ABSTRACT
Microalgae are key ecological players with a complex evolutionary history. Genomic diversity, in addition to limited availability of high-quality genomes, challenge studies that aim to elucidate molecular mechanisms underlying microalgal ecophysiology. Here, we present a novel and comprehensive transcriptomic hybrid approach to generate a reference for genetic analyses and resolve the microalgal gene landscape at the strain level. The approach is demonstrated for a strain of the coccolithophore microalga Emiliania huxleyi, which is a species complex with considerable genome variability. The investigated strain is commonly studied as a model for algal-bacterial interactions and was therefore sequenced in the presence of bacteria to elicit the expression of interaction-relevant genes. We applied complementary PacBio Iso-Seq full-length cDNA and poly(A)-independent Illumina total RNA sequencing, which resulted in a de novo-assembled, near-complete hybrid transcriptome. In particular, hybrid sequencing improved the reconstruction of long transcripts and increased the recovery of full-length transcript isoforms. To use the resulting hybrid transcriptome as a reference for genetic analyses, we demonstrate a method that collapses the transcriptome into a genome-like data set, termed “synthetic genome” (sGenome). We used the sGenome as a reference to visually confirm the robustness of the CCMP3266 gene assembly, to conduct differential gene expression analysis, and to characterize novel E. huxleyi genes. The newly identified genes contribute to our understanding of E. huxleyi genome diversification and are predicted to play a role in microbial interactions. Our transcriptomic toolkit can be implemented in various microalgae to facilitate mechanistic studies on microalgal diversity and ecology.
IMPORTANCE Microalgae are key players in the ecology and biogeochemistry of our oceans. Efforts to implement genomic and transcriptomic tools in laboratory studies involving microalgae suffer from the lack of published genomes. In the case of coccolithophore microalgae, the problem has long been recognized; the model species Emiliania huxleyi is a species complex with genomes composed of a core and a large variable portion. To study the role of the variable portion in niche adaptation, and specifically in microbial interactions, strain-specific genetic information is required. Here, we present a novel transcriptomic hybrid approach, and generated strain-specific genome-like information. We demonstrate our approach on an E. huxleyi strain that is cocultivated with bacteria. By constructing a “synthetic genome,” we generated comprehensive gene annotations that enabled accurate analyses of gene expression patterns. Importantly, we unveiled novel genes in the variable portion of E. huxleyi that play putative roles in microbial interactions.
KEYWORDS: differential gene expression, Emiliania huxleyi (coccolithophore, haptophyte), full-length cDNA, genome variability, hybrid transcriptome assembly, microbial interactions, sulfur and DMSP metabolism, whole transcriptome
INTRODUCTION
The ocean is rich in phytoplankton, namely, microalgae, which play key roles in global biogeochemical cycles and marine food webs (1–3). A well-studied microalgal representative is Emiliania huxleyi (4, 5); a coccolithophore species that creates vast annual blooms, covering hundreds of thousands of square kilometers of ocean surface (6–9). The algal blooms collapse in a sudden process that is largely attributed to viral infection (10–13); however, mounting evidence suggests that bacteria also play pivotal roles in the sudden demise of E. huxleyi (14–19).
To mechanistically study the influence of algal-bacterial interactions on E. huxleyi physiology and bloom dynamics, well-characterized E. huxleyi model systems are required. An emerging model is E. huxleyi CCMP3266 (derived from xenic culture RCC1216 [20]), a diploid and calcifying strain that resembles the predominant state of E. huxleyi under natural bloom conditions (21). Strain CCMP3266 can be cultured in the absence of bacteria (axenic) (20), allowing for controlled addition of selected bacteria in cocultivation experiments. Initial studies found that cocultivation of E. huxleyi CCMP3266 with the bacterium Phaeobacter inhibens, an ecologically relevant member of the Roseobacter group, changes the algal physiology and results in altered alkenone lipid metabolism (22). Furthermore, algal exudates were found to be modified by P. inhibens into growth-stimulating and protective agents (14, 23). Over time, interactions of E. huxleyi CCMP3266 with P. inhibens trigger a programmed cell death-like response in the alga (14, 19). This response was also observed for other pairs of E. huxleyi and bacteria (17) and resembles the sudden demise of E. huxleyi under natural bloom conditions (9, 12, 24). Yet, the mechanisms underlying E. huxleyi-bacterial interactions are still poorly understood.
Mechanistic studies on E. huxleyi face a tremendous challenge; E. huxleyi is a morphotypically and phenotypically diverse species complex with genomes composed of a core and a relatively large variable portion (25–27). The variable portion appears to contribute to the global success of E. huxleyi, which thrives under a wide variety of environmental conditions (25). Algal genome variability may also reflect a history of algal-bacterial interactions, as indicated by strain-specific responses of E. huxleyi towards bacterial cues (15, 17, 28, 29). Despite the complexity of E. huxleyi, to date only a single deep-sequenced, assembled, annotated, and partially curated draft genome is publicly available, generated for E. huxleyi CCMP1516 (25). Genome sequencing is still costly to conduct and, in the case of E. huxleyi, it is further complicated by gene duplications, repetitive sequences, ploidy, a high GC content, and possibly overlooked bacterial contaminations (25). As a result, genetic analyses that rely on the genome as sole reference cannot resolve the contribution of variable genes to the life and death of E. huxleyi strains other than CCMP1516.
There is a pressing need to generate accurate gene information for individual E. huxleyi strains. In the absence of strain-specific genomes, transcriptomes can be used as an alternative. Indeed, 17 E. huxleyi transcriptomes are currently available at the DDBJ/EMBL/GenBank Transcriptome Shotgun Assembly Sequence Database. Among them, 16 were generated as part of the Marine Microbial Eukaryotic Transcriptome Project (30; BioProject accession numbers PRJEB37159 and PRJEB37164), and one was created in order to study E. huxleyi CCMP371 calcium metabolism (31). However, transcriptomes have several known caveats: the genes of interest may not be transcribed under the applied laboratory conditions, resulting in their absence from the final transcriptome. In addition, transcriptomes may contain incomplete transcripts or partially fragmented transcript isoforms. Transcript isoforms originate from single genes that are differentially processed, involving, e.g., the utilization of alternative start/stop sites and splice sites (32). Transcript isoforms are usually assembled from short Illumina RNA sequencing reads, a computational approach that falls short of reconstructing accurate, full-length sequences from differentially processed transcripts (33, 34). The PacBio long-read sequencing platform overcomes this shortcoming; it produces assembly-free, full-length transcript isoforms, but at the expense of a lower sequencing depth that hampers the detection of low-abundance RNA (35). Novel hybrid approaches integrate reads from complementary short-read and long-read sequencing platforms, thereby increasing transcriptome depth, accuracy, and transcript isoform reconstruction efficiency (36–41). Transcriptome complexity can be further increased by applying total RNA sequencing, also termed whole-transcriptome sequencing. Total RNA sequencing is poly(A) independent and detects biologically important RNA molecules, which may be derived from chloroplasts and mitochondria (42), protein-coding circular RNA (circRNA [43]), or noncoding RNA with regulatory functions (44). However, total RNA sequencing is not yet commonly applied for the construction of hybrid transcriptomes.
Here, we report on the construction of an E. huxleyi CCMP3266 hybrid transcriptome, while addressing the caveats of generating transcript-based reference gene information. To elicit the expression of genes relevant for microbial interactions, we harvested RNA from cocultures of E. huxleyi CCMP3266 with the bacterium P. inhibens DSM17395. To resolve full-length transcript isoforms and increase transcriptome complexity, we applied two complementary sequencing approaches: (i) PacBio Iso-Seq cDNA sequencing and (ii) poly(A)-independent, stranded Illumina total RNA sequencing. Sequencing data were de novo assembled into a hybrid transcriptome, using the novel hybrid rnaSPAdes algorithm (40). To evaluate the quality of the assembled transcripts, we compared the hybrid transcriptome with three nonhybrid control transcriptomes, using a tailored analysis based on conserved E. huxleyi reference genes. The evaluation indicates that the hybrid transcriptome was nearly complete and contained comparatively long transcripts and more full-length transcript isoforms. Finally, we demonstrate a novel approach to use strain-specific transcriptomes as a reference for genetic analyses. To this end, we generated a “synthetic genome” (sGenome), which is based on the published E. huxleyi CCMP1516 reference genome but that also includes CCMP3266-specific gene constructs. This approach allowed us to cluster de novo assembled E. huxleyi CCMP3266 transcript isoforms into CCMP3266 gene loci in a genome-guided manner (using TAMA [45]), without discarding CCMP3266 transcripts that were absent from or dissimilar to the published reference genome.
The sGenome approach was applied to visually compare CCMP1516 and CCMP3266 gene annotations, to accurately conduct differential gene expression analyses, to identify gaps in the reference genome, and to gain novel insights into the functioning of E. huxleyi variable genes. The latter analysis indicates an involvement of E. huxleyi variable genes in microbial interactions. With the growing need for accurate microalgal genetic information, the approach we describe can be applied to resolve the strain-specific gene landscape of various microalgae.
RESULTS AND DISCUSSION
Sampling the algal transcriptome during dynamic microbial interactions.
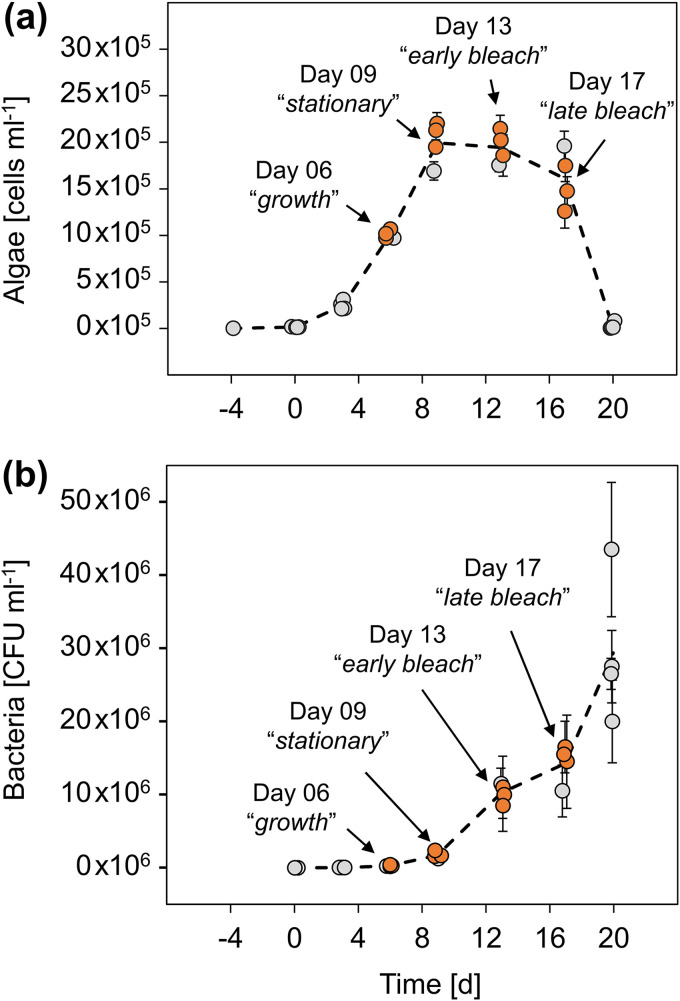
A transcriptome can be highly dynamic. Groups of genes may be transcribed only at specific life phases or under specific environmental conditions (46, 47). To capture the complete E. huxleyi CCMP3266 transcriptome during algal-bacterial interactions, we sampled algal-bacterial cocultures at different phases of their interaction. Therefore, axenic E. huxleyi CCMP3266 algal cultures were inoculated with the bacterium P. inhibens. This algal-bacterial pair was previously shown to engage in a dynamic interaction of ecological relevance (14). Briefly, it was shown that bacterial growth depends on metabolites exuded by the algae. However, as the culture ages, bacteria trigger an algal death, which is observed as bleaching of the culture. In the present study, cocultures largely followed the algal-bacterial dynamics that were previously described (Fig. 1). By closely monitoring algal and bacterial growth in cocultures, we identified four time points that represent key phases in the algal-bacterial interaction (Fig. 1). Cultures were sampled for RNA extraction during the algal exponential growth phase (day 6) and the algal stationary phase (day 9), as well as in the early and late bleaching phases (days 13 and 17). Sampled RNA extracts (see Fig. S1 in the supplemental material) were subjected to sequencing and used for transcriptome construction and gene expression analysis.
FIG 1.
Algal and bacterial growth during cocultivation. For each time point, four replicate cocultivation flasks were sampled for growth monitoring. (a and b) Algal growth was determined using three technical replicates per coculture (a), and bacterial growth was measured using technical duplicates per coculture (b). The data are presented as the means of the technical replicates. Error bars indicate the standard deviations. RNA was extracted from three cocultures at four time points during the algal-bacterial interaction (orange). A pool of RNA extracts from all four time points was used for PacBio Iso-Seq sequencing, while individual Illumina RNA sequencing libraries were prepared for each of the 12 RNA samples to allow time-resolved differential gene expression analyses.
Sequencing the transcriptome with two complementary methods.
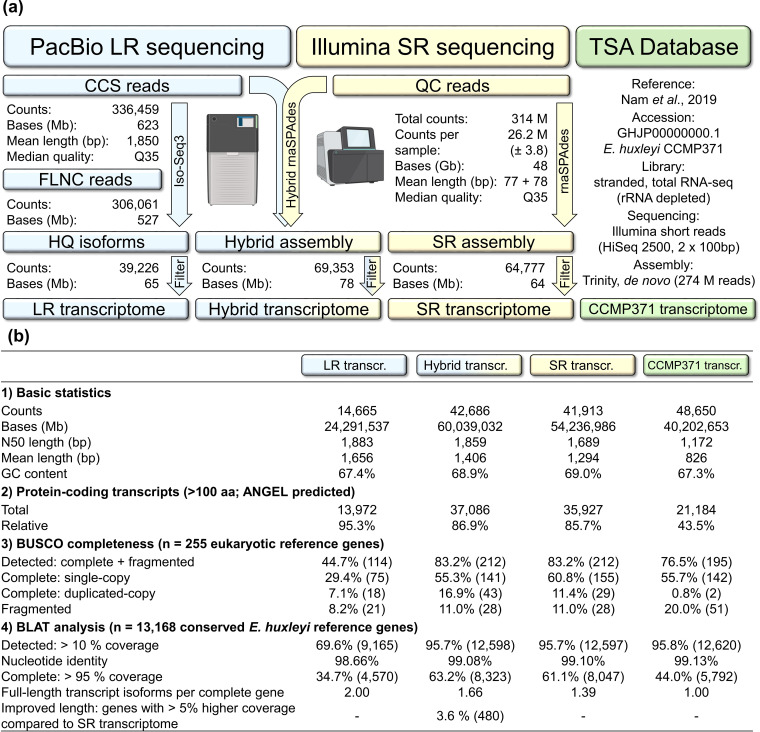
Our sequencing approach was designed with the aim to generate a comprehensive and accurate transcriptome of E. huxleyi CCMP3266. First, the PacBio Sequel I platform was used to sequence the mRNA with the Iso-Seq protocol. The protocol produces long sequencing reads that are suitable to detect full-length cDNA transcript isoforms, thereby reducing downstream assembly errors during transcript isoform reconstruction. Second, the Illumina NextSeq 500 platform was used to sequence rRNA-depleted total RNA, producing short sequencing reads with high sequencing depth, thereby improving the detection of genes with low expression levels. In contrast to the first protocol, the latter protocol also targets nonpolyadenylated RNA. Detailed information about the PacBio and Illumina sequencing results are provided in the supplemental material (see Table S1 and Fig. S2, as well as Table S2, respectively). Raw sequencing data from both platforms were processed into PacBio circular consensus sequence (CCS) reads and Illumina quality-controlled (QC) reads, as described in Materials and Methods, and are summarized in Fig. 2a. The PacBio method yielded 336,459 CCS reads (623 Mb), with a mean length of 1,850 bp. The Illumina method yielded 314 M QC reads (48 Gb), with a mean length of 155 bp (77 + 78-bp paired reads). As expected, the PacBio method produced less reads (930-fold), which, however, were significantly longer (12-fold). The processed reads from both platforms were characterized by an equally high median quality of Q35 (probability of 3.2 errors per 10,000 bp). In summary, sequencing yields and read qualities were within the upper boundaries of the manufacturers’ instrument specifications.
FIG 2.
Complementary long- and short-read sequencing of E. huxleyi CCMP3266 resulted in a hybrid transcriptome with improved transcript length, and full-length transcript isoform recovery. (a) PacBio long reads (LR) and Illumina total RNA short reads (SR) were used to construct a hybrid transcriptome, and two nonhybrid LR and SR control transcriptomes. A third nonhybrid E. huxleyi CCMP371 control transcriptome was downloaded from the Transcriptome Shotgun Assembly (TSA) database (31). (b) The quality of the hybrid transcriptome was evaluated by comparing it to the three nonhybrid control transcriptomes.
Constructing a hybrid transcriptome.
The generated PacBio CCS and Illumina QC reads were used to construct a hybrid transcriptome (Fig. 2a). For this purpose, we applied the hybrid rnaSPAdes algorithm, which de novo assembles short RNA sequencing reads into contiguous sequences (contigs), and integrates exon structure information of long-reads (40). The hybrid rnaSPAdes software produced an assembly with 69,353 contigs (Fig. 2a). We further implemented filtering steps, which removed 3,579 P. inhibens contigs (no other, possibly contaminating transcripts were detected; see Fig. S3), 11,073 redundant contigs (based on CD-HIT-EST clustering), and 1,502 erroneously assembled contigs derived from rRNA (see Fig. S4). In addition, 10,513 poorly supported contigs were removed, which were identified by multimapping Illumina QC reads to the hybrid assembly (<5 read support). The final hybrid transcriptome contained 42,686 contigs. Of note, we refrained from removing short, partially unassembled sequences (e.g., 200-bp cutoff). These sequences are potential artifacts; however, they may also include short RNAs with biological functions, such as previously described RNAs in E. huxleyi extracellular vesicles (48, 49). The final hybrid transcriptome, including functional transcript annotations are given in the supplement (see Data Sets S1 and S2, respectively).
Acquiring control transcriptomes to assess transcript qualities.
To assess the quality of the hybrid transcriptome, we generated two nonhybrid control transcriptomes (Fig. 2a). First, a long-read (LR) transcriptome was constructed, based on PacBio CCS reads that were processed into high-quality consensus transcript isoforms, using the default PacBio Iso-Seq3 pipeline. Second, a short-read (SR) transcriptome was constructed, based on Illumina QC reads that were processed with the hybrid transcriptome pipeline, while omitting the CCS read integration step. It is noteworthy that the hybrid transcriptome, as well as the two control transcriptomes, was filtered for technical redundancy using CD-HIT-EST with identical clustering options (95% nucleotide identity cutoff). This step ensured the presence of biologically relevant transcript isoforms, and allowed to compare the efficiency of transcript isoform reconstruction between the transcriptomes. In addition, a third E. huxleyi Illumina short-read transcriptome of strain CCMP371 was downloaded from the Transcriptome Shotgun Assembly database (TSA; Fig. 2a). The CCMP371 control transcriptome was generated with a similar, stranded, total RNA sequencing protocol as used for CCMP3266 total RNA sequencing (31), making it a suitable reference to evaluate the Illumina RNA sequencing and assembly strategy that we applied. The quality of the transcriptomes was assessed as described in “Assessment of transcriptome quality” in Materials and Methods. Briefly, we collected basic assembly statistics and quantified the number of protein-coding transcripts. In addition, transcriptome completeness and transcript isoform reconstruction efficiency were assessed with BUSCO (50), using a set of conserved eukaryotic reference genes (n = 255), as well as with a tailored BLAT-based method, using a newly compiled set of conserved E. huxleyi reference genes (n = 13,168; see Data Set S3).
Long-reads resolve transcript isoforms but lack complexity.
Using the above analyses, we found that the LR transcriptome contained the longest transcripts (1,656-bp mean length), with the highest relative number of protein-coding sequences (95.3%), and the highest number of full-length transcript isoforms per complete E. huxleyi reference gene (2.0; Fig. 2b). However, the LR transcriptome also contained the lowest number of detected eukaryotic and E. huxleyi reference genes (44.7% eukaryotic genes; 69.6% E. huxleyi genes), and the lowest number of complete E. huxleyi genes (34.7%). The low complexity of the LR transcriptome arises from the lower PacBio sequencing depth and/or the absence of nonpolyadenylated transcripts in the PacBio sequencing library. In conclusion, the PacBio long-read sequencing method generates valuable, full-length transcript isoform information but was insufficient in resolving a complex E. huxleyi CCMP3266 transcriptome when used as a stand-alone method.
Short-reads increase transcriptome complexity.
In comparison, all three other transcriptomes, which were assembled from total RNA Illumina short-reads, were found to be near complete (95.7 to 95.8% detected E. huxleyi genes; Fig. 2b). However, we found marked differences in the quality of the transcriptomes. To analyze the differences, we first compared the SR transcriptome with the CCMP371 transcriptome (Fig. 2b), which were both generated with a similar poly(A)-independent sequencing approach, and did not integrate PacBio long-reads. The SR transcriptome contained longer transcripts (a 1,294-bp versus 826-bp mean length) and a higher relative number of protein-coding sequences (85.7% versus 43.5%). The SR transcriptome also contained more detected eukaryotic reference genes (83.2% versus 76.5%), fewer fragmented eukaryotic genes (11.0% versus 20.0%), and more full-length transcript isoforms per complete eukaryotic gene (11.4% versus 0.8% duplicated copies). Likewise, the SR transcriptome contained more complete E. huxleyi reference genes (61.1% versus 44.0%) and more full-length transcript isoforms per complete E. huxleyi gene (1.39 versus 1.00). The results indicate that the Illumina total RNA sequencing strategy we applied was suitable for generating a short-read transcriptome of high quality.
The hybrid transcriptome harbors transcripts with improved length.
To assess the outcome of integrating long-reads during short-read assembly, we compared the hybrid transcriptome with the SR transcriptome (Fig. 2b). Both transcriptomes were similarly complex and contained the same amount of detected reference genes (83.2% versus 83.2% eukaryotic genes; 95.7% versus 95.7% E. huxleyi genes). However, the hybrid transcriptome contained more transcripts (42,686 versus 41,913), which were longer (a 1,406-bp versus 1,294-bp mean length), and encoded a slightly higher relative number of proteins (86.9% versus 85.7%). The hybrid transcriptome also contained more full-length transcript isoforms per complete eukaryotic reference gene (16.9% versus 11.4% duplicated copies). Furthermore, the hybrid transcriptome contained more complete E. huxleyi reference genes (63.2% versus 61.1%) and more full-length transcript isoforms per complete E. huxleyi gene (1.66 versus 1.39). We further assessed the number of E. huxleyi reference genes that were represented by longer transcripts in the hybrid transcriptome (>5% higher coverage). This analysis revealed that long-read integration improved the transcript length for 480 E. huxleyi reference genes. Noteworthy, the 480 improved E. huxleyi genes were 1.6-fold longer, compared to the total of 13,168 E. huxleyi reference genes (a 2,424-bp versus 1,488-bp median length; see Fig. S5). This highlights that long-read integration helps in reconstructing transcripts that originate from long genes. In summary, we found that the hybrid transcriptome approach did not notably alter the overall transcriptome complexity but gave rise to more complete transcripts and was advantageous in reconstructing full-length transcript isoforms. Complete transcripts are required to generate uninterrupted protein sequences and accurate functional annotations. In addition, full-length transcript isoforms are required to analyze differential transcript splicing—a mechanism that controls cellular differentiation in multicellular eukaryotes but is understudied in single-celled microalgae (51).
Generating a “synthetic genome” reference for genetic analyses.
Transcriptomes contain redundant transcript isoforms, which should be clustered into nonredundant gene loci for accurate downstream genetic analyses (52). Therefore, we used the hybrid transcriptome to generate a “synthetic genome” (sGenome), which is a genome-like data set that is based on the existing CCMP1516 reference genome assembly but also includes CCMP3266-specific genes. For sGenome construction, we first identified CCMP3266-specific transcripts in the hybrid transcriptome that had low similarity to the CCMP1516 reference genome (<95% nucleotide identity, <95% coverage) or which were completely absent from the genome. This step identified 19,537 CCMP3266-specific transcripts. Next, CCMP3266-specific transcripts were processed with COGENT to reconstruct the originating genes (53). COGENT clusters similar transcripts into families and then attempts to reconstruct the gene for each family based on shared exons and alternative introns. This step collapsed 19,537 CCMP3266-specific transcripts into 16,416 COGENT sequences. An example of a reconstructed COGENT gene sequence is presented in Fig. S6. Since COGENT cannot reconstruct single genes for each family, we additionally applied CD-HIT-EST filtering with a low nucleotide identity threshold (80% identify cutoff). This step selects a single representative sequence for clusters of similar sequences. The combination of COGENT with CD-HIT-EST resulted in 13,264 less-redundant sequences, which represent CCMP3266-specific genes. The representative CCMP3266-specific gene sequences were appended to the CCMP1516 genome (including mitochondrion and plastid), resulting in the preliminary sGenome. Finally, non-CCMP3266 regions were removed by mapping the hybrid transcriptome to the preliminary sGenome and masking unmapped regions (see Data Set S4). The mapping coordinates were also used to define CCMP3266 gene loci with TAMA (45; see also Data Set S5). As a result, 42,686 E. huxleyi CCMP3266 transcripts in the hybrid transcriptome were collapsed into 30,553 CCMP3266 genes. In comparison, the E. huxleyi CCMP1516 reference genome contains 38,554 NCBI genes and 33,341 JGI genes. In summary, the sGenome is a set of E. huxleyi CCMP3266 genes, which are based on transcriptional evidence and that harbor transcript isoform information.
Controlling for the robustness of E. huxleyi CCMP3266 genes.
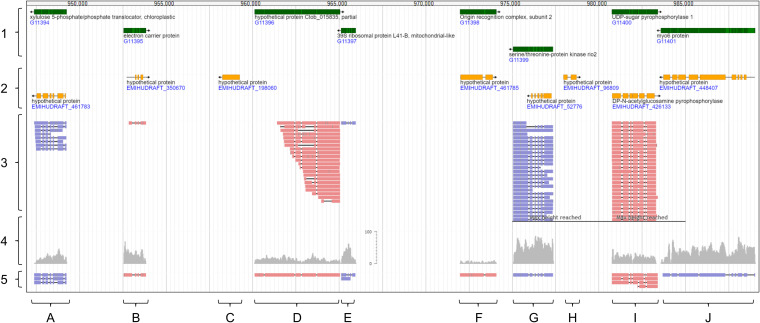
An advantage of the synthetic genome approach is that the resulting CCMP3266 sGenome has the same “chromosomal” structure as the CCMP1516 reference genome (scaffold names and lengths are preserved). This allows to visually compare the newly generated E. huxleyi CCMP3266 gene annotations, with those publicly available for CCMP1516. A representative region of the CCMP3266 sGenome is shown in Fig. 3. This region includes two novel E. huxleyi CCMP3266 genes, which were not annotated in the CCMP1516 genome (Fig. 3, loci D and E). Vice versa, the region includes two CCMP1516 genes that were not supported by CCMP3266 expression data (Fig. 3, loci C and H). Those unsupported CCMP1516 genes are possible pseudogenes and require experimental validation. The visualization further shows that CCMP3266 gene length and complexity was improved by combining PacBio long reads with poly(A)-independent Illumina short reads (compare to Fig. 2b). For example, one CCMP3266 gene presented in the region was incompletely covered by partially 5′-end degraded PacBio long reads (transcription start site) but extended by Illumina short reads (Fig. 3, locus D). In addition, two CCMP3266 genes presented in the region were only detected by short-read sequencing (Fig. 3, loci F and J). In these cases, the E. huxleyi transcript abundance was either below the PacBio detection threshold (Fig. 3, locus F), or transcripts were inefficiently detected by poly(A)-dependent long-read sequencing (Fig. 3, locus J), as indicated by relative differences in short-read coverages (Fig. 3, track 4). The visualization further exemplifies how E. huxleyi genes, which were described as “hypothetical” in the CCMP1516 genome, were now assigned with a function in the CCMP3266 sGenome. Taken together, the visualization supports the robustness of the generated E. huxleyi CCMP3266 genes.
FIG 3.
E. huxleyi CCMP3266 genes were generated by clustering transcript isoforms into gene loci. Visualized is a representative region of the E. huxleyi CCMP3266 sGenome (CCMP1516 reference genome plus CCMP3266-specific gene constructs). The visualization includes newly generated CCMP3266 gene annotations (left, track 1) and CCMP1516 gene annotations downloaded from NCBI (track 2). Bottom characters indicate gene loci discussed in the main text. The visualization includes alignments generated for PacBio CCS reads (track 3), Illumina QC reads (track 4), and contigs of the hybrid transcriptome (track 5). Track 4 is depicted as coverage track (light gray) with numbers on the scale indicating how often a read mapped to a specific position. Tracks 3 and 5 show individual transcripts (light red, + strand; light blue, – strand). The visualization was done with JBrowse (69), using the E. huxleyi CCMP3266 sGenome as reference (aee Data Sets S4 and S5).
Using the sGenome as a reference to track gene expression of known processes.
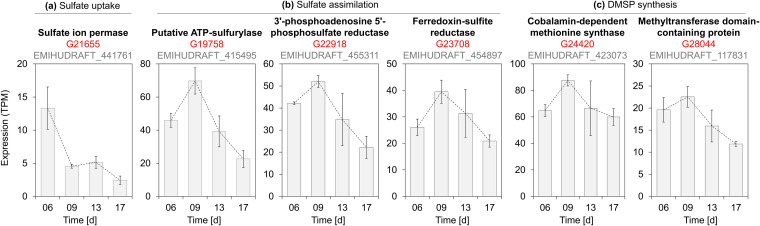
To assess the applicability of the E. huxleyi CCMP3266 sGenome as a reference for gene expression analyses, we mapped the Illumina QC reads to the sGenome and quantified CCMP3266 gene expression during cocultivation (Fig. 1). Using these expression data, we first conducted a targeted analysis to investigate the presence and temporal expression of genes involved in algal sulfur uptake and dimethylsulfoniopropionate (DMSP) biosynthesis. These cellular functions are well studied in E. huxleyi, are of major ecological importance, and have well-characterized expression patterns (54, 55). Our analysis indicated that key genes involved in sulfate uptake, reductive sulfate assimilation, and DMSP synthesis present in the E. huxleyi CCMP3266 sGenome were correctly annotated and, indeed, exhibited predicted expression patterns (Fig. 4). Examination of expression levels indicated that an ion permease involved in sulfate uptake exhibited the highest expression during growth (day 6), which decreased towards the stationary phase (day 9). In contrast, the expression levels of genes involved in reductive sulfate assimilation and DMSP synthesis were highest at the stationary phase (day 9) and decreased with the onset of bleaching (day 13). These data suggest that sulfate uptake takes place in dividing algal cells, while sulfur assimilation and DMSP synthesis are more pronounced in nondividing cells. These observations are in accordance with recent reports on high DMSP concentrations in stationary E. huxleyi cultures (17). It should be noted that a gene associated with the cleavage of DMSP to DMS (DMSP lyase, termed Alma1 [56]) was not found in strain CCMP3266, which corroborates reports on E. huxleyi strain variability regarding DMS production (57).
FIG 4.
Analysis of sulfur metabolism in E. huxleyi CCMP3266. (a) A sulfate ion permease gene exhibited high transcript abundance in dividing cells (day 06) and decreased as cultures aged. (b and c) Genes involved in reductive sulfate assimilation (b) and DMSP synthesis (c) exhibited the highest expression at stationary phase (day 9). Gene G28044 was identified as DSYB (methylthiohydroxybutryate methyltransferase), which catalyzes the last step of DMSP synthesis (99.7% amino acid identity), with the E. huxleyi CCMP1516 DSYB sequence reported by Curson et al (103). Information provided above the graphs include strain CCMP3266 gene annotations (bold), GeneID (red), and locus tag names of CCMP1516 homologues genes (gray). CCMP3266 GeneID, transcript IDs, and functional annotations are listed in Data Set S2; nucleotide sequences can be retrieved from Data Set S1, using the respective GeneID and/or transcript IDs.
Applying differential expression analysis to unravel biologically relevant genes.
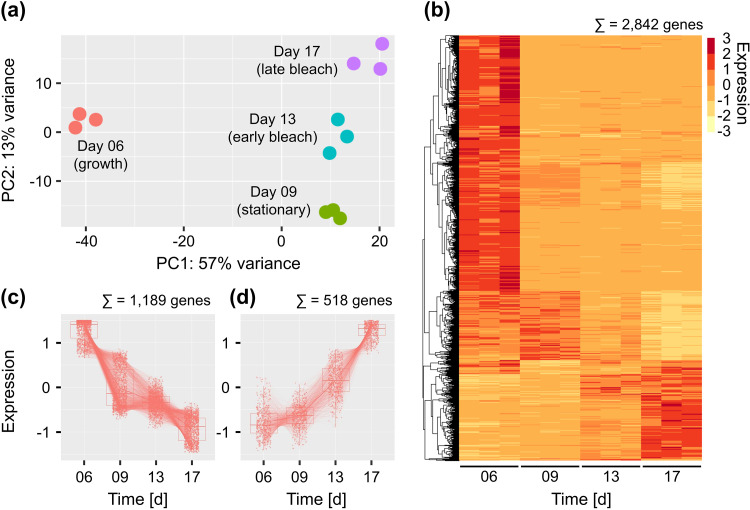
We further wished to gain an overview of algal genes and processes that were regulated during the cocultivation with bacteria. First, a principal component analysis was conducted, which shows that biological triplicates, which were collected for four different time points (Fig. 1), clustered closely together (Fig. 5a). The observed patterns in the PCA plot further suggest that E. huxleyi underwent major transcriptional rearrangements when transitioning from growth to stationary phase (day 6 to day 9). Fewer transcriptional changes were associated with the transition from stationary phase to algal demise (days 9 to 17). A likelihood-ratio test identified 2,842 algal genes that were differentially expressed (DE) during the four sampled time points (see Data Set S2, column 11). Gene expression is visualized as a heatmap (Fig. 5b), exhibiting clusters of genes that were characteristic for the different life phases of E. huxleyi (Fig. 1). A cluster of 1,189 downregulated, and 518 upregulated DE genes were extracted (Fig. 5c and d; see Data Set S2, columns 12 and 13) and analyzed for enrichment of biological processes based on Gene Ontology (GO) annotations (Table 1).
FIG 5.
Changes in E. huxleyi CCMP3266 gene expression patterns are most pronounced during the transition from growth to stationary phase. (a) A principal component analysis plot of algal gene expression in the different samples indicates that biological triplicates cluster closely together. The transition from growth (day 6) to stationary phase (day 9) is accompanied by global changes in gene expression, illustrated by the distance between the time points on the PC1 axis. Smaller differences in gene expression are observed during the transition from stationary phase to the early and late bleaching phase (days 9 to 17). (b) A heatmap with 2,842 differentially expressed genes (DEG) reveals clusters of E. huxleyi CCMP3266 genes that are characteristic for the different algal life phases. (c and d) Clusters of 1,189 downregulated (c) and 518 upregulated (d) DEGs were identified for subsequent GO enrichment analysis. Gene expression values are shown as z-scores. DEGs and down- and upregulated genes are indicated in Data Set S2 (columns 11 to 13).
TABLE 1.
E. huxleyi CCMP3266 gene expression in cocultures exhibits down- and upregulation of key biological processes during the transition from exponential growth to algal demise (Fig. 1)a
| Process no. | GO iD | Term | Annotated | Significant | Expected | P |
|---|---|---|---|---|---|---|
| Downregulated | ||||||
| 1 | GO:0006412 | Translation | 683 | 51 | 31.83 | 3.5E–04 |
| 2 | GO:0009765 | Photosynthesis, light harvesting | 79 | 29 | 3.68 | 3.4E–17 |
| 3 | GO:0044419 | Interspecies interaction between organisms | 535 | 28 | 24.93 | 1.4E–04 |
| 4 | GO:0006633 | Fatty acid biosynthetic process | 172 | 26 | 8.02 | 4.4E–07 |
| 5 | GO:0002181 | Cytoplasmic translation | 183 | 22 | 8.53 | 6.5E–05 |
| 6 | GO:0046034 | ATP metabolic process | 162 | 22 | 7.55 | 4.4E–03 |
| 7 | GO:0018298 | Protein-chromophore linkage | 49 | 19 | 2.28 | 1.9E–13 |
| 8 | GO:0006413 | Translational initiation | 177 | 15 | 8.25 | 4.2E–03 |
| 9 | GO:0006084 | Acetyl-CoA metabolic process | 91 | 13 | 4.24 | 1.4E–04 |
| 10 | GO:0010142 | Farnesyl diphosphate biosynthetic process, mevalonate pathway | 59 | 11 | 2.75 | 7.4E–05 |
| 11 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 65 | 11 | 3.03 | 1.8E–04 |
| 12 | GO:0000184 | Nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 87 | 11 | 4.05 | 2.2E–03 |
| 13 | GO:0006352 | DNA-templated transcription, initiation | 117 | 9 | 5.45 | 3.2E–03 |
| 14 | GO:0034975 | Protein folding in endoplasmic reticulum | 31 | 7 | 1.44 | 4.5E–04 |
| 15 | GO:0016126 | Sterol biosynthetic process | 34 | 7 | 1.58 | 2.4E–03 |
| Upregulated | ||||||
| 1 | GO:0006633 | Fatty acid biosynthetic process | 172 | 10 | 2.86 | 1.8E–03 |
| 2 | GO:0006986 | Response to unfolded protein | 85 | 7 | 1.42 | 6.3E–05 |
| 3 | GO:0051085 | Chaperone cofactor-dependent protein refolding | 21 | 6 | 0.35 | 8.7E–07 |
| 4 | GO:0006099 | Tricarboxylic acid cycle | 30 | 6 | 0.5 | 8.4E–06 |
| 5 | GO:0008340 | Determination of adult lifespan | 92 | 6 | 1.53 | 4.3E–03 |
| 6 | GO:0042026 | Protein refolding | 29 | 5 | 0.48 | 8.2E–04 |
| 7 | GO:0010142 | Farnesyl diphosphate biosynthetic process, mevalonate pathway | 59 | 5 | 0.98 | 2.9E–03 |
| 8 | GO:0006101 | Citrate metabolic process | 22 | 4 | 0.37 | 4.3E–04 |
| 9 | GO:0098630 | Aggregation of unicellular organisms | 40 | 4 | 0.67 | 4.3E–03 |
| 10 | GO:0042823 | Pyridoxal phosphate biosynthetic process | 10 | 3 | 0.17 | 5.0E–04 |
| 11 | GO:0051131 | Chaperone-mediated protein complex assembly | 17 | 3 | 0.28 | 2.6E–03 |
| 12 | GO:0000722 | Telomere maintenance via recombination | 19 | 3 | 0.32 | 3.6E–03 |
| 13 | GO:0009647 | Skotomorphogenesis | 4 | 2 | 0.07 | 1.6E–03 |
| 14 | GO:0034461 | Uropod retraction | 5 | 2 | 0.08 | 2.7E–03 |
| 15 | GO:0097552 | Mitochondrial double-strand break repair via homologous recombination | 6 | 2 | 0.1 | 4.0E–03 |
The “Annotated” column indicates the number of E. huxleyi CCMP3266 genes that were specified with the respective GO iD (see Data Set S2, column 21). The “Significant” column indicates the number of genes that were down- or upregulated. The “Expected” column indicates the theoretical number of down- and upregulated genes in the absence of an enrichment.
Processes that were downregulated during the transition from exponential growth (Fig. 1, day 6) to algal demise (Fig. 1, day 17) were associated with photosynthesis, ATP metabolism, and lipid biosynthesis, as well as DNA transcription and translation (Table 1). The identified processes are central for cell division, and their downregulation over time appears to be consistent with algal decline. Some downregulated DE genes were annotated with the GO term “interspecies interaction” and are therefore promising candidates for future studies on algal-bacterial interactions. We also found a rewiring of isoprenoid biosynthesis (i.e., the mevalonate pathway, which includes down- and upregulated genes; Table 1), which was previously reported to play an essential role in E. huxleyi during viral infections (58).
Next, we analyzed processes that were upregulated during algal demise. Upregulated processes were associated with DNA repair and protein folding, as well as with the tricarboxylic acid cycle involved in metabolite recycling and respiration. The upregulated genes are related to aging, and correspond to a general stress response in algae (46, 59). The increased expression of genes involved in DNA double-strand breaks during algal demise is in agreement with previous reports on programmed cell death in E. huxleyi (14, 60). Double-strand breaks could also be involved in meiosis, a cellular process that was shown to be triggered in E. huxleyi under viral infection (61). Noteworthy, the GO enrichment analysis identified genes involved in cell aggregation, a process that is extensively described in E. huxleyi (62), but for which no associated genes have yet been reported (63). Using the sGenome, we identified E. huxleyi candidate genes that are of biological and possibly ecological importance.
Expanding the E. huxleyi gene landscape.
Genomic diversity is assumed to contribute to the global success of E. huxleyi; however, the molecular functions encoded by variably distributed genes are poorly understood (25). To gain a better understanding of E. huxleyi variable genes, we sought to identify novel E. huxleyi genes that are expressed in CCMP3266 but absent from the E. huxleyi CCMP1516 reference genome. Therefore, we mapped the CCMP3266 genes (using the longest transcript per gene; see Data Set S2, column 7) to the genome of CCMP1516 (including mitochondrion and plastid). With this approach, we found that 6.3% of CCMP3266 genes (1,931 of 30,553) were not present in the CCMP1516 genome. Similarly, Read et al. (25) reported that 4.5 to 6.6% of CCMP1516 genes were absent from three other deep-sequenced E. huxleyi strains. The CCMP3266 genes absent in CCMP1516 were further filtered for those that contain functional annotations, resulting in a set 420 novel E. huxleyi genes (see Data Set S6).
Validating the novelty of the 420 new genes.
It is possible that part of the 420 novel E. huxleyi genes (see Data Set S6) are missing from the CCMP1516 reference genome due to technical constrains. To explore this possibility, we searched for the presence of the 420 novel genes in 17 publicly available E. huxleyi transcriptomes using BLAT (>10% coverage). Our analysis found that 142 of the 420 novel genes were present in all 17 transcriptomes (see Data Set S6, column 7). The common occurrence of these 142 genes suggests that they are missing from the CCMP1516 reference genome due to technical reasons. Indeed, among the 142 missing genes, we found a highly expressed nitrate reductase (GeneID G26296; see Data Set S6). The absence of this gene from the reference genome was previously noted (64), and was surprising due to its centrality in E. huxleyi nitrogen assimilation (65). In addition, we identified a gene encoding a decarboxylating phosphogluconate dehydrogenase (EC 1.1.1.44; GeneID G28049; see Data Set S6). This enzyme is required for the oxidative branch of the pentose phosphate pathway, a key biosynthetic pathway that produces essential 5-carbon sugars. In addition, an aminomethyl-transferring glycine dehydrogenase gene was detected (EC 1.4.4.2; GeneID G28787; see Data Set S6), which is necessary for photorespiration in mitochondria. In accordance with our findings, the three above-described genes are not represented in the public E. huxleyi KEGG metabolic pathway maps (66). In conclusion, the rediscovery of key metabolic E. huxleyi genes illustrates the importance of deep transcriptome sequencing from different growth stages and conditions, as well as the use of complementary long- and short-read sequencing methods in gene discovery and genome refinement.
E. huxleyi variable genes are potentially involved in microbial interactions.
Microalgae and bacteria have evolved together, and it appears plausible that E. huxleyi genome variability is influenced by microbial interactions (67). To address this hypothesis, we screened the list of 420 novel E. huxleyi genes (see Data Set S6) for genes that are variably distributed among E. huxleyi strains. Therefore, we filtered the list of 420 novel E. huxleyi genes for those that were detected in less than 50% of all 17 E. huxleyi transcriptomes (see Data Set S6, column 7). We further removed genes that lack PacBio long-read support (see Data Set S6, column 6) to increase the confidence of the functional annotations. The filtering resulted in a set of 86 variably distributed E. huxleyi CCMP3266 genes.
The set of 86 variable E. huxleyi genes was screened for functional annotations that indicate an involvement in algal-bacterial interactions. First, we searched for functions related to sensing bacteria. We found two genes that encode leucine-rich repeats (LRRs; G2551 and G5223; see Data Set S6 and Fig. S7) and which are part of a well-established plant-bacterium recognition mechanism (68). LRRs could play similar roles in algal-bacterial recognition.
Next, we searched for functions that alter the algal sugar profile. Algal sugars are important growth substrates for P. inhibens (69), and alterations in algal sugar compositions affect the assemblages of surrounding bacteria (70). Algal sugars are also found attached to the outer cell membrane, where they mediate interspecies communication (71). We identified several E. huxleyi CCMP3266 variable genes that are putatively involved in depolymerizing, modifying, and transporting sugars (see Fig. S6). These identified genes encode an alpha-amylase (G851; EC 3.2.1.1; see Data Set S6 and Fig. S7), which hydrolyses hexose polysaccharides, as well as an α-l-arabinofuranosidase (G548; EC 3.2.1.55; see Data Set S6 and Fig. S7), which hydrolyzes pentose polysaccharides. Two additional variable genes were found that can modify sugars by adding sulfur groups, including a galactose-3-O-sulfotransferase gene (G3328; EC 2.8.2.11; see Data Set S6 and Fig. S7) and a [heparan sulfate]-glucosamine 3-O-sulfotransferase gene (G1190; EC 2.8.2.30; see Data Set S6 and Fig. S7). In addition, a fucose/H+ symporter gene was found, which participates in transporting hexoses across cell membranes (G333; see Data Set S6 and Fig. S7).
Finally, we searched for E. huxleyi CCMP3266 variable genes that can modify algal lipids composition. Bacteria were previously shown to influence the abundance of lipid bodies in algal populations (22). Lipid bodies are also known to facilitate host defense in plants (67). In addition, virus-infected E. huxleyi cells were shown to accumulate lipid bodies that contain triacylglyerols (TAGs) and that can be secreted via extracellular vesicles (48, 72). Our search identified two variable CCMP3266 genes that are putatively involved in lipid modifications. First, we found a TAG-producing diacylglycerol acyltransferase gene, which was upregulated during algal demise (G26748; EC 2.3.1.20; see Data Set S6 and Fig. S7). Second, we identified a phospholipase A2 gene that can produce fatty acids and lysolipids and that was similarly upregulated during algal demise (G2440; see Data Set S6 and Fig. S7). Interestingly, the production and oxidation of fatty acids creates polyunsaturated aldehydes, known to be involved in grazer-defense of diatoms (73). Furthermore, lysolipids have known antimicrobial activity and were previously shown to protect certain protists against competing bacteria (74).
In conclusion, we identified E. huxleyi CCMP3266 genes that are variably distributed among E. huxleyi strains and that are promising candidates to be involved in microbial interactions. Our findings suggest that E. huxleyi genomic variability can be in part influenced by coevolution with bacteria. Our data further highlight the need to generate accurate gene information for individual E. huxleyi strains due to genomic variability between closely related strains. The pipeline we describe for hybrid transcriptome sequencing and analysis can facilitate further studies aimed at revealing strain-specific adaptations, which could provide broad insights into microalgal speciation and diversity.
MATERIALS AND METHODS
Strains and culture conditions.
The algal strain E. huxleyi CCMP3266 was purchased as an axenic culture from the National Center for Marine Algae and Microbiota (Bigelow Laboratory for Ocean Sciences, East Boothbay, ME). Algal axenic cultures were regularly controlled for the absence of bacteria, as described previously (14). To allow for cultivation under defined and reproducible conditions, cultures were grown in artificial seawater (ASW) prepared according to Goyet and Poisson (75). ASW contained mineral salts (NaCl, 409.41 mM; Na2SO4, 28.22 mM; KCl, 9.08 mM; KBr, 0.82 mM; NaF, 0.07 mM; Na2CO3, 0.20 mM; NaHCO3, 2 mM; MgCl2, 50.66 mM; SrCl2, 0.09 mM), L1 vitamins (thiamine HCl, 100 μg/L; biotin, 0.5 μg/L; vitamin B12, 0.5 μg/L), F/2 trace elements (FeCl3 · 6H2O, 3.15 mg/L; Na2EDTA · 2H2O, 4.36 mg/L; CuSO4 · 5H2O, 9.8 μg/L; Na2MoO4 · 2H2O, 6.3 μg/L; ZnSO4 · 7H2O, 22 μg/L; CoCl2 · 6H2O, 10 μg/L; MnCl2 · 4H2O, 180 μg/L), and L1 nutrients (NaNO3, 882 μM; NaH2PO4, 36.22 μM). ASW was adjusted to pH 8 using HCl. Algal cultivation was conducted at 18°C, with a light/dark cycle of 18/6 h and an illumination intensity of 130-μm photons m−2 s−1.
The bacterial strain Phaeobacter inhibens DSM17395 was purchased from the German collection of microorganisms and cell cultures (DSMZ, Braunschweig, Germany) and stored at −80°C with 20% glycerol. Cultivation of P. inhibens was conducted in ASW medium with additional carbon (glucose, 2 mM), nitrogen (NH4Cl, 5 mM), phosphorous (KH2PO4, 2 mM), and sulfur (Na2SO4, 33 mM), referred to as ASW+CNPS. Bacteria were cultivated at 30°C under constant shaking at 130 rpm.
Cocultivation.
E. huxleyi was cocultivated with P. inhibens to elicit the expression of “microbial interaction” genes. To avoid altering growth dynamics in cocultures during repetitive sampling, 24 replicate cocultures were setup in parallel flasks (1-L Erlenmeyer flasks, 150 ml of ASW). All 24 flasks were inoculated with 50,000 exponentially growing axenic E. huxleyi cells (day −4) and acclimated for 4 days. Cocultivation commenced by adding 300 CFU of a P. inhibens pure culture to all 24 flasks (day 0). To prevent glucose carryover, P. inhibens was precultivated for 48 h in ASW+CNPS until reaching stationary phase, while exhausting the glucose in the medium. Starting from day 4, four cocultivation flasks were randomly sampled per time point and then discarded after sampling (Fig. 1).
Growth monitoring.
Algal growth was determined for each sampled flask with three technical replicates by using a CellStream flow cytometer (Merck, Darmstadt, Germany). Bacterial growth was determined for each sampled flask with technical duplicates, by CFU counting on 1/2 YTSS agar plates, as previously described (14). Algal and bacterial growth is depicted in Fig. 1.
RNA sampling and quality control.
To capture E. huxleyi transcripts from different physiological states and different phases of the algal-bacterial interaction, four characteristic time points were chosen for RNA extraction (Fig. 1). At each of the four time points, RNA was extracted from three selected flasks, resulting in a total of 12 RNA samples (Fig. 1, orange dots). For RNA extraction, the sampled flasks were harvested (2 h after the beginning of the light cycle) by centrifugation of a 50-ml coculture at 18°C for 5 min at 3,220 relative centrifugal force using a 5810 R swing-out centrifuge (Eppendorf, Hamburg, Germany). The supernatant was removed with vacuum, and cell pellets were resuspended in 450 μl of RLT lysis buffer (Qiagen, Hilden, Germany). Cells were immediately disrupted by bead beating at room temperature with 300 mg of 150- to 220-μm acid-washed glass beads for 5 min at 30 s−1 in a mixer mill MM 400 (Retsch, Haan, Germany) and then directly subjected to RNA extraction. The RNA extraction was conducted using an RNeasy Plant minikit in combination with the RNase-Free DNase Set (Qiagen) for on-column DNase treatment. The final elution volume was 60 μl. RNA integrity was assessed using a 4150 TapeStation instrument with RNA ScreenTapes (Agilent, Santa Clara, CA; see Fig. S1 in the supplemental material). RNA purity was assessed using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA; see Fig. S1). The RNA quantity was assessed using a Qubit 2.0 Fluorometer with a Qubit RNA HS assay kit (Thermo Fisher Scientific; see Fig. S1). RNA extracts were stored at −80°C.
PacBio library preparation, sequencing, and raw read processing.
For PacBio long-read full-length cDNA sequencing, the non-size-selected, poly(A)-targeted PacBio Iso-Seq library protocol v06 was used (Pacific Biosciences, Menlo Park, CA). We constructed a single PacBio sequencing library with the intention to capture all E. huxleyi RNA transcripts expressed at the four sampled time points (Fig. 1, orange dots). Therefore, all 12 RNA samples were pooled (1 μl per sample; see Fig. S1), and the RNA pool used as the template for reverse transcription with the SMARTer PCR cDNA synthesis kit (two parallel reactions; 1 μg of pooled RNA per reaction; TaKaRa, Mountain View, CA). The resulting cDNA was amplified with 12 PCR cycles (see Fig. S8), and the product was used for SMRTbell template preparation as described in the protocol. The resulting SMRTbell template (see Fig. S9) was annealed to the sequencing polymerase, using a Sequel binding kit 3.0. Sequencing was conducted with a 8 pM on-plate concentration using a Sequel I System in CCS mode and a single SMRT Cell (1M; PacBio). PacBio raw reads (polymerase reads) were processed into circular consensus sequence (CCS) reads (≥Q20; also termed HiFi reads), using the SMRT Link software (v8.0). SMRT Link outputs that summarize sequencing productivity and quality are listed in Table S1. Diagnostic plots generated during CCS read generation are provided in Fig. S2.
Illumina library preparation, sequencing, and raw read processing.
For total RNA sequencing, a poly(A)-independent, stranded library preparation protocol was used. The protocol was adapted from Avraham et al. (76) (see Fig. S10), with minor modifications. As input, 100 ng of RNA extract was used for each of the samples collected from 12 cocultures (Fig. 1; see also Fig. S1). Fragmented RNA samples were individually tagged by 3′ ligation with the barcoded adapters 1 to 12 (desalted; Merck/Millipore/Sigma, Israel) to allow for downstream temporal gene expression analyses. Tagged RNA samples were pooled, and rRNA depleted with a RiboMinus kit using a combination of universal rRNA depletion probes for plants and bacteria (Invitrogen, Carlsbad, CA). The depleted RNA was purified using an RNA Clean & Concentrator-5 column (Zymo Research, Irvine, CA). First-strand cDNA synthesis was conducted with a SuperScript IV reverse transcriptase (Invitrogen) and an AR2 primer, which binds to the barcoded adapter region. The cDNA was 3′ ligated with adapter 3Tr3 and amplified with the adapter-binding primers P5 and P7_tagged_880 using an optimized eight-cycle PCR (see Fig. S11a). The PCR product was purified using 0.6× and 0.75× RNAClean XP SPRI beads for right- and left-sided cleanup, respectively (see Fig. S11b). Sequencing was conducted using an Illumina NextSeq 500 instrument in paired-end mode (88 + 78 cycles), a high-output v2.5 sequencing cassette (400 M), and 2 pM of prepared library with 1% PhiX as an internal control. Sequencing yields and quality are summarized in Table S2. Illumina raw reads were demultiplexed with fastq-multx (v1.3.1 [77]), allowing one mismatch per barcode. The demultiplexed reads were Q20 quality filtered with cutadapt (--nextseq-trim=20 [78]). Sequencing adapters and poly(A) tails were trimmed, and reads of <30 bp were discarded, resulting in processed raw reads, referred to as Illumina QC reads. Illumina read statistics were generated with FastQC (79) and MultiQC (80).
Transcriptome construction.
A hybrid transcriptome was constructed by integrating PacBio and Illumina sequencing data. To this end, PacBio CCS reads and Illumina QC reads were de novo assembled with hybrid rnaSPAdes (v3.14.0; --pe --ss rf --pacbio; 40). The resulting hybrid assembly was filtered, using the following steps in the given order: P. inhibens transcripts were identified and removed by mapping the hybrid assembly to the genome of P. inhibens DSM17395 (accession number GCF_000154765.2) with GMAP (v2019-02-26; --nosplicing [81]). Redundant transcripts were removed with CD-HIT-EST (-c 0.95 -n 10 [82]). Erroneously assembled rRNA fragments were curated by mapping the hybrid assembly to three manually constructed nucleus-, plastid-, and mitochondrion-encoded rRNA consensus sequences (nucleotide sequences of rRNA constructs are given in Table S3). The mapping was conducted with GMAP (v2019-02-26; -n 1 -z sense_force --canonical-mode 0 --no-chimeras --split-large-introns --totallength 5000 --max-intronlength-middle 5000 --cross-species [81]). Mapped fragments were removed and replaced by the curated rRNA consensus sequences (see Table S3 and Fig. S4). Contigs with poor expression support were identified by multimapping the Illumina QC reads (allowing up to 100 alignments per read) to the hybrid assembly with STAR (v2.7.5c; --outSAMattributes All --alignIntronMax 5000 --alignMatesGapMax 5000 --outSAMtype BAM SortedByCoordinate --outFilterMultimapNmax 100 --outSAMmultNmax 100 [83]). Mapped reads were counted with featureCounts (v2.0.0; -p -C -F SAF -M [84]), and contigs with <5 mapped reads were discarded. Finally, N-characters introduced by rnaSPAdes were removed with SeqKit (85), resulting in the final hybrid transcriptome (see Data Set S1). Functional annotations were created for each transcript by conducting a blastx search (blastx-v2.7.1; -db nr -soft_masking “true” -strand “plus” -num_alignments 10 -max_hsps 10 -evalue 1e-5 -outfmt 14 [86]) and using blast2GO, InterProScan, and EggNOG (v5.0) implemented in the OmicsBox (v1.3.11) (87–89). The functional transcript annotations are given in Data Set S2.
Two additional, nonhybrid transcriptomes were constructed as controls. First, a PacBio long-read transcriptome was constructed (LR transcriptome), by processing the PacBio CCS reads with the Iso-Seq3 pipeline, using the SMRT Link software (v8.0). As part of the pipeline, PacBio CCS reads that contain both cDNA primers and a poly(A) tail were trimmed at the ends, and artificial concatemers were removed (reads with additional cDNA primers in the middle of the sequence), resulting in full-length nonconcatemer reads (PacBio FLNC reads; see Table S1). FLNC reads with a minimum of seven sub-reads (inserts that were passed ≥7 times by the polymerase), at least two representative sequences (FLNC reads that differ by <100 bp on the 5′ end and by <30 bp on the 3′ end and have no internal gaps of >10 bp) and >0.99 accuracy (>Q20; less than 1 in 100 incorrectly called bases) were clustered and collapsed into consensus transcripts, referred to as PacBio HQ isoforms. Possible P. inhibens contaminations were removed by genome mapping, and redundant transcripts were discarded with CD-HIT-EST, as described above for the hybrid transcriptome, resulting in the final LR transcriptome. As a second nonhybrid control, an Illumina short-read transcriptome was constructed (SR transcriptome). The SR transcriptome was constructed with the same pipeline as described above for the hybrid transcriptome, including all filtering steps, but by using rnaSPAdes without PacBio CCS read integration (omitting the --pacbio option). An overview of the steps involved in transcriptome construction is given in Fig. 2a.
Assessment of transcriptome quality.
Basic fasta file statistics were collected for the transcriptomes with assembly_stats (v0.1.4 [90]). The number of transcripts with protein-coding sequences (≥100 amino acids) were quantified by predicting open reading frames with ANGEL (v2.7; dumb_predict.py --min_aa_length 100; angel_make_training_set.py --random; angel_train.py; angel_predict.py --output_mode=best --min_angel_aa_length 100 --min_dumb_aa_length 100 [91]). Transcriptome completeness was assessed with BUSCO v4 (Benchmarking Universal Single-Copy Orthologs [50]), using a eukaryotic reference data set with 255 conserved genes (eukaryota_odb10.2019-11-20). In addition, transcriptome completeness and transcript isoform reconstruction efficiency were assessed using a tailored analysis, which is conceptually similar to the BUSCO (50) and rnaQUAST (92) methods but relies on a newly compiled set of conserved E. huxleyi reference genes. For this analysis, a complete list of 33,341 E. huxleyi CCMP1516 genes was downloaded from the JGI PhycoCosm hub (25, 93) (reduced “haploid” gene model set). The gene list was filtered for those that are part of the E. huxleyi core genome (n = 20,055), and which were supported by expressed sequence tags (ESTs), based on data given by Read et al. (25). The nucleotide sequences of the genes were extracted from the E. huxleyi CCMP1516 genome assembly (Emihu1_scaffolds.fasta), using the associated annotation file (Emihu1_reduced_genes.gff). The result was a set of 13,168 conserved E. huxleyi CCMP1516 reference genes, which include intron and exon features (see Data Set S3). To identify which of these E. huxleyi reference genes were present in the analyzed transcriptomes, we ran a BLAT search (v35 [94]) using the reference genes as the database and the transcriptomes as queries (-tileSize=10 -stepSize=5 -extendThroughN). The BLAT output table was analyzed with basic Linux bash programs (awk, sort, and uniq), and the results are summarized in Fig. 2b. The coverage of an E. huxleyi reference gene was calculated based on the BLAT output table, using the columns for gene length (T size), target start (T start), and target end (T end), to include gaps. We considered an E. huxleyi reference gene to be detected in a transcriptome query if the gene exhibited at least 10% coverage by a matching transcript. Nucleotide identities between CCMP1516 reference genes and CCMP3266 matching transcripts were calculated based on the match and mismatch columns. A reference gene was considered to be complete if it exhibited at least 95% coverage by a matching transcript (= full-length transcript). To assess the efficiency of full-length transcript isoform reconstruction for each of the transcriptomes, we calculated the average number of full-length transcripts found per complete E. huxleyi reference gene. We also evaluated the outcome of hybrid transcriptome sequencing by quantifying the number of E. huxleyi reference genes that exhibited at least 5% higher coverage by the best matching transcript in the hybrid transcriptome compared to the best-matching transcript in the SR transcriptome (= reference genes with “improved length”; Fig. 2b).
Synthetic genome construction.
To generate a genome-like data set, a “synthetic genome” (sGenome) was constructed. The hybrid transcriptome was first mapped to the E. huxleyi CCMP1516 reference genome consisting of the main genome assembly (GCF_000372725.1), the plastid (JN022705.1), and the mitochondrion (JN022704.1). The mapping was conducted with GMAP (v2019-02-26; -n 1 -z sense_force --canonical-mode 0 --no-chimeras --split-large-introns --totallength 5000 --max-intronlength-middle 5000 --cross-species [81]). Unmapped transcripts and transcripts with <95% identity and <95% coverage alignment accuracy were identified with TAMA (tama_collapse.py -x no_cap -icm ident_map -c 95 -i 95 [45]) and subjected to reference-free transcript clustering and gene reconstruction with COGENT (v8.0.0; default options [53]). The COGENT output, which includes reconstructed genes and unassigned contigs, was used with CD-HIT-EST (v4.8.1; -c 0.80 -n 5 -d 0 -g 1 [82]) to reduce redundancy. The CD-HIT-EST output, which represents E. huxleyi CCMP3266 specific genes, was appended to the E. huxleyi CCMP1516 reference genome (including plastid and mitochondrion), resulting in a preliminary sGenome. For final sGenome generation (see Data Set S4), the hybrid transcriptome was mapped to the preliminary sGenome with GMAP (v2019-02-26; options as above [81]), and unmapped regions in the preliminary sGenome were masked with bedtools (v2.26.0 [95]). The GMAP mapping coordinates were further used as input for TAMA to assemble the mapped transcripts into genes (-x no_cap -icm ident_map -c 10 -i 10 [45]) and to generate a CCMP3266 gene annotation file. The annotation file was modified with AGAT to convert it into GFF3 format and to add additional feature attributes (96) (see Data Set S5). The E. huxleyi CCMP3266 sGenome (see Data Set S4) was used together with the gene annotation file (see Data Set S5) as a reference for visualization with JBrowse (v1.16.6 [97]; Fig. 3). The visualization includes expression data generated by mapping PacBio CCS reads (GMAP v2019-02-26, with options as described above [81]) and Illumina QC reads (STAR v2.7.5c, with options as described above [83]) to the sGenome.
Differential gene expression analysis.
E. huxleyi CCMP3266 gene expression analysis was conducted by mapping Illumina QC reads to the sGenome with STAR (v2.7.5c; --outSAMattributes All --alignIntronMax 5000 --alignMatesGapMax 5000 --outSAMtype BAM SortedByCoordinate --outFilterMultimapNmax 30 --outSAMmultNmax 1 [83]). The alignment output was used with the above generated GFF3 annotation file to count mapped reads per gene with featureCounts (-p -C -O --fraction; v2.0.0 [84]). For compatibility with featureCounts, the NH:i attribute of the STAR output was changed to “1,” using SAMtools (98) and Linux sed (STAR reports the number of alternative alignments in the NH:I attribute, which interferes with featureCounts fractional counting). The resulting count table was normalized to transcripts per million to account for differences in sample size and gene length. In addition, the nonnormalized count table was used as input for differential gene expression analysis with the DESeq2 package (v1.28.1 [34]) for R (v4.0.3). Sample variability was analyzed using the DESeq2 rlog transformation (blind=TRUE) and plotPCA function (ntop=2000), shown in Fig. 5a. Since the RNA sequencing data were from a time-course experiment, we identified differentially expressed genes across all samples by using the DESeq2 likelihood ratio test (adjusted P cutoff = 0.001), following recommendations of the Harvard Chan Bioinformatics Core Unit (99). The DESeq2 normalized counts for the DEGs were extracted, and then scaled (z-score) and plotted with pheatmap (100) (Fig. 5b). To extract groups of downregulated and upregulated genes, DESeq2 rlog-transformed counts were used as input, together with the list of DEGs, for the DEGReport package (101). The extracted DEG groups (Fig. 5c) were used as input for GO-term enrichment analysis with topGO, using the weight01 algorithm and Fisher test statistics (v2.40.0 [102]; Table 1).
Identification of novel E. huxleyi genes.
To identify CCMP3266 genes that are absent from the CCMP1516 reference genome, the CCMP3266 gene annotation file (see Data Set S5) was used with AGAT (96) to extract the longest representative transcript for each gene. The representative transcripts were mapped to the genome assembly of E. huxleyi CCMP1516 (including mitochondria and plastid) using GMAP (v2019-02-26; options as above [81]). Unmapped transcripts were used as query with BLAT (v35; -tileSize=10 -stepSize=5 -extendThroughN [94]) to search for homologues sequences in 17 E. huxleyi transcriptomes downloaded from the TSA database (accession numbers GHJP00000000, HBIR00000000, HBNU00000000, HBOB00000000, HBOC00000000, HBPW00000000, HBQI00000000, HBTF00000000, HBTI00000000, HBTL00000000, HBTO00000000, HBTP00000000, HBTT00000000, HBTU00000000, HBTV00000000, HBTW00000000, and HBTX00000000). The BLAT output table was filtered for matches with at least 10% coverage, as described in Materials and Methods under “Assessment of transcriptome quality.”
Data availability.
PacBio CCS reads, Illumina QC reads, and the hybrid transcriptome were deposited under the BioProject ID PRJNA698293. PacBio CCS reads and Illumina QC reads were uploaded to the Sequence Read Archive (SRA; accession numbers SRR13590462 to SRR13590474). The hybrid transcriptome was deposited at the Transcriptome Shotgun Assembly Sequence Database (TSA; master record, GIZZ00000000; assembly version, GIZZ01000000). The hybrid transcriptome described in the manuscript (see Data Set S1) slightly differs from the uploaded version; upload to the TSA database required the removal of 1,043 short transcripts (<200 bp), 14 contaminating transcripts, and the trimming of putative sequencing adapters for 164 transcripts (∼20 bp). The transcript IDs generated by hybrid rnaSPAdes were renamed for TSA submission (see Data Set S2, columns 3 and 4). A BLAST search of the E. huxleyi CCMP3266 hybrid transcriptome can be conducted from the NCBI webpage upon selecting the TSA Database. Supplemental Data Sets S1 to S6 can be downloaded from Zenodo (https://zenodo.org/record/5702921).
ACKNOWLEDGMENTS
We are grateful to Ester Feldmesser and Bareket Dassa for help with Illumina raw read processing and for fruitful discussions. We thank Hadas Keren-Shaul and her team from the Weizmann Institute of Science Sandbox Core facility for their guidance with PacBio Iso-Seq library preparation and for operating the sequencing instruments. Finally, we thank all members of the Segev lab for insightful discussions.
M.S. was funded by the Dean of Faculty fellowship and the Sir Charles Clore Postdoctoral Fellowship. E.S. was funded by the Israeli Science Foundation (ISF 947/18), the Peter and Patricia Gruber Foundation, and the Minerva Foundation, with funding from the Federal German Ministry for Education and Research.
E.S. and M.S. designed the research and wrote the manuscript. M.S. performed the research and collected the data. Data analyses and interpretation were conducted by M.S. and D.Y.
Footnotes
Supplemental material is available online only.
Contributor Information
Martin Sperfeld, Email: Martin.Sperfeld@weizmann.ac.il.
Einat Segev, Email: Einat.Segev@weizmann.ac.il.
Isaac Cann, University of Illinois at Urbana-Champaign.
REFERENCES
- 1.Falkowski PG, Barber RT, Smetacek V. 1998. Biogeochemical controls and feedbacks on ocean primary production. Science 281:200–206. 10.1126/science.281.5374.200. [DOI] [PubMed] [Google Scholar]
- 2.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281:237–240. 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 3.Moran MA, Kujawinski EB, Stubbins A, Fatland R, Aluwihare LI, Buchan A, Crump BC, Dorrestein PC, Dyhrman ST, Hess NJ, Howe B, Longnecker K, Medeiros PM, Niggemann J, Obernosterer I, Repeta DJ, Waldbauer JR. 2016. Deciphering ocean carbon in a changing world. Proc Natl Acad Sci USA 113:3143–3151. 10.1073/pnas.1514645113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lohmann H. 1908. Über die Beziehungen zwischen den pelagischen Ablagerungen und dem Plankton des Meeres. Int Revue Ges Hydrobiol Hydrogr 1:309–323. 10.1002/iroh.19080010302. [DOI] [Google Scholar]
- 5.Paasche E. 2001. A review of the coccolithophorid Emiliania huxleyi (Prymnesiophyceae), with particular reference to growth, coccolith formation, and calcification-photosynthesis interactions. Phycologia 40:503–529. 10.2216/i0031-8884-40-6-503.1. [DOI] [Google Scholar]
- 6.Holligan PM, Viollier M, Harbour DS, Camus P, Champagne-Philippe M. 1983. Satellite and ship studies of coccolithophore production along a continental shelf edge. Nature 304:339–342. 10.1038/304339a0. [DOI] [Google Scholar]
- 7.Balch WM, Holligan PM, Ackleson SG, Voss KJ. 1991. Biological and optical properties of mesoscale coccolithophore blooms in the Gulf of Maine. Limnol Oceanogr 36:629–643. 10.4319/lo.1991.36.4.0629. [DOI] [Google Scholar]
- 8.Thierstein HR, Young JR. 2013. Coccolithophores: from molecular processes to global impact. Springer Science & Business Media, New York, NY. [Google Scholar]
- 9.Behrenfeld MJ, Boss ES. 2014. Resurrecting the ecological underpinnings of ocean plankton blooms. Annu Rev Mar Sci 6:167–194. 10.1146/annurev-marine-052913-021325. [DOI] [PubMed] [Google Scholar]
- 10.Bratbak G, Egge JK, Heldal M. 1993. Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar Ecol Prog Ser 93:39–48. 10.3354/meps093039. [DOI] [Google Scholar]
- 11.Vardi A, Haramaty L, Van Mooy BA, Fredricks HF, Kimmance SA, Larsen A, Bidle KD. 2012. Host-virus dynamics and subcellular controls of cell fate in a natural coccolithophore population. Proc Natl Acad Sci USA 109:19327–19332. 10.1073/pnas.1208895109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehahn Y, Koren I, Schatz D, Frada M, Sheyn U, Boss E, Efrati S, Rudich Y, Trainic M, Sharoni S, Laber C, DiTullio GR, Coolen MJL, Martins AM, Van Mooy BAS, Bidle KD, Vardi A. 2014. Decoupling physical from biological processes to assess the impact of viruses on a mesoscale algal bloom. Curr Biol 24:2041–2046. 10.1016/j.cub.2014.07.046. [DOI] [PubMed] [Google Scholar]
- 13.Ku C, Sheyn U, Sebé-Pedrós A, Ben-Dor S, Schatz D, Tanay A, Rosenwasser S, Vardi A. 2020. A single-cell view on alga-virus interactions reveals sequential transcriptional programs and infection states. Sci Adv 6:eaba4137. 10.1126/sciadv.aba4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segev E, Wyche TP, Kim KH, Petersen J, Ellebrandt C, Vlamakis H, Barteneva N, Paulson JN, Chai L, Clardy J, Kolter R. 2016. Dynamic metabolic exchange governs a marine algal-bacterial interaction. Elife 5:e17473. 10.7554/eLife.17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harvey EL, Deering RW, Rowley DC, El Gamal A, Schorn M, Moore BS, Johnson MD, Mincer TJ, Whalen KE. 2016. A bacterial quorum-sensing precursor induces mortality in the marine coccolithophore Emiliania huxleyi. Front Microbiol 7:759. 10.3389/fmicb.2016.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayali X. 2018. Editorial: metabolic interactions between bacteria and phytoplankton. Front Microbiol 9:727. 10.3389/fmicb.2018.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barak-Gavish N, Frada MJ, Ku C, Lee PA, DiTullio GR, Malitsky S, Aharoni A, Green SJ, Rotkopf R, Kartvelishvily E, Sheyn U, Schatz D, Vardi A. 2018. Bacterial virulence against an oceanic bloom-forming phytoplankter is mediated by algal DMSP. Sci Adv 4:eaau5716. 10.1126/sciadv.aau5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whalen KE, Kirby C, Nicholson RM, O’Reilly M, Moore BS, Harvey EL. 2018. The chemical cue tetrabromopyrrole induces rapid cellular stress and mortality in phytoplankton. Sci Rep 8:15498. 10.1038/s41598-018-33945-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bramucci AR, Case RJ. 2019. Phaeobacter inhibens induces apoptosis-like programmed cell death in calcifying Emiliania huxleyi. Sci Rep 9:5215. 10.1038/s41598-018-36847-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Dassow P, Ogata H, Probert I, Wincker P, Da Silva C, Audic S, Claverie J-M, de Vargas C. 2009. Transcriptome analysis of functional differentiation between haploid and diploid cells of Emiliania huxleyi, a globally significant photosynthetic calcifying cell. Genome Biol 10:R114. 10.1186/gb-2009-10-10-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frada MJ, Bidle KD, Probert I, de Vargas C. 2012. In situ survey of life cycle phases of the coccolithophore Emiliania huxleyi (Haptophyta). Environ Microbiol 14:1558–1569. 10.1111/j.1462-2920.2012.02745.x. [DOI] [PubMed] [Google Scholar]
- 22.Segev E, Castañeda IS, Sikes EL, Vlamakis H, Kolter R. 2016. Bacterial influence on alkenones in live microalgae. J Phycol 52:125–130. 10.1111/jpy.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. 2011. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat Chem 3:331–335. 10.1038/nchem.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyrrell T, Merico A. 2004. Emiliania huxleyi: bloom observations and the conditions that induce them, p 75–97. In Thierstein HR, Young JR (ed), Coccolithophores: from molecular processes to global impact. Springer, Berlin, Germany. 10.1007/978-3-662-06278-4_4. [DOI] [Google Scholar]
- 25.Read BA, Kegel J, Klute MJ, Kuo A, Lefebvre SC, Maumus F, Mayer C, Miller J, Monier A, Salamov A, Young J, Aguilar M, Claverie JM, Frickenhaus S, Gonzalez K, Herman EK, Lin YC, Napier J, Ogata H, Sarno AF, Shmutz J, Schroeder D, de Vargas C, Verret F, von Dassow P, Valentin K, Van de Peer Y, Wheeler G, Dacks JB, Delwiche CF, Dyhrman ST, Glöckner G, John U, Richards T, Worden AZ, Zhang X, Grigoriev IV, Emiliania huxleyi Annotation Consortium . 2013. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499:209–213. 10.1038/nature12221. [DOI] [PubMed] [Google Scholar]
- 26.Kegel JU, John U, Valentin K, Frickenhaus S. 2013. Genome variations associated with viral susceptibility and calcification in Emiliania huxleyi. PLoS One 8:e80684. 10.1371/journal.pone.0080684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Dassow P, John U, Ogata H, Probert I, Bendif EM, Kegel JU, Audic S, Wincker P, Da Silva C, Claverie J-M, Doney S, Glover DM, Flores DM, Herrera Y, Lescot M, Garet-Delmas M-J, de Vargas C. 2015. Life-cycle modification in open oceans accounts for genome variability in a cosmopolitan phytoplankton. ISME J 9:1365–1377. 10.1038/ismej.2014.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labeeuw L, Khey J, Bramucci AR, Atwal H, de la Mata AP, Harynuk J, Case RJ. 2016. Indole-3-Acetic Acid Is Produced by Emiliania huxleyi Coccolith-Bearing Cells and Triggers a Physiological Response in Bald Cells. Front Microbiol 7. 10.3389/fmicb.2016.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayers TJ, Bramucci AR, Yakimovich KM, Case RJ. 2016. A bacterial pathogen displaying temperature-enhanced virulence of the microalga Emiliania huxleyi. Front Microbiol 7:892. 10.3389/fmicb.2016.00892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, Beszteri B, Bidle KD, Cameron CT, Campbell L, Caron DA, Cattolico RA, Collier JL, Coyne K, Davy SK, Deschamps P, Dyhrman ST, Edvardsen B, Gates RD, Gobler CJ, Greenwood SJ, Guida SM, Jacobi JL, Jakobsen KS, James ER, Jenkins B, John U, et al. 2014. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol 12:e1001889. 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nam O, Park J-M, Lee H, Jin E. 2019. De novo transcriptome profile of coccolithophorid alga Emiliania huxleyi CCMP371 at different calcium concentrations with proteome analysis. PLoS One 14:e0221938. 10.1371/journal.pone.0221938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keren H, Lev-Maor G, Ast G. 2010. Alternative splicing and evolution: diversification, exon definition, and function. Nat Rev Genet 11:345–355. 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 33.Steijger T, Abril JF, Engström PG, Kokocinski F, Hubbard TJ, Guigó R, Harrow J, Bertone P, RGASP Consortium . 2013. Assessment of transcript reconstruction methods for RNA-seq. Nat Methods 10:1177–1184. 10.1038/nmeth.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hölzer M, Marz M. 2019. De novo transcriptome assembly: a comprehensive cross-species comparison of short-read RNA-Seq assemblers. Gigascience 8:giz039. 10.1093/gigascience/giz039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stark R, Grzelak M, Hadfield J. 2019. RNA sequencing: the teenage years. Nat Rev Genet 20:631–656. 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- 36.Minio A, Massonnet M, Figueroa-Balderas R, Vondras AM, Blanco-Ulate B, Cantu D. 2019. Iso-Seq allows genome-independent transcriptome profiling of grape berry development. G3 (Bethesda) 9:755–767. 10.1534/g3.118.201008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guerrero-Sanchez VM, Maldonado-Alconada AM, Amil-Ruiz F, Verardi A, Jorrín-Novo JV, Rey M-D. 2019. Ion Torrent and Illumina, two complementary RNA-seq platforms for constructing the holm oak (Quercus ilex) transcriptome. PLoS One 14:e0210356. 10.1371/journal.pone.0210356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Liu C, He M, Xiang Z, Yin Y, Liu S, Zhuang Z. 2019. A full-length transcriptome of Sepia esculenta using a combination of single-molecule long-read (SMRT) and Illumina sequencing. Mar Genomics 43:54–57. 10.1016/j.margen.2018.08.008. [DOI] [Google Scholar]
- 39.Bråte J, Fuss J, Mehrota S, Jakobsen KS, Klaveness D. 2019. Draft genome assembly and transcriptome sequencing of the golden algae Hydrurus foetidus (Chrysophyceae). F1000Res 8:401. 10.12688/f1000research.16734.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prjibelski AD, Puglia GD, Antipov D, Bushmanova E, Giordano D, Mikheenko A, Vitale D, Lapidus A. 2020. Extending rnaSPAdes functionality for hybrid transcriptome assembly. BMC Bioinformatics 21:302. 10.1186/s12859-020-03614-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puglia GD, Prjibelski AD, Vitale D, Bushmanova E, Schmid KJ, Raccuia SA. 2020. Hybrid transcriptome sequencing approach improved assembly and gene annotation in Cynara cardunculus (L.). BMC Genomics 21:317. 10.1186/s12864-020-6670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stern DB, Goldschmidt-Clermont M, Hanson MR. 2010. Chloroplast RNA metabolism. Annu Rev Plant Biol 61:125–155. 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- 43.Chen D, Du Y, Fan X, Zhu Z, Jiang H, Wang J, Fan Y, Chen H, Zhou D, Xiong C, Zheng Y, Xu X, Luo Q, Guo R. 2020. Reconstruction and functional annotation of Ascosphaera apis full-length transcriptome utilizing PacBio long reads combined with Illumina short reads. J Invertebr Pathol 176:107475. 10.1016/j.jip.2020.107475. [DOI] [PubMed] [Google Scholar]
- 44.Statello L, Guo C-J, Chen L-L, Huarte M. 2021. Gene regulation by long noncoding RNAs and its biological functions. Nat Rev Mol Cell Biol 22:96–118. 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuo RI, Cheng Y, Zhang R, Brown JWS, Smith J, Archibald AL, Burt DW. 2020. Illuminating the dark side of the human transcriptome with long read transcript sequencing. BMC Genomics 21:751. 10.1186/s12864-020-07123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caron DA, Alexander H, Allen AE, Archibald JM, Armbrust EV, Bachy C, Bell CJ, Bharti A, Dyhrman ST, Guida SM, Heidelberg KB, Kaye JZ, Metzner J, Smith SR, Worden AZ. 2017. Probing the evolution, ecology and physiology of marine protists using transcriptomics. Nat Rev Microbiol 15:6–20. 10.1038/nrmicro.2016.160. [DOI] [PubMed] [Google Scholar]
- 47.Buccitelli C, Selbach M. 2020. mRNAs, proteins, and the emerging principles of gene expression control. Nat Rev Genet 21:630–644. 10.1038/s41576-020-0258-4. [DOI] [PubMed] [Google Scholar]
- 48.Schatz D, Rosenwasser S, Malitsky S, Wolf SG, Feldmesser E, Vardi A. 2017. Communication via extracellular vesicles enhances viral infection of a cosmopolitan alga. Nat Microbiol 2:1485–1492. 10.1038/s41564-017-0024-3. [DOI] [PubMed] [Google Scholar]
- 49.Schatz D, Schleyer G, Saltvedt MR, Sandaa R-A, Feldmesser E, Vardi A. 2021. Ecological significance of extracellular vesicles in modulating host-virus interactions during algal blooms. ISME J 10.1038/s41396-021-01018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 51.Schaefke B, Sun W, Li Y-S, Fang L, Chen W. 2018. The evolution of posttranscriptional regulation. Wiley Interdiscip Rev RNA 9:e1485. 10.1002/wrna.1485. [DOI] [PubMed] [Google Scholar]
- 52.Vijay N, Poelstra JW, Künstner A, Wolf JBW. 2013. Challenges and strategies in transcriptome assembly and differential gene expression quantification: a comprehensive in silico assessment of RNA-seq experiments. Mol Ecol 22:620–634. 10.1111/mec.12014. [DOI] [PubMed] [Google Scholar]
- 53.Tseng E. 2020. Cogent: COding GENome reconstruction tool. https://github.com/Magdoll/Cogent.
- 54.Stefels J. 2000. Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J Sea Res 43:183–197. 10.1016/S1385-1101(00)00030-7. [DOI] [Google Scholar]
- 55.Bochenek M, Etherington GJ, Koprivova A, Mugford ST, Bell TG, Malin G, Kopriva S. 2013. Transcriptome analysis of the sulfate deficiency response in the marine microalga Emiliania huxleyi. New Phytol 199:650–662. 10.1111/nph.12303. [DOI] [PubMed] [Google Scholar]
- 56.Alcolombri U, Ben-Dor S, Feldmesser E, Levin Y, Tawfik DS, Vardi A. 2015. Identification of the algal dimethyl sulfide-releasing enzyme: a missing link in the marine sulfur cycle. Science 348:1466–1469. 10.1126/science.aab1586. [DOI] [PubMed] [Google Scholar]
- 57.Steinke M, Wolfe G, Kirst G. 1998. Partial characterization of dimethylsulfoniopropionate (DMSP) lyase isozymes in 6 strains of Emiliania huxleyi. Mar Ecol Prog Ser 175:215–225. 10.3354/meps175215. [DOI] [Google Scholar]
- 58.Rosenwasser S, Mausz MA, Schatz D, Sheyn U, Malitsky S, Aharoni A, Weinstock E, Tzfadia O, Ben-Dor S, Feldmesser E, Pohnert G, Vardi A. 2014. Rewiring host lipid metabolism by large viruses determines the fate of Emiliania huxleyi, a bloom-forming alga in the ocean. Plant Cell 26:2689–2707. 10.1105/tpc.114.125641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blaby IK, Blaby-Haas CE, Pérez-Pérez ME, Schmollinger S, Fitz-Gibbon S, Lemaire SD, Merchant SS. 2015. Genome-wide analysis on Chlamydomonas reinhardtii reveals the impact of hydrogen peroxide on protein stress responses and overlap with other stress transcriptomes. Plant J 84:974–988. 10.1111/tpj.13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bidle KD, Haramaty L, Barcelos e Ramos J, Falkowski P. 2007. Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi. Proc Natl Acad Sci USA 104:6049–6054. 10.1073/pnas.0701240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frada M, Probert I, Allen MJ, Wilson WH, de Vargas C. 2008. The “Cheshire Cat” escape strategy of the coccolithophore Emiliania huxleyi in response to viral infection. Proc Natl Acad Sci USA 105:15944–15949. 10.1073/pnas.0807707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abada A, Segev E. 2018. Multicellular Features of Phytoplankton. Front Mar Sci 5:144. 10.3389/fmars.2018.00144. [DOI] [Google Scholar]
- 63.Brunet T, King N. 2017. The origin of animal multicellularity and cell differentiation. Dev Cell 43:124–140. 10.1016/j.devcel.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bruhn A, LaRoche J, Richardson K. 2010. Emiliania huxleyi (Prymnesiophyceae): nitrogen-metabolism genes and their expression in response to external nitrogen sources. J Phycol 46:266–277. 10.1111/j.1529-8817.2010.00809.x. [DOI] [Google Scholar]
- 65.Iwamoto K, Shiraiwa Y. 2003. Characterization of NADH:nitrate reductase from the coccolithophorid Emiliania huxleyi (Lohman) Hay & Mohler (Haptophyceae). Mar Biotechnol (NY) 5:20–26. 10.1007/s10126-002-0051-8. [DOI] [PubMed] [Google Scholar]
- 66.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delaux P-M, Schornack S. 2021. Plant evolution driven by interactions with symbiotic and pathogenic microbes. Science 371:eaba6605. 10.1126/science.aba6605. [DOI] [PubMed] [Google Scholar]
- 68.Steinbrenner AD. 2020. The evolving landscape of cell surface pattern recognition across plant immune networks. Curr Opin Plant Biol 56:135–146. 10.1016/j.pbi.2020.05.001. [DOI] [PubMed] [Google Scholar]
- 69.Wiegmann K, Hensler M, Wöhlbrand L, Ulbrich M, Schomburg D, Rabus R. 2014. Carbohydrate catabolism in Phaeobacter inhibens DSM 17395, a member of the marine Roseobacter clade. Appl Environ Microbiol 80:4725–4737. 10.1128/AEM.00719-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fu H, Uchimiya M, Gore J, Moran MA. 2020. Ecological drivers of bacterial community assembly in synthetic phycospheres. Proc Natl Acad Sci USA 117:3656–3662. 10.1073/pnas.1917265117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wanke A, Malisic M, Wawra S, Zuccaro A. 2021. Unraveling the sugar code: the role of microbial extracellular glycans in plant-microbe interactions. J Exp Bot 72:15–35. 10.1093/jxb/eraa414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Malitsky S, Ziv C, Rosenwasser S, Zheng S, Schatz D, Porat Z, Ben‐Dor S, Aharoni A, Vardi A. 2016. Viral infection of the marine alga Emiliania huxleyi triggers lipidome remodeling and induces the production of highly saturated triacylglycerol. New Phytol 210:88–96. 10.1111/nph.13852. [DOI] [PubMed] [Google Scholar]
- 73.Pohnert G. 2002. Phospholipase A2 activity triggers the wound-activated chemical defense in the diatom Thalassiosira rotula. Plant Physiol 129:103–111. 10.1104/pp.010974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ishibashi Y, Aoki K, Okino N, Hayashi M, Ito M. 2019. A thraustochytrid-specific lipase/phospholipase with unique positional specificity contributes to microbial competition and fatty acid acquisition from the environment. Sci Rep 9:16357. 10.1038/s41598-019-52854-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goyet C, Poisson A. 1989. New determination of carbonic acid dissociation constants in seawater as a function of temperature and salinity. Deep Sea Res A 36:1635–1654. 10.1016/0198-0149(89)90064-2. [DOI] [Google Scholar]
- 76.Avraham R, Haseley N, Fan A, Bloom-Ackermann Z, Livny J, Hung DT. 2016. A highly multiplexed and sensitive RNA-seq protocol for simultaneous analysis of host and pathogen transcriptomes. Nat Protoc 11:1477–1491. 10.1038/nprot.2016.090. [DOI] [PubMed] [Google Scholar]
- 77.Aronesty E. 2013. Comparison of sequencing utility programs. TOBIOIJ 7:1–8. 10.2174/1875036201307010001. [DOI] [Google Scholar]
- 78.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j 17:10. 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 79.Andrew S. 2010. FastQC: a quality control tool for high throughput sequence data. https://github.com/s-andrews/FastQC.
- 80.Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu TD, Watanabe CK. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21:1859–1875. 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 82.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 85.Shen W, Le S, Li Y, Hu F. 2016. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One 11:e0163962. 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 87.Götz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435. 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, von Mering C, Bork P. 2019. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Blum M, Chang H-Y, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, Richardson L, Salazar GA, Williams L, Bork P, Bridge A, Gough J, Haft DH, Letunic I, Marchler-Bauer A, Mi H, Natale DA, Necci M, Orengo CA, Pandurangan AP, Rivoire C, Sigrist CJA, Sillitoe I, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Bateman A, Finn RD. 2021. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49:D344–D354. 10.1093/nar/gkaa977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Trizna M. 2020. assembly_stats 0.1.4. Zenodo. 10.5281/zenodo.3968775. [DOI] [Google Scholar]
- 91.Tseng E. 2021. ANGEL: robust open reading frame prediction. https://github.com/PacificBiosciences/ANGEL.
- 92.Bushmanova E, Antipov D, Lapidus A, Suvorov V, Prjibelski AD. 2016. rnaQUAST: a quality assessment tool for de novo transcriptome assemblies. Bioinformatics 32:2210–2212. 10.1093/bioinformatics/btw218. [DOI] [PubMed] [Google Scholar]
- 93.Grigoriev IV, Hayes RD, Calhoun S, Kamel B, Wang A, Ahrendt S, Dusheyko S, Nikitin R, Mondo SJ, Salamov A, Shabalov I, Kuo A. 2021. PhycoCosm, a comparative algal genomics resource. Nucleic Acids Res 49:D1004–D1011. 10.1093/nar/gkaa898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kent WJ. 2002. BLAT: the BLAST-Like Alignment Tool. Genome Res 12:656–664. 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dainat J, Hereñú D, Pucholt P. 2020. NBISweden/AGAT: AGAT-v0.5.1. Zenodo 10.5281/zenodo.4205393. [DOI] [Google Scholar]
- 97.Buels R, Yao E, Diesh CM, Hayes RD, Munoz-Torres M, Helt G, Goodstein DM, Elsik CG, Lewis SE, Stein L, Holmes IH. 2016. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol 17:66. 10.1186/s13059-016-0924-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kolde R. 2019. pheatmap: pretty heatmaps. R package version 1.0.12. https://CRAN.R-project.org/package=pheatmap.
- 101.Pantano L. 2020. DEGreport: report of DEG analysis. R package version 1.24.1. http://lpantano.github.io/DEGreport/.
- 102.Alexa A, Rahnenführer J, Lengauer T. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22:1600–1607. 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 103.Curson ARJ, Williams BT, Pinchbeck BJ, Sims LP, Martínez AB, Rivera PPL, Kumaresan D, Mercadé E, Spurgin LG, Carrión O, Moxon S, Cattolico RA, Kuzhiumparambil U, Guagliardo P, Clode PL, Raina J-B, Todd JD. 2018. DSYB catalyzes the key step of dimethylsulfoniopropionate biosynthesis in many phytoplankton. Nat Microbiol 3:430–439. 10.1038/s41564-018-0119-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S11, Tables S1 to S3, links and descriptions for Data Sets S1 to S6. Download AEM.01418-21-s0001.pdf, PDF file, 2.6 MB (1.6MB, pdf)
Data Availability Statement
PacBio CCS reads, Illumina QC reads, and the hybrid transcriptome were deposited under the BioProject ID PRJNA698293. PacBio CCS reads and Illumina QC reads were uploaded to the Sequence Read Archive (SRA; accession numbers SRR13590462 to SRR13590474). The hybrid transcriptome was deposited at the Transcriptome Shotgun Assembly Sequence Database (TSA; master record, GIZZ00000000; assembly version, GIZZ01000000). The hybrid transcriptome described in the manuscript (see Data Set S1) slightly differs from the uploaded version; upload to the TSA database required the removal of 1,043 short transcripts (<200 bp), 14 contaminating transcripts, and the trimming of putative sequencing adapters for 164 transcripts (∼20 bp). The transcript IDs generated by hybrid rnaSPAdes were renamed for TSA submission (see Data Set S2, columns 3 and 4). A BLAST search of the E. huxleyi CCMP3266 hybrid transcriptome can be conducted from the NCBI webpage upon selecting the TSA Database. Supplemental Data Sets S1 to S6 can be downloaded from Zenodo (https://zenodo.org/record/5702921).