ABSTRACT
The type VI secretion system (T6SS) is an important translocation apparatus that is widely employed by Gram-negative bacteria to deliver toxic effectors into eukaryotic and prokaryotic target cells, causing host damage and providing competitive advantages in polymicrobial environments. The genome of Pseudomonas aeruginosa harbors three T6SS clusters (H1-T6SS, H2-T6SS, H3-T6SS). Activities of these systems are tightly regulated by a complicated signaling network which remains largely elusive. In this study, we focused on a previously characterized two-component system FleS/FleR, and performed comparative transcriptome analysis between the PAO1 wild-type strain and its isogenic ΔfleR mutant, which revealed the important role of FleS/FleR in regulating multiple physiological pathways including T6SS. Gene expression and bacterial killing assays showed that the expression and activity of H1-T6SS are repressed in the wild-type strain owing to the high intracellular c-di-GMP content. Further explorations demonstrated that c-di-GMP relies on the transcription factor FleQ to repress H1-T6SS and its synthesis is controlled by a global regulator AmrZ which is induced by the active FleS/FleR. Interestingly, repression of H1-T6SS by FleS/FleR in PAO1 is independent of RetS which is known to regulate H1-T6SS by controlling the central post-transcriptional factor RsmA. Together, our results identified a novel regulator of H1-T6SS and provided detailed mechanisms of this signaling pathway in PAO1.
IMPORTANCE Pseudomonas aeruginosa is an opportunistic human pathogen distributed widely in the environment. The genome of this pathogen contains three T6SS clusters which contribute significantly to its virulence. Understanding the complex regulatory network that controls the activity of T6SS is essential for the development of effective therapeutic treatments for P. aeruginosa infections. In this study, transcriptome analysis led to the identification of a novel regulator FleS/FleR which inversely regulates H1-T6SS and H2-T6SS in P. aeruginosa PAO1. We further revealed a detailed FleS/FleR-mediated regulatory pathway of H1-T6SS in PAO1 which involves two additional transcriptional regulators AmrZ and FleQ and the second messenger c-di-GMP, providing important implications to develop novel anti-infective strategies and antimicrobial drugs.
KEYWORDS: Pseudomonas aeruginosa, two-component system, FleS/FleR, T6SS, c-di-GMP
INTRODUCTION
The type VI secretion system (T6SS), first discovered in 2006, is an important virulence determinant distributed in more than 200 Gram-negative bacteria such as Pseudomonas aeruginosa, Escherichia coli, Burkholderia thailandensis, Vibrio cholerae, Serratia marcescens, and so forth (1–5). This system is composed of a set of core conserved genes including TssA-TssM, Hcp, VgrG, and ClpV to form its key structure which shares high similarity with the puncturing device of tailed bacteriophages (6). T6SS is known as a versatile secretion system. In addition to its function as a contractile molecular syringe to deliver toxins into neighboring competitors or translocate protein effectors into host cells which results in growth inhibition or death of the target cells, T6SS is involved in multiple other physiological processes such as biofilm formation and metal acquisition (7–9).
P. aeruginosa is an important opportunistic Gram-negative pathogen which can cause a variety of acute and chronic infections. Three independent T6SS clusters, namely, H1-T6SS, H2-T6SS, and H3-T6SS, have been identified in the genome of P. aeruginosa so far. H1-T6SS is the first discovered and well-characterized T6SS machinery displaying antibacterial activity by delivering at least seven different protein effectors, e.g., Tse1 to Tse7, providing a fitness advantage for P. aeruginosa in competition with other bacteria and contributing significantly to its infections in hosts (10–12). For example, Tse1 exhibits strong peptidoglycan-degrading activity which endows P. aeruginosa with a great ability to lyse bacterial competitors (13). Different from the H1-T6SS, the H2-T6SS and H3-T6SS can target both prokaryotic and eukaryotic cells using the effectors such as PldA and PldB (14–16).
T6SS activity in P. aeruginosa is tightly regulated at (post)transcriptional and (post)translational levels. Accumulating evidence has shown that T6SS is regulated by many factors such as the quorum sensing system (QS), copper-responsive regulator CueR, RNase YbeY, global regulator AmrZ, endogenous membrane stress, and the threonine phosphorylation pathway (TPP) (7, 17–22). Despite these progresses, the dominant factor controlling T6SS activity in P. aeruginosa is still regarded as the RNA binding protein RsmA which represses T6SS at the post-transcriptional level (22). RsmA is located downstream of the Gac/Rsm cascade which is responsible for the transitions of acute and chronic phases of P. aeruginosa infections (23). In this cascade, GacS phosphorylates GacA leading to the production of two small RNAs rsmY and rsmZ which are capable of sequestering RsmA and consequently de-repress the translation of T6SS mRNAs (24). Activity of the Gac/Rsm pathway is inversely controlled by two additional histidine kinases LadS and RetS which stimulates and represses the activity of the signaling pathway, respectively (25, 26). During in vitro growth, it has been reported that T6SS is poorly assembled in the strains with ordinary expression of retS and deletion of retS is required to obtain a fully active T6SS in P. aeruginosa (12, 27). However, whether additional regulatory factors exist to modulate T6SS activity remains largely unknown.
Two-component systems (TCSs) represent a group of important bacterial regulatory factors that control expression profiles of many genes or pathways in response to changing environments (28). In P. aeruginosa, a TCS FleS/FleR was previously characterized to mainly regulate bacterial motility and biofilm formation (29, 30). While interestingly, unlike canonical TCSs which consist of a transmembrane sensor histidine kinase, the histidine kinase FleS was found not containing a transmembrane domain (29). In order to comprehensively understand the biological roles of this peculiar TCS, in this study, we performed transcriptome analysis and discovered that FleS/FleR is a novel regulator of T6SS. We further investigated the detailed molecular mechanisms of how FleS/FleR controlled H1-T6SS activity in PAO1 and showed a RetS-independent signaling cascade that was composed of AmrZ, c-di-GMP, and FleQ. These findings expanded our understanding in the complexity of T6SS regulation and provided significant implications for therapeutic treatments for P. aeruginosa infections.
RESULTS
FleS/FleR regulates multiple physiological pathways in PAO1.
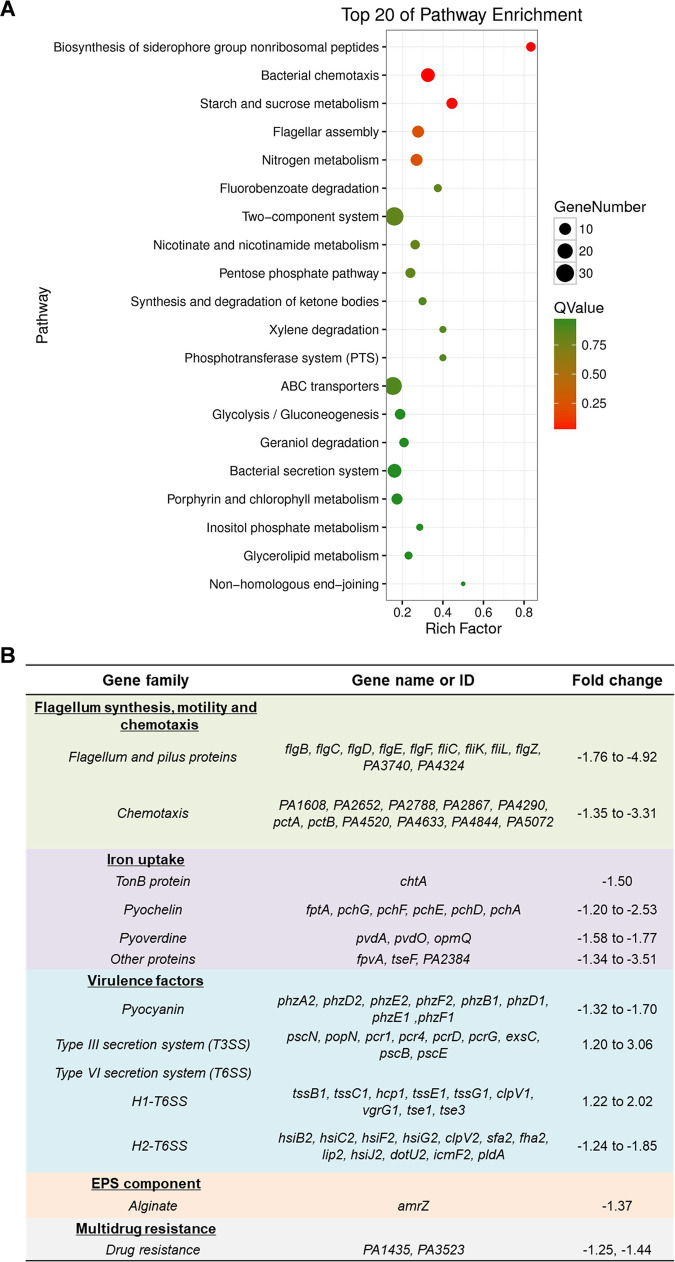
The TCS FleS/FleR plays important roles in regulating biofilm formation and motility in P. aeruginosa (29, 30). To further investigate whether this TCS is involved in regulating other physiological pathways in P. aeruginosa, we deleted fleR in PAO1 and conducted RNA-sequencing (RNA-seq) to compare its transcriptomic profiles with the wild-type PAO1 strain (PAO1 WT). A total of 440 differentially expressed genes with more than 1.2-log2fold changes were identified between these two strains (Table S1). Specifically, 121 genes were downregulated and 319 genes were upregulated in the fleR deletion mutant. To summarize the differentially expressed genes into interpretable pathways, enrichment analysis was performed for these genes based on the KEGG pathways (31), which showed that these genes were enriched in 20 pathways such as bacterial chemotaxis, flagellar assembly, two-component system, ABC transporters, and bacterial secretion system, etc. (Fig. 1A). As listed in Fig. 1B, a number of these genes were found to be associated with bacterial virulence and antibiotic resistance. Some of them were related to flagellar synthesis, motility, and chemotaxis, and this was in accordance with the previously reported regulatory functions of FleS/FleR in biofilm formation and motility (29).
FIG 1.
FleS/FleR regulates multiple physiological pathways in PAO1. (A) KEGG-enrichment of differentially expressed genes identified in the RNA-seq analysis. The y axis represents the names of the pathways. The x axis represents the rich factor which refers to the ratio of the number of differentially expressed genes to the total number of genes in the pathway. The size of the dot represents the number of differentially expressed genes in the pathways, and the color of the dot represents different Q values. The higher the value of rich factor represents the greater the enrichment degree. The smaller the Q value represents the more significant the enrichment. (B) Selected gene families related to bacterial virulence and drug resistance with more than 1.2-log2fold changes owing to the deletion of fleR in PAO1 (ΔfleR versus PAO1 WT).
FleR differentially regulates the expression of three T6SS clusters in PAO1 and repressed H1-T6SS is the major reason causing reduced antibacterial capacity.
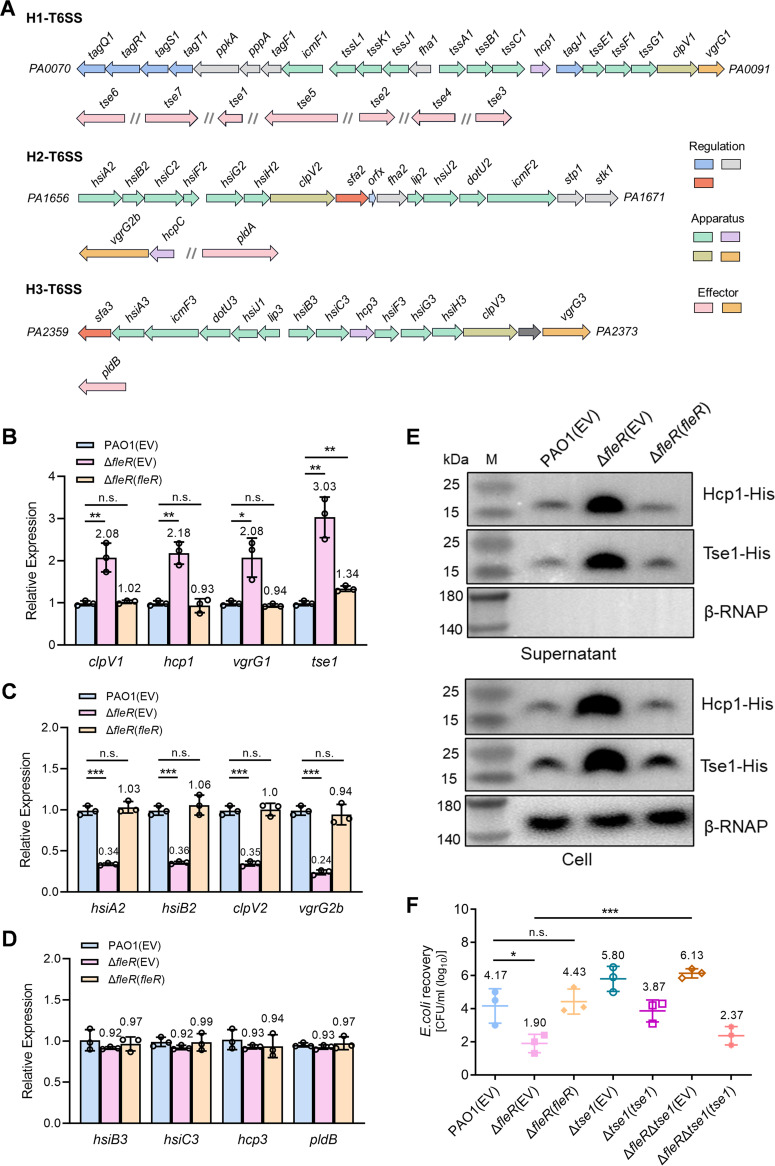
PAO1 contains three T6SS gene clusters in its genome which are named as H1-T6SS, H2-T6SS, and H3-T6SS, respectively (Fig. 2A) (5). It was interesting to notice that the expression of H1-T6SS and H2-T6SS were inversely regulated in the absence of fleR and expression changes of H3-T6SS genes were not detected (Fig. 1B), suggesting that FleS/FleR could be a novel regulator of T6SS in P. aeruginosa which displayed different regulatory mechanisms on different T6SS clusters. To verify the RNA-seq result, we selected four genes from each T6SS locus and performed RT-qPCR to examine their expression. Consistent with the RNA-seq result, deletion of fleR in PAO1 resulted in significantly upregulated expression of H1-T6SS genes, downregulated expression of H2-T6SS genes and undetectable expression changes of H3-T6SS genes (Fig. 2B to D), which indicated that FleR negatively regulated H1-T6SS and positively regulated H2-T6SS. Given that H1-T6SS in PAO1 is the first discovered and the most well-studied system (10, 12), in the present study, we decided to focus on H1-T6SS and aimed to reveal the molecular mechanisms underlying the regulation of FleS/FleR on this system.
FIG 2.
FleR differentially regulates the expression of three T6SS clusters in PAO1 and repressed H1-T6SS is the major reason causing reduced anti-bacterial capacity of PAO1. (A) A diagram showing the three T6SS gene clusters in P. aeruginosa PAO1. (B) Relative expression of H1-T6SS genes clpV1, hcp1, vgrG1, and tse1 measured by RT-qPCR in the strains of PAO1, ΔfleR and ΔfleR with ectopic expression of fleR (ΔfleR(fleR)). (C) Relative expression of H2-T6SS genes hsiA2, hsiB2, clpV2, and vgrG2b measured by RT-qPCR in PAO1, ΔfleR, and ΔfleR(fleR) strains. (D) Relative expression of H3-T6SS genes hsiB3, hsiC3, hcp3, and pldB measured by RT-qPCR in PAO1, ΔfleR, and ΔfleR(fleR) strains. (E) Western blot analysis of Hcp1-His and Tse1-His in the cell-associated and concentrated supernatant protein fractions from PAO1, ΔfleR, and ΔfleR(fleR) strains. The RNA polymerase β subunit (β-RNAP) was selected as an internal control for the assay. (F) Bacterial killing assay between the PAO1 strains and the E. coli prey. Data are represented as the mean ± SD (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus the indicated group based on paired Student's t test. n.s., not significant; EV, empty vector for the control.
To further confirm whether H1-T6SS is repressed by FleR, we selected a structural gene hcp1 and an effector-encoding gene tse1 from H1-T6SS and constructed the chromosomal His-tagged hcp1 and tse1 in PAO1 WT and ΔfleR strains to examine their protein productions by conducting the Western blot assay. Consistent with the RT-qPCR result, it showed that productions of both Hcp1 and Tse1 proteins were substantially increased in the ΔfleR strain which could be completely abolished with the ectopic expression of FleR (Fig. 2E). Considering that H1-T6SS is essential for the antibacterial activity of P. aeruginosa and Tse1 plays an important role in cell lysis of the competing bacterial cells (10), we moved to investigate whether increased expression of the H1-T6SS in ΔfleR could enhance its anti-bacterial ability. We assessed the antibacterial abilities of PAO1 WT and ΔfleR by performing a bacterial killing assay using E. coli as the prey. The survival rate of E. coli was significantly reduced when it was cocultured with ΔfleR compared with the PAO1 WT (Fig. 2F). The survival rate of E. coli was recovered when it was cocultured with ΔfleR which was complemented with an FleR-expressing vector (Fig. 2F), demonstrating that FleR repressed the antibacterial activity of H1-T6SS. We also found that Tse1 was the key antibacterial effector regulated by FleR because further deletion of tse1 in PAO1 ΔfleR completely abolished its killing ability (Fig. 2F). These results demonstrated that FleR inhibited the antibacterial activity of PAO1 by repressing the expression of H1-T6SS and its main effector Tse1.
FleR regulates H1-T6SS in a c-di-GMP-dependent manner.
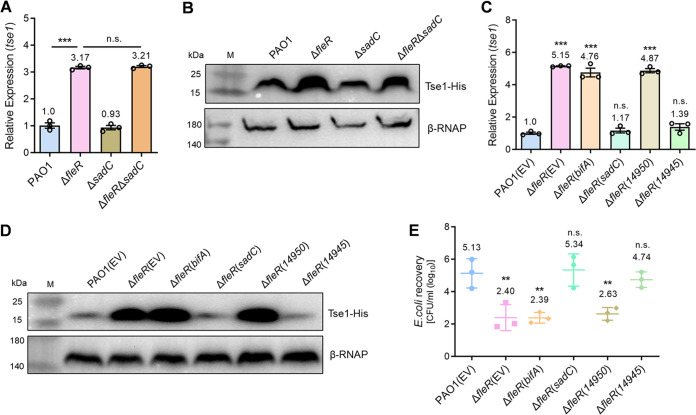
Interestingly, RNA-seq and RT-qPCR results showed that a group of diguanylate cyclase (DGCs) genes responsible for c-di-GMP synthesis including siaD, gcbA, sadC, mucR, and PA4929 were significantly downregulated in the ΔfleR strain (Table S1 and Fig. S1A); however, phosphodiesterases (PDEs) genes such as bifA, PA2071, and PA3825 responsible for c-di-GMP degradation were not influenced (Fig. S1B). Based on these observations in combination with a previous report that c-di-GMP inactivates T6SS activity in Agrobacterium tumefaciens (32), we speculated that FleR regulated H1-T6SS by modulating intracellular c-di-GMP contents. To examine this speculation, we first evaluated the intracellular c-di-GMP contents in PAO1 WT and ΔfleR strains using the expression of cdrA which is directly responsive to changing cellular c-di-GMP levels as an indicator (33–35). It was shown that deletion of fleR led to a significantly reduced expression of cdrA and such reduction could be fully restored by ectopic expression of FleR (Fig. S1C), indicating that FleR acted as an activator of c-di-GMP synthesis and higher intracellular c-di-GMP content potentially inhibited the H1-T6SS activity. We next examined the regulatory role of c-di-GMP in ΔfleR by either deleting or ectopically expressing a DGC gene sadC. Further deletion of sadC in ΔfleR did not cause obvious changes of hcp1 and tse1 expression (Fig. 3A and B, Fig. S2A), which was reasonable because sadC was already repressed to a low expression level in the absence of fleR. Interestingly, ectopic expression of sadC in ΔfleR to increase the c-di-GMP content completely abolished the elevated expression of hcp1 and tse1 (Fig. 3C and D, Fig. S2B). Consistently, the enhanced antibacterial activity of ΔfleR was abolished with the ectopic expression of sadC (Fig. 3E). In contrast, ectopic expression of a PDE gene bifA in ΔfleR did not show any differences on the hcp1 and tse1 expression as well as the antibacterial activity of this mutant (Fig. 3C to E, Fig. S2B). These results suggested that c-di-GMP mediated the regulation of FleR on H1-T6SS in PAO1.
FIG 3.
FleR regulates H1-T6SS in a c-di-GMP-dependent manner. (A) Relative expression of tse1 measured by RT-qPCR in the PAO1, ΔfleR, ΔsadC, and ΔfleR ΔsadC strains. (B) Western blot analysis of Tse1-His in the cell-associated protein fractions from PAO1, ΔfleR, ΔsadC, and ΔfleR ΔsadC strains. (C) Relative expression of tse1 measured by RT-qPCR in the strains of PAO1, ΔfleR, and ΔfleR with ectopic expression of DGCs and PDEs. (D) Western blot analysis of Tse1-His in the cell-associated protein fractions from the PAO1, ΔfleR, and ΔfleR strains with ectopic expression of DGCs and PDEs. (E) Bacterial killing assay between the indicated PAO1 strains and the E. coli prey. Data are represented as the mean ± SD (n = 3). **, P < 0.01; ***, P < 0.001 versus the indicated group based on paired Student's t test (A) or versus PAO1(EV) based on one-way ANOVA (C, E). n.s., not significant; EV, empty vector for the control.
Given that ectopic expression of the native DGC and PDE genes might coordinate other cellular activities other than to solely increase the c-di-GMP content as we expected, we chose two more genes (a DGC gene W909_14945 and a PDE gene W909_14950) involved in c-di-GMP metabolism from Dickeya zeae EC1 (36) and expressed them in ΔfleR, respectively. Consistent with the native genes sadC and bifA, expression of the heterologous DGC gene W909_14945 totally abolished the elevated expression of hcp1 and tse1 as well as the antibacterial activity of ΔfleR while W909_14950 did not generate any differences (Fig. 3C to E, Fig. S2B). These results confirmed that FleR functions to activate c-di-GMP synthesis which consequently inhibits the H1-T6SS activity.
FleQ represses H1-T6SS activity in response to c-di-GMP.
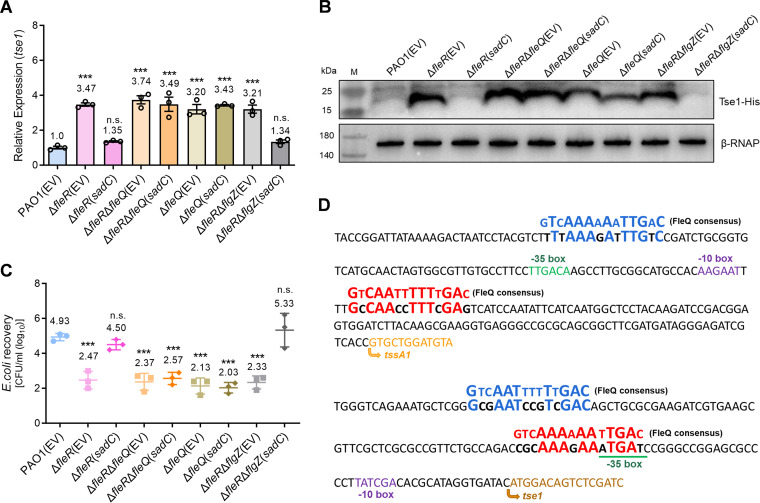
It is known that c-di-GMP exerts its regulatory roles through interacting with effector proteins or RNAs (37). Reported effectors include proteins containing the PilZ domain, degenerate GGDEF or EAL domains, transcriptional regulators, and mRNA riboswitches (38–40). In P. aeruginosa, proteins such as FleQ and FlgZ have been identified as c-di-GMP effectors to regulate a variety of physiological processes including bacterial chemotaxis, biofilm formation, motility, etc. (38, 39, 41, 42). We therefore moved to investigate whether the activity of H1-T6SS regulated by c-di-GMP was mediated by the effectors FleQ or FlgZ. We deleted FleQ and FlgZ in the ΔfleR mutant, respectively, and then introduced plasmids to ectopically express the DGC gene sadC. RT-qPCR showed that ectopic expression of sadC to increase intracellular c-di-GMP was incapable of restoring the hcp1 and tse1 expression in ΔfleR to the wild-type level when FleQ was absent (Fig. 4A, Fig. S3). Western blot examination and bacterial killing assay further confirmed that FleQ was indispensable to repress H1-T6SS activity under the c-di-GMP overproducing condition (Fig. 4B and C). In contrast, further deletion of flgZ in ΔfleR had no effect on the reduced tse1 expression and antibacterial activity during ectopic expression of sadC (Fig. 4A to C). Moreover, in the PAO1 WT which possessed an active FleR and contained relatively high c-di-GMP level, deletion of FleQ also increased the hcp1 and tse1 expression as well as the antibacterial activity (Fig. 4A to C, Fig. S3). These results demonstrated that FleQ was the key effector that responded to c-di-GMP and played an essential role in mediating the regulation of H1-T6SS activity by FleR.
FIG 4.
FleQ is the effector that responds to intracellular c-di-GMP and modulates the activity of H1-T6SS. (A) Relative expression of tse1 measured by RT-qPCR in the indicated PAO1 strains. (B) Western blot analysis of Tse1-His in the cell-associated protein fractions from the indicated PAO1 strains. (C) Bacterial killing assay between the indicated PAO1 strains and the E. coli prey. (D) Prediction of FleQ binding sites in the promoters of the gene cluster tssA1B1C1 and the effector gene tse1 by searching for the FleQ consensus sites (in blue and in red). Data are represented as the mean ± SD (n = 3). ***, P < 0.001 versus PAO1(EV) based on one-way ANOVA. n.s., not significant. EV, empty vector for the control.
It has been demonstrated that FleQ regulates expression of flagellar and biofilm genes by directly binding to the conserved sequence in the promoters of genes such as flhA, filE, fliL, psl, pel, etc. (41, 43). Therefore, we searched potential FleQ binding sites in promoters of H1-T6SS genes by aligning the promoter sequences with the consensus DNA binding sequence for FleQ. Sequence alignment identified two putative FleQ binding sites in the promoter regions of the H1-T6SS structural gene cluster tssA1B1C1 and the effector gene tse1, respectively, which shared high similarities with the FleQ DNA binding consensus sequence (Fig. 4D). This analysis suggested that FleQ might regulate H1-T6SS activity by directly interacting with the T6SS promoters.
The regulation of c-di-GMP synthesis and H1-T6SS activity by FleR is mediated by AmrZ.
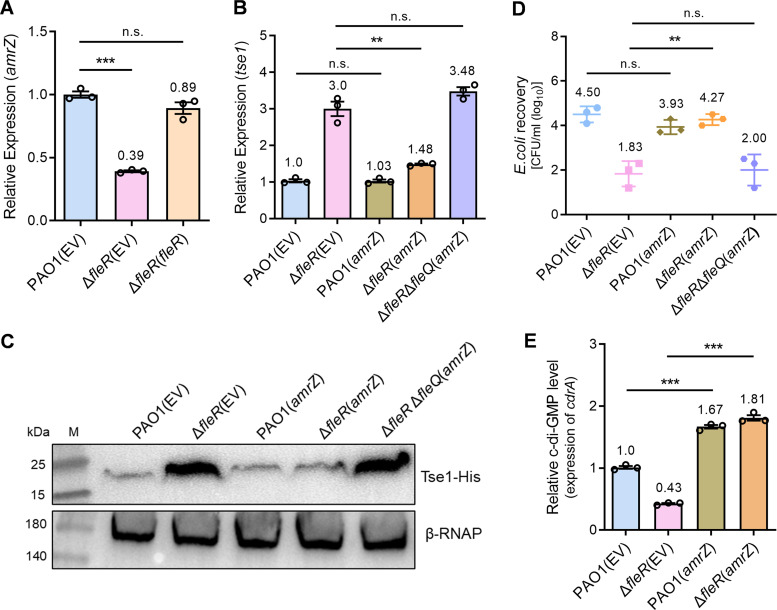
To understand how FleR induced intracellular c-di-GMP contents, we next purified FleR protein and conducted electrophoretic mobility shift assay (EMSA) to examine whether FleR could bind to the promoters of DGC genes to trigger their transcription. However, none of the five DGC genes promoters siaD, gcbA, sadC, mucR, or PA4929 showed interaction with FleR (Fig. S4), implying that FleR did not induce the expression of these DGC genes directly. Interestingly, our RNA-seq data in combination with RT-qPCR verification identified a global regulator gene amrZ which was significantly downregulated in the ΔfleR mutant (Fig. 1B and 5A). AmrZ is conserved in pseudomonads and has been reported as a major determinant of intracellular c-di-GMP levels by controlling the expression of a complex network of DGC and PDE genes in Pseudomonas fluorescens F113 and regulates bacterial motility in a c-di-GMP-dependent manner in P. aeruginosa PA14 (44, 45). Therefore, we supposed that AmrZ could be the primary target of FleR which subsequently modulated the c-di-GMP content and H1-T6SS activity in PAO1. To test this hypothesis, we expressed amrZ in the ΔfleR mutant and monitored the hcp1 and tse1 expression. RT-qPCR showed that ectopic expression of amrZ significantly reduced the expression of both genes in ΔfleR to the level similar as that in the PAO1 WT while ectopic expression of amrZ in PAO1 WT did not show obvious expression changes compared with the vector control (Fig. 5B, Fig. S5). Moreover, Western blot analysis and bacterial killing assay displayed that ectopic expression of amrZ in ΔfleR significantly reduced its Tse1 production and antibacterial activity, respectively (Fig. 5C and D), confirming that AmrZ mediated the regulation of H1-T6SS.
FIG 5.
The regulation of c-di-GMP synthesis and H1-T6SS activity by FleR is mediated by AmrZ. (A) Relative expression of amrZ measured by RT-qPCR in PAO1, ΔfleR, and ΔfleR(fleR) strains. (B) Relative expression of tse1 measured by RT-qPCR in the strains of PAO1 and ΔfleR with ectopic expression of amrZ. (C) Western blot analysis of Tse1-His in the cell-associated protein fractions from the strains of PAO1 and ΔfleR with ectopic expression of amrZ. (D) Bacterial killing assay between the indicated PAO1 strains and the E. coli prey. (E) Relative intracellular c-di-GMP levels in the indicated PAO1 strains were determined by measuring the relative expression of cdrA. Data are represented as the mean ± SD (n = 3). **, P < 0.01; ***, P < 0.001 versus the indicated group based on paired Student's t test. n.s., not significant; EV, empty vector for the control.
We next assessed the intracellular c-di-GMP content in ΔfleR with the ectopic expression of amrZ, which showed a significant increase of the c-di-GMP content in this strain relative to the ΔfleR strain containing the vector control (Fig. 5E). Moreover, we deleted fleQ in the ΔfleR strain with the ectopic expression of amrZ and examined the expression of hcp1 and tse1 as well as the antibacterial activity to further ensure the role of amrZ in H1-T6SS regulation was mediated by c-di-GMP and FleQ. As shown in Fig. 5B to D and Fig. S5, deletion of FleQ completely blocked the AmrZ-mediated repression of H1-T6SS. These results demonstrated that AmrZ transduced the signal from FleR to increase intracellular c-di-GMP synthesis and thereafter promoted H1-T6SS activity.
FleR regulates H1-T6SS in a novel pathway which is independent of RetS.
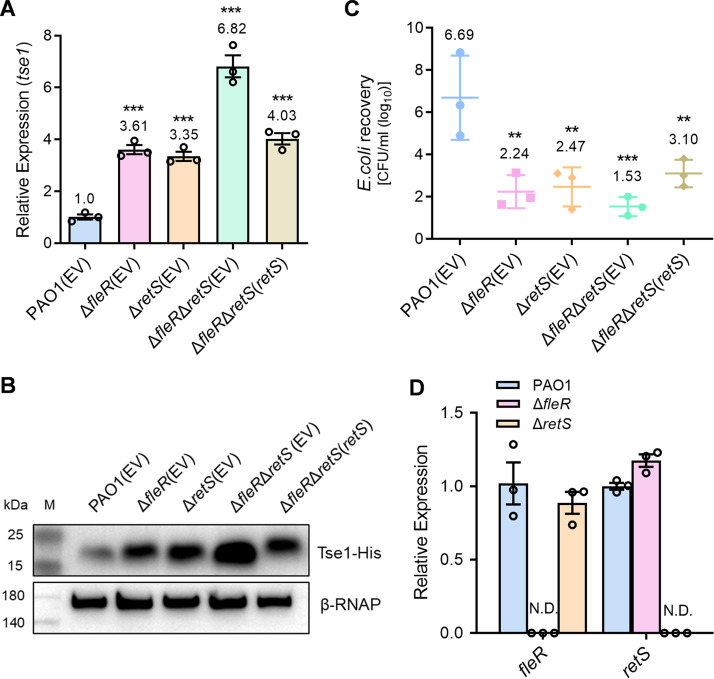
RetS is an important sensor kinase that represses H1-T6SS in P. aeruginosa via the Gac/Rsm pathway where RsmA is suggested as a central regulator silencing the translation of T6SS mRNAs (26). It has been well demonstrated that mutation of RetS is necessary to achieve a fully active H1-T6SS in P. aeruginosa under laboratory conditions (46). Moreover, it was reported that RetS switches expression of T3SS and H1-T6SS in a c-di-GMP-dependent manner in the P. aeruginosa PAK strain (47). Here, whether FleR interplayed with RetS in regulating H1-T6SS was not known. To clarify this, we further constructed a retS deletion mutant ΔretS and a fleR retS double deletion mutant ΔfleR ΔretS, and measured hcp1 and tse1 expression and antibacterial activity of these mutants. ΔfleR and ΔretS single deletion mutants displayed similar levels of the hcp1 and tse1 expression as well as the antibacterial activity, which were significantly higher than the PAO1 WT (Fig. 6A to C, Fig. S6). Intriguingly, double deletion of fleR and retS resulted in an additive effect of two single deletion mutants on the hcp1 and tse1 expression and the antibacterial activity (Fig. 6A to C, Fig. S6), suggesting that FleR and RetS pathways were independent in regulating H1-T6SS. We also determined FleR expression in ΔretS and RetS expression in ΔfleR, which showed no differences in both cases (Fig. 6D), further indicating that these two pathways were independent.
FIG 6.
FleR and RetS repress the activity of H1-T6SS independently. (A) Relative expression of tse1 measured by RT-qPCR in the PAO1, ΔfleR, ΔretS, and ΔfleR ΔretS strains. (B) Western blot analysis of Tse1-His in the cell-associated protein fractions from the strains of PAO1, ΔfleR, ΔretS, and ΔfleR ΔretS. (C) Bacterial killing assay between the indicated PAO1 strains and the E. coli prey. (D) Relative expression of fleR and retS in the PAO1, ΔfleR, and ΔretS strains. Data are represented as the mean ± SD (n = 3). **, P < 0.01; ***, P < 0.001 versus PAO1(EV) based on one-way ANOVA. N.D., not detected; EV, empty vector for the control.
DISCUSSION
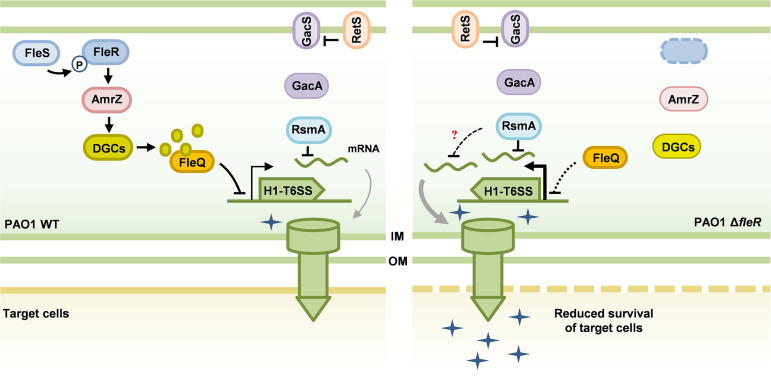
The T6SS is a powerful weapon employed by many Gram-negative bacteria to subvert or kill neighboring prey cells via injecting toxins and protein effectors (48). Understanding the regulation of T6SS in pathogens is important owing to the great contributions of T6SS in causing host infectious diseases and maintaining competitive advantages in polymicrobial communities (12). P. aeruginosa is a notorious opportunistic human pathogen equipped with three T6SS gene clusters and represents a huge threat of lifelong infections. Despite increasing efforts that have been made to elucidate the structure, biogenesis, and secreted effectors of T6SS in recent years, regulation of T6SS activity is still largely unclear. Facilitated by high-throughput RNA-seq analysis, we discovered the two-component system FleS/FleR is a novel regulator of T6SS. Interestingly, FleS/FleR regulates three T6SS clusters in PAO1 in completely different manners, e.g., downregulation of H1-T6SS, upregulation of H2-T6SS and no changed expression of H3-T6SS. In this study, we focused on the elucidation of the regulatory mechanism of H1-T6SS and demonstrated that FleR can induce the intracellular c-di-GMP content in an AmrZ-dependent manner and the elevated c-di-GMP further represses the H1-T6SS expression and antibacterial activity of PAO1 through the transcription regulator FleQ. Moreover, this regulatory circuit showed independency of the RetS pathway (Fig. 7).
FIG 7.
A model illustrating the regulation of H1-T6SS expression by FleS/FleR in PAO1. H1-T6SS is poorly expressed in PAO1 WT under laboratory conditions, which is mainly achieved by the RetS/Gac/Rsm pathway. The RetS/Gac/Rsm pathway regulates H1-T6SS at post-transcriptional level by release or sequester the central factor RsmA. In this study, we revealed a novel pathway under the control of the two-component system FleS/FleR which is independent of the RetS/Gac/Rsm pathway. FleS/FleR activates AmrZ to induce the synthesis of intracellular c-di-GMP, which subsequently represses the expression and activity of the H1-T6SS through FleQ. In the ΔfleR mutant, reduced c-di-GMP content leads to the de-repression and activation of H1-T6SS regardless of the presence of RetS and RsmA with unknown mechanisms.
FleS/FleR was previously identified to control biofilm formation and motility in P. aeruginosa (49). In this study, our RNA-seq result further revealed that FleS/FleR are also involved in regulating many other physiological pathways, including those associated with bacterial virulence and antibiotic resistance, highlighting the versatile regulatory roles of FleS/FleR in host adaptation. Unfortunately, physiologically relevant signals of FleS/FleR have not yet been identified, so it is still unclear how FleS/FleR is activated or repressed in responsive to the host environment. Because deletion of fleR significantly downregulated virulence traits related to acute infections such as motility, chemotaxis, iron uptake, and pyocyanin production, it seems that FleS/FleR is probably activated at the early stage of infection and functions predominantly to establish acute infections. Although we found that FleS/FleR represses the activity of H1-T6SS, which is a typic response during acute infection, it is still confusing how H2-T6SS is induced by FleS/FleR and what is the physiological function of the induced H2-T6SS.
AmrZ is a conserved global regulator in pseudomonads which binds hundreds of genomic regions to control many physiological pathways implicated in environmental adaptation (50, 51). A previous study demonstrated that AmrZ represses H2-T6SS and activates H1-T6SS and H3-T6SS through directly binding to T6SS promoters in PA14 (22). However, in PAO1, we showed that AmrZ inactivates H1-T6SS under the regulation of FleR, indicating the different regulatory patterns among P. aeruginosa isolates. Moreover, the regulation of H1-T6SS in this case seems more possibly dependent on the intracellular c-di-GMP contents rather than directly controlled by AmrZ. This is supported by the strong negative correlation between the H1-T6SS activity and the intracellular c-di-GMP content and the strong positive correlation between the intracellular c-di-GMP content and the AmrZ expression level we observed. We examined the expression of five major DGC genes to look for the potential target of AmrZ. Although we found three genes siaD, sadC, and PA4929, were upregulated significantly in ΔfleR with the ectopic expression of amrZ, all only showed a slight fold change (∼2 fold) (Fig. S7). A comprehensive evaluation of all the DGCs and PDEs is required to determine the role of AmrZ in regulating intracellular c-di-GMP contents. On the other hand, we showed that AmrZ is a key component bridging FleS/FleR and H1-T6SS, but how AmrZ is controlled by FleR requires further investigation. In this study, EMSA examination excluded the possibility of direct binding of FleR to the AmrZ promoter (Fig. S4) and RNA-seq analysis did not show significant expression changes of sigma factor genes such as algU, which was known to control the AmrZ expression (data not shown) (52).
Although T6SS expression is regulated by a complicated signaling network in P. aeruginosa, RsmA is regarded as the dominant regulator repressing T6SS activity by blocking the translation of all the T6SS mRNAs. Therefore, de-repression of T6SS by sequestering RsmA is a prerequisite for investigations on T6SS activity, which is commonly achieved by the deletion of retS. Interestingly, we found that deletion of FleR is adequate to induce the activity of H1-T6SS at both transcriptional and translational levels in PAO1 as evidenced by the RT-qPCR and Western blot examinations. This induction is independent of RetS, and simultaneous deletion of fleR and retS led to an additive induction of the H1-T6SS activity. However, it is still unknown how the signaling bypasses the RetS/Gac/Rsm pathway in PAO1 because it has been shown that all T6SS mRNAs are silenced by RsmA in spite of the regulation of T6SS transcripts by AmrZ in PA14 (22). Nonetheless, these results further highlight the complexity of the regulation of T6SS and the discrepancies of signaling pathways among P. aeruginosa isolates (53). Notably, H1-T6SS and H2-T6SS components were detected at 25°C in PA14 even in the presence of RetS and RsmA (22), which indicated that expression of H1-T6SS does not require the sequestration of RsmA under some peculiar conditions. Thus, it will be interesting to figure out whether additional factors exist to receive signals from FleS/FleR and regulate H1-T6SS activity in PAO1.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and culture conditions.
Strains and plasmids used in this study are summarized in Table 1. Primers used in this study are listed in Table 2. The plasmid pK18mobsacB was used for gene deletion in PAO1. The plasmids pBBR1-MCS5 and pUCP18 (for bacterial killing assays) were used for gene complementation. Unless indicated otherwise, PAO1 and its isogenic mutants were cultured at 37°C in Luria-Bertani (LB) broth (Tryptone 10g/L, Yeast extract 5g/L, NaCl 10g/L). Antibiotics were added in the medium when necessary: carbenicillin, 400 μg/mL for P. aeruginosa and 100 μg/mL for E. coli; gentamicin, 50 μg/mL for P. aeruginosa and E. coli; kanamycin, 50 μg/mL for E. coli; ampicillin, 100 μg/mL for E. coli.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains or plasmids | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| P. aeruginosa strains | ||
| PAO1 | Wild-type strain | Lab collection |
| ΔfleR | PAO1 with the deletion of the fleR gene | This study |
| ΔfleR(EV) | ΔfleR containing the empty plasmids pBBR1-MCS5 or pUCP18 | This study |
| ΔfleR(fleR) | ΔfleR containing the expression constructs pBBR1-MCS5-fleR or pUCP18-fleR | This study |
| ΔfleR(sadC) | ΔfleR containing the expression constructs pBBR1-MCS5-sadC or pUCP18-sadC | This study |
| ΔfleR(bifA) | ΔfleR containing the expression constructs pBBR1-MCS5-bifA or pUCP18-bifA | This study |
| ΔfleR(14945) | ΔfleR containing the expression constructs pBBR1-MCS5-W909_14945 or pUCP18-W909_14945 | This study |
| ΔfleR(14950) | ΔfleR containing the expression constructs pBBR1-MCS5-W909_14950 or pUCP18-W909_14950 | This study |
| ΔfleQ | PAO1 with the deletion of the fleQ gene | This study |
| ΔfleQ(EV) | ΔfleQ containing the empty plasmids pBBR1-MCS5 or pUCP18 | This study |
| ΔfleQ(sadC) | ΔfleQ containing the expression construct pBBR1-MCS5-sadC | This study |
| ΔfleRΔfleQ | PAO1 with the deletion of the fleR and fleQ genes | This study |
| ΔfleRΔfleQ(EV) | ΔfleRΔfleQ containing the empty plasmids pBBR1-MCS5 or pUCP18 | This study |
| ΔfleRΔfleQ(sadC) | ΔfleRΔfleQ containing the expression constructs pBBR1-MCS5-sadC or pUCP18-sadC | This study |
| Δtse1 | PAO1 with the deletion of the tse1 gene | This study |
| Δtse1(EV) | Δtse1 containing the empty plasmids pBBR1-MCS5 or pUCP18 | This study |
| Δtse1(tse1) | Δtse1 containing the expression construct pUCP18-tse1 | This study |
| ΔfleRΔtse1 | PAO1 with the deletion of the fleR and tse1 genes | This study |
| ΔfleRΔtse1(tse1) | ΔfleRΔtse1 containing the expression construct pUCP18-tse1 | This study |
| ΔsadC | PAO1 with the deletion of the sadC gene | This study |
| ΔfleRΔsadC | PAO1 with the deletion of the fleR and sadC genes | This study |
| ΔfleRΔflgZ | PAO1 with the deletion of the fleR and flgZ genes | This study |
| ΔfleRΔflgZ(sadC) | ΔfleRΔflgZ containing the expression constructs pBBR1-MCS5-sadC or pUCP18-sadC | This study |
| PAO1(amrZ) | PAO1 containing the expression constructs pBBR1-MCS5-amrZ or pUCP18-amrZ | This study |
| ΔfleR(amrZ) | ΔfleR containing the expression constructs pBBR1-MCS5-amrZ or pUCP18-amrZ | This study |
| ΔfleRΔfleQ(amrZ) | ΔfleRΔfleQ containing the expression constructs pBBR1-MCS5-amrZ or pUCP18-amrZ | This study |
| ΔretS | PAO1 with the deletion of the retS gene | This study |
| ΔretS(EV) | ΔretS containing the empty plasmids pBBR1-MCS5 or pUCP18 | This study |
| ΔfleRΔretS | PAO1 with the deletion of the fleR and retS genes | This study |
| ΔfleRΔretS(EV) | ΔfleRΔretS containing the empty plasmids pBBR1-MCS5 or pUCP18 | This study |
| ΔfleRΔretS(retS) | ΔfleRΔretS containing the expression constructs pBBR1-MCS5-retS or pUCP18-retS | This study |
| E. coli strains | ||
| DH5α | spuE44 ΔlacU169(φ80lacZΔM15) hsdR17λpir recA1 endA1 gyrA96 thi-1 relA1 | Lab collection |
| pRK2013 | Tra+, Mob-, ColE1-replicon, Kanr, Sper | Lab collection |
| Plasmids | ||
| pBBR1-MCS5 | Broad-host-range cloning vector; Gmr | Lab collection |
| pUCP18 | E. coli-P. aeruginosa shuttle expression vector with Plac, Ampr, Carr | Lab collection |
| pBBR1-MCS5-fleR | pBBR1-MCS5 containing fleR under the control of Plac | This study |
| pUCP18-fleR | pUCP18 containing fleR under the control of Plac | This study |
| pBBR1-MCS5-sadC | pBBR1-MCS5 containing sadC under the control of Plac | This study |
| pUCP18-sadC | pUCP18 containing sadC under the control of Plac | This study |
| pBBR1-MCS5-bifA | pBBR1-MCS5 containing bifA under the control of Plac | This study |
| pUCP18-bifA | pUCP18 containing bifA under the control of Plac | This study |
| pUCP18-tse1 | pUCP18 containing tse1 under the control of Plac | This study |
| pBBR1-MCS5-W909_14945 | pBBR1-MCS5 containing W909_14945 under the control of Plac | This study |
| pUCP18-W909_14945 | pUCP18 containing W909_14945 under the control of Plac | This study |
| pBBR1-MCS5-W909_14950 | pBBR1-MCS5 containing W909_14950 under the control of Plac | This study |
| pUCP18-W909_14950 | pUCP18 containing W909_14950 under the control of Plac | This study |
| pBBR1-MCS5-retS | pBBR1-MCS5 containing retS under the control of Plac | This study |
| pUCP18-retS | pUCP18 containing retS under the control of Plac | This study |
| pBBR1-MCS5-amrZ | pBBR1-MCS5 containing amrZ under the control of Plac | This study |
| pUCP18-amrZ | pUCP18 containing amrZ under the control of Plac | This study |
| pK18mobsacB | Broad-host-range sucrose counter-selection allelic exchange vector, sacB, Gmr | Lab collection |
| pK18-fleR | pK18 containing fleR flanking regions for generation of fleR in-frame deletion | This study |
| pK18-tse1 | pK18 containing tse1 flanking regions for generation of tse1 in-frame deletion | This study |
| pK18-retS | pK18 containing retS flanking regions for generation of retS in-frame deletion | This study |
| pK18-fleQ | pK18 containing fleQ flanking regions for generation of fleQ in-frame deletion | This study |
| pK18-flgZ | pK18 containing flgZ flanking regions for generation of flgZ in-frame deletion | This study |
| pK18-sadC | pK18 containing sadC flanking regions for generation of sadC in-frame deletion | This study |
| pK18-tse1-his | pK18 containing tse1. Six repeated CAC was added at the C-terminal. | This study |
| pK18-hcp1-his | pK18 containing hcp1. Six repeated CAC was added at the C-terminal. | This study |
TABLE 2.
PCR primers used in this study
| Primers | Sequence (5′ to 3′) | Description |
|---|---|---|
| For construction | ||
| fleR-Up-F | gagctcggtacccggggatccGCTGGTGTTCGCCCGCGG | For amplification of the 5′-region of fleR |
| fleR-Up-R | agcacggggttactcctgaaTCGCAG | |
| fleR-Dn-F | ttcaggagtaaccccgtgctCGCCATGTTCCCC | For amplification of the 3′-region of fleR |
| fleR-Dn-R | acgacggccagtgccaagcttACGCTGGCCTTCTGGCTG | |
| fleR-FC-F | gtcgacggtatcgataagcttTGCGGGCCCGAACTGCGC | For construction of pBBR1-MCS5-fleR |
| fleR-FC-R | cgctctagaactagtggatccGCGGACGCAAAAGGCCCG | |
| fleR-PC-F | gagctcggtacccggggatccTGCGGGCCCGAACTGCGC | For construction of pUCP18-fleR |
| fleR-PC-R | acgacggccagtgccaagcttGGACGCAAAAGGCCCGCA | |
| tse1-Up-F | agctcggtacccggggatccGACTGTACTTTCACCCAGCT | For amplification of the 5′-region of tse1 |
| tse1-Up-R | acggcctgaagtatcacctaTGCGTGTCGATAAGG | |
| tse1-Dn-F | taggtgatacttcaggccgtGCTGCGAATG | For amplification of the 3′-region of tse1 |
| tse1-Dn-R | cgacggccagtgccaagcttTCTCGATGGCCTGGATCACG | |
| tse1-PC-F | gagctcggtacccggggatccACGCATAGGTGATACATGGA | For construction of pUCP18-tse1 |
| tse1-PC-R | acgacggccagtgccaagcttATTCGCAGCACGGCCTGAA | |
| retS-Up-F | gagctcggtacccggggatccCATGGTCCGCCTGGAGTCC | For amplification of the 5′-region of retS |
| retS-Up-R | ggcgaagtcccttcgaagg | |
| retS-Dn-F | ccttcgaagggacttcgccGGGCAGCGACGTGCTCCG | For amplification of the 3′-region of retS |
| retS-Dn-R | cgacggccagtgccaagcttATAGAGCACCAGCATCTTCA | |
| retS-FC-F | gtcgacggtatcgataagcttGGCACCGCGCTGAAGGAT | For construction of pBBR1-MCS5-retS |
| retS-FC-R | cgctctagaactagtggatccATCCGCCGTGGCGGAGGC | |
| retS-PC-F | gagctcggtacccggggatccGGCACCGCGCTGAAGGAT | For construction of pUCP18-retS |
| retS-PC-R | acgacggccagtgccaagcttATCCGCCGTGGCGGAGGC | |
| fleQ-Up-F | gagctcggtacccggggatccATCGGTGAGCTGGATCAGGTC | For amplification of the 5′-region of fleQ |
| fleQ-Up-R | atccgattcgcgccacatttTGATC | |
| fleQ-Dn-F | aaatgtggcgcgaatcggatGATTGACAGGTCGTT | For amplification of the 3′-region of fleQ |
| fleQ-Dn-R | acgacggccagtgccaagcttCCTCGCGCGGAGCGAAGC | |
| flgZ-Up-F | gagctcggtacccggggatccTATCGGCCACGCCAACCA | For amplification of the 5′-region of flgZ |
| flgZ-Up-R | aacagttcgtcgaacgggttGGGCACCTT | |
| flgZ-Dn-F | aacccgttcgacgaactgttCTGAGTTTCACAGG | For amplification of the 3′-region of flgZ |
| flgZ-Dn-R | acgacggccagtgccaagcttTGCTGGAACGCGCCCTGC | |
| sadC-Up-F | gagctcggtacccggggatccTCCTGCTCTGGCTGGCGC | For amplification of the 5′-region of sadC |
| sadC-Up-R | actggtgacctcccacgtgtCCTGGTGCGCCTG | |
| sadC-Dn-F | acacgtgggaggtcaccagtGCCT | For amplification of the 3′-region of sadC |
| sadC-Dn-R | acgacggccagtgccaagcttGGTCGCAGAACGCGCCGG | |
| sadC-FC-F | gtcgacggtatcgataagcttCGGGTCGGCCAGGATCGA | For construction of pBBR1-MCS5-sadC |
| sadC-FC-R | cgctctagaactagtggatccAGGTTGCTGCCGGCGGCG | |
| sadC-PC-F | gagctcggtacccggggatccCGGGTCGGCCAGGATCGA | For construction of pUCP18-sadC |
| sadC-PC-R | acgacggccagtgccaagcttAGGTTGCTGCCGGCGGCG | |
| bifA-FC-F | gtcgacggtatcgataagcttCGACGTCTGGGAACACGC | For construction of pBBR1-MCS5-bifA |
| bifA-FC-R | cgctctagaactagtggatccCTGGGCAGCGCGCTATTG | |
| bifA-PC-F | gagctcggtacccggggatccCGACGTCTGGGAACACGC | For construction of pUCP18-bifA |
| bifA-PC-R | acgacggccagtgccaagcttCTGGGCAGCGCGCTATTG | |
| amrZ-FC-F | gtcgacggtatcgataagcttGGAGACTGTGTCAGCCCGG | For construction of pBBR1-MCS5-amrZ |
| amrZ-FC-R | cgctctagaactagtggatccAGCCGGCGAATGCCGGCC | |
| amrZ-PC-F | gagctcggtacccggggatccGGAGACTGTGTCAGCCCGG | For construction of pUCP18-amrZ |
| amrZ-PC-R | acgacggccagtgccaagcttAGCCGGCGAATGCCGGCC | |
| W909_14945-FC-F | gtcgacggtatcgataagcttAAATATTAGCCAGGCTTATGTCTATAACG | For construction of pBBR1-MCS5-W909_14945 |
| W909_14945-FC-R | cgctctagaactagtggatccTACGACAGGCCCGGATGG | |
| W909_14945-PC-F | gagctcggtacccggggatccAAATATTAGCCAGGCTTATGTC | For construction of pUCP18-W909_14945 |
| W909_14945-PC-R | acgacggccagtgccaagcttTACGACAGGCCCGGATGG | |
| W909_14950-FC-F | gtcgacggtatcgataagcttTACACAGCGTGGTTATGGTATAAACA | For construction of pBBR1-MCS5-W909_14950 |
| W909_14950-FC-R | cgctctagaactagtggatccCTCTAATCAAATAAGCAGTAGTGACTCAGG | |
| W909_14950-PC-F | gagctcggtacccggggatccTACACAGCGTGGTTATGGTATA | For construction of pUCP18-W909_14950 |
| W909_14950-PC-R | acgacggccagtgccaagcttCTCTAATCAAATAAGCAGTAGTG | |
| tse1-his-Up-F | gagctcggtacccggggatccGGGCCGGAGCGCCCCTTA | For amplification of the 5′-region of tse1. Six repeated CAC was added at the C-terminal. |
| tse1-his-Up-R | agcacggcctgaatcagtggtggtggtggtggtgactgGCCCTGGGCAGGCT | |
| tse1-his-Dn-F | cagtcaccaccaccaccaccacTGATTCAGGCCGTGCTGC | For amplification of the 3′-region of tse1. Six repeated CAC was added at the C-terminal. |
| tse1-his-Dn-R | acgacggccagtgccaagcttCGATGGCCTGGATCACGTC | |
| hcp1-his-Up-F | agctcggtacccggggatccCGGGAGGAAAGATGGCTGTT | For amplification of the 5′-region of hcp1. Six repeated CAC was added at the C-terminal. |
| hcp1-his-Up-R | tcatcagtggtggtggtggtggtgggcCTGCACGTTCTGG | |
| hcp1-his-Dn-F | gcccaccaccaccaccaccactgatgaGCCGGCTGCCGGTCA | For amplification of the 3′-region of hcp1. Six repeated CAC was added at the C-terminal. |
| hcp1-his-Dn-R | acgacggccagtgccaagcttCACCGGCGTCAGGCGCCC | |
| Sequences in lower case indicate the homology arms for recombination. | ||
| For qPCR: | ||
| q-clpV1-F | GTCTACGGCAGCCTGATGTT | |
| q-clpV1-R | GACCTTGAGCTTGGCGAA | |
| q-hcp1-F | TCCAAGGACAAGACTCACGC | |
| q-hcp1-R | CTTGGTGAACGACAGGTCCT | |
| q-vgrG1-F | CCGCATCTTCCAGAACCAG | |
| q-vgrG1-R | TGCACGCAGTATTCCCACTC | |
| q-tse1-F | CACCCCGAACAAGGACAAC | |
| q-tse1-R | TTGGTCCAGCTCTGCTCCA | |
| q-hsiA2-F | AAGCTCTCCTCGCATTATCTGG | |
| q-hsiA2-R | TTGCCCAGCTCATTTTCCAG | |
| q-hsiB2-F | TGACGCTGAGCGTACCCAAT | |
| q-hsiB2-R | CATCGCGCAACTCCATCAG | |
| q-clpV2-F | CATTGATCCTGGCCCTGCTA | |
| q-clpV2-R | GAGAGCGCGAAATCCTTCAAC | |
| q-vgrG2b-F | CGCATCTACCATGAGCACCT | |
| q-vgrG2b-R | CGAAGCGGAAGTAGTAGACCAG | |
| q-hsiB3-F | TCAACTATTTCGCCAACGGC | |
| q-hsiB3-R | GTAGCTGGCCGTTCAGGTAG | |
| q-hsiC3-F | ACGAAGAGGAATACGGCACC | |
| q-hsiC3-R | ACGTTCGACAGCTTCTCCAG | |
| q-hcp3-F | CTCAACGAGTATTGCTGCGC | |
| q-hcp3-R | TTGGAGAGCACCACGTTGTT | |
| q-pldB-F | AGTTGTGGGAGCTACATGCC | |
| q-pldB-R | TGTCACGACATCCCAGAAGC | |
| q-amrZ-F | AACTCCTACCTACTCCAGCCGT | |
| q-amrZ-R | GAGTTCATGCTGCGGTGATG | |
| q-cdrA-F | CCAGTTCAACCCCAACGAGA | |
| q-cdrA-R | GTCGAAGCCCTTCCAGTTGA | |
| q-fleR-F | GCCTGATCCGTACACGCTAC | |
| q-fleR-R | GAACGGCTTGACCAGGTAGTC | |
| q-retS-F | TGATCCAGCAGCTCAACCTG | |
| q-retS-R | GCTGATCTTGGCCAGGAACT | |
| q-siaD-F | CAGGGAGGAGAACGAACGCT | |
| q-siaD-R | TATTCGCGTAGCTCGGACTCC | |
| q-gcbA-F | CCGAATTGGCCAAGGTGAT | |
| q-gcbA-R | ATCGGCTTGGTGAGGAAGTC | |
| q-sadC-F | CGAACTCACCGGTCTGTTCA | |
| q-sadC-R | CCGCTTGAAGTGATCGAGGT | |
| q-mucR-F | AGATCGACCGTGGCTTCATC | |
| q-mucR-R | GGCGACGATCTTCAGGTTCA | |
| q-PA4929-F | ATCTGAAGCAGGAAAGGGCC | |
| q-PA4929-R | CCTCGCTCAACTCGTTGGTA | |
| q-bifA-F | GAAGATCACCCTGGACACCG | |
| q-bifA-R | GGTAGACCAGGAACAGCACC | |
| q-PA2072-F | TTCTACGTGGAAAAGGCGCT | |
| q-PA2072-R | CGACCCGGTTTCCCAGATAG | |
| q-PA3825-F | TTCGAACGGATGCTCGACAA | |
| q-PA3825-R | CGTATTGTCGCGCGTCAAAT | |
| rplU-F | GCAGCACAAAGTCACCGAAG | Internal control |
| rplU-R | CCGATTTTCACGTCTTCGCC | |
| For EMSA: | ||
| EMSA-fleSR-F | AAGGCCTGGACCTCAAGGAC | |
| EMSA-fleSR-R | GCTGGTTGCATTGCGTTTC | |
| EMSA-siaA-F | AAGACGTGCTGCCGCTCGAA | |
| EMSA-siaA-R | GCCATGGCTATCCCTATCAGT | |
| EMSA-gcbA-F | CAGCTCGATAGATGGGGGATTG | |
| EMSA-gcbA-R | GACGCGCTTCTTTCGTGGTC | |
| EMSA-sadC-F | GTGTTGTCCTTGGTGTTCTTCCG | |
| EMSA-sadC-R | TCCTACTACCACCCGGTCGAT | |
| EMSA-mucR-F | CACCTCCTGTCGAGACATTCAGA | |
| EMSA-mucR-R | GAAAGCGTCCGAACGGGATAG | |
| EMSA-PA4929-F | CCGAGCTTCATGAAGTCGTCC | |
| EMSA-PA4929-R | AGTGAAAACGTCGGAATTGCTC | |
| EMSA-amrZ-R | CAATCGGTTGCACGAAGACG | |
| EMSA-amrZ -R | AGTTGCCTGTTTCAGTGGGC | |
Construction of PAO1 mutants and gene complementation assay.
A SacB-based strategy was employed to achieve gene deletion in PAO1 according to a previous description (54). Briefly, 500-bp upstream and 500-bp downstream sequences of the target gene were amplified by PCR with Pfu DNA polymerase (Vazyme, China). The PCR products were assembled into the suicide plasmid pK18mobsacB which was predigested with BamHI and HindIII. The constructed plasmid pK18-fleR was introduced into PAO1 using the helper plasmid pRK2013 by triparental mating. Desired mutants were counter-selected using LB plates containing 10% sucrose and verified by PCR and DNA sequencing. For gene complementation, the open reading frames (ORF) together with its native promoter was amplified by PCR using primers listed in Table 2. PCR fragments were ligated into the downstream of the lac promoter of pBBR1-MCS5 or pUCP18 between the HindIII and BamHI sites or BamHI and HindIII sites, respectively. The plasmids were verified by PCR and DNA sequencing and introduced into PAO1 strains by tri-parental mating. Successful plasmid delivery into the PAO1 strains was confirmed by PCR.
RT-qPCR.
Bacterial cells were grown in LB medium and harvested at OD600 of 1.5 by centrifugation. Total RNA was isolated using the RNeasy minikit (Qiagen, Germany) according to the manufacturer’s instructions. The cDNA samples were synthesized from the isolated total RNA using SuperScript II reverse transcriptase (Invitrogen, USA) and random primers (Invitrogen, USA). qPCR was performed using the QuantiTect SYBR green PCR kit (Qiagen, Germany) on the ABI QuantStudioTM6 Flex system (Roche, Switzerland) according to the manufacturer’s instructions. The 50S ribosomal protein gene rplU was selected as an internal control (55). The relative gene expression level was calculated by using the 2−ΔΔCT method and presented as the mean of three independent biological isolates.
RNA-seq.
The enriched mRNA was fragmented as 200 nt to 700 nt and reverse transcribed into cDNA with random primers. Second-strand cDNA was synthesized by DNA polymerase I, RNase H, and dNTP. The cDNA fragments were then purified with QiaQuick PCR extraction kit with end repaired and poly(A) added and ligated to Illumina sequencing adapters. The ligation products were size selected by agarose gel electrophoresis, followed by PCR amplification, and sequencing by Illumina HiSeq TM 2500 (Gene Denovo Biotechnology Co., China). Differentially expressed genes with ≥1.2-log2fold changes were identified at a false discovery rate (FDR) of ≤0.05, and analyzed using the major public pathway-related database KEGG (56). The formula for calculating the P value is
where N is the number of all genes that with annotation in database, n is the number of differentially expressed genes in N, M is the number of all genes annotated to specific pathways, and m is the number of differentially expressed genes in M. The calculated P value has gone through FDR correction, taking FDR of ≤0.05 as a threshold. Q value is the P value underwent multiple hypothesis test corrections. The value ranges from 0 to 1 with more significant when it is closer to 0.
Protein purification and Western blot analysis.
Overnight bacterial cultures were 1:1,000 diluted into 10-mL fresh LB medium and incubated until OD600 reached 1.5. Bacterial cultures were chilled on ice for 20 min. For each sample, bacterial pellet was collected by centrifugation and lysed by radio immunoprecipitation assay (RIPA) lysis buffer (Biosharp, China). The supernatant was purified with a 0.22-μm filter and treated with trichloroacetic acid (TCA) at a final concentration of 10%. The precipitate was pelleted by centrifugation and resuspended in SDS loading buffer after washing with ice-cold acetone for three times. Western blot analysis was performed according to the method described previously (57). Proteins were separated by SDS-PAGE and then transferred to a polyvinylidene difluoride (PVDF) membrane by electroblotting. The PVDF membrane was blocked with 5% (wt/vol) skim milk in PBST buffer (PBS supplemental with 0.05% Tween 20) for 1 h, followed by immunoblotting using anti-His antibody (Abbkine, USA) and horseradish peroxidase-conjugated goat anti-mouse antibody (Abbkine, USA). Proteins were detected using the ECL kit (Abbkine, USA) according to the manufacture’s protocol. The RNA polymerase β subunit (β-RNAP) was selected as an internal control for Western blot assays.
Electrophoretic gel mobility shift assay.
The DNA probes used for EMSA were prepared by PCR using the primer pairs listed in Table 2. The purified PCR products were 3′-end labeled with biotin following the manufacturer’s instruction (Thermo Fisher Scientific, USA). The DNA-protein binding reactions were performed according to the manufacturer’s instructions (Thermo Fisher Scientific, USA). The 4% polyacrylamide gel was used to separate the DNA-protein complexes. After UV cross-linking, the biotin-labeled probes were detected using a biotin luminescent detection kit (Thermo Fisher Scientific, USA).
Bacterial killing assay.
The antibacterial activity of P. aeruginosa was examined following the method as described by Han et al. (7). E. coli cells containing the pBBR1-MCS5 plasmid (gentamicin resistance) served as the prey. Overnight cultures of P. aeruginosa and E. coli were washed three times with fresh LB and diluted to OD600 = 2.0 and 0.4, respectively. Then the two bacteria were 1:1 mixed and 5 μl of the mixture was spotted on a 0.22-μm nitrocellulose membrane and placed on an LB agar plate. After incubation at 37°C for 12 h, bacterial cells were re-suspended in 500 μl LB broth. The cell suspension was serially diluted and spread on LB agar plates containing gentamicin to select for E. coli cells. The number of recovered E. coli cells was counted.
ACKNOWLEDGMENTS
This work was supported by the Key Projects of Guangzhou Science and Technology Plan (Grant No. 201804020066), National Natural Science Foundation of China (Grant No. 31330002, Grant No. 32100020), and Key Realm R&D Program of Guangdong Province (Project No. 2018B020205003, Project No. 2020B0202090001).
We declare that they have no conflicts of interest with the contents of this article.
T.Z., Z.X., and L.Z. designed the study. T.Z., J.H., Z.L., and Q.L. performed experiments. T.Z., J.H., and Z.X. analyzed the results. T.Z., Z.X., and L.Z. drafted and revised the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Zeling Xu, Email: zelingxu@scau.edu.cn.
Lian-Hui Zhang, Email: lhzhang01@scau.edu.cn.
Gladys Alexandre, University of Tennessee at Knoxville.
REFERENCE
- 1.Navarro-Garcia F, Ruiz-Perez F, Cataldi Á, Larzábal M. 2019. Type VI secretion system in pathogenic Escherichia coli: structure, role in virulence, and acquisition. Front Microbiol 10:1965. 10.3389/fmicb.2019.01965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lennings J, West TE, Schwarz S. 2018. The Burkholderia type VI secretion system 5: composition, regulation and role in virulence. Front Microbiol 9:3339. 10.3389/fmicb.2018.03339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crisan CV, Hammer BK. 2020. The Vibrio cholerae type VI secretion system: toxins, regulators and consequences. Environ Microbiol 22:4112–4122. 10.1111/1462-2920.14976. [DOI] [PubMed] [Google Scholar]
- 4.Mariano G, Trunk K, Williams DJ, Monlezun L, Strahl H, Pitt SJ, Coulthurst SJ. 2019. A family of type VI secretion system effector proteins that form ion-selective pores. Nat Commun 10:5484. 10.1038/s41467-019-13439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L, Zou Y, She P, Wu Y. 2015. Composition, function, and regulation of T6SS in Pseudomonas aeruginosa. Microbiol Res 172:19–25. 10.1016/j.micres.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Coulthurst S. 2019. The type VI secretion system: a versatile bacterial weapon. Microbiology (Reading) 165:503–515. 10.1099/mic.0.000789. [DOI] [PubMed] [Google Scholar]
- 7.Han Y, Wang T, Chen G, Pu Q, Liu Q, Zhang Y, Xu L, Wu M, Liang H. 2019. A Pseudomonas aeruginosa type VI secretion system regulated by CueR facilitates copper acquisition. PLoS Pathog 15:e1008198. 10.1371/journal.ppat.1008198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J, Zhang W, Cheng J, Yang X, Zhu K, Wang Y, Wei G, Qian PY, Luo ZQ, Shen X. 2017. A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun 8:14888. 10.1038/ncomms14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Southey-Pillig CJ, Davies DG, Sauer K. 2005. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J Bacteriol 187:8114–8126. 10.1128/JB.187.23.8114-8126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347. 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sana TG, Berni B, Bleves S. 2016. The T6SSs of Pseudomonas aeruginosa strain PAO1 and their effectors: beyond bacterial-cell targeting. Front Cell Infect Microbiol 6:61. 10.3389/fcimb.2016.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530. 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeRoux M, De Leon JA, Kuwada NJ, Russell AB, Pinto-Santini D, Hood RD, Agnello DM, Robertson SM, Wiggins PA, Mougous JD. 2012. Quantitative single-cell characterization of bacterial interactions reveals type VI secretion is a double-edged sword. Proc Natl Acad Sci USA 109:19804–19809. 10.1073/pnas.1213963109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD. 2013. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496:508–512. 10.1038/nature12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang F, Wang X, Wang B, Chen L, Zhao Z, Waterfield NR, Yang G, Jin Q. 2016. The Pseudomonas aeruginosa type VI secretion PGAP1-like effector induces host autophagy by activating endoplasmic reticulum stress. Cell Rep 16:1502–1509. 10.1016/j.celrep.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 16.Jiang F, Waterfield NR, Yang J, Yang G, Jin Q. 2014. A Pseudomonas aeruginosa Type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe 15:600–610. 10.1016/j.chom.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 17.Hsu F, Schwarz S, Mougous JD. 2009. TagR promotes PpkA-catalysed type VI secretion activation in Pseudomonas aeruginosa. Mol Microbiol 72:1111–1125. 10.1111/j.1365-2958.2009.06701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. 2007. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol 9:797–803. 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 19.Stolle AS, Meader BT, Toska J, Mekalanos JJ. 2021. Endogenous membrane stress induces T6SS activity in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 118:e2018365118. 10.1073/pnas.2018365118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lesic B, Starkey M, He J, Hazan R, Rahme LG. 2009. Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology (Reading) 155:2845–2855. 10.1099/mic.0.029082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia YS, Xu CJ, Wang D, Weng YD, Jin YX, Bai F, Cheng ZH, Kuipers OP, Wu WH. 2021. YbeY controls the type III and type VI secretion systems and biofilm formation through RetS in Pseudomonas aeruginosa. Appl Environ Microbiol 87:e02171-20. 10.1128/AEM.02171-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allsopp LP, Wood TE, Howard SA, Maggiorelli F, Nolan LM, Wettstadt S, Filloux A. 2017. RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 114:7707–7712. 10.1073/pnas.1700286114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodman AL, Merighi M, Hyodo M, Ventre I, Filloux A, Lory S. 2009. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev 23:249–259. 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janssen KH, Diaz MR, Golden M, Graham JW, Sanders W, Wolfgang MC, Yahr TL, DiRita VJ. 2018. Functional analyses of the RsmY and RsmZ small noncoding regulatory RNAs in Pseudomonas aeruginosa. J Bacteriol 200:e00736-17. 10.1128/JB.00736-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol 73:434–445. 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broder UN, Jaeger T, Jenal U. 2016. LadS is a calcium-responsive kinase that induces acute-to-chronic virulence switch in Pseudomonas aeruginosa. Nat Microbiol 2:16184. 10.1038/nmicrobiol.2016.184. [DOI] [PubMed] [Google Scholar]
- 27.Basler M, Ho Brian T, Mekalanos JJ. 2013. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152:884–894. 10.1016/j.cell.2013.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 29.Zhou T, Huang JH, Liu ZQ, Xu ZL, Zhang LH. 2021. Molecular mechanisms underlying the regulation of biofilm formation and swimming motility by FleS/FleR in Pseudomonas aeruginosa. Front Microbiol 12:707711. 10.3389/fmicb.2021.707711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kollaran AM, Joge S, Kotian HS, Badal D, Prakash D, Mishra A, Varma M, Singh V. 2019. Context-specific requirement of forty-four two-component loci in Pseudomonas aeruginosa swarming. iScience 13:305–317. 10.1016/j.isci.2019.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang ML, Zhang SL, Dong LH, Kou YJ, Lin CX, Dai WJ, Zhang LH, Deng YZ. 2018. Label-free quantitative proteomics of lysine acetylome identifies substrates of Gcn5 in Magnaporthe oryzae autophagy and epigenetic regulation. mSystems 3:e00270-18. 10.1128/mSystems.00270-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarthy RR, Yu M, Eilers K, Wang YC, Lai EM, Filloux A. 2019. Cyclic di-GMP inactivates T6SS and T4SS activity in Agrobacterium tumefaciens. Mol Microbiol 112:632–648. 10.1111/mmi.14279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duan XK, Pan YR, Cai Z, Liu YM, Zhang YD, Liu MX, Liu Y, Wang K, Zhang LH, Yang L. 2021. rpoS-mutation variants are selected in Pseudomonas aeruginosa biofilms under imipenem pressure. Cell Biosci 11:138. 10.1186/s13578-021-00655-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moscoso JA, Jaeger T, Valentini M, Hui K, Jenal U, Filloux A. 2014. The diguanylate cyclase SadC is a central player in Gac/Rsm-mediated biofilm formation in Pseudomonas aeruginosa. J Bacteriol 196:4081–4088. 10.1128/JB.01850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75:827–842. 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen YF, Zhou JN, Lv MF, Liang ZB, Parsek MR, Zhang LH. 2020. Systematic analysis of c-di-GMP signaling mechanisms and biological functions in Dickeya zeae EC1. mBio 11:e02993-20. 10.1128/mBio.02993-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colley B, Dederer V, Carnell M, Kjelleberg S, Rice SA, Klebensberger J. 2016. SiaA/D interconnects c-di-GMP and RsmA signaling to coordinate cellular aggregation of Pseudomonas aeruginosa in response to environmental conditions. Front Microbiol 7:179. 10.3389/fmicb.2016.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6. 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 39.Baraquet C, Harwood CS. 2013. Cyclic diguanosine monophosphate represses bacterial flagella synthesis by interacting with the Walker A motif of the enhancer-binding protein FleQ. Proc Natl Acad Sci USA 110:18478–18483. 10.1073/pnas.1318972110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou H, Zheng C, Su JM, Chen B, Fu Y, Xie YQ, Tang Q, Chou SH, He J. 2016. Characterization of a natural triple-tandem c-di-GMP riboswitch and application of the riboswitch-based dual-fluorescence reporter. Sci Rep 6:20871. 10.1038/srep20871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baraquet C, Murakami K, Parsek MR, Harwood CS. 2012. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res 40:7207–7218. 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jyot J, Dasgupta N, Ramphal R. 2002. FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J Bacteriol 184:5251–5260. 10.1128/JB.184.19.5251-5260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muriel C, Arrebola E, Redondo-Nieto M, Martínez-Granero F, Jalvo B, Pfeilmeier S, Blanco-Romero E, Baena I, Malone JG, Rivilla R, Martín M. 2018. AmrZ is a major determinant of c-di-GMP levels in Pseudomonas fluorescens F113. Sci Rep 8:1979. 10.1038/s41598-018-20419-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hou L, Debru A, Chen Q, Bao Q, Li K. 2019. AmrZ regulates swarming motility through cyclic di-GMP-dependent motility inhibition and controlling Pel polysaccharide production in Pseudomonas aeruginosa PA14. Front Microbiol 10:1847. 10.3389/fmicb.2019.01847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hachani A, Lossi NS, Hamilton A, Jones C, Bleves S, Albesa-Jové D, Filloux A. 2011. Type VI secretion system in Pseudomonas aeruginosa: secretion and multimerization of VgrG proteins. J Biol Chem 286:12317–12327. 10.1074/jbc.M110.193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. 2011. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ Microbiol 13:3128–3138. 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]
- 48.Hachani A, Wood TE, Filloux A. 2016. Type VI secretion and anti-host effectors. Curr Opin Microbiol 29:81–93. 10.1016/j.mib.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 49.Ritchings BW, Almira EC, Lory S, Ramphal R. 1995. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect Immun 63:4868–4876. 10.1128/iai.63.12.4868-4876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones CJ, Newsom D, Kelly B, Irie Y, Jennings LK, Xu B, Limoli DH, Harrison JJ, Parsek MR, White P, Wozniak DJ. 2014. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog 10:e1003984. 10.1371/journal.ppat.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martínez-Granero F, Redondo-Nieto M, Vesga P, Martín M, Rivilla R. 2014. AmrZ is a global transcriptional regulator implicated in iron uptake and environmental adaption in P fluorescens F113. BMC Genomics 15:237. 10.1186/1471-2164-15-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wozniak DJ, Sprinkle AB, Baynham PJ. 2003. Control of Pseudomonas aeruginosa algZ expression by the alternative sigma factor AlgT. J Bacteriol 185:7297–7300. 10.1128/JB.185.24.7297-7300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mikkelsen H, McMullan R, Filloux A. 2011. The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS One 6:e29113. 10.1371/journal.pone.0029113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi KH, Schweizer HP. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol 5:30. 10.1186/1471-2180-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O’Toole GA. 2007. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol 189:8165–8178. 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484. 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu ZL, Wang PC, Wang HB, Yu ZH, Au-Yeung HY, Hirayama T, Sun HZ, Yan AX. 2019. Zinc excess increases cellular demand for iron and decreases tolerance to copper in Escherichia coli. J Biol Chem 294:16978–16991. 10.1074/jbc.RA119.010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S7, Table S1. Download AEM.01655-21-s0001.pdf, PDF file, 0.8 MB (854.1KB, pdf)