Abstract
Carotenoid pigments confer photoprotection and visual attraction and serve as precursors for many important signaling molecules. Herein, the orange-fruited phenotype of a tomato elite inbred line resulting from sharply reduced carotenoid levels and an increased β-carotene-to-lycopene ratio in fruit was shown to be controlled by a single recessive gene, oft3. BSA-Seq combined with fine mapping delimited the oft3 gene to a 71.23 kb interval on chromosome 4, including eight genes. Finally, the oft3 candidate gene SlIDI1, harboring a 116 bp deletion mutation, was identified by genome sequence analysis. Further functional complementation and CRISPR–Cas9 knockout experiments confirmed that SlIDI1 was the gene underlying the oft3 locus. qRT–PCR analysis revealed that the expression of SlIDI1 was highest in flowers and fruit and increased with fruit ripening or flower maturation. SlIDI1 simultaneously produced long and short transcripts by alternative transcription initiation and alternative splicing. Green fluorescent protein fusion expression revealed that the long isoform was mainly localized in plastids and that an N-terminal 59-amino acid extension sequence was responsible for plastid targeting. Short transcripts were identified in leaves and fruit by 5’ RACE and in fruit by 3’ RACE, which produced corresponding proteins lacking transit peptides and/or putative peroxisome targeting sequences, respectively. In SlIDI1 mutant fruit, SlBCH1 transcription involved in β-carotenoid catabolism was obviously suppressed, which may be responsible for the higher β-carotene-to-lycopene ratio and suggested potential feedback regulatory mechanisms involved in carotenoid pathway flux.
Introduction
Carotenoids are derived from isoprenoids and represent the largest class of natural pigments in plants. They are involved in plant growth and development in many aspects. Carotenoids are responsible for light harvesting and photoprotection during photosynthesis [1]. Carotenoids also contribute to red, yellow, and orange colors in mature flowers and ripe fruit that attract pollinators and promote seed dispersal [2]. In addition, carotenoids serve as precursors in the biosynthetic pathways of phytohormones and other signaling molecules that are very important for plant development and stress responses [3]. Some carotenoid derivatives are also associated with nutrition and flavor in human diets, which are important quality traits in crop plants [1, 4].
As the tomato is used as a model plant for studying fruit ripening and color formation, much knowledge regarding the regulation of carotenoid metabolism has been obtained based on the identification of relevant tomato mutants. Carotenoid biosynthesis occurs in plastids, starting with the condensation of two geranylgeranyl pyrophosphate (GGPP) molecules into phytoene by phytoene synthase (SlPSY), which is the committed step in the entire pathway [5]. Then, phytoene desaturase (SlPDS), carotene isomerase (SlZISO), carotene desaturase (SlZDS), and carotenoid isomerase (SlCRTISO) catalyze the following consecutive steps to produce trans-lycopene [6–8]. Next, the cyclization of trans-lycopene by lycopene ε-cyclase (SlLCY-E) or β-cyclase (SlLCY-B1 and SlLCY-B2) gives rise to α- or β-carotene, respectively [2, 9, 10]. Subsequent hydroxylation by the β-carotene hydroxylases SlBCH1 (CrtR-B1) and SlBCH2 [11] (CrtR-B2) and further oxygenation steps lead to the production of various xanthophylls, including lutein, zeaxanthin, violaxanthin and neoxanthin.
Carotenoid synthesis depends on isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP), synthesized from the terminal step in the plastidial 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway [12]. IPP and DMAPP are simultaneously produced from 1-hydroxy-2-methyl-2-butenyl-4-diphosphate (HMBPP) by HMBPP reductase (HDR) at a fixed ratio of 85:15 [13]. However, different ratios of DMAPP/IPP are required to serve as building blocks for the synthesis of various isoprenoids [14]. For monoterpenes, the ratio was 1:1; for sesquiterpenes and sterols, the ratio was 2:1; and for carotenoids, the ratio was 3:1. Reversible interconversion between IPP and DMAPP is catalyzed by isopentenyl diphosphate isomerase (IDI), which plays an important role in maintaining a dynamic optimal balance between them.
Two or more IDI isoforms have been discovered in many plant species. They have been found to be responsible for cytoplasmic and plastidial reactions. Plastidial IDI mediates the interconversion of IPP and DMAPP in the MEP pathway in plastids. Several plastidial IDI genes have been reported to date. In rice, plastid-localized OsIDI2 (OsIPPI2) was found to be expressed most highly in leaves but was not dispensable for chlorophyll or carotenoid accumulation in de-etiolated leaves [15]. In tobacco, NtIDI1 (NtIPP1) underwent transcription in response to high-salt and high-light stresses, and its encoded protein showed a longer N-terminal extension than cytosolic NtIDI2 (NtIPP2) and was found to target chloroplasts [16]. AtIDI1 in Arabidopsis thaliana was expressed mainly in the plastid but exhibited moderate peroxisome- and cytosol-localized activity with a lower efficiency. However, the AtIDI1 mutant was found to display only subtle morphological or chemical differences from the wild type [17].
In tomato, two IDI isoforms have been discovered [18]. SlIDI1 encodes a predicted protein of 294 amino acids, in which the main difference relative to its SlIDI2 isoform is a 59-amino acid extension sequence at the N-terminus. Both isoforms have been proven to be functional genes. SlIDI1 is predicted to target chloroplasts, while SlIDI2 is likely to be localized in the cytoplasm.
In this study, we focused on color formation in an orange-fruited tomato inbred line originally developed in our lab that has been assigned the name orange-fruited tomato3 (oft3). oft3 showed sharply reduced levels of carotenoids but an increased ratio of β-carotene to lycopene in fruit. BSA-seq and fine mapping based on segregating F2 and F3 populations identified SlIDI1, harboring a 116 bp deletion mutation, as the candidate gene for the oft3 locus. Further genetic functional complementation and CRISPR–Cas9 knockout assays confirmed that this mutation was responsible for the orange-fruited phenotype. SlIDI1 was proven to mainly target plastids, and the N-terminal 59-amino acid extension that was lacking in the SlIDI2 isoform was found to be indispensable for plastid localization. Rapid amplification of cDNA ends (RACE)-PCR experiments revealed the expression of truncated transcripts identified in leaves and fruit by 5’ RACE and in fruit by 3’ RACE, which produced corresponding proteins lacking transit peptides and/or putative peroxisome targeting sequences, respectively. Temporal and spatial expression assays conducted by qRT–PCR showed that SlIDI1 was transcribed at the highest abundance in carotenoid-rich flowers and fruit and was increasingly expressed during fruit ripening or flower maturation. SlBCH1, which is involved in β-carotenoid catabolism, was found to be obviously downregulated in SlIDI1 mutants, which may be the cause of the relatively high β-carotenoid content and orange-fruited phenotype.
Results
Phenotypic characterization and genetic analysis of the oft3 mutant
TB0735, an elite cherry tomato inbred line, is characterized by fruit with an orange color similar to that of tangerine (t) (Fig. 1a). Crosses between TB0735 and orange-fruited TB0017 (homozygous for mutated SlCRTISO, t) or yellow-fruited TB0040 (homozygous for mutated SlPSY1, r) produced F1 plants that all displayed a complementary red-fruited phenotype, which suggested that the TB0735 mutant does not present impairment at these two genetic loci. Here, we designate this mutant oft3.
Figure 1.
Fruit color development and genetic analysis of oft3. a Fruit color development. The tomato inbred lines oft3, the tangerine (t) mutant TB0017, and the r mutant TB0040 were originally developed in our lab. Fruit color (top), cross-sections (middle) and longitudinal sections (bottom) of these lines are shown at three different ripening stages. DPA: days post-anthesis. b Genetic analysis of oft3 conducted in the F2 and BC1F1 populations. Emasculated oft3 flowers were pollinated using AC (WT) pollen to produce the F1 generation. The F2 population was obtained from F1 self-pollinated plants, and the BC1F1 population was obtained by backcrossing using oft3 as a recurrent parent. The fruit color of each plant in the F2 and BC1F1 populations was confirmed when the fruit ripened.
High-performance liquid chromatography (HPLC) analysis showed no overaccumulated intermediates but an obviously reduced carotenoid content in oft3 fruit during ripening (Table 1), similar to the findings in the r mutant. Intriguingly, although there was a sharply decreased lycopene content (only 2.47% of that in the wild-type tomato cultivar Ailsa Craig (AC)), the β-carotene content of oft3 declined only slightly, resulting in an apparently higher β-carotene/lycopene ratio.
Table 1.
Carotenoid contents in oft3 fruit at three developmental stages
| Genotype | Phytoene | Lycopene | β-carotene | ζ-carotene | Lutein | Violaxanthin | Neoxanthin | Zeaxanthin | Total | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AC (WT) | 36 DPA | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.76 ± 0.09 b | 0.14 ± 0.03 a | 2.96 ± 0.36 a | 0.19 ± 0.05 a | 0.95 ± 0.23 a | 0.12 ± 0.03 a | 5.31 ± 0.79 a | |||||||||
| 40 DPA | 1.26 ± 0.17 a | 20.69 ± 2.41 a | 3.68 ± 0.40 a | 0.43 ± 0.08 a | 2.81 ± 0.33 a | 0.19 ± 0.03 a | 0.34 ± 0.08 a | 0.05 ± 0.02 b | 29.65 ± 2.61 a | ||||||||||
| 42 DPA | 5.27 ± 0.51 a | 59.05 ± 4.96 a | 5.37 ± 0.55 a | 1.20 ± 0.19 a | 2.20 ± 0.28 a | 0.09 ± 0.02 a | 0.01 ± 0.01 b | 0.04 ± 0.01 a | 73.32 ± 5.29 a | ||||||||||
| oft3 | 36 DPA | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 1.02 ± 0.16 a | 0.08 ± 0.01 b | 2.77 ± 0.34 a | 0.09 ± 0.03 b | 0.79 ± 0.10 a | 0.21 ± 0.08 a | 5.04 ± 0.63 a | |||||||||
| 40 DPA | 0.00 ± 0.00 b | 0.82 ± 0.16 b | 2.53 ± 0.30 b | 0.23 ± 0.06 b | 2.46 ± 0.44 a | 0.12 ± 0.02 b | 0.23 ± 0.05 ab | 0.11 ± 0.03 a | 6.63 ± 1.00 b | ||||||||||
| 42 DPA | 0.00 ± 0.00 b | 1.62 ± 0.22 b | 3.54 ± 0.33 b | 0.21 ± 0.05 b | 1.96 ± 0.26 a | 0.10 ± 0.04 a | 0.05 ± 0.02 a | 0.06 ± 0.05 a | 7.64 ± 1.03 b | ||||||||||
| TB0040 (r) | 36 DPA | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.79 ± 0.10 ab | 0.07 ± 0.01 b | 2.02 ± 0.33 b | 0.16 ± 0.04 a | 1.12 ± 0.23 a | 0.17 ± 0.06 a | 4.49 ± 0.74 a | |||||||||
| 40 DPA | 0.00 ± 0.00 b | 0.29 ± 0.04 c | 0.32 ± 0.06 c | 0.04 ± 0.03 c | 1.47 ± 0.18 b | 0.04 ± 0.01 c | 0.22 ± 0.03 b | 0.11 ± 0.03 a | 2.53 ± 0.32 c | ||||||||||
| 42 DPA | 0.00 ± 0.00 b | 0.65 ± 0.05 c | 0.08 ± 0.02 c | 0.01 ± 0.00 c | 1.16 ± 0.18 b | 0.02 ± 0.01 b | 0.03 ± 0.02 ab | 0.05 ± 0.02 a | 2.01 ± 0.25 c |
Note: The data represent the mean (μg g−1 fresh weight) ± SD from five biological replicates. Carotenoid contents were determined at three different ripening stages. The mean values were compared between genotypes at the same ripening stage, with different lowercase letters indicating significant differences (P < 0.05, Student’s t-test). The tomato inbred lines oft3 and the r mutant TB0040 were originally developed in our lab.
Genetic analysis was conducted by crossing oft3 with AC (WT) (Fig. 1b). All F1 plants showed a red-fruited phenotype. Color segregation (52 red/48 orange = 1.08) in the F1 × oft3 backcross progeny (BC1F1) corresponded approximately to a 1:1 Mendelian segregation ratio (χ2 = 0.08, P = 0.777). In the F2 population, color segregation (1101 red/355 orange = 3.10) matched the expected 3:1 Mendelian segregation ratio (χ2 = 0.15, P = 0.699). These results suggested that oft3 was controlled by a single recessive gene.
Mapping of oft3
BSA-Seq was employed to define the oft3 locus based on the generated F2 population derived from the crossing of oft3 with wild-type AC. A red-fruited pool (RF pool) and an orange-fruited pool (OF pool) were constructed for Illumina high-throughput sequencing. A total of 160 093 663 and 174 983 251 clean reads were obtained from the RF and OF pools, respectively. These clean reads were then mapped onto the Heinz 1706 reference genome (SL3.0), resulting in 89.99% and 89.93% coverage with at least a 50× depth.
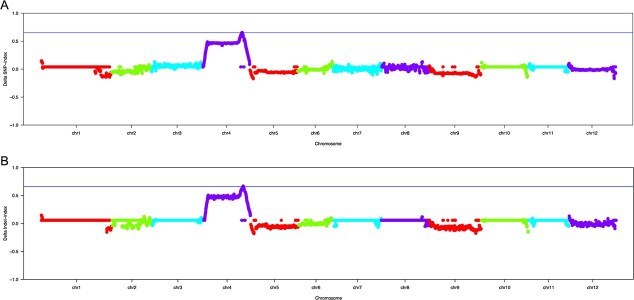
A total of 437 126 SNPs and 33 006 InDels were identified as differential between the RF pool and OF pool groups. The SNP/InDel index was computed within a 1 Mb interval using a 10 kb sliding window, and the ΔSNP/InDel index was generated by subtracting the SNP/InDel index of the OF pool from that of the RF pool. Subsequent correlation analysis based on the ΔSNP index revealed one candidate region (53 750 491 ~ 55 496 918 bp) on chromosome 4 (Fig. 2a, Table S1). Correlation analysis based on the ΔInDel index identified a candidate region on the same chromosome encompassing the 53 761 380 ~ 55 483 734 bp genomic positions (Fig. 2b, Table S2). By combining these two results, oft3 was mapped to a 1.70 Mb interval containing 136 genes (Table S3).
Figure 2.
Preliminary mapping of oft3 by using BSA-seq analysis. a Identification of the oft3 candidate region through the ΔSNP index association analysis method. b Identification of the oft3 candidate region through the ΔInDel index association analysis method. The X-axis represents the positions of twelve tomato chromosomes, and the Y-axis represents the Δ index. The colored dots represent the index value of every SNP/InDel locus. The blue imaginary lines indicate the association threshold of the Δ index.
Ten competitive allele-specific PCR (KASP) markers were developed based on the SNPs identified in the preliminary mapping interval (Table S4). By genotyping 1456 F2 plants for polymorphism analysis, the results from 36 recombinant individuals were shown to delimit the candidate area of oft3 to a 584.1 kb interval between markers OFT_Kp56 and OFT_Kp53. To further delimit the oft3 locus, 1033 F3 individuals derived from a single recombinant F2 plant were genotyped for recombinant screening using the other eight KASP markers. Finally, twelve informative recombinants allowed the oft3 locus to be narrowed to a 71.23 kb interval (54 083 697 ~ 54 154 931 bp) between markers OFT_Kp46 and OFT_Kp47 (Fig. 3). A total of eight genes were located in this region with diverse functional annotations, including a predicted lipid droplet biogenesis protein seipin-1 (Solyc04g056380), an isopentenyl diphosphate isomerase SlIDI1 (Solyc04g056390), a predicted chromatin remodeling protein (Solyc04g056410), a predicted peroxidase (Solyc04g056420) and four other unknown proteins (Solyc04g056370, Solyc04g056430, Solyc04g056440, Solyc04g056450) (Table S5). We proposed that the most likely candidate was SlIDI1, which is involved in reversible interconversion between IPP and DMAPP in the MEP pathway that acts upstream of carotenoid synthesis.
Figure 3.
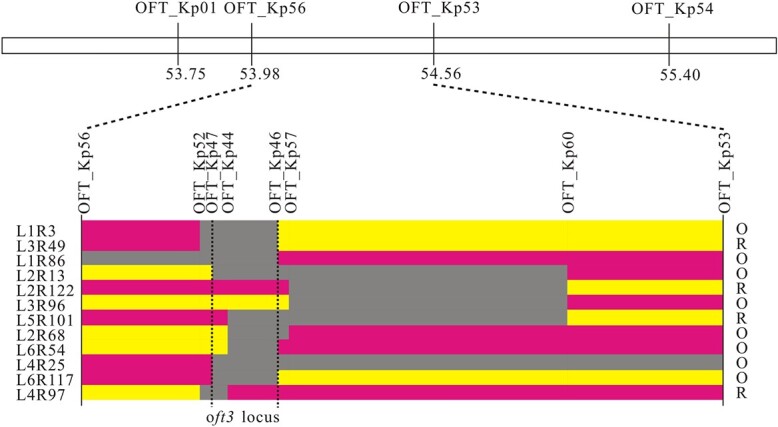
Fine mapping of oft3. The ten KASP markers shown above were used to screen recombinant individuals from the F2 population and the F3 population. The number of each individual recombinant plant is indicated to the left of the figure, and the fruit color phenotype is indicated to the right. Yellow blocks represent homozygosity, red blocks represent heterozygosity, and gray blocks represent the interval where crossover took place. The oft3 locus was refined to a 71.23 kb interval (54 083 697 ~ 54 154 931 bp) between markers OFT_Kp46 and OFT_Kp47 on chromosome 4. R: red-fruited phenotype; O: orange-fruited phenotype.
oft3 is a mutant allele of SlIDI1 with a 116 bp deletion
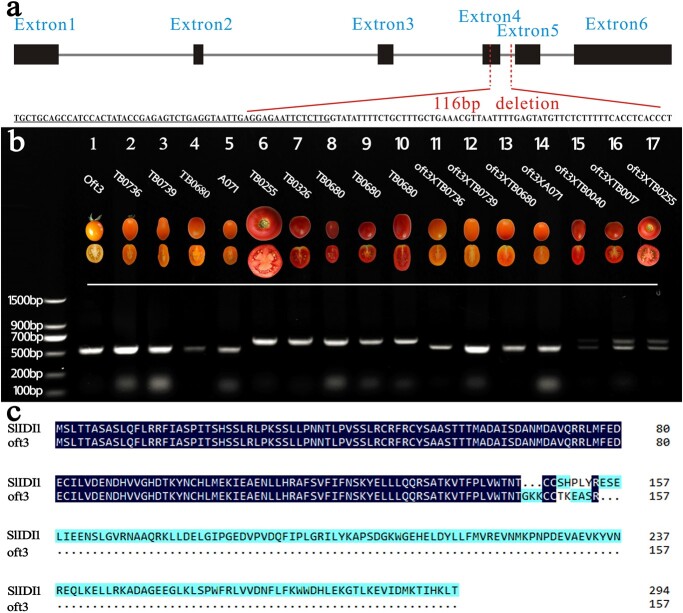
SlIDI1 in the Heinz 1706 reference genome (SL3.0) is 3770 bp in length, and no SNP or InDel mutations were found between oft3 and AC (WT) according to BSA-seq analysis. The genomic DNA sequences of SlIDI1 and the 1500 bp fragment upstream of the translation start codon were amplified by PCR from oft3 and AC (WT). Subsequent comparative analysis led to the identification of a long-fragment deletion in oft3. The deletion spanned an intron/exon junction encompassing 55 bp in exon 5 and 61 bp in intron 5, resulting in premature translation termination (Figs. 4a, c and S1).
Figure 4.
Sequence analysis of oft3. a The genomic DNA structure of SlIDI1 and the deleted fragment in oft3. Black boxes and gray lines represent the exons and introns of SlIDI1, respectively. The deleted fragment encompassed 55 bp in exon 5 (underlined) and 61 bp in intron 5. b Genotyping of tomato varieties using the markers developed based on the SlIDI1 deletion mutation of oft3. One 520 bp band was amplified by PCR in five orange-fruited inbred lines (Lanes 1–5), and their corresponding F1 hybrids (Lanes 11–14) were obtained by crossing with oft3, whereas a 636 bp band was amplified in five wild-type red-fruited inbred lines (Lanes 6–10). The two bands were produced by PCR in the three F1 hybrids between oft3 and the r mutant TB0040 (Lane 15), t mutant TB0017 (Lane 16) and wild-type AC (Lane 17). c Deduced protein sequence of SlIDI1 in oft3. SlIDI1 in oft3 was deduced to produce a truncated protein (157 aa) terminated by the premature translational stop signal resulting from the deletion mutation.
Genetic analysis through crossing experiments showed that four other inbred lines (TB0736, TB0739, TB0680 and A071) exhibiting the same orange-fruited phenotype as oft3 all carried impaired oft3 alleles. Genomic sequencing and comparative analysis revealed that the same 116 bp deletion mutation occurred in SlIDI1 in these lines (Fig. 4b).
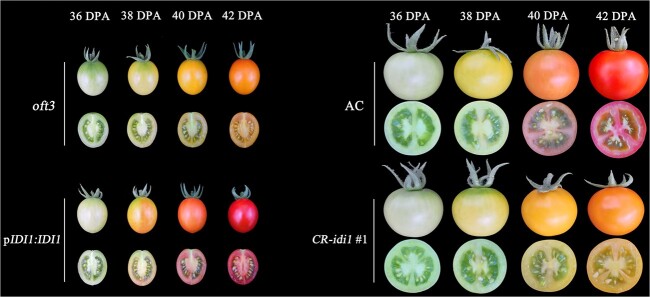
To obtain conclusive proof that SlIDI1 is the gene underlying the oft3 locus, transgenic complementation analysis was conducted. A construct pIDI1:IDI1-EGFP (enhanced green fluorescent protein) containing the CDS of SlIDI1 from AC (WT) under the control of its native promoter was generated and transformed into oft3 plants via Agrobacterium tumefaciens EHA105-mediated genetic transformation. Finally, a total of twelve independent transformants (pIDI1:IDI1 #1 ~ 12) were obtained, and all of these transformants restored the orange-fruited phenotype of oft3 to a red-fruited phenotype during ripening (Fig. S2, Fig. 5).
Figure 5.
Functional complementation and knockout analysis of oft3. Functional complementation analysis was conducted in oft3 by transforming oft3 plants with SlIDI1 driven by its native promoter, and all the transformants (pIDI:IDI1 #1 ~ 12) were restored to the red-fruited phenotype, as observed in wild-type AC (left). CR-idi1 #1 was generated by knocking out SlIDI1 in wild-type AC using the CRISPR–Cas9 system. CR-idi1 #1 plants from the T1 generation that were shown to be homozygous for mutated SlIDI1 by genotyping showed an orange-fruited phenotype, similar to that of oft3 (right). Fruit color and longitudinal sections from four different ripening stages are shown. DPA: days post-anthesis.
The knockout of SlIDI1 by using CRISPR–Cas9 technology was also conducted. A CRISPR/Cas9 construct with two synthetic gRNAs designed to target exon 1 and exon 2 was produced and introduced into AC (WT). Five of ten mutant plants (CR-idi1 #1 ~ 5) were screened by sequencing in the T0 generation (Fig. S3). T1 progenies generated from the five self-pollinated T0 mutants that were shown to be homozygous for mutated SlIDI1 and untransgenic by genotyping were further phenotyped while ripening. As expected, all CRISPR–Cas9-derived mutants displayed 1orange flesh, similar to oft3 (Fig. S2, Fig. 5).
SlIDI1 is involved in carotenoid synthesis in petals and anthers
Tomato flowers also accumulate high concentrations of carotenoids when they mature, among which yellow xanthophylls are the principal component. When compared with the parental line AC by visual inspection, the CRISPR–Cas9-generated SlIDI1 mutant exhibited a paler yellow color of the petals and anthers (Fig. S4). HPLC analysis revealed that SlIDI1 knockout led to apparently reduced xanthophyll contents in both petals and anthers (Table 2).
Table 2.
Carotenoid contents of the flowers of AC (WT) and the CRISPR–Cas9-generated SlIDI1 mutant
| Tissue | Genotype | Phytoene | β-carotene | Lutein | Violaxanthin | Neoxanthin | Zeaxanthin | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| petal | AC (WT) | 0.96 ± 0.26 a | 12.53 ± 1.15 a | 17.66 ± 2.28 a | 98.36 ± 8.98 a | 262.56 ± 17.91 a | 7.62 ± 0.88 a | 399.13 ± 29.48 a | |||||||
| CR-Slidi1 #1 | 0.78 ± 0.32 a | 16.54 ± 2.93 b | 11.96 ± 1.26 b | 53.31 ± 6.34 b | 113.39 ± 9.07 b | 4.36 ± 0.95 b | 200.79 ± 17.83 b | ||||||||
| anther | AC (WT) | 0.79 ± 0.18 a | 10.32 ± 1.89 a | 15.66 ± 2.28 a | 114.36 ± 18.92 a | 182.56 ± 20.93 a | 5.62 ± 1.84 a | 329.96 ± 34.46 a | |||||||
| CR-Slidi1 #1 | 0.85 ± 0.10 a | 13.24 ± 1.95 a | 8.96 ± 1.26 b | 63.31 ± 6.34 b | 88.39 ± 8.01 b | 4.36 ± 0.66 a | 177.46 ± 16.32 b |
Note: The data represent the mean (μg g−1 fresh weight) ± SD from five biological replicates. The mean values were compared between wild-type AC and the CR-Slidi1 #1 mutant, with different lowercase letters indicating significant differences (P < 0.05, Student’s t-test)
SlIDI1 expression increases are accompanied by carotenoid accumulation in fruit and flowers during maturation
To determine the expression pattern of SlIDI1, mRNA levels in different tissues and different stages of AC (WT) tissue development were measured by qRT–PCR. Transcripts of SlIDI1 were detected in all tissues but were expressed at the highest levels in flowers and fruit (Fig. 6).
Figure 6.
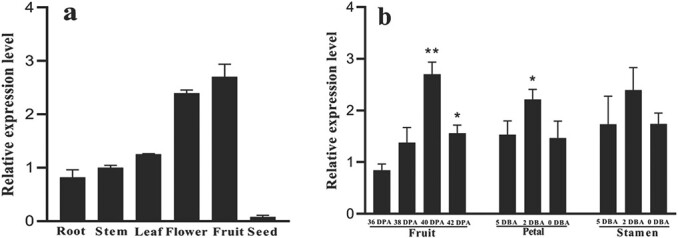
Spatiotemporal specific expression analysis of SlIDI1 in wild-type AC by qRT–PCR. a Tissue-specific expression analysis of SlIDI1 in different tissues of wild-type AC. b Stage-specific expression analysis of SlIDI1 in the fruit, petals and stamens of wild-type AC at different ripening or maturation stages. Values are the means of four biological replicates ± SD. For stage-specific expression analysis, values were compared among different stages of each tissue, and asterisks denote significance by Student’s t-test (*P < 0.05, **P < 0.01). DPA: days post-anthesis, DBA: days before anthesis.
In the fruit, SlIDI1 transcripts showed continuously increased expression throughout the ripening period and rose approximately threefold from 36 DPA to 40 DPA, in parallel with increased carotenoid production (Fig. 6a). Likewise, SlIDI1 transcripts in petals and anthers were upregulated as the flowers matured and carotenoids accumulated and reached a peak at 2 days after anthesis (2 DAA) (Fig. 6b). These results suggested that the expression pattern of SlIDI1 coincided well with its role in carotenoid synthesis in tomatoes.
Alternative transcripts and subcellular location of SlIDI1
In A. thaliana and Catharan thusroseus, IDI can be expressed as one long isoform and one short isoform through alternative transcription initiation to target different organelles [17, 19, 20]. RACE-PCR experiments were conducted for SlIDI1 in the fruit, petals, stamens and leaves of AC (WT). Finally, 5’ RACE-PCR and 3’ RACE-PCR both produced long and short products (Fig. 7). The long products were obtained from all the tissues. Sequencing and assembly gave rise to various long transcripts, with the only differences occurring in the length of the 3’-UTR or 5’-UTR (Figs. S5 and S6). The short 5’RACE product, designated SlIDI1–5’S, was identified from fruit and leaves. Sequence analysis revealed that in SlIDI1–5’S, the entire exon 1 and part of exon 2 were deleted, which may result in the translation of a shorter SlIDI1 isoform lacking 102 amino acids at the 5′ end. The short 3’ RACE product, designated SlIDI1–3’S, was identified from fruit, and sequence analysis showed that its entire exon 6 sequence was replaced with 165 bp of intron 5, resulting in a deduced truncated SlIDI1 isoform lacking 84 amino acids at the 3′ end. These results suggested that long and short isoforms (at least two isoforms) of SlIDI1 also exist in tomatoes, although we could not confirm whether SlIDI1–5’S and SlIDI1–3’S were derived from the same transcript.
Figure 7.
Alternative transcripts of SlIDI1 identified by RACE-PCR. a RACE-PCR products amplified from the leaves, fruit, petals and anthers of wild-type AC. For 5’ RACE-PCR (left), two identical short products were amplified from leaves and fruit. For 3’ RACE-PCR (right), one short product was amplified from fruit. b Schematic representation of the structures of DNA, the full-length cDNA of SlIDI1 (SlIDI1-L) and two short RACE-PCR products (SlIDI1–5’S and SlIDI1–3’S) identified by 5’ RACE-PCR and 3’ RACE-PCR. SlIDI1-L: long transcripts with the longest 5’-UTR (44 bp) and 3’-UTR (338 bp). Exons and introns are represented by boxes and lines. The 5′ and 3’ UTR are represented by gray boxes. The two specific primers used for 5’-RACE and 3’-RACE (5’RACE-GSP and 3’RACE-GSP) are represented by red boxes. In alternative transcription-generated SlIDI1–5’S, a 274 bp deletion occurred in the CDS region, including exon 1 and part of exon 2. In alternative-splicing-generated SlIDI1–3’S, 254 bp of exon 6 was replaced by 165 bp of intron 5 (white box). c Aligned sequences of the full-length cDNA of SlIDI1 (SlIDI1-L) and two short RACE-PCR products (SlIDI1–5’S and SlIDI1–3’S). The putative initiation codons and stop codons are indicated in red. The retained 165 bp of intron 5 in SlIDI1–3’S is boxed. d Aligned protein sequences deduced from SlIDI1-L, SlIDI1–5’S and SlIDI1–3’S. SlIDI1 is 294 amino acids in length. The putative CTP and type 1 peroxisome targeting sequence (PTS1) are underlined.
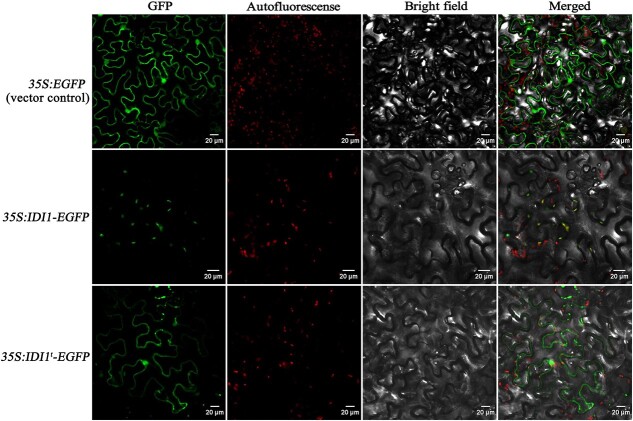
Subsequently, the localization of SlIDI1 was studied. The CDNA sequence of the SlIDI1-L long transcript was fused to the N-terminus of EGFP, and the corresponding 35S:SlIDI1-EGFP construct was then transiently expressed in tobacco leaves. Fluorescence microscopy associated with image overlay techniques demonstrated that, in contrast to the discrete fluorescence signals of 35S:EGFP (vector control), which were distributed evenly throughout the cytoplasm, most of the fluorescence signals from the 35S:SlIDI1-EGFP fusion protein coincided with chromoplast autofluorescence, indicating obvious plastid-localized characteristics of SlIDI1 (Fig. 8).
Figure 8.
Subcellular location of SlIDI1. 35S:SlIDI1-EGFP, with the full-length SlIDI1 CDS, and 35S:SlIDI1t-EGFP, with a truncated SlIDI1 CDS lacking the 59-amino acid extension sequence at the N-terminus, were agroinfiltrated into tobacco leaves, and the agroinfiltrated leaf epidermal cells were examined under a confocal microscope. 35S:EGFP served as the vector control
A 59-amino acid sequence at the N-terminus of SlIDI1 was predicted to be a chloroplast transit peptide (CTP) [18]. For confirmation, the 35S:IDI1t-EGFP construct expressing EGFP-fused truncated SlIDI1 that lacked the N-terminal 59-amino acid sequence was agroinfiltrated into tobacco leaves. In contrast to 35S:IDI1-EGFP, green fluorescence from 35S:IDI1t-EGFP did not coincide with plastid autofluorescence but produced discrete fluorescence signals (Fig. 8), which suggested that the truncated SlIDI1 failed to target plastids and that the 59-amino acid sequence is indispensable for the plastid localization of SlIDI1.
SlIDI1 suppresses the expression of SlBCH1 in fruit
Tomato fruit color is determined by the ratio of β-carotene to lycopene, which are the two most abundant carotenoids in tomatoes [7]. SlIDI1 acts upstream of SlPSY1 (Fig. S7); however, its disruption did not result in a yellow-flesh phenotype, as observed in the SlPSY1 mutant, but instead led to an orange-fruited phenotype owing to a relatively higher β-carotene level or higher ratio of β-carotene to lycopene (Table 1). To gain insight into this peculiar phenomenon, the transcription of key genes involved in the carotenoid pathway was examined during fruit ripening stages using qRT–PCR. Two wild-type individuals (WT-1 and WT-2) and two oft3 genotyped individuals (oft3–1 and oft3–2) used for expression measurements were selected from the segregating BC1F2 population on the basis of DNA marker analysis to minimize the genetic background differences. In oft3–1 and oft3–2 fruit, the expression levels of SlPSY1, SlPDS, SlCRTISO and SlLCY-B1 increased, but those of SlLCY-B2 and SlLCY-E decreased as the fruit ripened, showing similar expression patterns to those found in WT-1 and WT-2. None of these carotene biosynthetic genes displayed significant alterations in transcript abundance throughout the four ripening stages when the two oft3 genotyped plants were compared to the two wild-type BC1F2 plants (Fig. S8).
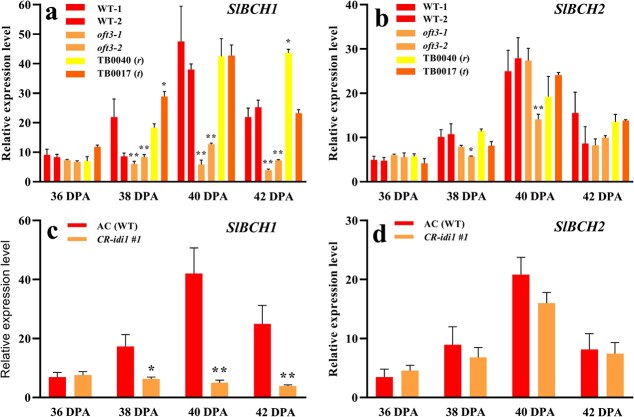
The expression levels of two carotene catabolism genes, SlBCH1 and SlBCH2, responsible for the conversion of β-carotene to zeaxanthin [11], were examined in fruit at the same time (Fig. 9a, b). SlBCH2 expression levels showed no obvious difference. Surprisingly, SlBCH1 transcription was found to be markedly inhibited in oft3–1 and oft3–2 fruit relative to WT-1 and WT-2 fruit. However, in the r mutant TB0040 and the t mutant TB0017, SlBCH1 expression was not affected. To obtain further confirmation, SlBCH1 and SlBCH2 transcripts were examined in the CRISPR–Cas9-generated mutant CR-idi1 #1 (Fig. 9c, d). Likewise, clearly distinguishable depressed expression of SlBCH1, but not SlBCH2, was observed in CR-idi1 #1 fruit relative to the parental line AC, with decreases of 3.5-, 10.4- and 8.7-fold being observed at 38 DPA, 40 DPA and 42 DPA, respectively.
Figure 9.
Expression analysis of SlBCH1 and SlBCH2 by qRT–PCR in the fruit of the SlIDI1 mutant at four ripening stages. a and b Expression analysis of SlBCH1 and SlBCH2 in two wild-type (WT-1 and WT-2) and two oft3 genotyped individuals (oft3–1 and oft3–2) from the segregating BC1F2 population. c and d Expression analysis of SlBCH1 and SlBCH2 in the CRISPR–Cas9-generated SlIDI1 mutant CR-idi1 #1 compared with its parental line AC. Values are the means of four biological replicates ± SD and were compared between genotypes at the same ripening stage. Asterisks denote significance by Student’s t-test (*P < 0.05, **P < 0.01). DPA: days post-anthesis.
Discussion
SlIDI1 is involved in carotenoids synthesis in tomato
Carotenoid synthesis is very important in plants. The 40-carbon hydrocarbon compounds produced in this pathway provide plants with orange, red, and yellow pigments and participate in a wide range of physiological processes, such as photosynthesis, plant development, and responses to environmental stimuli. Carotenoids mostly exist in nongreen tissues and are specifically synthesized in chromoplasts. Since the discovery of PSY1 in the carotenoid biosynthetic pathway, a set of key biosynthetic enzymes have been identified through extensive study, which has resulted in great progress in understanding the reactions involved in carotenoid biosynthesis (Fig. S7). In this study, we characterized a single major locus for carotenoid deficiency in the fruit and flowers of the tomato mutant oft3. A naturally occurring 116 bp deletion mutation in SlIDI1 was identified by map-based cloning. The absence of detectable transcripts, probably caused by nonsense-mediated mRNA decay, suggested that the oft3 mutant phenotype resulted from a complete loss-of-function mutation in SlIDI1 (data not shown). SlIDI1 was previously identified as a strong candidate gene for carotenoid deficiency in fruit of ethyl methanesulfonate (EMS)-induced tomato fcd1 (FRUIT CAROTENOID DEFICIENT 1) mutants [21]. Here, robust evidence was presented to confirm the involvement of SlIDI1 in carotenoid synthesis: 1) genetic complementation analysis combined with CRISPR–Cas9-mediated knockout analysis proved that the identified novel SlIDI1 deletion mutation was responsible for carotenoid deficiency in oft3 (Fig. 5); 2) temporal expression analysis showed that SlIDI1 was transcribed at the highest abundance in ripened fruit and matured flowers, which reflected the accumulation of carotenoids well during the fruit ripening and flower maturation processes (Fig. 6); and 3) subcellular location analysis indicated that SlIDI1 mainly targeted the plastid, where carotenoids are produced (Fig. 8).
In higher plants, carotenoid metabolism relies on the supply of the building blocks IDP and its isomer DMAPP, which both serve as substrates for the synthesis of the carotenoid precursor GGPP. IDP and DMAPP in plastids are produced simultaneously by HDR at a ratio of 85:15. The IDI enzyme catalyzes the transformation of IDP into DMAPP to produce the required stoichiometric ratio of 3:1 for GGPP synthesis [12, 13]. It can be speculated that markedly reduced carotenoid levels in fruit, petals and anthers all result from SlIDI1 deficiency-causing shortage of DMAPP.
A distinctive etiolated phenotype was discovered in the cotyledons of oft3- and CRISPR–Cas9-generated mutants, especially when cultured under a lower light intensity (Fig. S4). Interestingly, no color difference was observed in true leaves. These results were consistent with those found in the fcd1 mutant, which is allelic to oft3 [21]. SlIDI1 acts upstream in the chlorophyll synthetic pathway (Fig. S7), and SlIDI1 deficiency may block chlorophyll production in cotyledons.
SlIDI1 could be targeted to plastids and other organelles by alternative transcription initiation and alternative splicing
In higher plants, IDP and DMAPP can be synthesized via two independent pathways: the mevalonic acid (MVA) pathway in the cytoplasm and the MEP pathway in plastids [12]. Two or more IDI isoforms, produced via the MVA and/or MEP pathways, have been shown to target different subcellular compartments in many plants, such as Arabidopsis, Catharanthusroseus, tobacco and rice. In tomatoes, two IDI isoforms have been identified, and SlIDI1 exhibits a 59-amino acid extension sequence at its N-terminus compared to cytoplasm-localized SlIDI2 [18]. SlIDI1 has been predicted to target plastids, and the N-terminal 59-amino acid extension sequence is speculated to serve as a CTP. However, these hypotheses have yet to be confirmed by credible evidence. In this study, the evaluation of the subcellular localization of SlIDI1 by using a constitutively expressed protein fused with a GFP signal in tobacco leaves proved that SlIDI1 was mainly targeted to plastids. Moreover, a construct expressing an EGFP-fused truncated SlIDI1 that lacked the N-terminal 59-amino acid sequence was agroinfiltrated into tobacco leaves, and deletion/fusion experiments showed that the truncated SlIDI1 failed to be targeted to plastids. This further suggested that the 59-residue N-terminal extension is indispensable for plastid targeting.
It is worth noting that a small quantity of noncoincident fluorescence signals could be seen in the SlIDI1 subcellular localization analysis, suggesting the possible targeting of SlIDI1 to multiple subcellular compartments. According to studies in A. thaliana and Catharan thusroseus, IDI can be targeted to different organelles through alternative transcription. In A. thaliana [17, 19], AtIDI1 was found to be expressed in plastids but also in the cytosol due to the translation of one shorter transcript lacking the transit peptide (TP). In Catharanthus roseus [20], CrIDI1 is targeted to both plastids and mitochondria with a similar efficiency and is simultaneously targeted to peroxisomes as a short isoform that lacks the N-terminal sequence, including the transit peptide. To determine whether SlIDI1 also shows this mechanism, RACE-PCR experiments were conducted in tomato leaves, flowers and fruit. Interestingly, we not only discovered similar alternative transcription initiation at the 5′ end to that found in A. thaliana and Catharanthusroseus but also identified a novel alternative splicing event at the 3′ end that was not previously reported for IDI. Sequencing and alignment analysis showed that the shorter 5’RACE product SlIDI1–5’S, generated in leaves and fruit by alternative transcription initiation, was predicted to encode a truncated protein lacking the 59-amino acid transit peptide at the N-terminus. The shorter 3’RACE product SlIDI1–3’S, generated by alternative splicing in fruit, was found to lack the last exon and, thus, the putative type I peroxisome targeting (PST1) sequence at the C-terminus of the deduced protein sequence. These results were in accordance with the results of the SlIDI1 subcellular localization experiment that showed nonplastid-localized GFP signals. They further suggested a potentially similar mechanism of SlIDI1 to that found in A. thaliana and Catharanthusroseus; i.e. SlIDI1 could be targeted to multiple organelles via the expression of different isoforms. However, whether SlIDI1–5’S and SlIDI1–3’S were derived from the same transcript was not confirmed in this study. In addition, whether SlIDI1 could be targeted to mitochondria or peroxisomes was not confirmed. Further studies will be needed to address this question.
Relatively higher β-carotene content and repressed transcription of the carotenogenic gene SlBCH1 in response to SlIDI1 impairment suggested a novel feedback regulation mechanism in carotenoid synthesis
IDI1 functions in the MEP pathway upstream of PSY1, which encodes the first committed enzyme involved in carotenoid flux (Fig. S7). However, tomato carrying a mutated SlIDI1 gene showed an orange-fruited phenotype and not a yellow color, as observed in the r (SlPsy1) mutant. As revealed by HPLC analysis, oft3 fruit did not overaccumulate intermediates as did the orange-fruited t mutant. Compared with wild-type AC fruit, it showed sharply reduced levels of carotenoids but an increased ratio of β-carotene to lycopene due to a slight decrease in β-carotene accompanied by a sharp reduction in lycopene. In contrast, the yellow-fruited r mutant TB0040 exhibited lower levels of lycopene, β-carotene and total carotenoid content in fruit than oft3 (Table 1). This peculiar phenomenon raised the intriguing question of what the underlying mechanism may be. Transcript-level control is thought to be the major mechanism that determines the flux of carotenoid biosynthesis. Thus, the analysis of expression levels by qRT–PCR was conducted for the key carotenoid biosynthetic genes (SlPSY1, SlPDS, SlCRTISO, SlLCY-B1, SlLCY-B2 and SlLCY-E) in two oft3 genotyped BC1F2 individuals. However, no significant change was observed in any of these genes relative to the results in the wild-type counterparts (Fig. S8). Interestingly, the measurement of the expression level of SlBCH1, which is reported to encode β-carotene hydroxylase, responsible for catalyzing the transformation of β-carotene into other xanthophylls [11], revealed serious transcriptional repression. This result was further confirmed in CRISPR–Cas9-generated mutants. Thus, we speculated that SlIDI1 deficiency blocked carotenoid synthesis and that the decrease in SlBCH1 transcripts delayed β-carotene catabolism, which resulted in less obviously reduced β-carotene accumulation and contributed to an orange-fruited phenotype in the Slidi1 mutant. On the other hand, feedback regulation of carotenoid production in tomatoes has been reported previously [22, 23], and this result indicated a potential novel feedback loop involved in carotenoid pathway flux.
Materials and methods
Plant materials and growth conditions
Seeds of the tomato cultivar AC were provided by the Tomato Genetic Resource. All of the tomato inbred lines, including TB0735 (oft3), TB0040 (r), and TB0017 (t), were originally developed by us. Seedlings were cultivated in a mixture of peat and vermiculite (3:1, V/V) under controlled conditions (16 h light (200 μE m−2 s−1) at 25°C and 16 h dark at 18°C, 60% relative humidity) and then transplanted to tunnel greenhouses at the Tongzhou farm in Beijing, China. All of the cultivars and/or populations derived from crossing between them were grown side by side from March to July every year from 2018 to 2020 with regular cultivation.
Carotenoid extraction and analysis
Tomato flesh was sampled at three ripening stages: 36 DPA, 40 DPA and 42 DPA. In addition, petal and anther were sampled at the anthesis stage. Carotenoids were extracted from these samples and further analyzed on a Shimadzu Nexera HPLC system (Japan) using the method described previously [24], with five biological replicates. Commercial carotenoids (Sigma–Aldrich, USA) were used as standards.
Genetic analysis of oft3
oft3 flowers were emasculated and then pollinated with AC (WT) pollen to generate F1 seeds. F1 plants were then self-pollinated to give rise to the F2 population. The BC1F1 generation was obtained by backcrossing using oft3 as the recurrent parent. A total of 1456 F2 and 100 BC1F1 plants were grown with AC (WT), oft3 and F1 plants, and the fruit color of each plant was confirmed when the fruit ripened to identify the genetic basis of oft3 via the χ2 test.
DNA extraction
Young leaves from tomato plants were collected, quickly put into liquid nitrogen for pro-freezing, and then stored in a freezer at −80°C. DNA extraction was performed using the CTAB method [25]. DNA quality was examined by using a NanoDrop 2000 spectrophotometer (Thermo Fisher, USA) and agarose gel electrophoresis.
BSA-seq
Equimolar concentrations of DNA were pooled from 50 red-fruited F2 individuals (R-pool), 50 orange-fruited F2 individuals (O-pool) and the two parental lines. Libraries with mixtures of 300 ~ 500 bp DNA fragments for all of the DNA pools were prepared according to the manufacturer’s protocols and subjected to paired-end sequencing on an Illumina High-seq 2500 sequencing platform by the Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China). Raw reads were assigned to individual samples according to their nucleotide barcodes using the Axe package.
Low-quality reads, which were defined as more than 10% nucleotides with a quality value lower than 30, were filtered out with fastp software [26]. The Heinz 1706 genome (SL3.0) was used as the reference for qualified read mapping using BWA MEM software [27]. SNPs and InDels were called across all samples using Haplotype Caller in GATK 4.0.11.0 [28] and then filtered using standard hard filtering parameters according to the GATK Best Practices pipeline (mapping quality >37, quality depth > 24).
Genetic markers were identified using a modified QTL-seq method [29]. The SNP/InDel index was calculated based on read depth information for the homozygous SNPs/InDels in the R-pool and O-pool using each parental line as a reference. The SNP/InDel index of the base sites was defined as the ratio of the number of different reads to the total number of reads. The SNP/InDel index of the whole genome was calculated using sliding window methods, with a window size of 1 Mb and a step size of 10 kb as the default settings. The average of all SNP/InDel indices for the R-pool or O-pool in each window was used as the SNP/InDel index for that window, and the SNP/InDel index difference between the two pools was calculated as the ΔSNP/InDel index. ΔSNP/InDel index graphs were plotted separately, with the statistical confidence intervals calculated under the null hypothesis of no QTLs [21].
Fine mapping
According to the SNPs identified between the parental lines AC and oft3 by BSA-seq in the preliminary mapping interval, a total of ten KASP markers were successfully developed (Table S6). Recombination analysis was conducted in the F2 population based on genotyping and phenotyping. F3 populations were generated from the self-pollination of recombinant F2 individuals for further recombination analysis and fine mapping. Finally, the genes in the identified mapping region were assessed by the examination of the gene functional annotations to obtain the gene candidate(s) for oft3.
Genomic sequence analysis of SlIDI1 in oft3
For the candidate gene SlIDI1, the 1500-bp promoter sequences upstream of the translation start codon as well as the CDSs from AC (WT) and oft3 corresponding to that from the Heinz 1706 reference genome (SL3.0) were determined and compared. PCR amplification was conducted using the primers listed in Table S6. The obtained PCR products were purified and sequenced by Tianyi Huiyuan Bioscience & Technology, Inc. (Beijing, China). Sequence assembly and comparative analyses were conducted using DNAMAN (Version 5.0).
Functional complementation and knockout analysis
For the functional complementation experiment, we prepared a transformation construct with the SlIDI1 CDS under the control of its native promoter. The full-length CDS and the upstream 1500 bp genomic sequence of SlIDI1 from AC (WT) were artificially synthesized by GenScript Biotechnology Co., Ltd. (Nanjing, China). The synthesized SlIDI1 CDS was inserted into the PYBA-1332 vector, and CaMV35S in the recombinant vector was subsequently replaced with the synthesized promoter sequence. The produced construct was introduced into A. tumefaciens EHA105 for further transformation into oft3. Transformants were screened by checking the presence of transgenes using PCR.
The CRISPR–Cas9 system described by Deng et al. [30] was used to conduct the knockout experiment. The vector containing two sgRNAs targeting exon 1 and exon 2 was introduced into EHA105 cells for subsequent transformation into AC (WT) cells. Mutation analysis was conducted in the generated T0 plants by sequencing the PCR products amplified with primers that flanked the gRNA targets or potential off-targets (Tianyi Huiyuan Bioscience & Technology, Inc., Beijing, China). T1 progenies derived from the self-pollination of T0 mutants were screened for the absence of transgenes to obtain Cas9-free homozygous individuals.
The transformants, CRISPR-derived mutants obtained above and their corresponding wild-type plants were grown side by side for genotyping at the fruit ripening stage. All primers used are listed in Table S6.
Amplification of full-length cDNA by RACE-PCR
RACE-PCR experiments were conducted in the leaves, flowers and fruit of AC (WT) using the 5’-Full RACE and 3’-Full RACE Core Set (TaKaRa) to obtain the full-length cDNA of SlIDI1 in these tissues. The primers for 5′ or 3’-RACE were developed based on the Heinz 1706 reference genome (SL3.0) and are listed in Table S6. The resulting PCR products in each tissue were separated, purified and then inserted into pMD18-T simple vectors separately for sequencing (Tianyi Huiyuan Bioscience & Technology, Inc., Beijing, China). Finally, alignments were conducted using DNAMAN (Version 5.0) via the ClustalW method following sequence assembly.
Subcellular localization
Two constructs were produced to conduct subcellular localization analysis using the primers listed in Table S6. To obtain the 35S:SlIDI1-EGFP construct, the artificially synthesized full-length SlIDI1 CDS (Nanjing GenScript Biotechnology Co., Ltd) was cloned as an in-frame C-terminal fusion to EGFP in the PYBA-1332 vector under the control of the CaMV35S promoter. Likewise, the artificially synthesized truncated SlIDI1 CDS (lacking 177 bp at the 5′ end) was cloned into the PYBA-1332 vector to generate the construct 35S:SlIDI1t-EEFP. Three-week-old Nicotiana benthamiana leaves were infiltrated with A. tumefaciens EHA105 transformed with the constructs. After 36 h of incubation, a Zeiss LSM 780 confocal microscope (Germany) was used to observe the GFP fluorescence signals.
Gene expression analysis
Samples of young leaves, stems, roots, floral tissues at different developmental stages (petals and stamens) and fruit were collected for total RNA extraction using TRIzol reagent (Invitrogen, USA). Total RNA quality was examined by using a NanoDrop 2000 spectrophotometer (Thermo Fisher, USA). Then, mRNA was purified from total RNA and used as a template to synthesize double-stranded cDNA using a RT Reagent Kit (TaKaRa, Japan). qRT–PCR analysis was performed as described before [31], with the primers listed in Table S6. All experiments were performed with three biological replicates. The expression levels of target genes were normalized against that of SlACTIN2.
Supplementary Material
Acknowledgments
This work was supported by the Beijing Academy of Agricultural and Forestry Sciences (QNJJ201733, KJCX20200113), the Key-Area Research and Development Program of Guangdong Province (2018B020202006), and the Beijing Municipal Science and Technology Project (D171100007617001).
Author contributions
CB. L. and CY. L conceived and designed the research. M. Z. performed the experiments. L. D., SG. G. and GL. Y. analyzed the data. M. Z. wrote the manuscript. All authors read and approved the manuscript.
Data availability
The raw bulk sequencing data were deposited in the Genome Sequence Archive (GSA) at the Beijing Institute of Genomics (BIG) Data Center (https://bigd.big.ac.cn/gsa/browse/CRA004692), with the accession number CRA004692.
Conflict of interest
The authors declare that they have no conflicts of interest.
Supplementary data
Supplementary data is available at Horticulture Research online.
Statistical analyses
All data are expressed as the mean value ± standard deviation (SD) of biological replicates. Statistical significance was determined using Student’s t-test.
References
- 1. Demmig-Adams B, Adams WW 3rd.. Antioxidants in photosynthesis and human nutrition. Science. 2002;298:2149–53. [DOI] [PubMed] [Google Scholar]
- 2. Ronen G, Carmel-Goren L, Zamir Det al. An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proc Natl Acad Sci U S A. 2000;97:11102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schwartz SH, Qin X, Zeevaart JA. Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 2003;131:1591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Story EN, Kopec RE, Schwartz SJet al. An update on the health effects of tomato lycopene. Annu Rev Food Sci Technol. 2010;1:189–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fray RG, Grierson D. Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol Biol. 1993;22:589–602. [DOI] [PubMed] [Google Scholar]
- 6. Beltrán J, Kloss B, Holser JPet al. Control of carotenoid biosynthesis through a heme-based cis-trans isomerase. Nat Chem Biol. 2015;11:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Isaacson T, Ronen G, Zamir Det al. Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of beta-carotene and xanthophylls in plants. Plant Cell. 2002;14:333–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu L, Shao Z, Zhang Met al. Regulation of carotenoid metabolism in tomato. Mol Plant. 2015;8:28–39. [DOI] [PubMed] [Google Scholar]
- 9. Pecker I, Gabbay R, Cunningham FX Jret al. Cloning and characterization of the cDNA for lycopene beta-cyclase from tomato reveals decrease in its expression during fruit ripening. Plant Mol Biol. 1996;30:807–19. [DOI] [PubMed] [Google Scholar]
- 10. Ronen G, Cohen M, Zamir Det al. Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J. 1999;17:341–51. [DOI] [PubMed] [Google Scholar]
- 11. Galpaz N, Ronen G, Khalfa Zet al. A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell. 2006;18:1947–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eisenreich W, Rohdich F, Bacher A. Deoxyxylulose phosphate pathway to terpenoids. Trends Plant Sci. 2001;6:78–84. [DOI] [PubMed] [Google Scholar]
- 13. Tritsch D, Hemmerlin A, Bach TJet al. Plant isoprenoid biosynthesis via the MEP pathway: in vivo IPP/DMAPP ratio produced by (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase in tobacco BY-2 cell cultures. FEBS Lett. 2010;584:129–34. [DOI] [PubMed] [Google Scholar]
- 14. Gershenzon J, Kreis W. Biochemistry of Plant Secondary Metabolism. In: Wink M, ed. Annual Plant Reviews. Sheffield Academic Press, CRC Press, 1999,222–99. [Google Scholar]
- 15. Jin X, Baysal C, Gao Let al. The subcellular localization of two isopentenyl diphosphate isomerases in rice suggests a role for the endoplasmic reticulum in isoprenoid biosynthesis. Plant Cell Rep. 2020;39:119–33. [DOI] [PubMed] [Google Scholar]
- 16. Nakamura A, Shimada H, Masuda Tet al. Two distinct isopentenyl diphosphate isomerases in cytosol and plastid are differentially induced by environmental stresses in tobacco. FEBS Lett. 2001;506:61–4. [DOI] [PubMed] [Google Scholar]
- 17. Phillips MA, D'Auria JC, Gershenzon Jet al. The arabidopsis thaliana type I isopentenyl diphosphate isomerases are targeted to multiple subcellular compartments and have overlapping functions in isoprenoid biosynthesis. Plant Cell. 2008;20:677–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun J, Zhang YY, Liu Het al. A novel cytoplasmic isopentenyl diphosphate isomerase gene from tomato (solanum lycopersicum): cloning, expression, and color complementation. Plant Mol Biol Report. 2010;28:473–80. [Google Scholar]
- 19. Sapir-Mir M, Mett A, Belausov Eet al. Peroxisomal localization of Arabidopsis isopentenyl diphosphate isomerases suggests that part of the plant isoprenoid mevalonic acid pathway is compartmentalized to peroxisomes. Plant Physiol. 2008;148:1219–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guirimand G, Guihur A, Phillips MAet al. A single gene encodes isopentenyl diphosphate isomerase isoforms targeted to plastids, mitochondria and peroxisomes in Catharanthus roseus. Plant Mol Biol. 2012;79:443–59. [DOI] [PubMed] [Google Scholar]
- 21. Pankratov I, McQuinn R, Schwartz Jet al. Fruit carotenoid-deficient mutants in tomato reveal a function of the plastidial isopentenyl diphosphate isomerase (IDI1) in carotenoid biosynthesis. Plant J. 2016;88:82–94. [DOI] [PubMed] [Google Scholar]
- 22. McQuinn RP, Wong B, Giovannoni JJ. AtPDS overexpression in tomato: exposing unique patterns of carotenoid self-regulation and an alternative strategy for the enhancement of fruit carotenoid content. Plant Biotechnol J. 2018;16:482–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kachanovsky DE, Filler S, Isaacson Tet al. Epistasis in tomato color mutations involves regulation of phytoene synthase 1 expression by cis-carotenoids. Proc Natl Acad Sci U S A. 2012;109:19021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bang H, Davis AR, Kim Set al. Flesh color inheritance and gene interactions among canary yellow, pale yellow, and red watermelon. J Amer Soc Hort Sci. 2010;135:362–8. [Google Scholar]
- 25. Doyle J. Molecular Techniques in TaxonomyVol. 57. In: Hewitt G, Johnston AB, Young JP, eds. Springer, 1991,283–93. [Google Scholar]
- 26. Chen S, Zhou Y, Chen Yet al. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McKenna A, Hanna M, Banks Eet al. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takagi H, Abe A, Yoshida Ket al. QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013;74:174–83. [DOI] [PubMed] [Google Scholar]
- 30. Deng L, Wang H, Sun Cet al. Efficient generation of pink-fruited tomatoes using CRISPR/Cas9 system. J Genet Genomics. 2018;45:51–4. [DOI] [PubMed] [Google Scholar]
- 31. Zhou M, Guo S, Tian Set al. Overexpression of the watermelon ethylene response factor ClERF069 in transgenic tomato resulted in delayed fruit ripening. Hortic Plant J. 2020;6:247–56. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw bulk sequencing data were deposited in the Genome Sequence Archive (GSA) at the Beijing Institute of Genomics (BIG) Data Center (https://bigd.big.ac.cn/gsa/browse/CRA004692), with the accession number CRA004692.