ABSTRACT
The human skin is our outermost layer and serves as a protective barrier against external insults. Advances in next-generation sequencing have enabled the discoveries of a rich and diverse community of microbes—bacteria, fungi, and viruses that are residents of this surface. The genomes of these microbes also revealed the presence of many secretory enzymes. In particular, proteases which are hydrolytic enzymes capable of protein cleavage and degradation are of special interest in the skin environment, which is enriched in proteins and lipids. In this minireview, we will focus on the roles of these skin-relevant microbial secreted proteases, in terms of both their widely studied roles as pathogenic agents in tissue invasion and host immune inactivation and their recently discovered roles in intermicrobial interactions and modulation of virulence factors. From these studies, it has become apparent that while microbial proteases are capable of a wide range of functions, their expression is tightly regulated and highly responsive to the environments the microbes are in. With the introduction of new biochemical and bioinformatics tools to study protease functions, it will be important to understand the roles played by skin microbial secretory proteases in cutaneous health, especially the less studied commensal microbes with an emphasis on contextual relevance.
KEYWORDS: Candida, Cutibacterium, dermatophytes, Malassezia, proteases, skin microbiology, Staphylococcus, Streptococcus
INTRODUCTION
SKIN MICROBIOME
The skin is our outermost layer that interfaces with the external environment (1) and is also the site of residence of a rich and diverse microbial community composed of bacteria, viruses, and fungi (2). The major advance in understanding the community composition of skin microbes came in the early 2000s with the adoption of culture-independent techniques (3) together with next-generation sequencing which allows direct ecological profiling of the skin microbiome (4–6). This is a tremendous step forward in studying human skin microbes because culture-independent profiling of microbes greatly reduces the growth bias associated with laboratory culture conditions, providing significantly more accurate microbial composition analysis.
The human skin is divided into 3 main subtypes: oily/sebaceous, moist, and dry (7). Each type of skin site is associated with a particular microbial composition signature (6). The adult skin microbiome is very stable over a long period, as shown by the minimal changes to the composition especially at the sebaceous sites (8). This is fairly surprising, given the skin is constantly exposed to environmental perturbations and in contact with opportunistic pathogens. This stability underlines how skin microbes are masters of their environment—the microbial genes and associated products enable these microbes to thrive in this nutrient-deprived environment (9).
The skin environment.
The human skin is a stratified epithelium consisting of dividing basal layers of keratinocytes which differentiates into corneocytes in the uppermost layer of the skin (1). This layer, known as the stratum corneum, was thought to be the major residence site of microbes. Recent studies have shown the presence of a rich microbial community in skin invaginations and hair follicles, especially microbes that thrive in a more anaerobic environment (10, 11). In healthy individuals, the skin forms a formidable barrier against external insults through tight junctions in the stratum granulosum and the stratum corneum, which consists of highly keratinized cells in a lipid matrix (12). Compared to the gut environment that is enriched in carbohydrates, the main components of the skin surface are proteins and lipids (9). It is perhaps not surprising that skin microbes, the superior survivors in this dry, acidic environment, correspondingly harbor a repertoire of hydrolytic enzymes involved in protein and lipid metabolism. In this review, we will focus on how microbes use enzymes of protein metabolism to facilitate their growth, survival, and/or invasion on the skin surface.
Proteases and protein metabolism.
Proteins are one of the two major components of the stratum corneum (13). In order to utilize this resource, proteins need to be broken down into peptides and amino acids for uptake by the microbes. This catabolic process is performed by a class of hydrolytic enzymes called proteases (or proteinases) (14, 15). Proteases are divided into 7 main classes based on the catalytic mechanism: serine, metallo-, cysteine, aspartate, threonine, asparagine, and glutamate proteases (16). Serine, metallo- and cysteine proteases account for the majority of the proteases in bacteria and fungi, while aspartyl proteases are prevalent in many fungal genomes (17).
Secretory proteases are especially intriguing because microbial cells need to expend energy in synthesizing and secreting proteins, and this has to be carefully regulated under nutrient-deprived conditions (18). The conventional understanding is that secretory proteases, such as the Cutibacterium acnes proteases that release arginine from skin proteins (19), mainly function in nutrient acquisition (20). However, the roles of proteases in mediating processes beyond general catabolism have been increasingly evident, especially in mammalian systems (21). Through catalyzing irreversible peptide bond hydrolysis, proteases can precisely mediate biological events crucial for intermicrobial and host-microbial interactions. Most importantly, the repertoire of secretory enzymes is optimized for the environment where they reside (22); expression and activities of these enzymes are dynamic and change as the skin environment is altered.
The roles of secretory skin microbial proteases have been extensively studied, though the focus in early studies is mostly on cutaneous infection and invasion. With our renewed understanding of the human skin microbiome, many recent studies have revealed novel roles that microbial proteases play in regulating key biological processes beyond tissue invasion. In this minireview, we will outline the functional roles of these secretory proteases with relevance to the skin environment, with an emphasis on recent discoveries on both the pathogenic and potentially beneficial roles of these microbial proteases (Fig. 1).
FIG 1.
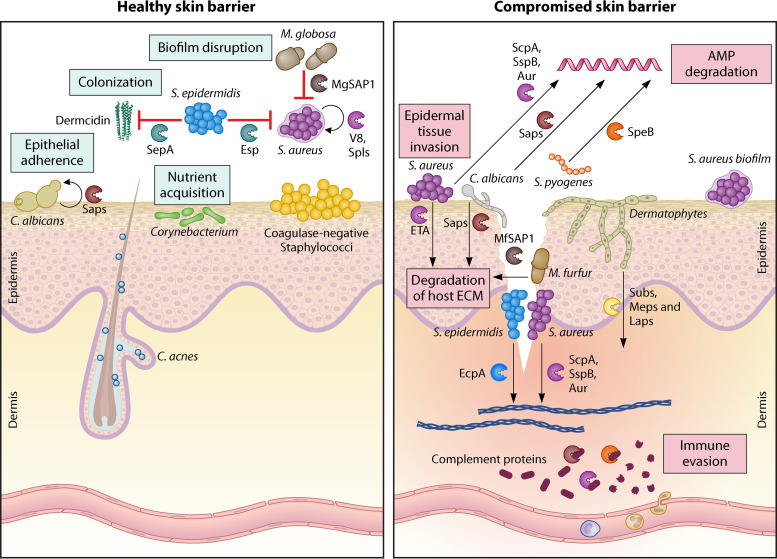
The role of skin-resident microbial secretory proteases in healthy and diseased skin. When the epidermal barrier is intact, the microbes and their secreted proteases are localized to the stratum corneum, the topmost layer of the epidermis, and epidermal invaginations. When the epidermal barrier is breached or compromised, these secretory proteases can reach the deeper layers of the epidermis and dermis, resulting in skin tissue damage and inflammation. The functions of the proteases depend on the skin environment—the same protease can have different roles depending on whether the skin barrier is intact. Only key secreted proteases from each species are shown. The color of the protease icons corresponds to the color of the microbe. Figure not drawn to scale. AMP, antimicrobial peptide; ECM, extracellular matrix.
SKIN FUNGAL SECRETORY PROTEASES
Historically, fungal secretory proteases are regarded as key virulence agents in skin and systemic infections. The most prominent members associated with skin infections are the dermatophytes, Candida and Aspergillus spp. However, next-generation sequencing studies of skin samples from healthy individuals in recent years revealed that aside from the feet, the skin is overwhelmingly populated by a single genus—Malassezia (23). This has led to a renewed understanding of fungal growth on human skin—colonization of commensals (growth without host tissue degradation) and infection by pathogens (involving host tissue destruction and penetration) (24). Recent studies on secretory proteases are increasing our understanding of how these proteases can be essential elements that facilitate colonization and infection (Table 1).
TABLE 1.
Fungal proteases and their associated targets and functions
| Organism | Protease | Target(s) | Process(es) involved | Key reference(s) | |
|---|---|---|---|---|---|
| Candida albicans | Secreted aspartyl proteases (Saps) | Sap1 to -3 | Complement C3b, C4b, C5; α1-protease inhibitor; His5; LL-37 (Sap2 only): α2-macroglobulin; cystatin A; cytokeratin; collagen; factor H; FHR-1 receptor; kininogens; macrophage factor-H receptors CR3, CR4; mucin; vimentin |
Colonization; epidermal barrier invasion; immune modulation and evasion | 31, 33, 38, 40, 42, 46 |
| Sap4 | α1-Protease inhibitor; His5; LL-37 | Immune modulation | 33, 42 | ||
| Sap5 | E-cadherin; His5 | Degradation of connective tissue; biofilm formation | 48, 49 | ||
| Sap6 | Unknown | Biofilm formation | 48 | ||
| Sap7 | His5 | Colonization; (Sap9 only) biofilm formation | 43 | ||
| Sap8 | His5; LL-37 | ||||
| Sap9 | α1-Protease inhibitor; His5; LL-37 | 29, 50 | |||
| Sap10 | His5 | ||||
| Candida parapsilosis | Secreted aspartyl proteases (SAPPs) | SAPP1 | IgA; complement 3b, 4b | Immune modulation and evasion | 54, 56 |
| SAPP2 | Complement 3b, 4b; factor H-related protein 5 | ||||
| Dermatophytes | Metalloprotease carboxypeptidases (Mcps), leucine aminopeptidases (Laps), deuterolysins (Npls) | McpA and -B; Lap1 and -2; NpIIA and -B | Keratin fragments | Epidermal barrier invasion | 75, 76 |
| Metalloprotease fungalysins (Meps) | Mep1, -3, -4 | Keratin | Epidermal barrier invasion | 71, 74, 75 | |
| Serine carboxypeptidases | CpyA, ScpA and -B | Keratin fragments | Epidermal barrier invasion | 75, 76 | |
| Serine dipeptidyl-peptidases (Dpps) | DppIV and -V (Tri 4) | Keratin fragments | Epidermal barrier invasion | 74, 75 | |
| Serine protease subtilisins (Subs) | Sub1 to -7 | Keratin; pro-Mcps | Epidermal barrier invasion; (Sub3 and -4) activation of pro-Mcps | 70, 76, 79, 85 | |
| Malassezia globosa | Aspartyl protease | MgSAP1 | S. aureus SpA | Reduce S. aureus biofilm | 61 |
| Malassezia furfur | Aspartyl protease | MfSAP1 | Cytokeratins; denatured collagen I/IV; fibronectin; thrombospondin-1; vitronectin | Epidermal barrier invasion | 66 |
Candida.
Candida is a genus of yeasts in which many species are commensals or symbionts of the human skin and gut. Candida spp. are often opportunistic pathogens, causing disease when the host skin or mucosal barriers are disrupted or when the host is immunocompromised (25). Candida albicans is the main species of Candida found on human skin and is linked to many common superficial infections (26). Candida spp. are only a minor component of the healthy skin microbiome. Despite this, Candida secreted aspartyl proteases (Saps) are perhaps the most well-studied among the human fungal skin microbiome (see reference 27 for a more extensive review). Candida albicans secretes 10 Saps belonging to 2 main families of aspartyl proteases—the candidapepsins (Sap1 to -8) and the yapsins which are bound to the fungal cell wall and membrane via a glycosylphosphatidylinositol anchor (Sap9 and -10) (28–30). The large number of protease-encoding genes is likely due to the need for specialized proteases during the various stages of host invasion as well as the specific location of infection, as these Saps have various pH optima and substrate specificities (30–32). Expression of Sap1 to -3 is often associated with C. albicans yeasts, while Sap4 to -6 are associated with hyphal growth (33); the morphological association of Sap7 to -10 has not been confirmed (30). In both the commensal and infection states, Sap9 is the most highly expressed protease (29, 34).
Saps are involved in both colonization and infection of epithelial surfaces; these enzymes are considered to be significant virulence factors in C. albicans invasion of host tissues (31, 35). First, Saps are involved in colonization by processing adhesion proteins, and increased Sap activity was correlated with stronger adherence of C. albicans to host cells (36). Saps1 to -4 and -9 are further able to degrade the human α1-protease inhibitor, which results in an increase in human neutrophil elastase activity, leading to further tissue damage and colonization by C. albicans (37). Second, Saps can degrade a wide range of human structural proteins in vitro, including cytokeratin, collagen, and vimentin (31), which enable epithelial cavitation and penetration to establish deeper infection. Third, Saps can modulate host immune responses in multiple ways. Saps contribute to Candida evasion of the host’s immune response by degrading complement system proteins C3b, C4b, and C5 (38). Sap2 degrades factor H, a complement regulator which promotes host immune cell recognition and binding to C. albicans (39). This degradation results in a lower host fungicidal response, facilitating fungal cell survival during infection. However, Sap2 or Sap5 overexpression in a hypofilamentous strain is insufficient to cause immunopathology and likely works in combination with various hypha-associated factors (40). Saps are further able to efficiently degrade some host antimicrobial peptides (AMPs), which promotes fungal survival in the host. AMPs are usually short, cationic, and amphiphilic peptides secreted by various cell types in the skin (41). Saps can inactivate human LL-37 (active fragment of cathelicidin), a key epidermal innate immunity AMP (42); the salivary AMP histatin5 (His5) (43); and AMPs liberated from kininogens by host proteases (44). Finally, these aspartyl proteases also elicit proinflammatory responses independent of their proteolytic activity—Sap1 to -3 stimulate macrophages, increasing interleukin-1β (IL-1β) and tumor necrosis factor alpha (TNF-α) production, while Sap1 and Sap3 also increase IL-6 production (45, 46).
Beyond host protein degradation, Saps are also involved in biofilm formation—a process that can reduce C. albicans susceptibility to current therapeutic agents. Global protease substrate profiling utilizing synthetic pooled peptides (47) revealed that Sap5 and Sap6 are upregulated and highly specific to C. albicans biofilms. C. albicans mutants harboring sap5 and/or sap6 deletions exhibit reduced biofilm formation both in vitro and in a rat catheter biofilm model (48). However, the precise substrates of Sap5 and Sap6 are undefined. The cell wall-associated Sap9 is also involved in this process; sap9 mRNA expression is upregulated in C. albicans biofilms (49), and a knockout mutant of sap9 displays flatter biofilm structure (50). This is likely attributed to Sap9’s role in the proteolytic processing of cell wall proteins, such as Eap1, involved in surface adhesion and biofilm formation (50). This role of Saps in biofilm formation has been targeted as a potential enhancer of antifungals. Several aspartyl protease inhibitors when used in combination with commonly prescribed antifungals, which alone have little to no inhibitory effect on biofilms, resulted in reduced formation of C. albicans biofilms. Lopinavir, the HIV aspartyl protease inhibitor, was able to both inhibit biofilm formation and disrupt mature biofilm when used in combination with the antifungal caspofungin (51). Overall, these studies demonstrate that aspartyl proteases are important for the formation and maintenance of the C. albicans biofilm structures.
Other skin-resident Candida species are also known to possess and express genes coding for Sap homologues (31). Candida parapsilosis, Candida tropicalis, and Candida dubliniensis possess SapP1 to -3, SapT1 to -4 (52, 53), and SapCD1 to -4, respectively. Of the 3 species, C. parapsilosis is the most studied, with SapP1 to -3 observed to degrade host proteins and activate proteolytic cascades in a similar fashion as C. albicans Saps (54–56).
Malassezia.
Malassezia (formerly Pityrosporum) is a genus of commensal fungi commonly found on human and animal skin. Currently, there are 17 known species of Malassezia (57). Malassezia species dominate the skin mycobiome in healthy individuals but are also associated with several skin conditions, such as pityriasis versicolor, seborrheic dermatitis, and atopic dermatitis (AD) (58). Common anthropophilic Malassezia species include Malassezia globosa, Malassezia restricta, Malassezia sympodialis, and Malassezia furfur (59). Functional genomic analysis of these recently sequenced Malassezia species has revealed that these Malassezia species possess many aspartyl proteases (Table 2) (60).
TABLE 2.
Predicted secreted proteases in common Malassezia species
| Protease class | No. of proteases in strain: |
||||
|---|---|---|---|---|---|
| Malassezia furfur CBS14141 | Malassezia globosa CBS7966 | Malassezia pachydermatis CBS1879 | Malassezia restricta CBS7877 | Malassezia sympodialis ATCC 42132 | |
| Aspartate | 5 | 14 | 6 | 12 | 6 |
| Metallo- | 2 | 1 | 0 | 1 | 3 |
| Serine | 7 | 2 | 9 | 4 | 6 |
| Total | 14 | 17 | 15 | 17 | 15 |
Malassezia globosa.
The M. globosa genome possesses 17 predicted secretory proteases, of which 14 are aspartyl proteases. In M. globosa culture, MgSAP1 is the secretory protease that dominates the extracellular proteolytic landscape of this yeast (61). MgSAP1 is ubiquitously expressed by M. globosa on healthy skin at the RNA level. Using an unbiased, mass spectrometry-based technique (47), we determined that MgSAP1 has a strong preference for positively charged residues at the P1 position. Interestingly, this cleavage preference is also observed in various fungal aspartyl proteases such as C. albicans Saps (48) and is likely due to the presence of an aspartate residue in the catalytic flap that confers S1 (binding pocket next to the cleavage site) specificity (62). MgSAP1 coincubation with Staphylococcus aureus inhibits biofilm formation without affecting planktonic growth, and this effect is likely attributable to the M. globosa protease cleaving Staphylococcus aureus protein A (SpA), an extracellular protein involved in S. aureus biofilm formation (63). Since Malassezia spp. and S. aureus share several common skin niches such as the anterior nares (6), this finding suggests that Malassezia could play a potentially beneficial role by preventing the formation of S. aureus biofilms which can be reservoirs for pathogenic dissemination (Fig. 1) (64).
Malassezia furfur.
M. furfur is the Malassezia species most often associated with pityriasis versicolor and systemic infections (65). M. furfur harbors 14 secretory proteases, in which 5 are aspartyl proteases. MfSAP1 is the homologue of MgSAP1 and is also the main extracellular aspartyl protease secreted by M. furfur (66). MfSAP1 cleaves many key extracellular matrix (ECM) proteins associated with the human skin, such as collagens I and IV, fibronectin, cytokeratins, thrombospondin-1, and vitronectin. Compared to MgSAP1 (61), this protease has similar preferences for substrate cleavage as determined by a synthetic fluorogenic substrate (66). However, MfSAP1 is a more catalytically efficient enzyme than MgSAP1 (66). This highlights that even though M. furfur is much less abundant than M. globosa on human skin, the high enzymatic activity of MfSAP1 makes this species functionally relevant. This is important considering that a high concentration of MfSAP1 can interfere with wound healing, as assessed in a three-dimensional (3D) skin wound model (66).
Dermatophytes.
The dermatophytes are a broad group of fungi comprising 3 genera—Microsporum, Trichophyton, and Epidermophyton (67). These fungi are largely pathogenic in nature, unlike Malassezia and Candida, which are mostly commensals. Dermatophyte infections are typically superficial, but in certain immunocompromised patients, deeper dermal infections can occur (68). Trichophyton rubrum is the most common anthropophilic dermatophyte.
Genes coding for the various secreted proteases are highly conserved across all dermatophyte species studied (69–71). Unlike Malassezia and Candida, where the secretory proteases are dominated by aspartyl proteases, dermatophytes’ secretory protease repertoire consists of serine and metalloproteases. The dermatophyte extracellular serine protease families include the subtilisins (Subs) (72), dipeptidyl peptidases (Dpps) (73, 74), carboxypeptidases (75, 76), and sedolysins (77). The secretory metalloprotease families consist of the metalloprotease M36 (MEPs) family (72), leucine aminopeptidases (Laps) (73), and deuterolysins (75). The expression of these proteases is regulated by the pH of the environment (78). At alkaline pH, expression is raised for the serine protease Sub3 and metalloprotease Lap1, whereas at acidic pH, secretion of aspartic protease Pep1 increases greatly, which may be relevant in the context of skin and nail infections (75).
The dermatophyte secretory proteases play important roles in degrading skin barrier and structural proteins for efficient epidermal colonization and invasion. These proteases are essential to dermatophyte colonization of host skin and nail tissues, where keratin is the major nutrient source, thereby playing a role in dermatophyte virulence and invasion of host tissues (79). It is, however, important to note that the keratinase activities of dermatophytes are likely relevant only in the presence of sulfite secretion by these fungi (80). The secretion of sulfites results in reduction of the keratin disulfide cross-linkages, allowing the proteases to access the compact keratin for proteolytic degradation (81). Degradation of keratin, which is a hallmark of dermatophytes, proceeds first by endopeptidic cleavage of reduced keratin by the Subs and fungalysins, followed by the degradation of the resulting fragments by the exoproteases Laps, Dpps, and carboxypeptidases to amino acids and small peptides for assimilation and utilization (71). Importantly, expression and secretion of these enzymes are highly regulated and influenced by the external environment (79, 82, 83). A recent study has shown that the protease expression profile of dermatophytes when grown in vitro and during infection varies considerably (84, 85). Many proteases secreted by T. rubrum when grown on keratin-based media in vitro are not found in infection samples and vice versa (86, 87). This emphasizes that potential virulence factors identified in vitro should be verified for expression in physiologically relevant human environments.
SKIN BACTERIUM SECRETORY PROTEASES
The skin bacterial community consists of 4 main phyla—Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria (6), where the Gram-positive Corynebacterium, Cutibacterium, and Staphylococcus account for over half of the skin bacteria (88). Most of the commensal bacterial strains possess few extracellular protease genes compared to the opportunistic pathogens (89). The study of skin bacterial proteases was one of the earliest investigations into the roles of microbial proteases on skin, and most studies have focused on the pathogenic bacteria utilizing proteases for tissue invasion and immune dysregulation (90). While many of these bacterial secretory proteases can serve as facile agents of invasion, recent investigations have shown that the expression of these proteases is tightly controlled and plays a complex role in modulating other virulence factors during bacterial pathogenesis (Table 3). In this minireview, we will focus on the secreted proteases produced by Gram-positive bacteria, which account for the majority of the skin bacterial community.
TABLE 3.
Bacterial proteases and their associated targets and functions
| Organism | Protease | Target(s) | Process(es) involved | Key reference(s) | |
|---|---|---|---|---|---|
| Cutibacterium acnes | Several unidentified proteases | Unknown | Keratinocyte protease activated receptor 2 | Immune modulation | 98, 100 |
| Staphylococcus aureus | Cysteine protease (staphopains) | ScpA | Collagen; CXCR2; complement C3, C5; fibrinogen; host cystatins; LL-37 | Epidermal barrier invasion; immune modulation and evasion; biofilm formation | 122, 127, 135, 162 |
| SspB | Collagen; CD31; complement C5; chemerin; fibrinogen; fibronectin; galectin; host cystatins | ||||
| Metalloprotease | Aureolysin | Collagen; complement C3, C5; LL-37; pro-MMP-9 | Epidermal barrier invasion; immune modulation and evasion; pro-V8 activation | 121, 124, 128 | |
| Serine proteases | Exfoliative toxin A and B | Desmoglein-1 | Epidermal barrier invasion | 116 | |
| V8 (SspA) | Complement C3, C5; IgG; kininogen; S. aureus FnBP | Immune modulation; colonization; biofilm formation; pro-SspB activation | 129, 131, 137, 139 | ||
| Serine protease-like family Spl | SplA to -F | IgG4; human mucin | Biofilm formation | 142 | |
| Staphylococcus epidermidis | Cysteine protease | EcpA | Collagen; elastin; fibronectin; fibrinogen; α1 protease inhibitor; IgA, IgM | Epidermal barrier invasion; immune modulation | 162 – 165 |
| Metalloprotease | SepA | Dermcidin; S. epidermidis Aap | Colonization; biofilm formation | 161, 170 | |
| Serine protease | Esp | Various S. aureus extracellular proteins; fibronectin; fibrinogen; vitronectin | Regulates S. aureus colonization and biofilm formation; epidermal barrier invasion | 166, 168, 169 | |
| Streptococcus pyogenes | Cysteine proteases | IdeS | IgG | Evasion of host phagocytes | 194, 195 |
| Streptopain (SpeB) | Chemokines; complement C3b desmogleins; fibronectin; vitronectin; various immunoglobulins; staphylococcal SdrC | Epidermal barrier invasion; immune modulation and evasion; biofilm formation | 180, 184, 186, 190, 196, 198 | ||
| Serine protease | ScpC | IL-8 | Evasion of host neutrophils | 187, 188 | |
| Streptococcal commensals | Metalloprotease | IgA1 | Human IgA1 | Immune modulation | 174 |
Cutibacterium (formerly Propionibacterium).
Cutibacterium species are Gram-positive, aerotolerant anaerobic bacteria that include the skin-dwelling Cutibacterium acnes, Cutibacterium avidum, and Cutibacterium granulosum (91, 92). They are prevalent across all skin sites and are the most abundant genus of bacteria in sebaceous skin sites due to their affinity for sebum as a nutrient source (23). As anaerobes, they are well adapted for survival in the oxygen-depleted skin invaginations, such as hair follicles and the pilosebaceous unit (93). The most well-studied species is Cutibacterium acnes, which is often associated with acne pathogenesis through biofilm formation which promotes bacterial adhesion to corneocytes, resulting in formation of comedones (94–96).
Early studies demonstrated that several Cutibacterium species have extracellular proteolytic activity when grown in synthetic media (97). Recent proteomic studies of C. acnes secretome (98) and biofilm (99) confirmed the presence and expression of secretory proteases including a putative subtilisin-like protease. While the exact substrates of these proteases are yet to be identified, studies have shown that C. acnes interacts with the host by activating host keratinocyte protease activated receptor 2 (PAR2), suggesting that C. acnes secretes one or more exogenous proteases (100). This activity is raised in acne lesions, which stimulate host expression of cytokines including gamma interferon (IFN-γ), IL-1α, IL-8, IL-17, and TNF-α and antimicrobial peptides human β-defensin 2 and LL-37 (101, 102). Metagenomic analysis of C. acnes in follicular microbiomes of acne patients and healthy individuals (103) revealed the presence of a CAAX amino protease, a membrane-bound metalloprotease involved in bacteriocin self-immunity in acne patients (104). These studies demonstrate that C. acnes can produce secretory proteases that have important effects on the host, but the exact proteases, their associated substrates, and molecular functions of the enzymes need further definition.
Staphylococcus.
Staphylococcus is arguably the most extensively studied genus of skin microbes. Traditionally, staphylococci are classified as coagulase positive or negative depending on the presence of coagulase that clots blood plasma (105). The skin harbors a wide range of the coagulase-negative Staphylococcus (CoNS) species, including Staphylococcus epidermidis, Staphylococcus capitis, Staphylococcus hominis, and Staphylococcus warneri (7). The coagulase-positive Staphylococcus aureus is widely regarded as an opportunistic pathogen capable of producing an arsenal of virulence factors such as extracellular enzymes involved in host epidermal invasion (Fig. 1).
Staphylococcus aureus.
In healthy individuals, S. aureus is harbored mostly in the anterior nares of about one-third of the adult population, while skin colonization is very low (106). S. aureus in the anterior nares can form biofilm that serves as a reservoir for the dissemination of this opportunistic pathogen (107). This coagulase-positive bacterium is historically associated with many skin disorders, ranging from inflammatory dermatological conditions such as atopic dermatitis (AD) to potentially life-threatening skin and soft tissue infections (108–111). A hallmark of S. aureus is its repertoire of secretory virulence factors that facilitate its survival and host invasion (112). These secretory pathogenic factors include allergens, toxins, adhesion factors, and extracellular enzymes including proteases that can facilitate superficial and invasive infections (113). The most well-characterized proteases include cysteine proteases staphopain A (ScpA) and staphopain B (SspB), metalloprotease aureolysin (Aur), serine protease V8 (SspA), and the serine protease-like family (SpIA to -F) (114). Aureolysin, which is activated in an autocatalytic manner, cleaves the proenzyme of V8, which then activates SspB (115). The exfoliative toxins A and B (ETA and ETB, respectively) are serine proteases that degrade desmoglein, a key desmosome protein that maintains the barrier integrity but are present only in a few S. aureus strains (116–118). The functions and involvement in infection of these proteases have been reviewed extensively (90, 119, 120), and we will highlight here the recent findings on the involvement of these proteases in host-microbial interactions.
(i) Degradation of host epidermis-associated proteins.
Host structural proteins such as the extracellular matrix (ECM) proteins and tight and gap junction proteins are involved in maintaining epidermal integrity. Many of these are targets of S. aureus extracellular proteases (90). Collagen, a major structural protein of the human dermal ECM, is a common substrate for S. aureus ScpA, SspB, and Aur (121, 122). A recent study using S. aureus aur gene knockout demonstrated that beyond its own ability to cleave collagen, Aur is able to further promote collagen degradation through its activation of host metalloprotease MMP-9 (121). While collagen is much more abundant in the dermal layer, the effect of these proteases becomes especially significant when the dermis is accessible to the bacteria, such as in wounds or skin abscesses. Interestingly, ScpA was also shown to be involved in inducing cell death when S. aureus propagates intracellularly, and this is likely due to its role in epithelial tissue destruction, which could facilitate bacterial exit from the host cell (123).
S. aureus ScpA, SspB, and Aur can degrade the cationic AMP LL-37 (124, 125), protecting S. aureus from the antimicrobial effect of cathelicidin. In a study done by Sonesson et al. (125), LL-37 fragments generated from staphopain degradation were shown to have immunomodulating effects on the host. Furthermore, the expression of ScpA and SspB was detected directly in skin biopsy specimens, demonstrating that these cysteine proteases are present in physiologically relevant environments (125).
(ii) Interference with host immune pathways.
S. aureus secreted proteases interfere with host immune pathways in multiple ways. SspB degrades multiple immune cell surface receptors such as CD11b, CD16, and CD31 crucial for induction of phagocytosis, enabling S. aureus to evade phagocytosis (126, 127). ScpA, SspB, V8, and Aur are effective inhibitors of the host complement pathway (128, 129), where the proteases act upon various complement components including C3 and C5. S. aureus isogenic mutants lacking SspB, V8, or Aur showed decreased survival in human blood (129). Galectin, an immunomodulating lectin produced by epithelial cells, is a newly discovered substrate of SspB. In a murine subcutaneous infection model, galectin knockout mice infected with S. aureus showed smaller lesions than wild-type mice. As galectin activates neutrophils, SspB enhances S. aureus virulence through cleavage of galectin that abolishes its ability to activate neutrophils (130). A recent study by Frey et al. utilized N-terminal degradomics to unravel new human serum protein substrates of V8 protease (131). This work highlights the role of V8 in interfering with host inflammatory signaling pathways through degradation of components of the complement pathway and host protease inhibitors such as SERPINs.
While most studies have focused on the proteolytic activity of S. aureus proteases in affecting the host immune response, a study by Stentzel et al. (132) observed increased IgE binding to the Spl serine proteases (A to F) in asthmatic patients. This suggests that the Spl proteases, which are encoded on a single operon, can be potential allergens (132).
(iii) Self-modulating effects.
S. aureus can form biofilms which have been associated with poor healing outcome in wounds (133) and potentially contribute to inflammation in AD (134). The staphopains ScpA and SspB were found to have an inhibitory effect on biofilm formation and maintenance (135). V8 and Aur were also reported to inhibit S. aureus biofilm formation (136), where V8 is known to cleave S. aureus adhesin FnBP (fibronectin-binding protein), a protein mediating cellular adhesion to host ECM substrates (137).
While S. aureus extracellular proteases are generally regarded as virulence factors, recent studies have shown that these enzymes can modulate virulence indirectly. A mutant lacking all 10 of the extracellular enzymes had dramatically reduced penetration into the deeper skin tissues (138) and decreased skin abscess formation but was found to be hypervirulent (139), likely due to the roles of these proteases in controlling the stability of secreted toxins (139–141) and surface-associated adhesion proteins (142, 143). Using a combination of protease deletion mutants, Gimza et al. identified Aur and ScpA as the two keystone proteases contributing to virulence through controlling abundance of S. aureus virulence factors (144). These studies highlight that the functions of S. aureus secreted proteases go beyond direct invasion and include modulation of other extracellular factors important to establish infection.
(iv) Importance of strain variation in pathogenicity of S. aureus.
In studying S. aureus secretory proteases, it has become clear in recent studies that it is important to consider the pathogenicity differences among S. aureus strains. This is relevant for both the presence of protease genes and the complex of regulatory elements that control protease gene expression (145), of which the agr operon (146, 147) and sarA are two key regulators (148–150). In a study involving 6-month-old infants, mutations in the Agr-quorum sensing system were more frequently observed in subjects who did not develop AD (151). Comparative genomics of diabetic foot ulcer S. aureus strains associated with different healing outcomes further revealed disparities in the presence of virulence-associated genes between the strains (152). Overall, it is crucial to consider the strain-associated heterogenicity leading to varied expression of S. aureus extracellular proteases as this could contribute to differences in disease severity.
Staphylococcus epidermidis.
The CoNS species represent one of the most abundant bacterial communities on the human skin and are especially prevalent on moist and sebaceous skin sites (7). Species of CoNS are generally considered to be commensal or even beneficial members of the skin microbiome as they utilize several mechanisms to limit growth of pathogens on the skin (23, 153). CoNS also possess fewer extracellular toxins capable of direct tissue invasion (154, 155). Functional annotations of the CoNS reveal that these species do possess secretory proteases, but the functions and molecular substrates of these enzymes are poorly defined. The most prominent member of the skin CoNS is S. epidermidis (156). Several studies have demonstrated S. epidermidis’s role as a beneficial microbe through increasing innate skin barrier immunity by tuning T cells (157, 158). However, S. epidermidis can cause opportunistic infections, such as when introduced into the body through medical implants and devices (159, 160), but such occurrences are rare compared to S. aureus. Overall, S. epidermidis favors persistence on the skin rather than host invasion and tissue destruction (160).
The secreted proteases of S. epidermidis include the serine protease Esp, metalloprotease SepA, and cysteine protease EcpA (89). While Esp has low homology to other staphylococcal proteases, SepA is a homologue of the S. aureus metalloprotease Aur (161) and EcpA shares substantial homology to the S. aureus staphopains ScpA and SspB (162).
(i) Degradation of host epidermis-associated proteins.
EcpA is expressed as part of the ecpAB operon upstream of its endogenous inhibitor EcpB. The cysteine protease EcpA can be found attached to the cell surface of S. epidermidis or secreted into the environment, and the proportions of these two forms differ between strains (163). Williams et al. reported that several cultured S. epidermidis strains isolated from a patient suffering from the rare Netherton syndrome (monogenic dermatological disease from loss of the human protease inhibitor LEKTI-1) had EcpA activity (162). In a recent study on AD patients, Cau et al. (164) observed that when S. epidermidis is present at high density, such as on AD lesional sites, this can induce expression of ecpA. This protease expression is under quorum sensing control, and high density of S. epidermidis is correlated with increased ecpA expression (164). Furthermore, this cysteine protease can degrade skin structural proteins such as collagen, elastin, fibronectin, and desmoglein-1 (164), the AMP LL-37, and various blood plasma-associated endogenous protease inhibitors (165) in vitro. In a mouse epicutaneous exposure model, only S. epidermidis wild type, but not the ecpA knockout strain, was able to elicit skin barrier damage and inflammation (164). In addition, S. epidermidis serine protease Esp can also degrade fibronectin, fibrinogen, and vitronectin in vitro (166), while the metalloprotease SepA is simultaneously upregulated by and degrades the sweat-associated anionic AMP dermicidin (161). Overall, the degradation of these host proteins indicates that S. epidermis has pathogenic potential, but further studies are needed to decipher what are the factors controlling expression and activities of these proteases as S. epidermis is prevalent and abundant on healthy human skin (2). One emerging theory, similar to S. aureus, is the expansion of particular S. epidermidis agr types that control and increase expression of the proteases at high density in skin diseases (167).
(ii) Interactions with other microbial communities.
The S. epidermidis serine protease Esp inhibits S. aureus colonization by blocking S. aureus biofilm formation and destroying preformed S. aureus biofilms (168, 169). Examination of S. aureus biofilm disassembly revealed the ability of Esp to degrade several S. aureus proteins involved in colonization and biofilm formation (166). Autolysin (Atl), extracellular matrix protein (Emp), FnBP, and SpA are some examples of S. aureus proteins targeted by Esp (166). The metalloprotease SepA also plays a role in biofilm formation by processing the S. epidermidis cell-wall-anchored Aap protein to form an adhesin that facilitates biofilm accumulation (170). Other than S. aureus biofilm inhibition, Esp was found to augment the bactericidal effect of human keratinocyte AMP β-defensin 2 against S. aureus (169). However, a follow-up study that assessed the expression of esp in S. epidermidis isolates from the nose of healthy adolescents using semiquantitative PCR did not find any correlation between esp expression and biofilm inhibition (171). As the gene expression analysis was done for only 9 strains, follow-up studies with a larger number of S. epidermidis strains are needed to decipher the relationship between esp expression and biofilm formation.
Streptococcus.
The Streptococcus genus is a group of Gram-positive bacteria that are commonly found in the oral and nasopharyngeal microflora of healthy individuals (172). Several Streptococcus species including Streptococcus mitis, Streptococcus oralis, Streptococcus pseudopneumoniae, and Streptococcus sanguinis are prevalent at the dry skin sites in healthy individuals (2). The most well-characterized Streptococcus is the pathogen group A Streptococcus (GAS) Streptococcus pyogenes. Though relatively uncommon in the healthy skin microbiome, S. pyogenes is implicated in a wide range of skin and soft tissue infections such as impetigo, ecthyma, cellulitis, and necrotizing fasciitis (111, 173).
Streptococcal commensals.
The skin commensals S. mitis, S. oralis, and S. sanguinis produce the extracellular metalloprotease IgA1 protease, which cleaves the human IgA1 at the hinge region of the heavy chain which connects the constant fragment (Fc) region to the antigen binding region (Fab) (174). The IgA1 proteases are large proteins (around 130 to 200 kDa) that possess a Gram-positive cell wall anchoring motif and can be secreted or cell wall associated. In particular, the iga genes in S. sanguinis and S. oralis share high homology while S. mitis and Streptococcus pneumoniae iga genes have moderate similarities (175). The precise functions of these Streptococcus IgA1 proteases are unclear, and most studies have focused on their roles as a virulence factor in S. pneumoniae, a pathogen that causes bacterial pneumonia and meningitis (172). The cleavage of the IgA1 hinge region leaves the Fab region intact, and these Fab fragments can in turn mask the bacterial surface epitopes without eliciting a downstream immune response as the Fc region is disconnected. Furthermore, IgA protease can inhibit phagocytic killing of S. pneumoniae by targeting the capsule-specific human IgA1 monoclonal antibody (176). However, the expression and functions of these IgA proteases, especially those secreted by the common streptococcal skin commensals, have yet to be investigated in detail.
Streptococcus pyogenes.
In contrast to the other Streptococcus skin commensals, S. pyogenes has high pathogenicity and causes infection on skin and mucosal surfaces. Similarly to S. aureus, S. pyogenes expresses a variety of extracellular virulence factors (173) including the secretory cysteine protease streptococcal pyrogenic exotoxin B (SpeB, also called streptopain), cysteine protease IdeS (IgG-degrading enzyme of S. pyogenes), and serine protease ScpC (also called SpyCEP).
(i) Degradation of host epidermis-associated proteins.
The cutaneous barrier is composed of layers of differentiated keratinocytes joined together by intercellular junctions such as the desmosomes and tight junctions (177, 178). SpeB was found to cleave these intercellular junction proteins including desmoglein-1, desmoglein-3, E-cadherin, and occludin, facilitating the dissemination of and invasion by S. pyogenes in skin infections (179, 180). Host ECM proteins fibronectin and vitronectin are also targets of SpeB (181). An S. pyogenes insertional mutant of SpeB which lacks expression of this protease (182) results in a smaller skin abscess and lesions in murine subcutaneous infection (183). Furthermore, a mutant strain that constitutively expressed SpeB in a murine subcutaneous infection model resulted in increased lesion size compared to the wild-type control (184). However, SpeB expression and protease activity were higher in clinical isolates from nonsevere infections compared to those isolated from severe infections (185). These studies demonstrate once again that it is important to consider contextual expression of these bacterial proteases, and similar to S. aureus, these extracellular S. pyogenes proteases can play a regulatory role in controlling other virulence factors important for infection to establish.
(ii) Interference with host immune pathways.
One of the best-characterized roles of S. pyogenes secretory proteases is their ability to degrade host immune factors ranging from cytokines (186–189) to complement proteins (190–193) and immunoglobulins (Igs). One crucial pathway that this pathogen has to overcome is the adaptive immune response facilitated by Igs. As antibodies can activate phagocytic cells and complement pathways, S. pyogenes gains resistance to antibody-mediated opsonophagocytosis by degrading these proteins (194, 195). Early studies uncovered the role of SpeB in degrading multiple Ig classes (196); IdeS, on the other hand, has specificity for cleavage of both circulating and Fab-bound IgG (194, 195) and was demonstrated to be a more efficient enzyme than SpeB in degrading IgG (197). Recent work by Persson et al. brought into question whether the degradation of multiple classes of Igs by SpeB is physiologically relevant as this cysteine protease can cleave the heavy chain of the Ig only when these substrates are in the reduced, semimonomeric form (197).
(iii) Interaction with other microbial communities.
Most of the attention on S. pyogenes secreted proteases has been on their effect on cleavage of host proteins, but less is understood about their role in interaction with other microbes, especially in the context of a mixed microbial community. In a recent study, Carothers et al. (198) demonstrated by using an isogenic SpeB mutant of a skin-tropic S. pyogenes strain and recombinant SpeB that this protease is involved in attenuation of S. aureus biofilm formation. SdrC, a cell-wall-anchored S. aureus adhesin, was shown to be a substrate of SpeB, and its degradation leads to disruption of the biofilm (198). This study highlights the importance of understanding the roles of these secretory proteases in the context of the mixed microbial communities present at different sites.
CONCLUSION/PERSPECTIVE
The roles of skin microbial secretory proteases have long intrigued many, and historically these proteases are regarded as virulent agents in microbial pathogenesis. Recent studies have revealed that many factors need to be taken into consideration when studying microbial protease functions. First, secreted proteases from microbes involved in infection can both serve as direct agents of invasion and indirectly modulate the abundance and stability of other virulence factors. Second, expression of the same proteases can differ dramatically within specific strains of the same species due to differences in the regulatory elements. Finally, it cannot be emphasized enough that skin host phenotype and environmental context are critical factors resulting in dynamic regulation of these secretory enzymes. The presence of potentially virulent genes does not automatically translate to constitutive expression and emphasizes the importance to go beyond metagenomic studies to validate expression in situ. Furthermore, the same protease could have beneficial roles when the skin barrier is intact but become an agent of pathogenesis when the skin barrier is compromised due to wounds or skin diseases (Fig. 1). It is therefore essential to consider the context and environment where the protease is expressed when elucidating the functional roles of the secreted proteases.
Many advanced analytical tools in studying microbial phenotype and protease activities have been introduced in recent years. These include large-scale culturomics studies (199, 200), mass spectrometry-based degradomics (201), and chemical probes to monitor protease activities (202, 203). Degradomics, the comprehensive functional analysis of proteases and their associated substrates (21), is a particularly powerful tool to identify novel protease substrates and quantify the extent of substrate degradation (204). The application of these techniques, together with the available database of microbe sequence databases, will advance our understanding of the molecular functions of skin microbial proteases in skin health, especially the understudied skin commensals that account for the vast majority of the skin microbiome.
ACKNOWLEDGMENTS
H.L. acknowledges support from the MOE AcRF Tier 1 grant (R-143-000-B79-114), Singapore Ministry of Health’s National Medical Research Council (MOH-000612-00), and Wound Care Innovations for the Tropics Program, A*STAR Industry Alignment Fund-Pre-Positioning grant (H19/01/a0/0GG9).
Biographies

Wisely Chua completed his undergraduate degree in Pharmacy at the National University of Singapore in 2019. He is currently working as a research assistant in the Institute of Molecular and Cell Biology at the Agency for Science, Technology and Research, Singapore. He is focused on characterizing Malassezia and bacterial secreted proteases and investigating their diagnostic and therapeutic potential for chronic wound infections.

Si En Poh graduated from Nanyang Technological University, Singapore, in 2017, with a B.Sc. in Biological Sciences. She is currently working as a research assistant at the Institute of Molecular and Cell Biology at the Agency for Science, Technology and Research, Singapore. Her current interest includes designing sequencing assays to determine expression of skin microbial secreted enzymes.

Hao Li, Ph.D., is an assistant professor at the Department of Chemistry at the National University of Singapore and research scientist at the Institute of Molecular and Cell Biology at the Agency for Science, Technology and Research, Singapore. She graduated with a B.Sc. (Hons) in Biochemistry at the University of Wisconsin-Madison and obtained her Ph.D. in Chemical and Systems Biology at Stanford University. Her laboratory’s research focuses on deciphering at a molecular level how microbial enzymes and their associated molecules structure the human environments where they reside. She is particularly interested in studying secretory enzymes of the human microbiome.
Contributor Information
Hao Li, Email: chmlihao@nus.edu.sg.
Anthony R. Richardson, University of Pittsburgh
REFERENCES
- 1.Proksch E, Brandner JM, Jensen JM. 2008. The skin: an indispensable barrier. Exp Dermatol 17:1063–1072. 10.1111/j.1600-0625.2008.00786.x. [DOI] [PubMed] [Google Scholar]
- 2.Byrd AL, Belkaid Y, Segre JA. 2018. The human skin microbiome. Nat Rev Microbiol 16:143–155. 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- 3.Gao Z, Perez-Perez GI, Chen Y, Blaser MJ. 2010. Quantitation of major human cutaneous bacterial and fungal populations. J Clin Microbiol 48:3575–3581. 10.1128/JCM.00597-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, NISC Comparative Sequencing Program, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192. 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, NIH Intramural Sequencing Center Comparative Sequencing Program, Kong HH, Segre JA. 2013. Topographic diversity of fungal and bacterial communities in human skin. Nature 498:367–370. 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh J, Byrd AL, Deming C, Conlan S, NISC Comparative Sequencing Program, Kong HH, Segre JA. 2014. Biogeography and individuality shape function in the human skin metagenome. Nature 514:59–64. 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belkaid Y, Segre JA. 2014. Dialogue between skin microbiota and immunity. Science 346:954–959. 10.1126/science.1260144. [DOI] [PubMed] [Google Scholar]
- 8.Oh J, Byrd AL, Park M, NISC Comparative Sequencing Program, Kong HH, Segre JA. 2016. Temporal stability of the human skin microbiome. Cell 165:854–866. 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen YE, Fischbach MA, Belkaid Y. 2018. Skin microbiota-host interactions. Nature 553:427–436. 10.1038/nature25177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo RL. 2013. The microbiome extends to subepidermal compartments of normal skin. Nat Commun 4:1431–1438. 10.1038/ncomms2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallo RL. 2017. Human skin is the largest epithelial surface for interaction with microbes. J Invest Dermatol 137:1213–1214. 10.1016/j.jid.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsui T, Amagai M. 2015. Dissecting the formation, structure and barrier function of the stratum corneum. Int Immunol 27:269–280. 10.1093/intimm/dxv013. [DOI] [PubMed] [Google Scholar]
- 13.Nemes Z, Steinert PM. 1999. Bricks and mortar of the epidermal barrier. Exp Mol Med 31:5–19. 10.1038/emm.1999.2. [DOI] [PubMed] [Google Scholar]
- 14.Drag M, Salvesen GS. 2010. Emerging principles in protease-based drug discovery. Nat Rev Drug Discov 9:690–701. 10.1038/nrd3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.López-Otín C, Bond JS. 2008. Proteases: multifunctional enzymes in life and disease. J Biol Chem 283:30433–30437. 10.1074/jbc.R800035200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein T, Eckhard U, Dufour A, Solis N, Overall CM. 2018. Proteolytic cleavage - mechanisms, function, and “omic” approaches for a near-ubiquitous posttranslational modification. Chem Rev 118:1137–1168. 10.1021/acs.chemrev.7b00120. [DOI] [PubMed] [Google Scholar]
- 17.Page MJ, Di Cera E. 2008. Evolution of peptidase diversity. J Biol Chem 283:30010–30014. 10.1074/jbc.M804650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cezairliyan B, Ausubel FM. 2017. Investment in secreted enzymes during nutrient-limited growth is utility dependent. Proc Natl Acad Sci USA 114:E7796–E7802. 10.1073/pnas.1708580114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holland KT, Greenman J, Cunliffe WJ. 1979. Growth of cutaneous propionibacteria on synthetic medium; growth yields and exoenzyme production. J Appl Bacteriol 10.1111/j.1365-2672.1979.tb01198.x. [DOI] [PubMed] [Google Scholar]
- 20.Allison SD, Vitousek PM. 2005. Responses of extracellular enzymes to simple and complex nutrient inputs. Soil Biol Biochem 37:937–944. 10.1016/j.soilbio.2004.09.014. [DOI] [Google Scholar]
- 21.López-Otín C, Overall CM. 2002. Protease degradomics: a new challenge for proteomics. Nat Rev Mol Cell Biol 3:509–519. 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen TTH, Myrold DD, Mueller RS. 2019. Distributions of extracellular peptidases across prokaryotic genomes reflect phylogeny and habitat. Front Microbiol 10:413. 10.3389/fmicb.2019.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flowers L, Grice EA. 2020. The skin microbiota: balancing risk and reward. Cell Host Microbe 28:190–200. 10.1016/j.chom.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Hoog S, Monod M, Dawson T, Boekhout T, Mayser P, Gräser Y. 2017. Skin fungi from colonization to infection. Microbiol Spectr 5(4). 10.1128/microbiolspec.FUNK-0049-2016. [DOI] [PubMed] [Google Scholar]
- 25.Patterson TF. 2005. Advances and challenges in management of invasive mycoses. Lancet 366:1013–1025. 10.1016/S0140-6736(05)67381-3. [DOI] [PubMed] [Google Scholar]
- 26.Sardi JCO, Scorzoni L, Bernardi T, Fusco-Almeida AM, Mendes Giannini MJS. 2013. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol 62:10–24. 10.1099/jmm.0.045054-0. [DOI] [PubMed] [Google Scholar]
- 27.Naglik J, Challacombe S, Hube B. 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev 67:400–428. 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krysan DJ, Ting EL, Abeijon C, Kroos L, Fuller RS. 2005. Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot Cell 4:1364–1374. 10.1128/EC.4.8.1364-1374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albrecht A, Felk A, Pichova I, Naglik JR, Schaller M, De Groot P, MacCallum D, Odds FC, Schäfer W, Klis F, Monod M, Hube B. 2006. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J Biol Chem 281:688–694. 10.1074/jbc.M509297200. [DOI] [PubMed] [Google Scholar]
- 30.Felk A, Kretschmar M, Albrecht A, Schaller M, Beinhauer S, Nichterlein T, Sanglard D, Korting HC, Schäfer W, Hube B. 2002. Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs. Infect Immun 70:3689–3700. 10.1128/IAI.70.7.3689-3700.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gogol M, Bochenska O, Zawrotniak M, Karkowska-Kuleta J, Zajac D, Rapala-Kozik M. 2017. Roles of Candida albicans aspartic proteases in host-pathogen interactions, p 353–380. In Chakraborti S, Dhalla N (ed), Pathophysiological aspects of proteases. Springer, Singapore. [Google Scholar]
- 32.Koelsch G, Tang J, Loy JA, Monod M, Jackson K, Foundling SI, Lin X. 2000. Enzymic characteristics of secreted aspartic proteases of Candida albicans. Biochim Biophys Acta 1480:117–131. 10.1016/S0167-4838(00)00068-6. [DOI] [PubMed] [Google Scholar]
- 33.Jackson BE, Wilhelmus KR, Hube B. 2007. The role of secreted aspartyl proteinases in Candida albicans keratitis. Invest Ophthalmol Vis Sci 48:3559–3565. 10.1167/iovs.07-0114. [DOI] [PubMed] [Google Scholar]
- 34.Naglik JR, Moyes D, Makwana J, Kanzaria P, Tsichlaki E, Weindl G, Tappuni AR, Rodgers CA, Woodman AJ, Challacombe SJ, Schaller M, Hube B. 2008. Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis. Microbiology (Reading) 154:3266–3280. 10.1099/mic.0.2008/022293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pereira R, Santos Fontenelle RO, Brito EHS, Morais SM. 2021. Biofilm of Candida albicans: formation, regulation and resistance. J Appl Microbiol 131:11–22. 10.1111/jam.14949. [DOI] [PubMed] [Google Scholar]
- 36.Ghannoum M, Elteen KA. 1986. Correlative relationship between proteinase production, adherence and pathogenicity of various strains of Candida albicans. J Med Vet Mycol 24:407–413. 10.1080/02681218680000621. [DOI] [PubMed] [Google Scholar]
- 37.Gogol M, Ostrowska D, Klaga K, Bochenska O, Wolak N, Aoki W, Ueda M, Kozik A, Rapala-Kozik M. 2016. Inactivation of α1-proteinase inhibitor by Candida albicans aspartic proteases favors the epithelial and endothelial cell colonization in the presence of neutrophil extracellular traps. Acta Biochim Pol 63:167–175. 10.18388/abp.2015_1163. [DOI] [PubMed] [Google Scholar]
- 38.Gropp K, Schild L, Schindler S, Hube B, Zipfel PF, Skerka C. 2009. The yeast Candida albicans evades human complement attack by secretion of aspartic proteases. Mol Immunol 47:465–475. 10.1016/j.molimm.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 39.Svoboda E, Schneider AE, Sándor N, Lermann U, Staib P, Kremlitzka M, Bajtay Z, Barz D, Erdei A, Józsi M. 2015. Secreted aspartic protease 2 of Candida albicans inactivates factor H and the macrophage factor H-receptors CR3 (CD11b/CD18) and CR4 (CD11c/CD18). Immunol Lett 168:13–21. 10.1016/j.imlet.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 40.Willems HME, Bruner WS, Barker KS, Liu J, Palmer GE, Peters BM. 2017. Overexpression of Candida albicans secreted aspartyl proteinase 2 or 5 is not sufficient for exacerbation of immunopathology in a murine model of vaginitis. Infect Immun 85:e00248-17. 10.1128/IAI.00248-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herman A, Herman AP. 2019. Antimicrobial peptides activity in the skin. Skin Res Technol 25:111–117. 10.1111/srt.12626. [DOI] [PubMed] [Google Scholar]
- 42.Rapala-Kozik M, Bochenska O, Zawrotniak M, Wolak N, Trebacz G, Gogol M, Ostrowska D, Aoki W, Ueda M, Kozik A. 2015. Inactivation of the antifungal and immunomodulatory properties of human cathelicidin LL-37 by aspartic proteases produced by the pathogenic yeast Candida albicans. Infect Immun 83:2518–2530. 10.1128/IAI.00023-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bochenska O, Rapala-Kozik M, Wolak N, Aoki W, Ueda M, Kozik A. 2016. The action of ten secreted aspartic proteases of pathogenic yeast Candida albicans on major human salivary antimicrobial peptide, histatin 5. Acta Biochim Pol 63:403–410. 10.18388/abp.2016_1318. [DOI] [PubMed] [Google Scholar]
- 44.Bochenska O, Rapala-Kozik M, Wolak N, Kamysz W, Grzywacz D, Aoki W, Ueda M, Kozik A. 2015. Inactivation of human kininogen-derived antimicrobial peptides by secreted aspartic proteases produced by the pathogenic yeast Candida albicans. Biol Chem 396:1369–1375. 10.1515/hsz-2015-0167. [DOI] [PubMed] [Google Scholar]
- 45.Pietrella D, Rachini A, Pandey N, Schild L, Netea M, Bistoni F, Hube B, Vecchiarelli A. 2010. The inflammatory response induced by aspartic proteases of Candida albicans is independent of proteolytic activity. Infect Immun 78:4754–4762. 10.1128/IAI.00789-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pietrella D, Pandey N, Gabrielli E, Pericolini E, Perito S, Kasper L, Bistoni F, Cassone A, Hube B, Vecchiarelli A. 2013. Secreted aspartic proteases of Candida albicans activate the NLRP3 inflammasome. Eur J Immunol 43:679–692. 10.1002/eji.201242691. [DOI] [PubMed] [Google Scholar]
- 47.O’Donoghue AJ, Eroy-Reveles AA, Knudsen GM, Ingram J, Zhou M, Statnekov JB, Greninger AL, Hostetter DR, Qu G, Maltby DA, Anderson MO, Derisi JL, McKerrow JH, Burlingame AL, Craik CS. 2012. Global identification of peptidase specificity by multiplex substrate profiling. Nat Methods 9:1095–1100. 10.1038/nmeth.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Winter MB, Salcedo EC, Lohse MB, Hartooni N, Gulati M, Sanchez H, Takagi J, Hube B, Andes DR, Johnson AD, Craik CS, Nobile CJ. 2016. Global identification of biofilm-specific proteolysis in Candida albicans. mBio 7:e01514-16. 10.1128/mBio.01514-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joo MY, Shin JH, Jang HC, Song ES, Kee SJ, Shin MG, Suh SP, Ryang DW. 2013. Expression of SAP5 and SAP9 in Candida albicans biofilms: comparison of bloodstream isolates with isolates from other sources. Med Mycol 51:892–896. 10.3109/13693786.2013.824623. [DOI] [PubMed] [Google Scholar]
- 50.Dutton LC, Jenkinson HF, Lamont RJ, Nobbs AH. 2016. Role of Candida albicans secreted aspartyl protease Sap9 in interkingdom biofilm formation. Pathog Dis 74:ftw005. 10.1093/femspd/ftw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lohse MB, Gulati M, Craik CS, Johnson AD, Nobile CJ. 2020. Combination of antifungal drugs and protease inhibitors prevent Candida albicans biofilm formation and disrupt mature biofilms. Front Microbiol 11:1027. 10.3389/fmicb.2020.01027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dabiri S, Shams-Ghahfarokhi M, Razzaghi-Abyaneh M. 2018. Comparative analysis of proteinase, phospholipase, hydrophobicity and biofilm forming ability in Candida species isolated from clinical specimens. J Mycol Med 28:437–442. 10.1016/j.mycmed.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 53.Silva S, Hooper SJ, Henriques M, Oliveira R, Azeredo J, Williams DW. 2011. The role of secreted aspartyl proteinases in Candida tropicalis invasion and damage of oral mucosa. Clin Microbiol Infect 17:264–272. 10.1111/j.1469-0691.2010.03248.x. [DOI] [PubMed] [Google Scholar]
- 54.Singh DK, Németh T, Papp A, Tóth R, Lukácsi S, Heidingsfeld O, Dostal J, Vágvölgyi C, Bajtay Z, Józsi M, Gácser A. 2019. Functional characterization of secreted aspartyl proteases in Candida parapsilosis. mSphere 4:e00484-19. 10.1128/mSphere.00484-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dostál J, Pecina A, Hrušková-Heidingsfeldová O, Marečková L, Pichová I, Řezáčová P, Lepšík M, Brynda J. 2015. Atomic resolution crystal structure of Sapp2p, a secreted aspartic protease from Candida parapsilosis. Acta Crystallogr D Biol Crystallogr 71:2494–2504. 10.1107/S1399004715019392. [DOI] [PubMed] [Google Scholar]
- 56.Hruskova-Heidingsfeldova O, Dostál J, Majer F, Havlíková J, Hradilek M, Pichová I. 2009. Two aspartic proteinases secreted by the pathogenic yeast Candida parapsilosis differ in expression pattern and catalytic properties. Biol Chem 390:259–268. 10.1515/BC.2009.034. [DOI] [PubMed] [Google Scholar]
- 57.Theelen B, Cafarchia C, Gaitanis G, Bassukas ID, Boekhout T, Dawson TL. 2018. Malassezia ecology, pathophysiology, and treatment. Med Mycol 56:S10–S25. 10.1093/mmy/myx134. [DOI] [PubMed] [Google Scholar]
- 58.Grice EA, Dawson TL. 2017. Host–microbe interactions: Malassezia and human skin. Curr Opin Microbiol 40:81–87. 10.1016/j.mib.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 59.Chandra SHV, Srinivas R, Dawson TL, Common JE. 2021. Cutaneous Malassezia: commensal, pathogen, or protector? Front Cell Infect Microbiol 10:614446. 10.3389/fcimb.2020.614446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu G, Zhao H, Li C, Rajapakse MP, Wong WC, Xu J, Saunders CW, Reeder NL, Reilman RA, Scheynius A, Sun S, Billmyre BR, Li W, Averette AF, Mieczkowski P, Heitman J, Theelen B, Schröder MS, De Sessions PF, Butler G, Maurer-Stroh S, Boekhout T, Nagarajan N, Dawson TL. 2015. Genus-wide comparative genomics of Malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genet 11:e1005614. 10.1371/journal.pgen.1005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Goh BN, Teh WK, Jiang Z, Goh JPZ, Goh A, Wu G, Hoon SS, Raida M, Camattari A, Yang L, O’Donoghue AJ, Dawson TL. 2018. Skin commensal Malassezia globosa secreted protease attenuates Staphylococcus aureus biofilm formation. J Invest Dermatol 138:1137–1145. 10.1016/j.jid.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 62.Rawlings ND, Salvesen G (ed). 2013. Handbook of proteolytic enzymes, 3rd ed. Academic Press, San Diego, CA. [Google Scholar]
- 63.Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, Calvo E, Lopez JA, Foster TJ, Penadés JR, Lasa I. 2009. Protein A-mediated multicellular behavior in Staphylococcus aureus. J Bacteriol 191:832–843. 10.1128/JB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.von Eiff C, Becker K, Machka K, Stammer H, Peters G. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N Engl J Med 344:11–16. 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 65.Prohic A, Jovovic Sadikovic T, Krupalija-Fazlic M, Kuskunovic-Vlahovljak S. 2016. Malassezia species in healthy skin and in dermatological conditions. Int J Dermatol 55:494–504. 10.1111/ijd.13116. [DOI] [PubMed] [Google Scholar]
- 66.Poh SE, Goh JPZ, Fan C, Chua W, Gan SQ, Lim PLK, Sharma B, Leavesley DI, Dawson TL, Li H. 2020. Identification of Malassezia furfur secreted aspartyl protease 1 (MfSAP1) and its role in extracellular matrix degradation. Front Cell Infect Microbiol 10:148. 10.3389/fcimb.2020.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Hoog GS, Dukik K, Monod M, Packeu A, Stubbe D, Hendrickx M, Kupsch C, Stielow JB, Freeke J, Göker M, Rezaei-Matehkolaei A, Mirhendi H, Gräser Y. 2017. Toward a novel multilocus phylogenetic taxonomy for the dermatophytes. Mycopathologia 182:5–31. 10.1007/s11046-016-0073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma Y, Wang X, Li R. 2021. Cutaneous and subcutaneous fungal infections: recent developments on host–fungus interactions. Curr Opin Microbiol 62:93–102. 10.1016/j.mib.2021.05.005. [DOI] [PubMed] [Google Scholar]
- 69.Jousson O, Léchenne B, Bontems O, Capoccia S, Mignon B, Barblan J, Quadroni M, Monod M. 2004. Multiplication of an ancestral gene encoding secreted fungalysin preceded species differentiation in the dermatophytes Trichophyton and Microsporum. Microbiology (Reading) 150:301–310. 10.1099/mic.0.26690-0. [DOI] [PubMed] [Google Scholar]
- 70.Jousson O, Léchenne B, Bontems O, Mignon B, Reichard U, Barblan J, Quadroni M, Monod M. 2004. Secreted subtilisin gene family in Trichophyton rubrum. Gene 339:79–88. 10.1016/j.gene.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 71.Monod M. 2008. Secreted proteases from dermatophytes. Mycopathologia 166:285–294. 10.1007/s11046-008-9105-4. [DOI] [PubMed] [Google Scholar]
- 72.Giddey K, Monod M, Barblan J, Potts A, Waridel P, Zaugg C, Quadroni M. 2007. Comprehensive analysis of proteins secreted by Trichophyton rubrum and Trichophyton violaceum under in vitro conditions. J Proteome Res 6:3081–3092. 10.1021/pr070153m. [DOI] [PubMed] [Google Scholar]
- 73.Monod M, Léchenne B, Jousson O, Grand D, Zaugg C, Stöcklin R, Grouzmann E. 2005. Aminopeptidases and dipeptidyl-peptidases secreted by the dermatophyte Trichophyton rubrum. Microbiology (Reading) 151:145–155. 10.1099/mic.0.27484-0. [DOI] [PubMed] [Google Scholar]
- 74.Kaufman G, Berdicevsky I, Woodfolk JA, Horwitz BA. 2005. Markers for host-induced gene expression in Trichophyton dermatophytosis. Infect Immun 73:6584–6590. 10.1128/IAI.73.10.6584-6590.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sriranganadane D, Waridel P, Salamin K, Feuermann M, Mignon B, Staib P, Neuhaus JM, Quadroni M, Monod M. 2011. Identification of novel secreted proteases during extracellular proteolysis by dermatophytes at acidic pH. Proteomics 11:4422–4433. 10.1002/pmic.201100234. [DOI] [PubMed] [Google Scholar]
- 76.Zaugg C, Jousson O, Léchenne B, Staib P, Monod M. 2008. Trichophyton rubrum secreted and membrane-associated carboxypeptidases. Int J Med Microbiol 298:669–682. 10.1016/j.ijmm.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 77.Burmester A, Shelest E, Glöckner G, Heddergott C, Schindler S, Staib P, Heidel A, Felder M, Petzold A, Szafranski K, Feuermann M, Pedruzzi I, Priebe S, Groth M, Winkler R, Li W, Kniemeyer O, Schroeckh V, Hertweck C, Hube B, White TC, Platzer M, Guthke R, Heitman J, Wöstemeyer J, Zipfel PF, Monod M, Brakhage AA. 2011. Comparative and functional genomics provide insights into the pathogenicity of dermatophytic fungi. Genome Biol 12:R7. 10.1186/gb-2011-12-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peres NTA, Sanches PR, Falcão JP, Silveira HCS, Paião FG, Maranhão FCA, Gras DE, Segato F, Cazzaniga RA, Mazucato M, Cursino-Santos JR, Aquino-Ferreira R, Rossi A, Martinez-Rossi NM. 2010. Transcriptional profiling reveals the expression of novel genes in response to various stimuli in the human dermatophyte Trichophyton rubrum. BMC Microbiol 10:39. 10.1186/1471-2180-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen J, Yi J, Liu L, Yin S, Chen R, Li M, Ye C, Zhang Y, Lai W. 2010. Substrate adaptation of Trichophyton rubrum secreted endoproteases. Microb Pathog 48:57–61. 10.1016/j.micpath.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 80.Mercer DK, Stewart CS. 2019. Keratin hydrolysis by dermatophytes. Med Mycol 57:13–22. 10.1093/mmy/myx160. [DOI] [PubMed] [Google Scholar]
- 81.Grumbt M, Monod M, Yamada T, Hertweck C, Kunert J, Staib P. 2013. Keratin degradation by dermatophytes relies on cysteine dioxygenase and a sulfite efflux pump. J Invest Dermatol 133:1550–1555. 10.1038/jid.2013.41. [DOI] [PubMed] [Google Scholar]
- 82.Apodaca G, McKerrow JH. 1989. Regulation of Trichophyton rubrum proteolytic activity. Infect Immun 57:3081–3090. 10.1128/iai.57.10.3081-3090.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maranhão FCA, Paião FG, Martinez-Rossi NM. 2007. Isolation of transcripts over-expressed in human pathogen Trichophyton rubrum during growth in keratin. Microb Pathog 43:166–172. 10.1016/j.micpath.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 84.Gräser Y, Monod M, Bouchara JP, Dukik K, Nenoff P, Kargl A, Kupsch C, Zhan P, Packeu A, Chaturvedi V, de Hoog S. 2018. New insights in dermatophyte research. Med Mycol 56(Suppl 1):2–9. 10.1093/mmy/myx141. [DOI] [PubMed] [Google Scholar]
- 85.Méhul B, Gu Z, Jomard A, Laffet G, Feuilhade M, Monod M. 2016. Sub6 (Tri r 2), an onychomycosis marker revealed by proteomics analysis of trichophyton rubrum secreted proteins in patient nail samples. J Invest Dermatol 136:331–333. 10.1038/JID.2015.367. [DOI] [PubMed] [Google Scholar]
- 86.Monod M, Méhul B. 2019. Recent findings in onychomycosis and their application for appropriate treatment. J Fungi 5:20. 10.3390/jof5010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu X, Liu T, Yang J, Chen L, Liu B, Wang L, Jin Q. 2018. The first whole-cell proteome- and lysine-acetylome-based comparison between Trichophyton rubrum conidial and mycelial stages. J Proteome Res 17:1436–1451. 10.1021/acs.jproteome.7b00793. [DOI] [PubMed] [Google Scholar]
- 88.Scharschmidt TC, Fischbach MA. 2013. What lives on our skin: ecology, genomics and therapeutic opportunities of the skin microbiome. Drug Discov Today Dis Mech 10:e83–e89. 10.1016/j.ddmec.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martínez-García S, Rodríguez-Martínez S, Cancino-Diaz ME, Cancino-Diaz JC. 2018. Extracellular proteases of Staphylococcus epidermidis: roles as virulence factors and their participation in biofilm. APMIS 126:177–185. 10.1111/apm.12805. [DOI] [PubMed] [Google Scholar]
- 90.Koziel J, Potempa J. 2013. Protease-armed bacteria in the skin. Cell Tissue Res 351:325–337. 10.1007/s00441-012-1355-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aubin GG, Portillo ME, Trampuz A, Corvec S. 2014. Propionibacterium acnes, an emerging pathogen: from acne to implant-infections, from phylotype to resistance. Med Mal Infect 44:241–250. 10.1016/j.medmal.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 92.Scholz CFP, Kilian M. 2016. The natural history of cutaneous propionibacteria, and reclassification of selected species within the genus Propionibacterium to the proposed novel genera Acidipropionibacterium gen. nov., Cutibacterium gen. nov. and Pseudopropionibacterium gen. nov. Int J Syst Evol Microbiol 66:4422–4432. 10.1099/ijsem.0.001367. [DOI] [PubMed] [Google Scholar]
- 93.Bojar RA, Holland KT. 2004. Acne and Propionibacterium acnes. Clin Dermatol 22:375–379. 10.1016/j.clindermatol.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 94.Achermann Y, Goldstein EJC, Coenye T, Shirtliff ME. 2014. Propionibacterium acnes: from commensal to opportunistic biofilm-associated implant pathogen. Clin Microbiol Rev 27:419–440. 10.1128/CMR.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Achermann Y, Tran B, Kang M, Harro JM, Shirtliff ME. 2015. Immunoproteomic identification of in vivo-produced Propionibacterium acnes proteins in a rabbit biofilm infection model. Clin Vaccine Immunol 22:467–476. 10.1128/CVI.00760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Beylot C, Auffret N, Poli F, Claudel JP, Leccia MT, Del Giudice P, Dreno B. 2014. Propionibacterium acnes: an update on its role in the pathogenesis of acne. J Eur Acad Dermatol Venereol 28:271–278. 10.1111/jdv.12224. [DOI] [PubMed] [Google Scholar]
- 97.Holland KT, Greenman J, Cunliffe WJ. 1979. Growth of cutaneous propionibacteria on synthetic medium; growth yields and exoenzyme production. J Appl Bacteriol 47:383–394. 10.1111/j.1365-2672.1979.tb01198.x. [DOI] [PubMed] [Google Scholar]
- 98.Holland C, Mak TN, Zimny-Arndt U, Schmid M, Meyer TF, Jungblut PR, Brüggemann H. 2010. Proteomic identification of secreted proteins of Propionibacterium acnes. BMC Microbiol 10:230. 10.1186/1471-2180-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gannesen AV, Zdorovenko EL, Botchkova EA, Hardouin J, Massier S, Kopitsyn DS, Gorbachevskii MV, Kadykova AA, Shashkov AS, Zhurina MV, Netrusov AI, Knirel YA, Plakunov VK, Feuilloley MGJ. 2019. Composition of the biofilm matrix of Cutibacterium acnes acneic strain RT5. Front Microbiol 10:1284. 10.3389/fmicb.2019.01284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee SE, Kim JM, Jeong SK, Jeon JE, Yoon HJ, Jeong MK, Lee SH. 2010. Protease-activated receptor-2 mediates the expression of inflammatory cytokines, antimicrobial peptides, and matrix metalloproteinases in keratinocytes in response to Propionibacterium acnes. Arch Dermatol Res 302:745–756. 10.1007/s00403-010-1074-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee SE, Kim J-M, Jeong SK, Choi EH, Zouboulis CC, Lee SH. 2015. Expression of protease-activated receptor-2 in SZ95 sebocytes and its role in sebaceous lipogenesis, inflammation, and innate immunity. J Invest Dermatol 135:2219–2227. 10.1038/jid.2015.151. [DOI] [PubMed] [Google Scholar]
- 102.Yu Y, Champer J, Agak GW, Kao S, Modlin RL, Kim J. 2016. Different Propionibacterium acnes phylotypes induce distinct immune responses and express unique surface and secreted proteomes. J Invest Dermatol 136:2221–2228. 10.1016/j.jid.2016.06.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Barnard E, Shi B, Kang D, Craft N, Li H. 2016. The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci Rep 6:39491. 10.1038/srep39491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kjos M, Snipen L, Salehian Z, Nes IF, Diep DB. 2010. The Abi proteins and their involvement in bacteriocin self-immunity. J Bacteriol 192:2068–2076. 10.1128/JB.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Foster T. 1996. Staphylococcus, Chapter 12. In Baron S (ed), Medical microbiology, 4th ed. University of Texas Medical Branch at Galveston, Galveston, TX. [Google Scholar]
- 106.Parlet CP, Brown MM, Horswill AR. 2019. Commensal staphylococci influence Staphylococcus aureus skin colonization and disease. Trends Microbiol 27:497–507. 10.1016/j.tim.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kluytmans J, van Belkum A, Verbrugh H. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev 10:505–520. 10.1128/CMR.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gong JQ, Lin L, Lin T, Hao F, Zeng FQ, Bi ZG, Yi D, Zhao B. 2006. Skin colonization by Staphylococcus aureus in patients with eczema and atopic dermatitis and relevant combined topical therapy: a double-blind multicentre randomized controlled trial. Br J Dermatol 155:680–687. 10.1111/j.1365-2133.2006.07410.x. [DOI] [PubMed] [Google Scholar]
- 109.Leung DYM. 2008. The role of Staphylococcus aureus in atopic eczema. Acta Derm Venereol 88:21–27. 10.2340/00015555-0388. [DOI] [Google Scholar]
- 110.Paller AS, Kong HH, Seed P, Naik S, Scharschmidt TC, Gallo RL, Luger T, Irvine AD. 2019. The microbiome in patients with atopic dermatitis. J Allergy Clin Immunol 143:26–35. 10.1016/j.jaci.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chiller K, Selkin BA, Murakawa GJ. 2001. Skin microflora and bacterial infections of the skin. J Invest Dermatol Symp Proc 6:170–174. 10.1046/j.0022-202x.2001.00043.x. [DOI] [PubMed] [Google Scholar]
- 112.Thammavongsa V, Kim HK, Missiakas D, Schneewind O. 2015. Staphylococcal manipulation of host immune responses. Nat Rev Microbiol 13:529–543. 10.1038/nrmicro3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Geoghegan JA, Irvine AD, Foster TJ. 2018. Staphylococcus aureus and atopic dermatitis: a complex and evolving relationship. Trends Microbiol 26:484–497. 10.1016/j.tim.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 114.Tam K, Torres VJ. 2019. Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol Spectr 7(2). 10.1128/microbiolspec.GPP3-0039-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nickerson NN, Prasad L, Jacob L, Delbaere LT, McGavin MJ. 2007. Activation of the SspA serine protease zymogen of Staphylococcus aureus proceeds through unique variations of a trypsinogen-like mechanism and is dependent on both autocatalytic and metalloprotease-specific processing. J Biol Chem 282:34129–34138. 10.1074/jbc.M705672200. [DOI] [PubMed] [Google Scholar]
- 116.Amagai M, Yamaguchi T, Hanakawa Y, Nishifuji K, Sugai M, Stanley JR. 2002. Staphylococcal exfoliative toxin B specifically cleaves desmoglein 1. J Invest Dermatol 118:845–850. 10.1046/j.1523-1747.2002.01751.x. [DOI] [PubMed] [Google Scholar]
- 117.Amagai M, Matsuyoshi N, Wang ZH, Andl C, Stanley JR. 2000. Toxin in bullous impetigo and staphylococcal scalded-skin syndrome targets desmoglein 1. Nat Med 6:1275–1277. 10.1038/81385. [DOI] [PubMed] [Google Scholar]
- 118.Jochmann A, Howser S, Davis S, Izakovic J, Plano LRW. 2013. Correlation of twenty virulence genes of Staphylococcus aureus with severity of atopic dermatitis in children as compared to healthy individuals. J Clin Exp Dermatol Res 4:174. [Google Scholar]
- 119.Kantyka T, Shaw LN, Potempa J. 2011. Papain-like proteases of Staphylococcus aureus. Adv Exp Med Biol 712:1–14. 10.1007/978-1-4419-8414-2_1. [DOI] [PubMed] [Google Scholar]
- 120.Potempa J, Pike RN. 2009. Corruption of innate immunity by bacterial proteases. J Innate Immun 1:70–87. 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lehman MK, Nuxoll AS, Yamada KJ, Kielian T, Carson SD, Fey PD. 2019. Protease-mediated growth of Staphylococcus aureus on host proteins is opp3 dependent. mBio 10:e02553-18. 10.1128/mBio.02553-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ohbayashi T, Irie A, Murakami Y, Nowak M, Potempa J, Nishimura Y, Shinohara M, Imamura T. 2011. Degradation of fibrinogen and collagen by staphopains, cysteine proteases released from Staphylococcus aureus. Microbiology (Reading) 157:786–792. 10.1099/mic.0.044503-0. [DOI] [PubMed] [Google Scholar]
- 123.Stelzner K, Boyny A, Hertlein T, Sroka A, Moldovan A, Paprotka K, Kessie D, Mehling H, Potempa J, Ohlsen K, Fraunholz MJ, Rudel T. 2021. Intracellular Staphylococcus aureus employs the cysteine protease staphopain A to induce host cell death in epithelial cells. PLoS Pathog 17:e1009874. 10.1371/journal.ppat.1009874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sieprawska-Lupa M, Mydel P, Krawczyk K, Wójcik K, Puklo M, Lupa B, Suder P, Silberring J, Reed M, Pohl J, Shafer W, McAleese F, Foster T, Travis J, Potempa J. 2004. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob Agents Chemother 48:4673–4679. 10.1128/AAC.48.12.4673-4679.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sonesson A, Przybyszewska K, Eriksson S, Mörgelin M, Kjellström S, Davies J, Potempa J, Schmidtchen A. 2017. Identification of bacterial biofilm and the Staphylococcus aureus derived protease, staphopain, on the skin surface of patients with atopic dermatitis. Sci Rep 7:8689. 10.1038/s41598-017-08046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Smagur J, Guzik K, Magiera L, Bzowska M, Gruca M, Thøgersen IB, Enghild JJ, Potempa J. 2009. A new pathway of staphylococcal pathogenesis: apoptosis-like death induced by staphopain B in human neutrophils and monocytes. J Innate Immun 1:98–108. 10.1159/000181014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Smagur J, Guzik K, Bzowska M, Kuzak M, Zarebski M, Kantyka T, Walski M, Gajkowska B, Potempa J. 2009. Staphylococcal cysteine protease staphopain B (SspB) induces rapid engulfment of human neutrophils and monocytes by macrophages. Biol Chem 390:361–371. 10.1515/BC.2009.042. [DOI] [PubMed] [Google Scholar]
- 128.Laarman AJ, Ruyken M, Malone CL, van Strijp JAG, Horswill AR, Rooijakkers SHM. 2011. Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J Immunol 186:6445–6453. 10.4049/jimmunol.1002948. [DOI] [PubMed] [Google Scholar]
- 129.Jusko M, Potempa J, Kantyka T, Bielecka E, Miller HK, Kalinska M, Dubin G, Garred P, Shaw LN, Blom AM. 2014. Staphylococcal proteases aid in evasion of the human complement system. J Innate Immun 6:31–46. 10.1159/000351458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Elmwall J, Kwiecinski J, Na M, Ali AA, Osla V, Shaw LN, Wang W, Sävman K, Josefsson E, Bylund J, Jin T, Welin A, Karlsson A. 2017. Galectin-3 is a target for proteases involved in the virulence of Staphylococcus aureus. Infect Immun 85:e00177-17. 10.1128/IAI.00177-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Frey AM, Chaput D, Shaw LN. 2021. Insight into the human pathodegradome of the V8 protease from Staphylococcus aureus. Cell Rep 35:108930. 10.1016/j.celrep.2021.108930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Stentzel S, Teufelberger A, Nordengrün M, Kolata J, Schmidt F, van Crombruggen K, Michalik S, Kumpfmüller J, Tischer S, Schweder T, Hecker M, Engelmann S, Völker U, Krysko O, Bachert C, Bröker BM. 2017. Staphylococcal serine protease–like proteins are pacemakers of allergic airway reactions to Staphylococcus aureus. J Allergy Clin Immunol 139:492–500.e8. 10.1016/j.jaci.2016.03.045. [DOI] [PubMed] [Google Scholar]
- 133.Roy S, Santra S, Das A, Dixith S, Sinha M, Ghatak S, Ghosh N, Banerjee P, Khanna S, Mathew-Steiner S, Das Ghatak P, Blackstone BN, Powell HM, Bergdall VK, Wozniak DJ, Sen CK. 2020. Staphylococcus aureus biofilm infection compromises wound healing by causing deficiencies in granulation tissue collagen. Ann Surg 271:1174–1185. 10.1097/SLA.0000000000003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Allen HB, Vaze ND, Choi C, Hailu T, Tulbert BH, Cusack CA, Joshi SG. 2014. The presence and impact of biofilm-producing staphylococci in atopic dermatitis. JAMA Dermatol 150:260–265. 10.1001/jamadermatol.2013.8627. [DOI] [PubMed] [Google Scholar]
- 135.Mootz JM, Malone CL, Shaw LN, Horswill AR. 2013. Staphopains modulate staphylococcus aureus biofilm integrity. Infect Immun 81:3227–3238. 10.1128/IAI.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Martí M, Trotonda MP, Tormo-Más MA, Vergara-Irigaray M, Cheung AL, Lasa I, Penadés JR. 2010. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect 12:55–64. 10.1016/j.micinf.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 137.McGavin MJ, Zahradka C, Rice K, Scott JE. 1997. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect Immun 65:2621–2628. 10.1128/iai.65.7.2621-2628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nakatsuji T, Chen TH, Two AM, Chun KA, Narala S, Geha RS, Hata TR, Gallo RL. 2016. Staphylococcus aureus exploits epidermal barrier defects in atopic dermatitis to trigger cytokine expression. J Invest Dermatol 136:2192–2200. 10.1016/j.jid.2016.05.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kolar SL, Ibarra JA, Rivera FE, Mootz JM, Davenport JE, Stevens SM, Horswill AR, Shaw LN. 2013. Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiologyopen 2:18–34. 10.1002/mbo3.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gonzalez DJ, Okumura CY, Hollands A, Kersten R, Akong-Moore K, Pence MA, Malone CL, Derieux J, Moore BS, Horswill AR, Dixon JE, Dorrestein PC, Nizet V. 2012. Novel phenol-soluble modulin derivatives in community-associated methicillin-resistant Staphylococcus aureus identified through imaging mass spectrometry. J Biol Chem 287:13889–13898. 10.1074/jbc.M112.349860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cassat JE, Hammer ND, Campbell JP, Benson MA, Perrien DS, Mrak LN, Smeltzer MS, Torres VJ, Skaar EP. 2013. A secreted bacterial protease tailors the Staphylococcus aureus virulence repertoire to modulate bone remodeling during osteomyelitis. Cell Host Microbe 13:759–772. 10.1016/j.chom.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Paharik AE, Salgado-Pabon W, Meyerholz DK, White MJ, Schlievert PM, Horswill AR. 2016. The Spl serine proteases modulate Staphylococcus aureus protein production and virulence in a rabbit model of pneumonia. mSphere 1:e00208-16. 10.1128/mSphere.00208-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Karlsson A, Saravia-Otten P, Tegmark K, Morfeldt E, Arvidson S. 2001. Decreased amounts of cell wall-associated protein a and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect Immun 69:4742–4748. 10.1128/IAI.69.8.4742-4748.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gimza BD, Jackson JK, Frey AM, Budny BG, Chaput D, Rizzo DN, Shaw LN. 2021. Unraveling the impact of secreted proteases on hypervirulence in staphylococcus aureus. mBio 12:e03288-20. 10.1128/mBio.03288-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gimza BD, Larias MI, Budny BG, Shaw LN. 2019. Mapping the global network of extracellular protease regulation in Staphylococcus aureus. mSphere 4:e00676-19. 10.1128/mSphere.00676-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cheung GYC, Wang R, Khan BA, Sturdevant DE, Otto M. 2011. Role of the Accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun 79:1927–1935. 10.1128/IAI.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Genetics 42:541–564. 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 148.Byrum SD, Loughran AJ, Beenken KE, Orr LM, Storey AJ, Mackintosh SG, Edmondson RD, Tackett AJ, Smeltzer MS. 2018. Label-free proteomic approach to characterize protease-dependent and -independent effects of sarA inactivation on the Staphylococcus aureus exoproteome. J Proteome Res 17:3384–3395. 10.1021/acs.jproteome.8b00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Alkam D, Jenjaroenpun P, Ramirez AM, Beenken KE, Spencer HJ, Smeltzer MS. 2021. The increased accumulation of Staphylococcus aureus virulence factors is maximized in a purR mutant by the increased production of SarA and decreased production of extracellular proteases. Infect Immun 89:e00718-20. 10.1128/IAI.00718-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zielinska AK, Beenken KE, Mrak LN, Spencer HJ, Post GR, Skinner RA, Tackett AJ, Horswill AR, Smeltzer MS. 2012. SarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol Microbiol 86:1183–1196. 10.1111/mmi.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nakamura Y, Takahashi H, Takaya A, Inoue Y, Katayama Y, Kusuya Y, Shoji T, Takada S, Nakagawa S, Oguma R, Saito N, Ozawa N, Nakano T, Yamaide F, Dissanayake E, Suzuki S, Villaruz A, Varadarajan S, Matsumoto M, Kobayashi T, Kono M, Sato Y, Akiyama M, Otto M, Matsue H, Núñez G, Shimojo N. 2020. Staphylococcus Agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci Transl Med 12:eaay4068. 10.1126/scitranslmed.aay4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kalan LR, Meisel JS, Loesche MA, Horwinski J, Soaita I, Chen X, Uberoi A, Gardner SE, Grice EA. 2019. Strain- and species-level variation in the microbiome of diabetic wounds is associated with clinical outcomes and therapeutic efficacy. Cell Host Microbe 25:641–655.e5. 10.1016/j.chom.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Heilmann C, Ziebuhr W, Becker K. 2019. Are coagulase-negative staphylococci virulent? Clin Microbiol Infect 25:1071–1080. 10.1016/j.cmi.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 154.Otto M. 2013. Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays 35:4–11. 10.1002/bies.201200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Michalik M, Samet A, Podbielska-Kubera A, Savini V, Międzobrodzki J, Kosecka-Strojek M. 2020. Coagulase-negative staphylococci (CoNS) as a significant etiological factor of laryngological infections: a review. Ann Clin Microbiol Antimicrob 19:26. 10.1186/s12941-020-00367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Otto M. 2009. Staphylococcus epidermidis - the “accidental” pathogen. Nat Rev Microbiol 7:555–567. 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, Spencer S, Hall JA, Dzutsev A, Kong H, Campbell DJ, Trinchieri G, Segre JA, Belkaid Y. 2012. Compartmentalized control of skin immunity by resident commensals. Science 337:1115–1119. 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Naik S, Bouladoux N, Linehan JL, Han S-J, Harrison OJ, Wilhelm C, Conlan S, Himmelfarb S, Byrd AL, Deming C, Quinones M, Brenchley JM, Kong HH, Tussiwand R, Murphy KM, Merad M, Segre JA, Belkaid Y. 2015. Commensal–dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520:104–108. 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Christensen GJM, Brüggemann H. 2014. Bacterial skin commensals and their role as host guardians. Benef Microbes 5:201–215. 10.3920/BM2012.0062. [DOI] [PubMed] [Google Scholar]
- 160.Otto M. 2010. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev Dermatol 5:183–195. 10.1586/edm.10.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lai Y, Villaruz AE, Li M, Cha DJ, Sturdevant DE, Otto M. 2007. The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in staphylococci. Mol Microbiol 63:497–506. 10.1111/j.1365-2958.2006.05540.x. [DOI] [PubMed] [Google Scholar]
- 162.Williams MR, Cau L, Wang Y, Kaul D, Sanford JA, Zaramela LS, Khalil S, Butcher AM, Zengler K, Horswill AR, Dupont CL, Hovnanian A, Gallo RL. 2020. Interplay of staphylococcal and host proteases promotes skin barrier disruption in Netherton syndrome. Cell Rep 30:2923–2933.e7. 10.1016/j.celrep.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Oleksy A, Golonka E, Bańbuła A, Szmyd G, Moon J, Kubica M, Greenbaum D, Bogyo M, Foster TJ, Travis J, Potempa J. 2004. Growth phase-dependent production of a cell wall-associated elastinolytic cysteine proteinase by Staphylococcus epidermidis. Biol Chem 385:525–535. 10.1515/BC.2004.062. [DOI] [PubMed] [Google Scholar]
- 164.Cau L, Williams MR, Butcher AM, Nakatsuji T, Kavanaugh JS, Cheng JY, Shafiq F, Higbee K, Hata TR, Horswill AR, Gallo RL. 2021. Staphylococcus epidermidis protease EcpA can be a deleterious component of the skin microbiome in atopic dermatitis. J Allergy Clin Immunol 147:955–966.e16. 10.1016/j.jaci.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dubin G, Chmiel D, Mak P, Rakwalska M, Rzychon M, Dubin A. 2001. Molecular cloning and biochemical characterization of proteases from Staphylococcus epidermidis. Biol Chem 382:1575–1582. 10.1515/BC.2001.192. [DOI] [PubMed] [Google Scholar]
- 166.Sugimoto S, Iwamoto T, Takada K, Okuda K-I, Tajima A, Iwase T, Mizunoe Y. 2013. Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host-pathogen interaction. J Bacteriol 195:1645–1655. 10.1128/JB.01672-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Brown MM, Horswill AR. 2020. Staphylococcus epidermidis—skin friend or foe? PLoS Pathog 16:e1009026. 10.1371/journal.ppat.1009026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen C, Krishnan V, Macon K, Manne K, Narayana SVL, Schneewind O. 2013. Secreted proteases control autolysin-mediated biofilm growth of Staphylococcus aureus. J Biol Chem 288:29440–29452. 10.1074/jbc.M113.502039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. 2010. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465:346–349. 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 170.Paharik AE, Kotasinska M, Both A, Hoang T-MN, Büttner H, Roy P, Fey PD, Horswill AR, Rohde H. 2017. The metalloprotease SepA governs processing of accumulation-associated protein and shapes intercellular adhesive surface properties in Staphylococcus epidermidis. Mol Microbiol 103:860–874. 10.1111/mmi.13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fredheim EGA, Flægstad T, Askarian F, Klingenberg C. 2015. Colonisation and interaction between S. epidermidis and S. aureus in the nose and throat of healthy adolescents. Eur J Clin Microbiol Infect Dis 34:123–129. 10.1007/s10096-014-2197-5. [DOI] [PubMed] [Google Scholar]
- 172.Patterson M. 1996. Streptococcus, Chapter 13. In Baron S (ed), Medical microbiology. 4th ed. University of Texas Medical Branch at Galveston, Galveston, TX. [Google Scholar]
- 173.Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V. 2014. Disease manifestations and pathogenic mechanisms of group A Streptococcus. Clin Microbiol Rev 27:264–301. 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang Z, Rahkola J, Redzic JS, Chi YC, Tran N, Holyoak T, Zheng H, Janoff E, Eisenmesser E. 2020. Mechanism and inhibition of Streptococcus pneumoniae IgA1 protease. Nat Commun 11:6063. 10.1038/s41467-020-19887-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Poulsen K, Reinholdt J, Jespersgaard C, Boye K, Brown TA, Hauge M, Kilian M. 1998. A comprehensive genetic study of streptococcal immunoglobulin A1 proteases: evidence for recombination within and between species. Infect Immun 66:181–190. 10.1128/IAI.66.1.181-190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Janoff EN, Rubins JB, Fasching C, Charboneau D, Rahkola JT, Plaut AG, Weiser JN. 2014. Pneumococcal IgA1 protease subverts specific protection by human IgA1. Mucosal Immunol 7:249–256. 10.1038/mi.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Niessen CM. 2007. Tight junctions/adherens junctions: basic structure and function. J Invest Dermatol 127:2525–2532. 10.1038/sj.jid.5700865. [DOI] [PubMed] [Google Scholar]
- 178.Schlüter H, Wepf R, Moll I, Franke WW. 2004. Sealing the live part of the skin: the integrated meshwork of desmosomes, tight junctions and curvilinear ridge structures in the cells of the uppermost granular layer of the human epidermis. Eur J Cell Biol 83:655–665. 10.1078/0171-9335-00434. [DOI] [PubMed] [Google Scholar]
- 179.Sumitomo T, Nakata M, Higashino M, Terao Y, Kawabata S. 2013. Group A streptococcal cysteine protease cleaves epithelial junctions and contributes to bacterial translocation. J Biol Chem 288:13317–13324. 10.1074/jbc.M113.459875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sumitomo T, Mori Y, Nakamura Y, Honda-Ogawa M, Nakagawa S, Yamaguchi M, Matsue H, Terao Y, Nakata M, Kawabata S. 2018. Streptococcal cysteine protease-mediated cleavage of desmogleins is involved in the pathogenesis of cutaneous infection. Front Cell Infect Microbiol 8:10. 10.3389/fcimb.2018.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kapur V, Topouzis S, Majesky MW, Li L-L, Hamrick MR, Hamill RJ, Patti JM, Musser JM. 1993. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb Pathog 15:327–346. 10.1006/mpat.1993.1083. [DOI] [PubMed] [Google Scholar]
- 182.Lukomski S, Sreevatsan S, Amberg C, Reichardt W, Woischnik M, Podbielski A, Musser JM. 1997. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J Clin Invest 99:2574–2580. 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lukomski S, Montgomery CA, Rurangirwa J, Geske RS, Barrish JP, Adams GJ, Musser JM. 1999. Extracellular cysteine protease produced by Streptococcus pyogenes participates in the pathogenesis of invasive skin infection and dissemination in mice. Infect Immun 67:1779–1788. 10.1128/.67.4.1779-1788.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Connolly KL, Roberts AL, Holder RC, Reid SD. 2011. Dispersal of Group A streptococcal biofilms by the cysteine protease SpeB leads to increased disease severity in a murine model. PLoS One 6:e18984. 10.1371/journal.pone.0018984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kansal RG, McGeer A, Low DE, Norrby-Teglund A, Kotb M. 2000. Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect Immun 68:6362–6369. 10.1128/IAI.68.11.6362-6369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Egesten A, Olin AI, Linge HM, Yadav M, Mörgelin M, Karlsson A, Collin M. 2009. SpeB of Streptococcus pyogenes differentially modulates antibacterial and receptor activating properties of human chemokines. PLoS One 4:e4769. 10.1371/journal.pone.0004769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kurupati P, Turner CE, Tziona I, Lawrenson RA, Alam FM, Nohadani M, Stamp GW, Zinkernagel AS, Nizet V, Edwards RJ, Sriskandan S. 2010. Chemokine-cleaving Streptococcus pyogenes protease SpyCEP is necessary and sufficient for bacterial dissemination within soft tissues and the respiratory tract. Mol Microbiol 76:1387–1397. 10.1111/j.1365-2958.2010.07065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sjölinder H, Lövkvist L, Plant L, Eriksson J, Aro H, Jones A, Jonsson AB. 2008. The ScpC protease of Streptococcus pyogenes affects the outcome of sepsis in a murine model. Infect Immun 76:3959–3966. 10.1128/IAI.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sumby P, Zhang S, Whitney AR, Falugi F, Grandi G, Graviss EA, Deleo FR, Musser JM. 2008. A chemokine-degrading extracellular protease made by group A Streptococcus alters pathogenesis by enhancing evasion of the innate immune response. Infect Immun 76:978–985. 10.1128/IAI.01354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Honda-Ogawa M, Ogawa T, Terao Y, Sumitomo T, Nakata M, Ikebe K, Maeda Y, Kawabata S. 2013. Cysteine proteinase from Streptococcus pyogenes enables evasion of innate immunity via degradation of complement factors. J Biol Chem 288:15854–15864. 10.1074/jbc.M113.469106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kuo C-F, Lin Y-S, Chuang W-J, Wu J-J, Tsao N. 2008. Degradation of complement 3 by streptococcal pyrogenic exotoxin B inhibits complement activation and neutrophil opsonophagocytosis. Infect Immun 76:1163–1169. 10.1128/IAI.01116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Terao Y, Mori Y, Yamaguchi M, Shimizu Y, Ooe K, Hamada S, Kawabata S. 2008. Group A streptococcal cysteine protease degrades C3 (C3b) and contributes to evasion of innate immunity. J Biol Chem 283:6253–6260. 10.1074/jbc.M704821200. [DOI] [PubMed] [Google Scholar]
- 193.Tsao N, Tsai W-H, Lin Y-S, Chuang W-J, Wang C-H, Kuo C-F. 2006. Streptococcal pyrogenic exotoxin B cleaves properdin and inhibits complement-mediated opsonophagocytosis. Biochem Biophys Res Commun 339:779–784. 10.1016/j.bbrc.2005.11.078. [DOI] [PubMed] [Google Scholar]
- 194.Su Y-F, Chuang W-J, Wang S-M, Chen W-Y, Chiang-Ni C, Lin Y-S, Wu J-J, Liu C-C. 2011. The deficient cleavage of M protein-bound IgG by IdeS: insight into the escape of Streptococcus pyogenes from antibody-mediated immunity. Mol Immunol 49:134–142. 10.1016/j.molimm.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 195.Von Pawel-Rammingen U, Johansson BP, Björck L. 2002. IdeS, a novel streptococcal cysteine proteinase with unique specificity for immunoglobulin G. EMBO J 21:1607–1615. 10.1093/emboj/21.7.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Collin M, Olsén A. 2001. Effect of SpeB and EndoS from Streptococcus pyogenes on human immunoglobulins. Infect Immun 69:7187–7189. 10.1128/IAI.69.11.7187-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Persson H, Vindebro R, Von Pawel-Rammingen U. 2013. The streptococcal cysteine protease SpeB is not a natural immunoglobulin-cleaving enzyme. Infect Immun 81:2236–2241. 10.1128/IAI.00168-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Carothers KE, Liang Z, Mayfield J, Donahue DL, Lee M, Boggess B, Ploplis VA, Castellino FJ, Lee SW. 2020. The streptococcal protease SpeB antagonizes the biofilms of the human pathogen Staphylococcus aureus USA300 through cleavage of the staphylococcal SdrC protein. J Bacteriol 202:e00008-20. 10.1128/JB.00008-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lagier J-C, Dubourg G, Million M, Cadoret F, Bilen M, Fenollar F, Levasseur A, Rolain J-M, Fournier P-E, Raoult D. 2018. Culturing the human microbiota and culturomics. Nat Rev Microbiol 16:540–550. 10.1038/s41579-018-0041-0. [DOI] [PubMed] [Google Scholar]
- 200.Lewis WH, Tahon G, Geesink P, Sousa DZ, Ettema TJG. 2021. Innovations to culturing the uncultured microbial majority. Nat Rev Microbiol 19:225–240. 10.1038/s41579-020-00458-8. [DOI] [PubMed] [Google Scholar]
- 201.Savickas S, Kastl P, Auf Dem Keller U. 2020. Combinatorial degradomics: precision tools to unveil proteolytic processes in biological systems. Biochim Biophys Acta Proteins Proteom 1868:140392. 10.1016/j.bbapap.2020.140392. [DOI] [PubMed] [Google Scholar]
- 202.Woehl JL, Kitamura S, Dillon N, Han Z, Edgar LJ, Nizet V, Wolan DW. 2020. An irreversible inhibitor to probe the role of Streptococcus pyogenes cysteine protease SpeB in evasion of host complement defenses. ACS Chem Biol 15:2060–2069. 10.1021/acschembio.0c00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Keller LJ, Babin BM, Lakemeyer M, Bogyo M. 2020. Activity-based protein profiling in bacteria: applications for identification of therapeutic targets and characterization of microbial communities. Curr Opin Chem Biol 54:45–53. 10.1016/j.cbpa.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Gordon MH, Anowai A, Young D, Das N, Campden RI, Sekhon H, Myers Z, Mainoli B, Chopra S, Thuy-Boun PS, Kizhakkedathu J, Bindra G, Jijon HB, Heitman S, Yates R, Wolan DW, Edgington-Mitchell LE, MacNaughton WK, Dufour A. 2019. N-Terminomics/TAILS profiling of proteases and their substrates in ulcerative colitis. ACS Chem Biol 14:2471–2483. 10.1021/acschembio.9b00608. [DOI] [PubMed] [Google Scholar]