Abstract
Nicotiana plumbaginifolia Vivianiis 1802 is an annual herb, native to Mexico and South America. It is one of the most widely distributed tobacco species. As a wild tobacco, N. plumbaginifolia has provided several economically important disease-resistance genes to cultivated tobacco. We assembled the complete chloroplast genome of N. plumbaginifolia. The chloroplast genome is 155,945 bp in length, which includes a large single copy region (86,621 bp), a small single copy region (18,528 bp) and two separated inverted repeat regions (25,398 bp). A total of 117 unique genes were annotated, consisting of 84 protein-coding genes, 29 tRNA genes and 4 rRNA genes. Based on chloroplast genomes of 17 Nicotiana species, phylogenetic analyses indicated that N. plumbaginifolia was closely related to N. suaveolens and N. amplexicaulis.
Keywords: Chloroplast genome, tobacco, Nicotiana plumbaginifolia
Species of the genus Nicotiana, including herbaceous plants and shrubs in the family Solanaceae, are used for smoking, ceremonial, or ornamental purposes (Lewis 2020). Nicotiana plumbaginifolia is commonly called Tex-Mex tobacco and wild tobacco and exhibits antimicrobial and antioxidant activities (Ajaib et al. 2016). It has been treated as an important donor of genes/beneficial alleles for tobacco breeding. A fair number of disease-resistance genes have been transferred to cultivated tobacco from N. plumbaginifolia, including black shank resistance and tobacco cyst nematode (Globodera tabacum) resistance (Johnson et al. 2009). Chloroplast genomes helps us understand the origin and evolution of plants. Previously, Nicotiana was divided into 13 sections based on multiple chloroplast markers (Clarkson et al. 2004). N. plumbaginifolia was divided into the section Alatae, which is considered as a monophyletic group (Kaczorowski et al. 2005). The species epithet ‘plumbaginifolia’ comes from the way in which the leaves resemble those of species in the genus Plumbago. Because of the complex development history of polyploidy and hybridization, N. plumbaginifolia and other Nicotiana species are also used as evolutionary model systems. Chloroplast genomes are also related with important crop traits such as yield, crop quality, resistance to disease and pest (Jin and Daniell 2015).
Here, we assembled the plastid genome of N. plumbaginifolia. The sample of N. plumbaginifolia was collected in Brazil, near Santa Catarina (27°3.462 S, 51°6.538 W) and deposited in the Herbarium of Zhejiang University (accession number: HZU60244006). Total genomic DNA was sequenced by the Illumina platform. After quality control with NGSQCToolkit v2.3 (Patel and Jain 2012), the high quality data was applied in de novo assembly by NOVOPlasty v3.6 (Dierckxsens et al. 2017) using the Nicotiana tabacum complete chloroplast genome (GenBank accession number: NC_001879) as a reference. Genome annotation was performed by the GeSeq online (Tillich et al. 2017). The assembled genome sequences and annotation information have been submitted to the DNA Data Bank of Japan under accession number LC649170.
The total length of N. plumbaginifolia chloroplast genome is 155,945 bp. Like most angiosperm chloroplast genomes, this genome exhibited a distinct quadripartite structure, including a pair of inverted repeats (IRa and IRb, 25,398 bp each), the large single-copy region (LSC, 86,621 bp) and the small single-copy region (SSC, 18,528 bp). The GC contents of the IR, LSC, and SSC regions are 42, 35, and 30%, respectively. A total of 117 unique genes were annotated. Among these, there are 84 protein-coding genes, 29 tRNA genes and 4 rRNA genes.
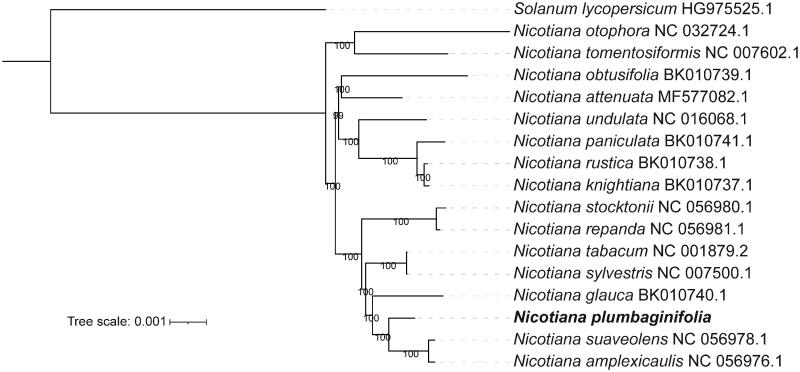
More recently, phylogeny inference based on complete chloroplast genomes provided insights into the phylogeny of certain families and genera (Amiryousefi et al. 2018). To investigate the evolutionary position of N. plumbaginifolia among Nicotiana species, we built a phylogenetic tree of 16 Nicotiana species based on complete chloroplast genome sequences downloaded from the NCBI GenBank database. Solanum lycopersicum was used as an outgroup. We first performed alignment by MAFFT v7.310 (Katoh et al. 2002) with the default parameter. Then, IQ-tree v1.6.12, an effective algorithm for estimating maximum-likelihood phylogenies, was used to construct a phylogenetic tree with recommended model TVM + F+R2 and 1000 bootstrap values (Nguyen et al. 2015). Finally, the tree was illustrated and modified using iTOL (Letunic and Bork 2019).
The phylogenic tree showed that N. plumbaginifolia first clustered with N. suaveolens and N. amplexicaulis forming as a monophyletic group (Figure 1). The young allotetraploid N. tabacum arose through the hybridization of the ancestral parents Nicotiana sylvestris and N. tomentosiformis (Edwards et al. 2017). The phylogenetic relationship between N. tabacum and N. sylvestris was quite close, which supported the assumption that N. sylvestris was the maternal genome donor (Figure 1). Our results provide basic information for further phylogenetic analysis on the genus Nicotiana.
Figure 1.
Maximum likelihood (ML) phylogenetic tree based on 16 Nicotiana species, using S. lycopersicum as an outgroup. The numbers on the node are the fast bootstrap value based on 1,000 replicates. The analyzed species and corresponding Genbank accession numbers are as follows: N. amplexicaulis NC_056976.1; N. attenuate NC_036467.1; N. debneyi NC_056977.1; N. glauca NC_056979.1; N. otophora NC_032724.1; N. repanda NC_056981.1; N. stocktonii NC_056980.1; N. suaveolens NC_056978.1; N. sylvestris NC_007500.1; N. tabacum NC_001879.2; N. tomentosiformis NC_007602.1; N. undulata NC_016068.1; N. knightiana BK010737; N. rustica BK010738; N. paniculate BK010741; N. obtusifolia BK010739; and S. lycopersicum HG975525.1.
Funding Statement
This work was supported by the Fundamental Research Program of Yunnan Province under [Grant 2018FB064]; Yunnan Tobacco Company under [Grant No. 2020530000241009]; the National Natural Science Foundation of China [NSFC, No.31860411]; and the Fundamental Research Funds for the Central Universities to E Shen.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data of this study is available in the DNA Data Bank of Japan (https://www.ddbj.nig.ac.jp/) under the accession number LC649170. The associated Bioproject, SRA, and Bio-Sample numbers are PRJNA763728, SAMN21448217, and SRR15927287, respectively.
References
- Ajaib M, Fatima S, Khan KM, Perveen S, Shah S.. 2016. Nicotiana plumbaginifolia: a rich antimicrobial and antioxidant source. J Chem Soc Pakistan. 38:143–149. [Google Scholar]
- Amiryousefi A, Hyvönen J, Poczai P.. 2018. The chloroplast genome sequence of bittersweet (Solanum dulcamara): plastid genome structure evolution in Solanaceae. PLOS One. 13(4):e0196069–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson JJ, Knapp S, Garcia VF, Olmstead RG, Leitch AR, Chase MW.. 2004. Phylogenetic relationships in Nicotiana (Solanaceae) inferred from multiple plastid DNA regions. Mol Phylogenet Evol. 33(1):75–90. [DOI] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KD, Fernandez-Pozo N, Drake-Stowe K, Humphry M, Evans AD, Bombarely A, Allen F, Hurst R, White B, Kernodle SP, et al. 2017. A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics. 18(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Daniell H.. 2015. The engineered chloroplast genome just got smarter. Trends Plant Sci. 20(10):622–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CS, Wernsman EA, Lamondia JA.. 2009. Effect of a chromosome segment marked by the Ph p gene for resistance to Phytophthora nicotianae on reproduction of tobacco cyst nematodes. Plant Dis. 93(3):309–315. [DOI] [PubMed] [Google Scholar]
- Kaczorowski RL, Gardener MC, Holtsford TP.. 2005. Nectar traits in Nicotiana section Alatae (Solanaceae) in relation to floral traits, pollinators, and mating system. Am J Bot. 92(8):1270–1283. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma KI, Miyata T.. 2002. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2019. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47:256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RS. 2020. Nicotiana tabacum L.: Tobacco. In Novak J. and Blüthner W.-D., eds. Medicinal, Aromatic and Stimulant Plants. Cham: Springer International Publishing, p. 345–375. [Google Scholar]
- Nguyen LT, Schmidt HA, Haeseler AV, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RK, Jain M.. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data of this study is available in the DNA Data Bank of Japan (https://www.ddbj.nig.ac.jp/) under the accession number LC649170. The associated Bioproject, SRA, and Bio-Sample numbers are PRJNA763728, SAMN21448217, and SRR15927287, respectively.