Abstract
Background
The association of cough with Mycoplasma hyopneumoniae (MHP) DNA detection in specimens was evaluated under conditions in which the MHP status of inoculated and contact-infected pen mates was closely monitored for 59 days post-inoculation (DPI).
Methods
Seven-week-old pigs (n = 39) were allocated to five rooms (with one pen). Rooms contained 9 pigs each, with 1, 3, 6, or 9 MHP-inoculated pigs, respectively, except Room 5 (three sham-inoculated pigs). Cough data (2 × week) and specimens, tracheal swabs (2 × week), oral fluids (daily), drinker wipes (~ 1 × week), and air samples (3 × week) were collected. At 59 DPI, pigs were euthanized, and lung and trachea were evaluated for gross and microscopic lesions. Predictive cough value to MHP DNA detection in drinker and oral fluid samples were estimated using mixed logistic regression.
Results
Following inoculation, MHP DNA was first detected in tracheal swabs from inoculated pigs (DPI 3), then oral fluids (DPI 8), air samples (DPI 10), and drinker wipes (21 DPI). MHP DNA was detected in oral fluids in 17 of 59 (Room 1) to 43 of 59 (Room 3) samples, drinker wipes in 4 of 8 (Rooms 2 and 3) to 5 of 8 (Rooms 1 and 4) samples, and air samples in 5 of 26 (Room 2) or 3 of 26 (Room 4) samples. Logistic regression showed that the frequency of coughing pigs in a pen was associated with the probability of MHP DNA detection in oral fluids (P < 0.01) and nearly associated with drinker wipes (P = 0.08). Pathology data revealed an association between the period when infection was first detected and the severity of gross lung lesions.
Conclusions
Dry, non-productive coughs suggest the presence of MHP, but laboratory testing and MHP DNA detection is required for confirmation. Based on the data from this study, oral fluids and drinker wipes may provide a convenient alternative for MHP DNA detection at the pen level when cough is present. This information may help practitioners in specimen selection for MHP surveillance.
Keywords: Mycoplasma hyopneumoniae, Enzootic pneumonia, Tracheal swabs, Oral fluids, Water samples, Air samples, Cough, Pathology
Background
Mycoplasma hyopneumoniae (MHP) is an impactful health challenge in swine [1], resulting in reduced daily weight gain and poor feed conversion [2]. MHP binds to ciliated epithelial cells of the respiratory tract by means of adhesins [3, 4], resulting in ciliostasis and diminished function of the mucociliary apparatus [5]. MHP virulence factors, i.e., adhesins and lipid associated membrane proteins, elicit a pro-inflammatory response, leading to infiltration and accumulation of immune cells, i.e., lymphocytes, plasma cells, and neutrophils, in the lumen and interstitium surrounding the conducting airways [6–8]. As the infection becomes chronic, marked hyperplasia of the bronchus-associated lymphoid tissues (BALT) and well-demarcated cranioventral pulmonary consolidation are identified as histologic and gross lesions, respectively, and are characteristic of porcine enzootic pneumonia (PEP). Due to bronchoconstriction resulting from the obstruction of airways, extensive cough may be observed in MHP-infected pigs [9–11].
Effective control measures are dependent on an accurate understanding of the limitations and benefits of sample techniques and diagnostic assays to assess herd status within specific clinical context [12]. Several different sample techniques and diagnostic tests have been developed to detect MHP nucleic acid, antigen or antibody, and are available for surveillance programs [13]. Yet, the selection of samples hinges on the diagnostic objectives, the level of disease in the herd, accuracy of the test, and cost.
Serum antibody based on enzyme-linked immunosorbent assay (ELISA) is commonly used to monitor swine herds. However, the utility of serological assays for MHP can be hindered by the limited correlation between a positive assay and disease, inability to differentiate natural infection from vaccination, the highly variable time lapse between infection and antibody production, variable diagnostic accuracy across commercially available ELISAs, and antibody cross-reactions with other mycoplasmas [13, 14]. Consequently, it is difficult to interpret results based on the individual or at the herd level.
In contrast, polymerase chain reaction (PCR) using specimens of the respiratory tract can determine the direct presence of the MHP deoxyribonucleic acid (DNA). The highest concentration of MHP DNA has been described in bronchoalveolar lavage fluid (BALF) over the course of infection [15]. Yet, BALF sampling is most commonly performed at post mortem examination or in sedated animals [16]. Alternatively, tracheal and tracheobronchial swabs for the detection and recovery of MHP have been described as the preferred sample for early detection in live pigs [17–19], followed by laryngeal swabs [20]. However, collection of such samples can be laborious given that it requires a specific number of trained personnel (two or three) and is stressful for the animals because of intensive restraint [17–19].
Several swine pathogens are present in and/or transmitted through oral fluids. Pen-based oral fluid is a non-invasive, cost-effective, aggregated sampling method to detect a number of swine pathogens. Pathogen surveillance for porcine circovirus 2 (PCV2), porcine reproductive and respiratory syndrome virus (PRRSV), and influenza A virus (IAV) through the detection of specific viral genes or antibodies in oral fluids has been successful [21]. The use of oral fluids for MHP DNA detection may be precluded due to the lower sensitivity than tracheal and laryngeal samples under field conditions [17]. Yet, there is very limited information concerning the agreement of oral fluid-based PCR for the detection of MHP DNA and other diagnostic samples with the predominate clinical sign of PEP, coughing [17, 22]. Establishing the diagnostic parameters of different sampling techniques of MHP and correlation between the PCR results of different sample types and clinical signs is critical for the strategic implementation of disease prevention and elimination/control strategies that will mitigate the economic and production losses incurred due to PEP. Therefore, this study investigated the predictive value of coughing for the detection of MHP DNA in aggregated specimens, such as pen-based oral fluids, pen-based drinker wipes, and air samples collected from pigs housed in rooms differing in within-pen MHP prevalence.
Materials and methods
Experimental design
Seven-week-old MHP-naïve pigs (n = 39) were blocked by litter (n = 19) and randomly assigned to one of five rooms: (Room 1) one MHP-inoculated pig commingled with eight uninoculated pigs (n = 9), (Room 2) three MHP-inoculated pigs with six uninoculated pigs (n = 9), (Room 3) six MHP-inoculated pigs with three uninoculated pigs (n = 9), (Room 4) nine MHP-inoculated pigs (n = 9), and (Room 5) 3 negative control pigs (n = 3). Clinical data collected, samples tested for MHP PCR, and pathologic evaluations are presented in Table 1. Twice weekly, coughing was assessed by individual pig and room by an observer blinded to inoculation status, and digital recording system captured cough daily. MHP DNA was tested by Real-Time qPCR testing in deep tracheal swabs, pen-based oral fluid, pen drinker, and pen air samples. On day post-inoculation (DPI) 59, pigs were humanely euthanized, and lung tissue samples were collected for post mortem examination. All procedures were conducted with the approval of the Iowa State University Office for Responsible Research and Institutional Animal Care and Use Committee.
Table 1.
Clinical data collected, samples tested for Mycoplasma hyopneumoniae (MHP) PCR, and pathologic evaluations throughout the study
| Samples | Frequency | Levela | No. of possible samples | No. of collected samples | Testing | Outcome |
|---|---|---|---|---|---|---|
| Coughs | ||||||
| Over 27-min | 2 × week | Individual | 702 | 621 | Blind observerb | Total coughs per pig (or per group) |
| Over 24-h | Real-time | Room | 355 | 303 | Digital recording systemc | Total coughs per group |
| Lung lesions | ||||||
| Gross | Necropsy | Individual | 39 | 37 | Blind observerb | % lung affected |
| Microscopic | Necropsy | Individual | 39 | 39 | Histopathologyb | Score 0, 1, 2, 3, 4 |
| Tracheal swab | 2 × week | Individual | 507 | 489 | PCR Protocol 1d | MHP DNA Pos or neg; Ct value |
| Oral fluid | Daily | Rooms (1–5) | 322 | 322 | PCR Protocol 1 | MHP DNA Pos or neg; Ct value |
| Drinker | 1 × week | Room (1–5) | 45 | 45 | PCR Protocol 1 | MHP DNA Pos or neg; Ct value |
| Air | 3 × week | Room (2, 4, and 5) | 84 | 84 | PCR Protocol 2e,f | MHP DNA Pos or neg; Ct value |
aRoom 1: One MHP-inoculated pig commingled with 8 uninoculated pigs. Room 2: 3 MHP-inoculated pigs with 6 uninoculated pigs. Room 3: 6 MHP-inoculated pigs with 3 uninoculated pigs. Room 4: 9 MHP-inoculated pigs. Room 5: 3 uninoculated pigs
bObserver blinded to the MHP inoculation pig status
cTotal number of coughs per day identified using a commercial digital quantitative cough recording system and software (SoundTalk®, SoundTalks NV, Leuven, Belgium)
dPCR Protocol 1: TaqMan® Fast Virus 1-Step Master Mix (Life Technologies, Carlsbad, CA USA) with AmpliTaq® 360DNA Polymerase (5U/uL) (Thermo Fisher Scientific, Inc., Waltham, MA USA)
ePCR Protocol 2: TaqMan® Fast Virus 1-Step Master Mix (Life Technologies)
fCollected from Room 2, 4, and 5
Animals, housing and groups
Seven-week-old MHP-, IAV-, and PRRSV-negative pigs (n = 39) were received into a BSL-2 livestock infectious disease isolation facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. At DPI -5, pigs were blocked by litter (n = 19) and randomly assigned (Microsoft Excel® random function) to one of five rooms: 9 pigs each in rooms 1 through 4 plus 3 pigs in the negative control room. Each room containing one pen was equipped with a single-pass non-recirculating ventilation system to prevent inadvertent aerosol transmission. On DPI -3, deep tracheal swabs were collected from all pigs and tested for MHP DNA (Real-Time qPCR). On DPI 0, after confirming all pigs to be negative for MHP infection, one randomly selected pig from Room 1, three pigs from Room 2, six pigs from Room 3, and nine pigs from Room 4 were intratracheally administrated a lung tissue homogenate (10 ml) containing MHP strain 232 [23] at concentration of ~ 1 × 105 color-changing units per ml (Lot 45, Iowa State University Veterinary Diagnostic Laboratory, ISU VDL). Crude lung homogenate was originally prepared in a specific-pathogen-free pig by inoculating it with MHP strain 11 [24] which was isolated at Iowa State University in the 1960s. The isolate from this pig, identified as MHP strain 232, has been serially passaged in specific-pathogen-free pigs for production of crude lung homogenates. The resultant pneumonic lungs have been harvested at 4 weeks post-inoculation. This inoculum is efficient in reproducing MHP infection under controlled and field conditions [14, 17, 19, 20, 25, 26]. All negative control pigs were administrated Friis broth (10 ml; ISU VDL Doc 9.6726). Intratracheal inoculation was done using a feeding tube catheter (Integral Funnel, Two Eyes, Rounded Closed Tip, 4.7 mm × 41 cm, COVIDEN™ Kendall™, Coviden, Mansfield, MA USA) introduced past the larynx. Thereafter, animals were observed for 59 days.
Sample collection
Deep tracheal swabs were taken at -3, 3, 7, 10, 14, 21, 24, 28, 35, 38, 45, 52, 59 DPIs from all pigs (Table 1). After restraining pigs with a snare and an oral speculum, a single-use artificial insemination catheter (IMV PCAI catheter, Nasco, Fort Atkinson, WI USA) was introduced into the distal part of the trachea and then a swab (100 mm, FLOQSwabs® 519CS01, Copan Diagnostics, Inc. Murrieta, CA USA) attached to the flexible inner tube was extended to collect mucus lining the trachea. To avoid contamination, the swab was retracted into the catheter before withdrawal from the larynx. After removal from the pig, the swab containing the collected material was severed from the inner rod and placed in a tube containing 1 mL sterile PBS. Pigs exhibiting signs of stress were administered a combination of tiletamine hydrochloride and zolazepam hydrochloride (500 mg; Telazol®, Zoetis Inc., Parsippany, NJ USA), xylazine (250 mg; XylaMed™, VetOne®, Boise, ID USA), and ketamine (250 mg; Zetamine™ Injection, VetOne®) at a dose of 1 mL per 4.4 kg of body weight prior to sample collection.
Aggregated samples included pen oral fluid samples, drinker wipe samples, and air samples. Pen-based oral fluid samples were collected as described elsewhere [21]. Beginning at DPI -4, pigs were provided daily access (25 min) to a 3-strand (1.6 cm diameter), 100% cotton rope (Web Rigging Supply, Lake Barrington, IL USA) suspended from a bracket fixed to the side of the pen (Table 1). Oral fluids were recovered by placing the wet portion of the rope inside a plastic bag and then passing the bag containing the wet rope through a wringer to express the fluid. Drinker wipe samples were collected on DPIs 0, 7, 14, 21, 24, 35, 45, 52, and 59 by wiping the surface (drinking nipple and water cup) with a paper towel and then recovering the absorbed liquid from the towel (Table 1). Air samples were collected three times per week in Rooms 2, 4, and 5 (negative control) by suspending an air sampler (Innovaprep prototype air collector, 200 L per min flow, Innovaprep, Drexel, MO USA) at ~ 25 cm above the pigs' heads in the center of the pen (Table 1). After a 60-min sampling period, collected air samples were eluted from the filter using 0.075% Tween 20/ PBS wet-foam (Innovaprep). Given the number of available air samplers for this study (n = 3), three rooms were selected to investigate the performance of the device in terms of specificity (Room 5, negative control pen) and in rooms highly MHP-infected (Room 4, e.g., with all 9 MHP-inoculated pigs) or undergoing transmission (Room 2, e.g., three MHP-inoculated pigs commingled with six uninoculated pigs). Following collection, all specimens were vortexed, transferred to 2 mL cryogenic tubes (Cryo.s™, Greiner Bio-One™), and then stored at −200980 °C until tested for MHP DNA.
Assays
MHP DNA was tested by Real-Time qPCR testing in individual pig deep tracheal swabs, pen oral fluids, drinker, and air samples. Nucleic acid extraction was done using the MagMAX™-96 Pathogen RNA/DNA kit (Applied Biosystems™, Carlsbad, CA USA) and the Kingfisher™ Flex Purification System (Thermo Fisher Scientific, Inc., Waltham, MA USA). Tracheal swabs, pen-based oral fluids, and drinker samples were tested using TaqMan® Fast Virus 1-Step Master Mix (Life Technologies, Carlsbad, CA) with primers and probes targeting the Mhp183 gene [27], primers and probes for internal positive control (IPC) [27], and AmpliTaq® 360DNA Polymerase (5U per µL) (Thermo Fisher Scientific, Inc.). Air samples were tested using TaqMan® Fast Virus 1-Step Master Mix (Life Technologies) with primers and probes targeting the Mhp183 gene [27] and primers and probes for IPC [27]. Amplification was done on the Applied Biosystems® 7500 Fast Real-Time PCR System (ThermoFisher Scientific, Inc.). PCR results were considered valid if the IPC cycle threshold (Ct) was < 36 and samples were considered positive for MHP DNA when the Ct < 37.
Clinical signs
Individual pigs were observed daily for general health. Dry, non-productive cough was measured two times per week, i.e., DPIs -3, 3, 7, 10, 14, 17, 21, 24, 31, 35, 38, 42, 45, 49, 52, 56, and 59. A single observer blinded to the pig inoculation status counted the number of coughs over a 27-min observation period (three minutes per pig) in the afternoon. At the room level, a digital quantitative cough recording system (SoundTalk®, SoundTalks NV, Leuven, Belgium) suspended in the center of each room directly over the pen counted coughs in real-time from DPI -7 through 63. The outputs provided by the system indicated the total number of coughs over a 24-h period and a respiratory distress index (RDI), which is given by a proprietary algorithm that generates warnings when the RDI is above a specified threshold [17]. The experiment was terminated on DPI 59, but the cough recording systems were left in the empty rooms to provide baseline data (DPIs 60 through 63).
Pathologic evaluation
On DPI 59, pigs (n = 39) were humanely euthanized using captive bolt followed by exsanguination [28]. A diagnostic pathologist certified by the American College of Veterinary Pathologists and blinded to pig MHP status performed the necropsy and postmortem assessment. Gross lesions were scored in terms of visible lung consolidation. The score was calculated as the total percent lung consolidation by accounting for the contribution of each lobe to the total lung volume as previously described [11, 29].
At necropsy, normal and lesioned lung tissue (3 × 3 cm) were placed in 10% neutral buffered formalin and processed for histopathologic examination. Formalin-fixed tissues were embedded in paraffin wax, sectioned at 4 µm, and then stained with hematoxylin and eosin (H&E) using routine procedures. Microscopic lung lesion scores were based on BALT hyperplasia adjacent to airways and the levels of peribronchiolar infiltration by lymphocytes (PIL). Microscopic lung lesions were classified as “marked BALT hyperplasia with distortion of airway and PIL” (Score 4), “moderate BALT and PIL” (Score 3), “minimal-to-mild BALT and mild PIL” (Score 2), “minimal-to-mild BALT or PIL” (Score 1), and “no lesions” (Score 0). Scores of 3 or 4 were considered consistent with a diagnosis of porcine enzootic pneumonia [10].
Statistical analysis
At both individual pig and room levels (Table 1), each analysis was performed based on data collected at the same time points. Analyses were performed in R (R program version 4.0.0, R core team 2020) with an alpha level ≤ 0.05 (P ≤ 0.05) used to establish statistical significance. The variation of PCR Ct values from MHP DNA positive samples over time in deep tracheal swabs, pen oral fluid, and pen drinker samples were analyzed using a linear mixed regression model and Tukey–Kramer adjustment for pairwise comparisons (lme4 and emmeans R packages). To account for temporal changes in cough over the course of MHP infection (initiation followed by resolution), the DPI was divided into four time periods of 15 days each (DPI < 15, 15 ≤ DPI < 30, 30 ≤ DPI < 45, and DPI ≥ 45). The PCR Ct was considered as the dependent variable, time period as an independent variable, and pig (tracheal swab analysis) or room (oral fluid and drinker sample analyses) as a random effect. Comparison of quantitative PCR results from MHP DNA positive samples across aggregated specimens (oral fluid, drinker, and air samples) was done using linear regression model and Tukey–Kramer adjustment for pairwise comparisons (emmeans R packages). PCR Ct was considered the dependent variable and specimen the independent variable.
Differences in the rate of cough over time measured by the blind observer or digital recording system by rooms were analyzed using negative binomial regression with Tukey–Kramer adjustment for pairwise comparisons (emmeans R package). For both models, the total number of coughs was considered the dependent variable, room, DPI, and their interaction as independent variables. Spearman’s rank coefficient was used to measure the correlation between coughs measured by the blind observer and digital recording system. As the digital recording system commonly spiked on tracheal swab sampling days, days on which sampling took place were excluded from the data set. Additionally, the data collected by the system the day before sampling was used to calculate the correlation between the blind observer and the digital recording system.
The predictive value of cough for MHP DNA detection in aggregated specimens was estimated using four mixed-logistic models (Model A–D). The MHP PCR binary response (positive or negative) from pen oral fluid (Model A), pen drinker samples (Model B), and pen air sample (Model C) was considered as the dependent variable, and the frequency of coughing pigs (number of coughing pigs / total pig in a pen by DPI) in the room (Model A–C), or documented cough data by digital system (Model D) as the independent variable, and room as the random effect. In these models, DPI was added as a covariate and then the effect of cough on the MHP DNA detection in different specimens was measured.
The association between time period of MHP infection (four intervals of 15 days each, i.e., DPI < 15, 15 ≤ DPI < 30, 30 ≤ DPI < 45, and DPI ≥ 45) and score for gross lung lesions (percent consolidation, log transformed) and microscopic lung scores (BALT and PIL) were evaluated using linear regression model with Bonferroni adjustment for pairwise comparisons (emmeans R package) and Kruskal–Wallis test and Dunn test for pairwise comparisons (FSA R package), respectively. Association between total number of coughs per pig (counted after DPI 45) with gross lung scores was done using linear mixed regression, using the natural logarithm of gross lung score as dependent variable, number of coughs per pig as the independent, and the period of MHP infection (four intervals of 15 days each) as random effect.
Normality and homoscedasticity assumptions of linear and linear mixed regression models were analyzed using Q-Q and residuals versus fitted plots (ggResidpanel R package), and model fitness following categorization of continuous variables (e.g. DPI divided into four time period) was evaluated based on Akaike information criteria. Over-dispersion and deviance assumptions of negative binomial regression were verified using simulated residuals versus fitted plots (R package DHARMa; P > 0.05). For logistic regressions, plots (logit versus continuous were used to evaluate the linear relationship between the logit of the outcome and continuous predictors, and model fitness was evaluated using Akaike information criteria and Hosmer–Lemeshow Test (P > 0.05).
Results
Pigs (n = 3) from Room 5 (negative control) were clinically normal throughout the study. One pig in Room 1 died acutely on DPI 10 and was diagnosed with chronic, fibrosing pericarditis and epicarditis. One pig in Room 4 became lame (DPI 28), was isolated and treated with ceftiofur (5.0 mg per kg; Zoetis Inc.), dexamethasone (2.0 mg per kg; MWI® Animal Health), and flunixin meglumine (2.2 mg per kg; Prevail™, MWI® Animal Health), but did not recover and was humanely euthanized on DPI 35.
MHP DNA detection in expected negative samples.
In Room 5 (negative control), all collected deep tracheal swabs (n = 39), pen-based oral fluid (n = 64), drinker wipes (n = 9), and air samples (n = 9) tested MHP PCR negative throughout the study. In Rooms 1–4, deep tracheal swabs (n = 9 per room), pen-based oral fluid (n = 5 per room), drinker wipes (n = 1 per room), and air samples (n = 1 per room) collected between DPI -4 through 0 were negative for MHP by PCR.
MHP DNA detection in deep tracheal swab samples
PCR results of deep tracheal swab samples by pig and room are presented in Fig. 1 and Table 2, respectively. Among deep tracheal swabs collected from inoculated pigs in Rooms 1–4, 15 of 19 MHP-inoculated pigs tested PCR positive on DPI 3, i.e., the one pig inoculated in Room 1, two of three pigs in Room 2, five of six pigs in Room 3, and seven of nine pigs in Room 4. The remaining inoculated pigs became MHP PCR positive via deep tracheal swab on DPI 7. Within-pen MHP transmission from pen inoculated pigs was observed through MHP DNA detection in deep tracheal swabs collected from uninoculated pigs in room groups. In Room 1 (1 MHP-inoculated: 8 uninoculated), the first MHP transmission event was observed in one pig on DPI 21, two pigs on DPI 24, two pigs on DPI 28, one pig on DPI 35, and the remaining pig on DPI 52, i.e., all uninoculated pigs except one pig that died on DPI 10 became MHP infected. In Room 2 (3 MHP-inoculated: 6 uninoculated), the first MHP transmission was observed in two pigs on DPI 10, one pig on DPI 21, one pig on DPI 24, and two pigs on DPI 28. In Room 3 (6 MHP-inoculated: 3 uninoculated), the first MHP transmission was observed in one pig on DPI 7 and the two remaining pigs on DPI 21.
Fig. 1.
Individual pig Mycoplasma hyopneumoniae (MHP) PCR results using deep tracheal swabs over time by room. Solid or dashed colored lines represent each MHP-inoculated or uninoculated pig in a room, respectively. Room 1: One MHP-inoculated pig commingled with 8 uninoculated pigs. Room 2: 3 MHP-inoculated pigs with 6 uninoculated pigs. Room 3: 6 MHP-inoculated pigs with 3 uninoculated pigs. Room 4: 9 MHP-inoculated pigs
Table 2.
Mycoplasma hyopneumoniae (MHP) DNA detection by specimen and room
| Specimen | MHP DNA detection | Room | |||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| 1 MHP-inoculated pig: 8 uninoculated pigs | 3 MHP-inoculated pigs: 6 uninoculated pigs | 6 MHP-inoculated pigs: 3 uninoculated pigs | 9 MHP-inoculated pigs | ||
| Tracheal swab | First detection | DPI 3 | DPI 3 | DPI 3 | DPI 3 |
| Pig-to-pig transmission detecteda | DPI 21 | DPI 10 | DPI 7 | NAb | |
| 100% detection in group | DPI 52 | DPI 38 | DPI 21 | DPI 7 | |
| MHP PCR positive of total samples | 47 of 99 | 69 of 107 | 94 of 104 | 98 of 103 | |
| MHP PCR overall mean ± se Ctc | 26.8 ± 0.7 | 27.6 ± 0.6 | 26.2 ± 0.4 | 26.2 ± 0.4 | |
| Highest MHP DNA concentration (DPI) | 23.4 (DPI 59) | 24.4 (DPI 45) | 23.6 (DPI 28) | 24.3 (DPI 28) | |
| Oral fluid | First detection | DPI 8 | DPI 9 | DPI 14 | DPI 8 |
| MHP PCR positive of total samplesd | 17 of 591 | 32 of 592 | 43 of 592 | 31 of 592 | |
| MHP PCR overall mean ± se Ct | 34.7 ± 0.3 | 33.3 ± 0.5 | 33.5 ± 0.3 | 33.4 ± 0.4 | |
| MHP PCR lowest Ct | 31.6 (DPI 51) | 25.7 (DPI 13) | 27.4 (DPI 45) | 26.1 (DPI 28) | |
| Drinker | First detection | DPI 21 | DPI 21 | DPI 21 | DPI 21 |
| MHP PCR positive of total samples | 5 of 8 | 4 of 8 | 4 of 8 | 5 of 8 | |
| MHP PCR overall mean ± se Ct | 35.3 ± 1.5 | 33.7 ± 1.9 | 34.7 ± 1.4 | 34.7 ± 1.6 | |
| MHP PCR lowest Ct | 34.5 (DPI 59) | 31.5 (DPI 21) | 33.3 (DPI 35) | 32.2 (DPI 24) | |
| Air | First detection | NDe | DPI 10 | NDe | DPI 17 |
| MHP PCR positive of total samples | NDe | 5 of 26 | NDe | 3 of 26 | |
| MHP PCR overall mean ± se Ct | 36.2 ± 0.4 | 36.3 ± 0.6 | |||
aFirst pig-to-pig transmission based on detection of MHP DNA in deep tracheal swab from commingled animal
bNA: not applicable
cNegative values (Ct ≥ 37) were excluded from calculations of statistical descriptive measures
dDifferent superscripted numbers (1, 2, 3, and 4) indicate statistical differences (P ≤ 0.05, logistic regression and Tukey–Kramer pairwise comparisons)
eND: not done
MHP DNA was consistently detected following first detection until the end of the study in deep tracheal swabs from inoculated pigs, with a total of 211 PCR positive deep tracheal swabs of 218 (97%). Following the first positive PCR result, MHP DNA was consistently detected until the end of the study in deep tracheal swabs in all three uninoculated pigs from Room 3. Within Room 1 and 2, all eight or six uninoculated pigs, respectively, were consistently PCR positive over the study, with exception of two pigs in each room. From those four pigs, in two or three time points, deep tracheal swabs resulted in MHP DNA negative following the first positive PCR result. A total of 97 positive PCR results of 195 collected deep tracheal swabs (50%) was observed in uninoculated pigs. The mean distribution of Ct values from MHP PCR positive tracheal swabs by room throughout the study is given in Table 2. Comparisons of individual pig tracheal swab PCR Ct values (least square means) by time period showed lower MHP DNA concentrations (P < 0.004, linear mixed model) at DPI < 15 (28.3; 95 CI% 27.1, 29.4) and DPI ≥ 45 (26.7; 95 CI% 25.7, 27.7) as compared to middle periods, i.e., 15 ≤ DPI < 30 (26.0; 95 CI% 24.8, 27.1), 30 ≤ DPI < 45 (26.3; 95 CI% 25.1, 27.2).
MHP DNA detection in pen oral fluid samples
After inoculation, 59 pen-based oral fluid samples were collected from each MHP-inoculated Room from DPI 1 through 59 (n = 236 samples). As shown in Table 2, MHP DNA was first detected in oral fluid samples from Rooms 1 and 4 on DPI 8, Room 2 on DPI 9, and Room 3 on DPI 14. Throughout the study, MHP DNA was detected in 17 of 59 pen oral fluids (29%) in Room 1, 32 of 59 (54%) in Room 2, 43 of 59 (73%) in Room 3, and 31 of 59 (53%) in Room 4. The rate of PCR positive oral fluid samples was lower in Room 1 vs Rooms 2, 3, and 4 (P < 0.001, logistic regression). The mean distribution of Ct values from MHP PCR positive oral fluids by room throughout the study is given in Table 2. Within room, there was no significant variation in oral fluid PCR Ct values (least square means) by time period (P > 0.05, linear mixed model).
MHP DNA detection in pen drinker samples
Following inoculation, eight drinker samples per MHP-inoculated room (n = 32 samples) were collected on DPIs 7, 14, 21, 24, 35, 45, 52, and 59. On DPI 21, MHP DNA was detected in all drinker samples. MHP DNA was detected in five of eight pen drinker samples (62.5%) in Room 1 and 4, and four of eight pen drinker samples (50%) in Rooms 2 and 3. The mean distribution of Ct values from MHP PCR positive drinker samples by room is given in Table 2. Within room, there was no significant variation in pen drinker PCR Ct values (least square means) by period (P > 0.05, linear mixed model).
MHP DNA detection in pen air samples
Air samples were collected three times per week from Rooms 2 and 4 (MHP-inoculated rooms), i.e., a total of 26 samples from each room following inoculation (2, 3, 7, 9, 10, 14, 16, 17, 21, 23, 24, 28, 30, 31, 35, 37, 38, 42, 44, 45, 49, 51, 52, 56, 58, and 59 DPIs). Among these, MHP DNA was detected in Room 2 air samples on DPIs 10, 14, 51, 52, and 59 and Room 4 on DPIs 17, 31, and 42. Throughout the study, the distribution of Ct values from MHP PCR positive pen air samples was 36.2 (standard error = 0.4) in Room 2, and 36.3 (standard error = 0.6) in Room 4 (Table 2). Due to the low number of positive PCR results for MHP DNA in air samples, the analysis of distribution of PCR Cts over DPIs was not done. No statistical difference among PCR Ct values from aggregated samples (pen oral fluids, pen drinker wipes, and pen air samples) was observed (P > 0.05, linear regression model).
Cough data based on four scenarios of within-pen MHP prevalence
Cough data per the observer or digital recording system by room are shown in Fig. 2 and Table 3. In Rooms 1–4, dry, non-productive coughs were first noted by the observer on DPI 10 (Rooms 2 and 4), DPI 14 (Room 3) and DPI 21 (Room 1). Thereafter, cough was continuously observed until the termination of the study in Rooms 1, 2, and 3, but not after DPI 45 in Room 4. The coughs rate was significantly higher in Rooms 3 and 4 compared to 1 and 2 (P < 0.001, negative binomial regression, Table 3). No dry, non-productive cough was observed in Room 5 (negative control).
Fig. 2.
Pattern of coughs counted based on a blinded observer to pig Mycoplasma hyopneumoniae (MHP)-inoculation status (27-min period) and digital recording system by rooms (24-h period, SoundTalk®, SoundTalks NV, Leuven, Belgium). Room 1: One MHP-inoculated pig commingled with 8 uninoculated pigs. Room 2: 3 MHP-inoculated pigs with 6 uninoculated pigs. Room 3: 6 MHP-inoculated pigs with 3 uninoculated pigs. Room 4: 9 MHP-inoculated pigs. Room 5: 3 uninoculated pigs
Table 3.
Cough indices and pathologic lesions associated with Mycoplasma hyopneumoniae (MHP) infection by room
| Clinical assessment and pathology | Rooma | ||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Cough counted per pig over 27-min (blind observer to MHP status)b | |||||
| First observation | DPI 21 | DPI 10 | DPI 14 | DPI 10 | - |
| Coughs per 27 min (mean ± se) | 5.8 ± 1.9 | 8.7 ± 2.1 | 18.5 ± 4.1 | 18.3 ± 6.7 | - |
| Coughing pigs per day (mean ± se) | 1.9 ± 0.8 | 1.8 ± 1.0 | 3.8 ± 2.0 | 4.6 ± 2.5 | 0 |
| Number of days with coughing pigs | 9 | 13 | 14 | 9 | 0 |
| Total number of coughs | 991 | 1471 | 3142 | 2922 | 0 |
| Maximum cough (number of coughs) | DPI 56 (26) | DPI 24 (30) | DPI 38 (50) | DPI 21 (79) | - |
| Coughs recorded over 24-h (SoundTalk®, SoundTalks NV, Leuven, Belgium)c | |||||
| Coughs per 24 h (mean ± se) | 23.4 ± 4.1 | 55.6 ± 6.6 | 33.1 ± 4.1 | 76.6 ± 11.2 | 23.1 ± 3.6 |
| Total number of coughs | 9591 | 2,2792,3 | 1,3901,2 | 3,2923 | 9921 |
| Maximum cough (number of coughs) | DPI 34 (139) | DPI 39 (152) | DPI 30 (108) | DPI 22 (232) | DPI 54 (83) |
| Respiratory distress index (mean ± se) | 0.02 ± 0.01 | 0.17 ± 0.03 | 0.04 ± 0.01 | 0.12 ± 0.02 | 0.01 ± 0.01 |
| Maximum respiratory distress index | DPI 34 (0.20) | DPI 31 (0.58) | DPI 30 (0.22) | DPI 35 (0.53) | DPI 3 (0.41) |
| Lung score | |||||
| Median gross lesion (min–max) | 2.9% (0.6–36) | 1.1% (0–12) | 1.1% (0–18) | 0.4% (0–3.8) | 0% (0–0) |
| Median microscopic score (min–max) | 4 (3–4) | 3 (1–4) | 4 (1–4) | 3 (2–4) | 1 (1–1) |
aRoom 1: One MHP-inoculated pig commingled with 8 uninoculated pigs. Room 2: 3 MHP-inoculated pigs with 6 uninoculated pigs. Room 3: 6 MHP-inoculated pigs with 3 uninoculated pigs. Room 4: 9 MHP-inoculated pigs. Room 5: 3 uninoculated pigs
bDifferent superscripted numbers (1 or 2) indicate statistical differences among Room groups (P ≤ 0.05, negative binomial regression and Tukey–Kramer pairwise comparisons)
cDifferent superscripted numbers (1, 2, and 3) indicate statistical differences among Room groups (P ≤ 0.05, negative binomial regression and Tukey–Kramer pairwise comparisons)
The digital recording system documented coughs and RDIs at various time points between -4 and 59 DPIs (n = 64 time points) in all Rooms (Fig. 2; Table 3). However, no warnings due to respiratory distress were generated. In Room 2, no RDI data was provided by the algorithm between DPIs 35 through 63 because of an internet connection issue. The first cough event in Rooms 3, 4 and 5 was noted at -3 DPI (the first day of tracheal sampling) and at 7 and 14 DPI for Rooms 1 and 2, respectively. The maximum number of coughs recorded by the system ranged from 83 for Room 5 at 54 DPI to 232 for Room 4 at 22 DPI. The cough rate documented by the digital recording system was statistical higher and similar in Rooms 4 and 2, and no statistically significant difference was detected among Rooms 1, 3, and 5 (P < 0.001, negative binomial regression). A moderate association (0.58; P < 0.001) was observed between the total number of coughs in a given DPI recorded by the blind observer and the digital system.
Predictive cough for MHP DNA in aggregated specimens
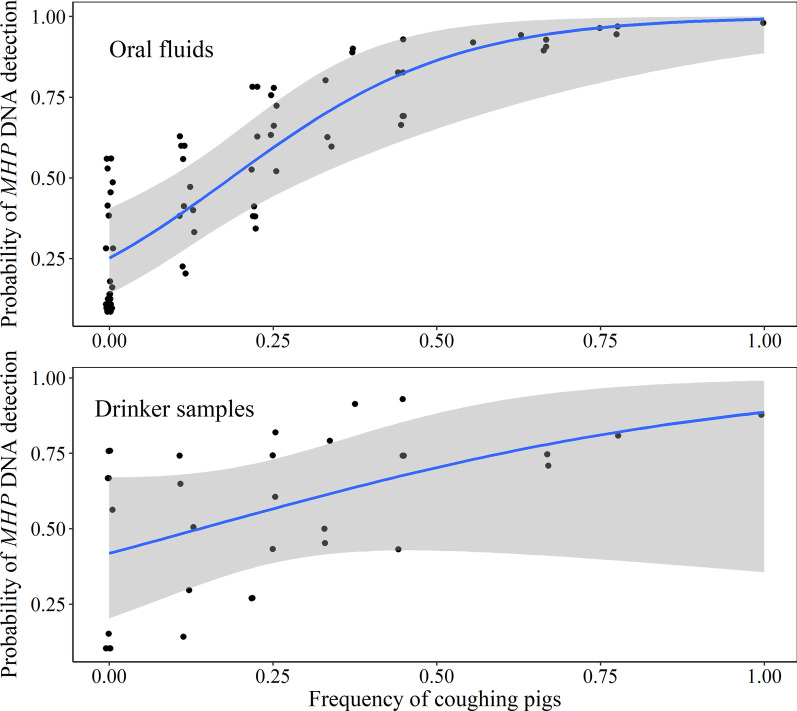
Figure 3 shows the cough pattern measured by the observer related to MHP DNA concentration by specimen. The frequency of coughing pigs, e.g., the number of coughing pigs divided by total number of pigs in pen given a DPI, as measured by the observer was a significant predictor for MHP DNA detection in pen oral fluid samples (beta coefficient = 4.85; SE = 1.59; P < 0.001; Model A) and nearly significant in pen drinker wipes (beta coefficient = 3.25; SE = 1.88; P = 0.08; Model B). As shown in Fig. 4, the probability of detecting MHP DNA in pen oral fluid and drinker samples increased as the frequency of coughing pigs increased. Likewise, the probability of MHP DNA detection increased by 1% as the cough data generated by the digital system increased by 1-unit (beta coefficient = 0.01; SE = 0.001; P < 0.001; Model D). The value of cough as a predictor for MHP DNA detection in air samples (Model C) was not done because of the limited number of positive PCR results in this study.
Fig. 3.
Number of coughs (box plot with median and interquartile range) related to Mycoplasma hyopneumoniae (MHP) DNA concentration (Adjusted PCR Ct) from deep tracheal swabs (TS; dark-green dots represent a pig), pen oral fluid (OF; red triangles represent a pen in a room), pen drinker (blue squares represent a pen in a room), and pen air samples (black diamonds represent a pen in a room). Room 1: One MHP-inoculated pig commingled with 8 uninoculated pigs. Room 2: 3 MHP-inoculated pigs with 6 uninoculated pigs. Room 3: 6 MHP-inoculated pigs with 3 uninoculated pigs. Room 4: 9 MHP-inoculated pigs
Fig. 4.
Overall probability of detection Mycoplasma hyopneumoniae DNA in oral fluid and drinker samples as a function of frequency of coughing pigs in a pen
Pathologic evaluation
Gross and microscopic lung lesions are given in Table 3. Gross lung lesions at necropsy were significantly higher in pigs that became infected ≥ DPI 15 versus those infected < DPI 15 (P < 0.002, linear regression). Microscopic lung scores did not differ between MHP-infected pigs by time period (P ≥ 0.05, Kruskal–Wallis test). The percent of lung consolidation was positively associated with total number of coughs per pig counted after DPI 45 (P < 0.001, linear mixed regression). That is, the median percent of lung consolidation increased when the number of coughs per pig increased by 7.6% (95 CI% 5.2%, 10.5%).
Discussion
The objective of this study was to evaluate the value of cough as a predictor for MHP DNA detection in different aggregated specimens. Cough due to MHP infection is triggered by the accumulated mucus and partial compression of airways by BALT hyperplasia [9–11]. The internal validity of the study was supported by the detection of MHP by PCR in tracheal swab samples from all pigs within MHP-positive rooms, gross and histologic lung lesions consistent with MHP disease, and the positive association between the percent of lung consolidation and total number of coughs per pig counted after DPI 45. In this study, microscopic lung score was not associated with time of MHP infection, but only a single section of lung was evaluated.
There was a significant association between the frequency of coughing pigs in a pen and MHP DNA detection in oral fluids, suggesting that pen-based oral fluid samples are an appropriate diagnostic specimen to evaluate for the presence of MHP by PCR in the presence of cough under the conditions of this study. Given that MHP is more likely to be found in the lower respiratory tract [15], cough may contribute to the presence of MHP in the oral cavity, increasing the likelihood of detecting MHP DNA in oral fluid samples. This has been suggested but not evaluated by Clavijo et al. [17] and Hernandez-Garcia et al. [22].
Pen drinker samples are not a common sample type submitted for molecular diagnostics. However, design of drinkers in modern production systems effectively flushes the oral cavity of each animal as it consumes water, providing a continuous, autonomous (labor-less), aggregated, non-invasive, population-based sample that could be assessed for various pathogens. In this study, analyses showed that PCR Ct values of drinker samples did not differ from the Ct values of oral fluid samples, but the model of cough as a predictor for MHP DNA detection in drinker samples was not significant (P = 0.08). This may have been impacted by the small sample size and limited sampling frequency in this study. The rate of detection of nucleic acid by PCR of water samples may be affected by PCR inhibitors [30]. In this study, PCR inhibitors in the water due to sample processing or nucleic acid extraction were monitored by the use of an internal positive control in every sample at the DNA extraction step [27]. The usefulness of pen drinker samples likely warrants further investigation.
In this study, air sampling resulted in occasional MHP PCR positive samples. Poeta Silva et al. did not detect MHP by PCR or culture using a similar cyclonic liquid impinger [26]. However, the duration of sampling (60 min versus 15 min), location of the air sampler (inside versus outside the pen) and PCR protocol differed. Under field conditions, Dee et al. [31], rarely (5%) detected MHP DNA by PCR near the exhaust fan in an MHP-positive herd using a cyclonic liquid impinger. These findings suggest that while detection of MHP in air samples is inconsistent that identification of MHP nucleic acid in this sample type could indicate numerous pigs are infected with MHP.
Due to the colonization site of MHP [17–19], MHP DNA was detected earliest in deep tracheal swabs, e.g., 15 of 19 MHP-inoculated pigs on DPI 3 with all commingled, non-inoculated pigs becoming positive at some point in the study (Table 2). Roos et al. [32] showed that a 100% within pen MHP infection prevalence was achieved with a ratio of six MHP-inoculated and four uninoculated pigs by DPI 28. While, in this study, a 100% within-pen MHP infection prevalence, based on MHP DNA detection in deep tracheal swabs, was achieved with a ratio of six MHP-inoculated and three uninoculated pigs on DPI 21 and three MHP-inoculated pigs and six uninoculated on DPI 38. That is, not surprisingly, transmission events based on MHP DNA detection by PCR via tracheal swabs occurred more quickly in pens with more MHP-infected pigs. As a general trend, MHP DNA concentration in tracheal swabs was lower through DPI 15, higher and steady from DPI 15 to 45, and lower after DPI 45, thus suggesting that the rate of MHP replication varied over time.
Analyses of the cough pattern measured by the observer demonstrated that cough rate was positively associated with within-pen MHP prevalence after DPI 15, as demonstrated by the higher cough observed in Room 3 (6 of 9 MHP-inoculated pigs) and 4 (9 of 9 MHP-inoculated pigs) (Fig. 2). Likewise, a higher cough incidence rate was observed in Rooms 2 and 4 using the digital recording system; however, there was no statistical difference among Rooms 1, 3, and 5 (negative control group). There was moderate correlation between cough measured by the observer and digital recording system. This moderate rather than strong correlation may be a result of multiple factors. First, it may reflect the difference in time used by each methodology (27 min vs 24 h). Second, based on the manufacturer’s instructions, these devices were designed to capture specific cough sounds in larger pig populations [33]. Therefore, in this study, the limited number of pigs in the room and/or the acoustics of the BSL-2 facility may have led to the absence of warnings and lower values of RDI by the digital recording system. This might have limited the use of the digital recording system under the conditions of this study, suggesting that the algorithm needs to be adapted to controlled environment. Clavijo et al. [17] reported at least seven warnings in a commercial wean-to-finish farm (1,250 pigs per room) undergoing MHP infection. Thus, their use under field conditions has been supported for MHP surveillance in larger pig populations.
Conclusion
The selection of specimens and testing will depend on the objectives of MHP surveillance. The data from this study suggests that in the presence of cough and high within-pen MHP prevalence, oral fluids and drinker samples may provide a more convenient (aggregate sampling with no pig handling) means to surveil the population for MHP infection than the more laborious tracheal sampling. Thus, these specimens may be utilized in herds with suggestive clinical signs of MHP infection.
Acknowledgements
We thank Chelsea Ruston, Justin Brown, Kristin Skoland, Locke Karriker, and Megan Nickel from the Swine Medicine Education Center Iowa State University (Ames, IA USA), and Aric McDaniel and Maria Merodio from the College of Veterinary Medicine Iowa State University for their assistance with pig sampling and necropsy.
Abbreviations
- BALT
Bronchus-associated lymphoid tissues
- DNA
Deoxyribonucleic acid
- DPI
Day post inoculation
- ELISA
Enzyme-linked immunosorbent assay
- H&E
Hematoxylin and eosin
- IAV
Influenza A virus
- ISU VDL
Iowa State University Veterinary Diagnostic Laboratory
- MHP
Mycoplasma hyopneumoniae
- PCR
Polymerase chain reaction
- PCV2
Porcine circovirus 2
- PEP
Porcine enzootic pneumonia
- PIL
Peribronchiolar infiltration by lymphocytes
- PRRSV
Porcine reproductive and respiratory syndrome virus
Authors' contributions
BA, MC, JZ, and RD designed the experiment. AS coordinated the animal study, data collection, statistical analyses, designed graphs, and wrote the manuscript. GS and FF assisted in sample collection. MZ and CW performed statistical analyses. EF and DP contributed with air samplers and digital recording systems for cough. BA, RD, MC, and JZ assisted with the animal study and edited the manuscript. All authors read and approved the final manuscript.
Funding
Funding provided by the Iowa Pork Producers Association (Clive, IA USA), grant no. #18-133 IPPA.
Availability of data and materials
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
All procedures were conducted with the approval of the Iowa State University Office for Responsible Research and Institutional Animal Care and Use Committee.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Swine Health Information Center. In: Swine bacterial disease matrix. Swine Health Information Center. 2021. https://www.swinehealth.org/swine-bacterial-disease-matrix/. Accessed 28 Jun 2021.
- 2.Maes D, Sibila M, Kuhnert P, Segalés J, Haesebrouck F, Pieters M. Update on Mycoplasma hyopneumoniae infections in pigs: knowledge gaps for improved disease control. Transbound Emerg Dis. 2018;65:110–124. doi: 10.1111/tbed.12677. [DOI] [PubMed] [Google Scholar]
- 3.DeBey MC, Ross RF. Ciliostasis and loss of cilia induced by Mycoplasma hyopneumoniae in porcine tracheal organ cultures. Infect Immun. 1994;62(12):5312. doi: 10.1128/iai.62.12.5312-5318.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leal Zimmer FM, Paes JA, Zaha A, Ferreira HB. Pathogenicity & virulence of Mycoplasma hyopneumoniae. Virulence. 2020;11(1):1600–1622. doi: 10.1080/21505594.2020.1842659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park SC, Yibchok-Anun S, Cheng H, Young TF, Thacker EL, Minion FC, Ross RF, Hsu WH. Mycoplasma hyopneumoniae increases intracellular calcium release in porcine ciliated tracheal cells. Infect Immun. 2002;70(5):2502. doi: 10.1128/IAI.70.5.2502-2506.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fourour S, Marois-Créhan C, Martelet L, Fablet C, Kempf I, Gottschalk M, Segura M. Intra-species and inter-species differences in cytokine production by porcine antigen-presenting cells stimulated by Mycoplasma hyopneumoniae, M. hyorhinis, and M. flocculare. Pathogens. 2019;8(1):34. doi: 10.3390/pathogens8010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siqueira FM, Gerber AL, Guedes RL, Almeida LG, Schrank IS, Vasconcelos AT, Zaha A. Unravelling the transcriptome profile of the swine respiratory tract mycoplasmas. PLoS ONE. 2014;9(10):e110327. doi: 10.1371/journal.pone.0110327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuo LL, Wu YM, You XX. Mycoplasma lipoproteins and Toll-like receptors. J Zhejiang Univ Sci B. 2009;10(1):67–76. doi: 10.1631/jzus.B0820256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Citti C, Nouvel LX, Baranowski E. Phase and antigenic variation in mycoplasmas. Future Microbiol. 2010;5(7):1073–1085. doi: 10.2217/fmb.10.71. [DOI] [PubMed] [Google Scholar]
- 10.Opriessnig T, Thacker EL, Yu S, Fenaux M, Meng XJ, Halbur PG. Experimental reproduction of postweaning multisystemic wasting syndrome in pigs by dual infection with Mycoplasma hyopneumoniae and porcine circovirus type 2. Vet Pathol. 2004;41(6):624–640. doi: 10.1354/vp.41-6-624. [DOI] [PubMed] [Google Scholar]
- 11.Thacker EL, Halbur PG, Ross RF, Thanawongnuwech R, Thacker BJ. Mycoplasma hyopneumoniae potentiation of porcine reproductive and respiratory syndrome virus-induced pneumonia. J Clin Microbiol. 1999;37(3):620–627. doi: 10.1128/JCM.37.3.620-627.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greiner M, Gardner IA. Epidemiologic issues in the validation of veterinary diagnostic tests. Prev Vet Med. 2000;45(1–2):3–22. doi: 10.1016/S0167-5877(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 13.Thacker EL. Diagnosis of Mycoplasma hyopneumoniae. Anim Health Res Rev. 2004;5(2):317–320. doi: 10.1079/AHR200491. [DOI] [PubMed] [Google Scholar]
- 14.Poeta Silva AP, Magtoto RL, Souza Almeida HM, McDaniel A, Magtoto PD, Derscheid RJ, Merodio MM, Matias Ferreyra FS, Gatto IR, Baum DH, Clavijo MJ. Performance of commercial Mycoplasma hyopneumoniae serum enzyme-linked immunosorbent assays under experimental and field conditions. J Clin Microbiol. 2020;58(12):e00485–e520. doi: 10.1128/JCM.00485-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Almeida HM, Mechler-Dreibi ML, Sonálio K, Ferreira MM, Martinelli PE, Gatto IR, Maes D, Montassier HJ, Oliveira LG. Dynamics and chronology of Mycoplasma hyopneumoniae strain 232 infection in experimentally inoculated swine. Porcine Health Manag. 2021;7(1):1–8. doi: 10.1186/s40813-020-00179-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumeister AK, Runge M, Ganter M, Feenstra AA, Delbeck F, Kirchhoff H. Detection of Mycoplasma hyopneumoniae in bronchoalveolar lavage fluids of pigs by PCR. J Clin Microbiol. 1998;36(7):1984–1988. doi: 10.1128/JCM.36.7.1984-1988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clavijo MJ, Hu D, Krantz S, Cano JP, Maróstica TP, Henao-Diaz A, Poeta Silva APS, Hemker D, Tapia E, Zimmerman S, Fano E. Mycoplasma hyopneumoniae surveillance in pig populations: establishing sampling guidelines for detection in growing pigs. J Clin Microbiol. 2021 doi: 10.1128/JCM.03051-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betlach AM, Valeris-Chacin R, Singer RS, Allerson M, Pieters M. Natural transmission and detection of Mycoplasma hyopneumoniae in a naïve gilt population. Vet Microbiol. 2020;248:108819. doi: 10.1016/j.vetmic.2020.108819. [DOI] [PubMed] [Google Scholar]
- 19.Sponheim A, Alvarez J, Fano E, Schmaling E, Dee S, Hanson D, Wetzell T, Pieters M. Comparison of the sensitivity of laryngeal swabs and deep tracheal catheters for detection of Mycoplasma hyopneumoniae in experimentally and naturally infected pigs early and late after infection. Vet Microbiol. 2020;241:108500. doi: 10.1016/j.vetmic.2019.108500. [DOI] [PubMed] [Google Scholar]
- 20.Pieters M, Daniels J, Rovira A. Comparison of sample types and diagnostic methods for in vivo detection of Mycoplasma hyopneumoniae during early stages of infection. Vet Microbiol. 2017;1(203):103–109. doi: 10.1016/j.vetmic.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Henao-Diaz A, Giménez-Lirola L, Baum DH, Zimmerman J. Guidelines for oral fluid-based surveillance of viral pathogens in swine. Porcine Health Manag. 2020;6(1):1–2. doi: 10.1186/s40813-020-00168-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernandez-Garcia J, Robben N, Magnée D, Eley T, Dennis I, Kayes SM, Thomson JR, Tucker AW. The use of oral fluids to monitor key pathogens in porcine respiratory disease complex. Porcine Health Manag. 2017;3(1):1–3. doi: 10.1186/s40813-017-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minion FC, Lefkowitz EJ, Madsen ML, Cleary BJ, Swartzell SM, Mahairas GG. The genome sequence of Mycoplasma hyopneumoniae strain 232, the agent of swine mycoplasmosis. J Bacteriol. 2004;186(21):7123–7133. doi: 10.1128/JB.186.21.7123-7133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zielinski GC, Ross RF. Effect of growth in cell cultures and strain on virulence of Mycoplasma hyopneumoniae for swine. Am J Vet Res. 1990;51(3):344–348. [PubMed] [Google Scholar]
- 25.Pieters M, Pijoan C, Fano E, Dee S. An assessment of the duration of Mycoplasma hyopneumoniae infection in an experimentally infected population of pigs. Vet Microbiol. 2009;134(3–4):261–266. doi: 10.1016/j.vetmic.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 26.Poeta Silva AP, Marostica TP, McDaniel A, Arruda BL, Alonso C, Derscheid R, Yeske P, Linhares DC, Giménez-Lirola L, Karriker L, Fano E. Comparison of Mycoplasma hyopneumoniae response to infection by route of exposure. Vet Microbiol. 2021;15:109118. doi: 10.1016/j.vetmic.2021.109118. [DOI] [PubMed] [Google Scholar]
- 27.Strait EL, Madsen ML, Minion FC, Christopher-Hennings J, Dammen M, Jones KR, Thacker EL. Real-time PCR assays to address genetic diversity among strains of Mycoplasma hyopneumoniae. J Clin Microbiol. 2008;46(8):2491–2498. doi: 10.1128/JCM.02366-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Underwood W, Anthony R. AVMA guidelines for the euthanasia of animals: 2020 edition. Retrieved on March 2020;2013(30):2020-1.
- 29.Halbur PG, Paul PS, Frey ML, Landgraf J, Eernisse K, Meng XJ, Lum MA, Andrews JJ, Rathje JA. Comparison of the pathogenicity of two US porcine reproductive and respiratory syndrome virus isolates with that of the Lelystad virus. Vet Pathol. 1995;32(6):648–660. doi: 10.1177/030098589503200606. [DOI] [PubMed] [Google Scholar]
- 30.Schrader C, Schielke A, Ellerbroek L, Johne R. PCR inhibitors–occurrence, properties and removal. J Appl Microbiol. 2012;113(5):1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 31.Dee S, Otake S, Oliveira S, Deen J. Evidence of long-distance airborne transport of porcine reproductive and respiratory syndrome virus and Mycoplasma hyopneumoniae. Vet Res. 2009;40(4):1–3. doi: 10.1051/vetres/2009022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roos LR, Fano E, Homwong N, Payne B, Pieters M. A model to investigate the optimal seeder-to-naïve ratio for successful natural Mycoplasma hyopneumoniae gilt exposure prior to entering the breeding herd. Vet Microbiol. 2016;29(184):51–58. doi: 10.1016/j.vetmic.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 33.Pessoa J, da Costa MR, Manzanilla EG, Norton T, McAloon C, Boyle L. Managing respiratory disease in finisher pigs: Combining quantitative assessments of clinical signs and the prevalence of lung lesions at slaughter. Prev Vet Med. 2021;186:105208. doi: 10.1016/j.prevetmed.2020.105208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.