Abstract
Coronavidae viruses, such as SARS-CoV, SARS-CoV-2, and MERS-CoV, cause severe lower respiratory tract infection, acute respiratory distress syndrome and extrapulmonary manifestations, such as diarrhea and fever, eventually leading to death. Fast, accurate, reproductible, and cost-effective SARS-CoV-2 identification can be achieved employing nano-biosensors, reinforcing conventional methodologies to avoid the spread of COVID-19 within and across communities. Nano-biosensors built using gold, silver, graphene, In2O3 nanowire and iron oxide nanoparticles, Quantum Dots and carbon nanofibers have been successfully employed to detect specific virus antigens – nucleic acid sequences and/or proteins –or host antibodies produced in response to viral infection. Biorecognition counterpart molecules have been immobilized on the surface of these nanomaterials, leading to selective virus detection by optical or electrochemical transducer systems. This systematic review assessed studies on described and tested immunonsensors and genosensors designed from distinct nanomaterials available at the Pubmed, Scopus, and Science Direct databases. Twenty-three nano biosensors were found suitable for unequivocal coronavirus detection in clinical samples. Nano-biosensors coupled to RT-LAMP/RT-PCR assays can optimize RNA extraction, reduce analysis times and/or eliminate sophisticated instrumentation. Although promising for the diagnosis of Coronavidae family members, further trials in large populations must be adequately and rigorously conducted to address nano-biosensor applicability in the clinical practice for early coronavirus infection detection.
Keywords: Nanodevices; Genosensors and imunnosensors; Signal transducers, minimal sample manipulation; Coronavirus detection; COVID-19 diagnosis
Graphical Abstract
1. Introduction
SARS‐CoV, the virus that causes the severe acute respiratory coronavirus syndrome SARS-CoV-2, and MERS-CoV, the causative agent of Middle East respiratory syndrome infection, have brought the world to attention in the last decades [1], [2], [3], [4].
The genome of these viruses presents high homology, with SARS-CoV-2 displaying a 79.5% homology to the SARS-CoV genome and 50% homology to the MERS-CoV genome. Besides the pathogenicity common to the members of this family, SARS-CoV-2, the etiologic agent of COVID-19 may cause severe infection and overall inflammation status, culminating with multiple organ failure, and is a highly transmissible virus. The SARS-CoV-2 genome comprises a 30 kb single positive-stranded RNA encoding 10 genes [5], [6], [7], [8], a spike (S) glycoprotein with affinity to the host cell receptor, known as angiotensin-converting enzyme 2 (ACE 2), the small envelope (E) and matrix (M) protein, both essential for virus assembly, the nucleocapsid (N) protein, and several other accessory proteins. SARS-CoV-2 can enter cells by recognizing the angiotensin-converting enzyme 2 (ACE2) through a viral receptor-binding domain located in the S protein [9], [10]. The M protein is responsible for transmembrane nutrient transport and virus envelope formation, and N and E proteins, as well as other several accessory proteins, obstruct the host immune response and present different functions [11].
The S and N proteins are among the most valuable antigen biomarkers for the diagnosis of COVID-19 [12], [13], [14]. The S protein displays the lowest sequence homology rate among the SARS-CoV and MERS virus genomes, indicating that it may be used as an antigen target in SARS-CoV-2 identification [6], [15], [16], [17].
Infected and asymptomatic individuals can quickly spread the disease, increasing the number of disease focal points and deaths [11], [18]. In November 2002, SARS-CoV emerged and culminated in over 8,000 cases, followed by MERS-CoV in January 2018, resulting in 2,143 confirmed cases, while SARS-CoV-2 has led to the current COVID-19 pandemic that began in December 2019 [19], [20], [21]. It is estimated that the number of confirmed COVID-19 cases and deaths have reached 167,492,769 and 3,482,907, respectively, on 26th May, 2021, according to the World Health Organization (WHO) [22].
SARS-CoV spread quickly and globally, resulting in large-scale outbreaks in different countries on all continents [23]. Early diagnosis of SARS-CoV-2-infected individuals is required to contain the COVID-19 pandemic and has been recognized as a successful strategy in several countries [17]. In the future, quick and easy SARS-CoV-2 diagnoses will represent the most powerful epidemiological tool and innovative methods must be developed to test and ensure the health status of large populations.
SARS-CoV-2 detection or quantification based on RNA sequences coding for viral proteins has been applied in the detection of the virus in clinical, biological, and environmental samples employing reverse transcriptase PCR (RT-PCR) [24] and quantitative RT-PCR (qRT-PCR) [25], [26], [27]. Immunological assays, such as lateral flow immunoassays (LFIA), enzyme-linked immunosorbent assays (ELISA), and chemiluminescence enzyme immunoassays (CLIA).have also been used to detect the presence of IgM and/or IgG immunoglobulins against SARS-CoV-2 in blood samples [28],[10].
A breakthrough in PCR detection systems comprises loop-mediated isothermal amplification (LAMP), allowing for coronavirus DNA detection in less than 60 min [29]. However, to prevent the transmission of infectious diseases in large populations, comprehensive but unequivocal and low-cost virus detection testing methods are required. In this sense, biosensors comprise portable, less expensive, simpler, and faster systems capable of detecting viruses in biological samples.
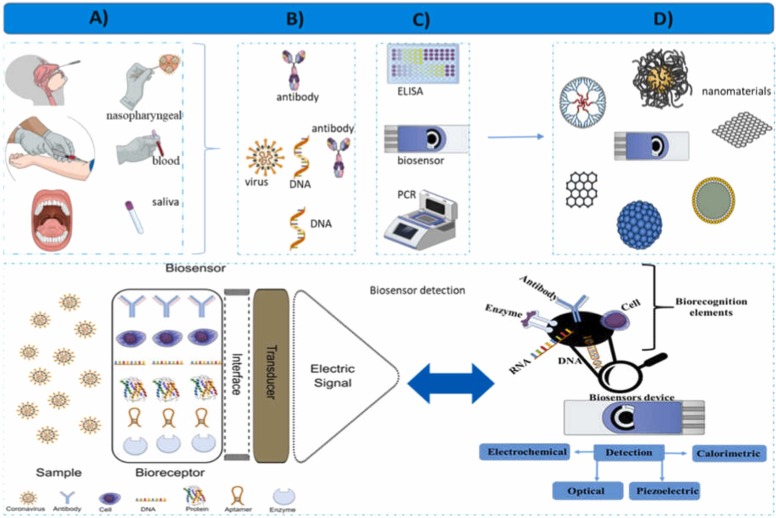
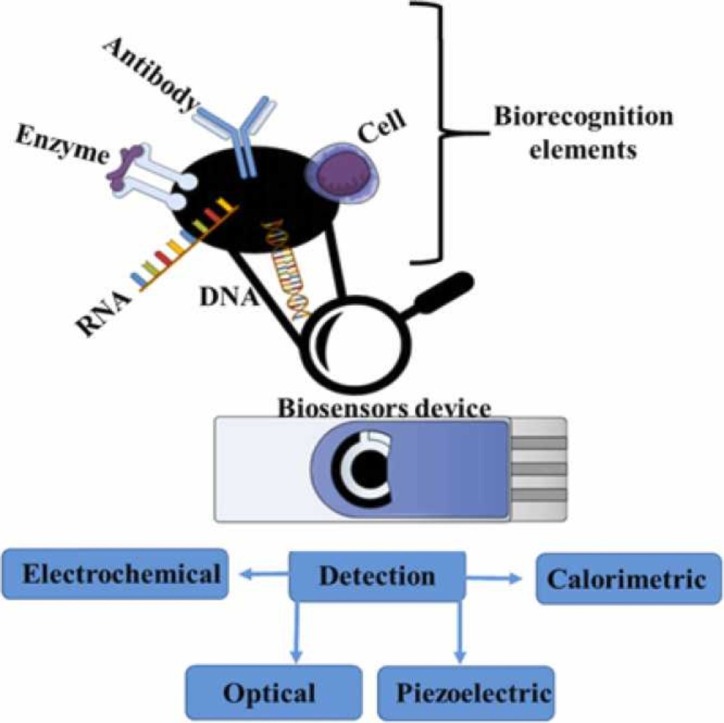
The possibilities concerning nano-analytical devices consisting of biological recognition apparatuses for the identification of coronavirus target molecules employing physical-chemical transducers to convert viral recognition into analytical signals are displayed in Fig. 1.
Fig. 1.
Nanodevice design for coronavirus detection, (A) sampling, (B) DNA and protein targets, (C) physicochemical coronavirus detection methods, (D) nanomaterials used in biosensor manufacturing, and (E) schematic biosensor components and biosensor detection.
Different nanomaterials can be used to manufacture nanodevices displaying unique and novel features, functionalities, and applicabilities [30]. The diagnosis and unequivocal biorecognition of COVID-19 employing virus biomarkers and biochemical indicators such as antigens, RNA sequences, proteins, or the entire virus itself can be performed by binding target molecules to these devices, and nano-biosensors carrying these biomarkers provide accurate, fast, and precise viral identification [31]. The combination of these elements makes it possible to design highly sensitive and selective nano-biosensor devices to detect SARS-CoV-2 in response to demands for early detection of infected individuals within large populations.
The present review provides a systematic analysis of innovative nano-biosensing systems applied or adapted to coronavirus detection. Different sensing strategies based on DNA receptors, aptamers, or antibodies immobilized onto nanoparticles coupled to transducers aiming at the detection of coronaviruses in clinical samples are presented and discussed. Nano-biosensors designed to detect SARS-CoV, SARS-CoV-2, and MERS-CoV viruses are discussed in terms of their effectiveness, simplicity, low costs, integration, and portability, allowing for their operation near patients by untrained people, dispensing sophisticated or expensive equipment.
2. Methodology
The search for studies concerning nano-biosensors was performed through the evaluation of keywords, titles, and abstracts. Studies that did not assess nano-biosensor applicability in the detection of coronaviruses and those employing biosensors and nanomaterials or aimed at the detection of other viruses that do not belong to the Coronavidae family were excluded. Only articles published in English from 2000 to 2021 were considered. The systematic review search and preparation followed the preferred reporting items for systematic review and meta-analyses statement (PRISMA) [32]. All available data obtained from the three scientific databases addressing biosensors employing nanoparticles for coronavirus detection were included.
2.1. Focus questions
Focus questions were prepared based on the problem, intervention, comparison, and outcome (PICO) method, established as follows: Which nanoparticle-based biosensors are used for coronavirus detection? What strategies have been applied the most concerning DNA-based biosensors or immuno-based biosensors for coronavirus detection? Which nano-biosensor exhibits the lowest limit of detection (LOD)? Which coupled transducers were used to build these biosensors?
2.2. Eligibility and selection criteria
Scientific data were collected from the PubMed, Scopus, and Science Direct databases. followed four main steps: (i) guiding question formulation; (ii) database searching; (iii); eligibility; and (iv) inclusion.
The selection and exclusion criteria listed in Table 1 are based on studies using nanomaterials and biosensors for the detection of coronaviruses and assessments regarding their LOD. This review considered only original research articles. The studies were selected based on the reading of titles and abstracts independently identified and, when these were not enough, the entire study was carefully read.
Table 1.
Inclusion/exclusion criteria presented in order of application.
| Inclusion criteria | Exclusion criterion |
|---|---|
| Articles in English | Non-English language articles |
| Biosensors using nanostructures | Short communications, reviews and thesis |
| Biosensors and coronavirus detection | Biosensors that do not employ nanomaterials |
| Immunosensors using nanostructures | Biosensors that were not applied for coronavirus detection |
| Nanostructures functionalized with aptasensors | – |
| Genosensor using nanostructures | – |
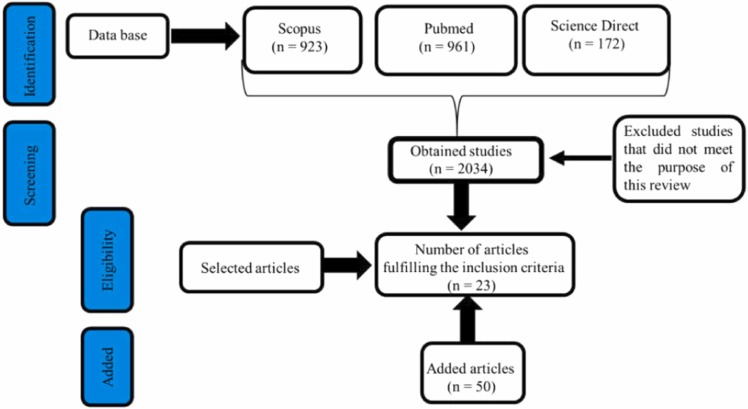
The flowchart of the study selection carried out herein is displayed in Fig. 2.
Fig. 2.
Flowchart concerning the selection of studies included in this systematic review following the PRISMA flow diagram.
2.3. Research strategy/database
This systematic review was conducted on November 23, 2020. A search was performed for articles published between 2000 and 2021 at the PubMed, Scopus, and Science Direct databases. The search strategy focused on the type of nanoparticles used for building biosensors designed for the detection of coronaviruses and was restricted to English-language articles. The inclusion criteria were as follows: (i) keyword identification; (ii) synonyms based on relevant studies on nanoparticle-based biosensors and coronavirus; and (iii) use of boolean operators, such as "AND", "OR" and " * ".
The search was performed using the following components for every database:
Search component 1 (SC1), including the key terms: Nanoparticle OR Nano OR Nanomaterial OR Nanocrystal AND Coronavirus.
Search component 2 (SC2), including the key terms: Biosensor OR Biosensing OR Biosens AND diagnosis OR detection.
2.4. Information sources
2.4.1. Data extraction and study quality
Data concerning the LOD, type of nanoparticles and transducers, coronavirus family member, linear range and type of clinical/biological sample(s) and studies revised by two independent reviewers were selected.
2.4.2. Risk of bias assessment
Bias sources may be related to the established study selection and exclusion criteria, such as the choice of articles written only in English, publication period, and the exclusion of reviews and editorials.
3. Results
3.1. Study selection
A total of 172 studies were found at the Science Direct database, 923 at the Scopus database, and 961 at the PubMed database, totaling 2,056 studies. After 22 duplicate or triplicate exclusions, a total of 2,034 studies remained. Study selection first comprised title and abstract readings, and the studies were or were not included according to the inclusion/exclusion criteria detailed in the methodology section. At the end of this process, 23 studies were selected based on the priority information defined as the use of nanoparticles in biosensor development and their use in coronaviruses detection. Other studies found outside the searching platforms were also included, as they added information on coronavirus biology and host transmission model. To better discuss the data from the selected articles, 50 additional studies were included, comprising both experimental and reviews, in order to provide theoretical and experimental support or complete information improving the discussion of data presented in the eligible papers, totaling 73 articles included in this review.
3.2. Conventional methods for coronavirus detection
Concerning COVID-19, the World Health Organization (WHO) recommends the collection of samples from the upper respiratory tract through nasopharyngeal swabs or from the lower respiratory tract employing endotracheal sputum aspiration [33]. Coronavirus mRNA can be detected by the amplification of specific sequences through the transcriptase reverse-polymerase chain reaction (RT-PCR) method, considered the gold standard for molecular diagnosis [34]. Although several improvements have been made to this methodology to reduce the analysis time, it can still take from 40 min to 3 h, and requires thorough sample handling, trained technicians, certified laboratories and mid-cost equipment. mRNA extraction is a crucial pre-analytical step for RT-PCR application, as it may generate false-negative results if not conducted appropriately. These limitations make it difficult to develop this methodology into a field application or for the screening of a significant number of samples as required during the current COVID-19 pandemic, which necessitates a quick but unequivocal diagnosis [35].
Serological tests based on enzyme-linked immunosorbent assay (ELISA) for IgM antibody detection can also be used for coronavirus diagnosis, but the period between infection and seroconversion analysis to carry out these tests make them unfeasible for early and large-scale diagnoses [14], [28], [36], [34]. Based on these considerations, the development of highly sensitive screening tests able to detect lower IgM antibody levels or lower viral loads may be useful in diagnosing the early stages of viral infection.
3.3. Nano-biosensor systems applied for coronavirus detection
Nanomaterials (NMs) less than 100 nm in size provide a greater surface volume, enhancing their intrinsic physicochemical properties and increasing their susceptibility to external influences when compared to the bulk material. Depending on NM composition, morphology, and size, these compounds may be used in very specialized applications, such as in special nanostructures made from iron or transition metals whose magnetic properties can be used for biosensing [30], [38], [39], [40].
NMs improve the performance of biosensors by enhancing signal amplification, thus increasing detection sensitivity. Nanostructured materials such as carbon nanotubes (CNTs), graphene, metal nanomaterials, magnetic nanoparticles (MNPs) and quantum dots (QDs) can be applied in biosensors combined to the electrochemical detection of viruses present in different samples [30]. NMs such as gold or lanthanide-doped polystyrene nanoparticles (NPs) and graphene iron oxides carrying biological recognition elements immobilized onto their surfaces and coupled to transducers can be used for coronavirus identification in clinical samples [13], [21], [41], [42], [43].
To this end, biorecognition must be attained in order to identify DNA (genosensors), aptamers (aptasensors) or antibodies (immunosensors) in viral particles. The detection of SARS-CoV-2 through different biorecognition molecules has been assessed, including sequences from ORF 1 ab, the nucleocapsid phosphoprotein gene (N gene), RNA-dependent RNA polymerase, (RdRp gene), the E gene or E protein, and the spike protein (S protein), or utilizing two targets, for example, protein N and ORF 1 ab, or E and S proteins, or S and RdRp, as well as other combinations [21], [44], [45], [46], [47].
The general design of nano-biosensors aimed at Coronavidae family member identification displayed in Fig. 1 demonstrates the numerous possibilities of nanobiosensors in detecting or measuring Coronavidade family member viral loads.
Biosensing systems can be applied for coronavirus screening of several clinical samples, such as saliva, nasopharyngeal swab specimens, endotracheal sputum aspirates, and blood [13], [35], [44]. The most common transducers used in biosensor systems comprise the combined effect of plasmonic photothermal (PPT) and localized surface plasmon resonance (LSPR), plasmonic effect based colorimetric biosensing, colorimetric, fluorescence, field-effect transistor-based amperometric and circle to circle amplification-based optomagnetics [13], [21], [35], [42], [43], [47].
3.4. Nano-biosensors for coronavirus screening and quantification in clinical samples: Exploring biorecognition molecules, aptasensors, genosensors and immunosensors
The results of this systematic review addressing nano-biosensors, biorecognition elements, nanomaterials, sensitivity, and detected Coronaviridae members family are presented in Table 2.
Table 2.
Nano-biosensor designs addressing the detection and quantification of Coronavidae family members considering types of clinical sample, nanomaterial, bioreceptor molecules, and transducers.
| Nano-material | Linear Range | Immobilized Viral Element | LOD | Transducer | References | |||
|---|---|---|---|---|---|---|---|---|
| IMMUNOSENSOR | MERS-CoV | AuNPs | 0.001–100ng/mL | Recombinant protein S | 1.0 pg/mL | Voltametric | [15] | |
| SARS-CoV | In2O3 nanowire | nd | Antibody mimic proteins (AMPs) to detect N protein | < 1 nM | FET | [54] | ||
| Gold binding polypeptides | nd | Fusion of polypetpides to coronavirus surphace antigen | 200ng/mL | SPR | [51] | |||
| SARS-CoV-2 | Graphene | nd | Specific antibody against S protein | 1.6 x 101 pfu/mL and 2.42 x 102 copies/mL | FET | [13] | ||
| AuNPs | 1 fM - 1 µM | Monoclonal antibody | 0.09 pM | Electrochemical | [44] | |||
| AuNPs | nd | Anti-human IgM and IgG antibodies | nd | Colorimetric | [35] | |||
| LNPs | nd | Recombinant N protein | nd | Optical | [42] | |||
| AuNPs | nd | Antibodies against surface proteins - S, E and M | Positive if Ct <35 at real-time PCR<< | Colorimetric | [53] | |||
| AuNPs | nd | N protein monoclonal antibodies | 103-104 virus copies/0.1mL corresponding to 10-18 M of N protein | Colorimetric | [55] | |||
| Carbon nanofiber-modified | 0.1 pg/mL - 1 μg/mL | N protein | 0.8 pg/mL | Electrochemical | [14] | |||
| Graphene | nd | S antibodies | 0.02 mg/mL S recombinant protein | Electrochemical | [17] | |||
| AuNRs AuNRs. |
nd | Recombinant N protein S antibodies |
F10 4E-5; F11 8.5E-5, and BK7-prism 7E-5 RIU* | SPP SPR |
[56] [57] |
|||
| GENOSENSOR | MERS-CoV | AuNPs | nd | 30 bp dsDNA from upE and ORF1a | 1 pmol/μL | Colorimetric- LSPR e |
[58] | |
| AgNPs | nd | acpcDNA | 1.53 nM | Colorimetric | [59] | |||
| SARS-CoV | QDs | nd | RNA aptamer | 0.1 pg/mL | Optical | [48] | ||
| AuNPs | 2.5 - 50 pmol/L | ssDNA | 2.5 pmol/L | Electrochemical | [49] | |||
| SARS-CoV-2 | AuNPs | 1 pM – 1 nM | Synthetic cDNA | 0.22 pM | PPT + LSPR | [47] | ||
| SPCnAuE | 2.5 – 50 pM | 30-mer oligonucleotide | 2.5pM | Voltametric | [60] | |||
| NPs | nd | FITC-labeled primers from ORF1ab and N gene | 12 copies/reaction | mRT-LAMP coupled toLFB h | [61] | |||
| pcMNPs (iron) | 10 - 105 copies/reaction | ORF l ab and N gene | 10 copies/reaction | RT-PCR i | [62] | |||
| Au-colloid | cDNA - RdRp gene | 0.05 ng RNA | Colorimetric | [46] | ||||
| AuNPs | 0.2 – 3 ng μL-1 | ASOS | 0.18 ng/μL | SPR-colorimetric | [21] | |||
| Iron oxide NPs | 10 – 100 pM | cDNA - RdRp gene | 0.4 fM | Optomagnetic | [43] | |||
| AuNPs | nd | ssDNA against N gene sequence | 6.9 copies/μL | Electrochemical | [63] | |||
LOD – minimum limit of detection (as included in the original articles); AuNPs - gold nanoparticles; FET - field-effect transistors; SPR - surface plasmon resonance; SPP - surface plasmon polaritons;; LSPR - surface plasmon resonance; PPT -plasmonic photothermal; mRT-LAMP - transcription loop-mediated isothermal amplification; LBF - lateral flow biosensor; RT-PCR - real-time reverse-transcription; C2CA - circle-to-circle amplification; ssDNA - single strand DNA; IgM - immunoglobulin M; IgG – immunoglobulin G; LNPs -lanthanide-doped polystirene nanoparticles; AuNRs - gold nanorods; Ct – cycle threshold; upE - E gene; ORF - open reading frames; RdRp - RNA-dependent RNA 34 polymerase; SA-DNPs -streptavidin-coated nanoparticles; pcMNPs – carboxil-coated magnetic nanoparticles; acpcDNA: pyrrolidinyl peptide nucleic acid; QDs -quantum dots; RNA aptamer – synthetic RNA probe from N protein gene; ssDNA: 45-nucleotide sequence from N protein gene; cDNA synthetized from RdRp and ORF1 ab; SPCnAuE: nanogold structured-screen-printed carbon electrode; 30-mer oligonucleotide; synthetic sequence from SARS genome comprising the 29218-29247 region; FITC (fluorescein)-/digoxin- and biotin-labeled primers; pcMNPs - poly (amino ester) with carboxyl groups (PC)-coated magnetic nanoparticles; ASOS - thiol-modified antisense oligonucleotides specific for N-gene; cDNA – complementary DNA synthetized from RNA sequence
Nucleotide-based biosensors or genosensors are designed by immobilizing a specific single-strand nucleotide employing a ligand, such as thiol or biotin, to the biosensor matrix. When samples contain viral RNA, hybridization occurs between the target sequence in the virus genome and the ssDNA probe coupled to the biosensor matrix, where the double-strand ssDNA probe forms a 3D double-strand nucleic molecule and becomes stable, allowing for signal amplification by a transducer [31]. Since the hybridization is very stringent, DNA-based sensors can be applied to detect SARS-CoV-2 in very complex microbiomes, as observed in clinical samples.
In nanodevices, DNA or RNA aptamer and nucleic acid sequences comprise biorecognition elements that bind to DNA or RNA targets with high specificity and affinity. Some advantages are noted for aptamer-based biosensors, such as full clinical applications in the detection of distinct pathogens and viruses due to the uniformity of different batches, allied to high stability and the possibility of adjusting the sequence to be synthesized according to the target. Aptamers comprise promising biorecognition effectiveness and are becoming increasingly important for coronavirus detection due to the superior stability of aptamer molecules, as well as their rapid response, low labor requirements and low cost, thus becoming superior to antibody detection [31], [48]. The broad use of aptamers for virus detection lies in the fact that they can be designed to target any virus molecule, such as genes, proteins, antigens, or even host antibodies synthesized during viral infection [37], [49].
Antibodies are highly versatile concerning immunobiosensor antibody antigen-binding interactions onto surfaces, and are, thus, capable of diagnosing SARS-CoV-2 in clinical samples. Immunosensors, genosensors or aptasensors can be classified according to the type of coupled transducer, although most are electrochemical or optical [44], [50].
3.5. Nanometric-scale immnunobiosensors for the screening and quantification of Coronaviridae family members
When employed in biosensors, NMs display the advantage of a high signal-to-noise ratio aimed at highly sensitive and selective diagnoses, as noted for the nanodevices included in Table 2. Some nano-biosensors have been developed for the analysis of clinical samples with no pre-treatment and results are obtained after only a few minutes. For example, in one special nanodevice, sample collection and detection tools have been joined into a single platform.
Immunobiosensors can detect the immunocomplex (Ab-Ag) with high specificity, where the antibodies are able recognize the entire SARS-CoV-2 virus or virus molecules such as protein or other viral compounds [51]. The recognition mechanism involves the bioaffinity between a monoclonal or polyclonal antibody (depending on the epitope) and the antigen, with very high affinity represented by an association constant (Ka = [Ab-Ag]/[Ab][Ag]) of up to 1015 [51]. A monoclonal antibody should be preferred for SARS-CoV-2, privileging a single epitope in order to decrease cross-reactivity and false-positives [51].
A carbon nanofiber-modified screen-printed electrode was functionalized with diazonium electrografting combined to the N protein coated with absorbing cotton-padding for swabbing the nasopharyngeal cavity of patients and tested against three positive RT-PCR samples. The quantification of the viral antigen was performed using a competitive assay with a fixed amount of N protein antibody, with an LOD of 0.8 pg mL−1. In addition to displaying high efficiency, the nanodevice was also highly selective, as no significant cross-reactivities were observed against influenza A and HCoV antigens [14].
An electrochemical immunosensor used for MERS-CoV diagnoses in artificially contaminated nasal samples was prepared from an array of carbon electrodes modified by the electrodeposition of gold nanoparticles functionalized with cysteamine to create a SAM and incubation with glutaraldehyde. A multiplex detection system was created by immobilizing both MERS-CoV S and human coronavirus (HCoV) recombinant protein antigens at 10 μg mL−1. The analytical performance of the immunobiosensor was conducted by square wave voltammetry (SWV), monitoring changes in the peak current at − 0.05 V vs. the Ag/AgCl electrode work potential and electrochemical measurements performed using a ferrocyanide/ferricyanide probe for 20 min, with linear responses from 0.001 to 100 ng.mL−1 for MERS-CoV and from 0.01 to 10,000 ng.mL−1 for HCoV detection in artificial nasal fluid samples. [15].
A combination of an optical surface plasmon resonance (SPR) platform to gold binding polypeptides (GBPs) and a SARS coronavirus surface antigen (SCVme) bound to gold-patterned chips prepared by photolithography has also been developed for rapid SARS-CoV diagnosis. The gold-micropatterned substrate was functionalized by immersion in GBP-E-SCVme fusion protein suspension. Anti-SCVme recognition formed a stable and specific sensing platform for the SARS antigen when tested on mouse IgG samples. Atomic force microscopy (AFM) and SPR imaging were both employed to assess anti-SCVme bonding between the protein and gold-micropatterned chips. This biosensor is simple, sensitive, selective, and able to detect up to 200 n g mL−1 anti-SCVme from SARS-CoV after 10 min [52].
A simple and rapid immunodiagnostic method using lanthanide-doped polystyrene nanoparticles (LNPs) directed towards a recombinant nucleocapsid protein (N) based on the lateral flow immunoassay (LFIA) has also been developed. Polystyrene nanoparticles were functionalized with mouse anti-human IgG antibody (M-HIgG) and rabbit IgG (RIgG) using a solution containing 1% of EDC and 1% of sulfo-NHS for 30 min. The functionalized LNPs were resuspended in blocking buffer followed by ultrasonication. Recombinant N protein was dispensed onto a nitrocellulose membrane to bind specific anti-SARS-CoV-2 IgG present in human serum samples diluted 1:1000-fold. The assay lasts 10 min and the results are consistent with RT-PCR applied to positive and negative clinical samples [42].
Graphene nano-biosensors coupled to optical and electrochemical transducers have also been created. The fabricated graphene-based device was functionalized with 1-pyrenebutanoic acid succinimidyl ester (PBASE) in methanol at room temperature by 1 h and coupled to 250 μg mL−1 of SARS-CoV-2 antibodies to the S protein (COVID-19 FET sensor) for the analysis of nasopharyngeal swabs samples and SARS-CoV-2 cultures. Graphene-FET biosensors were suitable for the detection of SARS-CoV-2 at 1.6 × 101 pfu/mL in culture samples and 2.42 × 102 copies/mL in clinical samples [13].
Gold NP biosensors can be very successful in detecting SARS-CoV-2 due to certain properties, such as signal amplification, biocompatibility, conductivity, and high surface volume, as well as the possibility of immobilizing distinct bioreceptors molecules on their surface, including, single or double-stranded DNAs, aptamers and antibodies [15], [21], [35], [44], [46], [47], [52], [53].
Another successful nano biosensor was developed in-house using a fluorine-doped tin oxide electrode (FTO) coated with gold NPs (AuNPs). The eCovSens-ultrasensitive device was functionalized with 90 μg anti-nCovid19 antibody mixed with 1 mL of AuNPs buffered solution. The functionalized AuNPs were drop-casted onto the FTO electrode to detect the presence of the nCovid-19 spike antigen. The FTO immunosensor and eCovSens were able to detect nCovid-19Ag in artificially contaminated saliva samples at concentrations ranging from 1 fM to 1 µM, with an LOD of 90 fM [44].
An electrochemical immunobiosensor comprising a graphene-working electrode functionalized with anti-spike antibodies covalently attached by carbodiimide coupled to the carboxylated surface was able to detect the viral S protein with a LOD of 20 µg mL−1 after 45 min of incubation with the target. The immunosensor was designed to detect absolute changes in [Fe(CN)6]3-/4- according to the range of spike protein concentrations on the immunosensor surface [17]. Another AuNP-based biosensor using a colorimetric transducer was designed to rapidly detect SARS-CoV-2 in nasal and oropharyngeal swab samples by targeting three surface SARS-CoV-2 proteins, namely the S, E, and M proteins. Briefly, 20 nm colloidal gold nanoparticles were functionalized with antibodies against one of each of the SARS-CoV-2 proteins by photochemical immobilization technique (PIT). Polyclonal antibodies were irradiated by Ab solution drop-casting in a colloidal suspension of AuNPs. Forty-five positive and 49 negative nasal and oropharyngeal swab samples previously tested by RT-PCR were confirmed. The immunobiosensors presented sensitivity and specificity thresholds of 96% and 98%, respectively, compared to RT-PCR at cycling (Ct) of 36.5 [54].
An interesting approach able to achieve a limit of detection of 0.6 nM of N protein from SARS-CoV-2 in clinical samples collected from COVID-19 patients was employed using antibody mimic proteins (fibronectin, Fn) as the biorecognition molecules, configured in In2O3 nanowire biosensors. The fibronectin probe molecules In2O3 were functionalized by the binding of the phosphonic acid groups to the In2O3 surface. The carboxylic acid functional groups on the In2O3 surface were then activated by EDC and allowing the activated COOHs to react with trifluoroacetic acid salt (BMPH). This cost-effective and rapid biosensor improves recognition and is claimed to be superior to those developed using antibodies and nucleotide aptamers, since the use of polypeptides and antibody mimic proteins (AMPs) displaying molecular masses < 10 kDa, can replace conventional antibodies, without compromising biosensor specificity and affinity, as demonstrated by the previously described immunological methods [55].
A forty-nanometer gold particle colloid conjugated with a recombinant antigen able to bind to the receptor domain of the SARS-CoV-2 spike protein has been used to detect IgM and IgG against SARS‐CoV‐2 in blood samples within 15 min in different host stage infections. The AuNP was functionalized in a buffered solution containing SARS-CoV-2 recombinant protein. In general, this biosensor exhibited an overall testing sensitivity of 88.66% and specificity of 90.63%. Venous and fingerstick blood samples were tested, indicating significant consistency among samples and demonstrating that this biosensor may be applied to the rapid screening of SARS‐CoV‐2 infection in both symptomatic and asymptomatic patients [35], [56].
The N protein from SARS CoV-2 N was directly attached to gold nanoparticles and used to develop another rapid test for the detection of total immunoglobulins G, M, and A against the nucleocapsid protein in the first seven days of infection using a multi-target lateral flow immunoassay platform. The immunoassay achieved a 100% diagnostic specificity (95.75–100, C.I. 95%, n = 85 healthy and individuals with other infections) and 94.6% sensitivity (84.9–98.9, C.I. 95%, n = 62 SARS CoV-2 infected subjects), reinforcing the accuracy and sensitivity of early viral infection detection in clinical samples [57].
In another approach, a 10 nm gold nanorod (Au NRs) sandwich plasmonic biosensor functionalized with SARS-CoV-2 spike (S) protein antibody was developed to monitor the detection of the COVID-19 SARS-CoV-2 spike protein through the refractive index. The introduction of Au NR into the plasmonic nanosheet-based system can lead to signal augmentation and a corresponding enhancement in sensitivity. The minimum refractive index achieved was 111.11 deg RIU−1 directed towards the spike protein of SARS-CoV-2 present in positive serum/nasopharyngeal swab samples [58].
Among the studies selected herein, several have employed antigens or antibodies as biological recognition elements for the detection of coronaviruses. Immunosensors have been shown to be suitable for this diagnosis, displaying high specificity when analyzing clinical samples if the dynamics of humoral immune responses following SARS-CoV-2 infection are observed. However, other nano-biosensor systems can be considered complementary to these, such as DNA-based sensors.
3.5.1. Genosensor and aptasensor nanoparticles designed for coronavirus infection detection: Viral load screening and quantification
The use of viral DNA bound to nano-biosensors for the diagnosis of COVID-19 has also been validated in the studies included in this systematic review, and genosensors have also been proven efficient for screening and detecting coronavirus family members in clinical samples.
A genosensor with an extended form of double-stranded DNA (dsDNA) bound to AuNPs was developed employing a platform using LSPR coupled to color change for the recognition and detection of complementary dsDNA upstream of the E gene (upE) and open reading frames (ORF) 1a, achieving MERS-CoV detection within 10 min, with a LOD of 1 pmol/μL of 30 bp MERS-CoV. The authors claimed that the genosensor designed for MERS-CoV identification could be adapted for the on-site detection of other viral infections as a cost-effective device [59] .
The development of a 5 min test at a low-cost was performed using a paper-based electrochemical sensor chip to enable the digital detection of SARS-CoV-2 in samples from Vero cells infected with SARS-CoV-2 virus and clinical samples. Highly specific antisense oligonucleotides (ssDNA) targeting viral N gene from SARS-CoV-2 were conjugated to AuNPs and immobilized on a paper-based electrochemical platform enabled the detection of SARS-CoV-2 genetic material in clinical samples. This genosensor provides a significant output signal improvement, with a LOD of 6.9 copies/μL and 100% reliability when compared to 22 COVID-19 positive patients and 26 healthy asymptomatic confirmed subjects pre-tested by an FDA-approved RT-PCR COVID-19 diagnostic kit. The characteristics and performance of the designed nano sensors make them feasible for large-scale diagnosis following the adjustment of ssDNA sequence immobilized onto AuNPs [60]. Pyrrolidinyl peptide nucleic acid (acpcPNA) probes can comprise a simple, quick, and sensitive alternative to DNA and RNA probes for the identification of MERS-CoV and related viruses. This biosensor induces AgNP aggregation in the presence of the target DNA, resulting in a detectable color change, with LODs of 1.53 nM for MERS-CoV, 1.27 nM for Mycobacterium tuberculosis (MTB), and 1.03 nM for human papillomavirus (HPV). It is important to emphasize that PNA probes are chemically and biologically stable since they cannot be degraded by nucleases, are easily synthesized, and hybridize efficiently with complementary DNA strands [61].
A quantum dot (QD)-conjugate RNA aptamer has also been considered an efficient platform based on optical signal changes, allowing for the detection of the SARS-CoV N protein at 0.1 pg mL−1 employing the chip system. The nanodevice designed for the visual detection of the SARS‐CoV N protein fulfills the parameters of ease to use, high sensitivity, suitability and specificity, and the ability to perform one‐spot monitoring [48].
A dual-functional plasmonic biosensor was developed for the diagnosis of COVID-19 by combining the plasmonic photothermal (PPT) effect and LSPR in the analysis of oropharyngeal swabs. Gold nanoislands (AuNIs) were functionalized with complementary multigene DNA, RdRp, ORF 1 ab and E gene sequences, exhibiting a high sensitivity of 0.22 pM for SARS-CoV-2. Even with high similarity among Coronavidae family members, devices functionalized with RdRp sequences can accurately discriminate between Coronavidade family members, allowing for the precise detection of the specific target in a multigene mixture, being very useful and reliable for the analysis of clinical samples [47].
A naked-eye colorimetric nanodevice composed of gold nanoparticles (AuNPs), capped with thiol-modified antisense oligonucleotides (ASOs) from N gene sequences has also been successfully developed. The agglomerate between the ASO-capped AuNPs and its target RNA causes a change in surface plasmon resonance. The precipitate can be visualized after RNA-DNA hybrid cleavage by RNAase treatment in positive samples, allowing for the detection of SARS-CoV-2 RNA at a minimum concentration of 0.18 ng μL−1. The selectivity of the assay was established by testing MERS-CoV viral RNA, with a limit of detection of 0.18 ng μL−1 of SARS-CoV-2 viral load RNA [21]. Another genosensor obtained from a gold nanostructured screen-printed carbon electrode (SPCnAuE) was also developed for the diagnosis of SARS-CoV infection through targeting viral RNA recognition. A 30-mer oligonucleotide with bases comprised between numbers 29,218 and 29,247 targeting the nucleocapside gene sequence was immobilized through thiol-gold interactions, leading to a biosensor displaying an LOD of 2.5 pmol and high specificity, with the ability to discriminate a 3-base mismatch from a complementary target strand using 25% formamide [62].
A multiplex reverse transcription loop-mediated isothermal amplification (mRT-LAMP) coupled with a nanoparticle-based lateral flow biosensor (LFB) assay (mRT-LAMP-LFB) was developed for the identification SARS-CoV-2 in oropharynx swab samples and is able to detect 12 copies of ORF 1 ab and N genes per reaction within 1 h. Two LAMP primer sets targeting the ORF1ab and N genes from SARS-CoV-2 generated numerous FITC-/digoxin- and biotin-attached duplex amplicons, which were determined by LFB and visualized without instrumentation. The association of a RT-PCR assay to a nano-biosensor guarantees the PCR specificity allied to a significant time assay reduction and free-instrumentation result interpretation [63].
The RdRp gene from SARS-CoV-2 was targeted by gold and optical sensing platform where the hybridization of the oligo probe (ssDNA) to target RNA promotes a salt-dependent aggregation and changes to the gold colloid color, which goes from pink to blue in the visible spectra. The affordable gold nanoparticles biosensor was able to detect infected individuals by testing nasopharyngeal samples in less than 30 min. The optimized method exhibited a LOD of 0.5 ng of SARS-CoV-2 mRNA [46].
Another important and useful application comprises the use of nano-biosensors in optimizing conventional techniques used for the diagnosis of COVID-19 patients, where their NPs can be employed to improve RNA extraction from clinical samples, reducing the chance of false negatives. The study reports that polyamino esters containing carboxyl groups coated by magnetic nanoparticles (pcMNPs) were efficiently used to lysis and extract viral RNA within 20 min and the resulting pcMNP-RNA complexes can be incorporated into RT-PCR reactions. RT-PCR assays were performed targeting ORFl ab and N gene sequences offering a 10-copy viral RNA sequence detection with a linear correlation between 10 and 105 copies [64] .
A homogeneous and isothermal nucleic acid amplification, the circle-to-circle amplification (C2CA) method, comprised by more than one round of probe ligation and rolling circle amplification generated amplicons that hybridize to probes grafted onto magnetic nanoparticles, forming clusters, can be detected by optomagnetic transducer measurements nucleic acid sequences within virus genome in real-time where the total assay time lasts around 100 min. The method was tested using a synthetic complementary DNA to the SARS-CoV-2 RdRp coding sequence, achieving a LOD of 0.4 fM [43].
The aforementioned methodologies join the high specificity of the gold standard RT-PCR methodology to detect SARS-CoV-2 in clinical samples, with the advantages of reducing the time-assay and/or simplifying RNA extraction and should be very useful in the early infection stages, where patients may present lower viral loads and/or are asymptomatic but still able to disseminate the virus. Another advantage comprises the unequivocal discrimination of distinct virus within the Coronavidade family, as well as new SARS-CoV-2 variants to obtain knowledge of circulating viral species in certain populations and offer these data to the public health authorities.
The most common material used for coronavirus diagnosis from clinical samples in genosensors comprises AuNPs functionalized as N gene detectors [59], [60], [61], [62], [47], [46], [21]. Sequences of nucleotides from each individual gene or multiple genes within the virus genome can be immobilized onto the surface of nanoparticles for coronavirus detection in clinical samples [65], [66], [67], [68], [69], [1].
Furthermore, genosensors can be designed or adapted to detect any SARS-CoV-2 virus variant, as they can target specific regions within the viral genome, resulting in specific nano biosensors to discriminate base substitutions in RNA sequences, for rapid tracking of recent mutations and revealing which specific variant is in circulation in a given area or population.
3.6. Transduction strategies in nano-biosensors for the detection of Coronavidae viruses in clinical samples
As mentioned previously, nano-biosensors display high specificity due to their nano-based structure and linked virus capsid or core molecules or antibodies, thus comprising nanodevices employed for the specific recognition of a particular virus. However, the target recognition must be transduced into a signal generated when the viral molecule interacts with the biorecognition element. The recognition signal can be detected by several transducers, giving rise to the broad applicability of these nanodevices. Electrical signal detectors can be generated from an electrochemical transducer, i.e., amperometric, potentiometric, conductometric, or impedimetric biosensors, while optical signals can also be detected, as in SPR, chemiluminescence, fluorescence or optical fiber or SPR transducers [31].
Optical transducers are probably the most applied for clinical analyses, due to their selectivity and sensitivity, allowing for broad use in COVID-19 diagnosis [70], [71]. Colorimetric detectors suitable for sample color change assessments by the naked eye and compound quantification through optical detectors are commonly applied in nanodevices, as well as in conventional methodologies [72]. Colorimetric detectors are considered the most advantageous, as there is no need for complex instruments, combining simplicity and low costs with the possibility of performing visual detections [21].
Among these biosensors, those employing electrochemical detectors comprise a critical class. An electrochemical immunosensor for MERS-CoV diagnosis using gold NP-modified carbon electrodes was developed based on the binding competition between the virus and the immobilized viral protein for a fixed antibody concentration added to the sample. Electrochemical measurements were used to assess the obtained voltametric responses, which were linear from 0.001 to 100 ng mL−1 and from 0.01 to 10,000 ng mL−1 for MERS-CoV and HCoV, respectively, achieving LODs of 0.4 pg mL−1 and 1.0 pg mL−1, respectively [15].
Early, low-cost, and rapid diagnoses make it possible to identify infected but asymptomatic individuals, minimizing the spread of the disease. In this sense, a plasma optic fiber absorption (P-FAB) biosensor was developed for the specific detection of proteins from the SARS-CoV-2 virus in saliva samples with minimal sample manipulation [56].
3.7. Biosensors advances compared to the gold standard diagnosis for COVID-19
Developed biosensors display several advantages in comparison to the gold standard methods of PCR and ELISA in the diagnosis of COVID-19. Genes targeted by RT-PCR are the same used in biosensing methods, including the genomic sequences encoding protein N, envelope protein (E), spike protein (S), RdRp, and ORF 1ab genes [73]. Due to COVID-19 pandemic, the detection of nucleic acid sequences from SARS-CoV-2 by RT-PCR has become the leading indicator for the diagnosis of this disease. However, RT-PCR is not fast (3–6 h) and requires several steps, making this method expensive and unfeasible in high demand in developing countries, which can contribute to increased non-testing or even false-negative results [47], [21], [14].
Currently, early diagnosis is indispensable to prevent COVID-19 transmission, and serological tests like ELISA are not reliable for early diagnosis, because of the time for seroconversion, making necessary to have sensitive methods but designed for the direct detection of the viral antigen. Among coronavirus antigens, the structural proteins N, S, M, and E, but particularly, the S and N proteins, are the most commonly used as biomarkers to distinguish coronavirus family members currently employed in biosensing devices [14].
The biosensor devices described herein display several advantages including being fast and accessible, employing no or minimal low-cost equipment, and displaying reliable detection. Several types of transducer, i.e., optical and electrochemical, have been reported for COVID-19 diagnosis and all of them presented good performance [14]. The designed biosensors were proven selective, with low LODs and able to provide accurate test results within minutes, making these innovative devices ideal and reliable solution for the clinical diagnosis of COVID-19 patients, privileging real-time detection and early diagnosis.
4. Conclusions
Among the most important points to refrain the current COVID-19 pandemic is the accurate diagnosis of infected individuals, mainly those asymptomatic, and/or in the early stages of the disease. The huge number of SARS-CoV-, SARS-CoV-2-, and MERS-CoV-infected or exposed individuals demands accurate, fast, easy and low-cost diagnostic technologies. Gold-standard molecular and serological virus detection methodologies should be reinforced by novel diagnostic strategies to enhance cost-effectiveness, reducing the analysis time, and avoiding the requirement of very qualified technicians or mid-expensive equipment while still reliable for screening large populations. Nano-biosensor technology can fulfill diagnosis gaps if they exceed their performance and overcome the disadvantages of the current methods in use for virus detection and diagnosis, with the possibility of being extended to target other virus families, particularly arboviruses that affect several populations in developing countries. Nano-biosensors should be designed using the same unequivocal recognition principles of conventional methods but must overcome their disadvantages and should certainly be considered promising epidemiological devices for clinical trials to diagnose viral infections during epidemics or local outbreaks, supporting public health authority decisions. Nano-biosensors proved they can achieve similar detection efficiency as the current gold-standard techniques in usage but at competitive prices, while also being fast and simple, increasing their reliability. In this review, several novel nano-biosensors were evaluated, and the approaches and challenges in their development and improvements in their applications for the diagnosis of SARS-CoV, SARS-CoV-2, and MERS-CoV in clinical samples were discussed. Nano-biosensors designed for the diagnosis of SARS-CoV-2 infection may, thus, become the best choice and most efficient tool for viral epidemiology to follow and control viral outbreaks in the near future if the nanodevices fulfill the general WHO-recommended criteria for screening tests, namely affordable, sensitive, specific, easy-to-use, quick and robust, with requirement of little or no additional equipment.
Among the studies included herein, biosensors are being developed to target different regions of the coronavirus genome and coronavirus capsid proteins in combination with nanoparticle matrices coupled to different types of transducers. Among the selected studies discussed herein, a greater use of ORF l ab genosensors was observed where the biorecognition element was bound to gold nanoparticles that generates a signal able to be detected by colorimetry. Considering immunosensors, the device used to recognize N protein from both SARS-CoV and SARS-CoV-2, bound to gold nanoparticles and coupled to an electrochemical transducer with colorimetric detection, seems to be the best alternative. Nanotechnology is currently being applied in several areas to minimize deaths caused by coronaviruses, including the use of nanoliposomes in coating pharmaceuticals, such as mRNA SARS-CoV-2 vaccines. The COVID-19 pandemic has increased the applications of nano-based materials from diagnosis to nucleic acid delivery for vaccination.
Novelty statement
A systematic review (SR) was developed based on the applications of the different nano biosensors for the SARS-CoV, SARS-CoV-2, and MERS-CoV diagnosis; moreover, few studies focused on the detection of these coronaviruses, and there is no systematic review addressing nano biosensor.
CRediT authorship contribution statement
A. A. conceived and constructed the study idea; L.L.G.T visualized the data and P.A.R.P, V.M.F.P and C.A.C.J edited and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
A.A, C.A.C.J and V.M.F.P are supported by Fundação de Amparo à Pesquisa Carlos Chagas Filho do Estado do Rio de Janeiro (FAPERJ), grants E-E26/203.187/2019, E-26/202.815/2018 and E‐26/010.000.984/2019 Nanotechnology Network. The authors are thankful to FAPESP (2016/01919-6), CNPq (423952/2018-8; 164569/2020-0; 150450/2020-6 and 311422/2016-0) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) Brazil - Finance Code [88887.518753/2020-00].
References
- 1.Lin S.C., Ho C.T., Chuo W.H., Li S., Wang T.T., Lin C.C. Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect. Dis. 2017;17:1–10. doi: 10.1186/s12879-017-2253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for 2019-nCoV based on SARS-CoV immunological studies. Viruses. 2020;12(254):1–5. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Rabia M.W., Alhakamy N.A., Ahmed O.A.A., Eljaaly K., Aloafi A.L., Mostafa A., Asfour H.Z., Aldarmahi A.A., Darwish K.M., Ibrahim T.S., et al. Repurposing of sitagliptin-melittin optimized nanoformula against sars-cov-2: antiviral screening and molecular docking studies. Pharmaceutics. 2021;13:1–19. doi: 10.3390/pharmaceutics13030307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nies A.T., König J., Hofmann U., Kölz C., Fromm M.F., Schwab M. Interaction of remdesivir with clinically relevant hepatic drug uptake transporters. Pharmaceutics. 2021;13:1–8. doi: 10.3390/pharmaceutics13030369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020;98:495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang X., Rayner S., Luo M.H. Does SARS-CoV-2 has a longer incubation period than SARS and MERS? J. Med. Virol. 2020;92:476–478. doi: 10.1002/jmv.25708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., de Meulder D., van Amerongen G., van den Brand J., Okba N.M.A., et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo J., Chen S., Tian S., Liu K., Ni J., Zhao M., Kang Y., Ma X., Guo J. 5G-enabled ultra-sensitive fluorescence sensor for proactive prognosis of COVID-19. Biosens. Bioelectron. 2021;181 doi: 10.1016/j.bios.2021.113160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yesudhas D., Srivastava A., Gromiha M.M. COVID-19 outbreak: history, mechanism, transmission, structural studies and therapeutics. Infection. 2021;49:199–213. doi: 10.1007/s15010-020-01516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58:1–7. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nal B., Chan C., Kien F., Siu L., Tse J., Chu K., Kam J., Staropoli I., Crescenzo-Chaigne B., Escriou N., van der Werf S., Yuen K.Y., Altmeyer R. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005;86:1423–1434. doi: 10.1099/vir.0.80671-0. [DOI] [PubMed] [Google Scholar]
- 12.Liu J., Han P., Wu J., Gong J., Tian D. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin. Chim. Acta. 2020;505:172–175. doi: 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., et al. Rapid Detection of COVID-19 Causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 14.Eissa S., Zourob M. Development of a low-cost cotton-tipped electrochemical immunosensor for the detection of SARS-CoV-2. Anal. Chem. 2021;93:1826–1833. doi: 10.1021/acs.analchem.0c04719. [DOI] [PubMed] [Google Scholar]
- 15.Layqah L.A., Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchim. Acta. 2019;186:224. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mavrikou S., Moschopoulou G., Tsekouras V., Kintzios S. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Sensors. 2020;20:3121. doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mojsoska B., Larsen S., Olsen D.A., Madsen J.S., Brandslund I., Alatraktchi F.A. Rapid SARS-CoV-2 detection using electrochemical immunosensor. Sens. (Switz. ) 2021;21:1–11. doi: 10.3390/s21020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu D.Y., Liu J.C., Liang S., Meng X.H., Greenbaum J., Xiao H.M., Tan L.J., Deng H.W. Drug repurposing for COVID-19 treatment by integrating network pharmacology and transcriptomics. Pharmaceutics. 2021;13:1–16. doi: 10.3390/pharmaceutics13040545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahn D.G., Jeon I.J., Kim J.D., Song M.S., Han S.R., Lee S.W., Jung H., Oh J.W. RNA aptamer-based sensitive detection of SARS coronavirus nucleocapsid protein. Analyst. 2009;134:1896–1901. doi: 10.1039/b906788d. [DOI] [PubMed] [Google Scholar]
- 20.Huang P., Wang H., Cao Z., Jin H., Chi H., Zhao J., Yu B., Yan F., Hu X., Wu F., et al. A rapid and specific assay for the detection of MERS-CoV. Front. Microbiol. 2018;9:1–9. doi: 10.3389/fmicb.2018.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moitra P., Alafeef M., Dighe K., Frieman M.B., Pan D. Selective naked-eye detection of SARS-CoV-2 mediated by N Gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.WHO. Novel coronavirus – COVID19. 〈https://covid19.who.int/(accessed〉 May 19, 2021).
- 23.Mccoy K., Gudapati S., He L., Horlander E., Kartchner D., Kulkarni S., Mehra N., Prakash J., Thenot H., Vanga S.V., et al. Biomedical text link prediction for drug discovery: a case study with COVID-19. Pharmaceutics. 2021;13:1–22. doi: 10.3390/pharmaceutics13060794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J.S., Ahn J.S., Yu B.S., Cho S.I., Kim M.J., Choi J.M., Seo S.H., Park S.S., Seong M.W. Evaluation of a real-time reverse transcription-PCR (RT-PCR) assay for detection of middle east respiratory syndrome coronavirus (MERS-COV) in clinical samples from an outbreak in South Korea in 2015. J. Clin. Microbiol. 2017;55:2554–2555. doi: 10.1128/JCM.00667-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fish E.J., Diniz P.P.V.P., Juan Y.C., Bossong F., Collisson E.W., Drechsler Y., Kaltenboeck B. Cross-sectional quantitative RT-PCR study of feline coronavirus viremia and replication in peripheral blood of healthy shelter cats in Southern California. J. Feline Med. Surg. 2018;20:295–301. doi: 10.1177/1098612X17705227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toptan T., Hoehl S., Westhaus S., Bojkova D., Berger A., Rotter B., Hoffmeier K., Cinatl Jr J., Ciesek S., Widera M. Optimized qRT-PCR approach for the detection of intra- and extra-cellular SARS-CoV-2 RNAs. Int J. Mol. Sci. 2020;21:1–11. doi: 10.3390/ijms21124396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jokerst J.C., Adkins J.A., Bisha B., Mentele M.M., Goodridge L.D., Henry C.S. Development of a paper-based analytical device for colorimetric detection of select foodborne pathogens. Anal. Chem. 2012;84:2900–2907. doi: 10.1021/ac203466y. [DOI] [PubMed] [Google Scholar]
- 28.Nicol T., Lefeuvre C., Serri O., Pivert A., Joubaud F., Dubée V., Kouatchet A., Ducancelle A., Lunel-Fabiani F., Le Guillou-Guillemette H. Assessment of SARS-CoV-2 serological tests for the diagnosis of COVID-19 through the evaluation of three immunoassays: two automated immunoassays (Euroimmun and Abbott) and one rapid lateral flow immunoassay (NG Biotech) J. Clin. Virol. 2020;129:1–7. doi: 10.1016/j.jcv.2020.104511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Estrela P.F.N., Mendes G., de M., de Oliveira K.G., Bailão A.M., Soares C.M., de A., Assunção N.A., Duarte G.R.M. Ten-minute direct detection of Zika virus in serum samples by RT-LAMP. J. Virol. Methods. 2019;271:1–5. doi: 10.1016/j.jviromet.2019.113675. [DOI] [PubMed] [Google Scholar]
- 30.Yatsunyk L.A., Mendoza O., Mergny J.L. "Nano-oddities": unusual nucleic acid assemblies for DNA-based nanostructures and nanodevices. Acc. Chem. Res. 2014;47:1836–1844. doi: 10.1021/ar500063x. [DOI] [PubMed] [Google Scholar]
- 31.Mujica M.L., Zhang Y., Bédioui F., Gutiérrez F., Rivas G. Label-free graphene oxide-based SPR genosensor for the quantification of microRNA21. Anal. Bioanal. Chem. 2020;412:3539–3546. doi: 10.1007/s00216-020-02593-w. [DOI] [PubMed] [Google Scholar]
- 32.Moher D., Liberati A., Tetzlaff J., Altman D.G., Altman D., Antes G., Atkins D., Barbour V., Barrowman N., Berlin J.A., et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:1–6. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thwe P.M., Ren P. Analysis of sputum/tracheal aspirate and nasopharyngeal samples for SARS-CoV-2 detection by laboratory-developed test and panther fusion system. Diagn. Microbiol. Infect. Dis. 2021;99:1–4. doi: 10.1016/j.diagmicrobio.2020.115228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kock R., Baselmans M., Scharnhorst V., Deiman B. Sensitive detection and quantification of SARS-CoV-2 by multiplex droplet digital RT-PCR. Eur. J. Clin. Microbiol Infect. Dis. 2020;40:807–813. doi: 10.1007/s10096-020-04076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W., et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020;92:1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beavis K.G., Matushek S.M., Abeleda A.P.F., Bethel C., Hunt C., Gillen S., Moran A., Tesic V. Evaluation of the EUROIMMUN Anti-SARS-CoV-2 ELISA Assay for detection of IgA and IgG antibodies. J. Clin. Virol. 2020;129:1–3. doi: 10.1016/j.jcv.2020.104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Percze K., Szakács Z., Scholz É., András J., Szeitner Z., Kieboom C.H., van den.; Ferwerda G., de Jonge M.I., Gyurcsányi R.E., Mészáros T. Aptamers for respiratory syncytial virus detection. Sci. Rep. 2017;7:1–11. doi: 10.1038/srep42794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melegari S.P., Fuzinatto C.F., Gonçalves R.A., Oscar B.V., Vicentini D.S., Matias W.G. Can the surface modification and/or morphology affect the ecotoxicityof zinc oxide nanomaterials? Chemosphere. 2019;224:237–246. doi: 10.1016/j.chemosphere.2019.02.093. [DOI] [PubMed] [Google Scholar]
- 39.Gomes L.P., Andrade C.T., Aguila E.M., Del, Alexander C., Paschoalin V.M.F. Assessing the antimicrobial activity of chitosan nanoparticles by fluorescence-labeling. World Acad. Sci. Eng. Technol. Int. J. Biotechnol. Bioeng. 2018;12:112–117. [Google Scholar]
- 40.Higashi S.L., Shibata A., Kitamura Y., Hirosawa K.M., Suzuki K.G.N., Matsuura K., Ikeda M. Hybrid soft nanomaterials composed of DNA microspheres and supramolecular nanostructures of semi-artificial glycopeptides. Chemistry. 2019;25:11955–11962. doi: 10.1002/chem.201902421. [DOI] [PubMed] [Google Scholar]
- 41.Hashemi S.A., Behbahan N.G.G., Bahrani S., Mousavi S.M., Gholami A., Ramakrishna S., et al. Ultra-sensitive viral glycoprotein detection NanoSystem toward accurate tracing SARS-CoV-2 in biological/non-biological media. Biosens. Bioelectron. 2021;171:1–8. doi: 10.1016/j.bios.2020.112731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Bian L., Li P., Yu L., Wu Y., et al. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal. Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 43.Tian B., Gao F., Fock J., Dufva M., Hansen M.F. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens. Bioelectron. 2020;165:1–7. doi: 10.1016/j.bios.2020.112356. [DOI] [PubMed] [Google Scholar]
- 44.Mahari, S.; Roberts, A.; Shahdeo, D.; Gandhi, S. eCovSens-Ultrasensitive Novel In-House Built Printed Circuit Board Based Electrochemical Device for Rapid Detection of nCovid-19 antigen, a spike protein domain 1 of SARS-CoV-2. 2020, https://doi.org/10.1101/2020.04.24.059204.
- 45.Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S., et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020;166:1–7. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar V., Mishra S., Sharma R., Agarwal J., Ghoshal U., Khanna T., Sharma L., Verma S.K., Mishra P., Tiwari S. Development of RNA-based assay for rapid detection of SARS-CoV-2 in clinical samples. bioRxiv. 2020 doi: 10.1101/2020.06.30.172833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 48.Roh C., Jo S.K. Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip. J. Chem. Technol. Biotechnol. 2011;86:1475–1479. doi: 10.1002/jctb.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho S.J., Woo H.M., Kim K.S., Oh J.W., Jeong Y.J. Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J. Biosci. Bioeng. 2011;112:535–540. doi: 10.1016/j.jbiosc.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martín-Yerga D., González-García M.B., Costa-García A. Electrochemical immunosensor for anti-tissue transglutaminase antibodies based on the in situ detection of quantum dots. Talanta. 2014;130:598–602. doi: 10.1016/j.talanta.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 51.Felix F.S., Angnes L. Electrochemical immunosensors - a powerful tool for analytical applications. Biosens. Bioelectron. 2018;102:470–478. doi: 10.1016/j.bios.2017.11.029. [DOI] [PubMed] [Google Scholar]
- 52.Park T.J., Hyun M.S., Lee H.J., Lee S.Y., Ko S. A self-assembled fusion protein-based surface plasmon resonance biosensor for rapid diagnosis of severe acute respiratory syndrome. Talanta. 2009;79:295–301. doi: 10.1016/j.talanta.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao H., Wei S., Chen Z., Cao L. Label-free electrochemical immunosensor based on gold and iron-oxide nanoparticle co-modified rGO-TEPA hybrid for sensitive detection of carcinoembryonic antigen. Electrocatalysis. 2021;11:513–521. [Google Scholar]
- 54.Ventura B., Della, Cennamo M., Minopoli A., Campanile R., Censi S.B., Terracciano D., Portella G., Velotta R. Colorimetric test for fast detection of SARS-COV-2 in nasal and throat swabs. ACS Sens. 2020;5:3043–3048. doi: 10.1021/acssensors.0c01742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ishikawa F.N., Chang H.K., Curreli M., Liao H.I., Olson C.A., Chen P.C., Zhang R., Roberts R.W., Sun R., Cote R.J., et al. Label-free, electrical detection of the SARS virus n-protein with nanowire biosensors utilizing antibody mimics as capture probes. ACS Nano. 2009;3:1219–1224. doi: 10.1021/nn900086c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murugan D., Bhatia H., Sai V.V.R., Satija J. P-FAB: a fiber-optic biosensor device for rapid detection of COVID-19. Trans. Indian Natl. Acad. Eng. 2020;5:211–215. doi: 10.1007/s41403-020-00122-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cavalera S., Colitti B., Rosati S., Ferrara G., Bertolotti L., Nogarol C., Guiotto C., Cagnazzo C., Denina M., Fagioli F., et al. A multi-target lateral flow immunoassay enabling the specific and sensitive detection of total antibodies to SARS COV-2. Tanta. 2020;223:1–8. doi: 10.1016/j.talanta.2020.121737. Talanta. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Das C.M., Guo Y., Yang G., Kang L., Xu G., Ho H.P., Yong K.T. Gold nanorod assisted enhanced plasmonic detection scheme of COVID-19 SARS-CoV-2 spike protein. Adv. Theory Simul. 2020;3:1–8. doi: 10.1002/adts.202000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim H., Park M., Hwang J., Kim J.H., Chung D.R., Lee K. sung, Kang M. Development of label-free colorimetric assay for MERS-CoV using gold nanoparticles. ACS Sens. 2019;4:1306–1312. doi: 10.1021/acssensors.9b00175. [DOI] [PubMed] [Google Scholar]
- 60.Alafeef M., Dighe K., Moitra P., Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teengam P., Siangproh W., Tuantranont A., Vilaivan T., Chailapakul O., Henry C.S. Multiplex paper-based colorimetric DNA sensor using pyrrolidinyl peptide nucleic acid-induced AgNPs aggregation for detecting MERS-CoV, MTB, and HPV Oligonucleotides. Anal. Chem. 2017;89:5428–5435. doi: 10.1021/acs.analchem.7b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martínez-Paredes G., González-García M.B., Costa-García A. Genosensor for SARS virus detection based on gold nanostructured screen-printed carbon electrodes. Electroanalysis. 2009;21:379–385. [Google Scholar]
- 63.Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S., et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020;166:1–7. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao Z., Cui H., Song W., Ru X., Zhou W., Yu X. A simple magnetic nanoparticles-based viral RNA extraction method for efficient detection of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.02.22.961268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan. China JAMA J. Am. Med. Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020;178:1–4. doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corman V.M., Ölschläger S., Wendtner C.M., Drexler J.F., Hess M., Drosten C. Performance and clinical validation of the RealStar® MERS-CoV Kit for detection of Middle East respiratory syndrome coronavirus RNA. J. Clin. Virol. 2014;60:168–171. doi: 10.1016/j.jcv.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim S.H., Chang S.Y., Sung M., Park J.H., Kim H., Bin, Lee H., Choi J.P., Choi W.S., Min J.Y. Extensive viable Middle East Respiratory Syndrome (MERS) coronavirus contamination in air and surrounding environment in MERS isolation wards. Clin. Infect. Dis. 2016;63:363–369. doi: 10.1093/cid/ciw239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tric M., Lederle M., Neuner L., Dolgowjasow I., Wiedemann P., Wölfl S., Werner T. Optical biosensor optimized for continuous in-line glucose monitoring in animal cell culture. Anal. Bioanal. Chem. 2017;409:5711–5721. doi: 10.1007/s00216-017-0511-7. [DOI] [PubMed] [Google Scholar]
- 71.Falohun T., McShane M.J. An optical urate biosensor based on urate oxidaseand long-lifetime metalloporphyrins. Sensors. 2010;20:1–15. doi: 10.3390/s20040959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mahato K., Chandra P. Paper-based miniaturized immunosensor for naked eye ALP detection based on digital image colorimetry integrated with smartphone. Biosens. Bioelectron. 2019;128:9–16. doi: 10.1016/j.bios.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 73.Attwood L.O., Francis M.J., Hamblin J., Korman T.M., Druce J., Graham M. Clinical evaluation of AusDiagnostics SARS-CoV-2 multiplex tandem PCR assay. J. Clin. Virol. J. 2020;128:1–4. doi: 10.1016/j.jcv.2020.104448. [DOI] [PMC free article] [PubMed] [Google Scholar]