Abstract
Object
Prallethrin is a pyrethroid-based insecticide, commonly used as a liquid vaporizer in household, schools, and offices to repel mosquitoes. Due to worldwide application, human beings are exposed to this compound via inhalation. Inhalation of prallethrin can expose lung surfactant proteins (SPs) to this compound. SPs such as SP-A and SP-D have anti-microbial activities, whereas SP-B and SP-C prevent alveolar collapse during exhalation by reducing surface pressure in alveolar walls. The present study aimed to investigate the binding affinities of prallethrin for the pulmonary SPs and the possible interactions involved in it.
Methods
In this study, molecular docking was performed using prallethrin as ligand and lung SPs as target molecules. The three-dimensional structure of prallethrin (PubChem CID: 9839306) was retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/), whereas the same for SPs were retrieved from RCSB Protein Data Bank (https://www.rcsb.org/). AutoDock 4.2 employing Lamarckian genetic algorithm was used to calculate binding affinities between the target protein and the ligand. Polar and nonpolar interactions between the amino acids of SPs and Prallethrin were studied utilizing Chimera X and Discovery Studio Visualizer.
Results
Results demonstrated that, prallethrin can bind with the four SPs using several interactions such as hydrogen bonds, alkyl bonds, Pi–Pi interaction, Van der Waals interaction and other. Prallethrin interacted with two binding pockets of SP-A and SP-C, whereas the prallethrin interacted with three binding pockets of SP-B and SP-D, respectively.
Conclusion
Findings of the study indicated that prallethrin can bind with the pulmonary SPs employing hydrogen and hydrophobic interactions. Such interactions could impair critical functions of SPs in lungs. This might increase susceptibility of lungs towards a range of respiratory illness, pathogenic infections, as well as malignancy.
Graphical abstract
Keywords: Prallethrin, Surfactant protein, Lungs, Insecticide, Molecular docking
Introduction
Prallethrin is a broad-spectrum insecticide widely used around the globe. Prallethrin is generally used as a liquid vaporizer to repel mosquitoes in household, offices, and academic institutions. It is also the primary insecticide in certain formulations for killing wasps and hornets. Pesticides can be toxic to non-target organisms [1–8]. Since human begins are regularly exposed to prallethrin chemical via oral and nasal routes, toxicity of this chemical has been assessed using several in vivo and in vitro studies. Neurotoxicity was observed throughout the database and is the most sensitive endpoint. Effects have been observed across species, sexes, and different routes of administration. The adverse outcome of prallethrin exposure may also include reduced motor activity and transient tremors. The weight of evidences considering mechanism of action and pharmacokinetics fuels concern regarding potential health hazards of prallethrin exposure in human beings.
Prallethrin is structurally similar to type I group of pyrethroid insecticides. Pyrethroid insecticides are broadly sprayed in farm lands, household, and various public places. Regardless of their world-wide use, there are few reports that directly explain the toxic effects of pyrethroids including prallethrin on human health. Pyrethroids are multiple times more poisonous to insects than vertebrates since insects have greater sodium channel affectability, small body size, and lower internal heat level. However, in human, ingestion of pyrethroids leads to sore throat, sickness, and stomach torment. There might be mouth ulceration and potentially dysphagia. Fundamental impacts are seen following 4–48 h of pyrethroid exposure. Unsteadiness, cerebral pain, and fatigue are normal; palpitation, chest pain, and obscured vision are less common; and extreme lethargies and seizures are the foremost hazardous features [9]. Moreover, ingestion of prallethrin causes convulsions, nausea, vomiting, and abdominal discomfort [10]. Respiratory ailments such as shortness of breath along with chest pain and acute respiratory distress syndrome have also been reported following prallethrin exposure [11]. Prallethrin is hepatotoxic in nature as it causes histopathological symptoms with indications of oxidative stress. In a study, Bhaskar et al. [12] observed that, exposure to prallethrin can disturb cardiac conduction by impacting sodium channels in the heart. Oral ingestion of prallethrin is also responsible for status epilepticus [13].
Pyrethrin is a concentrate derived from the blossom Chrysanthemum cinerarilifolium and is strongly toxic to insects. Exposure to prallethrin occurs through nasal and oral routes as it is used as a liquid vaporizer to repel insects in the residences, schools, and offices. Upon inhalation, lungs and its biomolecules are contaminated with the prallethrin. However, studies investigating the effects of prallethrin on lungs are rare.
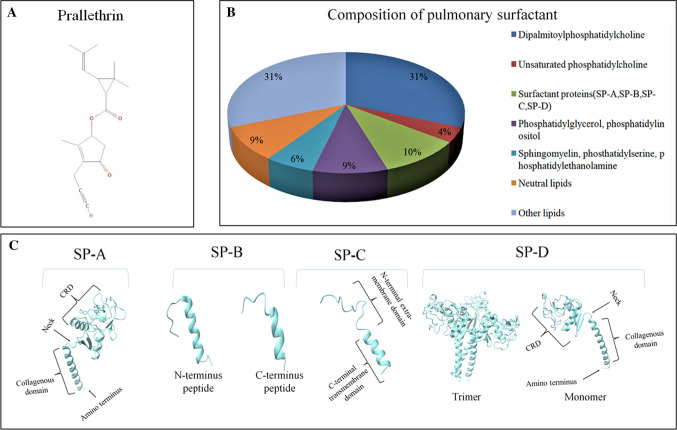
Respiratory cycle of lungs is regulated by respiratory surfactants. It is a lipoprotein complex secreted by type II alveolar cells. Pneumonic surfactants are known to reduce surface tension at the air/fluid interface within the alveoli. This prevents alveolar collapse during exhalation. Around 90% of this multi-molecular complex is made of phospholipids while 10% is comprised of surfactant proteins (SPs) (Fig. 1). SPs belong to either hydrophilic proteins (SP-A and SP-D) or hydrophobic proteins (SP-B and SP-C).
Fig. 1.
The figure represents a molecular structure of prallethrin, b bio-molecular composition of pulmonary surfactants, and c the three dimensional structure of surfactant proteins (SP-A, SP-B, SP-C, SP-D) as recovered from RCSB protein data bank
SP-A is the first recognized and most abundant surfactant protein in lungs [14]. It is an octadecameric peptide having an atomic mass of about 650 kDa. Eighteen monomers of SP-A in six trimeric units are assembled in a bouquet-like architecture. A human SP-A trimer is made of two monomers, viz. SP-A1 and SP-A2 [15]. Every monomer comprised of a calcium-based C-terminal carbohydrate recognition domain (CRD), a hydrophobic neck region, a collagenous domain, and an amino-terminal cysteine-rich domain. CRD is critical to most physiological functions of SP-A. CRD binds with lipopolysaccharides (LPS) of microbes and aggregate around them to impede infection. Additionally, SP-A lowers surface pressure in the alveolus and blocks the inhibitory impacts of plasma proteins involved in lung injury. SP-A is also found in human tracheal submucosal cells.
SP-B is a hydrophobic protein having atomic mass of about 8 kDa and made of 79 amino acids. Major amino acids in SP-B include leucine, isoleucine, phenylalanine, valine, alanine, and tryptophan. They are positively charged and hydrophobic in nature. Two monomers of SP-B form the homodimers in alveoli [16]. SP-B is needed for proper biophysical activity of the lungs. It enhances adsorption capability of phospholipids at the air–water interface [17]. Additionally, SP-B protects from oxygen-mediated lung injury and has healing property [18].
SP-C is a hydrophobic, 35–amino acid long, lipopeptide having two palmitates anchored at amino acids C5 and C6. The fragment between residues 13 and 28 is a hydrophobic α-helix including aliphatic amino acids, particularly valine. SP-C is solely produced by type II pneumocytes and secreted into alveolar spaces [19]. The α-helix of SP-C is very stable as transmembrane structure. Like SP-B, SP-C helps to stabilize alveolar surfactant film [20]. SP-C interacts with bacterial LPS and facilitates microbial degradation by phagocytes [21].
SP-D is a 43 kDa protein belonging to collectin family. SP-D has structural homology to SP-A. A mature SP-D molecule is a tetrameric complex made of four homotrimers. Each homotrimer is comprised of three subunits. Each SP-D subunit contains a N-terminal domain, a nucleating neck region, a collagenous domain and a C-terminal lectin region. SP-D is the collagenous group of proteins containing CRD. CRD helps SP-D to agglutinate microorganisms and apoptotic cells; and therefore facilitate their removal out of the lungs.
Surfactant proteins are critical to lung function. Since, prallethrin inhalation can expose surfactant proteins to this compound; hence the present in silico study was aimed to investigate the binding potential of prallethrin to respiratory surfactant proteins. This study will help to understand the potential impacts of prallethrin inhalation in lung physiology.
In silico studies using modern docking software for predicting toxicological endpoints, adverse effects, and metabolism of xenobiotics have become of high interest to the scientific community and the human society. Data obtained through in silico investigations accelerates the further toxicological investigations using in vivo and in vitro studies. In recent years, concerns regarding the use of animals in science research have risen exponentially, such that Regulatory Authorities had to reconsider the way of using animals within the scientific field. Notably, 3Rs core principles of Replacement, Reduction and Refinement for promoting ethical research, testing and education using animals, have been accepted globally leading to development and validation of alternative animal-free methods and approaches in science research. In silico study is a best fit for initial scientific investigations and analysis. This could also reduce the use of animals in preliminary investigations to overcome ethical issues and lengthy experimental procedures.
Results
Prallethrin–SP-A interaction
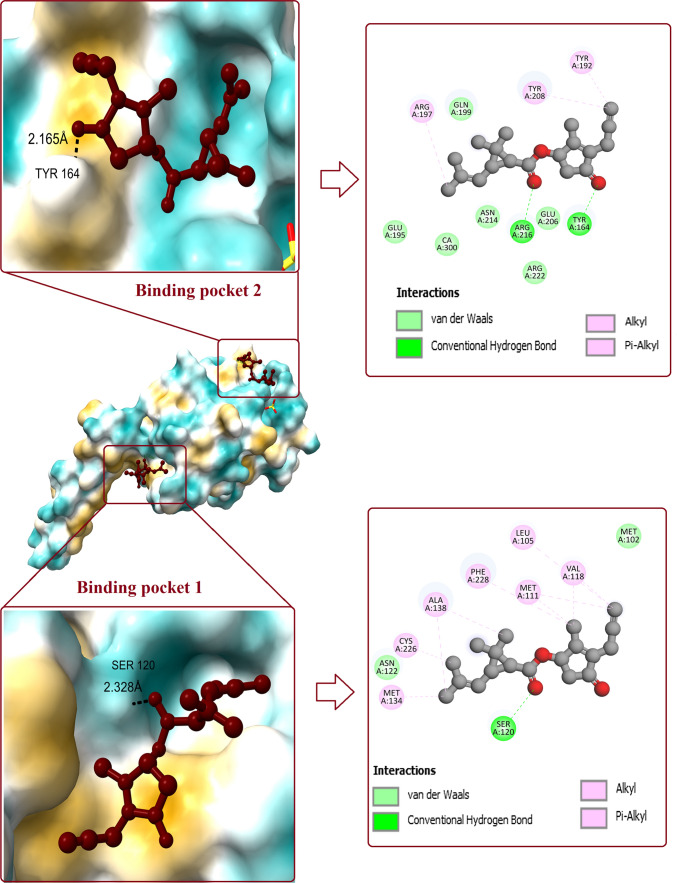
Molecular docking between prallethrin and SP-A has unraveled binding potential of prallethrin with SP-A (Fig. 2). At binding pocket 1 of CRD, prallethrin builds one H-bond with SER120 (bond length: 2.328 Å). Van der Waals interaction between prallethrin and SP-A involved two amino acid residues, viz. ASN122 and MET102. Other non-covalent interactions involved Alkyl and Pi-Alkyl established by MET134, CYS226, ALA138, PHE228, MET111, LEU105, and VAL118. The affinity for binding pocket 1 was recorded as -6.1 kcal/mol.
Fig. 2.
Interaction of prallethrin with two binding pockets of SP-A. Figure shows the binding pockets and the ligand bound to it. 2D diagram represents all the polar and non-polar interactions between different amino acid residues of protein and ligand
Binding of prallethrin with pocket 2 of CRD involved implication of two H-bonds. H-bond length between prallethrin and amino acid residues such as ARG216 and TYR164 were noted as 4.95 Å and 2.165 Å respectively. Other interactions such as Van der Waals (GLU195, GLN199, GLU206, ASN214, ARG222), and Pi-Alkyl interaction (ARG197, TYR208, TYR192) were additionally observed. The affinity for binding pocket 2 was recorded as -5.7 kcal/mol.
Prallethrin–SP-B interaction
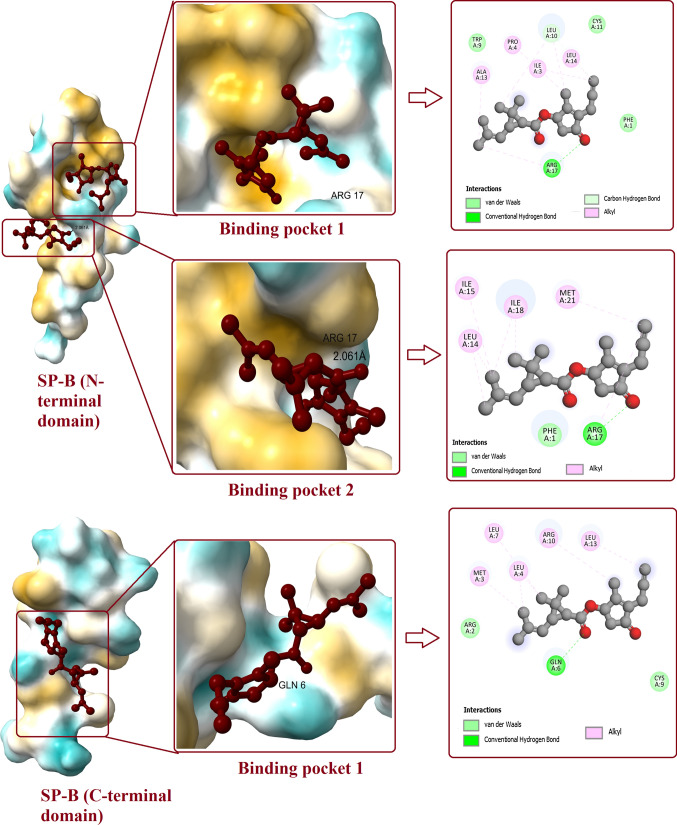
Molecular docking between prallethrin and SP-B has revealed that, prallethrin can interact with three binding pockets of SP-B (Fig. 3). Prallethrin interacted with binding pocket 1 of N-terminal peptide of SP-B with affinity of -5.2 kcal/mol. One H-bond was established between prallethrin and SP-B (ARG17) with bond length of 5.58 Å. Van der Waals interactions between prallethrin and SP-B involved amino acid residues viz. CYS11, PHE1, and TRP9. Other non-covalent interactions were established by amino acids ALA13, PRO4, ILE3, and LEU14.
Fig. 3.
Interaction of prallethrin with two binding pockets of SP-B. Figure shows the binding pockets in SP-B and the ligand bound to it. 2D diagram represents all the polar and non-polar interactions between different amino acid residues of protein and ligand
Prallethrin binds with N-terminal peptide of SP-B (binding pocket 2) with various bonds and interactions. Amino acid ARG17 formed H-bond with prallethrin, bond length was measured as 2.061 Å. Van der Waals interaction between prallethrin and SP-B was stabilized by PHE1. LEU14, ILE15, ILE18, and MET21 established other non-covalent interactions with prallethrin. Affinity for N-terminal peptide of SP-B at binding pocket 2 was recorded as -5.2 kcal/mol.
Prallethrin binds with C-terminal peptide of SP-B (binding pocket 1) by one H-bond (GLN6). Van der Waals interaction between prallethrin and C-terminal peptide of SP-B involved two amino acid residues, viz. CYS9 and ARG2. Alkyl interactions with prallethrin were established by MET3, LEU4, LEU7, ARG10, and LEU13. Affinity for C-terminal peptide of SP-B was recorded as -4.7 kcal/mol.
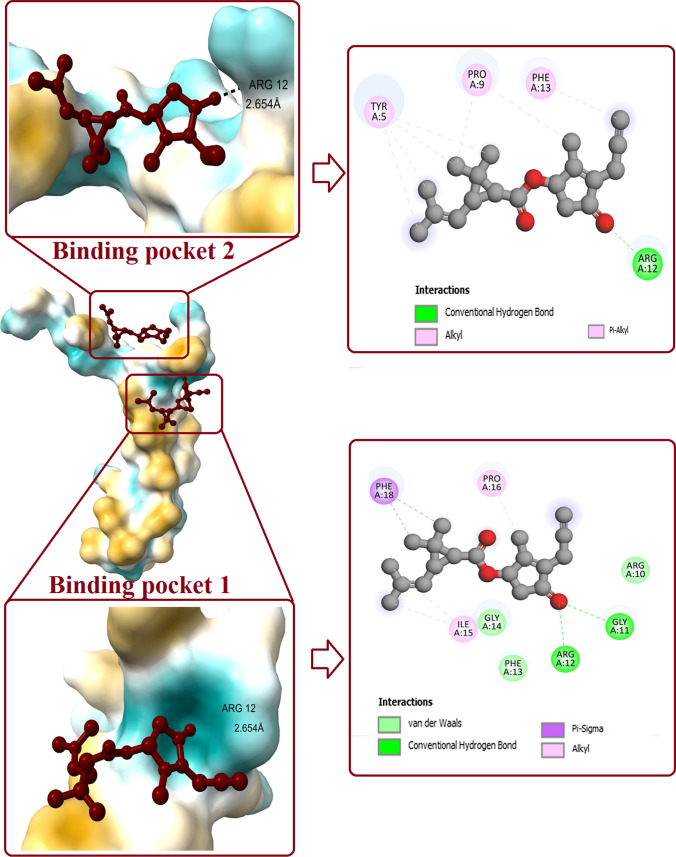
Prallethrin–SP-C interaction
Molecular docking between prallethrin and SP-C has revealed binding potential of prallethrin at two pockets of SP-C (Fig. 4). Prallethrin interacted with binding pocket 1 of SP-C with affinity of -5.5 kcal/mol. Two H-bonds were established between prallethrin and SP-C (GLY11, ARG12). Two Alkyl interactions were contributed by ILE15 and PRO16. Ligand established Van der Waals interaction using ARG10, PHE13, and GLY14. PHE18 formed Pi-Sigma bond with prallethrin.
Fig. 4.
Interaction of prallethrin with two binding pockets of SP-C. Figure shows the binding pockets in SP-C and the ligand bound to it. 2D diagram represents all the polar and non-polar interactions between different amino acid residues of protein and ligand
Interaction between binding pocket 2 of SP-C and prallethrin was established by one conventional H-bond (ARG12). Other interactions involved Pi-Alkyl and alkyl established byTYR5, PRO9, and PHE13. Affinity for binding pocket 2 was recorded as -4.7 kcal/mol.
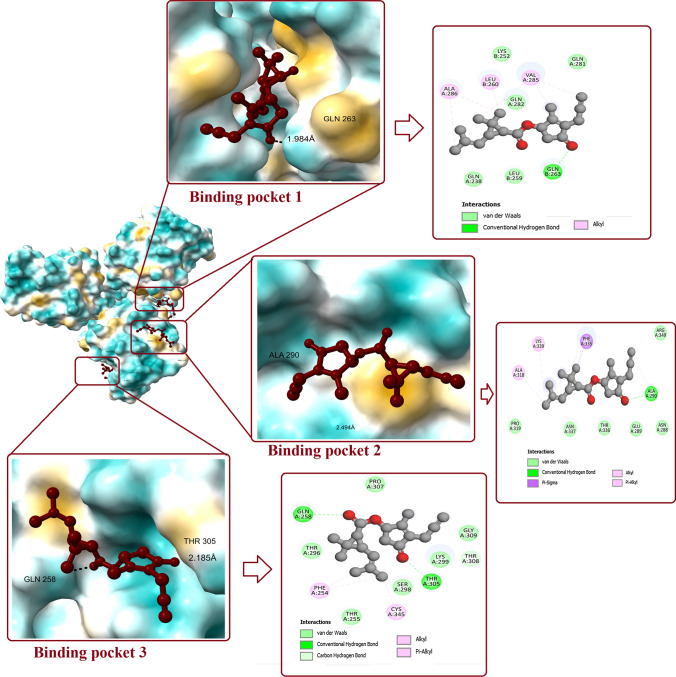
Prallethrin–SP-D interaction
Molecular docking between prallethrin and SP-D has revealed binding potential of prallethrin at multiple pockets on SP-D (Fig. 5). Interaction between prallethrin and binding pocket 1 of SP-D was stabilized by one conventional H-bond by GLN263 (bond length: 1.984 Å). Other interactions included Van der Waals (LYS252, LEU259, GLN238, GLN281, GLN282) and Alkyl interaction (LEU260, VAL285, ALA286). Affinity was measured as -5.5 kcal/mol (Fig. 6).
Fig. 5.
Interaction of prallethrin with two binding pockets of SP-D. Figure shows the binding pockets in SP-D and the ligand bound to it. 2D diagram represents all the polar and non-polar interactions between different amino acid residues of protein and ligand
Fig. 6.
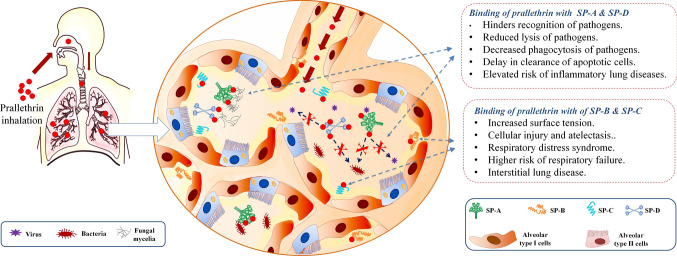
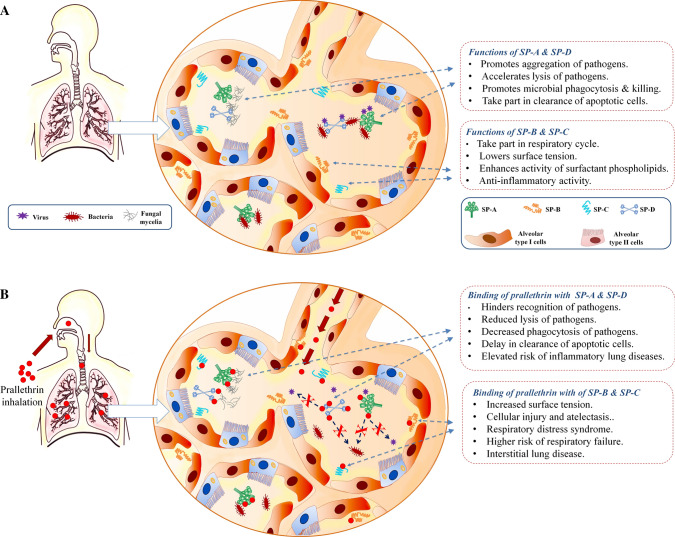
Brief summary of the study. a demonstrates the major surfactant proteins (SPs) and their functions. SPs are produced by alveolar type II cells in the form of lamellar bodies and later released into the alveolar lumen through exocytosis. CRD domain of SP-A and SP-D recognizes various pathogens like virus, bacteria, and fungi to facilitate microbial aggregation. This enhances opsonization and phagocytosis of microbes by macrophages. SP-A and SP-B also accelerate removal of apoptotic cells from alveolar environment. SP-B and SP-C are involved in maintenance of biophysical functions of lungs and inevitable for respiratory cycle. They can also impart anti-inflammatory function. b demonstrates that, inhalation of prallethrin can expose lung surfactant proteins to this insecticide. Prallethrin has potential to bind SPs through various interactions. Such interactions could disrupt several functions of alveolar SPs. Binding of prallethrin with SP-A and SP-D could impair recognition and opsonization of microbes in lungs. This might increase the risk of severe pathogenic infection/sepsis and inflammatory lung diseases. Binding of prallethrin with SP-B and SP-C could increase the surface tension at air/water interface in alveoli. This might fuel the onset of respiratory distress syndrome, forced breathing, risk of respiratory failure, and development of interstitial lung disease
Prallethrin interacted with binding pocket 2 of SP-D with affinity of -5.4 kcal/mol. One H-bond was established between prallethrin and SP-D (ALA290) with bond length of 2.494 Å. Van der Waals interaction between prallethrin and SP-D involved six amino acid residues, viz. ASN288, GLU289, PRO319, THR336, ASN337, and ARG349. One Pi-Sigma interaction was set up by PHE335 and two Pi-Alkyl interactions were established by ALA318 and LYS339.
At binding pocket 3, prallethrin established two H-bond with GLN258 and THR305. Van der Waals interaction between prallethrin and SP-D involved six amino acid residues, viz. THR255, THR296, SER298, LYS299, PRO307, and GLY309. Pi-Alkyl interaction was set up by PHE254 and CYS345. The affinity for binding pocket 3 was recorded as − 5.2 kcal/mol.
Discussion
The present in silico study has revealed binding potential of prallethrin at various sites of respiratory surfactant proteins.
Prallethrin was observed to bind with CRD of monomeric SP-A. Connections were stabilized by traditional H-bonds, Alkyl/Pi-Alkyl contacts and Van der Waals interaction between the ligand and the amino acid residues of SP-A. CRD is critical to most SP-A mediated biophysical activities of lungs required for support of appropriate respiratory cycle. Likewise, globular region of CRD associates with sugar or different ligands of microbial microorganisms in lungs and prevents infection. For example, CRD of SP-A binds with surface glycoprotein of Pneumocystis carinii, causative agent of pneumonia and promotes adherence to alveolar macrophages [22]. It helps in agglutination and phagocytosis of Pneumocystis carinii. SP-A also acts as an activity modulator of alveolar macrophage that is needed for phagocytosis and neutralization of Mycobacterium tuberculosis [23]. Respiratory syncytial virus (RSV) is the major reason of bronchitis in developing world. Trimeric units of SP-A successfully neutralizes RSV in human bronchial epithelial cells and subverts the level of disease [24]. In addition, SP-A by means of its sialic acid residues acts as an opsonin in the phagocytosis of influenza A virus (H1N1 and H3N2) by alveolar macrophages [25, 26]. Aspergillus is a contagious microbe that causes hypersensitive bronchopulmonary aspergillosis. Studies have shown that, SP-A binds with glycosylated antigens and allergens of Aspergillus fumigates and reduces harmful allergic reactions [27]. In a study, SP-A null mice was more susceptible to Histoplasma capsulatum than wild-type control mice [22]. It also protects surfactant phospholipids and macrophages from lipid peroxidation and oxidative cell injury [28]. Therefore, binding of prallethrin to CRD of SP-A can subvert anti-microbial and cytoprotective functions of SP-A. This could promote lung injury and respiratory troubles in prallethrin exposed individuals.
Prallethrin interacted with SP-B at N-terminal and C-terminal peptides. Protein–ligand binding was stabilized by conventional H-bonds, C-H bonds, Alkyl bonds, and Van der Waals interaction. Such protein–ligand interaction might block crucial functions of SP-B. SP-B is basically responsible for lowering the surface strain in alveolar lining [29]. SP-B malfunction is associated with respiratory ailments [30]. SP-B is associated with formation of lamellar bodies. Lamellar bodies contain surfactant phospholipids (and proteins) and associated with exocytosis of lung surfactants. Binding of prallethrin with SP-B might prevent the formation of lamellar bodies. Reduced level of lamellar bodies in type II alveolar cells is linked to severe respiratory illness [31].
SP-C is a solid hydrophobic peptide that can interact with inhaled prallethrin. In the present study, prallethrin interacted with SP-C at two binding pockets and the resultant ligand–protein complex was stabilized by conventional H-bonds, Pi-Sigma interaction, Alkyl/Pi-Alkyl bonds, and Van der Waals interactions. SP-C organizes phospholipids during respiratory cycle. It maintains the surface film and prevents the film breakdown in alveoli. Furthermore, synergistic activity of SP-B and SP-C controls the fluidity of phospholipid layers that is important to lung physiology [32]. Lack of alveolar SP-C has been connected to severe lung infections. Lack of SP-C enhances pSTAT3 activity in alveolar type 2 cells resulting in hyperinflammation with serious diffuse alveolar injury [33]. Intriguingly, recombinant SP-C has been utilized to treat intense respiratory trouble disorder [34]. SP-C deficient mice are sensitive to severe RSV infection [35]. In such cases, RSV exacerbated interstitial thickening, air space solidification, and hyperplasia. Hence, altered SP-C function following prallethrin binding could fuel diffuse lung injury.
In the present study, prallethrin made H-bonds, Pi-Sigma, Pi-Alkyl, and Van der Waals interactions with various pockets of SP-D. SP-D is responsible for maintaining lipid homeostasis. Besides, SP-D protects from detrimental effects of various allergens [36]. It down-regulates proinflammatory cytokines and prevents tissue injury [37]. CRD of SP-D binds with RSV glycoprotein in a calcium-dependent manner and blocks infection [38]. Flu virus is also opsonized by SP-D [39]. Intriguingly, recent study has shown that recombinant SP-D can bind with spike protein of SARS-CoV-2: the causative agent of coronavirus disease 19 (COVID-19) and hinders viral replication [40]. SP-D also restricts the activity of SARS-CoV-2 pseudotyped viral particles [41]. These studies suggest that, reduced function of SP-D could increase the susceptibility towards COVID-19. Additionally, CRD of SP-D can also bind with different fungal ligands, for example, 1,3-β-D-glucan, 1,6-β-D-glucan, galactosaminogalactan galactomannan, glucuronoxylomannan, and mannoprotein 1 of pathogenic fungi and hinder infection [42]. It agglutinates Aspergillus fumigatus conidia and enhances phagocytosis by alveolar macrophages and neutrophils [43]. SP-D additionally binds with a capsular type of cryptococci to promote their agglutination [44]. Pneumocystis carinii is a yeast-like fungus that causes severe pneumonia in immunocompromised patients. Studies have demonstrated that CRD of SP-D binds with surface glycoprotein A of Pneumocystis carinii and blocks the microbial activity [45]. Moreover, SP-D activates p53 and Fas-mediated apoptotic pathways to kill malignant cells [46, 47]. Hence, interaction of prallethrin with SP-D at multiple sites could dwarf anti-microbial and anti-cancerous activities of SP-D.
Materials & Method
Retrieval of prallethrin and preparation for molecular docking
The three dimensional structure of prallethrin (PubChem CID: 9839306) having molecular formula C19H24O3 was retrieved from PubChem (https://pubchem.ncbi.nlm.nih.gov/). Prallethrin was downloaded in.sdf arrangement and thereafter converted to .pdb file. Ligand was used for docking analysis utilizing AutoDock 4.2.
Retrieval of protein structures and preparation for molecular docking
The three-dimensional construction of four surfactant proteins, viz. SP-A (PDB ID: 1R13), SP-B (PDB ID: 1DFW), SP-C (PDB ID: 2ESY) and SP-D (PDB ID: 3IKP), was recovered in.pdb format from RCSB Protein Data Bank (https://www.rcsb.org/). To lower energy, water molecules and co-crystal ligands were removed from the SPs for molecular docking.
Molecular docking analysis
Molecular docking was performed utilizing prallethrin as ligand and pneumonic surfactant proteins as target molecules. AutoDock 4.2 employing Lamarckian genetic algorithm was utilized for the investigation. To lower energy, water molecules were discarded from crystalized SPs. In addition, polar hydrogen and Kollman charges were added to investigate binding affinity of prallethrin with SPs. Grid box was added separately to each target protein to cover the significant binding sites. Eventually, molecular docking was conducted utilizing AutoDock Vina [48] and results were saved in log.txt file. Additionally, prallethrin_output.pdbqt was also extracted for every docking analysis to observe binding interactions between prallethrin and target SP. Parameters like polar and non-polar interactions between amino acids of SPs and prallethrin were studied utilizing Chimera X and Discovery Studio Visualizer.
Conclusion
The current investigation has revealed that inhaled prallethrin can bind at multiple pockets of surfactant proteins in lungs. These surfactant proteins control the respiratory cycle of lungs by reducing surface tension at air–water interface. Besides, surfactant proteins like SP-A and SP-D provide first line of defense against a wide spectrum of bacterial, fungal, and viral infections. It is worth noting that SP-D has inhibitory action for COVID-19. Prallethrin has potential to bind surfactant proteins and therefore could impair their critical functions in lungs. This might increase susceptibility of lungs towards respiratory illness, pathogenic infections as well as malignancy.
Compliance with ethical standards
Conflict of interest
Siddhartha Ghanty, Moutushi Mandi, Abhratanu Ganguly, Kanchana Das, Anik Dutta, Sayantani Nanda, Gopal Biswas and Prem Rajak declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human subjects or animals performed by any of the authors.
Contributor Information
Siddhartha Ghanty, Email: siddharthaghanty20@gmail.com.
Moutushi Mandi, Email: mandimoutushi496@gmail.com.
Abhratanu Ganguly, Email: gangulyabhratanu@gmail.com.
Kanchana Das, Email: kdas151992@gmail.com.
Anik Dutta, Email: aniksep10@gmail.com.
Sayantani Nanda, Email: titli486@gmail.com.
Gopal Biswas, Email: biswasgopal329@gmail.com.
Prem Rajak, Email: prem.rjk@gmail.com.
References
- 1.Rajak P, Dutta M, Roy S. Altered differential hemocyte count in 3rd instar larvae of Drosophila melanogaster as a response to chronic exposure of Acephate. Interdiscip Toxicol. 2015;8(2):84–88. doi: 10.1515/intox-2015-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khatun S, Mandi M, Rajak P, Roy S. Interplay of ROS and behavioral pattern in fluoride exposed Drosophila melanogaster. Chemosphere. 2018;209:220–231. doi: 10.1016/j.chemosphere.2018.06.074. [DOI] [PubMed] [Google Scholar]
- 3.Rajak P, Khatun S, Dutta M, Mandi M, Roy S. Chronic exposure to acephate triggers ROS-mediated injuries at organismal and sub-organismal levels of Drosophila melanogaster. Toxicol Res (Camb) 2018;7(5):874–887. doi: 10.1039/c8tx00052b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajak P, Ganguly A, Sarkar S, Mandi M, Dutta M, Podder S, Khatun S, Roy S. Immunotoxic role of organophosphates: an unseen risk escalating SARS-CoV-2 pathogenicity. Food Chem Toxicol. 2021;149:112007. doi: 10.1016/j.fct.2021.112007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajak P, Roy S. Heat Shock Proteins and Pesticide Stress. In: Asea A, Kaur P, editors. Regulation of heat shock protein responses. Heat shock proteins. Cham: Springer; 2018. [Google Scholar]
- 6.Sarkar S, Rajak P, Roy S. Toxicological evaluation of a new lepidopteran insecticide, flubendiamide, in non-target Drosophila melanogaster meigen (Diptera: Drosophilidae) IJT. 2018;12(3):45–50. doi: 10.32598/IJT.12.3.477.1. [DOI] [Google Scholar]
- 7.Dutta M, Rajak P, Roy S. Determination of chronic median lethal concentration of sodium fluoride in Drosophila melanogaster and exploring effect of sub-lethal concentrations on differential hemocyte count. Proc Zool Soc. 2019;72:111–117. doi: 10.1007/s12595-017-0235-x. [DOI] [Google Scholar]
- 8.Rajak P, Sahana S, Roy S. Acephate-induced shortening of developmental duration and early adult emergence in a nontarget insect Drosophila melanogaster. Toxicol Environ Chem. 2013;95(8):1369–1379. doi: 10.1080/02772248.2014.880608. [DOI] [Google Scholar]
- 9.Bradberrry SM, Cage SA, Proudfoot AT, Vale JA. Poisoning due to pyrethroids. Toxicol Rev. 2005;24:93–106. doi: 10.2165/00139709-200524020-00003. [DOI] [PubMed] [Google Scholar]
- 10.Chandra A, Dixit MB, Banavaliker JN. Prallethrin poisoning: a diagnostic dilemma. J Anaesthesiol Clin Pharmacol. 2013;29(1):121–122. doi: 10.4103/0970-9185.105820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.George J, Malik R, Gogna A. Hypersensitivity reaction and acute respiratory distress syndrome in pyrethroid poisoning and role of steroid therapy. Asia Pac J Med Toxicol. 2015;4:91–93. [Google Scholar]
- 12.Bhaskar EM, Moorthy S, Ganeshwala G, Abraham G. Cardiac conduction disturbance due to prallethrin (pyrethroid) poisoning. J Med Toxicol. 2010;6:27–30. doi: 10.1007/s13181-010-0032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vijayaraju D, Ramkumar S, Subramanian SR. Prallethrin poisoning presenting as status epilepticus. Online J Health Allied Scs. 2013;12(4):11. [Google Scholar]
- 14.Weaver TE, Whitsett JA. Function and regulation of expression of pulmonary surfactant-associated proteins. Biochem J. 1991;273:249–264. doi: 10.1042/bj2730249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voss T, Melchers K, Scheirle G, Schafer KP. Structural comparison of recombinant pulmonary surfactant protein SP-A derived from two human coding sequences. Implications for the chain composition of natural human SP-A. Am J Respir Cell Mol Biol. 1991;4:88–94. doi: 10.1165/ajrcmb/4.1.88. [DOI] [PubMed] [Google Scholar]
- 16.Weaver TE, Conkright JJ. Functions of surfactant proteins B and C. Annu Rev Physiol. 2001;63:555–578. doi: 10.1146/annurev.physiol.63.1.555. [DOI] [PubMed] [Google Scholar]
- 17.Hawgood S. Surfactant protein B: structure and function. BiolNeonate. 2004;85(4):285–289. doi: 10.1159/000078169. [DOI] [PubMed] [Google Scholar]
- 18.Tokieda K, Ikegami M, Wert SE, Baatz JE, Zou Y, Whitsett JA. Surfactant protein B corrects oxygen-induced pulmonary dysfunction in heterozygous surfactant protein B-deficient mice. Pediatr Res. 1999;46:708–714. doi: 10.1203/00006450-199912000-00014. [DOI] [PubMed] [Google Scholar]
- 19.Ten Brinke A, Lambert MG, van Golde LG, Batenburg JJ. Palmitoylation and processing of the lipopeptide surfactant protein C. Biochim Biophys Acta. 2002;1583:253–265. doi: 10.1016/S1388-1981(02)00248-2. [DOI] [PubMed] [Google Scholar]
- 20.Qanbar R, Cheng S, Possmayer F, Schurch S. Role of the palmitoylation of surfactant-associated protein C in surfactant film formation and stability. Am J Physiol. 1996;271:L572–L580. doi: 10.1152/ajplung.1996.271.4.L572. [DOI] [PubMed] [Google Scholar]
- 21.Augusto LA, Li J, Synguelakis M, Johansson J, Chaby R. Structural basis for interactions between lung surfactant protein C and bacterial lipopolysaccharide. J Biol Chem. 2002;277:23484–23492. doi: 10.1074/jbc.M111925200. [DOI] [PubMed] [Google Scholar]
- 22.McCormack FX, Gibbons R, Ward SR, Kuzmenko A, Wu H, Deepe GS., Jr Macrophage-independent fungicidal action of the pulmonary collectins. J Biol Chem. 2003;278(38):36250–36256. doi: 10.1074/jbc.M303086200. [DOI] [PubMed] [Google Scholar]
- 23.Gaynor CD, McCormack FX, Voelker DR, McGowan SE, Schlesinger LS. Pulmonary surfactant protein A mediates enhanced phagocytosis of Mycobacterium tuberculosis by a direct interaction with human macrophages. J Immunol. 1995;155(11):5343–5351. [PubMed] [Google Scholar]
- 24.Watson A, Kronqvist N, Spalluto CM, Griffiths M, Staples KJ, Wilkinson T, Holmskov U, Sorensen GL, Rising A, Johansson J, Madsen J, Clark H. Novel expression of a functional trimeric fragment of human SP-A with efficacy in neutralisation of RSV. Immunobiology. 2017;222(2):111–118. doi: 10.1016/j.imbio.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benne CA, Benaissa-Trouw B, van Strijp JA, Kraaijeveld CA, van Iwaarden JF. Surfactant protein A, but not surfactant protein D, is an opsonin for influenza A virus phagocytosis by rat alveolar macrophages. Eur J Immunol. 1997;27(4):886–890. doi: 10.1002/eji.1830270413. [DOI] [PubMed] [Google Scholar]
- 26.Benne CA, Kraaijeveld CA, van Strijp JA, Brouwer E, Harmsen M, Verhoef J, van Golde LM, van Iwaarden JF. Interactions of surfactant protein A with influenza A viruses: binding and neutralization. J Infect Dis. 1995;171(2):335–341. doi: 10.1093/infdis/171.2.335. [DOI] [PubMed] [Google Scholar]
- 27.Madan T, Kishore U, Singh M, Strong P, Clark H, Hussain EM, Reid KB, Sarma PU. Surfactant proteins A and D protect mice against pulmonary hypersensitivity induced by Aspergillus fumigatus antigens and allergens. J Clin Invest. 2001;107(4):467–475. doi: 10.1172/JCI10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bridges JP, Davis HW, Damodarasamy M, Kuroki Y, Howles G, Hui DY, McCormack FX. Pulmonary surfactant proteins A and D are potent endogenous inhibitors of lipid peroxidation and oxidative cellular injury. J Biol Chem. 2000;275(49):38848–38855. doi: 10.1074/jbc.M005322200. [DOI] [PubMed] [Google Scholar]
- 29.Cochrane CG, Revak SD. Pulmonary surfactant protein B (SP-B): structure-function relationships. Science. 1991;254(5031):566–568. doi: 10.1126/science.1948032. [DOI] [PubMed] [Google Scholar]
- 30.Thompson MW. Surfactant protein B deficiency: insights into surfactant function through clinical surfactant protein deficiency. Am J Med Sci. 2001;321(1):26–32. doi: 10.1097/00000441-200101000-00005. [DOI] [PubMed] [Google Scholar]
- 31.Cutz E, Wert SE, Nogee LM, Moore AM. Deficiency of lamellar bodies in alveolar type II cells associated with fatal respiratory disease in a full-term infant. Am J Respir Crit Care Med. 2000;161(2 Pt 1):608–614. doi: 10.1164/ajrccm.161.2.9905062. [DOI] [PubMed] [Google Scholar]
- 32.Parra E, Moleiro LH, López-Montero I, Cruz A, Monroy F, Pérez-Gil J. A combined action of pulmonary surfactant proteins SP-B and SP-C modulates permeability and dynamics of phospholipid membranes. Biochem J. 2011;438(3):555–564. doi: 10.1042/BJ20110681. [DOI] [PubMed] [Google Scholar]
- 33.Jin H, Ciechanowicz AK, Kaplan AR, Wang L, Zhang PX, Lu YC, Tobin RE, Tobin BA, Cohn L, Zeiss CJ, Lee PJ, Bruscia EM, Krause DS. Surfactant protein C dampens inflammation by decreasing JAK/STAT activation during lung repair. Am J Physiol Lung Cell Mol Physiol. 2018;314(5):L882–L892. doi: 10.1152/ajplung.00418.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spragg RG, Lewis JF, Wurst W, Häfner D, Baughman RP, Wewers MD, Marsh JJ. Treatment of acute respiratory distress syndrome with recombinant surfactant protein C surfactant. Am J Respir Crit Care Med. 2003;167(11):1562–1566. doi: 10.1164/rccm.200207-782OC. [DOI] [PubMed] [Google Scholar]
- 35.Glasser SW, Witt TL, Senft AP, Baatz JE, Folger D, Maxfield MD, Akinbi HT, Newton DA, Prows DR, Korfhagen TR. Surfactant protein C-deficient mice are susceptible to respiratory syncytial virus infection. Am J Physiol Lung Cell Mol Physiol. 2009;297(1):L64–72. doi: 10.1152/ajplung.90640.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hawgood S, Brown C, Edmondson J. Pulmonary collectins modulate strain-specific influenza a virus infection and host responses. J Virol. 2004;78(16):8565–8572. doi: 10.1128/JVI.78.16.8565-8572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arroyo R, Kingma P. S. Surfactant protein D and bronchopulmonary dysplasia: a new way to approach an old problem. Respir Res. 2021;22(1):141. doi: 10.1186/s12931-021-01738-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LeVine AM, Elliott J, Whitsett JA. Surfactant protein-d enhances phagocytosis and pulmonary clearance of respiratory syncytial virus. Am J Respir Cell Mol Biol. 2004;31(2):193–199. doi: 10.1165/rcmb.2003-0107OC. [DOI] [PubMed] [Google Scholar]
- 39.White M, Kingma P, Tecle T, Kacak N, Linders B, Heuser J. Multimerization of surfactant protein D, but not its collagen domain, is required for antiviral and opsonic activities related to influenza virus. J Immunol. 2008;181:7936–7943. doi: 10.4049/jimmunol.181.11.7936. [DOI] [PubMed] [Google Scholar]
- 40.Madan T, Biswas B, Varghese PM, Subedi R, Pandit H, Idicula-Thomas S. A recombinant fragment of human surfactant protein D binds spike protein and inhibits infectivity and replication of SARS-CoV-2 in clinical samples. Am J Respir Cell Mol Biol. 2021 doi: 10.1165/rcmb.2021-0005OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsieh MH, Beirag N, Murugaiah V, Chou YC, Kuo WS, Kao HF. Human surfactant protein d binds spike protein and acts as an entry inhibitor of SARS-CoV-2 pseudotyped viral particles. Front Immunol. 2021;12:641360. doi: 10.3389/fimmu.2021.641360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madan T, Kishore U. Surfactant protein D recognizes multiple fungal ligands: a key step to initiate and intensify the anti-fungal host defense. Front Cell Infect Microbiol. 2020;10:229. doi: 10.3389/fcimb.2020.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madan T, Eggleton P, Kishore U, Strong P, Aggrawal SS, Sarma PU, et al. Binding of pulmonary surfactant proteins A and D to Aspergillus fumigatus conidia enhances phagocytosis and killing by human neutrophils and alveolar macrophages. Infect Immun. 1997;65:3171–3179. doi: 10.1128/iai.65.8.3171-3179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van de Wetering JK, Coenjaerts FE, Vaandrager AB, van Golde LM, Batenburg JJ. Aggregation of Cryptococcus neoformans by surfactant protein D is inhibited by its capsular component glucuronoxylomannan. Infect Immun. 2004;72:145–153. doi: 10.1128/IAI.72.1.145-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vuk-Pavlovic Z, Standing JE, Crouch EC, Limper AH. Carbohydrate recognition domain of surfactant protein D mediates interactions with Pneumocystis carinii glycoprotein A. Am J Respir Cell Mol Biol. 2001;24(4):475–484. doi: 10.1165/ajrcmb.24.4.3504. [DOI] [PubMed] [Google Scholar]
- 46.Kaur A, Riaz MS, Murugaiah V, Varghese PM, Singh SK, Kishore U. A recombinant fragment of human surfactant protein D induces apoptosis in pancreatic cancer cell lines via fas-mediated pathway. Front Immunol. 2018;9:1126. doi: 10.3389/fimmu.2018.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahajan L, Pandit H, Madan T, Gautam P, Yadav AK, Warke H, Sundaram CS, Sirdeshmukh R, Sarma PU, Kishore U, Surolia A. Human surfactant protein D alters oxidative stress and HMGA1 expression to induce p53 apoptotic pathway in eosinophil leukemic cell line. PLoS One. 2013;8(12):e85046. doi: 10.1371/journal.pone.0085046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]