Abstract
Biobanking in health care has evolved over the last few decades from simple biological sample repositories to complex and dynamic units with multi‐organizational infrastructure networks and has become an essential tool for modern medical research. Cardiovascular tissue biobanking provides a unique opportunity to utilize cardiac and vascular samples for translational research into heart failure and other related pathologies. Current techniques for diagnosis, classification, and treatment monitoring of cardiac disease relies primarily on interpretation of clinical signs, imaging, and blood biomarkers. Further research at the disease source (i.e. myocardium and blood vessels) has been limited by a relative lack of access to quality human cardiac tissue and the inherent shortcomings of most animal models of heart disease. In this review, we describe a model for cardiovascular tissue biobanking and databasing, and its potential to facilitate basic and translational research. We share techniques to procure endocardial samples from patients with hypertrophic cardiomyopathy, heart failure with reduced ejection fraction, and heart failure with preserved ejection fraction, in addition to aortic disease samples. We discuss some of the issues with respect to data collection, privacy, biobank consent, and the governance of tissue biobanking. The development of tissue biobanks as described here has significant scope to improve and facilitate translational research in multi‐omic fields such as genomics, transcriptomics, proteomics, and metabolomics. This research heralds an era of precision medicine, in which patients with cardiovascular pathology can be provided with optimized and personalized medical care for the treatment of their individual phenotype.
Keywords: Tissue biobanking, Translational research, Heart failure, Hypertrophic obstructive cardiomyopathy
Introduction
Biobanking in health care has evolved over the last few decades from simple biological sample repositories to complex and dynamic units with multi‐organizational infrastructure networks, as is the case in Europe. 1 , 2 , 3 They have become essential tools for modern medical research, including research into advanced treatments for cardiac disease, in particular heart failure.
Biobanks can be characterized as population‐based or disease‐oriented. The former contain epidemiological data collected from patients or volunteers without specific inclusion or exclusion criteria. 4 Disease‐oriented biobanks target a specific population with the disease in question, which would be true for many biobanks in the fields of oncology and cardiology.
There are comprehensive guidelines for tissue biobanking, which provide a framework for ethical and scientific governance. 5 Broadly speaking, the general features of a biobank, as outlined by the European Commission Joint Research Centre, include the following 6 :
The collection and storage of biological materials combined with medical and epidemiological data;
Dynamic development of biobanks involving continuous collection as a long‐term project;
Association with ongoing research projects;
Application of specimen anonymization for donor privacy; and
Implementation of governance standards and procedures for use of samples in future (not yet defined) research projects.
Cardiovascular biobanking represents a disease‐oriented biobank, providing unique opportunities to utilize cardiac and vascular samples for translational research into heart failure, valvular, and aortic diseases. 7 , 8 , 9 , 10 , 11 Current techniques for diagnosis, classification, and treatment of cardiovascular diseases rely primarily on interpretation of history and physical signs, imaging, and blood biomarkers. Further research into cardiovascular disease pathophysiology has been limited by a relative lack of access to quality human cardiac and aortic tissue, and the inherent shortcomings of most animal models of heart disease. 12 Indeed, the translation from animal (e.g. rodent) models of cardiovascular disease to humans can be less than 50%. 12
In this review, we describe a model for cardiovascular biobanking and databasing, and its potential to facilitate basic and translational research. The establishment of our cardiovascular biobank has been detailed previously 2 , 13 , 14 and is beyond the scope of this article. Herein, we share surgical techniques for procurement of biobank tissue samples, and we review processes and protocols for tissue transit and storage, the ethics and governance issues surrounding tissue biobanking, and detailed discussions for current and future uses of tissue to enhance cardiovascular research.
Patient identification and recruitment
The framework for cardiovascular biobanking includes procedures for patient recruitment and for donation, procurement and storage of tissue, and finally, the dissemination of samples for research, all within a predefined ethical framework and governance structure (Figure 1 ).
Figure 1.
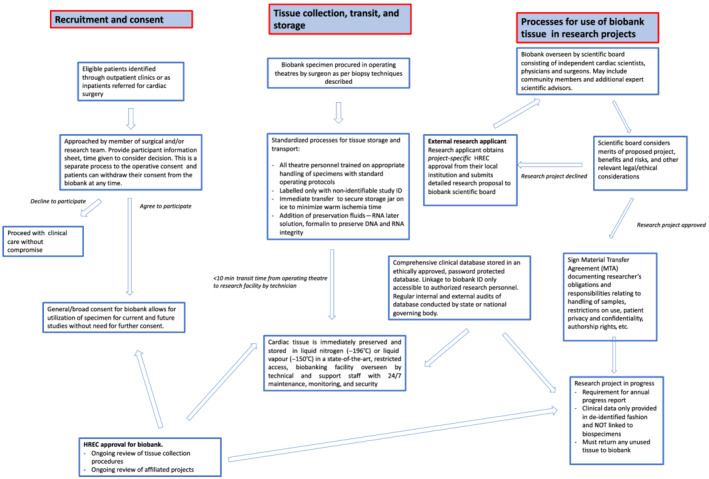
Our model of cardiovascular biobanking. HREC, Human Research Ethics Committee.
A multi‐stage process is followed in order to promote engagement with sample donors. Potential participants are initially identified as outpatients (in cardiology or cardiothoracic surgery clinics) or as inpatients who are clinically stable and referred for cardiac surgery. After identification, patients are reviewed by a member of our institutional research team for assessment and for the inclusion of tissue samples in the biobank. Relevant clinical and imaging data are reviewed on a standardized proforma and the initial interest of the patient to participate is ascertained.
Prior to surgery, eligible participants are approached by a senior member of the cardiothoracic surgical team to have the cardiovascular biobank explained in clear and non‐technical language and to provide them with an information sheet containing details pertinent to the biobank recruitment [this is specific for either hypertrophic cardiomyopathy (HCM), heart failure, or aortic tissue procurement]. The information sheets are operation‐specific as the procedures for sample procurement vary (as described in section below titled ‘techniques for cardiovascular sample collection’) and the inherent level of risk varies from no risk (when collecting aortic tissue which would otherwise be discarded) to low risk (when taking endocardial or atrial biopsies). The patients are given time to review this information independently and an opportunity to ask questions or discuss with their relatives or primary care physicians. Patients have until the day of surgery to provide or withdraw consent for study participation, which may be a period of months from the point of initial engagement. Those who agree to participate are requested to sign a detailed consent form.
Importantly, the biobank consent differs from the more traditional research consent in that, while it is operation‐specific, it also seeks a more general or broad consent, which allows for utilization of specimens for current studies, future projects, and experimental techniques (even those not yet conceived), without the need for future contact to seek additional consent from the patient. Hence, the biobank consent is operation‐specific but not project‐specific as is the case with more traditional research consents. Once patients are recruited, they are assigned a unique biobank ID so that tissue specimens are only labelled with this non‐identifiable ID. Patients retain the right to withdraw their consent at any time as explicitly stated in their patient information and consent forms.
Techniques for cardiovascular sample collection
All tissue samples are collected intraoperatively by the consultant surgeon (clinical experience is important here), and the surgical technique varies, according to the sample type being collected.
For HCM biobanking, the ventricular myocardium from the left ventricular outflow tract is resected as part of a septal myectomy procedure (Figure 2 ). Sample sizes for HCM biobanking are generous as HCM patients undergoing surgery have severe septal hypertrophy and this excess muscle mass is resected from the left ventricular outflow tract to achieve a satisfactory reduction of the outflow gradient. The risk of an iatrogenic ventricular septal defect is reduced if there is severe basal septal left ventricular hypertrophy (e.g. >18 mm) pre‐operatively.
Figure 2.
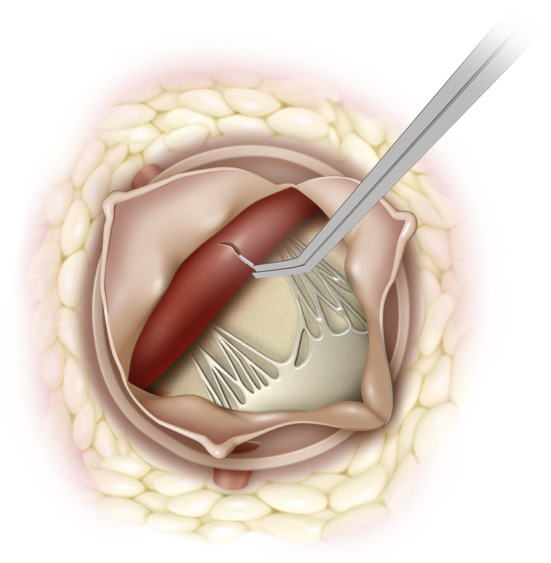
View of left ventricular outflow tract (LVOT) from aorta with aortic leaflets retracted. The ventricular septum can be biopsied with biopsy forceps or a blade under vision.
For heart failure biobanking, ventricular endocardial and atrial tissues are sampled from patients having operations such as mitral or aortic valve surgery. Essentially, these are cardiac procedures during which the atrial tissue (Figure 3 ) and left ventricular endocardium (Figures 2 and 4 ) are exposed as a routine part of the surgery. They are performed through a median full or hemi‐sternotomy with cardiopulmonary bypass support. The most common historic form of heart failure biobanking is from heart transplant patients, 2 but this can have limitations as it represents end‐stage disease only. The biobanking techniques described here offer an opportunity to conduct basic and translational research ‘up stream’ of severe heart failure, to include early systolic and diastolic dysfunction.
Figure 3.
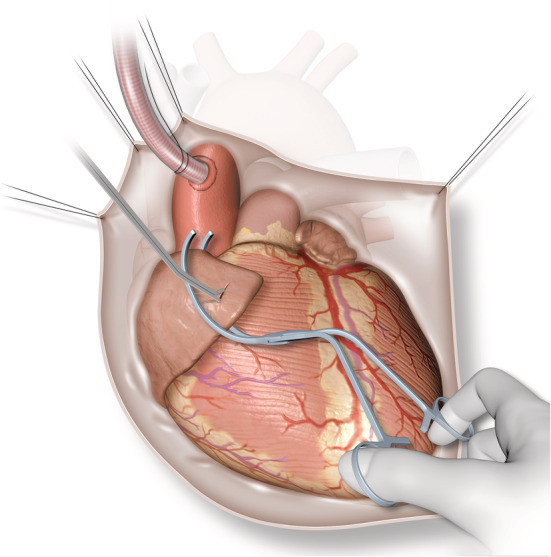
Biopsy of right atrium. Atraumatic clamp applied to tip of right atrial appendage and biopsy forceps used to take sample of atrial tissue.
Figure 4.
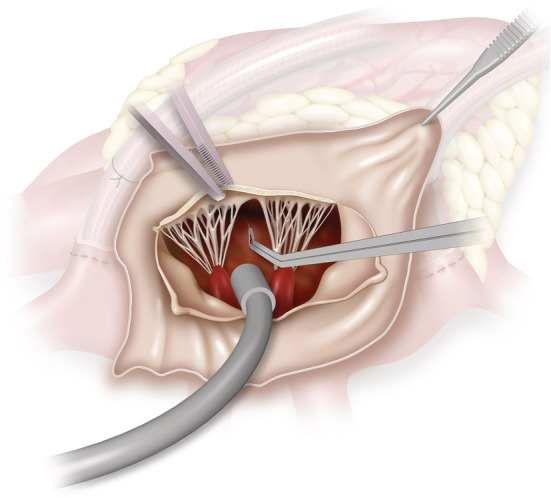
View of ventricular septum via left atriotomy, anterior mitral leaflets resected. The ventricular septum can be biopsied under direct vision.
Figure 3 illustrates the technique of atrial tissue sampling. To facilitate cardiopulmonary bypass, a 1–1.5 cm incision is usually made in the right atrial appendage to allow for the passage of a large cannula. Immediately prior to venous cannulation, an atraumatic clamp is applied to the tip of the right atrial appendage and a thin strip of atrial tissue (3–5 mm in size) is resected using Metzenbaum scissors or a surgical blade at the site of venous cannulation. This site is normally repaired at the end of cardiopulmonary bypass with a purse string suture (a circular stitch which cinches the tissue together to provide a haemostatic seal), and additional sutures can be applied as needed. The small quantity of tissue resected at this site causes minimal bleeding and the biopsy site is easily visible and accessible should further surgical repair be necessary.
Patients scheduled for left‐sided valvular surgery (e.g. aortic or mitral surgery) may be suitable for an endocardial biopsy. Figure 2 shows the surgical view of the left ventricular endocardium with the aortic leaflets retracted (or removed for aortic valve replacement), and Figure 4 illustrates the same left ventricular endocardium exposed through the mitral valve with a left atriotomy performed via Sondergaard's groove. In essence, any approach to the mitral valve (whether bi‐atrial, transeptal, or inter‐atrial) should provide adequate exposure of the left ventricular endocardium. Two small pieces of endocardial tissue (~5 mm each) are taken with biopsy forceps or a surgical blade under direct vision. This is generally in the region of the left ventricular outflow tract as the closest accessible point (if performed through a standard aortotomy), taking care to avoid other vital structures such as chordae or papillary muscles. The depth of this sample is under 3 mm (the length of the biopsy instrument tip used at our institution), much less than the average 6–10 mm thickness of the ventricular septum as measured by echocardiography. 15 Thus, the risk of an iatrogenic ventricular septal defect is low. As the endocardium faces directly into the left ventricle, there is no risk of blood loss from the biopsy site. Endomyocardial biopsies are also performed safely via transcatheter routes and major complications associated with left ventricular biopsy have an incidence of 0.33%. 16 Most of the described major complications from the transcatheter approach such as cardiac tamponade due to ventricular perforation or embolization to the brain are less problematic in a surgical setting during which the pericardium is open and the aorta has been cross‐clamped during myocardial arrest. To date, our institution has performed endocardial surgical biopsies in the manner described on a total of 24 patients without any clinical complications, supporting the safety of this technique in our early experience.
Thoracic aortic biobanking involves the collection of resected aortic tissue that would ordinarily be discarded as part of the surgery. The process for acquiring the aortic specimen is described here, but as it is not a specific biopsy technique, may vary considerably depending on the individual surgical plan (e.g. valve sparing vs. non‐valve sparing root reconstruction). Cardiopulmonary bypass for total body perfusion is instituted, and an aortic cross clamp is applied distal to the diseased portion of aorta and myocardial arrest achieved by delivery of hyperkaliaemic hypothermic cardioplegia solution. Once a bloodless and motionless operating field is achieved, the diseased aorta is resected using sharp dissecting scissors or bipolar diathermy, usually proximal to the cross clamp. Sometimes, the resection is completed under deep hypothermic circulatory arrest (14.1–20°C) depending on the extent of planned reconstruction. 17 The tissue specimen contains all three layers of aortic wall (adventitia, media, and intima) and may include portions of the ascending aorta, aortic root (with the sino‐tubular junction) or aortic arch. The tissue sample here is generous as these patients typically have dilated and diseased aortas (Marfan, Ehlers–Danlos syndrome, and other aortopathies), which are extensively resected prior to reconstruction.
Tissue transport and storage
Reliable procedures and processes for tissue transport and storage are vital to preserve sample quality. All specimen containers with preservatives and insulated coolers for transport are pre‐prepared by trained biobank personnel. Theatre personnel who handle the specimens intraoperatively (e.g. surgeons, nurses, and technical staff) are trained for the specific handling requirements of the biobank protocol, and they are provided with written instructions; the standard operating procedures and checklists utilized in our institution pertaining to intraoperative tissue collection are included in Data S1 . In addition, samples are only collected from patients who undergo surgery in a non‐emergency setting during working hours. This is to ensure adequate staff availability to procure, transport, and store/process the samples within the specified time frame. These procedures are important to minimize inter‐operator variability and maximize sample quality.
Once tissue samples are acquired, they are immediately placed into specimen containers on ice by the theatre nurse to minimize warm ischaemia time (the amount of time that tissue is at ambient room temperature after being resected and prior to stabilization by freezing or fixing in formalin at the time of biobanking). Aortic tissue specimens are further separated into three separate containers: one container with no preservation fluids for snap freezing with liquid nitrogen, the second containing RNAlater™ solution (ThermoFisher), and the third containing 10% neutral buffered formalin. Placing tissue immediately into RNAlater™ provides stabilization of the tissue RNA and enables downstream isolation of high‐quality RNA for gene expression analysis. Placing tissue in formalin enables fixation of the tissue in paraffin, which preserves tissue morphology and enables a wide range of downstream applications for immunohistochemical analysis. Other tissue samples (myectomy, endocardial, and atrial samples) are transported fresh for snap freezing. To protect donor anonymity and privacy, the samples are labelled only with the biobank ID and no other identifying details.
The specimen is then passed out of theatre to a waiting biobank technician who transports the specimens immediately to the research biobank where the fresh samples are cryopreserved in either liquid nitrogen (−196°C) or nitrogen vapour (−150°C) for storage, with the latter being ideal as it is safer. 18 The most important factor in maintaining quality of the tissue is time. This includes time from the body to being on ice, followed by time on ice to the laboratory bench, followed by time to be preserved at liquid nitrogen temperatures. The best practice arrangement is for the laboratory to be in close proximity to the operating theatre to minimize the transit time. In our institution, the laboratory is next door to the hospital with a transit time of around 2 min.
The next step is that of measuring quality control. This includes routine immunohistochemistry to examine the cellular architecture and to exclude obvious tissue damage such as disorganized sarcomere array, excess fibrosis. If there is any concern from this examination, then protein and RNA are extracted to check quality and yield at a molecular level. Once deemed to have satisfactory quality, the tissues are immediately preserved in ultra‐low (liquid nitrogen) temperature to maintain the integrity of proteins, DNA, RNA, and other cellular components. 18 Storage at ultra‐low temperatures allows biospecimens to be stored for years to decades (or beyond) without measurable tissue degradation. 19 , 20 Our model of immediate transport on ice and rapid cryopreservation (Figure 1 ) at ultra‐low temperatures is in line with best practice recommendations from the International Society for Biological and Environmental Repositories. 5
Principles of data collection and privacy
Patients undergoing cardiac surgery at the participating institution have their clinical data collected prospectively and stored in an ethically approved, password‐protected electronic database (Figure 1 ). The database is audited regularly (internal and/or external), and the audit findings are submitted at least annually, preferably quarterly to a state or national governing body (e.g. the ANZSCTS Cardiac Surgery Database, 21 the international equivalent of which is the European Association for Cardio‐Thoracic Surgery database).
In our institution, surgical patients have more than 130 clinical variables collected prospectively including demographic details, pre‐operative investigations, operative data, perfusion records, and post‐operative outcomes. Information such as genetic data, family history, echocardiographic parameters, medical therapy, laboratory or serum measurements, and previous surgical therapy (e.g. cardiac resynchronization therapy) are able to be linked with myocardial samples as they help to establish the clinical relevance of any molecular findings. Molecular characteristics are frequently the downstream outcomes of combined genetic, environmental, and iatrogenic influences. Examination of the clinical data is important to identify potential confounders, also noting the influences upstream of metabolomic or protein expression that may assist in the molecular pathway interpretation. For example, in one proteomic study of patients with HCM, the authors observed remarkably consistent alterations in the sarcomeric proteins of patients with HCM, despite distinct genotypes. This type of observation provides potential for molecular targets for drug therapies, which may benefit all patients with HCM. 11
As an adjunct to clinical data, blood samples are collected from patients in the anaesthetic bay immediately prior to surgery for the HCM and aortic biobanks. Work is in progress to add blood collection to the heart failure biobanking protocol. Matched serum samples can be used for proteomic, metabolomic, and genetic studies, and it is intended eventually to link into an extensive matched database of myocardial tissue, blood samples, cardiac magnetic resonance imaging, and other imaging. Perhaps the most impressive example of blood sampling in population studies is that of the UK Biobank 22 incorporating data regarding socio‐demographics, health status, blood sampling, and physical measurements, demonstrating the importance of linked data such as is being achieved in the tissue biobank described here.
The local institutional database is maintained in the cardiology or cardiothoracic surgery research offices of our institution and is accessible only to approved researchers and clinicians, according to the local ethics approval. All electronic access to the database is recorded and monitored. The clinical database is linked to the patient's biobank ID, and this is the ONLY identifying linkage between a patient's data and their biospecimen. The identifying link is housed only in a single hard copy format secured in a locked office within the local research precinct, where there is restricted and electronically monitored entry by ID pass to approved clinical and research personnel. The study consent provides for access to patients' clinical data, although this is provided in de‐identified format via approved and secure data storage platforms. Donor privacy is always maintained, but re‐identification of a patient is possible by specifically approved personnel to allow for incidental findings or specific conditions when clinically relevant information becomes known and can then be provided to the patient. 23
Ethics and governance of cardiovascular biobanking
With the development, expansion, and integration of large tissue biobanks, multiple ethical issues around consent, confidentiality, and governance have arisen. 24 While it is beyond the scope of this article to fully expand on these issues, we present here an example of governance standards and procedures in place for our collection, protection, and use of samples for cardiovascular surgery biobanking.
In line with best practice recommendations from International Society for Biological and Environmental Repositories, each biobank ‘subdivision’ of samples (from patients with HCM, heart failure, and aortic disease) needs to be approved separately by the institutional Human Research Ethics Committee, which also reviews the tissue collection procedures and affiliated projects annually. As described earlier, there is a procedure in place for recruitment and obtaining informed consent from participants of the biobank, including assurance that their clinical care will not be adversely affected if they decline to participate and that there are clearly outlined avenues for withdrawal of consent at any time. This ensures that consent is informed and given without coercion. In our institution, overall informed patient consent to the described research protocols is more than 90%.
The biobank itself is overseen by a committee including independent (not all employed by the institution that is the custodian of the biobank) cardiac scientists, cardiologists, and cardiac surgeons, representing experts in the fields of research, medicine, and surgery, respectively. Community (lay) members or patient advocates are also encouraged to participate. The primary role of the tissue access committee is to oversee the operation of the biobank and collectively evaluate the research project applications, including the scientific and logistical merit of the proposed projects. All researchers seeking to utilize the tissue samples and/or de‐identified patient clinical data are required to submit a detailed research proposal and a project‐specific human ethics approval from their own local institution. The proposal is then be assessed by the biobank committee to consider the scientific merit of the project, and whether the research is appropriate to the nature and composition of samples housed in the biobank, the availability of specimens being requested, benefits and risks of the proposed research, and other legal and ethical considerations as deemed important by our institution as the custodian of the biobank. With novel and more advanced technologies becoming available for tissue analysis, for example, single‐cell RNA sequencing and spatial transcriptomics, the scientific committee will evolve over time to include these experts to facilitate and advise on the most relevant use of the biobank tissue.
Once approved, all researchers sign a Material Transfer Agreement, which documents their obligations and responsibilities in matters such as proper handling and storage of samples, restrictions on sample use, maintenance of patient privacy and confidentiality, intellectual property, and authorship rights from resulting publications.
Basic and translational directions
We present here a model for biobanking of cardiovascular surgery tissue samples (including human myocardium and aortic specimens). Samples from septal myectomy procedures and aortic tissue samples are typically of significant quantity and represent high yield research specimens. They provide enough tissue for multiple analyses including mass spectrometry for metabolomics and proteomics, advanced microscopy, and cell culture with functional applications and other technologies as well. The endocardial and atrial biopsies provide enough tissue for microscopy (immunohistochemistry and immunofluorescence studies) and proteomics analysis (with as little as 2 mg of tissue being required).
Tissue from cardiovascular biobanks can be used by researchers in a multitude of projects including the study of cytoskeletal proteins, membrane proteins, contractile mechanics of the cardiomyocytes, protein expression in cardiomyopathies, myocardial regeneration after injury, and many others. To date, over 150 publications in peer reviewed journals have utilized tissue from the Sydney Heart Bank. A few recent examples include one where deep proteomics and metabolomics analyses were performed on left ventricular myocardium from patients with ischaemic and dilated cardiomyopathy to reveal significant alterations in X chromosome link proteins and mitochondrial substrates in those with heart failure. 10 Another performed extensive proteomic analysis to examine the protein landscape of tissue from patients with hypertrophic obstructive cardiomyopathy to further comprehensively characterize the hypertrophic obstructive cardiomyopathy phenotype on a molecular level. 11 More recently, RNA sequencing was performed on foetal and adult donor heart tissues, and this identified the progesterone receptor as a key mediator for sex‐dependent transcriptional programming during cardiomyocyte maturation. 25 The availability of high‐quality human heart tissue from biobanks such as the Sydney Heart Bank is a powerful research tool, particularly when combined with extensive information provided by publicly available protein databases such as the Human Protein Atlas. 14 In this way, tissue biobanks can be used to generate important insights into cardiovascular disease evolution.
The cardiovascular biobank described here is an extension of a larger biobank containing samples from patients with end‐stage cardiomyopathies as well as non‐diseased donor hearts, which have been described previously by us. 13 , 26 , 27 , 28 End‐stage heart failure biobanks are relatively common because these tissues would otherwise be discarded. The main sources of tissue in them are explanted hearts of transplant recipients with familial, idiopathic, ischaemic, restrictive, and hypertrophic cardiomyopathies. Samples from the left ventricle, right ventricle, interventricular septum, left and right atria, papillary muscles, and coronary arteries are usually collected 2–10 min after application of the aortic cross clamp. Best practice is for immediate snap freezing in liquid nitrogen.
Donor (non‐failing) samples are collected by the transplant team from healthy individuals after brain death. These hearts are assessed at the time of procurement as being eligible for transplant using standardized cardiac donor selection criteria, after visual inspection and manual palpation by the surgeon to assess atrial and ventricular size, presence of coronary calcification, any areas of transmural scarring, presence of mediastinal hematomas from chest compression or thoracic trauma and abnormalities of contractility. 29 Some of these hearts cannot be used for transplantation because there was no tissue and/or clinical match with an eligible patient waiting for a heart transplant, due to prohibitive logistical reasons or because the hearts do not fulfil the quality requirements for transplantation. When they are unable to be used as transplant hearts, donor hearts represent a collection of high‐quality human myocardium that can be used as a ‘control’ arm for failing hearts. They are particularly useful in high‐throughput multi‐omics analyses. 10 Of course, these samples do come from genetically diverse patients who have had varied iatrogenic and environmental exposures throughout their life. They are not controls in the purest sense of the word, especially when compared with animal control models, which are genetically modified, homogenous, and raised in a tightly controlled laboratory environment. 12 However, donor hearts do represent real‐life diversity, and this should be seen as an important factor in basic and translational research.
Cardiovascular tissue biobanks can overcome many of the limitations associated with animal models, given the significant differences in anatomical, functional, and molecular characteristics of the heart between species and the potential for poor applicability of animal models to the clinical setting. 30 , 31 Many therapies and drugs that perform well in animal studies fail in humans due to unacceptable safety profiles. The cardiovascular biobank procedures described here focus on sampling techniques that are upstream of end‐stage heart failure and the more traditional heart transplant samples of most cardiac biobanks. The goal is to establish a large repository of human tissue samples representative of the entire spectrum of heart disease, from non‐diseased to end‐stage disease.
Studies have used control samples procured from patients with a pathology different from the one being studied (e.g. left ventricular papillary muscle in patients undergoing mitral valve surgery were used as the control for end‐stage heart failure samples), 32 or another pathology with similar phenotype to serve as the ‘disease control’ in the absence of completely non‐diseased tissues (e.g. samples taken from patients with aortic stenosis for use as the disease control for familial HCM 33 ). This approach makes it easier for non‐transplant centres to use human tissue as their control group, but it has inherent limitations in the assumption that patients undergoing surgery for a different cardiac pathology are an appropriate control.
A limited number of studies have used myocardial samples procured from donor hearts as their control (Data S1 ). The donor hearts were available as they were not used for transplantation, sometimes due to logistical reasons 34 , 35 , 36 , 37 , 38 and sometimes because of quality issues such as palpable coronary calcification. 39 , 40 One was collected post‐mortem. 41 Another combined their analysis of donor hearts with explanted end‐stage heart failure hearts, 42 thus not using the donor as a control arm. There were variable levels of detail in describing the methods for tissue collection, transportation, storage, and quality control measures (Data S1 ). High‐quality donor hearts are ones which are not collected post‐mortem (this leads to degradation of protein epitopes and RNA) and where there is confirmation that these hearts are functionally and histologically normal. The Sydney Heart Bank addresses some of these limitations as all of the donor samples are collected from healthy individuals after brain death (not post‐mortem). Additionally, all donors have normal echocardiograms, no significant cardiovascular risk factors, have been assessed intraoperatively by the surgeon, and have undergone formal anatomical analysis to confirm the myocardium is not diseased. A large collaborative tissue biobank such as the one described here represents an important repository of high‐quality control human myocardial samples.
In addition to animal models and biobanked human tissues, pluripotent stem cells are increasingly being used as an alternative to study molecular mechanisms of disease. 43 Induced pluripotent stem cells and embryonic stem cells can differentiate into derivatives of all three germ cell layers (ectoderm, mesoderm, and endoderm) and renew indefinitely by mitosis. Cells from patients with a genetic variant causing a particular disorder can be isolated, induced into a pluripotent stem cell, programmed to become any other cell (a cardiomyocyte for example), and used as a model for the disease. They can be used for drug testing and to develop tailored treatments for patients based on their genetic mutation and risks. They can even be used to generate organoids and transplanted into damaged tissues to help regenerate them. The caveat is that these cardiomyocytes are relatively immature compared with adult ventricular cardiomyocytes.
Another particularly exciting field, but one which is in its infancy, is the use of artificial intelligence (AI) systems to manage and process clinical, imaging, and biospecimen data. AI systems use machine learning and natural language processing techniques to simultaneously and rapidly analyse large quantities of data and can learn from each incremental case to continually improve on their accuracy. 44 , 45 This technology is ideally suited to the type and volume of data within the biobank described here. For example, one could use an AI algorithm to define and measure the quality of tissue and DNA within samples and make recommendations for biobank quality control. AI systems can be used to identify links between imaging findings and biospecimens, helping in diagnosis and prognostication. The development and implementation of AI systems in cardiovascular biobanking represents a significant paradigm shift in the way that data are collected, managed, and processed.
Conclusions
There should be expansion in the future to include other subsets of surgical patients, providing even more diversity in the human cardiovascular tissues available for research. An opportunity is presented to link an imaging biobank that incorporates cardiac magnetic resonance imaging, and other imaging technologies such as ultrasound, computed tomography, and positron emission tomography to build a ‘super bank’ of linked clinical data, biospecimens and imaging data. The Genetically Triggered Thoracic Aortic Aneurysms and Cardiovascular Conditions Alliance is one such initiative that combines imaging, clinical, and some biospecimen data into one large repository. It is also proposed to make available some or all data generated from analysis of the biobank specimens to publicly accessible platforms in order to broaden researcher access to this information.
Human cardiac biobanking and the related basic and translational research heralds an era of precision medicine, in which patients with cardiovascular pathology can be provided with optimized and personalized medical care for the treatment of their individual phenotype. The goal of personalized medicine using genomic and proteomic analysis has already been realized in other areas of medicine such as oncology, 46 and this will become the standard of care for patients with cardiovascular diseases.
Supporting information
Data S1. Flowchart of cardiac tissue biobanking.
Data S2. Literature review of studies that utilise human donor hearts.
Supporting info item.
Zhu, Y. , Jackson, D. , Hunter, B. , Beattie, L. , Turner, L. , Hambly, B. D. , Jeremy, R. W. , Malecki, C. , Robertson, E. N. , Li, A. , dos Remedios, C. , Richmond, D. , Semsarian, C. , O'Sullivan, J. F. , Bannon, P. G. , and Lal, S. (2022) Models of cardiovascular surgery biobanking to facilitate translational research and precision medicine. ESC Heart Failure, 9: 21–30. 10.1002/ehf2.13768.
Paul G. Bannon and Sean Lal are co‐senior authors.
References
- 1. Yuille M, van Ommen GJ, Bréchot C, Cambon‐Thomsen A, Dagher G, Landegren U, Litton JE, Pasterk M, Peltonen L, Taussig M, Wichmann HE, Zatloukal K. Biobanking for Europe. Brief Bioinform 2008; 9: 14–24. [DOI] [PubMed] [Google Scholar]
- 2. Norlin L, Fransson M, Eaker S, Elinder G, Litton JE. Adapting research to the 21st century—the Swedish Biobank Register. Norsk Epidemiologi 2012; 21. [Google Scholar]
- 3. Zika E, Paci D, Schulte In Den Bäumen T, Braun A, Rijkers‐Defrasne S, Deschênes M, Fortier I, Laage‐Hellman JA, Scerri C, Ibarreta Ruiz D. Biobanks in Europe: Prospects for Harmonisation and Networking. Luxembourg (Luxembourg): Publications Office of the European Union; 2010. [Google Scholar]
- 4. Swede H, Stone CL, Norwood AR. National population‐based biobanks for genetic research. Genet Med 2007; 9: 141–149. [DOI] [PubMed] [Google Scholar]
- 5. Campbell LD, Jonas AJ, Judith GG, Rawley‐Payne M, Rush A, Sieffert N. Best practices: recommendations for repositories, Fourth Edition 2018 Available from: https://www.isber.org/page/BPR. Accessed May 2021.
- 6. Zika E, Paci D, Braun A, Rijkers‐Defrasne S, Deschênes M, Fortier I, Laage‐Hellman J, Scerri CA, Ibarreta D. A European survey on biobanks: trends and issues. Public Health Genomics 2011; 14: 96–103. [DOI] [PubMed] [Google Scholar]
- 7. Cao J, Koay YC, Quek LE, Parker B, Lal S, O'Sullivan JF. Myocardial substrate changes in advanced ischaemic and advanced dilated human heart failure. Eur J Heart Fail 2019; 21: 1042–1045. [DOI] [PubMed] [Google Scholar]
- 8. Lal S, Nguyen L, Tezone R, Ponten F, Odeberg J, Li A, dos Remedios C. Tissue microarray profiling in human heart failure. Proteomics 2016; 16: 2319–2326. [DOI] [PubMed] [Google Scholar]
- 9. Bagnall RD, Ingles J, Dinger ME, Cowley MJ, Ross SB, Minoche AE, Lal S, Turner C, Colley A, Rajagopalan S, Berman Y, Ronan A, Fatkin D, Semsarian C. Whole genome sequencing improves outcomes of genetic testing in patients with hypertrophic cardiomyopathy. J Am Coll Cardiol 2018; 72: 419–429. [DOI] [PubMed] [Google Scholar]
- 10. Li M, Parker BL, Pearson E, Hunter B, Cao J, Koay YC, Guneratne O, James DE, Yang J, Lal S, O'Sullivan JF. Core functional nodes and sex‐specific pathways in human ischaemic and dilated cardiomyopathy. Nat Commun 2020; 11: 2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tucholski T, Cai W, Gregorich ZR, Bayne EF, Mitchell SD, McIlwain SJ, de Lange WJ, Wrobbel M, Karp H, Hite Z, Vikhorev PG. Distinct hypertrophic cardiomyopathy genotypes result in convergent sarcomeric proteoform profiles revealed by top‐down proteomics. Proc Natl Acad Sci U S A 2020; 117: 24691–24700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Breckenridge R. Heart failure and mouse models. Dis Model Mech 2010; 3: 138–143. [DOI] [PubMed] [Google Scholar]
- 13. Lal S, Li A, Allen D, Allen PD, Bannon P, Cartmill T, Cooke R, Farnsworth A, Keogh A, dos Remedios C. Best practice biobanking of human heart tissue. Biophys Rev 2015; 7: 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li A, Estigoy C, Raftery M, Cameron D, Odeberg J, Pontén F, Lal S, dos Remedios CG. Heart research advances using database search engines, human protein atlas and the Sydney Heart Bank. Heart Lung Circ 2013; 22: 819–826. [DOI] [PubMed] [Google Scholar]
- 15. Lang RM, Badano LP, Mor‐Avi V, Afilalo J, Armstrong A, Ernande L, Flachskampf FA, Foster E, Goldstein SA, Kuznetsova T, Lancellotti P, Muraru D, Picard MH, Rietzschel ER, Rudski L, Spencer KT, Tsang W, Voigt JU. Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J Am Soc Echocardiogr 2015; 28: 1–39.e14. [DOI] [PubMed] [Google Scholar]
- 16. Chimenti C, Frustaci A. Contribution and risks of left ventricular endomyocardial biopsy in patients with cardiomyopathies: a retrospective study over a 28‐year period. Circulation 2013; 128: 1531–1541. [DOI] [PubMed] [Google Scholar]
- 17. Yan TD, Bannon PG, Bavaria J, Coselli JS, Elefteriades JA, Griepp RB, Hughes GC, LeMaire S, Kazui T, Kouchoukos NT, Misfeld M, Mohr FW, Oo A, Svensson LG, Tian DH. Consensus on hypothermia in aortic arch surgery. Annal Cardiothorac Surg 2013; 2: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shabihkhani M, Lucey GM, Wei B, Mareninov S, Lou JJ, Vinters HV, Singer EJ, Cloughesy TF, Yong WH. The procurement, storage, and quality assurance of frozen blood and tissue biospecimens in pathology, biorepository, and biobank settings. Clin Biochem 2014; 47: 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeisler R, Greenberg RR, Stone SF, Sullivan TM. Long‐term stability of the elemental composition in biological materials. Fresenius' Zeitschrift für analytische Chemie 1988; 332: 612–615. [Google Scholar]
- 20. Qualman SJ, France M, Grizzle WE, LiVolsi VA, Moskaluk CA, Ramirez NC, Washington MK. Establishing a tumour bank: banking, informatics and ethics. Br J Cancer 2004; 90: 1115–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tran L, Williams‐Spence J, Shardey G, Smith J, Reid C. The Australian and New Zealand Society of Cardiac and Thoracic Surgeons Database Program—two decades of quality assurance data. Heart Lung Circ 2019; 28: 1459–1462. [DOI] [PubMed] [Google Scholar]
- 22. Raisi‐Estabragh Z, Petersen SE. Cardiovascular research highlights from the UK biobank: opportunities and challenges. Cardiovasc Res 2019; 116: e12–e15. [DOI] [PubMed] [Google Scholar]
- 23. Wolf SM, Lawrenz FP, Nelson CA, Kahn JP, Cho MK, Clayton EW, Fletcher JG, Georgieff MK, Hammerschmidt D, Hudson K, Illes J, Kapur V, Keane MA, Koenig BA, LeRoy BS, McFarland EG, Paradise J, Parker LS, Terry SF, van Ness B, Wilfond BS. Managing incidental findings in human subjects research: analysis and recommendations. J Law Med Ethics 2008; 36: 219–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gottweis H, Zatloukal K. Biobank governance: trends and perspectives. Pathobiology 2007; 74: 206–211. [DOI] [PubMed] [Google Scholar]
- 25. Sim CB, Phipson B, Ziemann M, Rafehi H, Mills RJ, Watt KI, Abu‐Bonsrah KD, Kalathur RKR, Voges HK, Dinh DT, ter Huurne M, Vivien CJ, Kaspi A, Kaipananickal H, Hidalgo A, Delbridge LMD, Robker RL, Gregorevic P, dos Remedios CG, Lal S, Piers AT, Konstantinov IE, Elliott DA, el‐Osta A, Oshlack A, Hudson JE, Porrello ER. Sex‐specific control of human heart maturation by the progesterone receptor. Circulation 2021; 143: 1614–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dos Remedios CG, Li A, Lal S. Non‐sarcomeric causes of heart failure: a Sydney Heart Bank perspective. Biophys Rev 2018; 10: 949–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dos Remedios CG, Lal SP, Li A, McNamara J, Keogh A, Macdonald PS, Cooke R, Ehler E, Knöll R, Marston SB, Stelzer J, Granzier H, Bezzina C, van Dijk S, de Man F, Stienen GJM, Odeberg J, Pontén F, Linke WA, Linke W, van der Velden J. The Sydney Heart Bank: improving translational research while eliminating or reducing the use of animal models of human heart disease. Biophys Rev 2017; 9: 431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li A, Lal S, Dos Remedios CG. A step towards understanding the molecular nature of human heart failure: advances using the Sydney Heart Bank collection. Biophys Rev 2019; 11: 241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Copeland H, Hayanga JWA, Neyrinck A, MacDonald P, Dellgren G, Bertolotti A, Khuu T, Burrows F, Copeland JG, Gooch D, Hackmann A, Hormuth D, Kirk C, Linacre V, Lyster H, Marasco S, McGiffin D, Nair P, Rahmel A, Sasevich M, Schweiger M, Siddique A, Snyder TJ, Stansfield W, Tsui S, Orr Y, Uber P, Venkateswaran R, Kukreja J, Mulligan M. Donor heart and lung procurement: a consensus statement. J Heart Lung Transplant 2020; 39: 501–517. [DOI] [PubMed] [Google Scholar]
- 30. Sham JS, Hatem SN, Morad M. Species differences in the activity of the Na(+)‐Ca2+ exchanger in mammalian cardiac myocytes. J Physiol 1995; 488: 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guo G‐R, Chen L, Rao M, Chen K, Song J‐P, Hu S‐S. A modified method for isolation of human cardiomyocytes to model cardiac diseases. J Transl Med 2018; 16: 288. Available from: http://europepmc.org/abstract/MED/30348184 https://europepmc.org/articles/PMC6198433, https://europepmc.org/articles/PMC6198433?pdf=render. Accessed May 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sayour AA, Oláh A, Ruppert M, Barta BA, Horváth EM, Benke K, Pólos M, Hartyánszky I, Merkely B, Radovits T. Characterization of left ventricular myocardial sodium‐glucose cotransporter 1 expression in patients with end‐stage heart failure. Cardiovasc Diabetol 2020; 19: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Coats CJ, Heywood WE, Virasami A, Ashrafi N, Syrris P, dos Remedios C, Treibel TA, Moon JC, Lopes LR, McGregor CGA, Ashworth M, Sebire NJ, McKenna WJ, Mills K, Elliott PM. Proteomic analysis of the myocardium in hypertrophic obstructive cardiomyopathy. Circ: Genom Precis Med 2018; 11: e001974. [DOI] [PubMed] [Google Scholar]
- 34. Johnson EK, Matkovich SJ, Nerbonne JM. Regional differences in mRNA and lncRNA expression profiles in non‐failing human atria and ventricles. Sci Rep 2018; 8: 13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaynak B, von Heydebreck A, Mebus S, Seelow D, Hennig S, Vogel J, Sperling HP, Pregla R, Alexi‐Meskishvili V, Hetzer R, Lange PE. Genome‐wide array analysis of normal and malformed human hearts. Circulation 2003; 107: 2467–2474. [DOI] [PubMed] [Google Scholar]
- 36. Yang KC, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, Ewald GA, Mann DL, Nerbonne JM. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 2014; 129: 1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Di Salvo TG, Guo Y, Su YR, Clark T, Brittain E, Absi T, Maltais S, Hemnes A. Right ventricular long noncoding RNA expression in human heart failure. Pulm Circ 2015; 5: 135–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gaborit N, Le Bouter S, Szuts V, Varro A, Escande D, Nattel S, Demolombe S. Regional and tissue specific transcript signatures of ion channel genes in the non‐diseased human heart. J Physiol 2007; 582: 675–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kääb S, Barth AS, Margerie D, Dugas M, Gebauer M, Zwermann L, Merk S, Pfeufer A, Steinmeyer K, Bleich M, Kreuzer E. Global gene expression in human myocardium—oligonucleotide microarray analysis of regional diversity and transcriptional regulation in heart failure. J Mol Med 2004; 82: 308–316. [DOI] [PubMed] [Google Scholar]
- 40. Barth AS, Merk S, Arnoldi E, Zwermann L, Kloos P, Gebauer M, Steinmeyer K, Bleich M, Kääb S, Pfeufer A, Überfuhr P, Dugas M, Steinbeck G, Nabauer M. Functional profiling of human atrial and ventricular gene expression. Pflugers Arch 2005; 450: 201–208. [DOI] [PubMed] [Google Scholar]
- 41. Hsu J, Hanna P, Wagoner DRV, Barnard J, Serre D, Chung MK, Smith JD. Whole genome expression differences in human left and right atria ascertained by RNA sequencing. Circ Cardiovasc Genet 2012; 5: 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin H, Dolmatova E, Morley M, Lunetta K, McManus D, Magnani J, Margulies KB, Hakonarson H, del Monte F, Benjamin EJ, Cappola TP. Gene expression and genetic variation in human atria. Heart Rhythm 2013; 11: 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang C, Al‐Aama J, Stojkovic M, Keavney B, Trafford A, Lako M, Armstrong L. Concise review: cardiac disease modeling using induced pluripotent stem cells. Stem Cells 2015; 33: 2643–2651. [DOI] [PubMed] [Google Scholar]
- 44. Jiang F, Jiang Y, Zhi H, Dong Y, Li H, Ma S, Wang Y, Dong Q, Shen H, Wang Y. Artificial intelligence in healthcare: past, present and future. Stroke Vasc Neurol 2017; 2: 230–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buch VH, Ahmed I, Maruthappu M. Artificial intelligence in medicine: current trends and future possibilities. Br J Gen Pract 2018; 68: 143–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tyner JW. Functional genomics for personalized cancer therapy. Sci Transl Med 2014; 6(243): 243fs26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Flowchart of cardiac tissue biobanking.
Data S2. Literature review of studies that utilise human donor hearts.
Supporting info item.


