Abstract
The Streptomyces clavuligerus genome consists in a linear chromosome of about 6.7 Mb and four plasmids (pSCL1 to pSCL4), the latter one of 1.8 Mb. Deletion of pSCL4, results in viable mutants with high instability in the chromosome arms, which may lead to chromosome circularisation. Transcriptomic and proteomic studies comparing different mutants with the wild-type strain improved our knowledge on the biosynthesis and regulation of clavulanic acid, cephamycin C and holomycin. Additional knowledge has been obtained on the SARP-type CcaR activator and the network of connections with other regulators (Brp, AreB, AdpA, BldG, RelA) controlling ccaR expression. The transcriptional pattern of the cephamycin and clavulanic acid clusters is supported by the binding of CcaR to different promoters and confirmed that ClaR is a CcaR-dependent activator that controls the late steps of clavulanic biosynthesis. Metabolomic studies allowed the detection of new metabolites produced by S. clavuligerus such as naringenin, desferroxamines, several N-acyl tunicamycins, the terpenes carveol and cuminyl alcohol or bafilomycin J. Heterologous expression of S. clavuligerus terpene synthases resulted in the formation of no less than 15 different terpenes, although none of them was detected in S. clavuligerus culture broth. In summary, application of the Omic tools results in a better understanding of the molecular biology of S. clavuligerus, that allows the use of this strain as an industrial actinobacterial platform and helps to improve CA production.
Keywords: Streptomyces clavuligerus, Clavulanic acid; Cephamycin C; Clavams; Terpenes; Secondary metabolites
Streptomyces clavuligerus is the strain used industrially to produce clavulanic acid (CA). This compound is a potent β-lactamase inhibitor that binds the serine residue in the active centre of β-lactamases causing their irreversible inactivation and is used to potentiate the activity of classical β-lactam antibiotics against resistant bacteria. In addition, S. clavuligerus synthesises the β-lactam antibiotic cephamycin C and other compounds with antibacterial and antifungal activities. These facts made S. clavuligerus an interesting subject of study and numerous articles have been published on the physiology, nutrition, fermentation, and on enzymes and regulatory mechanisms involved in the formation of the metabolites produced by this species. The recent availability of powerful Omics tools provides new molecular genetic information on S. clavuligerus. The aim of this article is to analyse the impact of the Omics technology on the present knowledge on S. clavuligerus, especially in relation to secondary metabolite biosynthesis. The annotation of S. clavuligerus genes cited in this work is presented as in the Streptomyces database, StrepDB, and the equivalence to NCBI annotations is shown in Table S1.
Genomic of Streptomyces clavuligerus
S. clavuligerus, as all Streptomyces species, possess a linear chromosome. In addition, Netolitzky et al. (1995) using pulsed-field gel electrophoresis, found in the type strain S. clavuligerus NRRL 3585 three plasmids named pSCL1 (11.7 kb), pSCL2 (120 kb) and pSCL3 (430 kb). This research group also sequenced pSCL1 and partially pSCL2 (Wu & Roy, 1993; Wu et al., 2006).
The S. clavuligerus genome has been sequenced and analysed by several research groups. The Broad Institute (2008) was first to make public a very fragmented unannotated assembly of S. clavuligerus ATCC 27064 genome (Table 1). Medema et al. (2010) using Sanger sequencing of whole-genome shotgun libraries published the first S. clavuligerus ATCC 27064 genome assembly consisting of single scaffolds per replicon, and showed for first time the presence of a megaplasmid of 1,796 kb, named pSCL4, together with a chromosome of 6,760 kb (Table 1). The sequences described for pSCL2 were not found by these authors neither replicons of the sizes of pSCL2 and pSCL3. In parallel, Song et al. (2010) using a Sanger paired-end sequencing and 454 pyrosequencing published the S. clavuligerus NRRL 3585 genome (NRRL 3585 and ATCC 27064 refer to the type strain deposited in different culture collections) comprised by a 6.7-Mb chromosome and four linear replicons corresponding by size to the pSCL1, pSCL2, pSCL3 and pSCL4 plasmids. A refined genome of S. clavuligerus ATCC 27064 was obtained using a PacBio genome library and Illumina sequencing (Hwang et al., 2019). These authors published an assembly with single contigs for the chromosome (6.75 Mb) and pSCL4 (1.8 Mb), and annotated 7,163 genes including 6,880 protein-encoding genes, 196 pseudogenes, 66 tRNAs, 18 rRNAs and 3 ncRNAs; this assembly however does not contain any data on any of the other three plasmids. An additional genome assembly (GCA_015708605.1) has been recently made public from an S. clavuligerus ATCC 27064 isolate studied for more than 25 years in the Microbiology laboratory at the University of León in Spain (Gomez-Escribano et al., 2021). This is a refined sequence (200× PacBio coverage and optical mapping) in which plasmids pSCL1, pSCL2 and pSCL4 were sequenced close to completion but plasmid pSCL3 was not found. The authors, intrigued by the presence of pSCL3 only in the NRRL 3585 genome assembly from Song and colleagues (2010), further assessed the presence of plasmids in five different genuine type strain in culture collection by Illumina deep whole-genome sequencing, and found all four plasmids in all type strain deposits; this finding prompted the authors to suggest that pSCL3 might have been lost in the León isolate during subculturing of the original ATCC 27064 culture (Gomez-Escribano et al., 2021). Finally, the genomes of two industrial strains sequenced with PacBio/Ilumina have been described: S. clavuligerus F613-1 from which was reported a chromosome of 6.88 Mb and one plasmid of 0.7 Mb with sequence partially identical to pSCL4 (Cao et al., 2016) and S. clavuligerus F1D-5. The results obtained from these studies are summarised in Table 1.
Table 1.
Streptomyces clavuligerus Chromosome and Plasmid Sequences Available at NCBI
| Strain | Method used (Coverage) | Replicons | Size (bp) | CG (%) | Gene number | Reference and accesion number |
|---|---|---|---|---|---|---|
| S. clavuligerus NRRL 3585 | Sanger | Isolated plasmid pSCL1 | 11,696 | Wu and Roy, 1993 X54107 |
||
| S. clavuligerus NRRL 3585 | Sanger | Isolated plasmid pSCL2 | Partial sequence | Wu et al., 2006 AY392418 |
||
| S. clavuligerus ATCC 27064 | Unknown | 597 ungapped contigs (6,729,086 bp) | 6,941,740 | Broad Institute, 2008 GCA_000154925 |
||
| S. clavuligerus ATCC 27064 | Sanger (11,8×) | Chromosome plasmid pSCL4 | 6,760,392 1,799,650 |
7281 | Medema et al., 2010 ADGD01000000 |
|
| S. clavuligerus NRRL 3585 | Sanger/454 pyrosequencing (80,7×) | Chromosome Plasmid pSCL1 Plasmid pSCL2 Plasmid pSCL3 Plasmid pSCL4 |
6,736,473 10,266 149,326 442,792 1,796,117 |
72.7 71.9 70.0 70.7 71.8 |
7655 | Song et al., 2010 ADWJ01000000 |
| S. clavuligerus F613-1 | PacBio RSII/Illumina (60×) | Chromosome Plasmid pSCL4-like |
6,883,702 707,056 |
72.68 71.82 |
6340 | Cao et al., 2016 CP016559/CP016560 |
| S. clavuligerus F1D-5 | PacBio/Illumina (320×) | Chromosome Plasmid pSCL1 Plasmid pSCL2 |
6,900,908 1,051,520 106,697 |
6692 | Shandong Academy of Medical Science, 2018 GCA_003454 755 | |
| S. clavuligerus ATCC 27064 | PacBio/Ilumina (50×) | Chromosome-Plasmid pClaA1 | 6,784,591 1,795,495 |
7163 | Hwang et al., 2019 GSE128216 |
|
| S. clavuligerus ATCC 27064, León strain | PacBio/Ilumina (200×) | Chromosome Plasmid pSCL1 Plasmid pSCL2 Plasmid pSCL4 |
6,748,580 11,696 149,702 1,794,824 |
7227 | Gomez-Escribano et al., 2021 GCA_015708605 |
As usual in other Streptomyces species, S. clavuligerus linear chromosome has a conserved core region, rich in primary metabolism and housekeeping genes, and two chromosomal arms in which many secondary metabolites encoding clusters are located. The chromosome size is smaller than the average Streptomyces chromosomes probably due to the large 1.8-Mb pSCL4 plasmid. The differences in chromosome size and the number of plasmids found by different authors suggest that S. clavuligerus has a very dynamic genome due to plasmid loss and events of plasmid/chromosome recombination. In industrial strains the instability increases due to the effect of heavy mutagenesis as shown by the pSCL4 behaviour in strain S. clavuligerus F613 (Cao et al., 2016).
Plasmid pSCL4 does not have housekeeping genes except for some tRNA and rRNA genes which are duplicated in the chromosome but carries numerous gene clusters for secondary metabolites. A metabolic model for the growth of S. clavuligerus on several carbon and nitrogen sources, predicted in silico that removal of pSCL4 will not affect essentially the strain growth suggesting that this plasmid was not essential (Medema et al., 2010). This was experimentally confirmed by quantification of pSCL4 copy number in cultures of strain S. clavuligerus oppA2::aph, a strain that has about 25,000-fold lower pSCL4 copy number in relation to total DNA, than cultures of the parental strain (Álvarez-Álvarez et al., 2014a). A similar mechanistically unpredictable loss of pSCL4 was reported by Charusanti et al. (2012) due to selective pressure by co-culturing S. clavuligerus with Staphylococcus aureus.
Removal of pSCL4 by Deletion of the par Genes: Effect on Expression of Chromosomal Genes
A pSCL4-free strain, S. clavuligerus pSCL4–, was obtained by deletion of the pSCL4 parA-parB genes which differ 60–70% in sequence from the par genes required for chromosomal segregation. The pSCL4-free mutant fails to sporulate, grows more slowly than the parental strain and produced 20–30% less clavulanic acid. Colonies of the pSCL4-less strain have a spectacular yellow colour due to overproduction of the antibiotic holomycin, whose biosynthesis is deregulated in these mutants (Álvarez-Álvarez et al., 2014a, 2017). Comparative genomic and transcriptomic studies between the wild-type and the pSCL4-deleted strains reveal a high instability of S. clavuligerus genome. q-PCR quantification demonstrates that the lack of expression of genes in a 303-kb DNA fragment of S. clavuligerus pSCL4– chromosome right arm, was due to the absence of these genes. The chromosomal 303-kb DNA fragment, including the right telomere, probably was translocated to pSCL4, and lost in the plasmid-less mutant.
Recently CRISPR-cas9-mediated targeting of pSCL4 parB gene resulted in new pSCL4-free strains (Gomez-Escribano et al., 2021). Five pSCL4-cured isolates were sequenced and all of them showed deletions in one or both chromosomal arms, and other rearrangements, although they kept plasmids pSCL1 and pSCL2. For two of the mutants enough sequence data were obtained to show circularisation of the chromosome concomitant to the loss of around 100 kb of each end. The deletions are not site specific as they range from 88 kb to 303 kb (Álvarez-Álvarez et al., 2017; Gomez-Escribano et al., 2021) in the right arm of the chromosome, but probably chromosome circularisation relies on the deletion of fragments at both arm ends (Chen et al., 2002; Tidjani et al., 2020). From these results it can be concluded that pSCL4 is not essential for S. clavuligerus viability since the pSCL4-less mutant growth is only slightly delayed in relation to the wild-type strain; however, pSCL4 appears to be essential for the stability and maintenance of the linear chromosome, a genetic hallmark of streptomycetes, and it is still to be elucidated whether the lack of morphological development of pSCL4-cured strains is due to the loss of specific genes located in pSCL4 or the loss of chromosomal sequence.
Alvarez-Alvarez and colleagues found that deletion of pSCL4 resulted in a strong effect on the transcription of many chromosomal genes including genes for several regulators. The pSCL4-less strain upregulates the defence system for oxidative stress, the signal transduction system mediated by sigE-cseABC and downregulates the survival mechanisms mediated by the mce cluster (SCLAV_5177/_5193) (Álvarez-Álvarez et al., 2017). In addition, expression of genes for holomycin, tunicamycin and a type I polyketide compound (SCLAV_0446/_0497) biosynthesis were upregulated. Other gene clusters, as those for clavulanic acid, cephamycin C (SCLAV_4179/4217), desferrioxamine (SCLAV_1948/_1951), a lantibiotic-type compound (SCLAV_4387/_4392), an NRPS-type compound (SCLAV_5608/_5638) and for terpene biosynthesis (SCLAV_5670/_5674) were downregulated. Metabolomic studies confirmed that the products formed by S. clavuligerus pSCL4– differ significantly from those of the wild-type strain (see below). The requirement of a correct pSCL4 copy number for optimal clavulanic acid production is an important contribution of the genomic studies.
Transcriptomic and Proteomic Studies
Transcriptomic and proteomic studies in Streptomyces have been made by different authors using distinct strains and conditions: (a) studying the sequential expression of genes at different times of growth (Botas et al., 2018; Fergusson et al., 2016); (b) comparing gene expression in different culture media (Hwang et al., 2019; Pinilla et al., 2019); (c) studying the wild-type strain in relation to particular mutants (Álvarez-Álvarez et al., 2014b, 2017, 2018; Martínez-Burgo et al., 2015, 2019; Pérez-Redondo et al., 2012), and (d) comparing the wild-type strain with industrial strains (Medema et al., 2011; Ünsaldi et al., 2017). Transcriptomic studies provide a wealth of information that frequently is difficult to interpret and will require years of research to understand the up- and downexpression of hundreds of genes reported in the different studies. However, the differences in regulation of whole-gene clusters might be understood with the present available knowledge. In this article we will focus mostly on the transcriptomic studies of gene cluster expression, particularly of the clusters encoding secondary metabolites.
The genomic positions of transcription start sites (TSSs) have been determined in the complete S. clavuligerus genome, at four growth phases of the culture using an RNAseq library (Hwang et al., 2019). These authors found A/G-rich TSS in most S. clavuligerus genes. The −10 sites sequence, TANNNT, is conserved in about 90% of S. clavuligerus promoters and separated by an average distance of 18–19 nt from the less conserved, NTGAC, −35 sequence. The more abundant Shine–Dalgarno sequence in S. clavuligerus is RRGGAG. The S. clavuligerus regulons were proposed to be identified regarding their –35 promoter sequences (Hwang et al., 2019), which are recognised by specific sigma factors (Chauhan et al., 2016). Following this hypothesis Hwang et al. assumed that the SigF-binding RNA polymerase transcribes argR, and the ArgR regulator controls the arginine biosynthesis genes and the formation of clavulanic acid. However, experimental studies are required to confirm this claim. In a different study, these authors determined the 3′-end position (TEP) of 1,427 S. clavuligerus transcripts using a Term-seq library and classified the RNAs according to the free energy of the secondary structure of the TEP regions and the uridine chain downstream of the stem structure (Hwang et al., 2021). Specifically, the TSS and TEP structures of the 58 secondary metabolite gene clusters were studied and the transcripts of the CA and cephamycin gene clusters were confirmed.
The most complete S. clavuligerus proteome analysis reported 2,442 proteins, about 33% of the proteome, and compared them in the wild-type strain and two mutants deleted, respectively, in the regulatory genes bldA and bldG (Ferguson et al., 2016). The bldA gene encodes a leucine-specific tRNA that translates the rare TTA codon (Chandra & Chater, 2008) while bldG encodes an anti-anti-sigma factor (see below). Proteomic studies showed that the BglG factor is strongly downrepresented in the bldA mutant (Fergurson et al., 2016). Ünsaldi et al. (2017) compared 93 proteins in S. clavuligerus NRRL 3585 and the industrial clavulanic producer strain S. clavuligerus DEA. They found that proteins involved in CA formation (CeaS2, Bls2, Car) were overrepresented in the industrial strain but surprisingly the regulators CcaR, ClaR and the putative CagR regulator, were not. Partial proteome studies have been made also comparing S. clavuligerus ATCC 27064 and mutants in the ccaR, claR, oppA2 or argR genes (Nárdiz et al., 2011; Robles Reglero et al., 2013) and in the hom gene, encoding the homoserine dehydrogenase (Ünsaldi et al., 2021) as a way to understand the role of these genes in clavulanic acid and cephamycin C production.
An interesting fact observed in proteomic studies is the presence of multiple spots corresponding to the same protein. Sixteen proteins appeared in two or more spots in the two-dimensional gels reported by Ünsaldi et al. (2017) suggesting posttranslational modifications. As examples, the cystathionine-γ-synthase (SCLAV_5668) is represented by five spots and MetE by two isoforms. Also, the carboxyethylarginine synthetase (CeaS2) and the catalase of S. clavuligerus appear in several spots (Lorenzana & Liras, unpublished results). Although of interest, large-scale proteomic studies frequently are difficult to interpret and require parallel transcriptomic and physiological studies.
Organisation of the Genes of Cephamycin C, Clavulanic Acid and Clavams in Streptomyces clavuligerus Genome
Using AntiSMASH 4.0, Hwang et al. (2019) predicted that S. clavuligerus genome contains 58 gene clusters for secondary metabolites, 30 of them located in the chromosome and 28 in the megaplasmid pSCL4. This indicates that 21.7% of the genes in the genome are involved in secondary metabolite formation. A small number of these clusters have been thoroughly studied from the biochemical and genetical point of views. They are the clusters for the β-lactams (clavulanic acid, cephamycin C, clavams), for holomycin and for tunicamycin that will be considered in next section. In recent years additional metabolites produced by S. clavuligerus have been detected using mass spectrometry (MS) and nuclear magnetic resonance (NMR). These compounds and the possible clusters encoding them will be considered in a later section.
The Clavulanic Acid Pathway and Gene Cluster
From the pharmacological and industrial point of view, the most important compound produced by S. clavuligerus is clavulanic acid. The chemistry, biosynthetic pathway, enzymes and regulation of clavulanic acid formation have been reviewed previously (Baggaley et al., 1997; Hamed et al., 2013; Jensen & Paradkar, 1999; Liras & Rodríguez-García, 2000; Liras et al., 2008, 2011; Townsend, 2002). In addition, other reviews deal with the industrial production of clavulanic acid and its purification (López-Agudelo et al., 2021; Saudagar et al., 2008) or with strategies to obtain CA high-producing strains (Paradkar, 2013). Therefore, this article will focus only on the recent results on clavulanic acid biosynthesis and regulation, obtained mostly using Omics technology during the past 15 years.
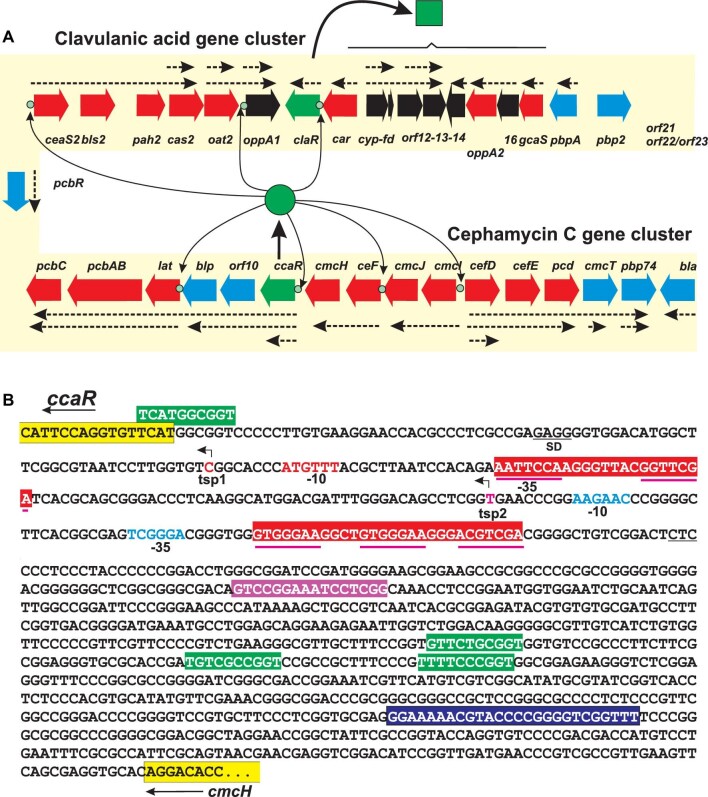
Cephamycin C, clavulanic and clavams are β-lactam antibiotics, containing a four-membered β-lactam ring. Moreover, the cephamycin C molecule has a six-membered dihydrothiazinic ring containing sulphur while clavulanic acid and the clavams possess a five-membered clavam ring containing oxygen. Although the initial steps of clavams and CA pathways are common, the stereochemical configuration of clavulanic acid is (3R, 5R) while the antifungal clavams have a (3S, 5S) stereochemistry and lack β-lactamase inhibitory properties. The biosynthetic pathway leading to cephamycin C is unrelated to that of CA; however, the cephamycin C and CA gene clusters in S. clavuligerus are adjacent forming a 60-kb supercluster (Fig. S1A). Expression of the genes in this supercluster is initiated by a common activator protein named CcaR encoded by ccaR, a gene located in the cephamycin C gene cluster. A second regulatory gene claR, located in the CA cluster encodes ClaR, an activator that controls specifically the late steps of clavulanic acid biosynthesis but not the formation of cephamycin C or clavams.
A similar supercluster organisation occurs in Streptomyces jumonjinensis and Streptomyces katsurahamanus (AbuSara et al., 2019; Nobary & Jensen, 2012; Ward & Hodgson, 1993) with small modifications in the order of some genes and the lack of genes blp, for a putative protein inhibitory of β-lactamases, and pcbR, confering β-lactam resistance. The three Streptomyces species produce equivalent amounts of cephamycin C, but clavulanic acid production in the S. clavuligerus wild type is about five- to sixfolds higher than in the other species. Other Streptomyces species contain only the genes for clavulanic acid production; the best studied are Streptomyces flavogriseus ATCC 33331 (renamed Streptomyces pratensis) and Saccharomonospora viridis DSM 43017 (Álvarez-Álvarez et al., 2013; Nobary & Jensen, 2012). These Streptomyces species have a silent CA cluster and, at difference of S. clavuligerus, carry the ccaR gene in the CA cluster. RT-PCR studies of S. flavogriseus show that five essential genes for CA biosynthesis are not expressed in this strain, even when the heterologous ccaR gene of S. clavuligerus, was expressed in S. flavogriseus. Probably S. flavogriseus has a poor conservation of nucleotides in the CcaR-binding sequences, since the S. flavogriseus CcaR protein is functional and able to activate the CA gene cluster of S. clavuligerus (Álvarez-Álvarez et al., 2013).
Clavulanic acid biosynthesis: New insights into genes and proteins
S. clavuligerus is a peculiar strain that does not use glucose because it lacks expression of the glucose permease gene, glP1 (Pérez-Redondo et al., 2010), however, it uses efficiently glycerol and amino acids as carbon sources. CA is formed from two precursors, glyceraldehyde-3-phosphate and arginine (Fig. S1B). Improved CA-producing strains have been obtained by increasing the glyceraldehyde-3-P pool through genetic engineering of the glycolytic pathway (Li & Townsend, 2006), by increasing the copy number of the glpK1D1 glycerol biosynthesis genes and by addition of glycerol to the cultures (Baños et al., 2009; Shin et al., 2021). The gene cluster for arginine and the arginine gene cluster regulation in Streptomyces have been thoroughly studied using genetic and transcriptomic tools and this might be useful to improve CA production (Botas et al., 2018; Pérez-Redondo et al., 2012; Rodríguez-García et al., 2000). The CA pathway starts with the condensation of glyceraldehyde-3-P and arginine by the carboxyethylarginine synthase (CeaS2), formation of a β-lactam ring by the β-lactam synthetase (Bls2), closing of the clavam ring by the multifunctional clavaminate synthase (Cas2), able to hydroxylate, cyclisise and desaturate its substrate in a single-binding cavity leading to the formation of (3S, 5S) clavaminic acid (Wojdyla & Borowski, 2021). This compound is the start point of a lateral branch of the CA pathway that results in the formation of (3S, 5S)-clavams (see below).
In the late steps of the CA pathway clavaminic acid is converted to (3S, 5S) N-glycylclavaminic acid by an ATP grass fold family protein (GcaS) that utilises glycine and ATP to introduce a glycyl group at the C9 amino group of clavaminic acid. Further steps in CA are poorly understood. However, mutations of several genes (oppA1, oppA2, cyp, orf12, 13, 14, 16) located in the CA cluster indicate that they are essential for CA formation suggesting the involvement of several additional enzymatic steps in the pathway, including the key change of (3S, 5S) to (3R, 5R) stereochemistry present in CA, which is essential for its β-lactamase inhibitory activity.
It was known that mutants deleted in oppA2, encoding an oligopeptide-binding protein, release large amounts of N-acetylglycylclavaminic acid (NAGC) to the broth (Jensen et al., 2004; Lorenzana et al., 2004). Formation of NAGC from N-glycylclavaminic acid requires an acetylation step, since the GcaS enzyme does not use N-acetylglycine as substrate (Arulanantham et al., 2006). Crystallised OppA2 protein is able to bind dipeptides or tripeptides containing arginine (MacKenzie et al., 2010); therefore, it was hypothesised that OppA2 could also bind NAGC, what might explain the release of NAGC by mutants lacking the OppA2 protein. Surprisingly, the co-culture of a CA non-producer oppA2 mutant with CA non-producer mutants blocked in genes for the early steps of the pathway (ceaS2 or bls2), results in CA formation by the mixed culture. This co-synthesis of CA does not occur when the oppA2-deleted mutant is co-cultured with mutants blocked in the cyp, orf14 or oppA1 genes, putatively involved in late steps of the pathway. Formation of CA is also observed in solid and liquid cultures when sterile broth of oppA2-null cultures, or pure N-acetylglycylclavaminic, is added to the cultures of ceaS2- or bls2-mutants, confirming that N-acetylglycylclavaminic is a real intermediate in the CA biosynthesis pathway (Álvarez-Álvarez et al., 2018). Transcriptomic studies show that only 233 genes changed their expression due to the lack of OppA2. They include genes for proteases and transport systems, but expression of all the CA biosynthesis genes was not significantly affected. This important advance in the knowledge of the CA pathway suggests that NAGC is formed by acetylation of N-glycylclavaminic acid. The release of N-acetylglycylclavaminic to the medium indicates that OppA2 binds this compound at the membrane level and presents it to the next enzyme in the pathway.
Expression of six additional genes in the CA gene cluster (oppA1, cyp-fd, orf12, orf13, orf14, orf16) is still required for CA production. No additional advances have been made on the mechanisms of action of the products of the genes: (1) cyp-fd, encoding a P450-ferredoxin complex (Li et al., 2000), (2) orf13, encoding a protein of the Eam family similar to the O-acetylserine/cysteine exporters, and (3) orf14/orf16 both of them encoding proteins with the GNAT motif of acetyltransferases. However, the proteins encoded by orf12 and orf14 have been crystallised. Protein Orf12 resembles type A β-lactamases and has a β-lactamase-like domain at the C-terminal end, although lacks in vitro β-lactamase activity (Valegard et al., 2013). Orf12 does not have amidase activity or the DD-carboxypeptidase activity characteristic of PBP proteins. Pure ORF12 is able to hydrolyse with low efficiency the 3′-O-acetyl side chain of cephalosporin C and also catalyses the reverse reaction, that is, the acetylation of deacetylcephalosporin C; therefore, this enzyme has been named Cpe for cephalosporin esterase (Srivastava et al., 2019). Disruption of the cpe gene does not affect cephamycin C production but completely blocks clavulanic acid formation although all the genes of the CA cluster are transcribed in this mutant. Although is a nucleophilic hydrolase, the Cpe protein lacks detectable peptide signal for secretion and Western blotting confirms that is a cytoplasmic protein suggesting that the cephalosporin C esterase/acetylase activities are not its real function. Pure Cpe protein has at the N-terminal end a domain isomerase/cyclase, characteristic of steroid isomerases, and is able to bind non-covalently clavulanic acid. For these reasons it has been suggested that Cpe may be involved in the opening of the clavam ring and its isomerisation to the 3R, 5R stereochemistry (Scrivastava et al., 2019). The isomerisation reaction has been also studied through a computational assisted approach (Gómez et al., 2020).
Orf14 possesses in the N- and C-terminal ends two motifs characteristic of the tandem GNAT-superfamily acetyl transferases. The crystal structure of Orf14 has an acetyl-CoA molecule deeply buried in the N-terminal domain, suggesting that this acetyl-CoA molecule has a structural rather than catalytic role. A second acetyl-CoA is bound by the protein, probably at the C-terminal domain (Iqbal et al., 2010). These authors propose that the C-terminal end is the catalytic site of the protein that may use acetyl-CoA or other acylated-CoA substrates. However, the involvement of Orf14 in N-acylglycylclavaminic acid formation has not been reported.
The final reaction of the CA pathway involves the action of the clavaldehyde dehydrogenase, Car, a tetrameric protein that uses NADPH to reduce the 3R, 5R clavaldehyde to 3R, 5R clavulanic acid (Fig. S1B).
RT-PCR analysis of the CA gene cluster polycistronic transcripts reveals a major transcript of the genes for the early steps of the pathway (ceaS2-bls2-pah2-cas2), large transcripts of genes for the late steps of the pathway (cyp-orf12-orf13 and orf16-oppA2) and genes transcribed individually (claR, car, orf14, gcaS) or transcribed both individually and forming part of larger transcripts (cas2, oppA1, cyp, orf12, orf13) (Hwang et al., 2021; Santamarta et al., 2011) (Fig. 1A).
Fig. 1.
Clusters of genes for clavulanic acid and cephamycin C biosynthesis and regulatory sites in the intergenic ccaR-cmcH region. (A) Cluster of genes for clavulanic acid biosynthesis (above) and for cephamycin C biosynthesis (below). The name of each gene is indicated. Notice that both clusters are linked through the pcbR gene (left side). Structural genes are shown in red colour and regulatory genes in green; genes for essential unknown functions are in black colour; genes for putative antibiotic resistance or transport are shown in blue. The known transcriptional units are indicated by dotted arrows. CcaR (green circle) binding sites are indicated as well as the putative sites for ClaR (green square) binding. (B) The sequence of the intergenic ccaR-cmcH is shown with the ends of the ccaR and cmcH genes boxed in yellow. The –35 site, –10 site, and the transcription start points are indicated with red lettering for tsp1 and with blue lettering for tsp2. Four putative Adp-binding sites are boxed in green. The double and tripled heptameric sequences for CcaR binding are boxed in red. The sequence for ARE binding (AREccaR) is boxed in dark blue. The putative sequence for PimM binding is shown in a purple box.
The Antifungal Clavams Biosynthesis Pathway: Gene Clusters and Regulation
Clavams are metabolites with a (3S, 5S) five-membered clavam ring containing oxygen (Fig. S2). They have antifungal activity but lack β-lactamase inhibitory properties. Two gene clusters for clavam biosynthesis are present in S. clavuligerus, the clavam gene cluster (cvm7-3-2-2-cas1-cvm4-5-6; SCLAV_2922 to _2929) and the CA paralogue gene cluster (orfDCBA-ceaS1-bls1-pah1-oat1-cvm6p-7p-snk-res1-res2; SCLAV_p1070 to _p1082), the latter located in plasmid pSCL4 (Tahlan et al., 2004, 2007). Studies using deleted mutants, elucidation of accumulated/or missing products of these strains, and comparison with the clavam clusters of other Streptomyces species, allowed to interpret the clavam biosynthetic pathway (Jensen, 2012; Tahlan et al., 2007). This pathway is branched and leads to the formation of alanylclavam and clavam-2-carboxylic acid, respectively; other previously reported clavams (2-formyloxymethylclavam, 2-hydroxymethylclavam) are intermediates in the pathway (Fig. S2). However, enzymatic studies to confirm the different steps and further research on the genes involved are still required. The snk-res1-res2-cvm7P genes, in the CA paralogue gene cluster, encode the clavam regulatory system (Kwong et al., 2012). Snk is a sensor kinase that is autophosphorylated and transfers the phosphate group to the two-component system Res1-Res2, in which Res1 is a truncated sensor and Res2 the response regulator. As detected by RT-PCR, deletion of snk and res2 results in lack of transcription of several essential genes for clavam biosynthesis, including cvm7p for a SARP-LAL-type regulator. Mutations of snk and res2 are complemented by transformation with the cvm7p parental allele expressed from a different promoter. In summary, Snk and the system Res1/Res2 control cvm7p transcription; in turn, Cvm7p activates expression of the clavam biosynthesis genes. The clavams and CA paralogue gene clusters are upregulated (1.5-fold) in a ccaR overexpressing, industrial strain (Medema et al., 2011); conversely, most of the genes in both clusters were downregulated in an S. clavuligerus ccaR-deleted strain (Álvarez-Álvarez et al., 2014b). The clavam activator gene cvm7p was weakly downregulated in this mutant while genes cas1 and orf8 were strongly upregulated suggesting that additional regulatory mechanisms control these clusters of genes (Álvarez-Álvarez et al., 2014b).
While the clavam gene clusters carry duplicated genes (cas2, ceaS2, bls2, pah2 and oat2) and might increase CA production, clavams utilise part of the valuable CA precursors and interfere with CA purification; moreover, some clavams, as clavam-2-carboxylate, have toxic properties. Therefore, the lack of production of clavams is an advantage in CA improved strains (Paradkar, 2013).
Cephamycin C Biosynthesis and Gene Cluster
The cephamycin C gene cluster (SCLAV_4198 to _4217) contains (a) genes (lat, pcd) to form the cephamycin precursor l-α-aminoadipic acid, (b) genes for biosynthetic enzymes leading to deacetylcephalosporin C formation (pcbAB, pcbC, cefD, cefE, cefF), and (c) genes for the late modifications of deacetylcephalosporin C to form cephamycin C (cmcI, cmcJ, cmcH) (Fig. S1C). In addition, some genes tentatively involved in transport (cmcT) and antibiotic resistance (bla, pcbR, pbp74) are located in the cephamycin gene cluster. The most interesting gene in this cluster is ccaR, located downstream of cmcH, which encodes the SARP-type activator that controls expression of the cephamycin C and clavulanic acid biosynthesis genes. The cephamycin C biosynthesis pathway, the pathway intermediates (Fig. S1C) and the mode of action of the enzymes involved have been extensively studied and reviewed (Liras, 1999; Liras & Demain, 2009) and will not be considered here.
One of the enzymes involved in l-α-aminoadipic acid formation is the piperideine-6-carboxylate dehydrogenase (P6C-DH), which belongs to the aldehyde dehydrogenase superfamily. This enzyme was purified to homogeneity and characterised years ago (Fuente et al., 1997) and has been recently crystallised (Hasse et al., 2019). Crystallographic studies indicated that this protein is a tetramer formed by a dimer-of-dimers (Hasse et al., 2019). Also, the methyl transferase CmcI involved in the C-7 methoxylation of the cephamycin nucleus has been crystallised (Öster et al., 2006). CmcI is a hexameric protein with a Rossmann domain at the C-terminal end that binds S-adenosylmethionine (SAM). The methyl group of SAM is transferred to the 7-α-OH cephalosporin in the final steps of formation of cephamycin C.
Expression of genes for cephamycin C biosynthesis proceeds through large transcripts expressing genes of the early-step enzymes (lat, pacAB, pcbC), the middle-step enzymes (cefD, cefE, pcd, cmcT, pbpA) and for enzymes acting late in the pathway (cmcI, cmJ, cefF, cmcH) (Fig. 1A). Some genes are transcribed individually or as bicistrons (pcbC, cefD-cefE, bla, pcbR). Finally, ccaR is expressed as a monocistronic unit and also coupled to orf10 and blp (Hwang et al., 2021; Santamarta et al., 2011). This pattern of transcription is important to understand the regulation of the gene cluster.
Due to its industrial interest the whole S. clavuligerus cephamycin C gene cluster has been transferred to Streptomyces coelicolor, Streptomyces albus and S. flavogriseus. Only the last strain is able to express, at a low level, the cephamycin biosynthetic genes. However, even under optimal conditions, the heterologous cephamycin C production, confirmed by LC-MS, was only 8% of that of S. clavuligerus ATCC 27064. The genes pcbA and pcbR for β-lactam resistance were expressed at almost the same levels as in S. clavuligerus, and transformants of the three Streptomyces strains showed an increased resistance to penicillin and cefoxitin (Martínez-Burgo et al., 2014). These results point out to an important limitation of cephamycin overproduction in heterologous Streptomyces species probably due to the difficulty in duplicating the regulatory network of S. clavuligerus in heterologous systems.
Regulation of Clavulanic Acid and Cephamycin C Biosynthesis: the CcaR and ClaR Activator Proteins
As indicated above CcaR is a SARP-type activator that controls cephamycin C and clavulanic acid gene cluster expression. Transcription of ccaR starts from two different TSSs: a major transcription site, tsp1, located 75-nt upstream of the translation ATG codon and a tsp2 site located at 173 nt (Wang et al., 2004). These tsp sites define two 5′-untranslated regions (UTR) that fit well with the average size of moderate and long UTRs described in S. clavuligerus (Hwang et al., 2019). RT-PCR studies show that a minor amount of ccaR transcript originates from the promoter of cmcH located upstream of ccaR (Kurt et al., 2013).
A second clavulanic acid regulatory gene, claR, is located in the CA cluster, downstream of car, encoding the clavaldehyde reductase. ClaR is a LysR-type regulator (Paradkar et al., 1998; Pérez-Redondo et al., 1998) with two helix-turn-helix DNA-binding motifs at the amino and carboxyl ends of the protein. The claR gene is transcribed from a tsp site 155-nt upstream from the claR ATG start codon and, in certain mutants, from an adenine located 107-nt upstream of the start codon. Mutants disrupted in claR accumulate clavaminic acid but do not produce clavulanic acid and amplification of claR copy number results in a threefold increase of CA production (Paradkar et al., 1998; Pérez-Redondo et al., 1998). Increasing the copy number of either claR, ccaR and/or ceaS2 genes has been used to obtain CA-overproducer strains (Kurt-Kizildoğan et al., 2017).
Regulatory cascades controlling clavulanic acid and cephamycin C biosynthesis
CcaR autoregulates its own expression as shown by binding of pure rCcaR protein to the ccaR upstream region. The CcaR-binding sites are duplicated/triplicated heptameric sequences characteristic of SARP-binding proteins that overlap with the tsp1 −35 promoter region, as shown by DNAse footprinting (Fig. 1B). The regulation, at the transcription level of the cephamycin and CA biosynthesis gene expression, has been studied by transcriptomic analysis, RT-PCR studies and DNA-binding gel shift comparing the wild-type strain with ccaR-null mutants (Álvarez-Álvarez et al., 2014b; Kurt et al., 2013; Santamarta et al., 2011).
CcaR also binds the promoters of cefD-cmcI, cefF and lat in the cephamycin cluster activating the transcription of these genes. Indeed, as shown by qRT-PCR, disruption of ccaR results in a transcription decrease of all the genes encoding cephamycin biosynthetic enzymes. The stronger effect is exerted on lat, encoding the first step in the pathway, which is 1,700-fold downregulated in a ccaR-disrupted mutant; also downregulated are the pcbAB gene (300-fold) for the early steps of the pathway, cefD and cefF (300–400-fold) for the medium steps and cmcI (550-fold) for the late modifications steps (Kurt et al., 2013; Santamarta et al., 2011). Genes involved in β-lactam resistance (pcbR, pcbA or bla), or transport (cmcT) are not affected by the lack of CcaR. These results were confirmed by microarray analysis of a different ccaR-deleted mutant which showed an almost null expression for lat, cmcI and cmcJ (Álvarez-Álvarez et al., 2014b). Enhancing the amount of CcaR protein either using strong promoters or increasing ccaR copy number (Kurt et al., 2013) results in higher levels of cephamycin C production, as also occurs in industrial strains (Medema et al., 2011). A similar upregulation of the cephamycin biosynthesis genes was also observed by growing S. clavuligerus in conditions of nutrient rich as compared to nutrient-depleted medium (Pinilla et al., 2019).
CcaR acts on the clavulanic acid gene cluster by binding the claR promoter region as shown by gel-shift experiments (Kurt et al., 2013; Santamarta et al., 2011). ccaR-null mutants showed a 400-fold lower expression of claR and 3,000-fold downregulation of ceaS2, encoding the first enzyme of the CA pathway. Binding was not studied for other CA gene promoters, but the transcription of cas2, encoding the multifunctional clavaminate synthase, was 10,000-fold downregulated (Álvarez-Álvarez et al., 2014b; Santamarta et al., 2011).
Recombinant-CcaR protein binds to a triple heptameric sequence TCGATTC-8-TTCAAGA-14-TTCAGCG located upstream of the claR promoter as shown by EMSA assays and DNAse footprinting (Santamarta et al., 2011); qRT-PCR studies indicate that claR is expressed 190-fold less in a ccaR-disrupted mutant, confirming early Northern transcriptional analysis (Paradkar et al., 1998). Transcriptomic studies of a claR-deleted mutant showed that, in relation to the wild-type strain, the genes for the late steps (oppA1, oppA2, car, cyp, orf12-13-14-16 and gcsA) were expressed at 1–8% level in relation to the wild-type strain while the genes for the early steps of the pathway were transcribed at 40–60% levels (Martínez-Burgo et al., 2015). In summary, ClaR is a CcaR-dependent activator that controls expression of genes for the late steps of CA biosynthesis but does not affect significantly expression of the early genes of the pathway (Paradkar et al., 1998) (Fig. 1A). The promoter sequences for ClaR-binding have not been yet reported.
Transcriptomic studies reveal additional global effects of CcaR and ClaR
Although CcaR appears to be a specific activator of cephamycin and clavulanic acid biosynthesis, transcriptomic analysis revealed that ccaR-deletion affects many additional genes for primary metabolism and secondary metabolite gene clusters (Álvarez-Álvarez et al., 2014b). Expression of genes related to energy production, carbon or nitrogen metabolism, phosphate metabolism, regulatory genes and genes for other physiological functions was affected at different degrees either in the exponential or stationary phase. Mutation of ccaR upregulated all the genes for holomycin biosynthesis an average value of 45-fold; downregulated an average of fourfold are all the arginine biosynthesis genes, and an average of 18-fold the genes for nitrogen metabolism (amtB, glnB, glnA2, glnA3) (Álvarez-Álvarez et al., 2013).
Similarly, transcriptomic studies of a claR-deleted mutant showed a weak upregulation of the cephamycin C genes at early times of the culture and, surprisingly, produced an upregulation of all the holomycin biosynthesis genes (3- to 572-fold higher expression). The lack of ClaR affects the expression level of more than 100 genes for primary metabolism and regulators. Downregulated are also all genes of clusters encoding enzymes for two NRPS-type metabolites, the cucumene synthase, and an unknown product, respectively (SCLAV_p1334/_p1341, SCLAV_p1471/_p1483, SCLAV_p1407/_p1415 and SCLAV_p1508/_p1510); also downregulated is the gene amfR that controls aerial mycelium formation (Martínez-Burgo et al., 2015). However, no significant effects on expression of the clavam and paralogous gene clusters were found in the claR-deleted strain. These facts indicate that ClaR exerts a direct or indirect effect on many genes other than CA biosynthesis genes.
Additional mechanisms of regulation of ccaR expression
Expression of ccaR is controlled additionally by several master regulators (i.e., Brp, Are, AdpA) that bind to the intergenic cmcH-ccaR region. We will consider two different regulatory cascades, a ‘proximal’ regulatory cascade close to the UTR region and a ‘distal’ cascade acting near the cmcH gene.
Regulation of ccaR expression by Brp and AdpA
The proximal cascade starts with the well-conserved butyrolactone receptor protein Brp which was found by two research groups using different strategies (Kim et al., 2004; Santamarta et al., 2005). Brp is encoded by the pSCL4-located brp gene (Nishida et al., 2007) and, in the absence of butyrolactones, it controls its own expression, as shown by binding of pure Brp to an autoregulatory sequence (AREbrp) upstream of brp (Santamarta et al., 2005).
Deletion of brp in S. clavuligerus results in an increase of clavulanic acid and cephamycin production (1.3- to 3-folds) in relation to the wild-type strain. Brp does not bind directly to the proximal UTR region of ccaR but its regulation is mediated through the AraC-type transcriptional regulator AdpA (López-García et al., 2010). Brp represses expression of adpA by binding to an AREadpA sequence upstream of adpA. The AdpA protein is a strong activator affecting ccaR expression; transcriptional analysis reveals that removal of adpA in S. clavuligerus results in a decrease expression of ccaR (7-fold) and drastically reduces the transcription of ceaS2/bls2 (50-fold) and cas2 (100-fold) in the CA cluster resulting in very low production of clavulanic acid and cephamycin C. Conversely, amplification of adpA copy number in S. clavuligerus results in more than twofold production of cephamycin C and CA in relation to the control strain. This proximal cascade is highly similar to that involved in the control of the streptomycin regulator StrR (Tomono et al., 2005). In the UTR close to the ccaR gene we have found four putative AdpA-binding sequences (Ohnishi et al., 2005). Three of them are located 380- to 450-pb upstream of tsp2 (Fig. 1B), one overlaps with the ATG codon of ccaR (Fig. 1B) and several putative, less conserved sites, are upstream of tsp1. Multiple AdpA-binding sites have been reported in the promoter of sanR encoding Streptomyces ansochromogenes activator of the nikkomycin cluster (Pan et al., 2009). In summary, all the available information indicates that the butyrolactone sensing system mediated by Brp controls the formation of AdpA which plays a major activator role in antibiotic production in S. clavuligerus.
In addition to the effect exerted through AdpA, Brp recognises an AREccaR sequence on the distant end of the intergenic cmcH-ccaR region. A regulatory protein of the IclR family, named AreB also binds to the AREccaR sequence, as shown by AREccaR-affinity capture method (Santamarta et al., 2007). AreB is an autoregulatory protein that recognises sequences upstream overlapping the −35 site in its promoter controlling also the expression of the leuCD genes, located tail to tail with the areB gene. The binding requires the presence of a small molecular weight ligand of still unknown structure, that may be leucine or a related branched chain amino acid (Santamarta et al., 2007; Yang et al., 2009). The significance of the Brp and AreB binding to this distal AREccaR sequence in cephamycin and clavulanic acid production is unclear since the ARE sequence is located almost 900-nt upstream of tsp1. Deletion of the areB gene produces a partial requirement of leucine for optimal growth but has no significant effect on biosynthesis of CA and cephamycin C.
Regulation by BldG
A regulatory protein that affects ccaR expression in a DNA-binding independent way is BldG, an anti-anti-sigma factor. Deletion of bldG results in mutants unable to differentiate and to produce cephamycin C and clavulanic acid due to lack of ccaR expression as shown by Northern analysis (Bignell et al., 2005). Complementation studies with the wild-type bldG allele resulted in the recovery of both β-lactam antibiotics formation. The bldG gene is located upstream of a gene encoding an anti-sigma factor. This organisation is identical to that of Bacillus subtilis in which the anti-sigma protein SpoIIAA binds the cognate sigmaFfactor preventing its association to the core of the RNA polymerase and the transcription of the genes targeted by the RNApol-sigmaF. The sigmaF-SpoIIAA association is released by the binding of an unphosphorylated anti-anti sigma factor. In S. clavuligerus a similar mechanism of action of BldG with the anti-sigma factor encoded by the bldG-adjacent gene has not been experimentally demonstrated. The null bldG mutant lacks also production of the antifungal clavams, which are not CcaR-dependent; this suggests that the sigma factor controlled by BldG, binds the RNA polymerase and expresses both ccaR and the genes for clavams.
Regulation of clavulanic acid and cephamycin C biosynthesis by relA
The relA gene in actinobacteria and Gram-negative bacteria encodes a guanosine tetraphosphate synthetase that forms the highly phosphorylated nucleotide ppGpp. Initially the ppGpp molecule was studied as an intermediate of the stringent response; however, its role is more complex, affecting many genes involved in growth, differentiation and secondary metabolite biosynthesis (Ochi, 1990). The S. clavuligerus relA gene was cloned independently by two research groups and is located upstream of apt, encoding an adenosine phosphoribosyl transferase. Notably, the intergenic region relA-apt was reported to be 29 nt long by Jin et al. (2004) and 176 nt long by Gomez-Escribano et al. (2008). S1 mapping revealed that the relA tsp is 43-nt upstream of the TTG start codon suggesting that the S. clavuligerus strain studied by Jin et al. (2004) suffered a rearrangement in this region that explains the different behaviour of both strains. The deleted mutant S. clavuligerus ΔrelA failed to produce aerial mycelium, spores or brown pigment in different solid media (Gomez-Escribano et al., 2008). Surprisingly, this mutant produced three- to fourfold more CA and 2.5-fold more cephamycin C than the parental strain. All these phenotypes were restored, and the antibiotics production decreased to wild-type levels, by complementing the ΔrelA mutant with the wild-type relA allele (Gomez-Escribano et al., 2008). Expression of both ccaR and claR were higher in the mutant, mainly at the stationary phase that resulted in a longer period of antibiotics production. Particularly high was the expression of ceaS2, catalysing the first step of CA biosynthesis, and of cefD for the medium steps of cephamycin biosynthesis. The differences in claR expression at the stationary phase parallel the activation of an additional transcription start point (tsp2), an adenine located 107-nt upstream of the ATG codon of claR. tsp2 is poorly active, and probably repressed by ppGpp, in the wild-type strain. This suggests that ppGpp acts as a negative effector in cephamycin C/clavulanic acid production in S. clavuligerus.
Other regulators
In a different study the pimM gene, encoding the Streptomyces natalensis PAS-LuxR-type transcriptional regulator of the pimaricin gene cluster, was integrated in S. clavuligerus genome (Martinez-Burgo et al., 2019). The pimM gene, when expressed in Streptomyces avermitilis, Streptomyces filipensis or Streptomyces nodosus activates expression of filipin and amphotericin B gene clusters and in Streptomyces albus that of the polyketide/non-ribosomal peptide antimycin (Olano et al., 2014). Heterologous expression of pimM in S. clavuligerus resulted in downregulation of all the genes in clusters for a not identified terpene and a clavam-like β-lactam (SCLAV_5670/_5674 and SCLAV_p1508/_1510, respectively); it causes also the upregulation of the cluster for non-characterised polyketide antibiotic (SCLAV_p0509/_p0520) and for the clavulanic and cephamycin C gene cluster. All the genes for CA biosynthesis were upregulated (1.4- to 8-fold) and the CA production increased 10-folds in relation to the control strain, while genes for clavam biosynthesis were not affected. A similar effect was observed on all the genes for cephamycin C biosynthesis, with the exception of pcd, and cephamycin C production increased sevenfold. Overproduction of both antibiotics coincides with an upregulation expression of ccaR (11-fold) and claR (2-fold) genes. Pure PimM binds to the intergenic ccaR-cmcH region resulting in gel shift in EMSA, and a bioinformatic analysis revealed in the intergenic region a sequence similar to the PimM-binding sequence, 5′-GTCCGGAAATCCTCGG-3′, with Schneider nucleotide conservation values, Ri, of 8.6 and 9.9 for the direct and reverse strands (Fig. 1B). This suggests that a PimM-like activator might exist in S. clavuligerus.
Three genes, orf21-orf22-orf23 (SCLAV_1477 to _1475), located in the CA gene cluster upstream of gcaS, have been studied as putative regulators of the cluster (Fu et al., 2019a; Song et al., 2009). Orf21 encodes an RNA polymerase sigma70-family factor, and orf22-orf23 (SCLAV_4176–4175) encode a two-component system (TCS) in which the sensor kinase orf22 contains a histidine residue (His283) for autophosphorylation and orf23 is the response regulator.
Transcription of the CA and cephamycin gene clusters, including ccaR, was not affected in the orf23-disrupted mutant, except for lat and claR transcripts, that were slightly downregulated. However, production of CA by the orf22 and orf23 mutants was decreased by 25% and 60%, while cephamycin C production was reduced by 40% and 60%, respectively. Only the orf23-null mutant showed delayed growth and poor sporulation (Song et al., 2009).
A double mutant, named cagRS, lacking both orf22 and orf23 has been obtained from the industrial strain S. clavuligerus F613-1 (Fu et al., 2019a). The cagRS mutant shows a strong downregulation of several genes for fatty acid degradation and a slight downregulation of glycerol metabolism genes. The mutation also resulted in downregulated expression of the arginine biosynthesis genes and of oat2, the CA cluster located gene encoding an ornithine aminotransferase. Recombinant CagR protein binds the argC and argG promoters as shown by EMSA gel shift, but the binding site or the relation with the ArgR repressor protein have not been studied. Expression of CA biosynthesis genes was not significantly affected in this mutant, although CagR was shown to bind the promoter of claR and the double mutant cagRS produced 25–60% less CA. Integration of these results in the current knowledge obtained from the wild-type strain is not easy. The study of industrial strains is important for the producer company; however, these are heavily mutagenised strains that are grown in industrial complex culture medium; therefore, their regulation might be different from that of the wild-type strain and the results obtained in industrial strains should be always confirmed in parallel with the wild-type strain. In summary, the proteins encoded by orf21, orf22, orf23 appear to have different effects on cell metabolism and may indirectly modulate the clavulanic acid and cephamycin C formation but should not be considered strictly regulators of the gene supercluster.
The effect on cephamycin production of a different TCS, named CepRS, (SCLAV_2959/2960), has been studied in the industrial strain S. clavuligerus F613-1 (Fu et al., 2019b). The mutant S. clavuligerus cepRS, deleted in cepR and cepS was found to produce normal levels of CA but only 30% of cephamycin C in relation to the parental strain. The CefR protein binds the intergenic cefD-cmcI region that controls genes encoding enzymes for the early, middle and late steps of the cephamycin pathway. The binding site appears to overlap with the three heptameric regions bound by CcaR, suggesting that CepR partially competes with CcaR, resulting in downregulation of the transcripts for the cephamycin biosynthesis genes. However, the cefD-cmcI region in strain F613-1 is slightly longer than in the ATCC 27064 strain and the binding site has not been clearly defined by footprinting.
Holomycin
Holomycin is a compound with dithiopyrrolone structure and yellow colour. Dithiopyrrolones are formed by two heterocyclic rings; one of them contains a disulphide bridge and the second has a nitrogen atom in the heterocycle (Fig. 2). The nitrogen-carrying heterocycle possesses an amino group, that is acetylated in holomycin, and propionylated in N-propionylholothin. Holomycin and related dithiopyrrolones are produced by Gram-negative bacteria and by actinomycetes, such as Saccharothrix species. More complex compounds containing the dithiopyrrolone chromophore (thiomarinols, xenorhabdins) are produced by enterobacteria that are mutualist or pathogens of invertebrates, and by marine bacteria (for reviews, see Li et al., 2014; Liras, 2014; Qin et al., 2013). S. clavuligerus IT1 was reported to produce a compound active against Micrococcus luteus that was not detectable in S. clavuligerus ATCC 27064 cultures; the compound was crystallised and identified as holomycin by 1H-NMR and MS spectrum (Kenig & Reading, 1979). Later, holomycin was confirmed to be produced by a mutant defective in the oppA2 gene. The yellow compound, is produced in increasing amounts by mutants blocked respectively in car, claR, orf12, oppA2 and cyp-fd, while mutants blocked in the early steps of the CA pathway were holomycin non-producers (Fuente et al., 2002; Lorenzana et al., 2004).
Fig. 2.
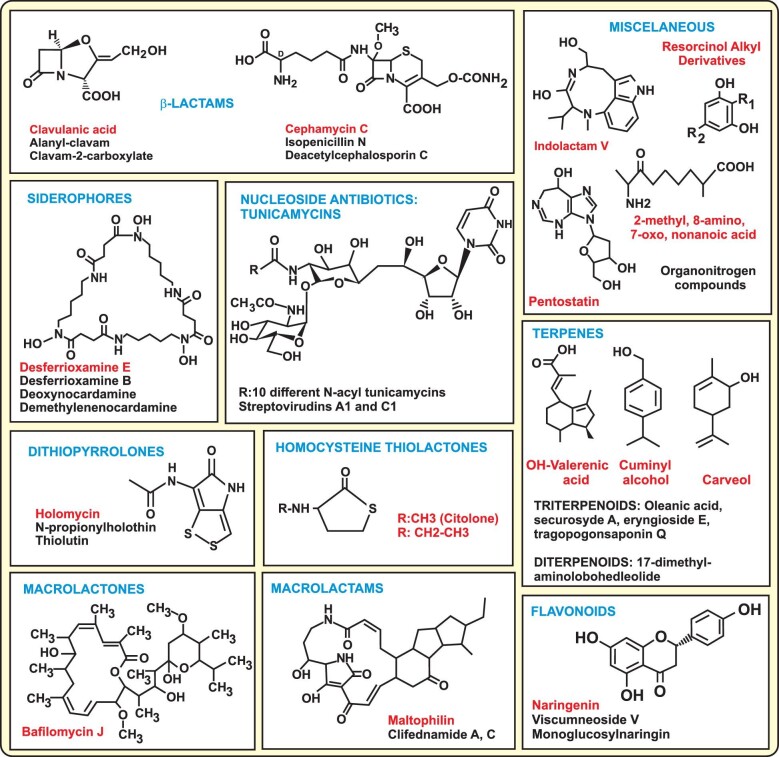
Chemical compounds produced by Streptomyces clavuligerus as demonstrated by HPLC LC/MS analysis. When more than one compound of each type is produced, the structure shown corresponds to the compound labelled with red letters.
The cluster of holomycin biosynthesis genes (Fig. S3A) was detected by bioinformatic analysis of the genes encoding the putative enzymes involved in holomycin biosynthesis (Huang et al., 2011; Li & Walsh, 2010) and heterologous expression of hlmE and hlmA. HlmE is a non-ribosomal peptide synthetase able to activate l-cysteine to form l-cys-AMP while HlmA is an acetyl transferase converting holothin into holomycin. Deletion of hlmE and of hlmF, encoding a lantibiotic-like decarboxylase, originated holomycin non-producer mutants (Li & Walsh, 2010). Independently, other research group observed S. clavuligerus oppA2::aph proteome intense protein spots corresponding to HlmD, HlmF and HlmG and a high transcription of genes located adjacent to hlmDFG which was not observed in the wild-type strain; holomycin production in S. coelicolor by heterologous expression of the hlm genes confirmed that this region corresponds to the holomycin biosynthesis gene cluster (Robles-Reglero et al., 2013). The hlm cluster has been expressed with low efficiency in S. albus and S. coelicolor (Huang et al., 2011; Robles-Reglero et al., 2013). A biosynthetic pathway for holomycin (Fig. S3B) has been proposed after purification and studies of the recombinant proteins HlmA, HlmC, HlmD, HlmE, HlmF and HlmI, and after deletion of the hlmF, hlmI and hlmE genes (Li & Walsh, 2010, 2011). A key enzyme in the pathway is HlmI, an oxygen-dependent dithiol oxidase, that oxidises the disulphide bond forming the heterologous ring and produces holomycin from reduced holomycin, a metal-chelating compound (Chan et al., 2017). Mutants in hlmI show lower resistance to holomycin, although additional genes conferring holomycin resistance cannot be excluded. Proteomic studies show that overproduction of holomycin correlates with an intense protein spot corresponding to a thiosulphate sulphur transferase, RhlA. This protein is essential for holomycin overproduction, although the rhlA gene is not located in the hlm gene cluster (Nárdiz et al., 2011).
Transcriptional studies in the oppA2-deleted mutant S. clavuligerus ΔoppA2::acc showed a twofold average increased expression of all the hlm genes at three sampling times in relation to the wild-type strain. However, comparison with cultures of S. clavuligerus pSCL4– indicated that the lack of plasmid pSCL4 affects the hlm genes expression more intensely (12.6-fold increase). This transcription deregulation of the pSCL– mutant holomycin cluster is independent of the oppA2 deletion and both effects are cumulative resulting in a 42.84-fold expression in a double mutant lacking oppA2 and the pSCL4 plasmid (Álvarez-Álvarez et al., 2018). The effect was especially high on genes hlmA to hlmI, while less affected were genes hlmK, hlmL and hlmM, the last one encoding a LuxR-like transcriptional regulator.
The overproduction of holomycin by S. clavuligerus pSCL4– might be explained if one of the genes located in pSCL4 encodes a repressor of the hlm gene cluster expression. In addition, pSCL4 contains several genes involved in sulphur, cysteine and methionine metabolism and their absence might channel sulphur-containing amino acids to produce dithiopyrrolones. However, the reasons for the overexpression of holomycin in clavulanic acid mutants blocked in the late, but not in the early steps of the CA pathway, is not understood.
Tunicamycin
Tunicamycins are a family of nucleoside antibiotics produced by several species of actinomycetes. They are bacterial cell wall inhibitors which target the translocase I involved in the first steps of peptidoglycan biosynthesis, affect the first step of protein glycosylation in eukaryotes, and also act as inhibitors of the palmitoyl transferase (Price & Tsvetanova, 2007). The tunicamycin structures are formed by a uracil molecule, N-acetylglucosamine and the rare sugar tunicamine (Fig. 2); members of the complex differ in the length of a lateral N-acyl chain (Tsvetanova & Price, 2001). Heterologous expression in Streptomyces lividans of a cosmid library of Streptomyces chartreusis, a tunicamycin producer, and HPLC analysis of the transformants culture broth revealed in an S. lividans transformant two novel peaks with antibiotic activity. LC/MS and MS/MS analysis and comparison with pure samples of tunicamycins identified the peaks as tunicamycin 15:1B and tunicamycin 16:1A (Chen et al., 2010). A biosynthetic pathway for tunicamycin has been proposed based on the characteristics of the S. chartreusis tun genes and in previous information (Chen et al., 2010; Tsvetanova & Price, 2001).
The S. clavuligerus IT1 strain reported to produce holomycin produces in addition a complex antibiotic, MM19290, formed by tunicamycins with N-acyl chains of 9–12 carbons (Kenig & Reading, 1979). Bioinformatic analysis of S. clavuligerus genome detected a cluster of genes (SCLAV_4275/_4287) almost identical in organisation to the tunicamycin cluster in S. chartreusis (Chen et al., 2010). This tun cluster is upregulated in S. clavuligerus pSCL4– lacking the megaplasmid pSCL4, showing overexpression of all the genes an average value of 6- to 25-fold in relation to the wild-type strain. The more upexpressed genes were tunA and tunB, encoding enzymes for the early steps of the pathway (Álvarez-Álvarez et al., 2017). This result suggests that a gene located in pSCL4 encodes a regulator that controls negatively the tunicamycin biosynthesis genes.
Transformation of S. clavuligerus with the pimM gene (see above) affects also tunicamycin production. Expression of pimM did not modify significantly the transcription of the tun genes; however, the transformant strain, but not of the control strain, showed bioactivity against yeasts. The LC-MS metabolic profile of S. clavuligerus::pimM cell extracts showed notable differences in metabolites in relation to the control strain, and two peaks with antifungal activity were shown to correspond to tunicamycins. Tunicamycin production in the transformant was 5.3-fold higher than in the control strain and included tunicamycins with N-acyl chains of 11, 12 and 18 carbons as shown by MS quantification Interestingly, the higher antifungal activity (4- to 16-fold) corresponded to tunicamycin with longer acyl chains (i.e., 17:1) that were shown for the first time to be more active than those of shorter acyl chains (Martínez-Burgo et al., 2019). Since no differences in expression of the tun biosynthesis genes was correlated with pimM expression it was proposed that the overproduction of tunicamycin was due to the effect of PimM on genes that might supply additional precursors for tunicamycin formation, that is, for new and longer acyl lateral chains.
Streptovirudins, aminoglycosides identical to tunicamycins that contains dihydrouracil instead of uracil, have been described to be produced by ribosome engineered strains of S. clavuligerus (Shaikh et al., 2021). These authors suggest that the tun cluster encodes also the streptovirudins, although the gene responsible for the dihydrouracil formation is not present in the tun cluster.
Naringenin
Comparative HPLC analysis of broths from S. clavuligerus cultures grown in complex TSB and SA media revealed a peak in the former cultures that was not present in SA-grown cells. The compound was purified and analysed by LC-MS and 1H-NMR. The substance was a flavonoid with a chemical structure corresponding to naringenin and differs from genistein in the lack of a double bond (Fig. 2). Naringenin is a typical plant compound not described previously to be produced naturally by microorganisms. The biosynthesis of naringenin in plants starts with the condensation of p-coumaroyl-CoA with three malonyl-CoA molecules by a chalcone synthase-type III. By comparison of genes/proteins in plants two clustered genes, ncyP and ncs (SCLAV_5491/5492), were located in S. clavuligerus genome (Álvarez-Álvarez et al., 2015). These genes encode, respectively, a cytochrome P450 oxygenase and a type III polyketide synthase with similarity to naringenin chalcone synthases. An additional gene, named tal (SCLAV_5457) was located 47-kb upstream of the cluster ncs-ncyP. The tal gene is similar to the encP gene of Streptomyces maritimus, which encodes a phenylalanine/tyrosine ammonia lyase (Tal) required for p-coumaroyl biosynthesis. Independent deletion of S. clavuligerus ncs, ncyP and tal genes resulted in strains unable to produce naringenin, and the production reverted to wild-type levels in each case by complementation with the deleted gene (Álvarez-Álvarez et al., 2015). The phenylalanine/tyrosine ammonia lyase protein has been purified to homogeneity; it has a prosthetic group methyl-idene-imidazol-5-one (MIO) giving the MIO characteristic spectrophotometric absorption; however, the Tal protein was very unstable which prevented enzymatic studies (Álvarez-Álvarez, personal communication). Supplementation of the cultures with fatty acids, that is, palmitic acid, and aromatic amino acids, as phenylalanine, improved naringenin production 3.7-fold. Under the optimal culture conditions studied S. clavuligerus ATCC 27064 produced 184 μg/ml naringenin. The fact that a natural compound is obtained from non-clustered genes, suggests that additional secondary metabolites might be encoded by Streptomyces species by distantly located (unlinked) genes.
Three flavonoid glycosides, including viscimneoside V and monoglucosyl-naringin, are produced by S. clavuligerus mutants (Shaikh et al., 2021). Adjacent to ncyP and ncs there are genes for a transketolase (SCLAV_5490) and a transglycosidase (SCLAV_5493); therefore, these authors suggest that these flavonoids are produced by the same gene cluster than naringenin.
Metabolomics
We have described above the compounds produced by S. clavuligerus from which enzymes and pathways are known, although in some of them, as is the case of tunicamycin, the knowledge partially relies on studies in other microorganisms.
Up to 58 secondary metabolite gene clusters have been described in S. clavuligerus genome (Hwang et al., 2019). Most of them are cryptic or silent clusters that have not been characterised, and the compounds derived from many of these clusters are still not known. Considering that, in any case, more than one compound might be encoded by every gene cluster the number of secondary metabolites potentially produced by S. clavuligerus is well over one hundred. Several strategies have been followed to characterise these secondary metabolites and the clusters responsible for their formation. This knowledge is important in order to delete the active clusters in industrial strains as a way to improve clavulanic acid production.
Clusters Characterised by the Similarity of Their Genes/Proteins to Those Present in Other Microorganisms
Tacrolimus. Tacrolimus (FK506) is a macrolide compound with inmunosupressant activity produced by Streptomyces tsukubaensis (Ordóñez-Robles et al., 2018). The cluster fkb for tacrolimus biosynthesis in S. tsukubaensis is formed by 16 ORFs and contains 3 genes for polyketide synthetases (fkbA to fkbC) and two genes for regulatory proteins (fkbN and fkbR). Mo et al. (2009) described the production of tacrolimus by S. clavuligerus CKD1119. Later Park et al. (2009) using LC/MS analysis reported the presence of tacrolimus and eight intermediates of the biosynthesis pathway in S. clavuligerus KCTC10561BP cultures.
In S. clavuligerus ATCC 27064 genome only genes SCLAV_0006 to _0010 have a similar organisation to the tacrolimus gene cluster as they encode PKSs with 45–50% identity to FkbA to FkbC. However, the proteins encoded by the surrounding genes have low identity to the Fkb proteins and the cluster lacks genes homologous to fkbO and fkbL. Moreover, the cluster SCLAV_0006 to _0017 has been found recently to be identical to that of the bafilomycin cluster of Streptomyces lohii (Zhang et al., 2013) and has been proposed that this cluster is responsible for bafilomycin production in S. clavuligerus (Shaikh et al., 2021). Therefore, S. clavuligerus ATCC 27064 does not appear to have a cluster for tacrolimus biosynthesis and was never reported to produce tacrolimus. It would be of interest to compare the genomes of the wild-type strain and the reported tacrolimus producers S. clavuligerus strains to clarify whether these strains obtained the tacrolimus cluster by horizontal transfer.
Moenomycins. Moenomycins are phosphoglycolipids acting on the glycosyltransferases involved in bacterial cell wall biosynthesis. These compounds are produced by Streptomyces ghanaensis (Ostash et al., 2009) while teichomycins, belonging to the moenomycin family, are produced by Actinoplanes teichomyceticus. A cluster of genes located in S. clavuligerus plasmid pSCL4 (SCLAV_p1274/_p1290) is similar to the moe and tchm clusters and encode proteins 47–82% identical, what suggests that this is a moenomycin gene cluster. Moenomycins could not be detected in fermentations of S. clavuligerus ATCC 27064 suggesting that this is a silent cluster under the studied conditions (Álvarez-Álvarez et al., 2017).
Compounds Characterised by HPLC-LC/Mass Spectrometry Analysis of Culture Broths
Comparison by LC/MS-MS analysis of S. clavuligerus ATCC 27064 culture broth and that of different mutants allowed the characterisation of several compounds (Álvarez-Álvarez et al., 2017; AbuSara et al., 2019; Martínez-Burgo et al., 2019).
The pSCL4-less mutant produces holomycin and N-propionylholothin, which are not detectable in S. clavuligerus ATCC 27064, in agreement with the upregulation of the holomycin biosynthesis genes in this mutant (Álvarez-Álvarez et al., 2017). The molecular mass of N-propionylholothin and thiolutin are identical but only N-propionylholothin has been detected by MS analysis to be produced by S. clavuligerus (Okamura et al., 1977). Two thiolactone compounds carrying sulphur in the molecule, N-acetyl and N-propionyl homocysteine, were also detected in the pSCL4-less strain. Nocardamine (desferrioxamine E) was the main desferrioxamine in both strains although small amounts of demethylenocardamine and deoxynocardamine were also present in the wild-type strain broth. The wild-type strain but not the pSCL4-less mutant produced resorcinol-derived compounds and methyl-7-oxononanoic acid (Álvarez-Álvarez et al., 2017) (Fig. 2).
A mixture of four tunicamycins, not completely characterised, was reported to be produced by S. clavuligerus (Kenig & Reading, 1979). Recently, the heterologous expression in S. clavuligerus of S. natalensis pimM gene (see Other regulators section) showed that the transformant produced eight members of the tunicamycin family with acyl chains of 11–18 carbons. They include two previously unreported compounds with acyl chains of 11 and 18 carbon atoms (Martínez-Burgo et al., 2019).
AbuSara et al. (2019) in a comparative study of the metabolome of S. clavuligerus confirmed the formation of desferrioxamine E (nocardamine), naringenin, holomycin and the tunicamycins by S. clavuligerus. In addition, using LC-MS and MS-MS these authors described the presence of several new compounds, as the terpenes carveol, cuminyl alcohol and hydroxyvalerenic acid, the linear siderophore desferrioxamine B, indolactamV and the nucleoside pentostatin (Fig. 2). In addition, this research group performed engineering of S. clavuligerus ribosomes overexpressing the rpsL for a ribosomal protein, the frr gene, for a recycling factor and obtaining by direct mutagenesis different mutants of rpsL. LC-MS/MS analysis of culture broth in these S. clavuligerus mutants revealed the presence of 28 new metabolites. They include nucleoside antibiotics as streptovirudins A1 and C1; macrolactone antibiotics as bafilomycin J; macrolactams as maltophilin and clifednamides A or B; flavonoids as viscumneoside V and monoglucosylnaringin; 10 different triterpenoids including oleanic acid and the saponins secursyde A, eryngioside E and tragopogonsaponin Q, three different diterpenoids, the hopanoids glochidone and 23-OH-hopen-1-one, and several organonitrogen compounds (Shaikh et al., 2021). Gene clusters for some of these compounds were suggested by the authors, but they still require confirmation.
Characterisation of Streptomyces clavuligerus Terpene Synthases by Heterologous Expression
Terpenoids used in the cosmetic and food industries as flavour and odour constituents have been isolated mostly from higher plants, and are rarely produced by microorganisms. Terpenoids derive from isopentenyl diphosphate (C5) and dimethylallyl diphosphate (C5), and by successive condensations form geranyl diphosphate (GPP, C10), farnesyl diphosphate (FPP, C15) and geranylgeranyl diphosphate (GGPP, C20). Cyclisation of these intermediates by terpene synthases/cyclases and further modifications result in the formation of different monoterpenes (C10), sesquiterpenes (C15) and diterpenes (C20).
Terpenoid platform vectors, containing inducible genes for the mevalonate pathway and/or for the precursors FPP, GPP or GGPP (fps, gps or ggs genes) expressed from strong promoters have been constructed in modified Escherichia coli strains (Hu et al., 2011; Yamada et al., 2016; Karuppiah et al., 2017) to produce terpenes.
More than 20 genes for terpene synthases have been found in S. clavuligerus genome (Hu et al., 2011). Most S. clavuligerus terpene synthase genes are located in plasmid pSCL4 and encode terpene synthases type I which contain the characteristic Mg2+-binding amino acid motifs (Yamada et al., 2016). About half of these genes have been expressed in E. coli and GC-MS/NMR of the transformants broth detected the production of monoterpene, sesquiterpene or diterpenes (Table 2).
Table 2.
Terpenes Characterised in Cultures of Heterologously Expressed Terpene Synthases of Streptomyces clavuligerus
| S. clavuligerus gene | Protein name | Substrate | Product | Reference |
|---|---|---|---|---|
| SCLAV_p0328 | FPP | (-)-δ-cadinene | Hu et al., 2011 | |
| SCLAV_p0068 | FPP | (+)-T-muurolol | Hu et al., 2011 | |
| SCLAV_p0068/SCLAV_00067 | (-)-drimenol | Yamada et al., 2015a | ||
| SCLAV_p0982 | CinS | GPP | 1,8 cineole, β-pinene, camphene | Nakano et al., 2011a; Yamada et al., 2015b. |
| SCLAV_p1185 | LinS | FPP y GPP | Linalool and nerolidol | Nakano et al., 2011b |
| SCLAV_p0765 | GPP | Hydropyrenol, hydropyrene, isoelisabethatriene B | Yamada et al., 2015a,b | |
| SCLAV_p1169 | GPP | Clavutrienes A and B, prenylgermacrene B, lobophytumin C | Yamada et al., 2015b | |
| SCLAV _1407 | FPP | (5S, 7S, 10R, 11S) cucumene (iso-hirsutene) | Yamada et al., 2015b | |
| Chow et al., 2015 | ||||
| SCLAV_p0490/_p0491 | GGPP | Labda 7,13(16), 14 triene | Yamada et al., 2016 |
Two genes, SCLAV_p0328 and SCLAV_p0068, encoding terpene synthases, flanked by genes for methyltransferases and cytochrome P450, probably involved in later modifications of the produced terpene, have been expressed in E. coli. The recombinant proteins incubated with FPP generated, respectively, a sesquiterpene (C15H24), a sesquiterpene alcohol (C15H26O) identified as (−)-δ-cadinene, and (+)-T-muurolol (Hu et al., 2011). In addition, the SCLAV-p0068 gene, when coexpressed with SCLAV_p0067, encoding a cytochrome P450, forms (−)-drimenol (Yamada et al., 2015a,b) (Fig. S4).
In parallel to the previous work, Nakano et al. (2011a) described that SCLAV_p0982 (cinS) encodes a type I monoterpene synthase, which uses only GPP as substrate and catalyses the synthesis of 1,8 cineole (eucalyptol), an essential oil used in the pharmaceutical and cosmetic industries, which is extracted normally from Eucalyptus polybractea. This gene has been described also as producer of β-pinene and camphene (Yamada et al., 2015b).
A different gene, SCLAV_p1185 (linS), encodes the linalool synthase. LinS uses FPP to produce nerolidol, and GPP to produce linalool (Nakano et al., 2011a, 2011b) as confirmed by GC-MS and comparison with pure compounds, but is unable to use GGPP. Linalool is a product used as fragrance in hygiene products; improved expression of the linalool synthase and strains with a better GPP assimilation resulted in a linalool yield of 1,027 mg/l in fed-batch cultures (Wang et al., 2020). However neither linalool, nerolidol or 1,8 cineole have been ever detected in S. clavuligerus cultures.
Labda 7,13(16), 14 triene, is formed in E. coli as product of the genes SCLAV_p0490/SCLAV_p0491. This is a novel triene (Fig. S4) which was not observed in S. clavuligerus cultures grown under different conditions. The cluster of genes SCLAV_p0484 to SCLAV_p0491, located in plasmid pSCL4, carry in addition to genes for terpene synthases, genes for a transcriptional regulator, a geranyl geranyl phosphate synthase (GGPPS) and a transport system (Yamada et al., 2016). Four different labda-type terpenes have been obtained by combined expression of SCLAV_p0490/_p0491 with genes for terpene cyclases of Streptomyces cyslabdanicus and Streptomyces anulatus (Yamada et al., 2016).
The S. clavuligerus cucumene synthase and terpene synthases LinS and CinS, have been crystalyzed (Blank et al., 2018; Karuppiah et al., 2017). These proteins are relatively unique since they lack the N-terminal alpha barrel domain present in the corresponding plant terpene synthases. The enzymatic reaction of several S. clavuligerus terpene synthases have been thoroughly analysed (Chow et al., 2015; Rinkel et al., 2016) since the modification of the key amino acid residues required for catalytic activity might allow to obtain modified monoterpenoids.
From all these results it can be concluded that S. clavuligerus has the ability to produce terpenes as shown by the detection of carveol, cumminyl alcohol or hydroxyvalerenic acid in culture broths (AbuSara et al., 2019); however, the wealth of terpenes obtained in heterologous expression platforms has never been observed in S. clavuligerus cultures even although a variety of culture conditions have been tested (Yamada et al., 2015a). In fact, although a gene (SCLAV_p0159) for geosmin biosynthesis, a sequiterpene responsible for the characteristic moist soil odour is present in the genome, this compound has never been detected in S. clavuligerus cultures (Yamada et al., 2015a).
The genes for terpene synthases are transcribed in S. clavuligerus as shown by downregulation of their expression in mutants lacking claR or pSCL4 (Álvarez-Álvarez et al., 2014a; Hwang et al., 2019; Martínez-Burgo et al., 2015), that is, they are not silent genes. The limited production of terpenes by S. clavuligerus cultures might be due to poor supply of precursors.
Conclusions
The advances in sequencing technology provides a high-coverage S. clavuligerus genome and provides an explanation for the instability found in plasmids and chromosome arms in different isolates of this species. The complexity of plasmid–chromosome relationships makes essential the confirmation of pSCL4 copy number and subsequent chromosome stability in every study. The genome sequence facilitates transcriptomic studies which still require a large amount of experimental research to connect the observations obtained by microarray analysis with the physiology and behaviour of S. clavuligerus in the different investigated conditions, especially in relation to primary metabolism.
Genomic studies provide also a wide view of the large number of clusters for secondary metabolism present in S. clavuligerus genome. Metabolomic studies, heterologous expression and comparison with other genomes permit to determine the compound encoded by different secondary metabolite clusters, but confirmation through gene disruption and biological and chemical analysis of the mutants is needed in most cases. In conclusion a new era is open to study the genetics and physiology of S. clavuligerus in order to convert this strain in an instrument for the efficient production of new compounds of industrial interest. These multi-omic studies open the way for the utilisation of S. clavuligerus has a platform for industrial production of secondary metabolites in addition to clavulanic acid.
Supplementary Material
Acknowledgments
This article is dedicated to the memory of Prof. Arnold L. Demain who was a very good friend and an excellent scientific teacher for the authors and to Dr. Jorge Romero that started the studies of clavulanic acid in our laboratory. J.F.M. and P.L. acknowledge the informatic support of the University of León (Spain) and the encouragement of Drs. Ruben Álvarez-Álvarez, Antonio Rodríguez-García and Juan P. Gomez-Escribano.
Contributor Information
Paloma Liras, Microbiology Section, Department of Molecular Biology, University of León, León 24071, Spain.
Juan F Martín, Microbiology Section, Department of Molecular Biology, University of León, León 24071, Spain.
Funding
None declared.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- AbuSara N. F., Piercey B. M., Moore M. A., Shaikh A. A., Nothias L. F., Srivastava S. K., Cruz-Morales P., Dorrestein P. C., Barona-Gómez F., Tahlan K. (2019). Comparative genomics and metabolomics analyses of clavulanic acid-producing Streptomyces species provides insight into specialized metabolism. Frontiers in Microbiology, 10, 2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Álvarez R., Martínez-Burgo Y., Pérez-Redondo R., Braña A. F., Martín J. F., Liras P. (2013). Expression of the endogenous and heterologous clavulanic acid cluster in Streptomyces flavogriseus: Why a silent cluster is sleeping. Applied Microbiology and Biotechnology, 97(21), 9451–9463. [DOI] [PubMed] [Google Scholar]
- Álvarez-Álvarez R., Rodríguez-García A., Martínez-Burgo Y., Robles-Reglero V., Santamarta I., Pérez-Redondo R., Martín J. F., Liras P. (2014a). A 1.8-Mb-reduced Streptomyces clavuligerus genome: Relevance for secondary metabolism and differentiation. Applied Microbiology and Biotechnology, 98(5), 2183–2195. [DOI] [PubMed] [Google Scholar]
- Álvarez-Álvarez R., Rodríguez-García A., Santamarta I., Pérez-Redondo R., Prieto-Domínguez A., Martínez-Burgo Y., Liras P. (2014b). Transcriptomic analysis of Streptomyces clavuligerus ΔccaR::tsr: Effects of the cephamycin C-clavulanic acid cluster regulator CcaR on global regulation. Microbial Biotechnology, 7(3), 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Álvarez R., Botas A., Albillos S. M., Rumbero A., Martín J. F., Liras P. (2015). Molecular genetics of naringenin biosynthesis, a typical plant secondary metabolite produced by Streptomyces clavuligerus. Microbial Cell Factories, 14(1), 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Álvarez R., Martínez-Burgo Y., Rodríguez-García A., Liras P. (2017). Discovering the potential of S. clavuligerus for bioactive compound production: Cross-talk between the chromosome and the pSCL4 megaplasmid. BMC Genomics [Electronic Resource], 18(1), 907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Álvarez R., Rodríguez-García A., Martínez-Burgo Y., Martín J. F., Liras P. (2018). Transcriptional Studies on a Streptomyces clavuligerus oppA2 deletion mutant: N-acetylglycyl-clavaminic acid is an intermediate of clavulanic acid biosynthesis. Applied and Environmental Microbiology, 84(22), e01701–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arulanantham H., Nadia J., Kershaw K., Hewitson S., Hughes C. E., Thirkettle J. E., Schofield C. J. (2006). ORF17 from the clavulanic acid biosynthesis gene cluster catalyzes the ATP-dependent formation of N-glycyl-clavaminic acid. Journal of Biological Chemistry, 281(1), 279–287. [DOI] [PubMed] [Google Scholar]
- Baggaley K. H., Brown A. G., Schofield C. J. (1997). Chemistry and biosynthesis of clavulanic acid and other clavams. Natural Product Reports, 14(4), 309–333. [DOI] [PubMed] [Google Scholar]
- Baños S., Pérez-Redondo R., Koekman B., Liras P. (2009). Glycerol utilization gene cluster in Streptomyces clavuligerus. Applied and Environmental Microbiology, 75(9), 2991–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell D. R. D., Tahlan K., Colvin K. R., Jensen S. E., Leskiw B. K. (2005). Expression of ccaR, encoding the positive activator of cephamycin C and clavulanic acid production in Streptomyces clavuligerus, is dependent on BldG. Antimicrobial Agents and Chemotherapy, 49(4), 1529–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank P. N., Pemberton T. A., Chow J.-Y., Poulter C. D., Christianson D. W.. . (2018). Crystal structure of cucumene synthase, a terpenoid cyclase that generates a linear triquinane sesquiterpene. Biochemistry, 57(44), 6326–6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botas A., Pérez-Redondo R., Rodríguez-García A., Álvarez-Álvarez R., Yagüe P., Manteca A., Liras P. (2018). ArgR of Streptomyces coelicolor is a pleiotropic transcriptional regulator: Effect on the transcriptome, antibiotic production, and differentiation in liquid cultures. Frontiers in Microbiology, 9, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G., Zhong C., Zong G., Fu J., Liu Z., Zhang G., Qin R. (2016). Complete genome sequence of Streptomyces clavuligerus F613-1, an industrial producer of clavulanic acid. Genome Announcements, 4(5), e01020–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A. N., Shiver A. L., Wever W. J., Razvi S. Z. A., Traxler M. F., Li B. (2017). Role for dithiolopyrrolones in disrupting bacterial metal homeostasis. Proceedings of the National Academy of Science USA, 114(10), 2717–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra G., Chater K. F. (2008). Evolutionary flux of potentially bldA-dependent Streptomyces genes containing the rare leucine codon TTA. Antonie van Leeuwenhoek, 94(1), 111–126. [DOI] [PubMed] [Google Scholar]
- Charusanti P., Fong N. L., Nagarajan H., Pereira A. R., Li H. J., Abate E. A., Su Y., Gerwick W. H., Palsson B. O. (2012). Exploiting adaptive laboratory evolution of Streptomyces clavuligerus for antibiotic discovery and overproduction. Plos One, 7(3), e33727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan R., Ravi J., Datta P., Chen T., Schnappinger D., Bassler K. E., Balázsi G., Gennaro M. L. (2016). Reconstruction and topological characterization of the sigma factor regulatory network of Mycobacterium tuberculosis. Nature Communications, 7(1), 11062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. W., Huang C. H., Lee H. H., Tsai H. H., Kirby R. (2002). Once the circle has been broken: Dynamics and evolution of Streptomyces chromosomes. Trends in Genetics, 18(10), 522–529. [DOI] [PubMed] [Google Scholar]
- Chen W., Qu D., Zhai L., Tao M., Wang Y., Lin S., Price N. P., Deng Z. (2010). Characterization of the tunicamycin gene cluster unveiling unique steps involved in its biosynthesis. Protein Cell, 1(12), 1093–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J. Y., Tian B. X., Ramamoorthy G., Hillerich B. S., Seidel R. D., Almo S. C., Jacobson M. P., Poulter C. D. (2015). Computational-guided discovery and characterization of a sesquiterpene synthase from Streptomyces clavuligerus. National Academy of Science USA, 112(18), 5661–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fergusson N. L., Peña-Castillo L., Moore M. A., Bignell D. R. D., Tahlan K. (2016). Proteomic analysis of global regulatory cascades involved in clavulanic acid production and morphological development in Streptomyces clavuligerus. Journal of Industrial Microbiology and Biotechnology, 43(4), 537–555. [DOI] [PubMed] [Google Scholar]
- Fu J., Qin R., Zong G., Liu C., Kang N., Zhong C., Cao G. (2019a). The CagRS two-component system regulates clavulanic acid metabolism via multiple pathways in Streptomyces clavuligerus F613-1. Frontiers in Microbiology, 10, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Qin R., Zong G., Zhong C., Zhang P., Kang N., Qi X., Cao G. (2019b). The two-component system CepRS regulates the cephamycin C biosynthesis in Streptomyces clavuligerus F613-1. Applied Microbiology and Biotechnology Express, 9, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuente A. de la., Lorenzana L. M., Martín J. F., Liras P. (2002). Mutants of Streptomyces clavuligerus with disruptions in different genes for clavulanic acid biosynthesis produce large amounts of holomycin: Possible cross-regulation of two unrelated secondary metabolic pathways. Journal of Bacteriology, 184(23), 6559–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuente J. L. de la., Rumbero A., Martín J. F., Liras P. (1997). Delta-1-piperideine-6-carboxylate dehydrogenase, a new enzyme that forms alpha-aminoadipate in Streptomyces clavuligerus and other cephamycin C-producing actinomycetes. Biochemical Journal, 327(1), 59–64. [PMC free article] [PubMed] [Google Scholar]
- Gómez S., Ramírez-Malule H., Cardona G. W., Osorio E., Restrepo A. (2020). Double-ring epimerization in the biosynthesis of clavulanic acid. The Journal of Physical Chemistry A, 124(45), 9413–9426. [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano J. P., Martín J. F., Hesketh A., Bibb M. J., Liras P. (2008). Streptomyces clavuligerus relA-null mutants overproduce clavulanic acid and cephamycin C: Negative regulation of secondary metabolism by (p)ppGpp. Microbiology (Reading), 154(3), 744–755. [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano J. P., Gallardo L. A., Bozhüyük K. A. J., Kendrew S. G., Huckle B. D., Crowhurst N. A., Bibb M. J., Collis A. J., Micklefield J., Herron P. R., Wilkinson B. (2021). Genome editing reveals that pSCL4 is required for chromosome linearity in Streptomyces clavuligerus. Microbial Genomics, (accepted for publication). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamed R. B., Gomez-Castellano J. R., Henry L., Ducho C., McDonough M. A., Schofield C. J. (2013). The enzymes of β-lactam biosynthesis. Natural Products Report, 30, 21–107. [DOI] [PubMed] [Google Scholar]
- Hasse D., Hülsemann J., Carlsson G. H., Valegård K., Andersson I. (2019). Structure and mechanism of piperideine-6-carboxylate dehydrogenase from Streptomyces clavuligerus. Acta Crystallography. D Structure Biology, 75(12), 1107–1118. [DOI] [PubMed] [Google Scholar]
- Hu Y., Chou W. K., Hopson R., Cane D. E. (2011). Genome mining in Streptomyces clavuligerus: expression and biochemical characterization of two new cryptic sesquiterpene synthases. Chemistry and Biology, 18(1), 32–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Zhao Y., Qin Z., Wang X., Onega M., Chen L., He J., Yu Y., Deng H. (2011). Identification and heterologous expression of the biosynthetic gene cluster for holomycin produced by Streptomyces clavuligerus. Process in Biochemistry, 46(3), 811–816. [Google Scholar]
- Hwang S., Lee N., Jeong Y., Lee Y., Kim W., Cho S., Palsson B. O., Cho B. K. (2019). Primary transcriptome and translatome analysis determines transcriptional and translational regulatory elements encoded in the Streptomyces clavuligerus genome. Nucleic Acids Research, 47(12), 6114–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S., Lee N., Choe D., Lee Y., Kim W., Jeong Y., Cho S., Palsson B. O., Cho B. K. (2021). Elucidating the regulatory elements for transcription termination and posttranscriptional processing in the Streptomyces clavuligerus genome. mSystems, 6(3), e01013–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal A., Arunlanantham H., Brown T. Jr., Chowdhury R., Clifton J. I., Kershaw N. J., Hewitson K. S., McDonough M. A., Schofield C. J. (2010). Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway. Proteins, 78(6), 1398–1407. [DOI] [PubMed] [Google Scholar]
- Jensen S. E., Paradkar A. S. (1999). Biosynthesis and molecular genetics of clavulanic acid. Antonie van Leeuwenhoek, 75(1/2), 125–133. [DOI] [PubMed] [Google Scholar]
- Jensen S. E., Paradkar A. S., Mosher R. H., Anders C., Beatty P. H., Brumlik M. J, Griffin A., Barton B. (2004). Five additional genes are involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. Antimicrobial Agents and Chemotherapy, 48(1), 192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S. E. (2012). Biosynthesis of clavam metabolites. Journal of Industrial Microbiology and Biotechnology, 39(10), 1407–1419. [DOI] [PubMed] [Google Scholar]
- Jin W., Ryu Y. G., Kang S. G., Kim S. K., Saito N., Ochi K., Lee S. H., Lee K. J. (2004). Two relA/spoT homologous genes are involved in the morphological and physiological differentiation of Streptomyces clavuligerus. Microbiology (Reading), 150(5), 1485–1493. [DOI] [PubMed] [Google Scholar]
- Karuppiah V., Ranaghan K. E., Leferink N. G. H., Johannissen L. O., Shanmugam M., Cheallaigh N. A., Bennett N. J., Kearsey L. J., Takano E., Gardiner J. M., van der Kamp M. W., Hay S., Mulholland A. J, Leys D., Scrutton N. S. (2017). Structural basis of catalysis in the bacterial monoterpene synthases linalool synthase and 1,8-cineole synthase. ACS Catalysis, 7(9), 6268–6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenig M., Reading C. (1979). Holomycin and an antibiotic (MM 19290) related to tunicamycin, metabolites of Streptomyces clavuligerus. Journal of Antibiotics (Tokyo), 32(6), 549–554. [DOI] [PubMed] [Google Scholar]
- Kim H.S., Lee Y. J., Lee C. K., Choi S. U., Yeo S-H., Hwang Y. I., Yu T. S, Kinoshita H., Nihira T. (2004). Cloning and characterization of a gene encoding the gamma-butyrolactone autoregulator receptor from Streptomyces clavuligerus. Archives in Microbiology, 182(1), 44–50. [DOI] [PubMed] [Google Scholar]
- Kurt A., Álvarez-Álvarez R., Liras P., Özcengiz G. (2013). Role of the cmcH-ccaR intergenic region and ccaR overexpression in cephamycin C biosynthesis in Streptomyces clavuligerus. Applied Microbiology and Biotechnology, 97(13), 5869–5880. [DOI] [PubMed] [Google Scholar]
- Kurt-Kizildoğan A., Jaccard V., Mutlu A., Sertdemir I., Özcengiz G. (2017). Genetic engineering of an industrial strain of Streptomyces clavuligerus for further enhancement of clavulanic acid production. Turkist Journal of Biology, 41, 342–353. [Google Scholar]
- Kwong T., Zelyas N. J., Cai H., Tahlan K., Wong A., Jensen S. E. (2012). 5S clavam biosynthesis is controlled by an atypical two-component regulatory system in Streptomyces clavuligerus. Antimicrobial Agents and Chemotherapy, 56(9), 4845–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Walsh C. T. (2010). Identification of the gene cluster for the dithiolopyrrolone antibiotic holomycin in Streptomyces clavuligerus. Proceeding of the National Academy of Science USA, 107(46), 19731–19735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Walsh C. T. (2011). Streptomyces clavuligerus HlmI is an intramolecular disulfide-forming dithiol oxidase in holomycin biosynthesis. Biochemistry, 50(21), 4615–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.-, Wever W. J., Walsh C. T., Bowers A. A. (2014). Dithiolopyrrolones: Biosynthesis, synthesis, and activity of a unique class of disulfide-containing antibiotics. Natural Product Reports, 31(7), 905–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Khaleeli N., Townsend C. A. (2000). Expansion of the clavulanic acid gene cluster: Identification and in vivo functional analysis of three new genes required for biosynthesis of clavulanic acid by Streptomyces clavuligerus. Journal of Bacteriology, 182(14), 4087–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Townsend C. A. (2006). Rational strain improvement for enhanced clavulanic acid production by genetic engineering of the glycolytic pathway in Streptomyces clavuligerus. Metabolic Engineering, 8(3), 240–252. [DOI] [PubMed] [Google Scholar]
- Liras P. (1999). Biosynthesis and molecular genetics of cephamycins. Antonie van Leeuwenhoek, 75(1/2), 109–124. [DOI] [PubMed] [Google Scholar]
- Liras P., Rodríguez-García A. (2000). Clavulanic acid, a beta-lactamase inhibitor: biosynthesis and molecular genetics. Applied Microbiology and Biotechnology, 54(4), 467–475. [DOI] [PubMed] [Google Scholar]
- Liras P., Gomez-Escribano J. P., Santamarta I. (2008). Regulatory mechanisms controlling antibiotic production in Streptomyces clavuligerus. Journal of Industrial Microbiology & Biotechnology, 35(7), 667–676. [DOI] [PubMed] [Google Scholar]
- Liras P., Demain A. L. (2009). Enzymology of β-lactam compounds with cephem structure produced by actinomycetes. In D. A. Hopwood (Ed.), Complex enzymes in microbial natural product biosynthesis, methods in enzymology (Vol. 458, pp. 401–429). Burlington Academic Press; [DOI] [PubMed] [Google Scholar]
- Liras P., Santamarta I., Pérez-Redondo R. (2011). Clavulanic acid and clavams biosynthesis and regulation. In Dyson P. (Ed.), Streptomyces: Molecular biology and biotechnology (pp. 167–178). Caister Acad. Press. [Google Scholar]
- Liras P. (2014). Holomycin, a dithiolopyrrolone compound produced by Streptomyces clavuligerus. Applied Microbiology and Biotechnology, 98(3), 1023–1030. [DOI] [PubMed] [Google Scholar]
- López-Agudelo V. A., Gómez-Ríos D., Ramírez-Malule H. (2021). Clavulanic acid production by Streptomyces clavuligerus: Insights from systems biology, strain engineering, and downstream processing. Antibiotics (Basel), 10(1), 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García M. T., Santamarta I., Liras P. (2010). Morphological differentiation and clavulanic acid formation are affected in a Streptomyces clavuligerus adpA-deleted mutant. Microbiology (Reading), 156(8), 2354–2365. [DOI] [PubMed] [Google Scholar]
- Lorenzana L. M., Pérez-Redondo R., Santamarta I., Martín J. F., Liras P. (2004). Two oligopeptide-permease-encoding genes in the clavulanic acid cluster of Streptomyces clavuligerus are essential for production of the beta-lactamase inhibitor. Journal of Bacteriology, 186(11), 3431–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie A. K., Valegård K., Iqbal A., Caines M. E. C., Kershaw N. J., Jensen S. E., Schofield C. J., Andersson I. (2010). Crystal structures of an oligopeptide-binding protein from the biosynthetic pathway of the beta-lactamase inhibitor clavulanic acid. Journal of Molecular Biology, 396(2), 332–344. [DOI] [PubMed] [Google Scholar]
- Martínez-Burgo Y., Álvarez-Álvarez R., Pérez-Redondo R., Liras P. (2014). Heterologous expression of Streptomyces clavuligerus ATCC 27064 cephamycin C gene cluster. Journal of Biotechnology, 186, 21–29. [DOI] [PubMed] [Google Scholar]
- Martínez-Burgo Y., Álvarez-Álvarez R., Rodríguez-García A., Liras P. (2015). The pathway-specific regulator ClaR of Streptomyces clavuligerus has a global effect on the expression of genes for secondary metabolism and differentiation. Applied and Environmental Microbiology, 81(19), 6637–6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Burgo Y., Santos-Aberturas J., Rodríguez-García A., Barreales E. G., Tormo J. R., Truman A. W., Reyes F., Aparicio J. F., Liras P. (2019). Activation of secondary metabolite gene clusters in Streptomyces clavuligerus by the PimM regulator of Streptomyces natalensis. Frontiers in Microbiology, 10, 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema M. H., Trefzer A., Kovalchuk A., van den Berg M., Müller U., Heijne W., Wu L., Alam M. T., Ronning C. M., Nierman W. C., Bovenberg R. A. L., Breitling R., Takano E. (2010). The sequence of a 1.8-Mb bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome Biology and Evolution, 2, 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema M. H., Alam M. T., Heijne W. H., van den Berg M. A., Müller U., Trefzer A., Bovenberg R. A. L., Breitling R., Takano E. (2011). Genome-wide gene expression changes in an industrial clavulanic acid overproduction strain of Streptomyces clavuligerus. Microbial Biotechnology, 4(2), 300–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo S., Ban Y. H., Park J. W., Yoo Y. J., Yoon Y. J. (2009). Enhanced FK506 production in Streptomyces clavuligerus CKD1119 by engineering the supply of methylmalonyl-CoA precursor. Journal of Industrial Microbiology & Biotechnology, 36(12), 1473–1482. [DOI] [PubMed] [Google Scholar]
- Nakano C., Hyo-Kyoung Kim H-K., Ohnishi Y. (2011a). Identification of the first bacterial monoterpene cyclase, a 1,8-cineole synthase, that catalyzes the direct conversion of geranyl diphosphate. Chembiochem, 12(13), 1988–1991. [DOI] [PubMed] [Google Scholar]
- Nakano C., Kim H. K., Ohnishi Y. (2011b). Identification and characterization of the linalool/nerolidol synthase from Streptomyces clavuligerus. Chembiochem, 12(16), 2403–2407. [DOI] [PubMed] [Google Scholar]
- Nárdiz N., Santamarta I., Lorenzana L. M., Martín J. F., Liras P. (2011). A rhodanese-like protein is highly overrepresented in the mutant S. clavuligerus oppA2::aph: Effect on holomycin and other secondary metabolites production. Microbial Biotechnology, 4(2), 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netolitzky D. J., Wu X., Jensen S. E., Roy K. L. (1995). Giant linear plasmids of beta-lactam antibiotic producing Streptomyces. FEMS Microbiology Letters, 131, 27–34. [DOI] [PubMed] [Google Scholar]
- Nishida H., Ohnishi Y., Beppu T., Horinouchi S. (2007). Evolution of gamma-butyrolactone synthases and receptors in Streptomyces. Environmental Microbiology, 9(8), 1986–1994. [DOI] [PubMed] [Google Scholar]
- Nobary G. S., Jensen S. E. (2012). A comparison of the clavam biosynthetic genec clusters in Streptomyces antibioticus Tu1718 and Streptomyces clavuligerus. Canadian Journal of Microbiology, 58(4), 413–425. [DOI] [PubMed] [Google Scholar]
- Ochi K. (1990). A relaxed (rel) mutant of Streptomyces coelicolor A3(2) with a missing ribosomal protein lacks the ability to accumulate ppGpp, A-factor and prodigiosin. Journal General of Microbiology, 136(12), 2405–2412. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y., Yamazaki H., Kato J. Y., Tomono A., Horinouchi S. (2005). AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Bioscience Biotechnology and Biochemistry, 69, 431–439. [DOI] [PubMed] [Google Scholar]
- Okamura K., Soga K., Shimauchi Y., Ishikura T., Lein J. (1977). Holomycin and N-propionylholothin, antibiotics produced by a cephamycin C producer. Journal of Antibiotics (Tokyo), 30(4), 334–336. [DOI] [PubMed] [Google Scholar]
- Olano C., García I., González A., Rodríguez M., Rozas D., Rubio J., Sánchez-Hidalg M., Braña A. F., Mendez C., Salas J. A. (2014). Activation and identification of five clusters for secondary metabolites in Streptomyces albus J1074. Microbial Biotechnology, 7(3), 242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordóñez-Robles M., Santos-Beneit F., Martín J. F. (2018). Unraveling nutritional regulation of tacrolimus biosynthesis in Streptomyces tsukubaensis through omic approaches. Antibiotics, 7(2), 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öster L. M., Lester D. R., Terwisscha van Scheltinga A., Svenda M., Lun van M., Généreux C., Andersson I. (2006). Insights into cephamycin biosynthesis: The crystal structure of CmcI from Streptomyces clavuligerus. Journal of Molecular Biology, 358(2), 546–558. [DOI] [PubMed] [Google Scholar]
- Ostash B., Doud E. H., Lin C., Ostash I., Perlstein D. L., Fuse S., Wolpert M., Kahne D., Walker S. (2009). Complete characterization of the seventeen step moenomycin biosynthetic pathway. Biochemistry, 48(37), 8830–8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Liu G., Yang H., Tian Y., Tan H. (2009). The pleiotropic regulator AdpA-L directly controls the pathway-specific activator of nikkomycin biosynthesis in Streptomyces ansochromogen. Molecular Microbiology, 72(3), 710–723. [DOI] [PubMed] [Google Scholar]
- Paradkar A. S., Aidoo K. A., Jensen S. E. (1998). A pathway-specific transcriptional activator regulates late steps of clavulanic acid biosynthesis in Streptomyces clavuligerus. Molecular Microbiology, 27(4), 831–843. [DOI] [PubMed] [Google Scholar]
- Paradkar A. (2013). Clavulanic acid production by Streptomyces clavuligerus: Biogenesis, regulation and strain improvement. Journal of Antibiotics, 66(7), 411–420. [DOI] [PubMed] [Google Scholar]
- Park J. W., Mo S-J., Park S. R., Ban Y. H., Yoo Y. J., Yoon Y. J. (2009). Liquid chromatography–mass spectrometry characterization of FK506 biosynthetic intermediates in Streptomyces clavuligerus KCTC 10561BP. Analytical Biochemistry, 393(1), 1–7. [DOI] [PubMed] [Google Scholar]
- Pérez-Redondo R., Santamarta I., Bovenberg R., Martín J. F., Liras P. (2010). The enigmatic lack of glucose utilization in Streptomyces clavuligerus is due to inefficient expression of the glucose permease gene. Microbiology (Reading), 156(5), 1527–1537. [DOI] [PubMed] [Google Scholar]
- Pérez-Redondo R., Rodríguez-García A., Botas A., Santamarta I., Martín J. F., Liras P. (2012). ArgR of Streptomyces coelicolor is a versatile regulator. Plos One, 7(3), e32697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Redondo R., Rodríguez-García A., Martín J. F, Liras P. (1998). The claR gene of Streptomyces clavuligerus, encoding a LysR-type regulatory protein controlling clavulanic acid biosynthesis, is linked to the clavulanate-9-aldehyde reductase (car) gene. Gene, 211(2), 311–321. [DOI] [PubMed] [Google Scholar]
- Pinilla L., Toro L. F., Laing E., Alzate J. F., Ríos-Estepa R. (2019). Comparative transcriptome analysis of Streptomyces clavuligerus in response to favorable and restrictive nutritional conditions. Antibiotics (Basel), 8(3), 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price N. P., Tsvetanova B. (2007). Biosynthesis of the tunicamycins: A review. J Antibiot (Tokyo), 60(8), 485–491. [DOI] [PubMed] [Google Scholar]
- Qin Z., Huang S., Yu Y., Deng H. (2013). Dithiolopyrrolone natural products: Isolation, synthesis and biosynthesis. Marine Drugs, 11(10), 3970–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkel J., Rabe P., Zur Horst L., Dickschat J. S. (2016). A detailed view on 1,8-cineol biosynthesis by Streptomyces clavuligerus. Beilstein Journal of Organic Chemistry, 12, 2317–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles-Reglero V., Santamarta I., Álvarez-Álvarez R., Martín J. F., Liras P. (2013). Transcriptional analysis and proteomics of the holomycin gene cluster in overproducer mutants of Streptomyces clavuligerus. Journal of Biotechnology, 163(1), 69–76. [DOI] [PubMed] [Google Scholar]
- Rodríguez-García A., Fuente de la A., Pérez-Redondo R., Martín J. F., Liras P. (2000). Characterization and expression of the arginine biosynthesis gene cluster of Streptomyces clavuligerus. Journal of Molecular Microbiology and Biotechnology, 2, 543–550. [PubMed] [Google Scholar]
- Santamarta I., Pérez-Redondo R., Lorenzana L. M., Martín J. F., Liras P. (2005). Different proteins bind to the butyrolactone receptor protein ARE sequence located upstream of the regulatory ccaR gene of Streptomyces clavuligerus. Molecular Microbiology, 56(3), 824–835. [DOI] [PubMed] [Google Scholar]
- Santamarta I., López-García M. T., Pérez-Redondo R., Koekman B., Martín J. F., Liras P. (2007). Connecting primary and secondary metabolism: AreB, an IclR-like protein, binds the AREccaR sequence of S. clavuligerus and modulates leucine biosynthesis and cephamycin C and clavulanic acid production. Molecular Microbiology, 66(2), 511–524. [DOI] [PubMed] [Google Scholar]
- Santamarta I., López-García M. T., Kurt A., Nárdiz N., Álvarez-Álvarez R., Pérez-Redondo R., Martín J. F., Liras P. (2011). Characterization of DNA-binding sequences for CcaR in the cephamycin-clavulanic acid supercluster of Streptomyces clavuligerus. Molecular Microbiology, 81(4), 968–981. [DOI] [PubMed] [Google Scholar]
- Saudagar P. S., Survase S. A., Singhal R. S. (2008). Clavulanic acid: A review. Biotechnology Advances, 26(4), 335–351. [DOI] [PubMed] [Google Scholar]
- Shaikh A. A., Nothias L. F., Srivastava S. K., Dorrestein P. C., Tahlan K. (2021). Specialized metabolites from ribosome engineered strains of S. clavuligerus. Metabolites, 11(4), 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C-H., Cho H. S., Won H-J., Kwon H. J., Kim C-W., Yoon Y. J. (2021). Enhanced production of clavulanic acid by improving glycerol utilization using reporter-guided mutagenesis of an industrail. Streptomyces clavuligerus strain. J. Ind. Microb. Biotechnol, 48, (3-4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. Y., Jeong H., Yu D. S., Fischbach M. A., Park H. S., Kim J. J., Seo J-S., Jensen S. E., Oh T. K., Lee K. J., Kim J. F. (2010). Draft genome sequence of Streptomyces clavuligerus NRRL 3585, a producer of diverse secondary metabolites. Journal of Bacteriology, 192(23), 6317–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. Y., Kim E. S., Kim D. W., Jensen S. E., Lee K. J. (2009). A gene located downstream of the clavulanic acid gene cluster in Streptomyces clavuligerus ATCC 27064 encodes a putative response regulator that affects clavulanic acid production. Journal of Industrial Microbiology and Biotechnology, 36(2), 301–311. [DOI] [PubMed] [Google Scholar]
- Srivastava S. K., King K. S., AbuSara N. F., Malayny C. J., Piercey B. M., Wilson J. A., Tahlan K. (2019). In vivo functional analysis of a class A β-lactamase-related protein essential for clavulanic acid biosynthesis in Streptomyces clavuligerus. Plos One, 14(4), e0215960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidjani A-R., Bontemps C., Leblond P. (2020). Telomeric and sub-telomeric regions undergo rapid turnover within a Streptomyces population. Scientific Reports, 10(1), 7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahlan K., Anders C., Wong A., Mosher R. H., Beatty P. H., Brumlik M. J., Griffin A., Hughes C., Griffin J., Barton B., Jensen S. E. (2007). 5S clavam biosynthetic genes are located in both the clavam and paralog gene clusters in Streptomyces clavuligerus. Chemistry and Biology, 14(2), 131–142. [DOI] [PubMed] [Google Scholar]
- Tahlan K., Anders C., Jensen S. E. (2004). The paralogous pairs of genes involved in clavulanic acid and clavam metabolite biosynthesis are differently regulated in Streptomyces clavuligerus. Journal of Bacteriology, 186(18), 6286–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetanova B. C., Price N. P. (2001). Liquid chromatography-electrospray mass spectrometry of tunicamycin-type antibiotics. Analytical Biochemistry, 289(2), 147–156. [DOI] [PubMed] [Google Scholar]
- Tomono A., Tsai Y., Yamazaki H., Ohnishi Y., Horinouchi S. (2005). Transcriptional control by A-factor of strR, the pathway-specific transcriptional activator for streptomycin biosynthesis in Streptomyces griseus. Journal of Bacteriology, 187(16), 5595–5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend C. A. (2002). New reactions in clavulanic acid biosynthesis. Current Opinion in Chemical Biology, 6(5), 583–589. [DOI] [PubMed] [Google Scholar]
- Tsvetanova B. C., Price N. P. (2001). Liquid chromatography-electrospray mass spectrometry of tunicamycin-type antibiotics. Analytical Biochemistry, 289(2), 147–156. [DOI] [PubMed] [Google Scholar]
- Ünsaldi E., Kurt-Kizildoğan A., Voigt B., Becher D., Özcengiz G. (2017). Proteoma-wide alterations in an industrial clavulanic acid strain of Streptomyces clavuligerus. Synthetic and Systems Biotechnology, 2(1), 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ünsaldi E., Kurt-Kizildoğan A., Özcan S., Becher D., Voigt B., Aktaş C., Özcengiz G. (2021). Proteomic analysis of a hom-disrupted, cephamycin C overproducing Streptomyces clavuligerus. Protein and Peptide Letters, 28(2), 205–220. [DOI] [PubMed] [Google Scholar]
- Valegård K., Iqbal A., Kershaw N. J., Ivison D., Généreux C., Dubus A., Blikstad C., Demetriades M., Hopkinson R. J., Lloyd A. J., Roper D. I., Scofield C. J., Andersson I., McDonough M. A. (2013). Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway. Acta Crystallography. D Biologica. Crystallography, 69(8), 1567–1579. [DOI] [PubMed] [Google Scholar]
- Wang L., Tahlan K., Kaziuk T. L., Alexander D. C., Jensen S. E. (2004). Transcriptional and translational analysis of the ccaR gene from Streptomyces clavuligerus. Microbiology (Reading), 150(12), 4137–4145. [DOI] [PubMed] [Google Scholar]
- Wang X., Wu J., Chen J., Xiao L., Zhang Y., Wang F., Li X. (2020). Efficient biosynthesis of R-(-) linalool through adjusting the expression strategy and increasing GPP supply in Escherichia coli. Journal of Agricultural Food Chemistry, 68(31), 8381–8390. [DOI] [PubMed] [Google Scholar]
- Ward J. M., Hodgson J. E. (1993). The biosynthetic genes for clavulanic acid and cephamycin production occur as a ‘super-cluster’ in three Streptomyces. FEMS Microbiology Letters, 110(2), 239–242. [DOI] [PubMed] [Google Scholar]
- Wojdyla Z., Borowski T. (2021). Enzyme multifunctionality by control of substrate positioning within the catalytic cycle-A QM/MM study of clavaminic acid synthase. Chemistry, 27(6), 2196–2211. [DOI] [PubMed] [Google Scholar]
- Wu X., Roy K. L. (1993). Complete nucleotide sequence of a linear plasmid from Streptomyces clavuligerus and characterization of its RNA transcripts. Journal of Bacteriology, 175(1), 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Leblanc S. K. D., Piktel J., Jensen S. E., Roy K. L. (2006). Prediction and functional analysis of the replication origin of the linear plasmid pSCL2 in Streptomyces clavuligerus. Canadian Journal of Microbiology, 52(4), 293–300. [DOI] [PubMed] [Google Scholar]
- Yamada Y., Kuzuyama T., Komatsu M., Shin-ya K., Omura S., Cane D. E., Ikeda H. (2015a). Terpene synthases and widely distributed in bacteria. Proceedings of the National Academy of Science USA, 112(3), 857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Arima S., Nagamitsu T., Johmoto K., Uekusa H., Eguchi T., Shin-ya K., Cane D. E., Ikeda H. (2015b). Novel terpenes generated by heterologous expression of bacterial terpene synthase genes in an engineered Streptomyces host. Journal of Antibiotics (Tokyo), 68(6), 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Komatsu M., Ikeda H. (2016). Chemical diversity of labdane-type bicyclic diterpene biosynthesis in actinomycetales microorganism. Antibiotics, 69(7), 515–523. [DOI] [PubMed] [Google Scholar]
- Yang Y-H., Song E., Kim E-J., Lee K., Kim W-S., Park S-S., Hahn J-S., Kim B-G. (2009). NdgR, an IclR-like regulator involved in amino-acid-dependent growth, quorum sensing, and antibiotic production in Streptomyces coelicolor. Applied Microbiology and Biotechnology, 82(3), 501–511. [DOI] [PubMed] [Google Scholar]
- Zhang W., Fortman J. L., Carlson J. C., Yan J., Liu Y., Bai F., Guan W., Jia J., Matainaho T., Sherman D. H., Li S. (2013). Characterization of the bafilomycin gene cluster from Streptomyces lohii. Chembiochem, 14(3), 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.