Abstract
Background
There are no validated biomarkers that can aid clinicians in selecting who would best benefit from anticytotoxic T lymphocyte-associated antigen 4 monotherapy versus combination checkpoint blockade in patients with advanced melanoma who have progressive disease after programmed death 1 (PD-1) blockade.
Methods
We conducted a randomized multicenter phase II trial in patients with advanced melanoma. Patients were randomly assigned to receive either 1 mg/kg of nivolumab plus 3 mg/kg of ipilimumab or 3 mg/kg of ipilimumab every 3 weeks for up to four doses. Patients were stratified by histological subtype and prior response to PD-1 therapy. The primary clinical objective was overall response rate by week 18. Translational biomarker analyses were conducted in patients with blood and tissue samples.
Results
Objective responses were seen in 5 of 9 patients in the ipilimumab arm and 2 of 10 patients in the ipilimumab+nivolumab arm; disease control rates (DCRs) (66.7% vs 60.0%) and rates of grade 3–4 adverse events (56% vs 50%) were comparable between arms. In a pooled analysis, patients with clinical benefit (CB), defined as Response Evaluation Criteria in Solid Tumors response or progression-free for 6 months, showed increased circulating CD4+ T cells with higher polyfunctionality and interferon gamma production following treatment. Tumor profiling revealed enrichment of NRAS mutations and activation of transcriptional programs associated with innate and adaptive immunity in patients with CB.
Conclusions
In patients with advanced melanoma that previously progressed on PD-1 blockade, objective responses were seen in both arms, with comparable DCRs. Findings from biomarker analyses provided hypothesis-generating signals for validation in future studies of larger patient cohorts.
Trial registration number
Keywords: melanoma, tumor microenvironment, immunotherapy
Introduction
Survival outcomes for patients with advanced melanoma have markedly improved over the past decade with the introduction and Food and Drug Administration (FDA) approval of immune checkpoint inhibitors (ICIs)1 and BRAF and MEK targeted inhibitors for patients with BRAF mutated tumors. ICIs enhance antitumor immunity by blocking negative regulators of T-cell function. There are three types of FDA-approved ICI for the treatment of advanced melanoma: the anticytotoxic T lymphocyte-associated antigen 4 (CTLA-4) antibody ipilimumab, the antiprogrammed death 1 (PD-1) antibodies pembrolizumab and nivolumab, and the antiprogrammed death ligand 1 (PD-L1) antibody, atezolizumab (given in combination with vemurafenib and cobimetinib). CTLA-4 is expressed during the early stages of T-cell activation and is maintained on a subset of activated T cells, acting as a negative regulator of T-cell receptor signaling and T-cell activation.2 3 PD-1 is a coinhibitory receptor that is expressed by effector T cells on activation.4 Cognate interactions between PD-1 and its ligands expressed on tumor cells (PD-L1) and tumor-infiltrating immune cells (PD-L1 and PD-L2) result in reduced T-cell proliferation,5 cytokine production6 7 and decreased survival.8 9 Mechanism of action studies have revealed that CTLA-4 and PD-1 blockade have complementary, non-overlapping effects on T cells,10 suggesting that combination therapies could result in additive or synergistic effects. Ipilimumab and PD-1 blocking antibodies have both demonstrated an overall survival (OS) benefit for patients with advanced melanoma.11–13 In untreated patients, the two drugs appear to have additional activity when combined, with a median OS of more than 60 months compared with 36.9 months for nivolumab alone and 19.9 months for ipilimumab alone.1 However, this comes at a cost of an increased rate of high-grade treatment-related adverse events (59% in patients treated with ipilimumab plus nivolumab vs 28% in patients treated with ipilimumab).1
Despite significant efforts to identify predictive biomarkers for clinical benefit (CB) for ICI treatment in melanoma, they remain imperfect. Proposed mechanisms of resistance to ICIs include exclusion of T cells in the tumor microenvironment (TME), high levels of immunosuppressive cells or factors, and tumor mutations that result in immune ignorance, such as disruption of antigen processing and presentation.14–17 Taken together, these studies suggest the necessity for applying broad peripheral immune and tumor profiling before and on treatment when interrogating biomarkers for ICI in the pretreated setting.
In this multicenter phase II study, we investigated the efficacy and safety of ipilimumab alone and in combination with nivolumab in patients with advanced melanoma with progressive disease after PD-1 blockade as monotherapy.
Patients and methods
Patients were eligible if they met the following criteria: had histologically confirmed, AJCC stage IV or inoperable stage III cutaneous, acral or mucosal melanoma; had received prior treatment with a PD-1/PD-L1 inhibitor in the adjuvant or metastatic setting with evidence of clinical or radiological progression; had measurable disease based on the revised Response Evaluation Criteria In Solid Tumors (RECIST) guidelines V.1.1; had Eastern Cooperative Oncology Group performance status score of 0–1; and had adequate renal, hepatic, and bone marrow function. There were no restrictions placed on time elapsed from the last PD-1/PD-L1 dose. Patients previously treated with an anti-CTLA-4 antibody, history of autoimmune disease, symptomatic or untreated brain metastases or leptomeningeal disease, or history of a grade 4 immune-related toxicity or grade 3 pneumonitis were ineligible. This study is registered at ClinicalTrials.gov.
Study design and treatment
This was a multicenter, randomized, open-label, phase II study. Patients were randomly allocated at study entry to receive either nivolumab 1 mg/kg of body weight plus ipilimumab 3 mg/kg every 3 weeks for up to four doses, or ipilimumab 3 mg/kg every 3 weeks for up to four doses. Centralized random assignment software was used, ensuring the concealment of the next patient allocation. Balanced 1:1 random assignment was stratified based on melanoma histological subtype, as well as prior response to PD-1 therapy; primary refractory patients were defined as those who had anti-PD-1 therapy within 2 months of study enrollment, whereas progressive disease was defined as those patients who received their last dose of PD-1 blocking antibody ≥2 months prior to enrollment. Both nivolumab and ipilimumab were administered by intravenous infusion. Treatment was discontinued at the onset of disease progression or the development of unacceptable toxic effects. Patients could receive up to four cycles of therapy and then were observed for up to 2 years. There was no nivolumab maintenance therapy mandated per protocol.
The primary objective was to determine the overall response rate (ORR) (either partial response (PR) or complete response (CR)) defined by RECIST V.1.1 criteria18 of combination ipilimumab plus nivolumab and ipilimumab monotherapy by week 18. Secondary endpoints included disease control rate (DCR), time-to-treatment failure (TTF), OS, and safety. Translational biomarker analyses were conducted as exploratory outcomes.
Assessments
Disease was assessed by CT or MRI of the chest, abdomen and pelvis within 28 days prior to study treatment, and then at weeks 12 and 18 according to RECIST V.1.1.18 Adverse events, as assessed according to the National Cancer Institute Common Terminology Criteria for Adverse Events (CTCAE) V.4.0, were monitored during each cycle. Guidelines for the management of adverse events were provided by the sponsor and were published previously.19 20
Sample collection and centralized processing
Peripheral blood was collected via venipuncture into sodium heparin vacutainer tubes at baseline, the first day of treatment cycles 1–4, and at 3 and 6 weeks following the final dose of the study drug. Paired tumor samples were obtained prior to the start of treatment and at day 14 following first drug administration (if safe and medically appropriate). Blood and tissue samples for exploratory analyses were collected at respective sites and shipped to a centralized repository under PICI BioTrust protocols.21
Peripheral blood immunophenotyping
Mass cytometry ((CyTOF)
Peripheral blood was collected via venipuncture into sodium heparin vacutainer tubes and PBMC samples were processed at baseline, the first day of treatment cycles 1–4, and at 3 and 6 weeks following the final dose of study drug. Details on the CyTOF panel for broad immune system profiling are displayed in online supplemental table S1. All samples were thawed, stained for viability and antibodies, and run under uniform protocols22 at Primity BiosciencesCellCarta (formerly Primity Biosciences, Fremont, California, USA).
jitc-2021-003853supp001.pdf (146KB, pdf)
Single-cell functional phenotyping of CD4+ T cells by IsoPlexis IsoCode
PBMCs were subjected to single-cell functional phenotyping of T cells by the single-cell 32-plex IsoCode chip as previously described.23–28 Protein secretions from ~1000 single cells were captured by the 32-plex antibody barcoded chip and analyzed by fluorescence ELISA-based assay. Polyfunctional T cells that cosecreted 2+ cytokines per cell were evaluated by the IsoSpeak software across the five functional groups: (1) effector: granzyme B, tumour necrosis factor (TNF)-α, interferon gamma (IFN-γ), MIP1α, perforin, and TNF-β; (2) stimulatory: GM-CSF, IL-2, IL-5, IL-7, IL-8, IL-9, IL-12, IL-15, and IL-21; (3) chemoattractive: CCL11, IP-10, MIP-1β, and RANTES; (4) regulatory: IL-4, IL-10, IL-13, IL-22, sCD137, sCD40L, transforming growth factor-β1 (TGF-β); and (5) inflammatory: IL-6, IL-17A, IL-17F, MCP-1, MCP-4, and IL-1β. The Polyfunctional Strength Index (PSI) of T-cell functional group products was computed using a prespecified formula, defined as the percentage of polyfunctional cells, multiplied by mean fluorescence intensity, which is in indirect measure of preformed proteins in the secretory pathway.
Tumor immune microenvironment analysis
Whole-exome and RNA sequencing with Personalis ImmunoID NeXT platform
For each patient, a single paired formalin-fixed, paraffin-embedded or fresh-frozen tumor and normal PBMC sample was collected and profiled using the ImmunoID NeXT platform (Personalis) for whole-exome and whole-transcriptome analyses. Whole-exome library preparation and sequencing were performed as previously described,29 and paired-end sequencing was performed on NovaSeq instrumentation (Illumina, San Diego, California, USA). The resulting data were analyzed for single nucleotide variants, gene expression quantification, neoantigen characterization, human leukocyte antigen (HLA) typing and allele-specific HLA loss of heterozygosity data (HLA LOH). Whole-transcriptome sequencing results were aligned using STAR30 and normalized expression values in transcripts per million calculated using Personalis’ ImmunoID NeXT tool, Expressionist.
Statistical analysis
This parallel-arm study was not intended for hypothesis testing between arms. An optimal Simon two-stage design31 was implemented for each arm. The study planned to enroll 12 patients per arm in the first stage. If 2 or more out of 12 patients responded, an additional 23 patients would be enrolled for a total accrual of 35 patients per arm. If at least six responses were observed in these 35 patients, the treatment would be considered promising for future investigation. Assuming 10% and 30% ORRs for the null and alternative hypotheses, respectively, this design yields type I and type II errors of 0.10.
Safety and efficacy were assessed in patients who received at least one dose of study drug. Patients were grouped by the treatment assigned at randomization for efficacy analyses and by the treatment regimen actually received for safety analyses. OS was defined as the time from treatment initiation to death from any cause. TTF was defined as the time from treatment initiation until the patient starts another line of systemic therapy or death, whichever occurs first. For translational endpoints, where appropriate, patients were pooled across the two arms and classified based on CB, defined as a CR or PR by RECIST, or progression-free for 6 months. Patients who were alive and had not started another therapy at the time of database lock were censored at the time of the last follow-up. CIs for ORR and DCR were calculated using the Clopper-Pearson method. OS and TTF were estimated by the Kaplan-Meier method.
Peripheral blood immune data (CyTOF and IsoPlexis cytokine profiling) were analyzed in multiple ways. For trends in systemic immune populations over time, values were normalized to the baseline measurement and plotted as mean fold change of the group with 95% CIs. To evaluate differences in circulating immune cell populations between CB and treatment groups at single timepoints, two-sided Wilcoxon rank-sum tests were employed at a 0.05 significance level. Gene expression signatures (online supplemental table S2) were calculated as the average of all the genes in each signature after standard normalization. Cell proportions in tissue were estimated from EPIC (Estimating the Proportions of Immune and Cancer cells) deconvolution32 run using default parameters. All clustering was hierarchical clustering using a Ward-D2 metric within the heatmap package V.1.0.12. All statistical analyses were performed using R.33
Results
Patients
Between June 2016 and May 2018, a total of 20 patients provided consent and were randomized from four institutions in the USA. One patient was randomized to the ipilimumab monotherapy arm but withdrew consent before starting treatment (online supplemental figure S1) and thus was not included in the efficacy or safety analyses. The trial was closed early due to slow accrual. Patient characteristics were generally balanced across the two treatment arms (table 1).
Table 1.
Baseline demographics and patient characteristics
| Characteristics | Ipilimumab (N=9) | Ipilimumab plus nivolumab (N=10) | Total (N=19) |
| Age (year), median (range) | 66 (35–83) | 56 (39–66) | 60 (35–83) |
| Sex, n (%) | |||
| Male | 6 (67) | 9 (90) | 15 (79) |
| Female | 3 (33) | 1 (10) | 4 (21) |
| Race, n (%) | |||
| Asian | 0 | 1 (10) | 1 (5) |
| White | 9 (100) | 7 (70) | 16 (84) |
| Other | 0 | 2 (20) | 2 (11) |
| Eastern Cooperative Oncology Group performance status score, n (%) | |||
| 0 | 6 (67) | 7 (70) | 13 (68) |
| 1 | 3 (33) | 3 (30) | 6 (32) |
| M stage, n (%) | |||
| M0 | 3 (33) | 1 (10) | 4 (21) |
| M1a | 1 (11) | 2 (20) | 3 (16) |
| M1b | 2 (22) | 3 (30) | 5 (26) |
| M1c without brain metastases | 3 (33) | 4 (40) | 7 (37) |
| Type of melanoma, n (%) | |||
| Acral | 1 (11) | 1 (10) | 2 (11) |
| Cutaneous | 7 (89) | 8 (90) | 17 (90) |
| Mucosal | 1 (11) | 1 (10) | 2 (11) |
| Lactate dehydrogenase (unit/L), median (range) | 208 (152–1800) | 214 (157–310) | 211.0 (152–1800) |
| Genomic driver | |||
| BRAF | 2 (22) | 3 (30) | 5 (26) |
| NRAS | 4 (44) | 2 (20) | 6 (32) |
| Other/unknown | 3 (33) | 5 (50) | 8 (42) |
| Prior treatment, n (%) | |||
| Anti-PD-1 | 9 (100) | 10 (100) | 19 (100) |
| Other* | 1 (11) | 3 (30) | 4 (21) |
| Time since last anti-PD-1 treatment (weeks), median (range) | 6.0 (3–55) | 4.3 (2–36) | 5.1 (2.0–54.7) |
| Best response to prior anti-PD-1 treatment, n (%) | |||
| Stable disease | 1 (11) | 0 | 1 (5) |
| Progressive disease | 6 (67) | 9 (90) | 15 (79) |
| Unknown | 2 (22) | 1 (10) | 3 (16) |
*Other prior treatments include talimogene laherparepvec (patient randomized to ipilimumab), high-dose interferon, dabrafenib plus trametinib, and vemurafenib plus cobimetinib.
PD-1, programmed death 1.
jitc-2021-003853supp002.pdf (193.5KB, pdf)
jitc-2021-003853supp003.pdf (54.5KB, pdf)
In the efficacy-evaluable population, 12 patients (63%) discontinued study treatment prematurely, most commonly for disease progression (n=5, 26%) or adverse events (n=5, 26%). The median number of treatment cycles was four (range 2–4) in the ipilimumab arm and three (range 1–4) in the ipilimumab plus nivolumab arm. All patients were followed up for a minimum of 7.6 months (median 12.2).
Efficacy
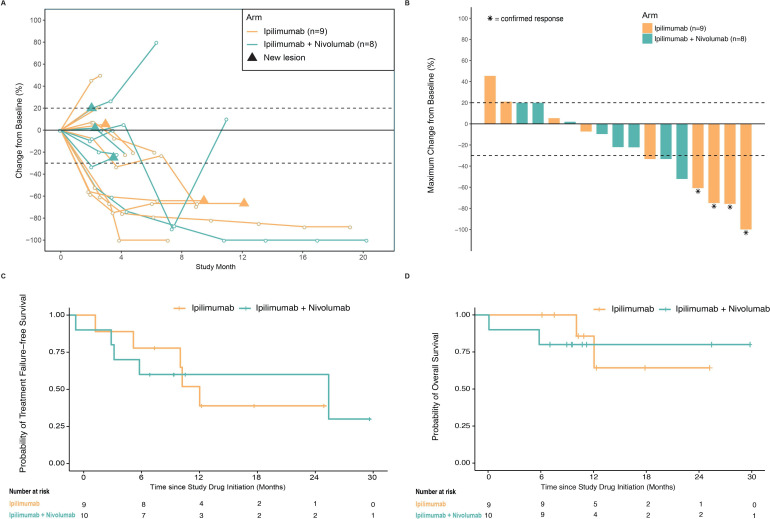
Objective responses were observed in 5 of 9 (56%, 95% CI 21% to 86%) evaluable patients in the ipilimumab arm and 2 of 10 (20%, 95% CI 3% to 56%) in the ipilimumab plus nivolumab arm (figure 1 and table 2) at week 18.
Figure 1.
Change in tumor burden, TTF, and overall survival. (A) Percentage change from baseline in the sum of the diameters of the target lesions in patients receiving ipilimumab (gold) or ipilimumab plus nivolumab (teal). Triangles denote the presence of a new lesion. Two patients in the ipilimumab plus nivolumab arm did not have postbaseline target lesion measurements and are not displayed on the plot (clinical progression, presence of a new lesion (counted as radiographic progressive disease) but no measurement of the target lesions). (B) Maximum percentage change from baseline in the sum of the diameters of the target lesions at week 18. Asterisks denote that the response (complete or partial) was confirmed by a second scan by week 18. Two patients, one in each arm, had a maximum change in the sum of target lesion diameters of less than 20% but had a best overall response of progressive disease due to the presence of a new lesion. (C) Kaplan-Meier curves for TTF. (D) Kaplan-Meier curves for overall survival. TTF, time-to-treatment failure.
Table 2.
Clinical activity of ipilimumab and ipilimumab plus nivolumab
| Ipilimumab (N=9) | Ipilimumab plus nivolumab (N=10) | Total (N=19) | |
| Best response at week 18, n (%) | |||
| Complete response | 1 (11) | 0 | 1 (5) |
| PR | 4 (44) | 2 (20) | 6 (32) |
| Stable disease | 1 (11) | 4 (40) | 5 (26) |
| Progressive disease | 3 (33) | 3 (30) | 6 (32) |
| Could not be evaluated* | 0 | 1 (10) | 1 (5) |
| Objective response rate at week 18 | |||
| n (%) | 5 (56) | 2 (20) | 7 (37) |
| 95% CI | 21 to 86 | 3 to 56 | 16 to 62 |
| Disease control rate at week 18 | |||
| n (%) | 6 (67) | 6 (60) | 12 (63) |
| 95% CI | 30 to 93 | 26 to 88 | 38 to 84 |
| CB rate† | |||
| n (%) | 6 (67) | 5 (50) | 11 (58) |
| 95% CI | 30 to 93 | 19 to 81 | 34 to 80 |
| Time to treatment failure (months) | |||
| Median | 13.6 | 26.9 | 13.6 |
| 95% CI | 2.8 to NE | 0.7 to NE | 6.7 to NE |
| Overall survival (months) | |||
| Median | NE | NE | NE |
| 95% CI | 11.5 to NE | 1.6 to NE | 13.5 to NE |
The best overall response was assessed by the investigator with the use of the Response Evaluation Criteria in Solid Tumors V.1.1. Patients are grouped according to the treatment assigned at randomization. Two patients had treatment different from the one assigned at randomization. The patient randomized to the ipilimumab arm who received ipilimumab plus nivolumab had a best overall response of PR. The patient randomized to the ipilimumab plus nivolumab arm who received ipilimumab monotherapy had a best overall response of PR.
*One patient in the ipilimumab plus nivolumab arm died due to clinical progression prior to the first scheduled on-treatment tumor assessment.
†CB is defined as a complete or PR at any time or a best response of stable disease with no evidence of radiographic progression within 6 months.
CB, clinical benefit; NE, not estimable; PR, partial response.
One patient in the ipilimumab arm achieved a best response of CR; four patients in the ipilimumab arm and two patients in the ipilimumab plus nivolumab arm achieved a PR. The DCR at week 18 was similar across the arms (67% and 60%, respectively). The median TTF was 13.6 months (95% CI 2.8 to not estimable) in the ipilimumab arm and 26.9 months (95% CI 0.7 to not estimable) in the ipilimumab plus nivolumab arm. The median OS was not reached in either arm; two deaths (22%) were observed in the ipilimumab arm and two deaths (20%) in the ipilimumab plus nivolumab arm; none were attributed to the study drug.
Safety
A total of 19 patients enrolled received at least one dose of study drug and therefore were evaluable for safety analysis (table 3). No fatal adverse events attributable to the drug were reported. TRAEs were generally consistent with previously reported adverse events. While the rate of high-grade TRAEs was comparable across the arms, there were four TRAEs that led to treatment withdrawal in the ipilimumab plus nivolumab arm, including two patients with diarrhea (grades 1 and 2), one patient with an elevated aspartate transminase (AST) and alanine transaminase (ALT) (grade 2), and one with hypophysitis (grade 2). This is compared with one patient in the ipilimumab arm who discontinued for treatment-related adrenal insufficiency and infection (both grade 3).
Table 3.
Selected TRAEs
| Event | Ipilimumab (N=9) | Ipilimumab plus nivolumab (N=10) | Total (N=19) | |||
| Any grade | Grade 3 or 4 | Any grade | Grade 3 or 4 | Any grade | Grade 3 or 4 | |
| Any TRAE | 9 (100) | 5 (56) | 9 (90) | 4 (40) | 18 (95) | 9 (47) |
| Pruritus | 5 (56) | 0 | 5 (50) | 0 | 10 (53) | 0 |
| Maculopapular rash | 3 (33) | 0 | 4 (40) | 0 | 7 (37) | 0 |
| Diarrhea | 3 (33) | 1 (11) | 6 (60) | 2 (20) | 9 (47) | 3 (16) |
| Colitis | 2 (22) | 2 (22) | 1 (10) | 0 | 3 (16) | 2 (11) |
| Alanine aminotransferase increased | 2 (22) | 0 | 5 (50) | 1 (10) | 7 (37) | 1 (5) |
| Aspartate aminotransferase increased | 1 (11) | 0 | 4 (40) | 1 (10) | 5 (26) | 1 (5) |
| Hyponatremia | 2 (22) | 1 (11) | 2 (20) | 0 | 4 (21) | 1 (5) |
| Hypokalemia | 2 (22) | 1 (11) | 1 (10) | 1 (10) | 3 (16) | 2 (11) |
| Arthralgia | 2 (22) | 0 | 1 (10) | 0 | 3 (16) | 0 |
| Hypophysitis | 1 (11) | 0 | 3 (30) | 0 | 4 (21) | 0 |
| Adrenal insufficiency | 1 (11) | 1 (11) | 0 | 0 | 1 (5) | 1 (5) |
| White blood cell count decreased | 2 (22) | 1 (11) | 1 (10) | 0 | 3 (16) | 1 (5) |
| Neutrophil count decreased | 1 (11) | 1 (11) | 1 (10) | 0 | 2 (11) | 1 (5) |
| Urinary tract infection | 1 (11) | 1 (11) | 0 | 0 | 1 (5) | 1 (5) |
| Hypotension | 0 | 0 | 2 (20) | 1 (10) | 2 (11) | 1 (5) |
| TRAE leading to discontinuation | 1 (11) | 1 (11) | 4 (40) | 0 | 5 (26) | 1 (5) |
TRAE, treatment-related adverse event.
Shown are treatment-related adverse events of any grade that occurred in more than 10% of patients in a treatment arm. Patients are grouped according to the treatment regimen actually received. The relatedness of the adverse event to treatment was determined by the investigator. The severity of adverse events was graded according to the National Cancer Institute CTCAE V.4.0.
Peripheral immune characteristics
In order to gain insight into the peripheral immune phenotypes that are associated with benefit from ipilimumab-based treatment (ipilimumab alone and in combination with nivolumab) after progression on PD-1 blockade, we performed multiple analyses on blood samples obtained over the course of therapy. Where appropriate, patients were pooled across the two arms. Given the small number of samples, for the purpose of translational analyses, patients were classified by CB, defined as those patients who achieved a CR or PR by RECIST, or were progression-free for at least 6 months (table 2).
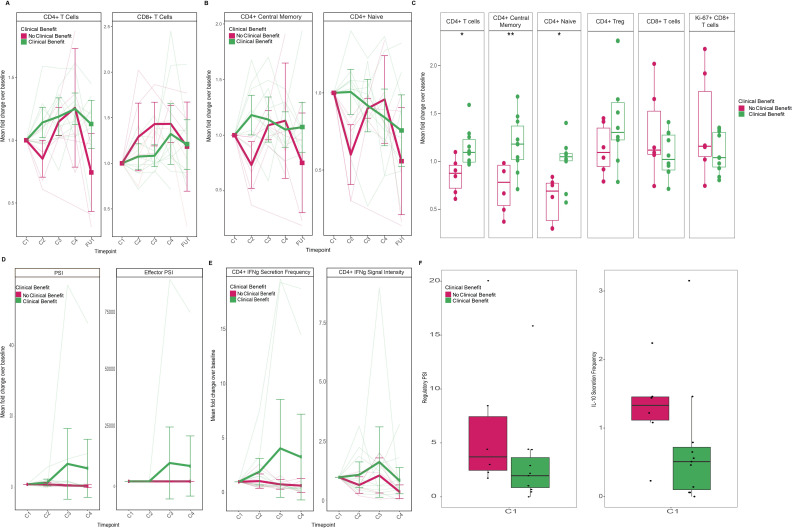
Longitudinal immune profiling by mass cytometry (CyTOF) by percent of total leukocytes showed increases in circulating CD4+, but not CD8+, T cells associated with CB to ipilimumab-based therapy (figure 2A). Specifically, CD4+ central memory and naïve T-cell populations (figure 2B), as opposed to CD4+ T regulatory cell populations (figure 2C and online supplemental figure S2), showed the largest disparities when comparing patients by CB classification, and differences in fold change in overall CD4+, CD4+ central memory and naïve immune cell subsets were most apparent following one dose of therapy (figure 2C; p<0.05, p<0.01, p<0.05, respectively), consistent with existing reports.34 Importantly, these immune T-cell changes were similar when comparing CB within treatment arms (online supplemental figure S2). Also in line with prior findings,10 CD4+ inducible T-cell costimulator was higher in the ipilimumab monotherapy patients early on-treatment, most dramatically in those with CB (online supplemental figure S3).
Figure 2.
Trends in peripheral immune characteristics and CB over the course of therapy. (A) Trends in CD4+ and CD8+ T cells by CB (pink denotes no CB, n=6; green denotes CB, n=10). Thick lines represent mean fold change±95% CI of group cell population over time as percent of total leukocytes; light-colored lines represent measurements of individual patients. (B) Trends in CD4+ T cells by CB. Thick lines represent mean fold change±95% CI of group cell population over time as percent of total leukocytes; light-colored lines represent measurements of individual patients. (C) Comparison of fold change from baseline in CD4+ and CD8+ T-cell populations by CB prior to cycle 2 of therapy. (D) Trends in CD4+ T-cell cytokine PSI and effector PSI by CB (pink denotes no CB, n=7; green denotes CB, n=11). Thick lines represent mean fold change±95% CI of group cell population over time as percent of total leukocytes; light-colored lines represent measurements of individual patients. (E) Trends in IFN-γ secretion frequency and signal intensity from CD4+ T cells by CB. Thick lines represent mean fold change±95% CI of group cell population over time as percent of total leukocytes; light-colored lines represent measurements of individual patients. (F) Differences in regulatory PSI and IL-10 secretion frequency and from CD4+ T cells at baseline by CB (pink denotes no CB; n=6; green denotes CB, n=11). *P<0.05, **P<0.01 by two-sided Wilcoxon rank-sum test. C1, baseline; C2, precycle 2 of therapy; C3, precycle 3 of therapy; C4, precycle 4 of therapy; CB, clinical benefit; FU1, after four cycles of therapy; IFN-γ, interferon gamma; IL, interleukin; PSI, Polyfunctional Strength Index.
jitc-2021-003853supp004.pdf (2.1MB, pdf)
jitc-2021-003853supp005.pdf (705.9KB, pdf)
Further interrogation of the potential role of CD4+ T cells in those patients who derived CB following progression on PD-1 also suggested differences in functionality. Characterizing PSI, which captures the percentage of single T cells secreting at least two cytokines and their respective signal intensities, we found that patients with CB demonstrated CD4+ T cells with increased PSI following initiation of therapy compared with patients who did not benefit, particularly among effector cytokines that are known to be associated with antitumor immunity (figure 2D). In examining contributions of individual effector cytokines secreted from CD4+ T cells that may promote immune response to therapy, IFN-γ was higher in the CB group compared with those without benefit over the course of therapy (figure 2E). Conversely, the immune suppressive cytokine, IL-10, was higher at baseline in patients without CB, and a similar association was seen in PSI of regulatory cytokines of these patients at the same timepoint (figure 2F). Similar effect patterns were observed when comparing CB within treatment arms, although trends were more moderate but consistent over time in the ipilimumab monotherapy arm and more dramatic but variable in the ipilimumab plus nivolumab arm (online supplemental figure S4).
jitc-2021-003853supp006.pdf (2.5MB, pdf)
Characteristics of the TME
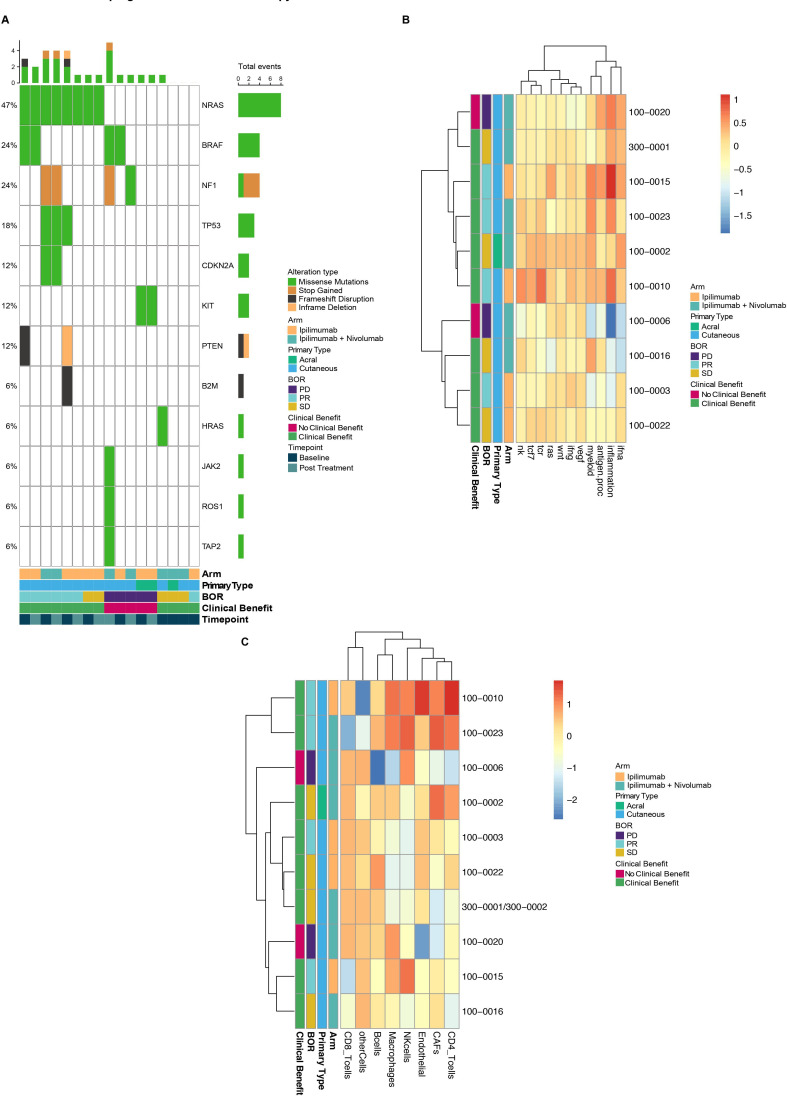
Profiling of the tumor was performed using multiple approaches. Tumor mutational burden (TMB) has been proposed and studied as a potential biomarker of response to ICIs in ICI-naïve patients.35–37 TMB was not associated with CB in response to an ipilimumab-containing regimen when examined at a single timepoint or in paired biopsies (online supplemental figure S5), though the sample size is small. Additional whole-exome sequencing (WES) analysis showed an enrichment of NRAS mutations in patients with CB (figure 3A), as 100% of patients with this alteration benefitted from therapy, consistent with prior findings in patients with melanoma treated with ICIs.38 Select patients had alterations in genes previously associated with PD-1 resistance and antigen presentation in melanoma including B2M, JAK2 and HLA loci,39 40 although it was not determined whether these alterations were loss of function.
Figure 3.
Features of the tumor microenvironment in patients with melanoma treated with ipilimumab or ipilimumab plus nivolumab. (A) Mutational landscape indicating melanoma and immunotherapy-related genes with alterations in patients (n=10) at all timepoints. (B) Heatmap of expression profiling of patient tumors (n=10) by RNA sequencing at the on-treatment timepoint. The list of genes comprising each signature (heatmap columns) is detailed in online supplemental table S2. (C) Heatmap showing immune population scores calculated by EPIC deconvolution of the gene expression data for all patients with on-treatment samples sent for RNA sequencing (n=10). BOR, best overall response; CAFs, cancer associated fibroblasts; PD, progressive disease; PR, partial response; SD, stable disease.
jitc-2021-003853supp007.pdf (94.9KB, pdf)
Analysis of RNA sequencing at multiple timepoints showed lack of convergence in terms of pathways associated with CB to ICIs following PD-1 resistance (figure 3B, online supplemental figure S6). Comparing multiple expression signatures important for melanoma and ICI response-related biology41 (online supplemental table S2) by CB and RECIST response showed an enrichment in patients experiencing CB in tumors displaying upregulation in active immune and myeloid cell signatures post-treatment (five out of six patients with upregulation of these signatures experienced CB; only three out of four experienced CB in samples without upregulation of these signatures; figure 3B). Deconvolution of RNA sequencing data by EPIC32 to extrapolate specific immune subsets associated with CB revealed the natural killer (NK) cell, macrophage and CD4+ T-cell pathways were most commonly increased post-treatment among patients with CB (figure 3C). Notably, peripheral NK cell populations decreased among patients with CB post treatment, potentially suggesting trafficking of NK cells out of the periphery and into the TME as a hypothesis for contributing to antitumor benefit in these patients (online supplemental figure S7). Lastly, HLA expression was also assessed in baseline and on-treatment tumor samples; there was no significant difference in HLA expression between those patients who derived CB and those that did not (online supplemental figure S8).
jitc-2021-003853supp008.pdf (117.1KB, pdf)
jitc-2021-003853supp009.pdf (812.7KB, pdf)
jitc-2021-003853supp010.pdf (1MB, pdf)
Discussion
We conducted a prospective randomized clinical trial with longitudinal biomarker sample collection to evaluate the clinical and biological activities of ipilimumab alone and in combination with nivolumab in patients with progression of disease on anti-PD-1 monotherapy. The trial was ended early due to poor accrual after 20 patients were enrolled out of a planned 24 in the first stage. Had the study remained open, sufficient responses were observed in the first 20 patients to warrant expanding the study and enrolling a total of 35 patients in both arms. A numerically higher ORR was observed in the ipilimumab arm (56%) than the ipilimumab plus nivolumab arm (20%); however, the study was not designed to test between arms and is limited by the small sample size. Due to truncated follow-up, the median OS was not reached in either group. The safety profile of ipilimumab with or without nivolumab is consistent with previously published data for both agents, and the rate of high-grade TRAEs was similar with monotherapy and combined ICIs.
There have been several studies to examine the management of advanced melanoma with progression of disease after PD-1 blockade. In a large retrospective study of 330 patients treated with ipilimumab alone or in combination, the ORR was 31% with combination ICIs compared with 12% with ipilimumab alone.42 In a small prospective study of pembrolizumab plus low-dose ipilimumab (1 mg/kg), the ORR was 29% in all enrolled patients.43 In our study, the DCRs at week 18 were comparable in patients treated with ipilimumab and ipilimumab plus nivolumab. Notably, the elapsed time interval between the last dose of anti-PD-1 monotherapy and the study drug was short in both arms: 6 weeks (range 3–55 weeks) in patients treated with ipilimumab arm and 4.3 (range 2–36 weeks) in patients treated with ipilimumab plus nivolumab. This raises the possibility that there may have been an anti-PD-1 antibody present in both groups at the time of ipilimumab administration. Moreover, it is notable that the majority of patients in our study were classified as ‘primary refractory’ to anti-PD-1 therapy based on the time interval since the last anti-PD-1/PD-L1 infusion; this may have adversely impacted the ORR to ipilimumab-based therapy. There is an ongoing randomized phase II study of ipilimumab versus ipilimumab plus nivolumab in patients with PD-1 refractory advanced melanoma (Clinicaltrials.gov identifier NCT03033576), which is likely to provide further evidence of whether combination ICIs is superior in this patient population.
Biomarker analyses for the trial were limited by the small sample size and the limited amount of tissue material available, highlighting the challenges of biomarker development. Despite the small sample size, potentially important trends were revealed in both longitudinal peripheral and tumor-specific studies, suggesting a direction for future work on PD-1 resistance.
In longitudinal peripheral immune profiling by CyTOF, we found that increased fold change in CD4+ immune cell subsets, but not CD8+ T cells, was associated with CB to ipilimumab-based therapy, and these effects were more consistent and sustained in the ipilimumab monotherapy arm than in combination with nivolumab. Similar observations have been reported in peripheral blood analysis at baseline, where increased frequencies of CD45RA− T cells were associated with response to ipilimumab.44 The longitudinal profiling in our study expands on a link between engagement of the CD4+ T-cell compartment and CB with ipilimumab-containing therapies. This is consistent with observations reported in murine studies of the TME, where anti-CTLA-4 monotherapy drives a T-helper type 1 (Th1) effector-like T-cell enrichment when compared with combination therapy.45 Additionally, in patients with melanoma, treatment with ipilimumab was associated with the specific enrichment of Th1-like CD4 effector T cells in the TME, an effect not observed in response to PD-1 monotherapy treatment.10 Cytotoxic CD8+ T cells directly lyse tumor cells, whereas CD4+ Th1 responses broadly engage innate and adaptive cell function, indirectly resulting in tumor cytotoxicity. These data suggest that successful eradication of metastatic tumors in patients that progress on PD-1 may require the concerted activation of both the CD4+ and CD8+ compartments of the adaptive immune response.
It is notable that in our study, all patients whose tumor harbored an NRAS mutation derived CB. This has been demonstrated in previous retrospective analyses38 and is of potential mechanistic interest, given NRAS mutations in metastatic melanoma are enriched in older patients with more chronic ultraviolet exposure.46 Future studies to evaluate if a specific CD4+ T cell reactive with this shared oncogenic mutation may inform T-cell response to this subset of melanomas and possibly design of T-cell therapies.47 In sum, these data suggest that the different genomic subpopulations should be prospectively evaluated in future clinical trials in the post-PD-1 progression setting. Conversely, in this small heterogenous cohort of PD-1 progressors, we found no association between TMB and CB to ipilimumab-based treatment. The exact role of TMB in predicting response to ICIs in patients with advanced melanoma has yet to be defined. In a study of 206 patients treated with anti-PD-1 monotherapy, of whom 44% had previously been treated with ipilimumab, researchers found that, when stratified by melanoma subtype, there was no positive correlation between TMB and benefit to ICIs.48 In contrast, in a meta-analysis of data from the CheckMate 066 and CheckMate 067 studies, which enrolled treatment-naïve patients, high TMB was associated with improved PFS and OS in patients treated both with nivolumab monotherapy, as well as ipilimumab plus nivolumab and ipilimumab alone.49 Additionally, the FDA has recently granted accelerated approval to pembrolizumab for the treatment of unresectable and metastatic tumors with TMB of at least 10 mutations per megabase.50 It may be that TMB plays a less substantial role in predicting response to ICIs in the PD-1-resistant setting.
Gene expression analysis of the tumors demonstrated immunological changes in the TME with treatment. Active immune and myeloid signatures containing genes associated with myeloid and T-cell activation (PD-L1, iNOS and arginase-1; online supplemental table S2) were most frequently upregulated and commonly associated with CB. CB was associated with increased peripheral CD4+ T-cell activation, which may be linked to the increase in immune activation signatures in the TME. Immune cell deconvolution of our gene expression data yielded similar trends; NK cells, macrophages, B cells and, to a lesser extent, CD4 T cells were more frequently increased in the TME with treatment than, for example, CD8 T cells. While our dataset is not powered to draw conclusions from the heterogeneous immune responses observed, it provides preliminary signals that suggest the melanoma microenvironment can undergo immunological remodeling with ipilimumab alone or in combination with PD-1 blockade in patients that progressed on PD-1 monotherapy. These mechanistic insights suggest that antitumor immunity in PD-1-resistant patients can be activated by CD4+ T cells and NK cells.
While active immune and myeloid signatures trended higher with treatment in a subset of tumor samples from patients with CB, two outliers were apparent. One patient with CB in the ipilimumab arm (100–0003) had a lower than median active immune signature, whereas a patient in the ipilimumab plus nivolumab arm with progressive disease (100–0020) demonstrated a higher than median active immune signature. Taken alone, the scenarios appear counterintuitive; however, peripheral blood analysis from these subjects trended with their respective groupings for CD4+ and CD8+ T-cell dynamics (online supplemental figure S9). Interestingly, the patient without CB and a high active immune signature also had a mutation in JAK2 (figure 3A), a signaling molecule for the type-II interferon receptor that has previously been described in a patient with PD-1 refractory melanoma.39 This deeper look at discordant patients exemplifies the need for multidimensional profiling of peripheral and tumor compartments over multiple therapeutic timepoints when investigating response and resistance to ICIs.
jitc-2021-003853supp011.pdf (734.9KB, pdf)
The role of antigen presentation and response to ICI has previously been explored in patients with treatment-naïve advanced melanoma, in whom it was demonstrated that major histocompatability complex (MHC) class I expression was associated with response to CTLA-4 blockade, whereas MHC class II was associated with response to PD-1 blockade.40 In addition, genes in the IFN-γ pathway predicted response to anti-PD1 but were less relevant in anti-CTLA-4 therapy.40 It is unclear whether these data can be extrapolated to this study, given the differing patient populations; however, we analyzed the WES data for mutations in the antigen-presentation pathway (notably B2M, JAK 1/2, and STAT) in pretreatment and post-treatment tumor samples, as well as analyzed IFN-γ signature via RNAseq. We did not observe a strong IFN-γ expression pattern associated with response nor was there a statistically significant correlation between HLA expression and CB. Further work may be needed to delineate the role of MHC and alterations in antigen presentation in a PD-1-resistant population.
In summary, objective responses were seen in PD-1-resistant patients treated with ipilimumab and ipilimumab plus nivolumab, with similar DCRs at week 18. Overall, these data suggest that ipilimumab-based therapies broadly engage the innate and adaptive arms of the immune response, resulting in CB. Further mechanistic studies and clinical biomarker discovery in this patient population are needed, especially those that longitudinally incorporate diverse immune cell and functional phenotypes from both the tumor and periphery. These studies will hopefully characterize biomarkers to aid clinicians when making treatment decisions following PD-1 progression, along with identifying potential targets and combinations for novel therapeutics to improve patient outcomes.
jitc-2021-003853supp012.pdf (5.5MB, pdf)
Acknowledgments
The authors thank all patients and families for participating in this study, and also Sean Mackay and Jing Zhou (IsoPlexis), Charles Abbott and Sean Boyle (Personalis), and Primity Biosciences for their efforts toward the lab work associated with this study.
Footnotes
Twitter: @HTawbi_MD
CFF and CS contributed equally.
Contributors: CFF: conceptualization, investigation, project administration, original draft, review, and editing, and serves as the guarantor for the overall content; CS: methodology, investigation, data curation, formal analysis, visualization, original draft, review, and editing; CRC: methodology, review, and editing; KSP: conceptualization, methodology, review, and editing; DKW: investigation, data curation, methodology, review, and editing; AR, KT, MP, AS, and PC: investigation, review and editing; JK: supervision, review, and editing; SB: supervision, methodology, review, and editing; PG: methodology, data curation, Investigation, review, and editing; TJH: investigation, formal analysis, visualization, review, and editing; ROC: methodology, data curation, formal analysis, review, and editing; MC: investigation, methodology, review, and editing; TLV and RI: project administration, review, and editing; JW: conceptualization, investigation, methodology, project administration, review, and editing.
Funding: The clinical trial was sponsored by the Parker Institute for Cancer Immunotherapy, with financial and drug support provided by Bristol Myers Squibb. The study was supported in part by the MSK Cancer Center (support grant P30 CA008748). This study was also supported by a 2016 SITC-Genentech Postdoctoral Cancer Immunotherapy Clinical Fellowship and a 2017 ASCO Young Investigator Award.
Competing interests: CFF reports personal/consultancy fees from AstraZeneca, as well as participation in steering committees (compensation waived) for Merck and Genentech; these are outside the scope of the submitted work. She also reports institutional research funding from Genentech, Merck, Bristol Myers Squibb, Daiichi, and AstraZeneca. AR reports personal/consultancy fees from Amgen, Chugai, Genentech, Jounce, Merck, Novartis, Nurix, Sanofi, and Vedanta; stock from prior consulting with Advaxis, CytomX, Five Prime, RAPT, IsoPlexis, and Kite-Gilead; being a member of the scientific advisory board and stockholder in 4C Biomed, Apricity, Arcus, Highlight, Compugen, ImaginAb, MapKure, Merus, Rgenix, Lutris, PACT Pharma, and Tango; and receiving research funding to the institution from Agilent and Bristol-Myers Squibb through a Stand Up to Cancer Catalyst grant. KKT reports institutional research funding from Array/Pfizer, Bristol Myers Squibb. Oncosec, Regeneron, and Replimune. HT reports personal/consultancy fees from Genentech, Merck, BMS, Novartis, Iovance, and Eisai and research funding to institution from Genentech, Merck, BMS, Novartis, Celgene, and GSK. TL discloses the following: LISCure Biosciences Scientific Advisory Board, June 2020 up to the present and stock ownership in AstraZeneca; consulting: TRex Bio, Grey Wolf Therapeutics, Exosis, and BiOne Cure; these are outside the scope of the submitted work. MP reports personal/consultancy fees from Array BioPharma, Aduro Biotech, Bristol Myers Squibb, Incyte, Merck, Newlink Genetics, Novartis, and Eisai; honoraria from Bristol Myers Squibb and Merck; research funding from Array BioPharma, AstraZeneca/Medimmune, Bristol Myers Squibb, Infinity Pharmaceuticals, Merck, Novartis, and RgenixA. NS reports personal/consultancy fees from Bristol-Myers Squibb, Immunocore, and Castle Biosciences; research funding to institution from BMS, Immunocore, Xcovery, and Novartis. MC reports institutional research support and employment of a family member by Bristol-Myers Squibb, and consulting, advisory, or speaking compensation for AstraZeneca/MedImmune, Incyte, Moderna, Immunocore and Merck. PC reports consulting/advisory/or speaking compensation from Immunocore, Merck, Cell Medica, Takeda Millenium, and Astra Zeneca; stock ownership in Rgenix; and research funding from Pfizer. DKW is a scientific founder of, holds equity in, and receives consulting fees from Immunai. RI discloses the following: scientific advisory board membership at Harpoon, BitBio, and Arcus; board of directors at Surface Oncology; non-compensated board membership at Lyell; non-compensated scientific advisory board membership at ImaginAb; advisor: IMV and Georgiamune. JK discloses the following: either scientific advisory board membership, stock ownership, or receiving consulting fees from AstraZeneca, Arcus Biosciences, Tizona Therapeutics, Trishula Therapeutics, Primevax, and Elucida Oncology. JW is a consultant for Amgen, Apricity, Ascentage Pharma, Arsenal IO, Astellas, AstraZeneca, Bayer, Bicara Therapeutics, Boehringer Ingelheim, Bristol Myers Squibb, Chugai, Daiichi Sankyo, Dragonfly, Eli Lilly, F Star, Georgiamune, Idera, Imvaq, Kyowa Hakko Kirin, Maverick Therapeutics, Merck, Neon Therapeutics, Psioxus, Recepta, Tizona, Trieza, Truvax, Trishula, Sellas, Surface Oncology, Syndax, Syntalogic, and Werewolf Therapeutics. JDW reports receiving grant/research support from Bristol Myers Squibb and Sephora; having equity in Tizona Pharmaceuticals, Adaptive Biotechnologies, Imvaq, Beigene, Linneaus, Apricity, Arsenal IO, Georgiamune, Trieza, Maverick, and Ascentage.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request. Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as online supplementary information. The corresponding author may be contacted with any requests.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human subjects and was approved by MSKCC Institutional Review Board (protocol ID #16-043). The subjects gave informed consent to participate in the study before taking part.
References
- 1.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med 2019;381:1535–46. 10.1056/NEJMoa1910836 [DOI] [PubMed] [Google Scholar]
- 2.Shrikant P, Khoruts A, Mescher MF. CTLA-4 blockade reverses CD8+ T cell tolerance to tumor by a CD4+ T cell- and IL-2-dependent mechanism. Immunity 1999;11:483–93. 10.1016/S1074-7613(00)80123-5 [DOI] [PubMed] [Google Scholar]
- 3.Krummel MF, Allison JP. CTLA-4 engagement inhibits IL-2 accumulation and cell cycle progression upon activation of resting T cells. J Exp Med 1996;183:2533–40. 10.1084/jem.183.6.2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agata Y, Kawasaki A, Nishimura H, et al. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int Immunol 1996;8:765–72. 10.1093/intimm/8.5.765 [DOI] [PubMed] [Google Scholar]
- 5.Parry RV, Chemnitz JM, Frauwirth KA, et al. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol Cell Biol 2005;25:9543–53. 10.1128/MCB.25.21.9543-9553.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett F, Luxenberg D, Ling V, et al. Program death-1 engagement upon TCR activation has distinct effects on costimulation and cytokine-driven proliferation: attenuation of ICOS, IL-4, and IL-21, but not CD28, IL-7, and IL-15 responses. J Immunol 2003;170:711–8. 10.4049/jimmunol.170.2.711 [DOI] [PubMed] [Google Scholar]
- 7.Saunders PA, Hendrycks VR, Lidinsky WA, et al. PD-L2:PD-1 involvement in T cell proliferation, cytokine production, and integrin-mediated adhesion. Eur J Immunol 2005;35:3561–9. 10.1002/eji.200526347 [DOI] [PubMed] [Google Scholar]
- 8.Chemnitz JM, Parry RV, Nichols KE, et al. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J Immunol 2004;173:945–54. 10.4049/jimmunol.173.2.945 [DOI] [PubMed] [Google Scholar]
- 9.Ishida Y, Agata Y, Shibahara K, et al. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. Embo J 1992;11:3887–95. 10.1002/j.1460-2075.1992.tb05481.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei SC, Levine JH, Cogdill AP, et al. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell 2017;170:1120–33. 10.1016/j.cell.2017.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodi FS, O'Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711–23. 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robert C, Ribas A, Schachter J, et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study. Lancet Oncol 2019;20:1239–51. 10.1016/S1470-2045(19)30388-2 [DOI] [PubMed] [Google Scholar]
- 13.Weber JS, D'Angelo SP, Minor D, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol 2015;16:375–84. 10.1016/S1470-2045(15)70076-8 [DOI] [PubMed] [Google Scholar]
- 14.Nowicki TS, Hu-Lieskovan S, Ribas A. Mechanisms of resistance to PD-1 and PD-L1 blockade. Cancer J 2018;24:47–53. 10.1097/PPO.0000000000000303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang AC, Orlowski RJ, Xu X, et al. A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat Med 2019;25:454–61. 10.1038/s41591-019-0357-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valpione S, Galvani E, Tweedy J, et al. Immune-awakening revealed by peripheral T cell dynamics after one cycle of immunotherapy. Nat Cancer 2020;1:210–21. 10.1038/s43018-019-0022-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zappasodi R, Budhu S, Hellmann MD, et al. Non-Conventional inhibitory CD4+Foxp3−PD-1hi T cells as a biomarker of immune checkpoint blockade activity. Cancer Cell 2018;33:1017–32. 10.1016/j.ccell.2018.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 19.Opdivo (R) [package insert]. Bristol-Myers squibb; initial approval 2011, 2020. Available: https://packageinserts.bms.com/pi/pi_opdivo.pdf [Accessed 23 Jul 2020].
- 20.Yervoy (R) [package insert]. Bristol-Myers Squibb; initial approval 2011, 2020. Available: http://packageinserts.bms.com/pi/pi_yervoy.pdf [Accessed 23 Jul 2020].
- 21.Spencer CN, Wells DK, LaVallee TM. It is a capital mistake to Theorize who to treat with checkpoint inhibitors before one has data. Trends Cancer 2019;5:79–82. 10.1016/j.trecan.2018.12.003 [DOI] [PubMed] [Google Scholar]
- 22.Hartmann FJ, Babdor J, Gherardini PF, et al. Comprehensive immune monitoring of clinical trials to advance human immunotherapy. Cell Rep 2019;28:819–31. 10.1016/j.celrep.2019.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossi J, Paczkowski P, Shen Y-W, et al. Preinfusion polyfunctional anti-CD19 chimeric antigen receptor T cells are associated with clinical outcomes in NHL. Blood 2018;132:804–14. 10.1182/blood-2018-01-828343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue Q, Bettini E, Paczkowski P, et al. Single-cell multiplexed cytokine profiling of CD19 CAR-T cells reveals a diverse landscape of polyfunctional antigen-specific response. J Immunother Cancer 2017;5:85. 10.1186/s40425-017-0293-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma C, Cheung AF, Chodon T, et al. Multifunctional T-cell analyses to study response and progression in adoptive cell transfer immunotherapy. Cancer Discov 2013;3:418–29. 10.1158/2159-8290.CD-12-0383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma C, Fan R, Ahmad H, et al. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat Med 2011;17:738–43. 10.1038/nm.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y, Xue Q, Eisele MR, et al. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc Natl Acad Sci U S A 2015;112:E607–15. 10.1073/pnas.1416756112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parisi G, Saco JD, Salazar FB, et al. Persistence of adoptively transferred T cells with a kinetically engineered IL-2 receptor agonist. Nat Commun 2020;11:660. 10.1038/s41467-019-12901-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patwardhan A, Harris J, Leng N, et al. Achieving high-sensitivity for clinical applications using augmented exome sequencing. Genome Med 2015;7:71. 10.1186/s13073-015-0197-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon R. Optimal two-stage designs for phase II clinical trials. Control Clin Trials 1989;10:1–10. 10.1016/0197-2456(89)90015-9 [DOI] [PubMed] [Google Scholar]
- 32.Racle J, de Jonge K, Baumgaertner P, et al. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife 2017;6 10.7554/eLife.26476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [computer program] 2014.
- 34.Riaz N, Havel JJ, Makarov V, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 2017;171:934–49. 10.1016/j.cell.2017.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hellmann MD, Ciuleanu T-E, Pluzanski A, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med Overseas Ed 2018;378:2093–104. 10.1056/NEJMoa1801946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rizvi H, Sanchez-Vega F, La K, et al. Molecular determinants of response to anti-programmed cell death (PD)-1 and anti-programmed death-ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol 2018;36:633–41. 10.1200/JCO.2017.75.3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 2017;377:2500–1. 10.1056/NEJMc1713444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson DB, Lovly CM, Flavin M, et al. Impact of NRAS mutations for patients with advanced melanoma treated with immune therapies. Cancer Immunol Res 2015;3:288–95. 10.1158/2326-6066.CIR-14-0207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med 2016;375:819–29. 10.1056/NEJMoa1604958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodig SJ, Gusenleitner D, Jackson DG, et al. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci Transl Med 2018;10 10.1126/scitranslmed.aar3342 [DOI] [PubMed] [Google Scholar]
- 41.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva PD, Ahmed T, Lo S. Ipilimumab (IPI) alone or in combination with anti-PD-1 (IPI+PD1) in patients (pts) with metastatic melanoma (MM) resistant to PD1 monotherapy. J Clin Oncol 2020;38. [Google Scholar]
- 43.Olson DJ, Eroglu Z, Brockstein B, et al. Pembrolizumab plus ipilimumab following Anti-PD-1/L1 failure in melanoma. J Clin Oncol 2021;39:2647–55. 10.1200/JCO.21.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subrahmanyam PB, Dong Z, Gusenleitner D, et al. Distinct predictive biomarker candidates for response to anti-CTLA-4 and anti-PD-1 immunotherapy in melanoma patients. j. immunotherapy cancer 2018;6:18. 10.1186/s40425-018-0328-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei SC, Anang N-AAS, Sharma R, et al. Combination anti-CTLA-4 plus anti-PD-1 checkpoint blockade utilizes cellular mechanisms partially distinct from monotherapies. Proc Natl Acad Sci U S A 2019;116:22699–709. 10.1073/pnas.1821218116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devitt B, Liu W, Salemi R, et al. Clinical outcome and pathological features associated with NRAS mutation in cutaneous melanoma. Pigment Cell Melanoma Res 2011;24:666–72. 10.1111/j.1755-148X.2011.00873.x [DOI] [PubMed] [Google Scholar]
- 47.Veatch JR, Jesernig BL, Kargl J, et al. Endogenous CD4+ T cells recognize neoantigens in lung cancer patients, including recurrent oncogenic KRAS and ERBB2 (Her2) driver mutations. Cancer Immunol Res 2019;7:910–22. 10.1158/2326-6066.CIR-18-0402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu D, Schilling B, Liu D, et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med 2019;25:1916–27. 10.1038/s41591-019-0654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hodi FS, Wolchok JD, Schadendorf D. Abstract CT037: genomic analyses and immunotherapy in advanced melanoma. Cancer Research 2019;79:CT037. [Google Scholar]
- 50.Marabelle A, Fakih M, Lopez J, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol 2020;21:1353–65. 10.1016/S1470-2045(20)30445-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-003853supp001.pdf (146KB, pdf)
jitc-2021-003853supp002.pdf (193.5KB, pdf)
jitc-2021-003853supp003.pdf (54.5KB, pdf)
jitc-2021-003853supp004.pdf (2.1MB, pdf)
jitc-2021-003853supp005.pdf (705.9KB, pdf)
jitc-2021-003853supp006.pdf (2.5MB, pdf)
jitc-2021-003853supp007.pdf (94.9KB, pdf)
jitc-2021-003853supp008.pdf (117.1KB, pdf)
jitc-2021-003853supp009.pdf (812.7KB, pdf)
jitc-2021-003853supp010.pdf (1MB, pdf)
jitc-2021-003853supp011.pdf (734.9KB, pdf)
jitc-2021-003853supp012.pdf (5.5MB, pdf)
Data Availability Statement
Data are available upon reasonable request. Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as online supplementary information. The corresponding author may be contacted with any requests.