Abstract
Chromatin is a supramolecular DNA-protein complex that compacts eukaryotic genomes and regulates their accessibility and functions. Dynamically disordered histone H3 N-terminal tails are among key chromatin regulatory components. Here, we used high-resolution-magic-angle-spinning NMR measurements of backbone amide 15N spin relaxation rates to investigate, with residue-specific detail, the dynamics and interactions of H3 tails in recombinant 13C,15N-enriched nucleosome arrays containing 15, 30 or 60 bp linker DNA between the nucleosome repeats. These measurements were compared to analogous data available for mononucleosomes devoid of linker DNA or containing two 20 bp DNA overhangs. The H3 tail dynamics in nucleosome arrays were found to be considerably attenuated compared to nucleosomes with or without linker DNA due to transient electrostatic interactions with the linker DNA segments and the structured chromatin environment. Remarkably, however, the H3 tail dynamics were not modulated by the specific linker DNA length within the 15–60 bp range investigated here.
Graphical Abstract
Chromatin is a dynamic supramolecular protein-DNA complex that compacts the genome and regulates DNA accessibility in eukaryotic cells. The building block of chromatin is the nucleosome, which is made up of ~147 base pairs (bp) of DNA double helix wrapped ~1.7 times around a histone octamer complex containing two copies each of highly conserved H2A, H2B, H3, and H4 histone proteins.1 Each histone contains an intrinsically disordered N-terminal tail domain that extends out of the nucleosome1 and retains considerable flexibility even in highly condensed mononucleosomes2,3 and nucleosome arrays.4 This conformational plasticity of the histone tails appears to play an important role in the regulation of chromatin structure and recruitment of regulatory protein complexes.5–7
The histone H3 N-terminal tail, spanning ~35 amino acid (aa) residues, is the longest among the histone tail domains and contains numerous post-translational modification (PTM) sites.8 For example, methylation of H3K4 and H3K36 are key transcription activating PTMs, while methylation of H3K9 and H3K27 are transcription silencing PTMs.9 Given that distinct parts of histone tails recruit specific regulatory complexes, changes in the local conformation and flexibility along the tails may influence their interactions with protein complexes that recognize histone PTMs, i.e. histone readers. Additionally, multiple studies point to the presence of transient interactions between the histone H3 N-terminal tails and DNA.10 These interactions, which likely involve both nucleosomal and linker DNA,11–14 are expected to compete with the interactions of H3 tails with chromatin regulators including chromatin remodeling complexes and transcription coactivators.15,16
Linker DNA, the segment of DNA that connects neighboring nucleosomes, significantly impacts chromatin structure since a change in length of 5 bp changes the orientation of adjacent nucleosomes by 180 degrees.17 Linker DNA length also appears to affect the phase separation properties of chromatin.18 The length of linker DNA is regulated and varies between ~10–90 bp for different cell types and organisms.19,20 Shorter linker lengths (~40 bp or less) are generally found in simpler eukaryotes and associated with active transcription and longer linker lengths (~50 bp or more) are typically associated with mature transcriptionally-inactive cells,21 probably in part due to the binding of linker histone H1 for longer linker DNA lengths.22 In addition, although the length of linker DNA does not extensively vary in a given cell type,17 it is quantized23 in different cell types and has a profound effect on chromatin compaction extent and pattern. This results in nucleosome spacings of 10n+5 (n is an integer), which is predominant in several cell types,24–28 being less compactable29 and associated with active transcription30 compared to linker lengths of 10n. While histone H3 tails and linker DNA both affect chromatin structure and function and appear to interact with one another, it is unclear whether the length of the linker DNA modulates this interaction or has an impact on the conformational flexibility of H3 tail domains in chromatin.
In terms of atomic level understanding of histone H3 tail dynamics and interactions in nucleosomes, Stützer et al.12 have carried out an elegant comparative study based on measurements of solution state nuclear magnetic resonance (NMR) backbone amide 1H and 15N chemical shift perturbations and 15N longitudinal (R1) and transverse (R2) spin relaxation rates for a free histone H3 peptide (H3 residues 1–44), H3 in nucleosomes reconstituted with 187 base pairs (bp) of double-stranded DNA containing the 147 bp Widom 601 nucleosome positioning sequence31 flanked by 20 bp of linker DNA on each side, as well as H3 in the 187 bp DNA nucleosomes above also containing bound linker histone H1. The 15N relaxation measurements, which were used to infer site-specific rotational correlation times (τc) for histone H3 tail residues in the different contexts, revealed an approximately 4-fold reduction in tail dynamics within 187 bp DNA nucleosomes relative to the free peptide, and a further reduction in dynamics within nucleosomes complexed with H1, stemming from transient interactions of the H3 tail residues with linker DNA (but not with the linker histone H1).12 This study also demonstrated that charge-modulating PTMs weaken the H3 tail-linker DNA interactions and increase H3 tail dynamics. In a complementary study, which included solution NMR measurements of 1H and 15N chemical shifts (but not 15N spin relaxation rates), Morrison et al.14 investigated the histone H3 tail conformation and interactions in “minimal” nucleosomes reconstituted with 147 bp Widom 601 DNA containing no linker DNA. They concluded that the H3 tails engage in considerable interactions with nucleosomal DNA even in the absence of linker DNA.
In order to explore how the above findings reporting on the conformational dynamics and interactions of histone H3 tails in nucleosomes translate to larger assemblies of nucleosomes, we carried out magic-angle spinning (MAS) NMR measurements of residue-specific resonance intensities and backbone amide 15N spin relaxation rates in concentrated recombinant nucleosome arrays, reconstituted with 13C,15N-labeled H3 and a DNA template containing 16 repeats of the Widom 601 nucleosome positioning sequence, which allows for the nucleosome spacing to be precisely controlled. Furthermore, to investigate any dependence of histone H3 conformational dynamics and interactions on the linker DNA length, measurements were performed on arrays containing 15, 30 or 60 bp DNA linkers between successive nucleosome units. Rotational correlation times for the H3 tail residues in the 16-mer nucleosome arrays, determined from the 15N R1 and R2 values, were compared with those obtained in analogous fashion for 147 bp DNA nucleosomes as part of the present study as well as those reported by Stützer et al.12 for 187 bp DNA nucleosomes containing two 20 bp linker DNA overhangs.
The 16-mer nucleosome arrays with 15, 30 or 60 bp linker DNA and the control 147 bp DNA nucleosome sample were reconstituted with 13C,15N-labeled histone H3 as described in the Experimental Section (Figure 1A,B, Supporting Information (SI) Figures S1 and S2). The nucleosome array purity and level of saturation with histone octamer were assessed by using composite gel electrophoresis, AvaI restriction enzyme digestion, and atomic force microscopy (AFM) (Figure 1C, SI Figure S2). Altogether, these assays indicate that the nucleosome array samples used for the NMR measurements are highly homogeneous and effectively saturated (>95%) with histone octamer, in line with earlier studies.4 The increase in the array length and separation between adjacent nucleosomes as a function of increasing linker DNA length is also clearly visible in the AFM images of the nucleosome arrays (Figure 1C).
Figure 1.
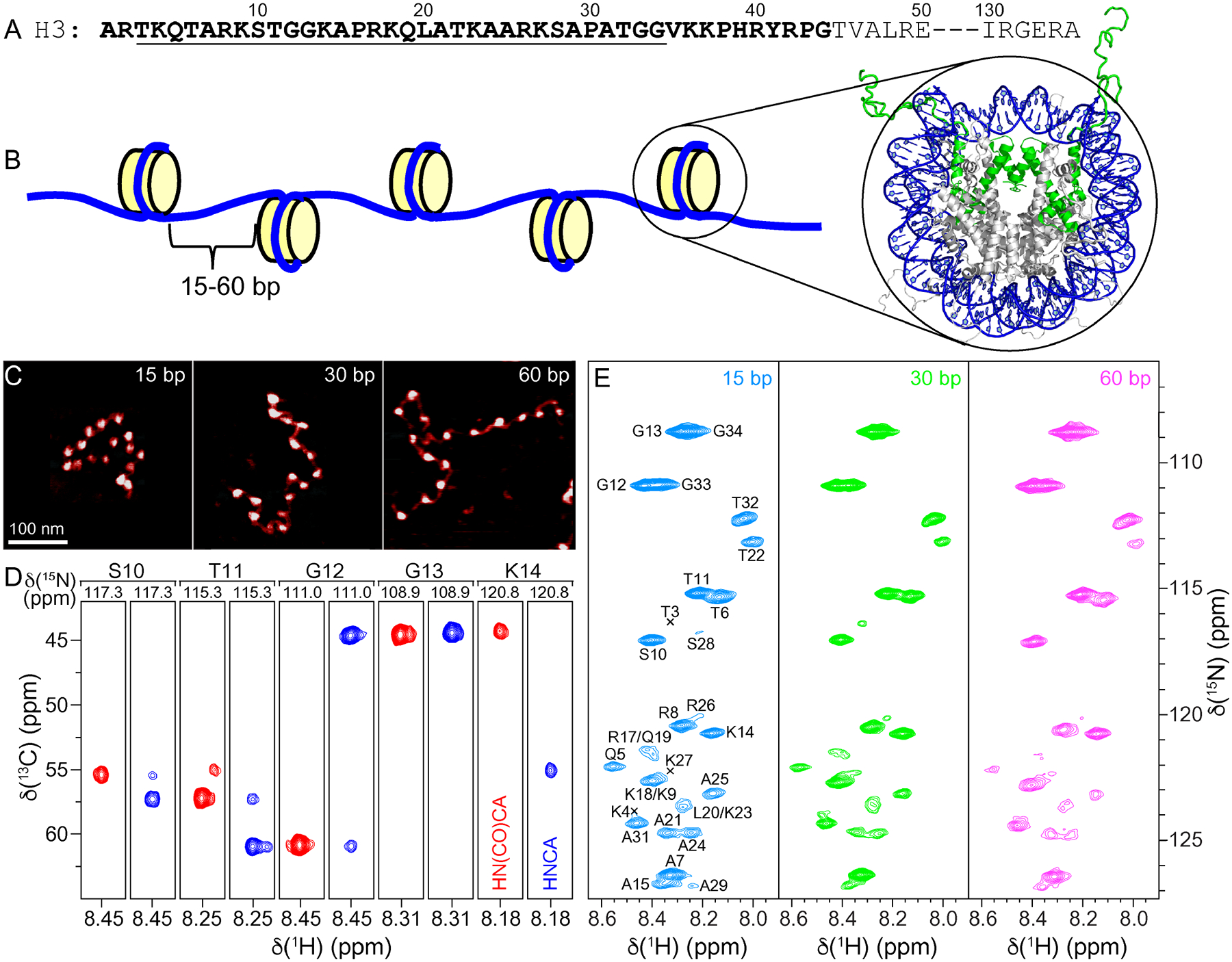
(A) Partial amino acid sequence of Xenopus laevis histone H3. Relatively unstructured residues based on the nucleosome core particle crystal structure are bold. Conformationally flexible residues detected in MAS NMR spectra of 16-mer nucleosome arrays in this study are underlined. (B) Schematic representation of the 16-mer nucleosome arrays with variable length (15, 30 or 60 bp) DNA linkers between nucleosome units. Also shown is the crystal structure of the nucleosome core particle (PDB entry 1KX5)32 with DNA and histone H3 colored blue and green, respectively, and histones H2A, H2B and H4 colored grey. (C) Representative AFM images of the 16-mer nucleosome arrays containing 15, 30 and 60 bp DNA linkers as indicated, showing the increase in internucleosome separation as a function of increasing linker DNA length. All images are shown on the same scale with the scale bar indicated in the leftmost panel. (D) Representative strips from 3D HNCA (blue contours) and HN(CO)CA (red contours) spectra of 16-mer nucleosome arrays with 15 bp DNA linkers reconstituted with 13C,15N-enriched histone H3, showing sequential connectivity for residues S10-K14. (E) 15N–1H HSQC spectra of 16-mer nucleosome arrays with 15 bp (blue), 30 bp (green), and 60 bp (magenta) DNA linkers, with the resonance assignments indicated. All spectra were recorded at 800 MHz 1H frequency, 10 kHz MAS rate, and sample temperature of ~35 °C.
Figure 1E shows INEPT-based 2D 15N-1H heteronuclear single quantum coherence (HSQC) MAS NMR spectra of histone H3 in the 16-mer nucleosome arrays with 15, 30 and 60 bp linker DNA length. These spectra each contain ~30 relatively well-resolved resonances, corresponding to the N-terminal H3 tail residues,2–4 and are nearly identical to one another in terms of the chemical shifts and relative resonance intensities irrespective of the linker DNA length. The latter finding suggests that the local environment of the histone H3 tail and its interactions with linker DNA are not significantly influenced by the precise linker DNA length within the 15–60 bp range investigated here.
To establish the sequential assignments of 15N-1H resonances for H3 tail residues in the nucleosome arrays, we recorded 3D HNCA and HN(CO)CA33 spectra under MAS for the 15 bp linker DNA array sample (Figure 1D). These spectra enabled unambiguous assignments to be obtained for 21 out of 30 non-proline residues in the H3 tail (aa 5–15, 20–25 and 31–34) detectable in the 15N-1H HSQC spectrum. The remaining non-proline H3 tail residues (aa 3–4, 17–19, 26–29), associated with particularly weak cross-peak intensities in the 15N-1H spectrum (Figure 2A) and not detected in the 3D HNCA and HN(CO)CA datasets for the nucleosome arrays, could be readily assigned by comparing the 15N-1H HSQC MAS NMR spectrum for the 15 bp linker DNA array sample to solution NMR 15N-1H HSQC spectra of nucleosomes reconstituted with isotope-enriched histone H3 reported previously in multiple independent studies2,12,14 and confirmed in the present study (not shown), for which the amide 1H and 15N chemical shifts of the different residues were generally within 0.1 and 1 ppm, respectively, of the corresponding ones in the array sample. In analogy to the 15N-1H spectra of histone H3 in nucleosome arrays recorded under MAS, the 15N-1H solution NMR spectra of H3 in nucleosomes contain exclusively correlations for the highly dynamically disordered tail residues (aa 3–34 and 3–36 for arrays and nucleosomes, respectively), with the two N-terminal residues not observed in neither arrays nor nucleosomes likely due to amide proton exchange.2,12,14
Figure 2.
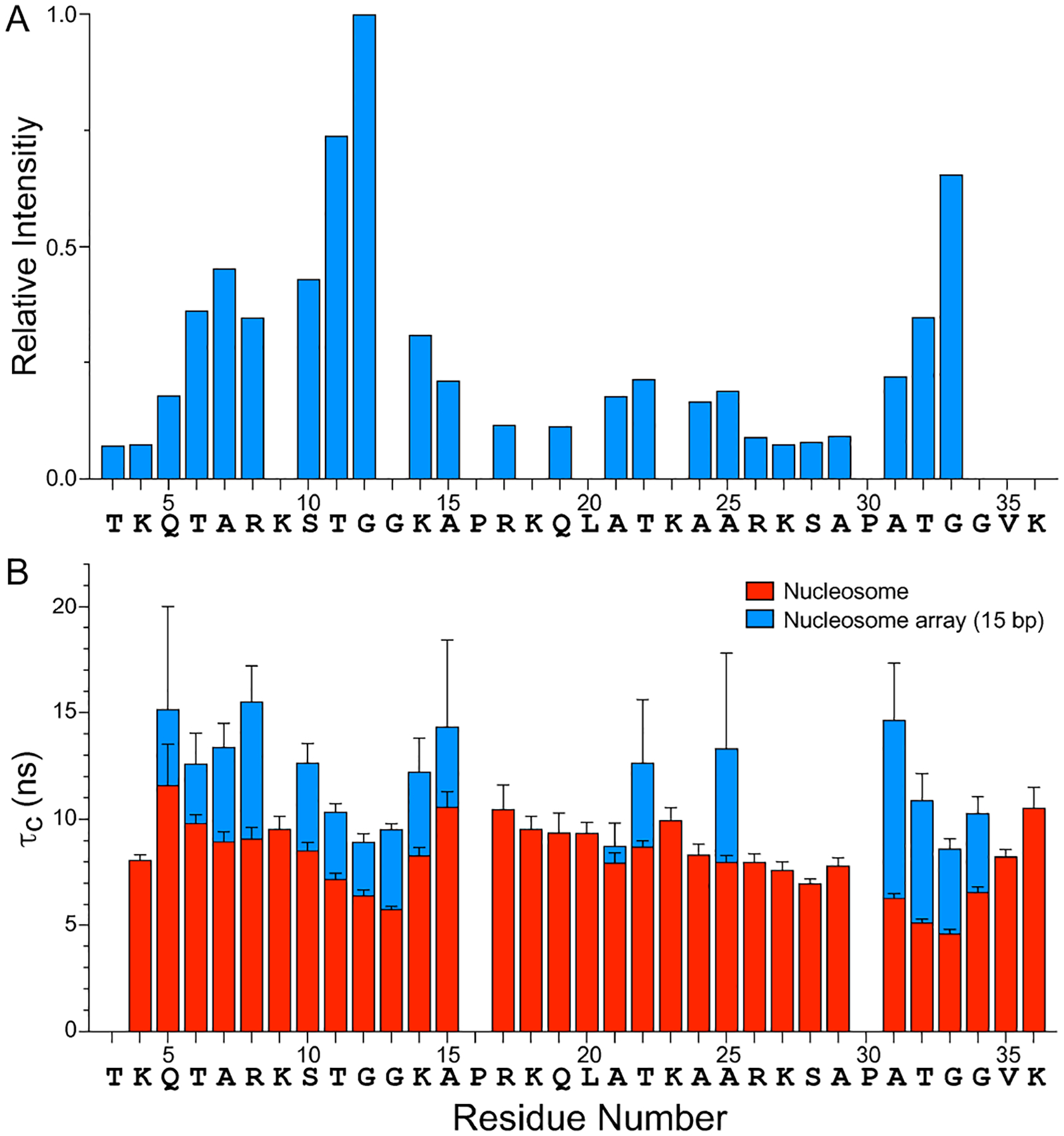
(A) Relative resonance intensities in the 15N–1H HSQC MAS NMR spectrum of histone H3 in 16-mer nucleosome arrays with 15 bp linker DNA (c.f., Figure 1E) as a function of residue number. The residue-specific resonance intensities are scaled according to the intensity for G12, and the intensities for residues K9, G13, K18, L20, K23 and G34 have been omitted due to spectral overlap which precluded the accurate extraction of resonance intensities. Nucleosome arrays with 30 and 60 bp linker DNA were found to exhibit nearly identical relative resonance intensity profiles (not shown). (B) Rotational correlation times, τc, for histone H3 tail residues in nucleosomes (red) and 16-mer nucleosome arrays with 15 bp linker DNA (blue) determined based on quantitative measurements of amide 15N R1 and R2 spin relaxation rates (SI Figure S3A,B). The τc vs. residue number profiles for nucleosome arrays with 30 and 60 bp linker DNA were found to be generally similar to the corresponding profile for the 15 bp linker DNA arrays (SI Figure S3C).
To obtain more detailed insights into the conformational flexibility and interactions of histone H3 tails in nucleosome arrays with different linker DNA lengths, we performed quantitative measurements of amide backbone 15N spin relaxation rate constants (SI Figure S3A,B) via series of 2D 15N-1H HSQC MAS NMR spectra;34,35 for reference, analogous measurements were carried out for histone H3 tail residues in 147 bp DNA nucleosomes in solution. Residue-specific rotational correlation times, τc, were calculated based on 15N R1 and R2 values according to Kay et al.36
| (1) |
where νN is the 15N resonance frequency in Hz.
Figure 2B shows plots of τc as a function of residue number for the H3 tails in 15 bp linker DNA nucleosome arrays and in nucleosomes. For the arrays, τc values for 17 of 30 non-proline H3 tail residues (aa 5–8, 10–15, 21–22, 25, and 31–34) could be determined, with data for the remaining residues not accessible due to resonance overlap and/or vanishing intensities in the 15N spin relaxation experiments, while for the nucleosomes τc values could be determined for all non-proline H3 tail residues detectable in the 15N-1H HSQC spectra (aa 3–36) except for T3. Comparison of these τc profiles reveals that all H3 tail residues in the nucleosome arrays consistently display higher τc values (by ~50% on average) relative to the corresponding residues in nucleosomes, indicative of a reduction in H3 tail dynamics in the arrays compared to nucleosomes. For the 15 bp linker DNA arrays, the average τc was found to be 12.0 ± 2.3 ns, with τc values for individual residues ranging from 8.6 to 15.5 ns, and for the nucleosomes the average τc was 8.3 ± 1.7 ns with τc values for individual residues ranging from 4.6 to 11.6 ns. Measurements of the 15N spin relaxation rates for the 30 bp and 60 bp linker DNA nucleosome array samples yielded τc profiles (SI Figure S3C) that, within experimental error, were overall effectively indistinguishable from that obtained for the 15 bp linker DNA arrays (average τc values of 11.0 ± 2.0 ns and 12.5 ± 2.8 ns for the 30 bp and 60 bp linker DNA arrays, respectively). These results are in line with the nearly identical 15N-1H correlation spectra obtained for all three 16-mer nucleosome array samples irrespective of the linker DNA length (Figure 1E). Note that the conformational dynamics of the H3 tails in nucleosomes and nucleosome arrays, which occur on the time scale of ~10 ns, exceed by more than an order of magnitude the overall rotational tumbling time of the nucleosome core particle (τrot ~ 150 ns)37–39 indicating that the observed differences in the τc profiles are not due to any differences in the global reorientation of nucleosomes vs. nucleosome arrays but rather reflect changes in the local dynamics of the H3 tail residues as a function of the chromatin environment.
The observed ~50% reduction in the H3 tail dynamics for the nucleosome arrays vs. 147 bp nucleosomes devoid of linker DNA points to enhanced transient interactions between the H3 tail residues and DNA due to the presence of the 15–60 bp linkers. As expected, and in line with earlier studies,12,14 the interactions of H3 tails with both nucleosomal and linker DNA are largely electrostatic in nature, evidenced by the fact that segments containing positively charged arginine and lysine residues generally show larger τc values indicative of relatively restricted motions while neutral segments such as the TGG motifs (aa 11–13 and 31–34) are associated with smaller τc values and hence more dynamic. In addition, the TGG motifs are likely to have higher inherent flexibility than the arginine/lysine rich regions, which we anticipate will also contribute to differences in flexibility along the H3 tail. Furthermore, the finding that no significant systematic differences are observed between the H3 tail τc profiles for nucleosome arrays with 15, 30 and 60 bp DNA linkers suggests that for the array samples the H3 tail domain interactions with additional DNA are confined to the DNA linker regions nearest to the nucleosome, involve ~15 bp or fewer, and are independent of nucleosome-nucleosome orientation. Since 15 bp corresponds to about 5 nm in length, these results are consistent with the H3 tail contour length of about 6 nm.32 In this context, it is also instructive to compare the H3 tail τc profiles obtained in the present study for the nucleosome arrays and “minimal” 147 bp DNA nucleosomes with the data reported by Stützer et al.12 for 187 bp DNA nucleosomes containing two 20 bp linker DNA overhangs. Specifically, inspection of the H3 tail τc profile presented for 187 bp DNA nucleosomes in Figure 2 of Stützer et al.12 reveals that, in terms of relative τc values for different residues along the H3 tail, it overall mirrors those presented here for the nucleosome arrays and 147 bp DNA mononucleosomes. Remarkably, the τc values for individual H3 tail residues in 187 bp DNA nucleosomes range from ~6 to ~13 ns with the average τc estimated to be in the ~10–11 ns regime. These τc values exceed those determined in this study for the 147 bp DNA nucleosomes (τc range of 4.6 to 11.6 ns and average τc of 8.3 ns) but fall below those determined for the nucleosome arrays (e.g., τc range of 8.6 to 15.5 ns and average τc 12.0 ns for the 15 bp linker DNA array sample). The successive reduction in H3 tail dynamics observed when going from 147 bp DNA nucleosomes to 187 bp DNA nucleosomes and finally to nucleosome arrays reinforces the notion that this reduction in histone tail mobility results from transient interactions between the H3 tail residues and linker DNA. These interactions appear to be enhanced in nucleosome arrays relative to a single nucleosome containing linker DNA overhangs, and we speculate that this is due at least in part to reduced conformational flexibility of the DNA linkers in the chromatin environment as well as possible interactions with non-adjacent linker and nucleosomal DNA. The finding that H3 tails in 187 bp DNA nucleosomes complexed with linker histone H1—which binds to and presumably immobilizes the linker DNA segments but does not interact directly with H3 tails—show reduced mobility (τc range of ~12 to ~20 ns and average τc of ~15 ns)12 lends additional support for this idea.
In summary, the conformational dynamics of histone H3 tail domains in “minimal” 147 bp Widom 601 DNA nucleosomes and oligonucleosome arrays containing 16 nucleosome repeats with intervening 15, 30 or 60 bp linker DNA segments were probed by using quantitative 15N NMR spin relaxation measurements. The ~35 N-terminal H3 residues are highly conformationally flexible overall in both nucleosomes and nucleosome arrays, in line with earlier studies,2–4,12,14 but the tail domain dynamics in the nucleosome arrays were found to be markedly attenuated compared to 147 bp DNA nucleosomes as well as 187 bp DNA nucleosomes containing two 20 bp linker DNA overhang regions.12 The reduced H3 tail mobility observed for the nucleosome arrays relative to mononucleosomes stems from transient electrostatic interactions between positively charged H3 residues and negatively charged linker DNA segments, which appear to be enhanced within the structured chromatin environment and can be modulated by PTMs at key sites that activate or suppress transcription. Remarkably, these transient interactions between H3 tail residues and linker DNA are not influenced by the precise length of the linker DNA regions within the 15 to 60 bp regime indicating that nucleosome-nucleosome orientation within chromatin does not influence the histone tail interactions with linker DNA.
EXPERIMENTAL SECTION
Preparation of DNA constructs.
For the nucleosome array samples, three DNA templates were prepared by ligating 16 tandem repeats of a 147 bp variant of the Widom 601 DNA high-affinity nucleosome positioning sequence (NPS).31 These DNA constructs were designed to contain 15, 30, or 60 bp of linker DNA between each nucleosome after array reconstitution (SI Figure S1A,B). The DNA templates were prepared using Qiagen Gigaprep kits and digested with DdeI (New England Biolabs). The digested DNA mixture (Figure S1C) contains a long linear double stranded DNA with the variable linker DNA length and seven shorter DNA fragments (653, 535, 421, 404, 245, 230, and 161 bp), which serve as buffering DNA in the process of nucleosome array reconstitution and help minimize non-specific aggregation of the arrays.40
For the nucleosome samples, a pJ201 plasmid containing 32-copies of the Widom 601 DNA variant was transformed in E. coli DH5α, amplified in Luria-Bertani rich medium, extracted by using a Qiagen Gigaprep kit, digested with EcoRV (New England Biolabs), and purified by PEG precipitation as described previously.41 Briefly, 4 M NaCl and 40% PEG 6000 were added to the EcoRV-digested plasmid in a 0.192:0.346:1 v/v/v ratio to precipitate the vector. The mixture was incubated on ice for 1 h, and centrifuged at 27,000 g and 4 °C for 20 min. The supernatant containing the 147 bp Widom 601 DNA sequence was collected and combined with 100% ice cold ethanol in a 1:2.5 v/v ratio. The precipitated DNA was centrifuged at 27,000 g and 4 °C for 40 min, the supernatant decanted, and the precipitate air-dried for ~10 min and dissolved in 1 × TE buffer (10 mM Tris pH 7.5, 1 mM EDTA pH 8.0).
Histone protein expression and purification.
Histones H2A, H2B, H3, and H4 (Xenopus laevis) were overexpressed in E. coli BL21(DE3)pLysS and purified as described previously42 using gel filtration and ion-exchange chromatography in 7 M urea followed by dialysis against a solution of 2 mM β-mercaptoethanol (BME) and lyophilized. Uniformly 13C,15N-labeled H3 was prepared by using a minimal medium with 13C glucose (3 g/L) and 15NH4Cl (1 g/L) (Cambridge Isotope Laboratories) as the sole carbon and nitrogen sources, respectively. Histone octamer containing 13C,15N-H3 was prepared by dissolving the four unfolded histone proteins (H2A, H2B, 13C,15N-labeled H3, and H4) at concentrations of ≤ 10 mg/ml in unfolding buffer (7 M guanidine hydrochloride, 20 mM Tris, 10 mM dithiothreitol, pH 7.5) in a H2A:H2B:H3:H4 molar ratio of 1.2:1.2:1:1, refolded by double dialysis against refolding buffer (1 × TE, 2 M NaCl, 5 mM BME, pH 8.0). After dialysis, the solution was removed from the dialysis bag, concentrated to < 5 mL by Amicon ultracentrifugal filters (30 kDa cutoff, MilliporeSigma) and purified by gel filtration chromatography in refolding Buffer (without BME) as described previously.42
Reconstitution of nucleosome arrays and nucleosomes.
Nucleosome array reconstitution was performed as follows. An aqueous solution was made in 0.5 × TE (5 mM Tris pH 7.5, 0.5 mM EDTA pH 8.0), 2 M NaCl, 1 mM BZA (benzamidine hydrochloride hydrate, MilliporeSigma) buffer, containing the DdeI digested DNA mixture ([DNA] ≤ 0.5 mg/ml) and histone octamer in a molar ratio of NPS:histone octamer of 1:1.5. This ensures an effectively complete saturation of all the nucleosome positioning sites, with excess histone octamer binding to the buffering DNA and avoiding precipitation. The nucleosomes were reconstituted as follows. DNA and histone octamer were combined in a molar ratio of DNA:histone octamer of 1:0.65, in an aqueous 0.5 × TE, 2 M NaCl and 1 mM BZA buffer. The NaCl was removed by double dialysis at 4 °C against 0.5 × TE, 1 mM BZA buffer.
Both nucleosome array and nucleosome samples were concentrated ~30-fold using Amicon ultracentrifugal filters (100 kDa and 30 kDa cut-off for arrays and nucleosomes, respectively). To separate pure nucleosomes and nucleosome arrays from free DNA and buffering DNA, the purification was followed by sucrose gradient centrifugation using sucrose gradients of 5–40% (for arrays) or 5–30% (for nucleosomes) in 0.5 × TE buffer. The fractions containing pure 16-mer nucleosome arrays or nucleosomes were combined and the sucrose was removed by exchanging into 0.5 × TE buffer using Amicon ultracentrifugal filters.
Electrophoretic mobility shift and atomic force microscopy assays.
The formation and purity of nucleosomes were confirmed by 5% native polyacrylamide gel electrophoresis (SI Figure S2A). The formation, purity, and level of saturation of the 16-mer nucleosome arrays with histone octamer were confirmed by using composite gel electrophoresis (1% agarose-2% polyacrylamide, 0.2 × TB running buffer: 18 mM Tris-borate) for as-prepared arrays and 5% polyacrylamide gel electrophoresis for AvaI-digested arrays (SI Figure S2B,C). This resulted in a single clear mononucleosome band which shows that the nucleosomes in the arrays contain full histone octamers and are well-positioned. The sucrose gradient-purified arrays were further analyzed by atomic force microscopy (AFM) as follows (SI Figure S2D). Freshly cleaved mica was rinsed with 2 × 200 μl of ultrapure water, followed by treatment with 50 μl of 10 ng/μl aqueous solution of poly-D-lysine (PL; MilliporeSigma) and an additional 2 × 200 μl ultrapure water rinse to remove any unbound PL, and finally, air-dried. The PL-treated mica was incubated for 5 min with 50 μl of a dilute (~0.2 nM) solution of nucleosome arrays in 0.1 × TE buffer, rinsed with 200 μl of ultrapure water and air-dried. The samples were imaged by a Bruker AXS Dimension Icon Atomic Force Microscope at a scan rate of 1 Hz.
NMR spectroscopy.
The 16-mer nucleosome array samples in 0.5 × TE buffer reconstituted with 13C,15N-labeled H3 were pelleted by ultracentrifugation for 16–24 h at 4 °C and ~400,000 g using a Beckman-Coulter TLA-100.3 rotor. The pellets were transferred to a Bruker Kel-F insert (~15 μl sample volume) and sealed using a plug and sealing screw to prevent sample dehydration during experiments. The insert was inserted into a Bruker 4 mm zirconia MAS rotor and capped with a Kel-F drive cap. The final NMR samples contained ~2–3 mg of nucleosome arrays with total protein:DNA ratios of ~1:1 (w/w) for each sample, corresponding to chromatin concentrations in the ~200 mg/ml regime. NMR spectra were recorded using a 800 MHz Bruker Avance III HD spectrometer equipped with a 4 mm high-resolution magic-angle spinning (HR-MAS) probe, and processed and analyzed using NMRPipe43, Sparky44 and nmrglue.45 The MAS rate and sample temperature during the experiments were actively controlled at 10 kHz and ca. 35 °C, respectively. Sequential assignments of the H3 tail residues were established by using conventional 3D HNCA and HN(CO)CA experiments,33 with chemical shift evolution periods of ca. 80, 10 and 5 ms in the 1H, 15N and 13C dimensions, respectively. 15N longitudinal relaxation, R1, and longitudinal relaxation in the rotating frame, R1ρ, rate constants were determined from series of 2D 15N-1H correlation spectra34,35 recorded with chemical shift evolution periods of ca. 80 and 60 ms in the 1H and 15N dimensions, respectively, and R1 and R1ρ relaxation delays of 40, 160, 320, 480, 640 and 960 ms and 0, 10, 20, 40, 60 and 80 ms, respectively. For the R1ρ experiments, the 15N spin-lock field strength was 2 kHz. Residue-specific 15N R2 relaxation rate constants were calculated using the corresponding R1 and R1ρ values according to: R2 = R1ρ/sin2θ − R1/tan2θ, where θ = tan−1(ω1/Ω), ω1 is the 15N spin-lock field strength and Ω is the resonance offset. Note that perdeuteration followed by back-exchange of amide protons was not required in the context of 15N spin relaxation measurements in the present study given the significant conformational flexibility of histone H3 tails.
Solution NMR experiments on nucleosomes reconstituted with 13C,15N-labeled H3, analogous to the MAS NMR experiments described above for the nucleosome arrays, were recorded using a 850 MHz Bruker Avance III HD spectrometer equipped with a TCI CryoProbe. The NMR sample consisted of nucleosomes at a concentration of ~35 μM in 0.5 × TE buffer (200 μl sample volume) in a 3 mm tube.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (R01GM118664 to C.P.J., R01GM123743 to C.P.J. and M.G.P. and S10OD012303 to C.P.J.) and the National Science Foundation (MCB-1715174 to C.P.J.). We thank Prof. Catherine Musselman for providing the pJ201 plasmid.
Footnotes
SUPPORTING INFORMATION
Figures with plasmid map, gel electrophoresis and atomic force microscopy assays, and NMR relaxation data (PDF).
References:
- (1).Luger K; Mäder AW; Richmond RK; Sargent DF; Richmond TJ Crystal Structure of the Nucleosome Core Particle at 2.8 Å Resolution. Nature 1997, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- (2).Zhou BR; Feng H; Ghirlando R; Kato H; Gruschus J; Bai Y Histone H4 K16Q Mutation, an Acetylation Mimic, Causes Structural Disorder of Its N-Terminal Basic Patch in the Nucleosome. J. Mol. Biol 2012, 421, 30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Xiang SQ; le Paige UB; Horn V; Houben K; Baldus M; van Ingen H Site-Specific Studies of Nucleosome Interactions by Solid-State NMR Spectroscopy. Angew. Chem. Int. Ed 2018, 57, 4571–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Gao M; Nadaud PS; Bernier MW; North JA; Hammel PC; Poirier MG; Jaroniec CP Histone H3 and H4 N-Terminal Tails in Nucleosome Arrays at Cellular Concentrations Probed by Magic Angle Spinning NMR Spectroscopy. J. Am. Chem. Soc 2013, 135, 15278–15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Pepenella S; Murphy KJ; Hayes JJ Intra- and Inter-Nucleosome Interactions of the Core Histone Tail Domains in Higher-Order Chromatin Structure. Chromosoma 2014, 123, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Taverna SD; Li H; Ruthenburg AJ; Allis CD; Patel DJ How Chromatin-Binding Modules Interpret Histone Modifications: Lessons from Professional Pocket Pickers. Nat. Struct. Mol. Biol 2007, 14, 1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Musselman CA; Khorasanizadeh S; Kutateladze TG Towards Understanding Methyllysine Readout. Biochim. Biophys. Acta 2014, 1839, 686–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Rothbart SB; Strahl BD Interpreting the Language of Histone and DNA Modifications. Biochim. Biophys. Acta - Gene Regul. Mech 2014, 1839, 627–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zentner GE; Henikoff S Regulation of Nucleosome Dynamics by Histone Modifications. Nat. Struct. Mol. Biol 2013, 20, 259–266. [DOI] [PubMed] [Google Scholar]
- (10).Kan P-Y; Lu X; Hansen JC; Hayes JJ The H3 Tail Domain Participates in Multiple Interactions during Folding and Self-Association of Nucleosome Arrays. Mol. Cell. Biol 2007, 27, 2084–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zheng C; Lu X; Hansen JC; Hayes JJ Salt-Dependent Intra- and Internucleosomal Interactions of the H3 Tail Domain in a Model Oligonucleosomal Array. J. Biol. Chem 2005, 280, 33552–33557. [DOI] [PubMed] [Google Scholar]
- (12).Stützer A; Liokatis S; Kiesel A; Schwarzer D; Sprangers R; Söding J; Selenko P; Fischle W Modulations of DNA Contacts by Linker Histones and Post-Translational Modifications Determine the Mobility and Modifiability of Nucleosomal H3 Tails. Mol. Cell 2016, 61, 247–259. [DOI] [PubMed] [Google Scholar]
- (13).Rhee HS; Bataille AR; Zhang L; Pugh BF Subnucleosomal Structures and Nucleosome Asymmetry across a Genome. Cell 2014, 159, 1377–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Morrison EA; Bowerman S; Sylvers KL; Wereszczynski J; Musselman CA The Conformation of the Histone H3 Tail Inhibits Association of the BPTF PHD Finger with the Nucleosome. Elife 2018, 7, e31481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Musselman CA; Ramiŕez J; Sims JK; Mansfield RE; Oliver SS; Denu JM; Mackay JP; Wade PA; Hagman J; Kutateladze TG Bivalent Recognition of Nucleosomes by the Tandem PHD Fingers of the CHD4 ATPase Is Required for CHD4-Mediated Repression. Proc. Natl. Acad. Sci. USA 2012, 109, 787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Gatchalian J; Wang X; Ikebe J; Cox KL; Tencer AH; Zhang Y; Burge NL; Di L; Gibson MD; Musselman CA; Poirier MG; Kono H; Hayes JJ; Kutateladze TG Accessibility of the Histone H3 Tail in the Nucleosome for Binding of Paired Readers. Nat. Commun 2017, 8, 1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Grigoryev SA Chromatin Higher-Order Folding: A Perspective with Linker DNA Angles. Biophys. J 2018, 114, 2290–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Gibson BA; Doolittle LK; Schneider MWG; Jensen LE; Gamarra N; Henry L; Gerlich DW; Redding S; Rosen MK Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 2019, 179, 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Lantermann AB; Straub T; Strålfors A; Yuan GC; Ekwall K; Korber P Schizosaccharomyces Pombe Genome-Wide Nucleosome Mapping Reveals Positioning Mechanisms Distinct from Those of Saccharomyces Cerevisiae. Nat. Struct. Mol. Biol 2010, 17, 251–257. [DOI] [PubMed] [Google Scholar]
- (20).Hughes AL; Rando OJ Mechanisms Underlying Nucleosome Positioning in Vivo. Annu. Rev. Biophys 2014, 43, 41–63. [DOI] [PubMed] [Google Scholar]
- (21).Perišić O; Collepardo-Guevara R; Schlick T Modeling Studies of Chromatin Fiber Structure as a Function of DNA Linker Length. J. Mol. Biol 2010, 403, 777–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Woodcock CL; Skoultchi AI; Fan Y Role of Linker Histone in Chromatin Structure and Function: H1 Stoichiometry and Nucleosome Repeat Length. Chromosom. Res 2006, 14, 17–25. [DOI] [PubMed] [Google Scholar]
- (23).Widom J A Relationship between the Helical Twist of DNA and the Ordered Positioning of Nucleosomes in All Eukaryotic Cells. Proc. Natl. Acad. Sci. USA 1992, 89, 1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Lohr DE Detailed Analysis of the Nucleosomal Organization of Transcribed DNA in Yeast Chromatin. Biochemistry 1981, 20, 5966–5972. [DOI] [PubMed] [Google Scholar]
- (25).Brogaard K; Xi L; Wang JP; Widom J A Map of Nucleosome Positions in Yeast at Base-Pair Resolution. Nature 2012, 486, 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Strauss F; Prunell A Nucleosome Spacing in Rat Liver Cluomatin. A Study with Exonuclease III. Nucleic Acids Res. 1982, 10, 2275–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lohr D The Salt Dependence of Chicken and Yeast Chromatin Structure. Effects on Internucleosomal Organization and Relation to Active Chromatin. J. Biol. Chem 1986, 261, 9904–9914. [PubMed] [Google Scholar]
- (28).Voong LN; Xi L; Sebeson AC; Xiong B; Wang JP; Wang X Insights into Nucleosome Organization in Mouse Embryonic Stem Cells through Chemical Mapping. Cell 2016, 167, 1555–1570.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Correll SJ; Schubert MH; Grigoryev SA Short Nucleosome Repeats Impose Rotational Modulations on Chromatin Fibre Folding. EMBO J. 2012, 31, 2416–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Norouzi D; Katebi A; Cui F; Zhurkin VB Topological Diversity of Chromatin Fibers: Interplay between Nucleosome Repeat Length, DNA Linking Number and the Level of Transcription. AIMS Biophys. 2015, 2, 613–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Lowary PT; Widom J New DNA Sequence Rules for High Affinity Binding to Histone Octamer and Sequence-Directed Nucleosome Positioning. J. Mol. Biol 1998, 276, 19–42. [DOI] [PubMed] [Google Scholar]
- (32).Davey CA; Sargent DF; Luger K; Maeder AW; Richmond TJ Solvent Mediated Interactions in the Structure of the Nucleosome Core Particle at 1.9 Å Resolution. J. Mol. Biol 2002, 319, 1097–1113. [DOI] [PubMed] [Google Scholar]
- (33).Grzesiek S; Bax A Improved 3D Triple-Resonance NMR Techniques Applied to a 31 kDa Protein. J. Magn. Reson 1992, 96, 432–440. [Google Scholar]
- (34).Farrow NA; Muhandiram R; Pascal SM; Kay LE; Singer AU; Forman-Kay JD; Kay CM; Gish G; Pawson T; Shoelson SE Backbone Dynamics of a Free and a Phosphopeptide-Complexed Src Homology 2 Domain Studied by 15N NMR Relaxation. Biochemistry 1994, 33, 5984–6003. [DOI] [PubMed] [Google Scholar]
- (35).Korzhnev DM; Skrynnikov NR; Millet O; Torchia DA; Kay LE An NMR Experiment for the Accurate Measurement of Heteronuclear Spin-Lock Relaxation Rates. J. Am. Chem. Soc 2002, 124, 10743–10753. [DOI] [PubMed] [Google Scholar]
- (36).Kay LE; Torchia DA; Bax A Backbone Dynamics of Proteins As Studied by 15N Inverse Detected Heteronuclear NMR Spectroscopy: Application to Staphylococcal Nuclease. Biochemistry 1989, 28, 8972–8979. [DOI] [PubMed] [Google Scholar]
- (37).Feigon J; Kearns DR 1H NMR Investigation of the Conformational States of DNA in Nucleosome Core Particles. Nucleic Acids Res. 1979, 6, 2327–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Brown DW; Libertini LJ; Small EW Fluorescence Anisotropy Decay of Ethidium Bound to Nucleosome Core Particles. 1. Rotational Diffusion Indicates an Extended Structure at Low Ionic Strength. Biochemistry 1991, 30, 5293–5303. [DOI] [PubMed] [Google Scholar]
- (39).Rabdano SO; Shannon MD; Izmailov SA; Gonzalez Salguero N; Zandian M; Purusottam RN; Poirier MG; Skrynnikov NR; Jaroniec CP Histone H4 Tails in Nucleosomes: A Fuzzy Interaction with DNA. Angew. Chem. Int. Ed 2021, 60, 6480–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Dorigo B; Schalch T; Bystricky K; Richmond TJ Chromatin Fiber Folding: Requirement for the Histone H4 N-Terminal Tail. J. Mol. Biol 2003, 327, 85–96. [DOI] [PubMed] [Google Scholar]
- (41).Dyer PN; Edayathumangalam RS; White CL; Bao Y; Chakravarthy S; Muthurajan UM; Luger K Reconstitution of Nucleosome Core Particles from Recombinant Histones and DNA. Methods Enzymol. 2003, 375, 23–44. [DOI] [PubMed] [Google Scholar]
- (42).Luger K; Rechsteiner TJ; Richmond TJ Preparation of Nucleosome Core Particle from Recombinant Histones. Methods Enzymol. 1999, 304, 3–19. [DOI] [PubMed] [Google Scholar]
- (43).Delaglio F; Grzesiek S; Vuister GW; Zhu G; Pfeifer J; Bax A NMRPipe: A Multidimensional Spectral Processing System Based on UNIX Pipes. J. Biomol. NMR 1995, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- (44).Goddard T; Kneller D SPARKY 3. University of Califronia: San Francisco: 2006. [Google Scholar]
- (45).Helmus JJ; Jaroniec CP Nmrglue: An Open Source Python Package for the Analysis of Multidimensional NMR Data. J. Biomol. NMR 2013, 55, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


