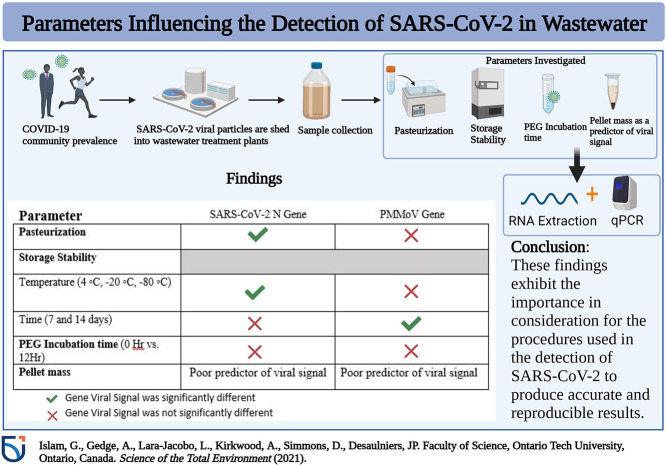
Abstract
The COVID-19 pandemic presents many public health challenges including the tracking of infected individuals from local to regional scales. Wastewater surveillance of viral RNA has emerged as a complementary approach to track and monitor the presence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus in a variety of communities of different land use and population size. In the present study, we investigate how five different parameters (pasteurization, storage temperature, storage time, polyethylene glycol (PEG) concentration, and pellet mass) affect the detection of the SARS-CoV-2 N gene and fecal abundance indicator pepper mild mottle virus (PMMoV) gene. Pre-treatment of 24-h composite wastewater samples (n = 14) by pasteurization at 60 °C resulted in a significant reduction of total RNA concentration and copies of the SARS-CoV-2 N gene copies/L (paired Student's t-test, P < 0.05). Comparing the wastewater samples collected from 6 wastewater treatment plants (WWTPs) for a storage period of 7 and 14 days at 4 °C, −20 °C and −80 °C, demonstrated a decrease in SARS-CoV-2 N gene copies/L when samples were stored for 14 days at −20 °C. Polyethylene glycol-NaCl for purification and concentration of viral particles from the wastewater samples demonstrated that a short PEG incubation of 2 h during centrifugation at 4 °C was sufficient for the consistent detection of the SARS-CoV-2 N gene from a 30 mL sample volume. Combined, this paper presents method recommendations for developing a reliable, accurate, sensitive, and reproducible estimation of the SARS-CoV-2 virus in diverse domestic wastewater samples.
Keywords: COVID-19, SARS-CoV-2, Wastewater, RT-qPCR, Pasteurization, Viral RNA concentration
Graphical abstract
1. Introduction
The ongoing COVID-19 pandemic has had an immeasurable impact on society, as evidenced by the loss of 5.32 million lives globally in over 81 affected countries as of December 2021 (WHO https://covid19.who.int/). It is particularly challenging for low- and middle-income countries to conduct mass testing of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in large populations due to several logistical challenges such as trained personnel, supply shortages, and reporting delays in reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) test results (Lopes-Júnior et al., 2020). In addition, current prevalence rates are largely based on symptomatic, clinically diagnosed cases, which excludes the existence of an estimated 45% of undiagnosed and asymptomatic individuals (Oran & Topal, 2020; Yang et al., 2020). Therefore, COVID-19 infections remain largely undetected when sufficient clinical testing does not occur. This necessitates the availability of alternative surveillance systems for the monitoring of the SARS-CoV-2 virus.
Clinical testing of the SARS-CoV-2 respiratory virus is mostly confirmed using nasopharyngeal mucosa, saliva, or sputum samples. However, recent studies have also revealed that 48 to 67% of COVID-19 infected individuals shed SARS-CoV-2 in their stool (Chan et al., 2020; Cheung et al., 2020; Parasa et al., 2020; Wölfel et al., 2020) and urine (Wu et al., 2020a; Wu et al., 2020b). The fecal and respiratory shedding of SARS CoV-2 follows a classic infection pattern (rapid buildup followed by a slow decline) (Sethuraman et al., 2020), but it is evident that shedding may occur ca. 3–5 days before the appearance of classic SARS-CoV-2 symptoms such as fever or diarrhea (Wang et al., 2020). It also remains detectable in fecal positive patients for a period of 9–14 days post-symptom onset date (Chen et al., 2020; Holshue et al., 2020; Ling et al., 2020; Pan et al., 2020). Both symptomatic and asymptomatic individuals may therefore shed the etiological agent of SARS-CoV-2 through saliva, sputum, feces, and urine (Jones et al., 2020; Lu et al., 2020; Van Vinh Chau et al., 2020), all of which are collected in domestic wastewater and consequently demonstrate the potential advantages of wastewater surveillance of SARS-CoV-2.
Wastewater based epidemiology (WBE) can provide a complementary surveillance system to measure the SARS-CoV-2 RNA in wastewater at the community level and may have the potential to be used as an early-detection system. As an example, in the Netherlands, SARS-CoV-2 RNA was detected in wastewater six days before the first clinical cases were reported (Medema et al., 2020). In Milan, Italy, SARS-CoV-2 RNA was detected in wastewater only a few days after the first Italian autochthonous SARS-CoV-2 positive case was reported (La Rosa et al., 2020), as well as in Brisbane, Australia in the early stages of the pandemic (Ahmed et al., 2020a). Despite the early successes, there are aspects of the current wastewater surveillance methodology that remain sub-optimized and thus further refinement is urgently needed for improved RNA detection. Sample site characteristics, primary viral concentration methods, sample volume, temperature and residence times en-route to sampling points can all influence accurate RNA detection and signal decay (Ahmed et al., 2020b). Current methods for the surveillance of SARS-CoV-2 in wastewater also vary widely and remain unstandardized. Some examples include: (1) Heat-based pasteurization to deactivate pathogens as a biosafety pre-treatment step (Pecson et al., 2021; Whitney et al., 2021; Wu et al., 2020a; Wu et al., 2020b); (2) Primary viral concentration involving one or a combination of ultrafiltration, PEG precipitation, or direct silica or skimmed milk flocculation extraction (Ahmed et al., 2020b; Casanova et al., 2009; Medema et al., 2020; Whitney et al., 2021); and (3) Varied sample storage times and temperatures (Ahmed et al., 2020c). Additionally, individual WWTPs will differ in sewershed size, flow, and source-community demographics, which can all affect wastewater sample characteristics including the variability of pellet sizes after ultracentrifugation (Whitney et al., 2021). Each of these parameters may contribute to variability in the detection and measurement of viral RNA targets such as SARS-CoV-2 and the fecal-normalizing-control pepper mild mottle virus (PMMoV). Due to these factors, there is a need to compare the performance of different protocols focused on RNA concentration, storage, extraction, and detection of the SARS-CoV-2 virus in wastewater to develop improved methodology that can be used consistently and with a high level of sensitivity.
The goal of the present study was to test some of the technical parameters and conditions that influence the measurement and detection of SARS-CoV-2 RNA and PMMoV viral particles in domestic wastewater samples. Specifically, this research examines: (1) the effect of pasteurizing wastewater influent samples prior to viral target detection; (2) the persistence of SARS-CoV-2 and PMMoV RNA at different storage temperatures (4 °C, −20 °C and −80 °C) for a duration of 7 and 14 days; (3) the efficacy of a PEG-based separation and viral concentration method under different incubation periods; and (4) the influence of pellet mass on the SARS-CoV-2 and PMMoV gene copies. This study provides guidance on these variables of sample processing methods and provides recommendations to estimate the SARS-CoV-2 virus more reliably in wastewater.
2. Methods
2.1. Aspects of Durham Region WWTPs
Raw influent samples containing 24-h time proportional composites were collected from four wastewater treatment plants, and two upstream pumping stations (PS) across the Durham Region in Ontario, Canada, over three months (January–March 2021). The residential population data and name of the plants are included in Table 1 . The sewershed of Durham region services approximately 640,000 residents and was one of the highly affected COVID-19 areas in Ontario, Canada with approximately 34,812 total confirmed cases reported on Dec 30th, 2021 (Durham Region Health Department, www.durham.ca).
Table 1.
Residential characteristics of the investigated WWTPs.
| Sampling site | Sample ID | Residential population |
|---|---|---|
| Corbett Creek | WWTP1 | 147,879 |
| Harmony Creek | WWTP2 | 37,485 |
| Ajax (Bayly PS) | WWTP3 | 155,734 |
| Pickering (Liverpool PS) | WWTP4 | 90,333 |
| Port Darlington | WWTP5 | 41,223 |
| Courtice | WWTP6 | 145,224 |
2.2. Wastewater processing and RNA analysis
2.2.1. Wastewater sample collection and PEG-NaCl viral concentration
Composite wastewater samples were collected from each plant three times per week using an autosampler. Each sample reflected hourly sub-samples of equal volume collected over a 24-h period, for a final sample volume of 500 mL collected and stored at 4 °C. Wastewater samples were transported in sterile and sealed 500 mL plastic containers at 4 °C to the laboratory at Ontario Tech University, Oshawa, Ontario, Canada. Upon arrival, samples were stored at 4 °C for up to 24 h until processing and analysis.
To precipitate the SARS-CoV-2 and PMMoV particles from wastewater, all samples were mixed thoroughly before 30 mL of wastewater was transferred to Nalgene™ Oak Ridge High-Speed PPCO Centrifuge Tubes containing 10 mL of 4× PEG-NaCl buffer (40% w/v PEG 8000 and 1.5 M NaCl), vortexed briefly and centrifuged using a SORVALL RC 6+ Ultracentrifuge (ThermoFisher Scientific, MA, USA) at 12,000 ×g for 2 h at 4 °C (Lewis & Metcalf, 1988; Wu et al., 2020a; Wu et al., 2020b). After discarding the supernatant, a second centrifugation step at 12,000 ×g for 10 min followed, to help solidify the pellet. The PEG-NaCl method was utilized for all experimental samples to concentrate the viral particles. The pellet mass for all samples was measured using a top loading balance (Sartorius, Goettingen, Germany), before RNA extraction.
2.2.2. Effect of pasteurization on RNA detection
To compare the effects of pasteurization on SARS-CoV-2 and PMMoV RNA detection, 14 raw influent wastewater composite samples collected in random from all wastewater treatment sites were split into duplicate aliquots in falcon tubes and placed into two groups. One group of 14 sample tubes was pasteurized by incubating at 60 °C in a pre-heated water bath for 1 h (Pecson et al., 2021). The pasteurized and unpasteurized samples (without heat treatment) were concentrated by using the same PEG processing method described above.
2.2.3. Effect of storage temperature on in-sample stability
To determine the effect of storage on viral RNA decay in the composite wastewater samples, duplicate samples collected from 4 different WWTPs and 2 upstream pumping stations in the region of Durham, were transferred into falcon tubes and stored in either a 4 °C refrigerator, a −20 °C or a −80 °C freezer for a period of 7 and 14 days (n = 72). Frozen aliquots of samples were thawed over a 1-h period in a room temperature water bath prior to processing using the PEG/NaCl ultracentrifugation and RNA extraction method, as previously described, for 7 and 14 days.
2.2.4. PEG incubation time
To precipitate the SARS-CoV-2 and PMMoV viral particles, 12 wastewater samples were selected from 4 WWTPs and one pumping station and were split into duplicate tubes by the addition of 30 mL of each wastewater sample to 10 mL of 4× polyethylene glycol (PEG)/NaCl buffer (40% w/v PEG 8000 and 1.5 M NaCl, pH = 7.2) in Nalgene™ Oak Ridge High-Speed PPCO Centrifuge Tubes (Thermo-Scientific, Waltham, Massachusetts, USA) and vortexed briefly (Lewis & Metcalf, 1988). To determine the efficacy of PEG incubation time, one group of duplicates were ultracentrifuged immediately after the addition of PEG/NaCl with no incubation period, and the other set of duplicates were incubated for 12 h at 4 °C without agitation prior to the ultracentrifugation step. All 24 samples were centrifuged using a SORVALL RC 6+ Ultracentrifuge (ThermoFisher Scientific, MA, USA) at 12,000 ×g for 2 h at 4 °C (Wu et al., 2020a; Wu et al., 2020b). To help solidify the pellet, after discarding the supernatant, a second centrifugation step at 12,000 ×g for 10 min followed.
2.2.5. RNA extraction
To determine extraction efficiencies for all wastewater samples, a viral surrogate control was carried out by adding 3 μL from a 1000 infectious units/mL of human coronavirus 229-E (HCoV-229E) viral standard stock (Chik et al., 2021) to the lysis buffer prior to RNA extraction. Recovery efficiency data was not included due to the significant loss of surrogate virus within the blank matrix (matrix), which severely affected the calculation of % Recovery of HCoV-229E and thus the estimated recovery of the target SARS-CoV-2.
Total RNA was extracted from the concentrated wastewater pellets using the RNeasy® PowerMicrobiome® Kit (Cat # 26000-50. Qiagen, Germantown, MD) with the following alterations from the recommended protocol: 100 μL of phenol-chloroform-isoamyl alcohol (25:24:1, pH 6.5–8) was added to each sample prior to the lysis step (ThermoFisher Scientific, MA, USA). The pellet was then resuspended with 650 μL of the lysis buffer and transferred to the PowerBead (glass, 0.1 mm) tubes (QIAGEN, Germantown, MD). The subsequent steps were performed following the recommended protocol from the manufacturer's kit. The total RNA was eluted from the kit spin column using 100 μL of RNAse-free water. Nucleic acid concentrations and RNA purity was measured using the Biodrop Nanospectrophotometer (BioDrop, UK) for the pasteurized and unpasteurized samples. The 260/280 ratios for all RNA samples used for analysis was ~1.9–2.0.
2.2.6. qRT-PCR
Quantification of SARS-CoV-2 viral nucleocapsid (N) gene and the PMMoV coat protein gene in the composite wastewater samples was performed using the Reliance One-Step Multiplex RT-qPCR Supermix (BioRad, Hercules, CA) utilizing a TaqMan (IDT, Iowa, USA) probe-based approach. The gene copy numbers of SARS-CoV-2 in wastewater were determined using the US CDC 2019-nCoV N1 Assay RUO primer/probe mix to target a region of the N gene. To determine the PMMoV gene copy numbers, PCR primers developed by Zhang et al. (Zhang et al., 2005) were employed to target a region of the PMMoV strain S genomic sequence. In order to determine RNA extraction efficiencies, the HCoV-229E gene was quantified using primers/probe designed by Dr. Lilly Pang (Alberta Provincial Lab, University of Alberta). All probe/primers used in this study and their sequences can be found in Table 2 .
Table 2.
Primers, probes and standards used in this study.
| Primer/probe | Sequence | Source |
|---|---|---|
| 2019-nCoV_N1 F | 5′-GACCCCAAAATCAGCGAAAT-3′ | (US Centers for Disease Control and Prevention, 2019) |
| 2019-nCoV_N1 R | 5′-TCTGGTTACTGCCAGTTGAATCTG-3′ | (US Centers for Disease Control and Prevention, 2019) |
| 2019-nCoV_N1 Probe | 5′-FAM-ACCCCGCATTACGTTTGGTGGACC-MGBNFQ-3′ | (US Centers for Disease Control and Prevention, 2019) |
| PMMV F | 5′-GAGTGGTTTGACCTTAACGTTGA-3′ | (Haramoto et al., 2013) |
| PMMV R | 5′-TTGTCGGTTGCAATGCAAGT-3′ | (Haramoto et al., 2013) |
| PMMV Probe | 5′-6FAM-CCTACCGAAGCAAATG-MGBNFQ-3′ | (Zhang et al., 2005) |
| HCoV-229E F | 5′-TTCCGACGTGCTCGAACTTT-3′ | Lilly Pang (APL) AB |
| HCoV-229ER | 5′-CCAACACGGTTGTGACAGTGA-3′ | Lilly Pang (APL) AB |
| HCoV-229 E Probe | 5′-FAM-TCCTGAGGTCAATGCA-MGBNFQ-3′ | Lilly Pang (APL) AB |
| HCoV-229E G-block | 5′-GTGTACCTGAAACAAAACCCATTGTAATTTTTCCGACGTGCTCGAACTTTTTTTCCTGAGG-3′ | Lilly Pang (APL) AB |
| N1 Standard | Custom IDT Twist RNA Control 2 | Twist Bioscience |
| PMMoV Standard | RNA N gene standard in vitro transcribed from an IDT gBlock DNA | IDT |
For each wastewater sample, technical replicates were run in triplicate, and serial dilutions of the viral RNA standard TWIST control 2 (Twist Bioscience, SF, USA) were run on every plate to quantify the gene copies of SARS-CoV-2 using the standard curve method. For PMMoV, a custom DNA PMMoV gBlock standard (IDT, Iowa, USA) was used to generate standard curves and quantify the PMMoV gene. Each reaction comprised of a mixture including 5 μL RNA template, 500 nM (N1) or 400 nM (PMMoV and H-CoV229E) of each forward and reverse primers, 125 nM (N1) or 200 nM (PMMoV and H-CoV229E) probe, and the Reliance mastermix, in a final reaction volume of 20 μL. Reactions were performed in a CFX Connect Real-Time PCR Detection System (BioRad, Hercules, CA) beginning with a reverse transcription (RT) step at 50 °C for 10 min, followed by a polymerase activation at 95 °C for 10 min, and then 45 cycles of denaturation and annealing/extension at 95 °C for 10 s, then 60 °C for 30 s.
The RT-qPCR analysis was validated with no-template controls (NTCs) using PCR grade water instead of RNA, no-reverse transcriptase controls (NRTs), and the VetMax™ Xeno™ kit (ThermoFisher Scientific, MA, USA) as internal inhibition control to monitor the presence of PCR inhibitors. All samples analyzed were quantified according to the MIQE recommendations (Bustin et al., 2009) using the standard curve method with a synthetic RNA standard (TWIST BioSciences Control 2) that contains our gene of interest, the nucleocapsid gene of SARS-CoV-2. A minimum 5-point standard curve with technical triplicates for each point was performed for every RT-qPCR experiment. Primer efficiencies for each target ranged from 95 to 110%. The R2 value was ≥0.98, and the slope of the standard curve was ~3.3–3.4. The limit of detection for the SARS-CoV-2 N gene with 95% coefficient of variation was 3333 copies/L of wastewater. Any crossing threshold values above 40 cycles were identified as negative reactions, assuming no amplification/detection occurred. The dynamic range of our linear standard curve was between 1 × 105 copies/μL and 1 × 101 copies/μL.
2.2.7. Data analysis
All experimental data were tested for normality using a Shapiro-Wilk test, and log-transformed when tests of normality failed. All statistical analyses were conducted using GraphPad Prism 9.0.2 software (La Jolla, CA) or SigmaPlot 14.0 software (San Jose, CA).
A paired Student's t-test (α = 0.05) was used to determine if there were statistical differences between pooled unpasteurized samples (control) and pasteurized samples (treatment) in (1) total RNA concentrations (n = 14), and (2) SARS-CoV 2 N gene copies/L (n = 14), and (3) PMMoV gene copies/L (n = 14).
To evaluate the effect of storage conditions, the data from all wastewater treatment plants were pooled in order to determine if three independent factors (WWTP, temperature and time (7 and 14 days)) had an effect on the quantification of both the SARS-CoV-2 and the PMMoV genes in wastewater (n = 72). The statistical output confirmed that the data passed normality and conditions of the equal variance test (Brown-Forsythe test) and a multi-factorial three-way analysis of variance (ANOVA) (balanced design, no interactions) was employed, using the Holm-Sidak method, where α = 0.05.
The percent change (% change) was calculated for all sites on day 7 and day 14 as % change = 100 × (control − treatment) / control, where control (day 0 samples directly extracted) and treatment (samples stored at 4 °C, −20 °C and −80 °C) are the associated measurements. A positive % change indicated that the treatment result was smaller than the control, whereas a negative % change indicated the treatment had higher concentrations than the control.
For the PEG incubation experimental data, a paired Student's t-test (α = 0.05) was also used to test for significant differences in SARS-CoV-2 and PMMoV gene copies/L between pooled samples with no incubation (control) and 12-h incubation times (treatment) (n = 12).
Lastly, a least-squares linear regression (goodness of fit) was used to assess pellet mass (wet weight in mg) as a predictor variable of gene signal of N and PMMoV gene copies/L (n = 120).
3. Results and discussion
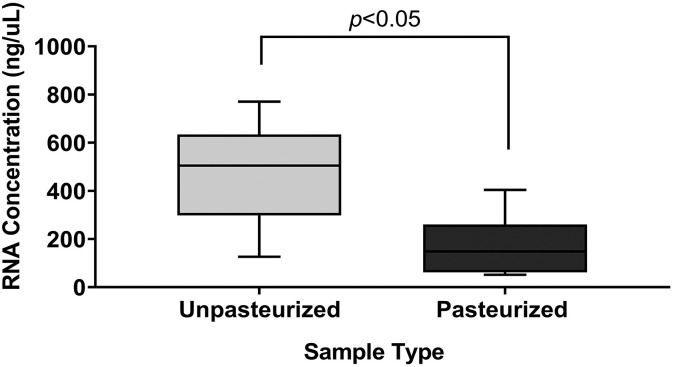
3.1. Effect of pasteurization on viral RNA detection in wastewater
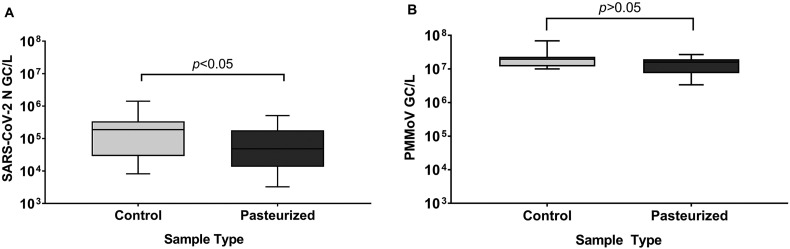
The analysis of total RNA from pasteurized and unpasteurized wastewater samples demonstrated that significantly higher total RNA concentrations (ng/μL) were recovered from unpasteurized wastewater in comparison to the pasteurized samples (paired Student's t-test, P < 0.05) (Fig. 1 ). Further investigation of the SARS-CoV-2 N gene copy numbers revealed that N gene copies/L were significantly higher in unpasteurized samples compared to pasteurized samples (paired Student's t-test, P < 0.05) (Fig. 2A), although this was not the case for PMMoV gene copies (paired Student's t-test, P > 0.05) (Fig. 2B). Using a 4S (silica and NaCl) method, Whitney et al. (Whitney et al., 2021) reported that heat pasteurization did not affect the concentration of SARS-CoV-2 gene copies and increased the concentration of PMMoV gene copies. In contrast, Steele et al. (Steele et al., 2021) observed that heat inactivation at 70 °C resulted in a significant reduction (1–3 Log10) in SARS-CoV-2 N gene concentrations using membrane concentrated samples. Palmer et al. (Palmer et al., 2021), also reported that pasteurization at 60 °C, resulted in an average decrease of approximately 50% in SARS-CoV-2 N gene concentrations. Differences in pasteurization temperature and duration may explain discrepancies between our results and others (Whitney et al., 2021), but there is a growing body of literature that implicates pasteurization in reduced SARS-CoV-2 concentration in wastewater (Palmer et al., 2021; Steele et al., 2021).
Fig. 1.
Total RNA concentration (ng/μL) in a 100 μL extraction elution volume comparing unpasteurized and pasteurized samples. Statistical significance was determined using a paired Student’s t-test (n = 14, P < 0.05).
Fig. 2.
(A) SARS-CoV-2 N gene copies/L comparing pasteurized and unpasteurized samples of raw influent. Data was log transformed and significance was determined using a paired Student’s t-test (P < 0.05, n = 14). (B) PMMoV gene copies/L comparing pasteurized and unpasteurized samples of raw influent. Data was log transformed and significance was determined using a paired Student’s t-test (P > 0.05, n = 14).
There were no effects of pasteurization on the PMMoV gene shown in this study (Fig. 2B), suggesting that PMMoV is more resistant to heat exposure and likely retains its encapsulated form that provides protection from degradation (Steele et al., 2021; Whitney et al., 2021). Sewage flow rate is not stable and may be impacted by factors such as inflows from storm events or winter-spring snow melts, that are prominent in Ontario, Canada. Thus, to correct for those sample-to-sample variations, PMMoV, which is a rod shaped, single-strand positive sense virus is used as an internal reference for the quantification of SARS-CoV-2 in wastewater. In the sewage system, PMMoV has shown remarkable stability and is highly abundant, which makes the PMMoV virus a suitable normalization target. However, since it is an entirely different virus with a unique morphology, it may show different partitioning characteristics and may not be as susceptible to heat-inactivation when compared to SARS-CoV-2. Therefore, PMMoV would not be suitable as an internal reference when samples are pasteurized before analysis.
The ability to detect low viral RNA concentration is particularly important for the surveillance of SARS-CoV-2 when active cases are low. Pasteurization may affect the sensitivity of the detection method by producing false-negative results when the RT-PCR assay is negative for the SARS-CoV-2 virus, yet trace amounts of the SARS-CoV-2 RNA may be present in the wastewater sample (Ahmed et al., 2021).
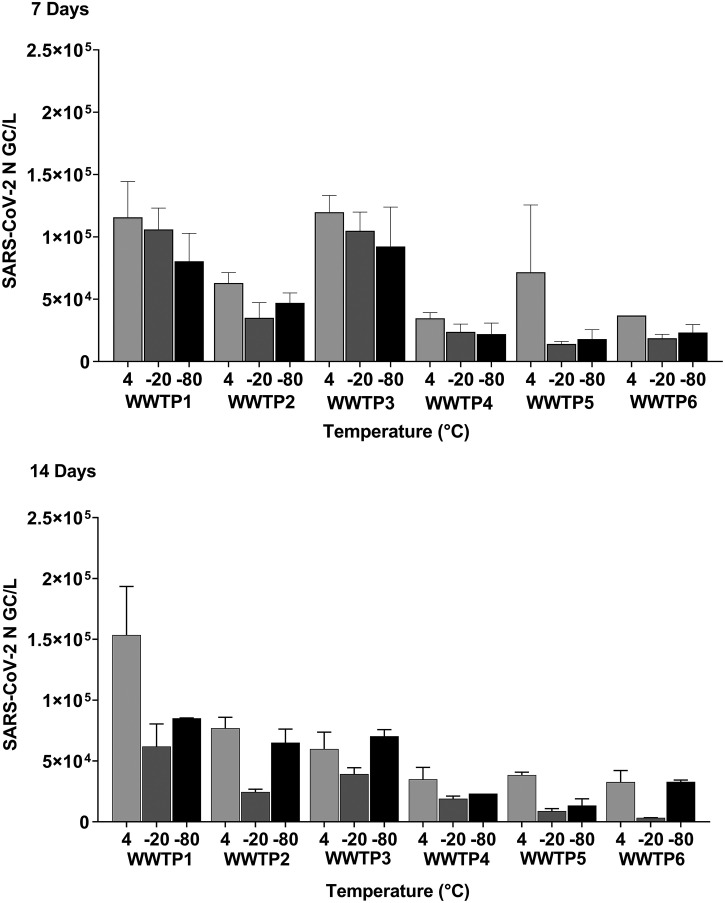
3.2. Effect of storage temperature on SARS-CoV-2 and PMMoV RNA stability
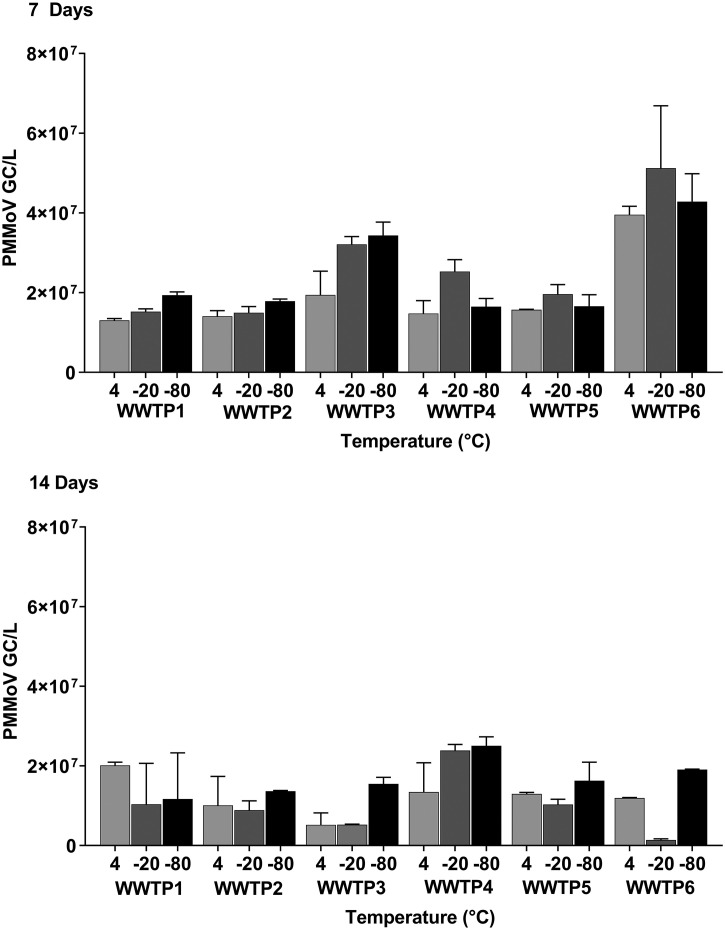
The stability of SARS-CoV-2 RNA in wastewater may be another potential contributing factor accounting for the loss of SARS-CoV-2 RNA signal. Comparing the SARS-CoV-2 N gene copy numbers of pooled sample data from all WWTP sites, we observed that storage temperature (F-Statistic 10.278, P < 0.001) and WWTP (F-Statistic: 20.352, P < 0.001) were the main contributing factors that resulted in significant differences of the SARS-CoV-2 N gene copies/L concentrations (Table 3 ). Analysis of the pair-wise comparison statistics (Holm-Sidak Method) of the Three-Way ANOVA (Table S1 in Supplementary Material) showed that wastewater samples stored at 4 °C had significantly different N gene copies/L from those stored at −20 °C (F-Statistic: 4.416, P < 0.001) and − 80 °C (F-Statistic: 3.099, P < 0.001). After 14 days, samples stored at −20 °C contained lower N gene copies than samples stored at 4 °C from pooled data which included all six WWTPs (Fig. 3 ). The % change for N gene copies for samples stored at 4 °C, −20 °C and −80 °C was 1.11%, 32.29% and 36.61% at 7 days and 11.23%, 65.12% and 35.44% after 14 days, demonstrating a greater loss of N gene concentrations when samples under-go freezing and thawing (Table 4A ).
Table 3.
Summary statistics for Three-Way Analysis of Variance (balanced design, no interactions) comparing the effects of wastewater treatment plant (WWTP), storage time, and storage temperature as independent variables and pooled quantified viral RNA copy number as the dependent variable. Results for both N and PMMoV are presented below, where statistical significance was set at α = 0.05, n = 72.
| Viral RNA target | Independent variable | F-statistic | P-value |
|---|---|---|---|
| N | WWTP (6 plants) | 20.352 | <0.001 |
| Storage time (7-day and 14-day) | 3.134 | 0.088 | |
| Storage temperature (4 °C, −20 °C, −80 °C) | 10.278 | <0.001 | |
| PMMoV | WWTP (6 plants) | 12.395 | <0.001 |
| Storage time (7-day and 14-day) | 26.377 | <0.001 | |
| Storage temperature (4 °C, −20 °C, −80 °C) | 2.628 | 0.091 |
Fig. 3.
SARS-CoV-2 N gene copies/L of raw influent wastewater stored at 4 °C, −20 °C, −80 °C for 7 and 14 days sampled from 6 different WWTP sites. A multi-factorial Three-Way Analysis of Variance was performed (balanced design, no interactions) on pooled data comparing the effects of WWTP, storage time, and storage temperature as independent variables and quantified viral RNA copy number as the dependent variable (n = 72). Statistical significance was set at α = 0.05. Error bars represent the standard error of the mean (SEM).
Table 4A.
Percent change in SARs-CoV-2 N gene viral signal at 4 °C, −20 °C, and −80 °C.⁎
| Time (days) | % change at 4 °C | % change at −20 °C | % change at −80 °C |
|---|---|---|---|
| 7 | 1.11 | 32.29 | 36.61 |
| 14 | 11.23 | 65.12 | 35.44 |
The percent change (% change) was calculated for all sites on day 7 and day 14 as % change = 100 × (control − treatment) / control, where control (day 0 samples directly extracted) and treatment (samples stored at 4 °C, −20 °C and − 80 °C) are the associated measurements. A positive % change indicated that the treatment result was smaller than the control, whereas a negative % change indicated the treatment had higher concentrations than the control.
For the normalization marker PMMoV, it was found that the gene copies were 2–3 orders of magnitude higher that SARS-CoV-2 and that storage temperature was not a significant factor affecting PMMoV gene copies/L concentrations (Table 3). However, when analyzing time as a factor, there was a significant difference in PMMoV gene copies/L (F-statistic: 26.377, P < 0.001, Table 3 and Fig. 4 ). PMMoV demonstrated an increase in gene copies/L at 7 days but a greater % change for PMMoV was observed at 14 days with 36.74% and 44.11% change when samples were stored at 4 °C and −20 °C (Table 4B ), compared to only 8.04% at −80 °C. Thus, PMMoV demonstrated higher stability at −80 °C.
Fig. 4.
Pepper Mild Mottle Virus (PMMoV) gene copies/L of raw influent wastewater stored at 4 °C, −20 °C, −80 °C for 7 and 14 days sampled from 6 different WWTP sites. A multi-factorial Three-Way Analysis of Variance was run on pooled data (balanced design, no interactions) comparing the effects of WWTP, storage time, and storage temperature as independent variables and quantified viral RNA copy number as the dependent variable (n = 72). Statistical significance was set at α = 0.05. Error bars represent the standard error of the mean (SEM).
Table 4B.
Percent change in PMMoV gene viral signal at 4 °C, −20 °C, and −80 °C.⁎
| Time (days) | % change at 4 °C | % change at −20 °C | % change at −80 °C |
|---|---|---|---|
| 7 | −0.07 | −36.09 | −26.67 |
| 14 | 36.74 | 44.11 | 8.04 |
The percent change (% change) was calculated for all sites on day 7 and day 14 as % change = 100 × (control − treatment) / control, where control (day 0 samples directly extracted) and treatment (samples stored at 4 °C, −20 °C and − 80 °C) are the associated measurements. A positive % change indicated that the treatment result was smaller than the control, whereas a negative % change indicated the treatment had higher concentrations than the control.
The storage of samples is critical when immediate processing cannot be achieved. Samples may need to be re-analyzed to ensure quality control assays are met or retroactively be investigated for the presence of known variants. Hokajärvi et al. (Hokajärvi et al., 2021) reported that in liquid influent, a linear decay of spiked SARS-CoV-2 RNA was observed at 4 °C, while no decay was visible within 58 days at the freezing temperatures of −20 °C and −80 °C. In contrast, Fernandez-Cassi et al. (Fernandez-Cassi et al., 2021) reported an extensive reduction in SARS-CoV-2 RNA concentrations measured in raw influent wastewater stored at −20 °C (1–2 orders of magnitude) after one month. Both freezing and thawing processes are known to be detrimental to microorganisms (Meyer & Calcott, 1980; Morley et al., 1983) by contributing to the degradation of nucleic acids via physical shearing (i.e., formation of ice crystals that cut the nucleic acid strands) (Röder et al., 2010) and oxidative damage caused by the release of metal ions and change in pH (Krajden et al., 1999).
For samples that were not processed immediately (24–48 h), several studies observed minimal differences in SARS-CoV-2 RNA concentrations for samples stored at 4 °C for up to 9 days (Markt et al., 2021; Simpson et al., 2021). Our results complement this finding by demonstrating that samples can be stored at 4 °C for a period of 14 days without a great reduction in viral signal. Thus, the consensus is to avoid freezing and thawing wastewater samples prior to testing to reduce damage to fragile RNA templates (Ahmed et al., 2021).
Comparing the most recent findings on the effect of storage conditions on SARS-CoV-2 detection in wastewater, it is evident that although the sample type (raw influent) and volume processed (30–70 mL) are similar in most experimental methods, ultracentrifugation was employed as a pre-treatment step to remove the particulate fraction of the wastewater (Fernandez-Cassi et al., 2021; Hokajärvi et al., 2021; Markt et al., 2021). Thus, these studies only utilized the liquid fraction of the wastewater samples for the analysis of SARS-CoV-2; whereas we utilized the solid fractions of wastewater which may have led to the differences in our observations. The solid fraction may provide higher affinity of an enveloped virus like SARS-CoV-2 to attach onto the particulate matter of wastewater and may be a more optimal choice for the analysis of enveloped viruses such as SARS-CoV-2 (Hokajärvi et al., 2021). Additionally, to compare our findings, it is also important to consider key differences in the experimental parameters such as whether the SARS-CoV-2 being targeted is spiked in or is within the matrix, volume of sample frozen/thawed, fraction of wastewater being frozen, and sample thawing procedures prior to analysis. This information may be crucial in comparing the effects of storage temperature on SARS-CoV-2 detection and should be made available in all future studies.
Since PMMoV is widely used as an internal process control to normalize SARS-CoV-2 RNA concentrations in domestic wastewater, the applicability of using it as normalizing factor should be re-considered when analyzing samples after long-term storage. Whitney et al. (Whitney et al., 2021) reported that the PMMoV gene signal remained stable throughout storage conditions at 4 °C for up to one month, and this is likely due to the non-enveloped morphology of the PMMoV, which contains a more robust capsid structure that allows it to persist in wastewater (Alonso et al., 1991; Kitajima et al., 2018). However, when observing the effects of freezing, (Simpson et al., 2021) concluded that since PMMoV was affected by storage in a similar manner to SARS-CoV-2, normalizing SARS-CoV-2 concentrations using PMMoV can correct for the effects of storage of samples from one WWTP site. Our sampling approach included samples from 6 different wastewater treatment sites to capture potential variations between properties of wastewater solids and observed that storage time (days) and not temperature may be resulting in differences in PMMoV concentrations. Further analysis is necessary to fully understand the effect of storage on PMMoV stability and its feasibility as an internal control when analyzing samples after long-term storage.
3.3. PEG incubation and relationship to pellet mass
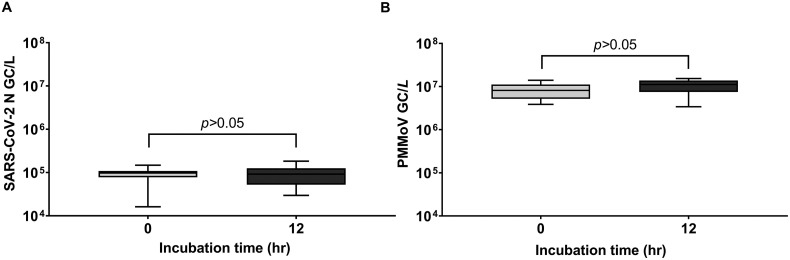
Due to the relatively low abundance of viral pathogens in wastewater, it is important to consider viral concentration when establishing a SARS-CoV-2 monitoring protocol. It is also imperative that a rapid turn-around time be incorporated into the methodology of a SARS CoV-2 wastewater surveillance program so that public health organizations can base their decisions on timely data. PEG-NaCl precipitation is a promising approach because it concentrates viruses from both the liquid and solid fractions of wastewater, is relatively inexpensive, and requires little technical expertise (Ahmed et al., 2020b). In this study, 30 mL sample aliquots of wastewater were precipitated with PEG-NaCl incubation times of 0 and 12 h, and the results demonstrated that there were no significant differences in viral copies between incubation times ((N) P > 0.05 and (PMMoV) P > 0.05) (Fig. 5 ). The extraction efficiencies for samples from all WWTPs was obtained using the no incubation method and ranged within 8–16%.
Fig. 5.
Comparing viral concentration after different incubation time frames for (A) SARS-CoV-2 N and (B) PMMoV gene copies/L of raw influent wastewater. Samples either had no incubation time with PEG (0 h) or a 12-h incubation time (12h) ((N) P > 0.05, n = 12 and (PMMoV) P > 0.05, n = 12).
Although several PEG precipitation methods have been used in the assessment of SARS-CoV-2 in untreated wastewater (Bar-Or et al., 2020; Kocamemi et al., 2020; Philo et al., 2021; Wu et al., 2020a; Wu et al., 2020b), many of these protocols recommend using large volumes (>200 mL) to concentrate the SARS-CoV-2 virus (Ahmed et al., 2020b), and include a pre-treatment such as pre-filtration (to dispose of cell debris and large particulates) with a lengthy incubation time (4–12 h) (Philo et al., 2021; Wu et al., 2020a; Wu et al., 2020b). The findings in this study demonstrated that the PEG/NaCl precipitation method can also detect SARS-CoV-2 and PMMoV viral RNA from a 30 mL volume of wastewater without the requirement of a pre-treatment or lengthy incubation step (Fig. 5). Due to the simplicity of PEG-NaCl precipitation, this rapid and effective method of viral concentration should be incorporated into the sample processing of SARS-CoV-2 wastewater surveillance programs.
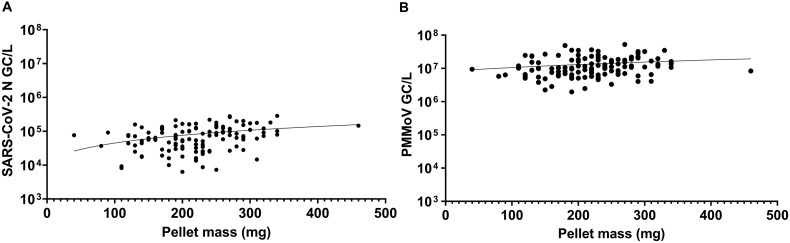
The differences in WWTP characteristics such as sewershed size and flow regime can impact not only the composition of the influent, but also the composition and size of the concentrated wastewater pellet obtained after processing. Several studies have implicated the partitioning of SARS CoV-2 to solid fractions compared to the liquid fractions (D'Aoust et al., 2021; Kim et al., 2021; Li et al., 2021). Thus, wastewater pellet mass (wet weight) was examined as a predictor variable for SARS-CoV-2 and PMMoV gene copy numbers obtained from a 30 mL fixed volume using RT-qPCR. Using a linear regression analysis model, it was found that for both the SARS CoV-2 N and PMMoV gene, pellet mass was observed to a be poor predictor variable for N and PMMoV gene copies (R2 = 0.113 for N), (R2 = 0.029 for PMMoV) (Fig. 6 ). These results indicate that the differences observed in SARS-CoV-2 and PMMoV gene copies between different samples is unlikely due to the size of the concentrated wastewater pellet (wet weight).
Fig. 6.
(A) SARS-CoV-2 N gene copies/L vs. wet weight (mg) (R2 = 0.113, n = 120), (B) PMMoV gene copies/L vs. wet weight (mg) (R2 = 0.029, n = 120).
4. Conclusions
-
•
We conclude that pasteurization of wastewater samples negatively impacted the detection of SARS-CoV-2 by decreasing the total RNA concentration and SARS-CoV-2 N gene copies/L measured, but not PMMoV gene copies.
-
•
SARS-CoV-2 gene copies/L were lower at −20 °C and −80 °C storage temperatures compared to 4 °C in 30 mL raw influent wastewater samples after a period of 7 and 14 days, having undergone a thawing process of 1 h in a room temperature water bath. This finding suggested that freezing/thawing of samples can be detrimental to SARS-CoV-2 RNA quantification, whereas storage time affected PMMoV gene quantity regardless of temperature.
-
•
PEG precipitation does not require an incubation period to be an effective method of viral concentration. Pellet wet-weight was a poor predictor variable of SARS-CoV-2 N and PMMoV gene copies/L.
The following is the supplementary data related to this article.
Pair-wise comparison statistics for Three-Way Analysis of Variance (balanced design, no interactions) comparing the effects of wastewater treatment plant (WWTP), storage time, and storage temperature as independent variables and quantified viral RNA copy number as the dependent variable. Results for both N1 and PMMoV are presented below, where statistical significance was set at α = 0.05, n = 72.
CRediT authorship contribution statement
Golam Islam: Conceptualization, Methodology, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing. Ashley Gedge: Methodology, Formal analysis, Investigation, Data curation, Visualization, Writing – original draft, Writing – review & editing. Linda Lara-Jacobo: Data curation, Writing – review & editing. Andrea Kirkwood: Formal analysis, Writing – review & editing, Supervision, Funding acquisition. Denina Simmons: Formal analysis, Writing – review & editing, Supervision, Funding acquisition. Jean-Paul Desaulniers: Writing – review & editing, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors wish to acknowledge that the financial support for the conduction of the research was supported by funds from the MECP (Ministry of the Environment, Conservation and Parks) and Province of Ontario's “Wastewater Surveillance Initiative” awarded to Dr. Jean-Paul Desaulniers, Dr. Denina Simmons and Dr. Andrea Kirkwood. In addition, funding was also provided by a post-doctoral fellowship awarded to Dr. Golam Islam by OCWA (Ontario Clean Water Agency), MITACS, and Cole Engineering Group Ltd. The authors also acknowledge the help and assistance from Ontario Tech University, The Regional Municipality of Durham's Health and Works Departments, and all their employees involved in the project during this study. Their time, facilities, resources, and thoughts provided throughout the study helped the authors greatly. Lastly, the authors would like to declare that views expressed in the publication are their own and do not necessarily reflect those of the Province of Ontario, and The Regional Municipality of Durham's Health and Works Departments. The funding source(s) was not involved in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Editor: Warish Ahmed
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., Mcminn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M., Simpson S.L., Tandukar S., Thomas K.V., Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimization and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020;17:82–93. doi: 10.1016/j.coesh.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Simpson S., Bertsch P., Bibby K., Bivins A., Blackall L., Bofill-Mas S., Bosch A., Brandao J., Choi P., Ciesielski M., Donner E., D'Souza N., Farnleitner A., Gerrity D., Gonzalez R., Griffith J., Gyawali P., Haas C., Shanks O. 2021. Minimizing Errors in RT-PCR Detection and Quantification of SARS-CoV-2 RNA for Wastewater Surveillance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso E., Garcia-Luque I., De La Cruz A., Wicke B., Avila-Rincon M.J., Serra M.T., Castresana C., Diaz-Ruiz J.R. Nucleotide sequence of the genomic RNA of pepper mild mottle virus, a resistance-breaking tobamovirus in pepper. J. Gen. Virol. 1991;72(12):2875–2884. doi: 10.1099/0022-1317-72-12-2875. [DOI] [PubMed] [Google Scholar]
- Bar-Or I., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E., Mannasse B., Shirazi R., Kramarsky-Winter E., Nir O., Abu-Ali H., Ronen Z., Rinott E., Lewis Y.E., Friedler E., Bitkover E., Paitan Y., Berchenko Y., Kushmaro A. 2020. Regressing SARS-CoV-2 Sewage Measurements Onto COVID-19 Burden in the Population: A Proof-of-concept for Quantitative Environmental Surveillance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43(7):1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan V.W.-S., Chiu P.K.-F., Yee C.-H., Yuan Y., Ng C.-F., Teoh J.Y.-C. A systematic review on COVID-19: urological manifestations, viral RNA detection and special considerations in urological conditions. World J. Urol. 2020 doi: 10.1007/s00345-020-03246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Lou J., Bai Y., Wang M. COVID-19 disease with positive fecal and negative pharyngeal and sputum viral tests. Am. J. Gastroenterol. 2020;115(5):790. doi: 10.14309/ajg.0000000000000610. 10.14309/ajg.0000000000000610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R., Ng Y.Y., Chu M.Y., Chung T.W.H., Tam A.R., Yip C.C.Y., Leung K.-H., Fung A.Y.-F., Zhang R.R., Lin Y., Cheng H.M., Zhang A.J.X., To K.K.W., Chan K.-H., Leung W.K.… Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159(1):81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chik A., Glier M.B., Servos M., Mangat C.S., Pang X.L., Qiu Y., D’Aoust P.M., Burnet J.B., Delatolla R., Dorner S., Geng Q., Giesy J.P., Jr., McKay R.M., Jr., Mulvey M.R., Jr., Prystajecky N., Jr., Srikanthan N., Jr., Xie Y., Jr., Conant B., Jr., Hrudey S.E., Jr., Canadian SARS-CoV-2 Inter-Laboratory Consortium Comparison of approaches to quantify SARS-CoV-2 in wastewater using RT-qPCR: results and implications from a collaborative inter-laboratory study in Canada. 2021;107:218–229. doi: 10.1016/j.jes.2021.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Mercier E., Montpetit D., Jia J.J., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Langlois M.A., Servos M.R., MacKenzie M., Figeys D., MacKenzie A.E., Graber T.E., Delatolla R. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188 doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Cassi X., Scheidegger A., Bänziger C., Cariti F., Tuñas Corzon A., Ganesanandamoorthy P., Lemaitre J.C., Ort C., Julian T.R., Kohn T. Wastewater monitoring outperforms case numbers as a tool to track COVID-19 incidence dynamics when test positivity rates are high. Water Res. 2021;200 doi: 10.1016/j.watres.2021.117252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Kishida N., Konno Y., Katayama H., Asami M., Akiba M. Occurrence of pepper mild mottle virus in drinking water sources in Japan. Appl. Environ. Microbiol. 2013;79(23):7413–7418. doi: 10.1128/AEM.02354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokajärvi A.-M., Rytkönen A., Tiwari A., Kauppinen A., Oikarinen S., Lehto K.-M., Kankaanpää A., Gunnar T., Al-Hello H., Blomqvist S., Miettinen I.T., Savolainen-Kopra C., Pitkänen T. The detection and stability of the SARS-CoV-2 RNA biomarkers in wastewater influent in Helsinki,Finland. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., Debolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pillai S.K.… First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382(10):929–936. doi: 10.1056/nejmoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.L., Baluja M.Q., Graham D.W., Corbishley A., McDonald J.E., Malham S.K., Hillary L.S., Connor T.R., Gaze W.H., Moura I.B., Wilcox M.H., Farkas K. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020;749 doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kennedy L.C., Wolfe M.K., Criddle C.S., Duong D.H., Topol A., White B.J., Kantor R.S., Nelson K.L., Steele J.A., Langlois K., Griffith J.F., Zimmer-Faust A.G., Mclellan S.L., Schussman M.K., Ammerman M., Wigginton K.R., Bakker K.M., Boehm A.B. 2021. SARS-CoV-2 RNA Is Enriched by Orders of Magnitude in Solid Relative to Liquid Wastewater at Publicly Owned Treatment Works. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Sassi H.P., Torrey J.R. Pepper mild mottle virus as a water quality indicator. Npj CleanWater. 2018;1(1) doi: 10.1038/s41545-018-0019-5. [DOI] [Google Scholar]
- Kocamemi B.A., Kurt H., Sait A., Sarac F., Saatci A.M., Pakdemirli B. 2020. SARS-CoV-2 Detection in Istanbul Wastewater Treatment Plant Sludges. [DOI] [Google Scholar]
- Krajden M., Minor J.M., Rifkin O., Comanor L. Effect of multiple freeze-thaw cycles on hepatitis B virus DNA and hepatitis C virus RNA quantification as measured with branched-DNA technology. J. Clin. Microbiol. 1999;37(6):1683–1686. doi: 10.1128/jcm.37.6.1683-1686.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Bonanno Ferraro G., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis G.D., Metcalf T.G. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 1988;54(8):1983–1988. doi: 10.1128/aem.54.8.1983-1988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Di D., Saingam P., Jeon M.K., Yan T. Fine-scale temporal dynamics of SARS-CoV-2 RNA abundance in wastewater during a COVID-19 lockdown. Water Res. 2021;197 doi: 10.1016/j.watres.2021.117093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y., Xu S.B., Lin Y.X., Tian D., Zhu Z.Q., Dai F.H., Wu F., Song Z.G., Huang W., Chen J., Hu B.J., Wang S., Mao E.Q., Zhu L., Zhang W.H., Lu H.Z. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. 2020;133(9):1039–1043. doi: 10.1097/CM9.0000000000000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes-Júnior L.C., Bomfim E., Silveira D.S.C.D., Pessanha R.M., Schuab S.I.P.C., Lima R.A.G. Effectiveness of mass testing for control of COVID-19: a systematic review protocol. BMJ Open. 2020;10(8) doi: 10.1136/bmjopen-2020-040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., du Plessis L., Liu Z., Hill V., Kang M., Lin H., Sun J., François S., Kraemer M., Faria N.R., McCrone J.T., Peng J., Xiong Q., Yuan R., Zeng L., Zhou P., Liang C., Yi L., Liu J., Xiao J., Ke C.… Genomic epidemiology of SARS-CoV-2 in Guangdong Province,China. Cell. 2020;181(5):997–1003.e9. doi: 10.1016/j.cell.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markt R., Mayr M., Peer E., Wagner A.O., Lackner N., Insam H. 2021. Detection and Stability of SARS-CoV-2 Fragments in Wastewater: Impact of Storage Temperature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Meyer H.W., Calcott P.H. Freezing and Thawing Microbes (Patterns of Progress, 14). 68 S., 12 Abb., 4 Tab. Shildon, Co. Durham 1978. Meadowfield Press Ltd. $ 8.40. 1980;20(4):295. doi: 10.1002/jobm.19800200410. [DOI] [Google Scholar]
- Morley C.R., Trofymow J.A., Coleman D.C., Cambardella C. Effects of freeze-thaw stress on bacterial populations in soil microcosms. Microb. Ecol. 1983;9(4):329–340. doi: 10.1007/bf02019022. [DOI] [PubMed] [Google Scholar]
- Oran D.P., Topal E.J. Prevalence of asymptomatic SARS-CoV-2 infection. Ann. Intern. Med. 2020;173(5):362–367. doi: 10.7326/m20-3012%m32491919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer E.J., Maestre J.P., Jarma D., Lu A., Willmann E., Kinney K.A., Kirisits M.J. Development of a reproducible method for monitoring SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;799 doi: 10.1016/j.scitotenv.2021.149405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/s1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasa S., Desai M., Thoguluva Chandrasekar V., Patel H.K., Kennedy K.F., Roesch T., Spadaccini M., Colombo M., Gabbiadini R., Artifon E.L.A., Repici A., Sharma P. Prevalence of gastrointestinal symptoms and fecal viral shedding in patients with coronavirus disease 2019. JAMA Netw. Open. 2020;3(6) doi: 10.1001/jamanetworkopen.2020.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson B.M., Darby E., Haas C.N., Amha Y.M., Bartolo M., Danielson R., Dearborn Y., Di Giovanni G., Ferguson C., Fevig S., Gaddis E., Gray D., Lukasik G., Mull B., Olivas L., Olivieri A., Qu Y., Sars-Cov-2 Interlaboratory Consortium Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the U.S. 2021;7(3):504–520. doi: 10.1039/d0ew00946f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo S.E., Keim E.K., Swanstrom R., Ong A., Burnor E.A., Kossik A.L., Harrison J.C., Demeke B.A., Zhou N.A., Beck N.K., Shirai J.H., Meschke J.S. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.144215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röder B., FrüHwirth K., Vogl C., Wagner M., Rossmanith P. Impact of long-term storage on stability of standard DNA for nucleic acid-based methods. J. Clin. Microbiol. 2010;48(11):4260–4262. doi: 10.1128/jcm.01230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323(22):2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- Simpson A., Topol A., White B.J., Wolfe M.K., Wigginton K.R., Boehm A.B. Effect of storage conditions on SARS-CoV-2 RNA quantification in wastewater solids. PeerJ. 2021;9 doi: 10.7717/peerj.11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele J.A., Zimmer-Faust A.G., Griffith J.F., Weisberg S.B. 2021. Sources of Variability in Methods for Processing, Storing, and Concentrating SARS-CoV-2 in Influent From Urban Wastewater Treatment Plants. [DOI] [Google Scholar]
- US Centers for Disease Control and Prevention 2019-Novel coronavirus (2019-nCoV) real-time rRT-PCR panel primers and probes. 2019. https://www.cdc.gov/coronavirus/2019-ncov/downloads/rt-pcr-panel-primer-probes.pdf
- Van Vinh Chau N., Lam V.T., Dung N.T., Yen L.M., Minh N., Hung L.M., Ngoc N.M., Dung N.T., Man D., Nguyet L.A., Nhat L., Nhu L., Ny N., Hong N., Kestelyn E., Dung N., Xuan T.C., Hien T.T., Phong N.T., Tu T., Oxford University Clinical Research Unit COVID-19 Research Group The natural history and transmission potential of asymptomatic severe acute respiratory syndrome coronavirus 2 infection. 2020;71(10):2679–2687. doi: 10.1093/cid/ciaa711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan,China. JAMA. 2020;323(11):1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney O.N., Kennedy L.C., Fan V.B., Hinkle A., Kantor R., Greenwald H., Crits-Christoph A., Al-Shayeb B., Chaplin M., Maurer A.C., Tjian R., Nelson K.L. Sewage, salt, silica, and SARS-CoV-2 (4S): an economical kit-free method for direct capture of SARS-CoV-2 RNA from wastewater. Environ. Sci. Technol. 2021;55(8):4880–4888. doi: 10.1021/acs.est.0c08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J., Gilbert J.A. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5(4):e00614–e00620. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Gui X., Xiong Y. Comparison of clinical characteristics of patients with asymptomatic vs symptomatic coronavirus disease 2019 in Wuhan,China. JAMA Netw. Open. 2020;3(5) doi: 10.1001/jamanetworkopen.2020.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Breitbart M., Lee W.H., Run J.-Q., Wei C.L., Soh S.W.L., Hibberd M.L., Liu E.T., Rohwer F., Ruan Y. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2005;4(1) doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pair-wise comparison statistics for Three-Way Analysis of Variance (balanced design, no interactions) comparing the effects of wastewater treatment plant (WWTP), storage time, and storage temperature as independent variables and quantified viral RNA copy number as the dependent variable. Results for both N1 and PMMoV are presented below, where statistical significance was set at α = 0.05, n = 72.