Abstract
Peroxisome proliferator-activated receptors γ (PPARγ) is a master regulator that controls energy metabolism and cell fate. PPARγ2, a PPARγ isoform, is highly expressed in the normal prostate but expressed at lower levels in prostate cancer tissues. In the present study, PC3 and LNCaP cells were used to examine the benefits of restoring PPARγ2 activity. PPARγ2 was overexpressed in PC3 and LNCaP cells, and cell proliferation and migration were detected. Hematoxylin and eosin (H&E) staining was used to detect pathological changes. The genes regulated by PPARγ2 overexpression were detected by microarray analysis. The restoration of PPARγ2 in PC3 and LNCaP cells inhibited cell proliferation and migration. PC3-PPARγ2 tissue recombinants showed necrosis in cancerous regions and leukocyte infiltration in the surrounding stroma by H&E staining. We found higher mixed lineage kinase domain-like (MLKL) and lower microtubule-associated protein 1 light chain 3 (LC3) expression in cancer tissues compared to controls by immunohistochemistry (IHC) staining. Microarray analysis showed that PPARγ2 gain of function in PC3 cells resulted in the reprogramming of lipid- and energy metabolism-associated signaling pathways. These data indicate that PPARγ2 exerts a crucial tumor-suppressive effect by triggering necrosis and an inflammatory reaction in human prostate cancer.
Keywords: inflammatory reaction, necrosis, PPARγ, prostate cancer, tissue recombination-xenografting
INTRODUCTION
Although screening and early detection are valuable strategies to decrease mortality, prostate cancer (PCa) remains the third leading cause of male cancer death in the United States.1 The mechanisms of PCa initiation and progression are not well understood. The advent of prostate-specific antigen as a screening tool and increased public awareness have resulted in a significant decrease in PCa stage at presentation, and patients are now presenting sooner with curable, organ-confined disease. However, up to 22% of newly diagnosed patients still present with advanced or metastatic disease.2 There is a need to develop new approaches to prevent and treat PCa to improve patient survival and decrease disease- and treatment-associated mortality.
The nuclear receptor peroxisome proliferator-activated receptors γ (PPARγ) is a transcription factor that plays a key role in lipid and energy metabolism,3 differentiation, and inflammation in many tissues and organs, including the prostate.4,5,6 The important role of PPARγ in carcinogenesis is highlighted by the ability of its ligands to affect cellular proliferation and differentiation or interfere in apoptosis and angiogenesis.7,8 Early studies reported the detection of high PPARγ expression in PCa.9 Conditional deletion of PPARγ in mouse prostatic luminal epithelial cells resulted in a range of phenotypes, including hyperplastic growth and cumulative high-grade prostatic intraepithelial neoplasia, suggesting the biological role of PPARγ signaling in regulating mouse prostate epithelial homeostasis.10 The identified phenotypes were associated with dysregulation of cell cycle control and metabolic signaling networks related to peroxisomal and lysosomal maturation, lipid oxidation, and degradation. Specifically, loss of the PPARγ2 isoform resulted in autophagocytosis in mouse prostate epithelial cells.11 Mouse prostatic glandular differentiation in tissue recombination was achieved by rescue with the PPARγ2 isoform but not with PPARγ1, and expression of the two isoforms resulted in differential reprogramming of lipid- and energy metabolism-associated signaling pathways and expression of tissue-specific differentiation markers.12
In the present study, we investigated the effects of genetic gain of function of the human-specific PPARγ2 isoform on specific end points in human PCa cells in vitro and in vivo using a translational human prostatic tissue recombination xenograft mouse model. The aim of the work was to confirm the pivotal role of PPARγ2 and to decipher the molecular mechanisms by which the PPARγ2 isoform elicited its effects in human PCa cells. Two advanced human PCa cell lines, PC3 and LNCaP cells, were used to study the consequences of gain of function of the PPARγ2 isoform, and one normal prostate cell line, RWPE cells, was used as a control.
MATERIALS AND METHODS
Reagents and cell lines
The set of primary antibodies used in the experiments is provided in Supplementary Table 1. Secondary antibodies, Cell Counting Kit-8 (CCK-8), and other chemicals used in this study were purchased from Beyotime Biotechnology (Haimen, China).
Supplementary Table 1.
Primary antibodies used in the experiments
| Name | Brand | Applications and dilutions | Article number |
|---|---|---|---|
| PPARγ | Santa Cruz Biotechology | 1:800 (WB) | sc-7273 |
| MLKL | Abcam | 1:400 (IHC) | ab184718 |
| LC3 | Cell Signaling Technology | 1:400 ( IHC) | 12741 |
| β-actin | Abcam | 1:1000 (WB) | ab8226 |
WB: western blot; IHC: immunohistochemistry; PPARγ: peroxisome proliferators-activated receptors γ; MLKL: mixed lineage kinase domain-like; LC3: light chain 3
RWPE cells purchased from iCell Bioscience Inc. (Shanghai, China) were cultured in 50%/50% Dulbecco's modified Eagle's medium (DMEM)/F12 (Gibco, Burlington, Canada) supplemented with 5% heat-inactivated fetal bovine serum (FBS; Gibco), 1% insulin-transferrin-selenium-X (ITS; Gibco), 0.4% bovine pituitary extract (BPE; Hammond Cell Tech, Shanghai. China), and 10 ng ml−1 epidermal growth factor (EGF; Sigma-Aldrich, Shanghai, China) with 1% penicillin (100 U ml−1; Gibco) and streptomycin (100 mg ml−1; Gibco) at 37°C in an incubator with a humidified atmosphere of 5% CO2. PC3 and LNCaP cells purchased from iCell Bioscience Inc. were routinely cultured in RPMI 1640 medium (HyClone, Logan, UT, USA) with 10% heat-inactivated FBS and 1% antibiotic/antimycotic solution (Gibco).
Retroviral constructs and establishment of a stable cell line
A retroviral construct containing full-length human PPARγ2 complementary DNA (cDNA) in pBABE was a gift from Professor C Ronald Kahn at the Joslin Diabetes Center (Department of Medicine, Harvard University Medical School, Boston, MA, USA). Stable puromycin-resistant cells were generated.
Cell proliferation and migration assay
Cell morphology was observed using an inverted microscope (Olympus Fluoview, Tokyo, Japan). Cell viability was determined using the CCK-8 assay. In brief, RWPE-empty vector (EV), RWPE-PPARγ2, PC3-EV, PC3-PPARγ2, LNCaP-EV, and LNCaP-PPARγ2 cells were plated in 96-well plates. CCK-8 solution (10 μl) was added to each well and incubated for an additional 2 h. Finally, the sample absorbance at 450 nm (650 nm as reference wavelength) was measured by an automatic microplate reader (ThermoFisher, Carlsbad, CA, USA) according to the manufacturer's protocol.
For the in vitro cell migration assay, 1 × 105 cells were detached and plated in a 24-well plate. After overnight incubation, two parallel wounds approximately 400 μm in length were made using a P100 pipette tip. After rinsing with phosphate-buffered saline (PBS), images were collected at 24 h after wounding at the same positions on the underside of the dish indicated with a marker. The cell migration distance was determined by measuring the width of the wound.
Protein extraction and western blot assays
Proteins were extracted with a protein extraction kit (Beyotime Biotechnology) according to the manufacturer's instructions. The protein content was measured by bicinchoninic acid (BCA) protein assay. PPARγ2 protein expression levels were determined by western blot analysis. Samples containing a total protein content of 50 μg were loaded in each lane of 12% polyacrylamide gel electrophoresis (PAGE) gel with Precision Plus protein molecular weight standards (Bio-Rad, Hercules, CA, USA) and separated with a Mini-PROTEAN Tetra system (Bio-Rad). The proteins were transferred to nitrocellulose membranes, which were then incubated with fat-free milk powder for 1 h at room temperature (RT) to block nonspecific binding. The membranes were incubated with primary antibody overnight at 4°C. The membrane was then washed and incubated with horseradish peroxidase (HRP)-labeled goat anti-rabbit IgG as a secondary antibody (ab6728, Cambridge, UK, Abcam), followed by visualization using enhanced chemiluminescence detection reagent (ThermoFisher). Bands were observed by scanning the blot with a computer, and relative intensities were determined by densitometry using Scion Image version 4.0.2 (Scion Corporation, Frederick, CA, USA). Negative control membranes were incubated with the appropriate preabsorbed primary antibody or blocking solution without primary antibody. Relative PPARγ2 protein expression levels were normalized to β-actin as a loading control.
Animal experiments
Adult male immunodeficient mice (BALB/c nude) used for PPARγ2 overexpression approach were purchased from Shanghai Silaike Experimental Animals Inc. (Shanghai, China). The animals were housed at four animals per cage in a temperature-controlled room (20°C–22°C) under a 12-h light/12-h dark cycle. Food and water were available ad libitum unless noted. All animal protocols used in this study had been reviewed and approved by Nantong University Medical School (Nantong, China; S20190323-312).
Tissue recombinant xenograft (TRX) modeling
Viable cells were counted using a hemacytometer. A total of 6 × 105 cultured cells were pelleted and resuspended in 50 μl of rat tail collagen gel (pre-equilibrated to pH 7.4). After polymerization, the collagen was overlaid with growth medium. After incubation at 37°C overnight, the tissue recombinants were grafted under the renal capsule of each intact nude mouse at two recombinants per kidney. Hosts were sacrificed at 12-week postgrafting. The kidneys with the grafts were removed and imaged before processing for histology.
Morphological detection and immunohistochemistry
The fixed grafts were embedded in paraffin blocks, sectioned perpendicular to the longest axis at a thickness of 6 μm, and stained with hematoxylin and eosin (H&E). The stained sections were mounted and examined by light microscopy (Olympus Fluoview).
Sections of the grafts were prepared for immunohistochemical analysis in a manner similar to a previously reported method13 with slight modulations. Briefly, the sections were deparaffinized with xylene and rehydrated in graded ethanol before being washed with twice-distilled water. To increase epitope exposure, the sections were heated for 15 min in sodium citrate buffer (0.01 mol l−1, pH 6.0) in a microwave oven. The sections were cooled, washed with PBS, and then blocked with 10% bovine serum albumin (BSA) in PBS for 1 h at RT. The sections were incubated overnight at 4°C with diluted antibodies. The antibodies were visualized with an streptavidin–biotin complex (SABC) Kit Elite (Zhongshanjinqiao Biotech, Beijing, China) and 0.05% 3,3-diaminobenzidine tetrachloride (Zhongshanjinqiao Biotech) in PBS containing 0.01% H2O2 for 2 min. The sections were counterstained with hematoxylin and mounted with coverslips. Antibody specificity was examined with the use of 1% BSA instead of primary antibody.
Globe gene expression profiling by microarray
The expression and functional profiles of genes in PC3 cells expressing PPARγ2 cDNA and empty vector were determined using an Agilent SurePrint G3 Human Gene Expression 8 × 60K version 2 Microarray (Agilent Technologies, Santa Clara, CA, USA) and compared. Analysis of functional PPARγ2-regulating gene profiles was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) based on the biological pathways from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Arrays underwent background correction, quantile normalization, and log transformation.
All microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) database (accession No. GSE108309) with a link for review: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108309.
Validation of the microarray data using real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR)
To verify the gene expression responses observed in the microarray, we performed qRT-PCR analysis of ten significant differentially expressed genes in each cell type from the microarray data. Primers for the target genes are listed in Supplementary Table 2. cDNA was produced from total RNA using the Prime Script RT Reagent Kit (TaKaRa Biotechnology, Dalian, China). qRT-PCR was conducted in 25 μl reaction volumes consisting of 12.5 μl of TB Green Premix Ex Taq (TaKaRa Biotechnology), 2 μl of first-strand cDNA (template), 0.5 μl of the PCR forward primer, 0.5 μl of the PCR reverse primer, and 9.5 μl of double distilled H2O. Three biological replicates were used for qRT-PCR experiments.
Supplementary Table 2.
Primers and details of the quantitative polymerase chain reaction analysis of mRNA levels
| Gene | Sequence (5′→3′) | Product size (bp) | 40 PCR cycles |
|---|---|---|---|
| ALDH1A3 (NM_000693.4) | F: GGAGACTTGCCTGGTGAA R: GCATTTGTGGTTGGGTCA |
195 | 95°C for 15 s 60°C for 30 s 72°C for 30 s |
| CD44 (NM_000610.4) | F: GTTCCTGGACTGATTTCTT R: AATTACTCTGCTGCGTTG |
186 | 95°C for 15 s 60°C for 30 s 72°C for 30 s |
| CDC25C (NM_001790.5) | F: CTTCCTTTACCGTCTGTC R: AAACCATTCGGAGTGCTA |
292 | 95°C for 15 s 60°C for 30 s 72°C for 30 s |
| CDKN1C (NM_000076.2) | F: GGGACCGTTCATGTAGCAGC R: CTTGGGACCAGTGTACCTTCT |
147 | 95°C for 15 s 60°C for 30 s 72°C for 30 s |
| FOS (NM_005252.4) | F: CAGTGCCAACTTCATTCC R: GGAGATAACTGTTCCACCTT |
234 | 95°C for 15 s 60°C for 30 s 72°C for 30 s |
| IL6 (NM_000600.5) | F: GGAGACTTGCCTGGTGAA R: GCATTTGTGGTTGGGTCA |
195 | 95°C for 15 s 60°C for 30 s 72°C for 30 s |
| RAC1 (NM_006908.5) | F: TAGGGATGATAAAGACACG R: GACAGGACCAAGAACGAG |
253 | 95°C for 15 s 60°C for 30 s 72°C for 30 s |
| ZHX2 (NM_001362797.2) | F: GTTGAGCAGCATCAGAGCG R: CATCATATCAGAGTGGGAGCA |
103 | 95°C for 15 s 60°C for 30 s 72°C for 30 s |
| GAPDH (NM_002046.6) | F: ATGAGAAGTATGACAACAGCC R: TTCAGCTCAGGGATGACCTT |
266 | 95°C for 15 s 60°C for 30 s 72°C for 30 s |
PCR: polymerase chain reaction; FOS: fos protein; ALDH1A3: aldehyde dehydrogenase 1 family member A3; CD44: cell surface glycoprotein 44; CDC25C: cell division cycle 25 homolog C; CDKN1C: cyclin dependent kinase inhibitor 1C; IL6: interleukin-6; RAC1: ras-related C3 botulinum toxin substrate 1
Statistical analyses
All data are presented as the mean ± standard error of the mean (s.e.m.) and were analyzed by one-way analysis of variance (ANOVA) followed by Bonferroni's multiple comparison test. Statistical analyses were performed with GraphPad Prism software (GraphPad Software, San Diego, CA, USA). Differences were considered statistically significant when P < 0.05.
RESULTS
Restoration of PPARγ2 isoform activity in PC3 and LNCaP cells suppressed cellular proliferation and migration in vitro
The advanced human PCa cell lines PC3 and LNCaP show low endogenous PPARγ2 expression.11 To explore the benefits of restoring PPARγ2 isoform activity in PC3 cellular growth and differentiation in vitro, we overexpressed the wild-type full-length cDNA for the human PPARγ2 isoform wild (pBABE-Puro-EV as a control) in the cells and analyzed resultant cell proliferation and migration as well as related gene expression profiles. High expression of the specific PPARγ2 isoform was validated by western blot analysis (Figure 1a and Supplementary Figure 1a (255.9KB, tif) ).
Figure 1.
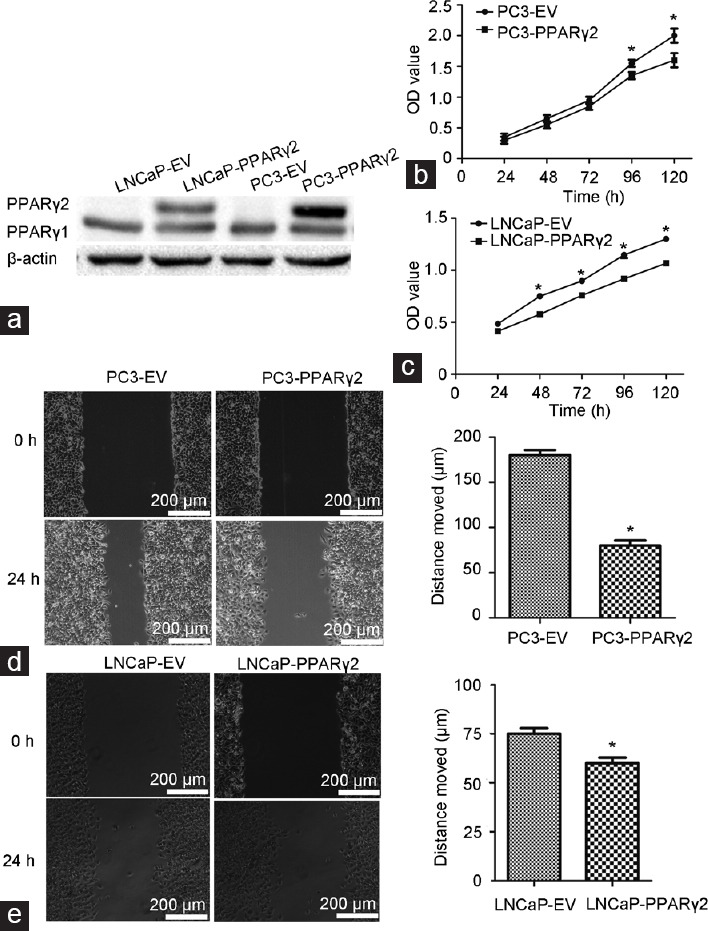
Restoration of PPARγ2 activity in PC3 and LNCaP cells inhibited cellular proliferation and migration in vitro. (a) Restoration of specific PPARγ2 isoform activity using WT full-length human PPARγ2 cDNA in PC3 and LNCaP cells by stable transfection was used to create a series of isogenic cell lines for genetic and functional comparisons. PPARγ2 protein expression was validated by western blot analysis. The proliferation of (b) PC3- and (c) LNCaP-PPARγ2 cDNA-restored cells was significantly inhibited compared with that of control cells. *P < 0.05, n = 6 for each group. The migration of (d) PC3- and (e) LNCaP-PPARγ2-restored cells was significantly inhibited compared with that of control cells. *P < 0.05, n = 6 for each group. WT: wild type; EV: empty vector; PPARγ2: peroxisome proliferators-activated receptors γ 2.
Restoration of PPARγ2 isoform activity in PC3 and LNCaP cells resulted in remarkable physiologic and biological changes. Although we did not observe major differences in cell morphology between the PC3- and LNCaP-PPARγ2-restored cells, the proliferation of PC3- and LNCaP-PPARγ2-restored cells was significantly suppressed (Figure 1b and 1c). Overexpression of PPARγ2 had no effect on RWPE proliferation (Supplementary Figure 1b (255.9KB, tif) ). In addition, the monolayer wound healing assay showed that the migration of PC3- and LNCaP-PPARγ2-restored cells was significantly suppressed (Figure 1d and 1e).
Restoration of PPARγ2 isoform activity in PC3 cells induced necrosis and leukocyte infiltration in vivo
To determine the effects of restoration of PPARγ2 activity on advanced human prostate cancer progression in vivo, tissue recombinants of PC3-PPARγ2-restored cells and control cells mixed with rat tail collagen gel were grafted into the kidney capsules of 8-week-old nude mice and remained for 3 months. In total, six to eight tissue recombinants were made for each group, and the PC3 tumor remodeling ratio was analyzed (Supplementary Table 3). H&E staining showed that PC3-EV cells regenerated a structure typical of solid prostate carcinoma showing decreased differentiation (Figure 2a, left). Histologically, restoration of PPARγ2 activity in PC3 tissue recombinants increased leukocyte infiltration in the surrounding stroma, accompanied by severe necroptosis in the reconstituted cancerous region (Figure 2a, right).
Supplementary Table 3.
Tissue recombinants were made using PC3-peroxisome proliferators-activated receptors γ 2 and control PC3-empty vector cells
| Tissue recombinants | PC3-EV | PC3-PPARγ2 |
|---|---|---|
| Total number | 6 | 8 |
| Regenerated tumor number | 6 | 6 |
| Regenerated tumor ratio (%) | 100 | 75% |
Recombinant tissues generated from PC3-PPARγ2 and control PC3-EV cells were grafted under the subrenal capsules of male nude mice and examined 3 months after grafting. The histology of the regenerated tumors was microscopically evaluated. PPARγ2: peroxisome proliferators-activated receptors γ 2; EV: empty vector
Figure 2.
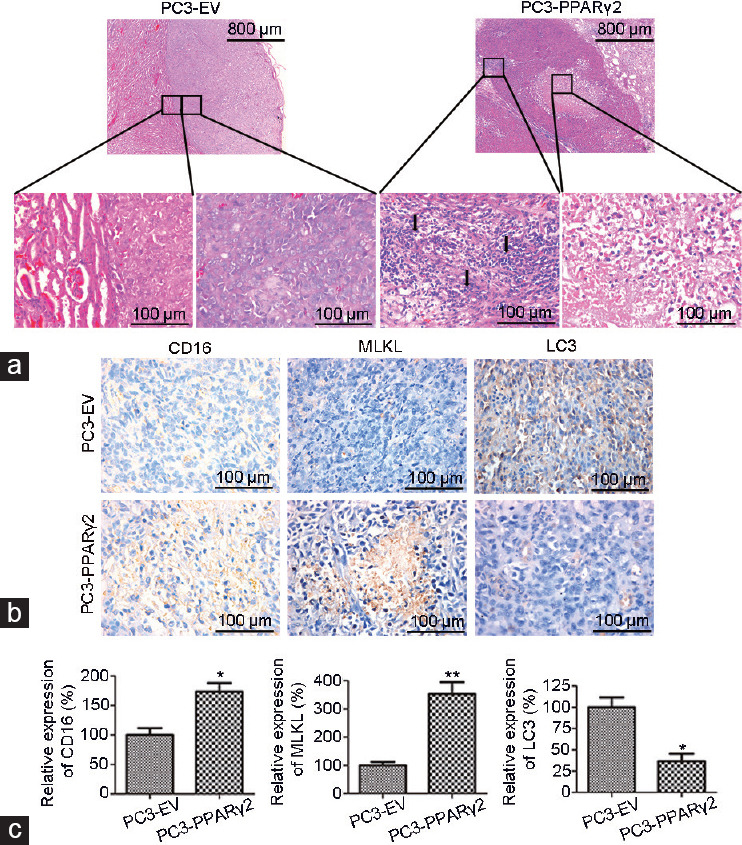
Restoration of PPARγ2 isoform activity in PC3 cells induced necrosis and leukocyte infiltration in vivo. (a) Histological analyses of human prostate cancer tissue recombinants indicate significant morphological differences between the PC3-EV and PC3-PPARγ2 groups at 3 months postgrafting. PC3-EV tissue recombinants showed no leukocyte accumulation in the stroma or solid tumors (left). PC3-PPARγ2 tissue recombinants presented stromal leukocyte infiltration and necrosis of cancerous areas (right). The arrow shows leukocyte infiltration. (b) IHC staining shows low protein expression of CD16 and MLKL and high protein expression of LC3 in cancerous regions of PC3-EV tissue recombinants and high CD16 and MLKL protein expression and low LC3 protein expression in PC3-PPARγ2 tissue recombinants. (c) The relative expression of CD16, MLKL, and LC3. *P < 0.05, **P < 0.01; n = 5 for each group. PPARγ2: peroxisome proliferators-activated receptors γ 2; EV: empty vector; MLKL: mixed lineage kinase domain-like; LC3: lower microtubule-associated protein 1 light chain 3; IHC: immunohistochemistry.
In addition, little signal for CD16 (a natural killer cell marker) and mixed lineage kinase domain-like (MLKL; a necroptotic marker) and a large signal for lower microtubule-associated protein 1 light chain 3 (LC3; an autophagic marker) were observed in cancerous regions regenerated from PC3-EV cells, while enhanced CD16 and MLKL protein expression and decreased LC3 expression were detected in cancerous regions regenerated by PC3-PPARγ2 cells (Figure 2b and 2c).
These data suggest that restoration of PPARγ2 isoform activity in PC3 tissue recombinants played an important tumor-suppressive role in the induction of cancer necrosis and the immune response or inflammation in the cancer microenvironment in vivo.
Messenger RNA (mRNA) expression was changed in PC3 cells with restored PPARγ2 activity
To explore the associated signaling pathways linked to the benefits of restored PPARγ2 activity in PC3 cells, we analyzed the global gene expression profiles of PC3-PPARγ2-restored and PC3-EV control cells in vitro using a microarray platform. Principal component analysis (PCA) was used to reduce the complexity of high-dimensional gene expression data. Three-dimensional plots were obtained by plotting differentially expressed genes in PC3-PPARγ2-restored cells and control cells (Figure 3a). The data from PC3-PPARγ2-restored cells were closely clustered, and a similar finding was observed in control cells (Figure 3a). The results from PCA suggested that global gene expression was altered by the restoration of PPARγ2 isoform activity. Then, a box plot was made to visualize the distributions of the data. Assessment of the box plot suggested that the data from six RNA gene chips were similarly distributed (Figure 3b). Variations in mRNA expression among samples were shown by scatter plotting (Figure 3c). Then, to distinguish differential gene expression patterns among samples, hierarchical clustering was conducted (Figure 3d). Thirty genes were upregulated, and 112 genes were downregulated upon the restoration of PPARγ2 activity in PC3 cells (Figure 3d). The top 20 more differentially expressed genes are listed in Supplementary Table 4 and 5.
Figure 3.
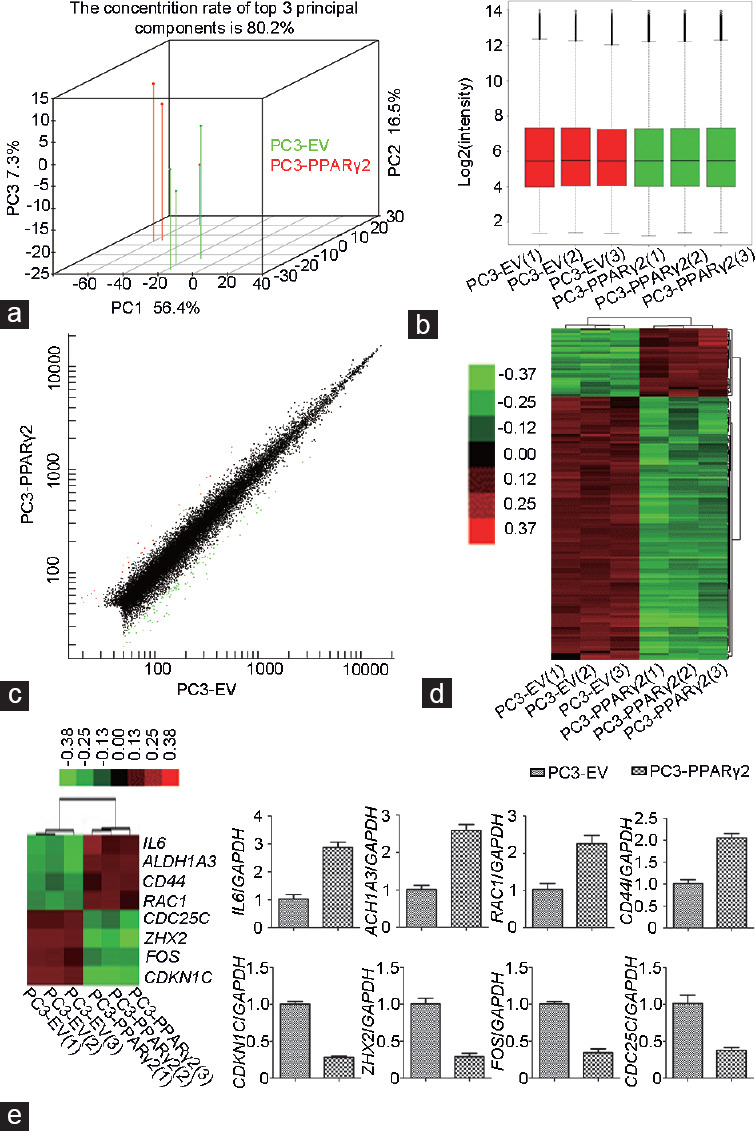
Global gene expression profiles were analyzed by microarray and validated by qRT-PCR in PC3 cells, in which PPARγ2 activity was restored and control PC3-EV cells. (a) Principal component analysis of PC3 cells in which PPARγ2 activity was restored and control cells. (b) Box and whisker plots of mRNA levels showing the distribution of their intensities in all samples. (c) Scatter plots showed variations in mRNA expression. (d) Hierarchical clustering showing gene expression profiles. (e) The expression of selected differentially expressed genes was validated by qRT-PCR. The numbers in brackets represent three samples for each group. PPARγ2: peroxisome proliferators-activated receptors γ 2; EV: empty vector; qRT-PCR: real-time quantitative reverse transcription polymerase chain reaction; IL6: interleukin-6; CDKN1C: cyclin-dependent kinase inhibitor 1C; ALDH1A3: aldehyde dehydrogenase 1 family member A3; CD44: cell surface glycoprotein 44; RAC1: ras-related C3 botulinum toxin substrate 1; FOS: fos protein; CDC25C: cell division cycle 25 homolog C; ZHX2: zinc fingers and homeoboxes 2; mRNA: messenger RNA; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
Supplementary Table 4.
The top 20 most upregulated mRNAs
| Gene number | Fold change | Score (d) | Gene title | Gene symbol |
|---|---|---|---|---|
| NM_198833.1 | 3.033 | 17.98284224 | SERPINB8 | Serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| NM_005556.3 | 2.9159 | 19.3799738 | KRT7 | Keratin 7 |
| NM_001126336.1 | 2.8959 | 9.723185182 | VCAN | Versican |
| NM_004598.3 | 2.7589 | 9.139741868 | SPOCK1 | Sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| X15998.1 | 2.6789 | 10.75276679 | VCAN | Versican |
| CR624632.1 | 2.631 | 7.903625006 | IL6 | Interleukin 6 |
| BX538027.1 | 2.6127 | 10.0784025 | ALDH1A3 | Aldehyde dehydrogenase 1 family, member A3 |
| NM_005949.3 | 2.6089 | 12.18734769 | MT1F | Metallothionein 1F |
| BM982921 | 2.5688 | 6.136702928 | LCN2 | Lipocalin 2 |
| AK302607.1 | 2.5537 | 10.73348507 | ALDH1A3 | Aldehyde dehydrogenase 1 family, member A3 |
| NM_001432.2 | 2.5459 | 6.302285608 | EREG | Epiregulin |
| AI077622 | 2.5359 | 5.22933458 | LOC100128551///ZDHHC14 | Uncharacterized LOC100128551///zinc finger, DHHC-type containing 14 |
| NM_001110503.1 | 2.4971 | 3.748435483 | TMEM87A | Transmembrane protein 87A |
| CX756248 | 2.485 | 9.90040295 | RHOB | Ras homolog family member B |
| NM_002638.3 | 2.3813 | 6.527071477 | PI3 | Peptidase inhibitor 3, skin-derived |
| AF136373.1 | 2.2166 | 8.396541344 | RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
| CD642045 | 2.1367 | 7.437555631 | CD44 | CD44 molecule (Indian blood group) |
| AK300391.1 | 2.1076 | 10.77529713 | SERPINB8 | Serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| NM_018697.3 | 2.0886 | 12.13180687 | LANCL2 | LanC lantibiotic synthetase component C-like 2 (bacterial) |
| NM_004385.3 | 2.0821 | 6.969702142 | VCAN | Versican |
Supplementary Table 5.
The top 20 most downregulated mRNAs
| Gene number | Fold change | Score (d) | Gene title | Gene symbol |
|---|---|---|---|---|
| NM_021623.1 | 0.3787 | −15.40164962 | PLEKHA2 | Pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| NM_004348.3 | 0.3781 | −7.732465628 | RUNX2 | Runt-related transcription factor 2 |
| NM_177987.1 | 0.3773 | −7.234198871 | TUBB8///TUBBP5 | Tubulin, beta 8 class VIII///tubulin, beta pseudogene 5 |
| AK126854.1 | 0.3759 | −11.11085267 | KDM4B | Lysine (K)-specific demethylase 4B |
| AK298659.1 | 0.3734 | −9.705048613 | FOS | FBJ murine osteosarcoma viral oncogene homolog |
| AB014766.1 | 0.3664 | −5.196096628 | – | – |
| NM_001206.2 | 0.3644 | −7.538694252 | KLF9 | Kruppel-like factor 9 |
| NM_015568.2 | 0.3634 | −11.21549651 | PPP1R16B | Protein phosphatase 1, regulatory subunit 16B |
| NM_177987.1 | 0.3543 | −8.912983074 | TUBB7P///TUBB8///TUBBP5 | Tubulin, beta 7, pseudogene///tubulin, beta 8 class VIII///tubulin, beta pseudogene 5 |
| NM_177987.1 | 0.346 | −11.01428081 | TUBB8///TUBBP5 | Tubulin, beta 8 class VIII///tubulin, beta pseudogene 5 |
| NM_181726.2 | 0.3365 | −12.13704369 | ANKRD37 | Ankyrin repeat domain 37 |
| NM_001005404.3 | 0.3334 | −9.24516789 | YPEL2 | Yippee-like 2 (Drosophila) |
| BU615833 | 0.3122 | −14.18372082 | LINC00341///SYNE3 | Long intergenic non-protein coding RNA 341///spectrin repeat containing, nuclear envelope family member 3 |
| CR627389.1 | 0.3069 | −12.24438793 | ETV1 | Ets variant 1 |
| CR605750.1 | 0.3056 | −21.59932672 | CDKN1C | Cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| NM_080616.3 | 0.3008 | −8.45570563 | NOL4L | Nucleolar protein 4-like |
| NM_014943.3 | 0.3005 | −10.18239551 | ZHX2 | Zinc fingers and homeoboxes 2 |
| H99290 | 0.2815 | −12.96901942 | – | – |
| U22398.1 | 0.2787 | −11.22869711 | CDKN1C | Cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| CR605750.1 | 0.2632 | −12.75239734 | CDKN1C | Cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
Confirmation of differentially expressed mRNAs by qRT-PCR
To examine the reliability of the microarray data, we selected eight mRNAs that were found to have differential gene expression by microarray analysis and confirmed their expression in PC3-PPARγ2-restored and PC3-EV control cells using qPCR. The variations in expression of these mRNAs assessed by qRT-PCR were consistent with the microarray data (Figure 3e).
To strengthen the conclusions of the microarray data, expression of the same mRNAs whose abundance in PC3 cells was tested was also validated in LNCaP cells. Although the results in LNCaP cells were slightly different from those in PC3 cells, changes in the expression of five genes in LNCaP cells were consistent with those in PC3 cells (Supplementary Figure 2 (304.5KB, tif) ). These results indicate that the function of PPARγ2 is universal in both cell lines.
Inflammatory response- and cell proliferation-associated signaling pathways were reprogrammed in PC3 cells with restored PPARγ2 activity
Functional classification of the differentially expressed genes was performed, revealing ontological gene groupings with high-fold enrichment and low false discovery rates. The biological processes most significantly enriched in these differentially expressed genes in PC3-PPARγ2 cells were cellular response to interleukin-1 (Gene Ontology [GO]: 0071347), regulation of peptidyl-tyrosine phosphorylation (GO: 0050730), positive regulation of peptidyl-tyrosine phosphorylation (GO: 0050731), regulation of epithelial cell proliferation (GO: 0050678), and cellular response to endogenous stimulus (GO: 0071495), as shown in Figure 4a. The cellular components most significantly enriched in the differentially expressed genes in PC3-PPARγ2 cells were anchoring junction (GO: 0070161), phagocytic cup (GO: 0001891), plasma membrane region (GO: 0098590), actin A complex (GO: 0048180), and macropinocytic cup (GO: 0070685), as shown in Figure 4b. The molecular functions most significantly enriched in the differentially expressed genes in PC3-PPARγ2 cells were single-stranded DNA-dependent ATPase activity (GO: 0043142), enzyme regulator activity (GO: 0030234), RNA polymerase II core promoter sequence-specific DNA binding (GO: 0000979), protein tyrosine kinase activator activity (GO: 0030296), and growth factor binding (GO: 0019838), as shown in Figure 4c.
Figure 4.
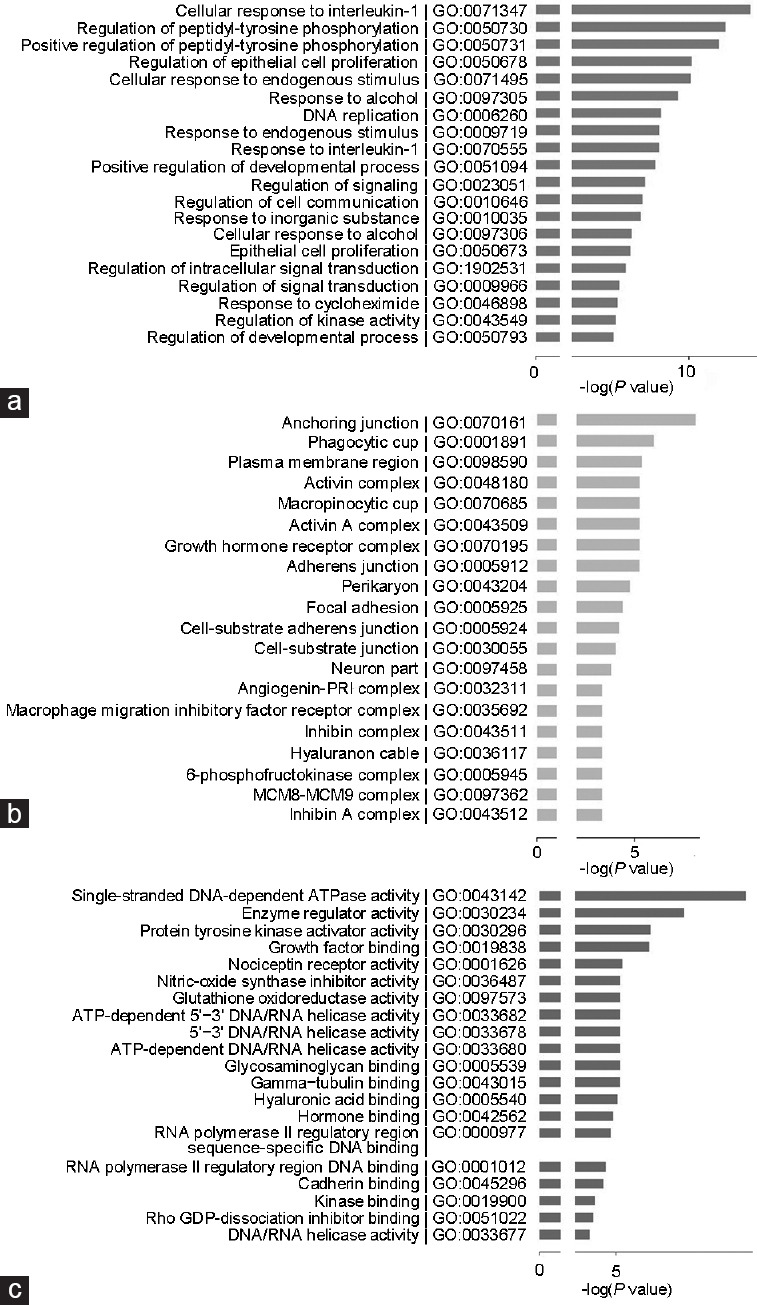
Results of GO analysis of the differentially expressed genes upon restoration of PPARγ2 isoform activity in PC3 cells. (a) Main biological processes enriched in the differentially expressed genes. (b) Main cellular components enriched in the differentially expressed genes. (c) Main molecular functions enriched in the differentially expressed genes. PPARγ2: peroxisome proliferators-activated receptors γ 2; GO: Gene Onotology; MCM8: minichromosome maintenance 8; MCM9: minichromosome maintenance 9; ATP: adenosine triphosphate; PRI: placental ribonuclease inhibitor.
KEGG analysis of diseases showed that the diseases related to differentially expressed genes were mainly cancer, cleidocranial dysplasia, camptodactyly, tall stature, scoliosis, and hearing loss (CATSHL) syndrome, achondroplasia, and glycogen storage disease (GSD; Figure 5a). Notably, PCa was also present among the top 30 associated diseases, although it was not the disease most relevant to the differentially expressed genes.
Figure 5.
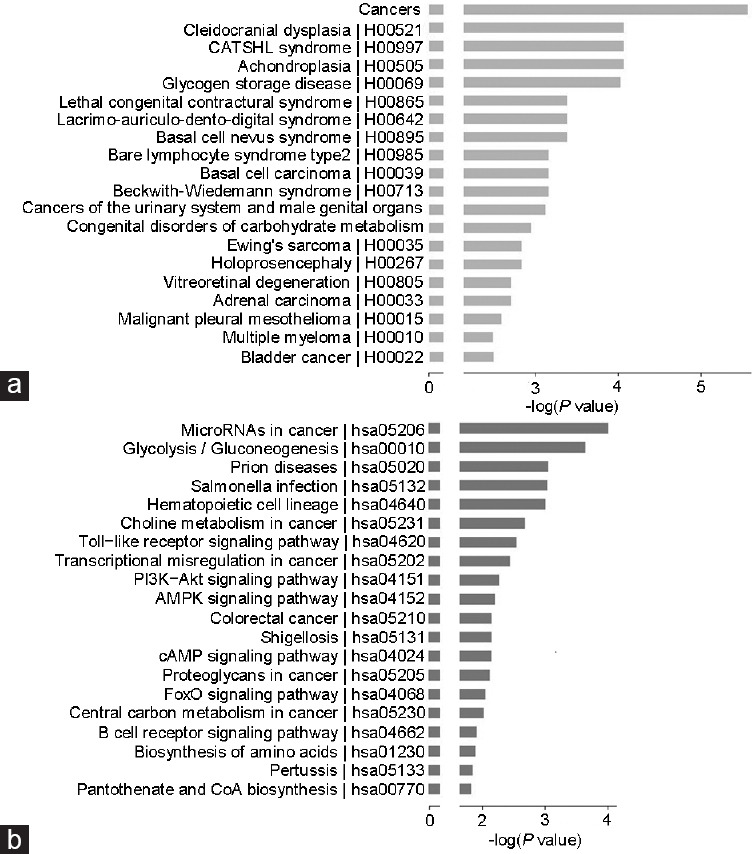
Results of pathway and disease analyses of the differentially expressed genes upon restoration of PPARγ2 isoform activity in PC3 cells. (a) Main pathways that involve the differentially expressed genes. (b) Main diseases that involve the differentially expressed genes. PPARγ2: peroxisome proliferators-activated receptors γ 2; PI3K: phosphatidylinositol 3-kinase; Akt: protein kinase B; AMPK: adenosine 5’-monophosphate-activated protein kinase; cAMP: cyclic adenosine monophosphate; FoxO: forkhead box O3; CoA: coenzyme A.
KEGG pathway analysis suggested that the differentially expressed genes in PC3-PPARγ2 cells are involved in microRNAs in cancer (hsa05206), glycolysis/gluconeogenesis (hsa00010), prion diseases (hsa05020), Salmonella infection (hsa05132), and the hematopoietic cell lineage (hsa04640), listed in Figure 5b and Supplementary Table 6.
Supplementary Table 6.
Genes differentially expressed in PC3-peroxisome proliferators-activated receptors γ 2 and control PC3-empty vector cells and their functions in ten representative pathways
| Description | Official symbol | Fold change |
|---|---|---|
| 1. MicroRNAs in cancer | ||
| Cell division cycle 25C | CDC25C | 0.4389 |
| Fibroblast growth factor receptor 3 | FGFR3 | 0.4977 |
| CD44 molecule (Indian blood group) | CD44 | 2.1367 |
| V-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 | ERBB3 | 0.4015 |
| Platelet-derived growth factor alpha polypeptide | PDGFA | 0.4007 |
| 2. Glycolysis/gluconeogenesis | ||
| Aldehyde dehydrogenase 1 family, member A3 | ALDH1A3 | 2.6127 |
| Enolase 2 (gamma, neuronal) | ENO2 | 0.4605 |
| Phosphofructokinase, muscle | PFKM | 0.4613 |
| 3. Prion diseases | ||
| Interleukin 6 | IL6 | 2.631 |
| Early growth response 1 | EGR1 | 0.4598 |
| 4. Salmonella infection | ||
| FBJ murine osteosarcoma viral oncogene homolog | FOS | 0.436 |
| Interleukin 6 | IL6 | 2.631 |
| Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | RAC1 | 2.2166 |
| 5. Hematopoietic cell lineage | ||
| CD24 molecule | CD24 | 0.4604 |
| Interleukin 6 | IL6 | 2.631 |
| CD44 molecule (Indian blood group) | CD44 | 2.1367 |
| 6. Choline metabolism in cancer | ||
| FBJ murine osteosarcoma viral oncogene homolog | FOS | 0.436 |
| Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | RAC1 | 2.2166 |
| Platelet-derived growth factor alpha polypeptide | PDGFA | 0.4007 |
| 7. Toll-like receptor signaling pathway | ||
| Interleukin 6 | IL6 | 2.631 |
| Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | RAC1 | 2.2166 |
| FBJ murine osteosarcoma viral oncogene homolog | FOS | 0.436 |
| 8. Transcriptional misregulation in cancer | ||
| Runt-related transcription factor 2 | RUNX2 | 0.3781 |
| Interleukin 6 | IL6 | 2.631 |
| Platelet-derived growth factor alpha polypeptide | PDGFA | 0.4007 |
| Ets variant 1 | ETV1 | 0.3069 |
| 9. PI3K-Akt signaling pathway | ||
| Fibroblast growth factor receptor 3 | FGFR3 | 0.4977 |
| Interleukin 6 | IL6 | 2.631 |
| Platelet-derived growth factor alpha polypeptide | PDGFA | 0.4007 |
| Glycogen synthase 1 (muscle) | GYS1 | 0.4811 |
| Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | RAC1 | 2.2166 |
| Growth hormone receptor | GHR | 0.4952 |
| 10. AMPK signaling pathway | ||
| Stearoyl-CoA desaturase 5 | SCD5 | 0.4453 |
| Phosphofructokinase, muscle | PFKM | 0.4613 |
| Glycogen synthase 1 (muscle) | GYS1 | 0.4811 |
CD44: cell surface glycoprotein 44
Ingenuity pathway analysis (IPA) can identify published direct binding partners, transcriptional regulators, and translational regulators of specific molecules. In our study, gene expression networks were revealed based on accepted databases of molecular interactions reported in the literature using IPA. Restoration of the PPARγ2 isoform affected the cyclin-dependent kinase inhibitor 1C (CDKN1C), aldehyde dehydrogenase 1 family member A3 (ALDH1A3), cell surface glycoprotein 44 (CD44), ras-related C3 botulinum toxin substrate 1 (RAC1), interleukin-6 (IL6), fos protein (FOS), and cell division cycle 25 homolog C (CDC25C) genes, which play important biological roles in human PCa development and progression (Supplementary Figure 3a (466.7KB, tif) –3e (466.7KB, tif) ).
DISCUSSION
Many tumors are characterized by a drastic change in cellular energy metabolism14 marked by altered glucose15 and lipid metabolism.16 PCa is characterized by the loss of expression of enzymes in the lipid metabolic pathways, which generate natural ligands for PPARγ, and the increases in the expression of enzymes in pathways that generate proinflammatory prostaglandins.17 The treatment of primary cultured human PCa cells with PPARγ agonists was shown to suppress cell growth and proliferation, induce apoptosis, and increase secondary lysosomes and neutral lipid droplets.18 Degenerative lysosomes and autophagocytosis were found to accumulate in PPARγ-deficient mouse prostate epithelial cells both in vitro and in vivo, resulting in mouse prostatic intraepithelial neoplasia,10,11 suggesting that PPARγ signaling plays an important role in maintaining cellular organelle differentiation and mouse prostate organization. However, the molecular mechanisms responsible for these effects remain incompletely elucidated.
Alterations in lipid metabolism resulting in the loss of PPARγ signaling have been suggested to predispose the prostate to premalignant or malignant changes;19 thus, PPARγ may be a key regulator in the maintenance of human prostate epithelial cell homeostasis.20 A previous study indicated that the dyslipidemic phenotypes associated with PPARγ function in mouse prostate epithelia may be linked to isoform-specific expression and function.10 To date, little has been done to describe the functions of the individual PPARγ isoforms in the human prostate, but the consensus seems to be that PPARγ2 in other tissues (adipose, muscle) is responsible for a reduction in lipotoxicity through the conversion of toxic-free fatty acids into less harmful triglycerides.21
In the present study, we found that advanced human PCa cells showed a low level of the endogenous PPARγ2 protein. Restoration of PPARγ2 isoform activity inhibited cell proliferation and migration in PC3 and LNCaP cells and resulted in the reprogramming of cellular lipid- and energy metabolism-related genes in PC3 cells. Tissue recombinants generated with PC3-PPARγ2-restored cells using the TRX mouse model showed the induction of differentiation and necrosis in cancerous regions and leukocyte infiltration in the surrounding stroma. These results indicate that the PPARγ2 isoform may play an important tumor-suppressive role in the development and progression of PCa. In previous studies, PPARγ was found to be activated in metastatic PCa.22 Taken together, the results in our published papers4,6,10,11 and current experimental results indicate that the activated PPARγ proteins localized in the cytoplasm of cancer cells in metastatic PCa are the phosphorylated PPARγ1 isotype, not PPARγ1 or PPARγ2.
The role of autophagy in cancer is complex.23,24 Autophagy may be protumorigenic and promote tumor cell survival.25,26,27 Alternatively, autophagy may represent either a barrier or an adaptive response to cancer.28,29 In the present study, immunohistochemical analysis of tissue recombinants showed that the expression of LC3 was decreased in PC3 cells with restored PPARγ2, which generated cancerous regions, indicating the protumorigenic function of autophagy.
Necroptosis, a form of regulated necrosis, is classically initiated by death receptors, such as tumor necrosis factor receptor 1.30 Necroptotic cell death is dependent on the phosphorylation of MLKL, an essential necroptosis-induced protein that migrates and localizes to the plasma membrane after its phosphorylation and is degraded, which in turn releases intracellular proinflammatory molecules.31,32,33 The evasion of programmed cell death is considered a hallmark of cancers, and this condition can facilitate tumor initiation, progression, and drug resistance.34 Necroptosis provides potential novel molecular targets and strategies for therapeutic intervention because its molecular mechanisms and signaling regulation are distinct from those of apoptosis.35 The expression of MLKL was increased in cancerous regions generated from PC3 cells, in which PPARγ2 was restored, which illustrates the tumor-suppressive function of restoration of PPARγ2.
In summary, the data presented here suggest that modulation of PPARγ2 signaling is a potential approach to inhibit the progression of PCa by triggering an inflammatory reaction- and energy metabolism-associated signaling pathways. Specific PPARγ2 isoform gain of function in PCa cells resulted in cancer cell necrosis with stromal lymphocyte infiltration. These findings have further deepened our understanding of the effects of specific PPARγ2 isoform signaling on human prostate epithelial cells. Such a scenario justifies the consideration of screening specific PPARγ2 agonists as chemopreventive agents to inhibit the pathogenesis of early-stage prostate carcinogenesis (Figure 6). An active surveillance program incorporating preventative agents could be applied in patients and would add an additional noninvasive approach for use in this large patient population. However, the functions of PPARγ, including effects on inflammatory cell recruitment, still require further exploration.
Figure 6.
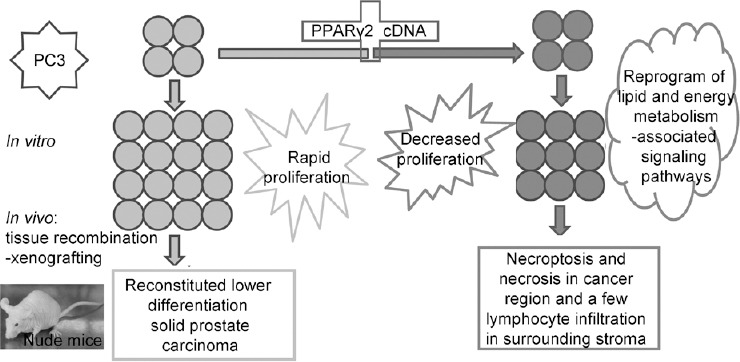
A schematic showing a proposed translational model for functional dissection of the PPARγ2 isoform in human prostate cancer initiation and progression in vitro and in vivo. Specific PPARγ2 isoform gain of function in PC3 advanced human prostate cancer cells resulted in increased cancer cell differentiation and severe necroptosis with stromal leukocyte infiltration. These findings deepen our understanding of the effects of the effects of the specific PPARγ2 isoform on human prostate epithelial cellular homeostasis and provide a novel strategy for preventing prostate carcinogenesis and blocking cancer progression. PPARγ2: peroxisome proliferators-activated receptors γ 2.
AUTHOR CONTRIBUTIONS
YHZ conceived the project. YHZ and FLD designed and performed the experiments and wrote the manuscript. FLD assisted with bioinformatic data analysis and interpretation. DML carried out the genetic studies and participated in the proteomic analysis. TTL, FL, CZ, and QE provided technical assistance. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declared no competing interests.
Restoration of PPARγ2 activity in RWPE cells had no effect on cellular proliferation or migration in vitro. (a) Restoration of specific PPARγ2 isoform activity using WT full-length human PPARγ2 cDNA in RWPE cells by stable transfection was used to create a series of isogenic cell lines for genetic and functional comparisons. PPARγ2 protein expression was validated by western blot analysis. (b) The proliferation of RWPE-PPARγ2-restored cells was not affected. PPARγ2: peroxisome proliferators-activated receptors γ 2.
Restoration of PPARγ2 activity in LNCaP cells has effects on gene expression similar to those in PC3 cells. PPARγ2: peroxisome proliferators-activated receptors γ 2.
Interrelated gene expression networks in PC3-PPARγ2 cells and PC3-EV cells were identified by IPA and compared. (a–e) Five important networks of interrelated target genes were identified in PC3-PPARγ2 cells and PC3-EV control cells by IPA and compared. IPA: ingenuity pathway analysis; PPARγ2: peroxisome proliferators-activated receptors γ 2.
ACKNOWLEDGMENTS
The authors thank Dr. Ming Jiang at Vanderbilt University Medical Center for his critical comments on the manuscript. The work was supported by the National Natural Science Foundation of China (NSFC; No. 81874171 and 81703259).
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Rathkopf DE, Antonarakis ES, Shore ND, Tutrone RF, Alumkal JJ, et al. Safety and antitumor activity of apalutamide (ARN-509) in metastatic castration-resistant prostate cancer with and without prior abiraterone acetate and prednisone. Clin Cancer Res. 2017;23:3544–51. doi: 10.1158/1078-0432.CCR-16-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubois V, Eeckhoute J, Lefebvre P, Staels B. Distinct but complementary contributions of PPAR isotypes to energy homeostasis. J Clin Invest. 2017;127:1202–14. doi: 10.1172/JCI88894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang M, Shappell SB, Hayward SW. Approaches to understanding the importance and clinical implications of peroxisome proliferator-activated receptor gamma (PPAR gamma) signaling in prostate cancer. J Cell Biochem. 2004;91:513–27. doi: 10.1002/jcb.10770. [DOI] [PubMed] [Google Scholar]
- 5.Metzger D, Imai T, Jiang M, Takukawa R, Desvergne B, et al. Functional role of RXRs and PPARgamma in mature adipocytes. Prostaglandins Leukot Essent Fatty Acids. 2005;73:51–8. doi: 10.1016/j.plefa.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Jiang M, Strand DW, Franco OE, Clark PE, Hayward SW. PPARγ: a molecular link between systemic metabolic disease and benign prostate hyperplasia. Differentiation. 2011;82:220–36. doi: 10.1016/j.diff.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byndloss MX, Olsan EE, Rivera-Chávez F, Tiffany CR, Cevallos SA, et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science. 2017;357:570–5. doi: 10.1126/science.aam9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Tan H, Xu D, Ma A, Zhang L, et al. The combinatory effects of PPAR-γ agonist and survivin inhibition on the cancer stem-like phenotype and cell proliferation in bladder cancer cells. Int J Mol Med. 2014;34:262–8. doi: 10.3892/ijmm.2014.1774. [DOI] [PubMed] [Google Scholar]
- 9.Jung Y, Cackowski FC, Yumoto K, Decker AM, Wang Y, et al. Abscisic acid regulates dormancy of prostate cancer disseminated tumor cells in the bone marrow. Neoplasia. 2021;23:102–11. doi: 10.1016/j.neo.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang M, Fernandez S, Jerome WG, He Y, Yu X, et al. Disruption of PPARgamma signaling results in mouse prostatic intraepithelial neoplasia involving active autophagy. Cell Death Differ. 2010;17:469–81. doi: 10.1038/cdd.2009.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang M, Jerome WG, Hayward SW. Autophagy in nuclear receptor PPARgamma-deficient mouse prostatic carcinogenesis. Autophagy. 2010;6:175–6. doi: 10.4161/auto.6.1.10700. [DOI] [PubMed] [Google Scholar]
- 12.Strand DW, Jiang M, Murphy TA, Yi Y, Konvinse KC, et al. PPARγ isoforms differentially regulate metabolic networks to mediate mouse prostatic epithelial differentiation. Cell Death Dis. 2012;3:e361. doi: 10.1038/cddis.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Cao Y, Wang F, Song M, Rui X, et al. 4-Nitrophenol induces activation of Nrf2 antioxidant pathway and apoptosis of the germ cells in rat testes. Environ Sci Pollut Res Int. 2016;23:13035–46. doi: 10.1007/s11356-016-6470-2. [DOI] [PubMed] [Google Scholar]
- 14.Lazar MA. Maturing of the nuclear receptor family. J Clin Invest. 2017;127:1123–5. doi: 10.1172/JCI92949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci. 2016;73:377–92. doi: 10.1007/s00018-015-2070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beyaz S, Mana MD, Roper J, Kedrin D, Saadatpour A, et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature. 2016;531:53–8. doi: 10.1038/nature17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernandez-Quiles M, Broekema MF, Kalkhoven E. PPARgamma in metabolism, immunity, and cancer: unified and diverse mechanisms of action. Front Endocrinol (Lausanne) 2021;12:624112. doi: 10.3389/fendo.2021.624112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galbraith LC, Mui E, Nixon C, Hedley A, Strachan D, et al. PPAR-gamma induced AKT3 expression increases levels of mitochondrial biogenesis driving prostate cancer. Oncogene. 2021;40:2355–66. doi: 10.1038/s41388-021-01707-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu RZ, Choi WS, Jain S, Dinakaran D, Xu X, et al. The FABP12/PPARγ pathway promotes metastatic transformation by inducing epithelial-to-mesenchymal transition and lipid-derived energy production in prostate cancer cells. Mol Oncol. 2020;14:3100–20. doi: 10.1002/1878-0261.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elix C, Pal SK, Jones JO. The role of peroxisome proliferator-activated receptor gamma in prostate cancer. Asian J Androl. 2018;20:238–43. doi: 10.4103/aja.aja_15_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schadinger SE, Bucher NL, Schreiber BM, Farmer SR. PPARgamma2 regulates lipogenesis and lipid accumulation in steatotic hepatocytes. Am J Physiol Endocrinol Metab. 2005;288:E1195–205. doi: 10.1152/ajpendo.00513.2004. [DOI] [PubMed] [Google Scholar]
- 22.Ahmad I, Mui E, Galbraith L, Patel R, Tan EH, et al. Sleeping beauty screen reveals Pparg activation in metastatic prostate cancer. Proc Natl Acad Sci U S A. 2016;113:8290–5. doi: 10.1073/pnas.1601571113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12(Suppl 2):1509–18. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 24.Bergmann A. Autophagy and cell death: no longer at odds. Cell. 2007;131:1032–4. doi: 10.1016/j.cell.2007.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 27.Galluzzi L, Bravo-San Pedro JM, Kroemer G. Defective autophagy initiates malignant transformation. Mol Cell. 2016;62:473–4. doi: 10.1016/j.molcel.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Liu J, Xu Y, Wu RL, Chen X, et al. Tumor heterogeneity in autophagy-dependent ferroptosis. Autophagy. 2021 doi: 10.1080/15548627.2021.1872241. Doi: 10.1080/15548627.2021.1872241. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan I, Yousif A, Chesnokov M, Hong L, Chefetz I. A decade of cell death studies: breathing new life into necroptosis. Pharmacol Ther. 2021;220:107717. doi: 10.1016/j.pharmthera.2020.107717. [DOI] [PubMed] [Google Scholar]
- 31.Dovey CM, Diep J, Clarke BP, Hale AT, McNamara DE, et al. MLKL requires the inositol phosphate code to execute necroptosis. Mol Cell. 2018;70:936–48.e7. doi: 10.1016/j.molcel.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samson AL, Zhang Y, Geoghegan ND, Gavin XJ, Davies KA, et al. MLKL trafficking and accumulation at the plasma membrane control the kinetics and threshold for necroptosis. Nat Commun. 2020;11:3151. doi: 10.1038/s41467-020-16887-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karunakaran D, Nguyen MA, Geoffrion M, Vreeken D, Lister Z, et al. RIPK1 expression associates with inflammation in early atherosclerosis in humans and can be therapeutically silenced to reduce NF-κB activation and atherogenesis in mice. Circulation. 2021;143:163–77. doi: 10.1161/CIRCULATIONAHA.118.038379. [DOI] [PubMed] [Google Scholar]
- 34.Baidya R, Gautheron J, Crawford DH, Wang H, Bridle KR. Inhibition of MLKL attenuates necroptotic cell death in a murine cell model of ischaemia injury. J Clin Med. 2021;10:212. doi: 10.3390/jcm10020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong Y, Fan Z, Luo G, Yang C, Huang Q, et al. The role of necroptosis in cancer biology and therapy. Mol Cancer. 2019;18:100. doi: 10.1186/s12943-019-1029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Restoration of PPARγ2 activity in RWPE cells had no effect on cellular proliferation or migration in vitro. (a) Restoration of specific PPARγ2 isoform activity using WT full-length human PPARγ2 cDNA in RWPE cells by stable transfection was used to create a series of isogenic cell lines for genetic and functional comparisons. PPARγ2 protein expression was validated by western blot analysis. (b) The proliferation of RWPE-PPARγ2-restored cells was not affected. PPARγ2: peroxisome proliferators-activated receptors γ 2.
Restoration of PPARγ2 activity in LNCaP cells has effects on gene expression similar to those in PC3 cells. PPARγ2: peroxisome proliferators-activated receptors γ 2.
Interrelated gene expression networks in PC3-PPARγ2 cells and PC3-EV cells were identified by IPA and compared. (a–e) Five important networks of interrelated target genes were identified in PC3-PPARγ2 cells and PC3-EV control cells by IPA and compared. IPA: ingenuity pathway analysis; PPARγ2: peroxisome proliferators-activated receptors γ 2.


