Abstract
We studied the human immunodeficiency virus type 1 phenotypic and genotypic profiles of a dual drug-resistant isolate (isolate 14aPost-DR) selected for zidovudine (ZDV) and lamivudine (3TC) resistance and then cultured in the presence of 3TC and a protease inhibitor: indinavir (IDV), ritonavir, or KNI-272. The IDV-treated virus was highly resistant to 3TC, ZDV, and IDV and accumulated protease mutations at positions M46I and V82F. A change from alanine to valine was observed in 4 of 10 clones in the P2 position of the p7-p1 Gag-protease cleavage site, linked to position M46I in the dominant viral quasispecies. Previous 3TC resistance did not impair the development of additional mutations in the protease and Gag-protease cleavage regions.
Current antiretroviral therapy includes the combination use of reverse transcriptase (RT) inhibitors and protease (PR) inhibitors (PIs) (6, 7, 19). Multiply targeted therapies with agents active against both PR and RT enzymes showed more benefits than monotherapy, but these clinical benefits have been limited due to drug resistance and particularly multidrug resistance (MDR) (1, 5, 8, 16, 22).
Sequential acquisition of mutations in the region that codes for viral PR induces increased levels of resistance, and cross-resistance could confer resistance to more than one other PI (3, 4, 13, 20, 27, 28). The overlap in the PI mutational pathway led to cross-resistance to inhibitors actually used (K. Hertogs, S. Kemp, S. Bloor, V. Miller, S. Steszewski, J. Mellors, C. Van den Eynde, F. Peeters, B. Larder, and R. Pauwels, 2nd Int. Workshop on HIV Drug Resistance and Treatment Strategies, Antivir. Ther. 3(Suppl. 1):49–50, 1998) when four amino acid substitutions were examined (M46I, L63P, V82T, I84V) (2).
The selective pressure of indinavir (IDV) induces the early appearance of primary mutations that confer resistance to this compound, i.e., mutation V82A (3, 4, 29), but the PI-specific “genetic barrier” implies that three or more mutations are necessary for the development of a high level of resistance (4). These resistant mutants often reveal a loss of fitness. The viral replication capacity is restored at the appearance of compensatory substitutions (L10I, M36I, M46I, L63P, or A71V), that, without interfering directly with PI binding, have been shown to increase the catalytic activity of PR (21). Such mutations in the PR nonactive site increase viral fitness as a result of primary mutations that elicit phenotypic resistance (2, 12). The resistant strains display an overlapping mutational pathway that results in cross-resistance to several PIs (3). Some compensatory mutations map in the substrate of the human immunodeficiency virus type 1 (HIV-1) PR, i.e., the Gag-Pol polyprotein precursor cleavage sites (9).
Thus, the initial therapy could have an important role in future therapeutic choices, while cross-resistance to additional compounds should be always considered (25). With the aim of clarifying the in vitro appearance of PI MDR, we have studied the evolution of HIV-1 PR genotypes and phenotypic changes during culture of an HIV-1 strain resistant to both zidovudine (ZDV) and lamivudine (3TC) either in the presence or in the absence of a single PI, e.g., IDV, ritonavir (RTV), or KNI-272. The third drug (a PI) was added in order to achieve a more active antiviral regimen.
In the study described here we examined the in vitro susceptibility of this viral isolate to RT inhibitors and PI, and its linkage to Gag-protease cleavage site modifications at different time points of the culture.
In vitro passage of a dual RT inhibitor-resistant HIV-1 isolate.
The HIV-1 strain used in this study was isolate 14aPost, which was used after in vitro selection of resistance to both ZDV and 3TC and which then had five amino acid substitutions: D67N, K70R, M184V, T215F, and K219Q (isolate 14aPost-DR) (23).
We cultivated this dual drug-resistant viral isolate under four conditions: in three different flasks we added 3TC as selective pressure to maintain 3TC resistance and one of each of the PIs (IDV, RTV, KNI-272); in the fourth flask isolate 14aPost-DR was cultivated without drug treatment as a control.
The culture flasks were kept by the method described by Johnson et al. (15). Cell-free supernatants were tested twice a week by a p24 antigen enzyme-linked immunosorbent assay (NEN Research Products, Boston, Mass.) and for syncytium induction (14). Viral titration was done in peripheral blood mononuclear cells (PBMCs), and the viral titer, measured as the 50% tissue culture infective dose per milliliter, was calculated by the method of Reed and Muench (10).
The pharmacological compounds were obtained from the respective companies. 3TC was used at its 25% inhibitory concentration IC25 (0.2 μM), while the PIs were added at their IC50s (0.5 μM for IDV, 0.3 μM for RTV and KNI-272).
The IDV-treated virus and the untreated virus presented similar patterns regarding p24 antigen production; in fact, both had growth increases after day 221. Under the other two flask conditions in which viruses were treated with RTV or KNI-272, viral replications were suppressed earlier (day 21) and the values for p24 antigen were negative.
Phenotypic pattern of MDR isolate.
Table 1 shows the drug susceptibilities of the viruses in IDV-treated (MDR) and control flasks. Susceptibility tests were done at day 60 and once a month after day 116. The drugs used were ZDV, 3TC, IDV, RTV, and nelfinavir (NFV). Drug susceptibility tests were carried out as described previously (23), and the cutoff values for the various inhibitors were derived from experiments with isolate 14a-Pre, which was obtained from an HIV-1-seropositive person before any antiretroviral treatment. In the presence of IDV the dual drug-resistant isolate had a high level of resistance to 3TC (IC50, >1 μM) at different time points, whereas it was moderately resistant to ZDV at day 0 (IC50, 0.563 μM) and was found to be resistant (IC50, >10 μM) at the subsequent time points. The susceptibility to IDV was tested at every time point, and the isolate was sensitive only at day 0 but became resistant from day 116 on (IC50, >0.1 μM).
TABLE 1.
MDR strain appearance on a genotypic and phenotypic background of ZDV-3TC resistancea
| Day | Drug IC50 (μM) in the following:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IDV-containing flask
|
Control flask
|
|||||||||
| ZDV | 3TC | IDV | RTV | NFV | ZDV | 3TC | IDV | RTV | NFV | |
| 0b | 0.563 | 8.408 | 0.010 | 0.010 | 0.010 | 0.298 | 0.170 | 0.010 | 0.010 | 0.010 |
| 59 | >1 | >1 | 0.020 | NDc | ND | <0.01 | <0.01 | 0.010 | ND | ND |
| 116 | >10 | >10 | >0.1 | ND | ND | 0.081 | 0.491 | 0.035 | ND | ND |
| 186 | >10 | >10 | 0.806 | ND | ND | <0.01 | 0.362 | 0.010 | ND | ND |
| 221 | >10 | >10 | >0.1 | >0.1 | >0.1 | 0.076 | 0.210 | 0.024 | 0.063 | 0.046 |
| 256 | >10 | >10 | >0.1 | >0.1 | >0.1 | 0.011 | 0.069 | 0.009 | 0.009 | 0.009 |
Drug pressure in the IDV-containing flask consisted of 3TC at its IC25 (0.2 μM), IDV at its IC50 (0.01 μM), and no ZDV. No drugs were included in the control flask.
Day 147 for the previous passage.
ND, not done.
Cross-resistance to the other PIs was evaluated at the two last time points: days 221 and 256. On these days, the IDV-treated virus was resistant to RTV and NFV (IC50s, >0.1 μM). The untreated control virus was sensitive to each antiretroviral compound, both PIs and RT inhibitors, for the entire duration of the experiment.
Reference IC50s for the standard strain (14aPre) were as follows: ZDV, 0.005 ± 0.003 μM; 3TC, 0.140 ± 0.073, μM; IDV, 0.030 ± 0.010 μM; RTV, 0.020 ± 0.010 μM; and NFV, 0.049 ± 0.020 μM.
Occurrence of gag-pol mutations in passaged MDR virus.
Table 2 shows the evolution of the genotypic pattern of the PR and RT regions both in the absence and in the presence of pharmacological pressure with IDV plus 3TC from day 28 after the start of culture in the presence of IDV and 3TC.
TABLE 2.
Evolution of 14aPost-DR genotype in vitro in presence of IDV plus 3TC and without drugs at sequential times of culturea
| Culture and day of passage | Amino acid at the following codonsb:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p7-p1 (P3)Q | p7-p1 (P2)A | p1-p6 (P1′)L | PR
|
RT
|
|||||||||||||
| 10 Leu | 35 Glu | 37 Asn | 46 Met | 63 Leu | 64 lle | 82 Val | 67 Asp | 70 Lys | 103 Lys | 184 Met | 211 Arg | 215 Thr | 219 Lys | ||||
| MDRd28 | |||||||||||||||||
| Bulk | — | Asp | Pro | — | Leu-Pro | Val | — | Asn | Arg | — | Ile | Lys | Phe | Gln | |||
| Clone 1 | — | — | Pro | — | Asp | Pro | — | Ala | Val | — | Asn | Arg | Glu | Val | Lys | Phe | Gln |
| Clone 2 | — | — | Pro | — | Asp | Pro | — | Ala | Val | — | Asn | Arg | — | Ile | Lys | Phe | Gln |
| Clone 3 | — | — | Pro | Pro | Asp | Pro | — | — | Val | — | Asn | Arg | — | Val | Lys | Phe | Gln |
| Clone 4 | — | — | Pro | — | Asp | Pro | — | — | Val | — | Asn | Arg | — | Ile | Lys | Phe | Gln |
| Clone 6 | — | — | Pro | — | Asp | Pro | — | Ser | Val | — | Asn | Arg | — | Val | Lys | Phe | Gln |
| Clone 7 | — | — | — | — | Asp | Pro | — | Ala | Val | — | Asn | — | — | Ile | Lys | Phe | Gln |
| MDRd88 (bulk) | — | Asp | Pro | — | Leu-Pro | Val | — | Asn | Arg | — | Val | Lys | Phe | Gln | |||
| MDRd116 (bulk) | — | Asp | Pro | Met-Ile | Ser-Pro | Val | — | Asn | Arg | — | Val | Lys | Phe | Gln | |||
| MDRd144 | |||||||||||||||||
| bulk | — | Asp | Pro | Met-Ile | Ser | Val | — | Asn | Arg | — | Val | Lys | Phe | Gln | |||
| Clone 1, 2, and 6 | — | — | Pro | — | Asp | Pro | Ile | Ser | Val | — | Asn | Arg | — | Val | Lys | Phe | Gln |
| Clones 3 and 8 | — | — | Pro | — | Asp | Pro | Ile | Ala | Val | — | Asn | Arg | — | Val | Lys | Phe | Gln |
| Clone 4 | — | Val | Pro | — | Asp | Pro | Ile | Pro | Val | — | Asn | Arg | — | Val | Lys | Phe | Gln |
| Clones 7 and 10 | — | Val | Pro | — | Asp | Pro | Ile | Ser | Val | — | Asn | Arg | — | Val | Lys | Phe | Gln |
| Clone 9 | — | Val | Pro | — | Asp | Pro | Ile | — | Val | — | Asn | Arg | — | Val | Lys | Phe | Gln |
| Clone 11 | — | — | Pro | — | Asp | Pro | Ile | Ser | Val | Phe | Asn | Arg | — | Val | Lys | Phe | Gln |
| MDR d186 (bulk) | — | Asp | Pro | Ile | Ser | Val | Phe | Asn | Arg | — | Val | Lys | Phe | Gln | |||
| MDR d221 (bulk) | — | Asp | Pro | Ile | Ser | Val | Phe | Asn | Arg | — | Val | Lys | Phe | Gln | |||
| MDRd256 (bulk) | — | Val | Pro | — | Asp | Pro | Ile | Ser | Val | Phe | Asn | Arg | — | Val | Lys | Phe | Gln |
| CTRLd28 (bulk) | — | Asp | Pro | — | Leu-Pro-Asp | Val | — | Asn | Arg-Lys | — | Val | Lys | Phe | Gln | |||
| CTRLd88 (bulk) | — | Asp | Pro | — | Leu-Pro | Val | — | Asn | Arg-Lys | — | Met-Val | Lys | Phe | Gln | |||
| CTRLd144 (bulk) | — | Asp | Pro | — | Leu-Pro | Val | — | Asn | Arg-Lys | — | — | Lys | Phe | Gln | |||
| CTRLd221 (bulk) | — | Asp | Pro | — | Leu-Pro-Asp | Val | — | Asn | — | — | — | Lys | Phe | Gln | |||
| CTRLd256 (bulk) | — | Asp | Pro | — | Pro | Val | — | Asn | — | — | — | Lys | Phe | Gln | |||
MDR, presence of IDV plus 3TC; CTRL, control (no drugs). Sequential times of culture were days 28, 88, 144, 221, and 256. Bulk indicates the genotype detected by direct sequencing of PCR products. When clonal analysis was done, PCR products were cloned into the pAMP vector and the clones were sequenced. Sequences of gag protease cleavage sites were available only for clones and bulk MDRd256.
Relative to the sequence of the reference strain, NL4-3. —, amino acid identical to that in reference strain sequence.
Cellular DNA in PBMCs in culture was extracted with a nucleic acid extraction kit (Elu-Quick isolation kit; Schleicher & Schuell, Dassel, Germany) according to the manufacturer's recommendation. The pol gene was amplified in the first step by using #1 and #2 as outer primers (25), while the primers used to amplify the two regions of the RT gene in the second step were #3-seq1 and Pia3-#4 (24), and the inner primers for the PR region were PRO-F (5′-TGTAAAACGACGGCCAGT-3′) and PRO-R (5′-CAGGAAACAGCTATGACC-3′).
The conditions of amplification were described previously (24, 25). The purified PCR products were directly sequenced by using internal primers and ABI sequencing kit reagents with dye-labeled dideoxy terminators (Applied Biosystems, Inc., Foster City, Calif.). An automatic DNA sequencer (ABI PRISM 377; Applied Biosystems, Inc.) was used. The sequences were edited with Factura software and aligned with the HIV-1 IIIB consensus sequence with the Sequence Navigator software program (Applied Biosystems, Inc.).
In the case of the IDV-treated flask, we observed a quite homogeneous mutational profile for the RT gene, meaning that the mutations conferring ZDV resistance (D67N, K70R, R211K, T215F, K219Q) were maintained at each time point considered. The clonal analysis of the RT sequence on day 28 revealed a mix of Ile and Val at codon 184 that was not detected by bulk sequencing. The mutant with the 184V mutation became dominant at the subsequent time points. The mutational pattern of the PR gene in the IDV-treated virus showed evolution, with accumulation of additional PR mutations over time. The PR gene profile showed the appearance of a viral mixture at position 46 (M46M/I) on day 116. The clonal analysis on day 144 showed that a minority of the viral population (1 of 10 clones) had V82F linked to M46I. On day 186 the V82F mutation became dominant and was detected by bulk sequencing. The mutants containing the 82F mutation rapidly outgrew the other viruses in the viral population, indicating that this variant gained a sufficient increase in fitness in the presence of IDV to dominate the population.
In the untreated virus, wild-type codons at positions 70 and 184 of the RT gene were reexpressed. This result suggests a selective advantage in the absence of drugs for HIV-1 variants that have the wild-type sequence at these codons. In the absence of IDV polymorphic changes in the PR gene (L63L/P, I64V) were maintained at different time points.
HIV-1 DNA extracted was amplified with primers RVA1607 and RVA4522 (0.4 μM each) as outer primers (18), with 1 mM magnesium acetate, each deoxynucleoside triphosphate at a concentration of 0.2 mM, and 2 U of XL rTth DNA polymerase (Perkin-Elmer). In the nested PCR inner primers 1811 and 4335 (18), modified from the previously published sequence by the addition of tails to allow cloning into pAMP, were used to amplify a 2,524-bp fragment including the 3′ end of Gag and all of PR. The thermal cycling conditions used for both amplifications were those described previously (18), except that an annealing temperature of 62°C was used for the nested PCR. DNA was extracted from healthy donor PBMCs and was amplified in all PCR assays along with cultured PBMC DNA as a negative control. PCR products were cloned into vector pAMP by using the UDG cloning system (CloneAmp; GIBCO-BRL) and transformed into electrocompetent DH10B cells (GIBCO-BRL), and recombinant clones were identified by restriction enzyme digestion of plasmid DNA minipreps (Qiagen). Molecular clone pol genotypes were determined with the BigDye terminator cycle sequencing ready reaction kit (PE Applied Biosystems) and electrophoresis in an automated sequencer (ABI 377; PE Applied Biosystems). Assembly and sequence editing were done with Sequencher, version 3.1 (Gene Codes Corp.), from the 3′ end of the gag region that includes p7-p1 and p1-p6 cleavage sites through codon 350 in the RT.
PI therapy can select for substitutions in the cleavage sites in the Gag and Gag-Pol polyproteins. In clones of isolate 14aPost-MDR d144 a mutation from alanine to valine was observed in 4 of 10 clones at position (P2) of the p7-p1 cleavage site, and this mutation was always linked to M46I in the PR. In the (P1′) of p1-p6 cleavage site, a proline was detected as a polymorphic change. No others mutations were observed.
Phylogenetic analysis of viral genotypes at different time points.
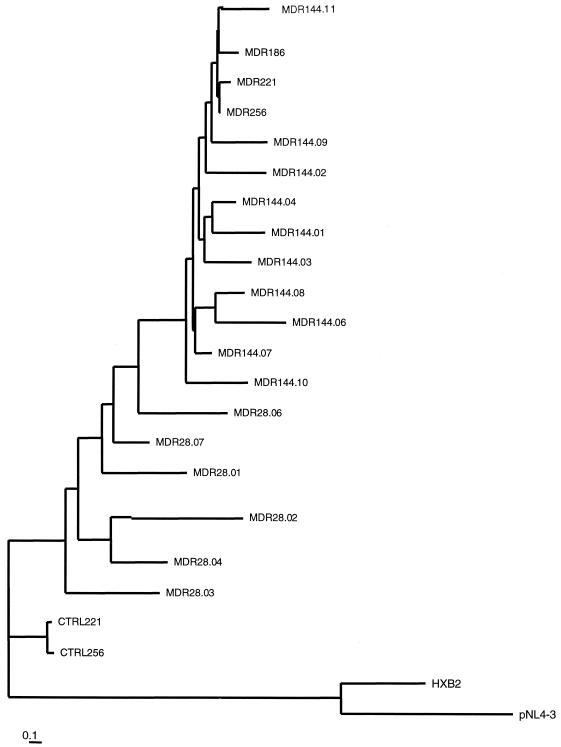
Phylogenetic analysis based on nucleotide alignment was performed to study the evolution of the pol gene during the in vitro treatment. Figure 1 shows the tree obtained by the neighbor-joining method. The bootstrapping with 1,000 replicates gave high values for all the branches, indicating that this tree is a good estimate of the true evolutionary relationships of the pol sequences. The analysis revealed the appearance of three new sequence clusters at day 144 that harbored the mutations 46I, 46I plus 82F, and p7-p1 (P2)V plus 46I, all of which were associated with IDV resistance. Variants that contained the 82F and 46I mutations rapidly dominated the viral population, which was highly homogeneous on days 186, 221, and 256. This is indicated by the presence of one major cluster in the tree (which included variants MDRd186, MDRd221, and MDRd256) with almost no genetic diversity. Sequences from the untreated flask (isolates CTRLd221 and CTRLd256) clustered separately. These controls were more related to the treated virus (isolate MDRd28) at the initial time point than to the treated virus sampled at the same time (isolates MDRd221 and MDRd256). This suggests that in the absence of selective drug pressure (and the absence of immunological pressure active in vivo but not in vitro), the evolutionary force dragging the genetic variability is lower.
FIG. 1.
Neighbor-joining phylogenetic tree of proviral sequences (996 bp) encoding PR and RT up to codon 233. The sequences of HXB2 and pNL4-3 from a database were used to root the tree. The distances between two sequences can be derived by comparing the length of the horizontal branches to the scale bar. The scale bar represents 1 nucleotide change per 100 nucleotides (1% divergence). Viruses grown in the presence of IDV and 3TC and in the absence of drugs are labeled MDR and CTRL, respectively, followed by the day of culture and the clone number (when cloning was performed). The corresponding genotypes are reported in Table 2.
Multiple alignments were done with the Wisconsin package (version 9.1; Genetics Computer Group, Madison, Wis.). Phylogenetic analysis was performed with the PHYLIP package, version 3.57c (11). Pairwise distances among sequences were computed with DNADIST and were calculated according to the two-parameter Kimura algorithm, with a transition-to-transversion ratio of 2 and maximum likelihood (DNAML, in PHYLIP). Neighbor-joining phylogenetic trees (NEIGHBOR) were constructed with TREEVIEW, version 1.5.3. Bootstrap analysis was performed with SEQBOOT (1,000 replicates). Phylogenetic analyses included pNL3-4 and HXB2 laboratory reference strains.
Evolution and linkage among mutations in the PR-coding region and the Gag-Pol cleavage site.
In the present study, the evolution of resistance in an HIV-1 strain resistant to two drugs (ZDV and 3TC) was characterized by gradual increases in the IC50 of IDV, accompanied by the emergence of variants that harbored the mutations 46I and 82F and A→V at the position (P2) of the p7-p1 cleavage site added to a viral background with polymorphisms at codons 35, 37, 63, and 64 in the PR gene and mutations at codons 67, 70, 184, 211, 215, and 219 in the RT gene. The 14aPost-MDR virus had a high level of resistance to IDV and was cross-resistant to RTV and NFV but did not accumulate other major mutations dominant in the viral population. To determine whether the mutations in the RT and PR genes occurred in the same HIV-1 genomes, we sequenced the 3′ termini of the gag and the pol regions of molecular clones from the viral population at the initial time and on day 144, when phenotypic IDV resistance became significant. The mutations that emerged in the PR gene were harbored in the same molecules linked to the RT M184V and ZDV resistance mutations.
It has been suggested that the M184V mutation resulted in an increase in the fidelity of polymerase for the RT and could affect the rate of mutation of mutant viruses (26). This study suggests that M184V in the 14aPost-MDR RT enzyme does not affect significantly the ability of the virus to accumulate mutations.
A resistance pathway can involve mutations in the viral gag and gag-pol polyprotein cleavage sites that can affect processing and viral maturation. Recent data suggest a very high frequency of gag-pol cleavage site mutations in PI-resistant clinical isolates (86% with at least one site mutated, 39% with two or more sites mutated), a significant prevalence of these mutations (15% of 300 plasma samples contained PI-resistant virus), and a high level of linkage (90% for the A(P2)V mutation in the p7-p1 cleavage site) to active-site mutations. (H. Cote, Z. Brumme, K. Hertogs, B. Larder, and P. R. Harrigan, 7th Conf. Retrovir. Opportunistic Infect. abstr. 722, 2000). Previous studies reported HIV-1 gag cleavage site changes associated with specific PR mutations in HIV-1 RNA in plasma in the case of A(P2)V in p1-p7 and L(P1′)F in p1-p6 associated with mutations at positions 82 and 84. Of note, these changes are not always retained in replication-competent PI-resistant recombinant viruses (B. A. Larder, S. Bloor, H. Hertogs, C. Van den Eynde, and R. Pauwels, 2nd Int. Workshop HIV Drug Resistance Treatment Strategies, Antivir. Ther. 3(Suppl. 1) abstr. 23, 1998). In our study, we have evidenced the emergence of a mutant which carries the M46I mutation along with A(P2)V in the p1-p7 cleavage site. The viral population that harbored IDV-selected mutations 46I and 82F and the polymorphic changes at codons 63 and 64 in the PR and that retained 67N, 70R, 211K, 215F, 219Q, and 184V in the RT rapidly became dominant, suggesting that this novel variant had gained increased fitness. Although cleavage site mutations do not induce drug resistance by themselves, they could restore fitness in PR mutants, as enhanced processing has been demonstrated (30), and since they do not affect drug binding they could contribute to cross-resistance to all PIs. Therefore, more studies should investigate the role of the gag-pol cleavage site mutations in the emergence of PR-resistant HIV-1 variants in order to clarify their clinical relevance. In our study the cross-resistance that resulted from the initial selection of resistance with IDV was sufficient to cause resistance to RTV and NFV. We also showed that the IDV-resistant virus displayed in vitro a reduced phenotypic susceptibility to the other two PIs. Interestingly, a recent study demonstrated in vivo cross-resistance between PIs during sequential therapy with the same class of compounds (8). This could be explained by the greater initial replicative fitness of mutant strains than wild-type virus in the presence of selective drug pressure (17).
The 14aPost-DR isolate in the present study exhibited a progressive accumulation of PI-related mutations over time following in vitro selection pressure with IDV, thus mimicking a frequent situation observed in individuals exposed to multiple treatments. Cross-resistance to the other two compounds (RTV, NFV) tested by susceptibility assay was supported by the sequence of the PR gene.
Our in vitro study supports resistance testing for the tailoring of antiretroviral therapy to individual subjects, taking into consideration that both genotyping and phenotyping were helpful in identifying the pattern of appearance of MDR to PIs and the maintenance of drug resistance to both PIs and nucleoside RT inhibitors.
It is important to determine the prospective efficacies of therapeutic regimens after the genotypic pattern is known, and the level of resistance and cross-resistance should be considered in order to establish the most effective combination. The gradual accumulation of mutations associated with reduced sensitivity to some antiretroviral compounds revealed that the choice of the initial therapy may be very important for the future therapeutic changes. After considering the potential occurrence of MDR HIV-1 strains to both PIs and nucleoside RT inhibitors, as modeled in our experiments, large prospective in vitro analyses are warranted to properly define future strategies for the treatment of HIV-1-infected persons.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in the present study have been submitted to GenBank and have been given accession numbers AF152964 to AF152995.
Acknowledgments
Support for this research was provided in part by the Progetto Terapia Antiretrovirale, Istituto Superiore di Sanità (Rome, Italy; grant 1997, 30A.01.43).
We thank Elizabeth L. Kaplan for manuscript editing, Matteo Romagnoni for precious collaboration, and Nicholas A. Kartsonis for helpful discussions and helping with the phylogenetic analysis.
Footnotes
This report is dedicated to the memory of Alessandro Caporali.
REFERENCES
- 1.Boden D, Hurley A, Zhang L, Cao Y, Guo Y, Jones E, Tsay J, Ip J, Farthing C, Limoli K, Parkin N, Markowitz M. HIV-1 drug resistance in newly infected individuals. JAMA. 1999;12:1135–1141. doi: 10.1001/jama.282.12.1135. [DOI] [PubMed] [Google Scholar]
- 2.Borman A, Paulous S, Clavel F. Resistance of HIV-1 to protease inhibitors: selection of resistance mutations in the presence and in the absence of the drug. J Gen Virol. 1996;77:419–426. doi: 10.1099/0022-1317-77-3-419. [DOI] [PubMed] [Google Scholar]
- 3.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, Titus D, Yang T, Squires K E, Deutch P J, Emini E A. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;274:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 4.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rodhes A, Robbins H L, Roth E, Shivaprekash M, Yang T, Chodakewits J A, Deutsch P J, Leavitt R Y, Massari F E, Mellors J W, Squires K E, Steigbigel R T, Teppler H, Emini E. Genetics correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitors. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condra J H. Virological and clinical implications of resistance to HIV-1 protease inhibitors. Drug Resist Updates. 1998;1:292–299. doi: 10.1016/s1368-7646(98)80045-9. [DOI] [PubMed] [Google Scholar]
- 6.Debouck C. The HIV-1 protease as a therapeutic target for AIDS. AIDS Res Hum Retrovir. 1992;8:153–164. doi: 10.1089/aid.1992.8.153. [DOI] [PubMed] [Google Scholar]
- 7.Deeks S G, Smith M, Holodniy M, Kahn J O. HIV-1 protease inhibitors: a review for clinicians. JAMA. 1997;277:145–153. [PubMed] [Google Scholar]
- 8.Delioust A, Paulous S, Guillemont L, Delavalle A-M, Boué F, Clavel F. Constrained evolution of human immunodeficiency virus type 1 protease during sequential therapy with two distinct protease inhibitors. J Virol. 1999;73:850–854. doi: 10.1128/jvi.73.1.850-854.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doyon L, Croteau G, Thibeault D, Poulin F, Pilote L, Lamarre D. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J Virol. 1996;70:3763–3769. doi: 10.1128/jvi.70.6.3763-3769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dulbecco R. Endpoint methods—measurements of the infectious titer of a viral sample. Virology and M. 1988;69:23–31. [Google Scholar]
- 11.Felsenstein J. Phylogeny inference package (PHYLIP). 3.57c ed. Seattle: Department of Genetics, University of Washington; 1995. [Google Scholar]
- 12.Ho D D, Toyoshima T, Mo H, Kempf D J, Norbeck D, Chen C M, Wideburg N E, Burt S K, Erickson J W, Singh M K. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J Virol. 1994;68:2016–2020. doi: 10.1128/jvi.68.3.2016-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobsen H, Hanggi M, Otto M, Duncan I B, Owen S, Andreoni M, Vella S, Mous J. In vivo resistance to a human immunodeficiency virus type 1 proteinase inhibitor: mutations, kinetics, and frequencies. J Infect Dis. 1996;173:1379–1387. doi: 10.1093/infdis/173.6.1379. [DOI] [PubMed] [Google Scholar]
- 14.Japour A J, Fiscus S A, Arduino J-M, Mayers D L, Reichelderfer P S, Kuritzkes D R. Standardized microtiter assay for the termination of syncytium-inducing phenotypes of clinical immunodeficiency virus type 1 isolates. J Clin Microbiol. 1994;32:2291–2294. doi: 10.1128/jcm.32.9.2291-2294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson V A, Merrill D P, Videler J A, Chou T-C, Byington R E, Eron J J, D'Aquila R T, Hirsch M S. Two-drug combinations of zidovudine, didanosine, and recombinant interferon-α inhibit replication of zidovudine resistant human immunodeficiency virus type 1 synergistically in vitro. J Infect Dis. 1991;164:646–655. doi: 10.1093/infdis/164.4.646. [DOI] [PubMed] [Google Scholar]
- 16.Markowitz M, Vesanen M, Tenner-Racz K, Cao Y, Binley J M, Talal A, Hurley A, Ji X, Chaudry M R, Yaman M, Frankel S, Heath-Chiozzi M, Leonard J M, Moore J P, Racz P, Nixon D F, Ho D D. The effect of commencing combination antiretroviral therapy soon after immunodeficiency virus type 1 infection on viral replication and antiviral immune response. J Infect Dis. 1999;179:525–537. doi: 10.1086/314628. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Picado J, Savara A V, Sutton L, D'Aquila R T. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J Virol. 1999;73:3744–3752. doi: 10.1128/jvi.73.5.3744-3752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Picado J, Sutton L, De Pasquale M P, Savara A V, D'Aquila R T. Human immunodeficiency virus type 1 cloning vectors for antiretroviral resistance testing. J Clin Microbiol. 1999;37:2943–2951. doi: 10.1128/jcm.37.9.2943-2951.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald C K, Kuritzkes D R. Human immunodeficiency virus type 1 protease inhibitors. Arch Intern Med. 1997;157:951–959. [PubMed] [Google Scholar]
- 20.Molla A, Korneyva M, Gao Q, Vasavanonda S, Schipper P J, Mo H-M, Markowitz M, Chernyavskiy T, Niu P, Lyons N, Hsu A, Granneman G R, Ho D D, Boucher C A B, Leonard J M, Norbeck D W, Kempf D J. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 21.Nijhuis M, Schuurman R, de Jong D, Erickson J, Gustchina E, Albert J, Schipper P, Gulnik S, Boucher C A B. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS. 1999;13:2349–2359. doi: 10.1097/00002030-199912030-00006. [DOI] [PubMed] [Google Scholar]
- 22.Richman D D. Drug resistance and its implications in the management of HIV infection. Antivir Ther. 1997;2(Suppl. 4):41–58. [Google Scholar]
- 23.Rusconi S, De Pasquale M P, Milazzo L, Bulgheroni E, Citterio P, Kurtagic S, d'Arminio-Monforte A, Galli M. In vitro effects of a continuous pressure with zidovudine (ZDV) and lamivudine on a ZDV-resistant HIV-1 isolate. AIDS. 1997;11:1406–1411. [PubMed] [Google Scholar]
- 24.Rusconi S, De Pasquale M P, Milazzo L, Moscatelli G, Bulgheroni E, Citterio P, d'Arminio-Monforte A, Moroni M, Galli M. Loss of antiviral effect owing to zidovudine and lamivudine double resistance in HIV-1-infected patients in an ongoing open-label trial. Antivir Ther. 1997;2:41–48. [PubMed] [Google Scholar]
- 25.Tisdale M, Myers R E, Maschera B, Parry N R, Oliver N M, Blair E D. Cross-resistance analysis of human immunodeficiency type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob Agents Chemoter. 1995;39:1704–1710. doi: 10.1128/aac.39.8.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wainberg M A, Drosopoulos W C, Salomon H, Hsu M, Borkow G, Parniak M, Gu Z, Song Q, Manne J, Islam S, Castriota G, Prasad V R. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science. 1996;271:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]
- 27.Winters M A, Schapiro J M, Lawrence J, Merigan T C. Human immunodeficiency virus type-1 protease genotypes and in vitro protease inhibitor susceptibilities of isolates from individuals who were switched to other protease inhibitor after long-term saquinavir treatment. J Virol. 1998;72:5303–5306. doi: 10.1128/jvi.72.6.5303-5306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young B, Johnson S, Bahktiari M, Shugarts D, Young R K, Allen M, Ramey II R R, Kuritzkes D R. Resistance mutations in protease and reverse transcriptase genes of human immunodeficiency virus type 1 isolates from patients with combination antiretroviral therapy failure. J Infect Dis. 1998;178:1497–1501. doi: 10.1086/314437. [DOI] [PubMed] [Google Scholar]
- 29.Zang Z Q, Schuler T, Cavert W, Notermans D W, Gebarth K, Herry K, Havlir D V, Gunthard H F, Wong J K, Little S, Feinberg M B, Polis M A, Schrager L H, Schacker T W, Richman D D, Corey L, Danner S A, Haase A T. Reversibility of the pathological changes in the follicular dendritic cell network with treatment of HIV-1 infection. Proc Natl Acad Sci USA. 1999;96:5169–5172. doi: 10.1073/pnas.96.9.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y M, Imamichi H, Imamichi T, Lane H C, Falloon J, Vasudevachari M B, Salzman N P. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J Virol. 1997;71:6662–6670. doi: 10.1128/jvi.71.9.6662-6670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]