ABSTRACT
Cyclic dimeric GMP (c-di-GMP) is a universal second messenger in bacteria. A large number of c-di-GMP-related diguanylate cyclases (DGCs), phosphodiesterases (PDEs), and effectors are responsible for the complexity and dynamics of c-di-GMP signaling. Some of these components employ various methods to avoid undesired cross talk to maintain signaling specificity. The synthesis of the antibiotic HSAF (heat-stable antifungal factor) in Lysobacter enzymogenes is regulated by a specific c-di-GMP signaling pathway that includes a PDE, LchP, and a c-di-GMP effector, Clp (also a transcriptional regulator). In the present study, from among 19 DGCs, we identified a diguanylate cyclase, LchD, that participates in this pathway. Subsequent investigation indicates that LchD and LchP physically interact and that the catalytic center of LchD is required for both the formation of the LchD-LchP complex and HSAF production. All the detected phenotypes support that LchD and LchP display local c-di-GMP signaling to regulate HSAF biosynthesis. Although direct evidence is lacking, our investigation, which shows that the interaction between a DGC and a PDE maintains the specificity of c-di-GMP signaling, suggests the possibility of the existence of local c-di-GMP pools in bacteria.
IMPORTANCE Cyclic dimeric GMP (c-di-GMP) is a universal second messenger in bacteria. The signaling of c-di-GMP is complex and dynamic, and it is mediated by a large number of components, including c-di-GMP synthases (diguanylate cyclases [DGCs]), c-di-GMP-degrading enzymes (phosphodiesterases [PDEs]), and c-di-GMP effectors. These components deploy various methods to avoid undesired cross talk to maintain signaling specificity. In the present study, we identified a DGC that interacted with a PDE to specifically regulate antibiotic biosynthesis in L. enzymogenes. We provide direct evidence to show that the DGC and PDE form a complex and also indirect evidence to argue that they may balance a local c-di-GMP pool to control antibiotic production. These results represent an important finding regarding the mechanism of a DGC and PDE pair to control the expression of specific c-di-GMP signaling pathways.
KEYWORDS: c-di-GMP, signaling specificity, local c-di-GMP signaling, HSAF, diguanylate cyclase
INTRODUCTION
Cyclic dimeric GMP (c-di-GMP), first discovered by Ross and colleagues in the mid-1980s (1), was recently shown to participate in biofilm formation, pathogenicity, the cell cycle, secondary metabolite biosynthesis, and other processes and is recognized as a ubiquitous nucleotide second messenger in bacteria (1–5). The synthesis and hydrolysis of c-di-GMP are carried out by diguanylate cyclases (DGCs) and c-di-GMP-specific phosphodiesterases (PDEs), respectively (3). DGCs, which possess a GGDEF domain, catalyze the condensation of two GTP molecules into a c-di-GMP molecule (6, 7). PDEs, which possess either an EAL or an HD-GYP domain, hydrolyze c-di-GMP into a 5′-pGpG molecule or two GMP molecules (8–11). Generally, the concentration of c-di-GMP is determined by DGCs and PDEs, and downstream effectors receive the c-di-GMP signal and generate a response. The c-di-GMP signaling pathway was considered simple until genomic analyses showed that numbers of DGCs and PDEs vary from a few to dozens in various bacteria (12). Additionally, the similarity between c-di-GMP effectors that have been identified so far, such as PilZ, GIL, and MshEN domains or inhibitory sites (I-sites) of the GGDEF domain (13–16), is limited, indicating that the c-di-GMP signaling network is large and complex.
Despite the number of c-di-GMP-related components at work, many researchers have found that several DGCs and PDEs seem to specifically participate in one or two c-di-GMP-dependent processes (17, 18). Meanwhile, more and more studies have demonstrated that physical interactions can occur among DGCs, PDEs, and particular c-di-GMP effectors (19–21). Paul and colleagues proposed that the local action of a DGC, PleD, might constitute a general regulatory principle in bacterial growth when they were studying the pole development and cell cycle of Caulobacter crescentus (7), and the proposal was then discussed in a review by Römling et al. (22), thus leading to the development of the concept of local c-di-GMP signaling or c-di-GMP signaling specificity.
Lysobacter enzymogenes is a soilborne, nonpathogenic, Gram-negative bacterium that can secrete the secondary metabolite HSAF (heat-stable antifungal factor), which has potent broad-spectrum antifungal activity targeting the sphingolipid biosynthesis pathway of filamentous pathogens (23–25). Therefore, we focused on the regulatory mechanism of the HSAF biosynthesis pathway, aiming to enhance the production of HSAF for use in fungal disease control as an antifungal antibiotic.
Our previous study showed that high levels of c-di-GMP inhibit HSAF production (26). A further study investigated the role of a specific c-di-GMP-dependent regulatory pathway in HSAF biosynthesis (27). In this signaling pathway, LchP, acting as a c-di-GMP PDE, and the transcriptional regulator Clp, acting as a c-di-GMP effector, physically interact (27). When LchP is activated via as-yet-unknown factors, it actively degrades c-di-GMP from the Clp–c-di-GMP complex, which leads to the release of Clp and the activation of the HSAF biosynthesis operon (27). The Clp-LchP interaction increases the PDE activity of LchP, providing positive feedback. However, our previous study described an unusual phenotype: lchP gene deletion resulted in only a modest increase in the intracellular c-di-GMP concentration but had a large effect on HSAF production (27). We thus aimed to investigate other factors in addition to the LchP-Clp interaction that may contribute to the maintenance of signal specificity.
In the present work, we used indirect experiments to demonstrate that LchP does not affect the global c-di-GMP concentration. Next, we identified a partner of LchP, the DGC LchD (Lysobacter c-di-GMP and HSAF-related diguanylate cyclase), from among 19 DGCs in L. enzymogenes OH11 using genetic methods. We further demonstrated that LchP and LchD physically interact and that the catalytic center of LchD is important for the LchP-LchD interaction and HSAF production. The present findings not only complement previous studies on the role of the specific c-di-GMP signaling pathway in regulating the secondary metabolite HSAF but also suggest the possibility of the existence of local c-di-GMP pools in bacteria.
RESULTS
LchP does not regulate the global c-di-GMP pool.
Our previous study reported LchP as a weak PDE that regulates HSAF biosynthesis (27). To fully characterize the function of LchP, we used transcriptome sequencing (RNA-seq) to identify differentially expressed genes in the lchP mutant strain. Only 16 genes were differentially expressed (log2 ratio of greater than 2 or less than −2) in the lchP mutant strain compared to the wild-type strain, including 1 upregulated gene and 15 downregulated genes (Fig. 1A; see also Table S1 in the supplemental material). Nine of these genes belong to the HSAF biosynthesis gene cluster, and 4 have unknown functions (Fig. 1A and B). To confirm the RNA-seq data, we used quantitative real-time PCR (qRT-PCR) to determine the transcription level of the key HSAF biosynthesis gene lafB (Fig. 1C). The qRT-PCR results were consistent with the RNA-seq data (Fig. 1D). These results support the previous finding that LchP specifically regulates HSAF biosynthesis (27). But previous RNA-seq data showed that 775 genes, belonging to 19 functional categories, were differentially expressed (showing a ≥2-fold change) between LchP’s downstream c-di-GMP effector Clp mutant strain and the wild-type strain (28), which indicates that LchP may not control the global c-di-GMP pool. Because LchP is an inner membrane-anchored protein and functions as a PDE, we hypothesized that a DGC may participate in the specific c-di-GMP signaling pathway to precisely balance the c-di-GMP concentration.
FIG 1.
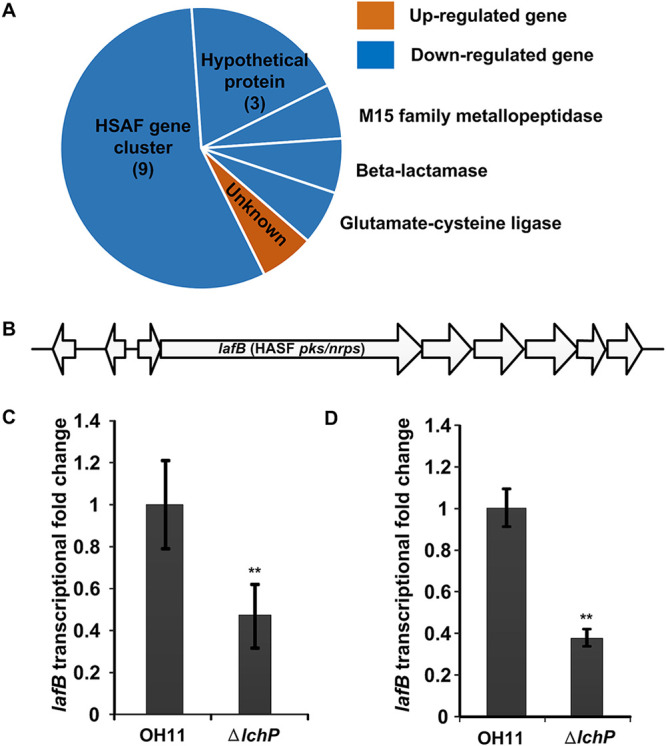
RNA-seq data indicate that LchP does not regulate the global c-di-GMP concentration. (A) Sixteen genes were differentially expressed in the lchP deletion mutant. Among these genes were 1 upregulated gene and 15 downregulated genes. (B) lafB, a key gene in the HSAF biosynthesis gene cluster, was downregulated in the lchP mutant. (C) qRT-PCR analyses of lafB mRNA levels in the wild-type and lchP mutant ΔlchP strains. The qRT-PCR data were obtained from three biological experiments with three technical replicates. The lafB mRNA level in the wild-type OH11 strain was assumed to be 1. Statistical comparisons were performed with GraphPad software (GraphPad, La Jolla, CA) using one-way ANOVA. The error bars indicate standard errors. **, P < 0.01 relative to wild-type OH11. (D) Transcription levels (RNA-seq data) of lafB in the ΔlchP and wild-type strains. The lafB mRNA level in the wild-type OH11 strain was assumed to be 1. Statistical comparisons were performed with GraphPad software (GraphPad, La Jolla, CA) using one-way ANOVA. The error bars indicate standard errors. **, P < 0.01 relative to wild-type OH11.
Identification of the partner DGC of LchP involved in regulating HSAF synthesis.
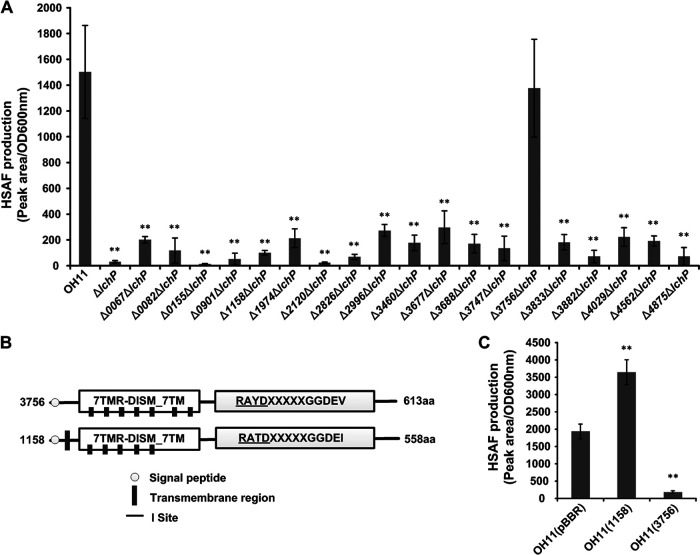
The numbers of DGCs in bacteria vary from several to dozens (12, 29). Studies have shown that some DGCs participate in only one or two processes and coordinate their functions without excessive cross talk (17, 18). L. enzymogenes OH11 has 14 single GGDEF domain-containing proteins and 6 GGDEF-EAL hybrid proteins (30). Here, we hypothesized that the deficiency in HSAF production in the lchP mutant is caused by an increase in the c-di-GMP concentration. Thus, we aimed to determine whether the c-di-GMP level would not be increased by halting c-di-GMP synthesis and whether this manipulation would restore HSAF production. We used a genetic approach to identify the DGC that cooperates with LchP in the regulation of HSAF production. We first deleted 19 DGC-encoding genes on the background of the lchP mutant to generate 19 double mutants and then measured HSAF production in these mutants. The results in Fig. 2A show that only the 3756 and lchP double mutant produced HSAF at a level identical to that in the wild-type strain, while knockouts of the other 18 genes did not restore HSAF production or had only a small amount of recovery. Notably, both 3756 and 1158 contain the 7TMR-DISM_7TM and GGDEF domains (Fig. 2B), and multiple-sequence alignment demonstrated that they share 55% amino acid sequence similarity (Fig. S1); however, mutation of 1158 did not restore HSAF production. Additionally, the 7TMR-DISM_7TM domain is a multi-intracellular signal receptor with 7 transmembrane regions (31), indicating that 3756 is also a membrane-anchored protein with a specific location. These data suggested that 3756 is probably the DGC that partners with LchP to regulate HSAF. In another experiment, when we overexpressed 3756 in the wild-type background, HSAF production was significantly reduced (Fig. 2C), confirming our hypothesis in which an elevated c-di-GMP concentration inhibits HSAF production. However, the overexpression of 1158 increased HSAF production, indicating that 1158 may employ other mechanisms to regulate HSAF production (Fig. 2C). We renamed 3756 LchD (Lysobacter c-di-GMP- and HSAF-related DGC).
FIG 2.
Identification of the partner DGC of LchP. (A) Quantification of HSAF production in the double mutants of 19 DGCs based on the lchP mutant. Statistical comparisons were performed with GraphPad software (GraphPad, La Jolla, CA) using one-way ANOVA. **, P < 0.01 relative to the wild-type OH11 strain. Biological experiments for each treatment were performed three times and assayed in triplicate. (B) Proteins 3756 and 1158 share the same domain organization. (C) Quantification of HSAF production by the overexpression of 3756 and 1158. The error bars indicate standard errors. Statistical comparisons were performed with GraphPad software (GraphPad, La Jolla, CA) using one-way ANOVA. **, P < 0.01 relative to the wild-type OH11 strain containing the original pBBR-MCS5 vector. Biological experiments for each treatment were performed three times and assayed in triplicate.
LchD physically interacts with LchP.
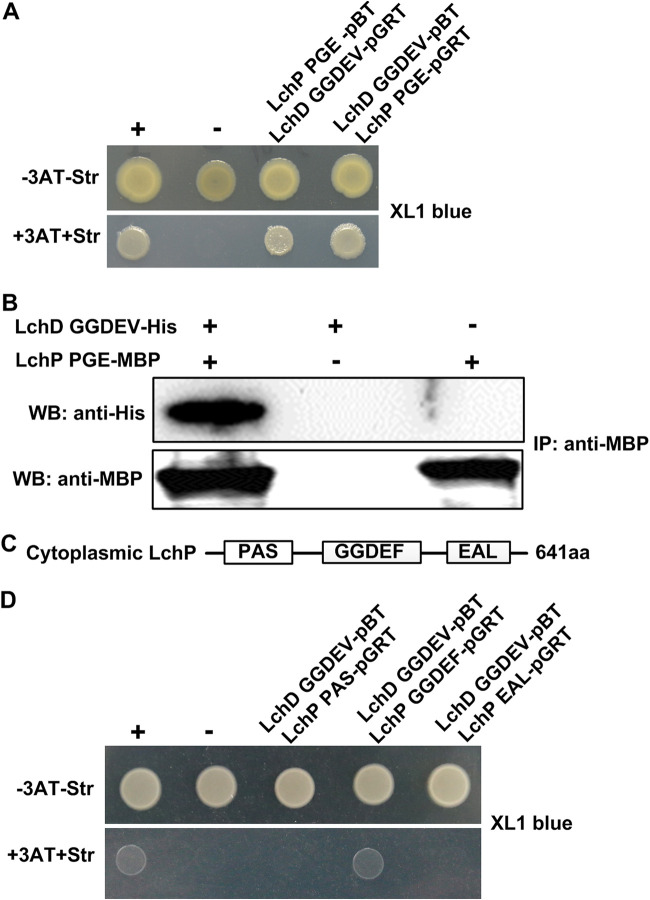
We next aimed to determine why LchD, but not the similar protein 1158, is the partner of LchP. Furthermore, we wondered whether there is a connection between LchP and LchD. Do these enzymes interact? To assess these questions, we used a bacterial two-hybrid (B2H) system. In this system, the interaction of two target proteins activates the transcription of the HIS3 reporter gene, which allows growth in the presence of 3-amino-1,2,4-triazole (3-AT), a competitive inhibitor of the His3 enzyme. Positive results are verified by using the aadA gene, which confers streptomycin (Str) resistance, as a secondary reporter (32). The results in Fig. 3A show that the cytoplasmic regions of LchP and LchD interact. This interaction was further confirmed via pulldown of LchD-GGDEV-His using purified LchP-PGE-maltose binding protein (MBP) (27), which produced a positive signal (Fig. 3B). Both experiments strongly suggested that the cytoplasmic regions of LchP and LchD physically interact. To evaluate whether c-di-GMP is involved in the LchP-LchD interaction, we first detected it by the B2H system in the XL1-Blue yhjH mutant, which is considered to be a high-c-di-GMP-level background strain (15), and obtained results similar to those shown in Fig. 3A (Fig. S2A). More directly, we used a pulldown assay to compare the interactions between LchP and LchD in the presence and absence of c-di-GMP and found no difference (Fig. S2B). Considering that LchP has 3 cytoplasmic domains (Fig. 3C), to determine the details of this interaction, we further investigated which domain of LchP is involved in binding LchD-GGDEV with the B2H system. As shown in Fig. 3D, the two GGDEF domains interact. We also used the B2H system to determine whether the cytoplasmic regions of LchP and 1158 interacted; however, a negative result was obtained (Fig. S3). These results indicate that the LchP-LchD interaction is specific and may explain why only LchD participates in HSAF production in the absence of LchP, as shown in Fig. 2A.
FIG 3.
LchD physically interacts with LchP. (A) An E. coli-based B2H assay shows that the cytoplasmic regions of LchD and LchP interact. +, positive control (GacS-pBT and GacS-pTRG); −, negative control (vectors pBT and pTRG). (B) Diagram of LchP fragments. (C) Confirmation of the LchD and LchP interaction by a pulldown assay. The immunoprecipitation (IP) assay was carried out using anti-MBP antibody. Western blotting (WB) was performed by using anti-His and anti-MBP antibodies. (D) The B2H assay shows that the GGDEF domains of LchP and LchD interact. +, positive control (GacS-pBT and GacS-pTRG); −, negative control (vectors pBT and pTRG). aa, amino acids.
LchD and LchP may regulate the local c-di-GMP concentration.
Our previous study reported a very modest increase in the c-di-GMP concentration in the lchP mutant compared to the wild-type strain (27). A somewhat similar result was obtained in the present study. We collected Lysobacter strains in HSAF production medium (1/10 tryptic soy broth [TSB]) at the logarithmic growth phase (27), which is the same growth phase as that in the RNA-seq experiment, to detect the c-di-GMP levels. The results in Fig. 4 show that the concentration of c-di-GMP in the lchD mutant was slightly decreased compared to that in the wild-type strain (Fig. 4). Moreover, we measured the c-di-GMP level in the lchD-lchP double mutant and found that it was maintained as the level in the wild-type strain. Taken together, the results described above support that LchD may cooperate with LchP to control the local, but not global, c-di-GMP concentration to regulate HSAF biosynthesis.
FIG 4.
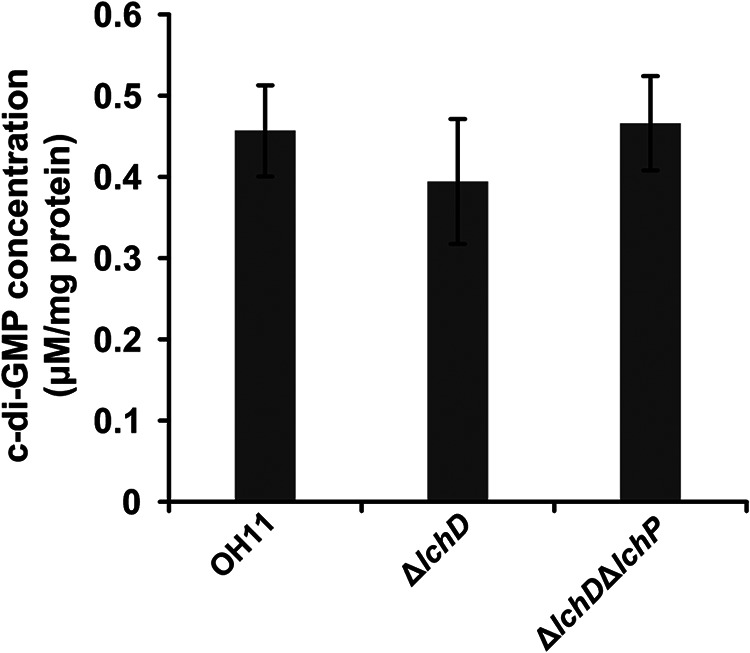
Quantification of intracellular c-di-GMP concentrations. The lchD mutant showed modestly decreased levels of intracellular c-di-GMP compared to those in the wild-type strain, while the lchD and lchP double mutant showed c-di-GMP levels restored to those detected in the wild-type strain. The data from three experiments are shown. Statistical comparisons were performed with GraphPad software (GraphPad, La Jolla, CA) using one-way ANOVA. The error bars indicate standard errors.
The GGDEV active site is necessary for the regulation of HSAF biosynthesis.
LchP contains both active GGDEF and EAL domains but utilizes its PDE activity to regulate HSAF biosynthesis (27). However, it is unclear whether the DGC activity of LchD is important for this process since the GGDEF domains of LchD and LchP interact (Fig. 3D).
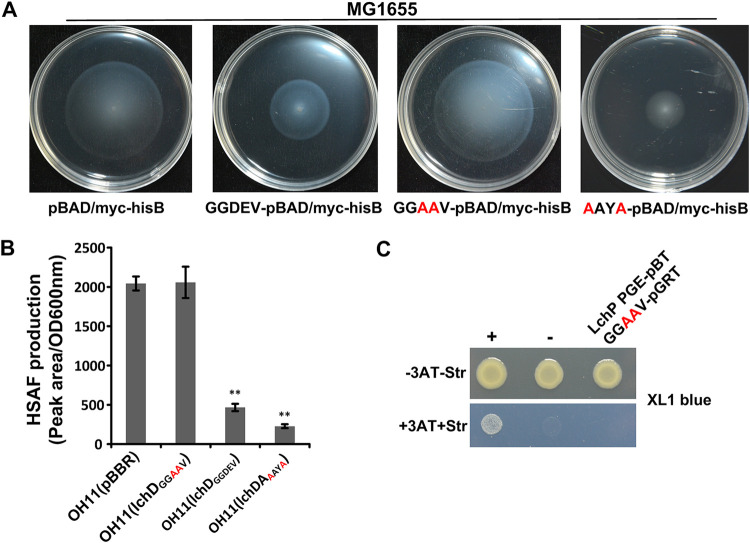
The GGDEF motif is the known catalytic center of a DGC. We aimed to determine whether the regulation of HSAF biosynthesis would be retained upon the inactivation of the DGC activity of LchD. Therefore, we mutated the GGDEV active site of LchD to GGAAV to generate catalytically inactive LchD and used an Escherichia coli-based motility assay to detect the DGC activity of the mutant protein. The results in Fig. 5A show that the expression of the mutant LchD protein did not change the swimming area of E. coli MG1655, but the expression of the wild-type LchD protein decreased the swimming area due to an increase in the c-di-GMP level, indicating that mutant LchD was deficient in the DGC activity. Next, we overexpressed inactive lchD in the wild-type background and measured HSAF production. As shown in Fig. 5B, inactive LchD lost the ability to regulate HSAF biosynthesis since similar amounts of HSAF were produced by the strain overexpressing the inactivated mutant and the wild-type strain containing the pBBR vector. Furthermore, B2H assay data (Fig. 5C) indicated that the catalytically inactive LchD protein no longer interacted with LchP. Thus, regardless of the DGC activity of LchD or its interaction with LchP, the GGDEV active site plays a key role in the regulation of HSAF production.
FIG 5.
The catalytic center of LchD is important for complex formation with LchP and HSAF production. (A) An E. coli-based swimming motility assay indicated that mutant LchD does not have DGC activity and that the I-site mutant has stronger DGC activity. (B) Quantification of HSAF production by a series of mutant strains. Statistical comparisons were performed with GraphPad software (GraphPad, La Jolla, CA) using one-way ANOVA. (C) A B2H assay showed that mutant LchD does not interact with LchP. +, positive control (GacS-pBT and GacS-pTRG); −, negative control (vectors pBT and pTRG).
Another structure that may contribute to LchD DGC activity is the conserved I-site RAYD (Fig. 2B). We constructed an RAYD inactive-site mutant, LchDAAYA, and found that it created a smaller swimming area than the wild type, indicating that the mutant of the I-site relieved the inhibition of its DGC activity. Next, we overexpressed LchDAAYA in wild-type OH11, measured its HSAF production, and found that it was lower than the overexpression of wild-type LchD (Fig. 5B). We then purified LchDAAYA and used a protein pulldown assay to detect its interaction with LchP. The results in Fig. S2C show that the mutant of the I-site of LchD did not influence its interaction with LchP. We also evaluated the addition of c-di-GMP to the interaction system and found that c-di-GMP did not show any influence (Fig. S2B), which was consistent with the B2H results (Fig. 3A and Fig. S2A).
DISCUSSION
Our previous study investigated a specific c-di-GMP pathway involved in the biosynthesis of the antibiotic HSAF in the potential biocontrol agent Lysobacter enzymogenes OH11. In this pathway, the c-di-GMP PDE LchP and the c-di-GMP receptor Clp (which is also a transcription regulator) form a complex to ensure the specificity of c-di-GMP-mediated signaling (27). Clp is an important transcriptional regulator that participates in many processes in L. enzymogenes (28), indicating that Clp should be distributed throughout the whole cell. In the present study, RNA-seq data indicated that LchP specifically regulates HSAF biosynthesis. The inconsistent effects of Clp and LchP suggested that LchP may be a local c-di-GMP-hydrolyzing enzyme but not a global modulator. Thus, we aimed to identify the DGC that acts on the same local c-di-GMP signaling pathway.
Like excess water spilling out of a container, excess c-di-GMP can interfere with other functions. Therefore, we constructed DGC mutants based on the lchP mutant, but not overexpression strains, to directly target the specific c-di-GMP pathway associated with LchP and found only one DGC, LchD, out of 19 DGCs in L. enzymogenes OH11 that may modulate the same c-di-GMP pathway with LchP (Fig. 2A). B2H and protein pulldown assays showed that LchP physically interacts with LchD but does not interact with the similar DGC 1158 (see Fig. S3 in the supplemental material), indicating that LchP and LchD have the same subcellular localization and confirming that LchP and LchD are partners that regulate the same c-di-GMP pathway.
An increasing number of recent studies have focused on local c-di-GMP signaling, and several mechanisms employed by some bacteria to maintain signaling specificity have been identified (22, 33). Because various types of N-terminal signal transduction domains are usually connected to a GGDEF or an EAL domain (5, 34), the activation of a given DGC or PDE by the desired signals at the desired time is a straightforward way to maintain c-di-GMP signaling specificity (35). The physical interaction among the c-di-GMP signaling components is another regulatory mechanism. An increasing number of examples have shown the interactions between DGCs, PDEs, and downstream receptors. Classic cases include but are not limited to the complexes formed by the PDE YciR and the DGC YdaM, the DGC DosC and the PDE DosP, and the DGC DgcC and the PDE PdeK from E. coli (19, 33, 36) and the DGC GcbC and the receptor LapD from Pseudomonas fluorescens (20). Physical interaction enables the delivery of c-di-GMP to a given target or the triggering of downstream targets in some cases. However, the mechanism by which signaling specificity is maintained by the formation of a complex between a DGC and a PDE remains unclear. Similar to LchP and LchD in the present study, deletion of only lchD (but not the 18 other DGCs) in the lchP mutant background restored HSAF production. Thus, this phenotype is consistent with the presence of a local c-di-GMP pool that is used to balance the concentration of c-di-GMP by LchP and LchD to trigger the downstream effector Clp, which activates HSAF biosynthesis to fight against filamentous fungi.
Although direct evidence showing the existence of a local c-di-GMP pool is lacking, there is a method that can measure the levels of another second messenger, cAMP, and cAMP gradients in real time with spatial accuracy in the nanometer range. Bock and colleagues used 8-FDA [2-(5(6)-carboxyfluoresceindiacetate)-aminoethylthio]-cAMP, the hydrolysis of which generates fluorescent 8-F-cAMP, which can be assayed by fluorescence fluctuation spectroscopy in combination with confocal microscopy (37). Using this method, the effects of binding sites on free cAMP concentrations in cells and on the spatial profile of cAMP gradients generated by PDE-mediated degradation could be analyzed. Those authors demonstrated that a low concentration of free cAMP enables PDEs to establish a local cAMP pool with a lower cAMP concentration below the activation threshold for the downstream effectors (37). Although cAMP signaling exists in eukaryotes, bacteria seem to employ an analogous strategy. We anticipate that a similar mechanism will be illustrated by bacteriologists in the near future.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Lysobacter strains were cultured in LB medium at 28°C unless stated otherwise. E. coli strains were cultured in LB medium at 37°C. Appropriate antibiotics (25 μg/mL kanamycin, 25 μg/mL gentamicin, 12.5 μg/mL tetracycline, 34 μg/mL chloramphenicol, and 8 μg/mL streptomycin [Str]) were used as needed.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| Lysobacter enzymogenes | ||
| OH11 | Wild type; Kmr | 46 |
| Δ3756 (ΔlchD) | le3756 in-frame deletion mutant | This study |
| Δ2762 (ΔlchP) | lchP in-frame deletion mutant | 27 |
| Δ0082 ΔlchP | le0082 and lchP double-deletion mutant | This study |
| Δ4875 ΔlchP | le4875and lchP double-deletion mutant | This study |
| Δ3882 ΔlchP | le3882 and lchP double-deletion mutant | This study |
| Δ0901 ΔlchP | le0901 and lchP double-deletion mutant | This study |
| Δ2826 ΔlchP | le2826 and lchP double-deletion mutant | This study |
| Δ0155 ΔlchP | le0155 and lchP double-deletion mutant | This study |
| Δ2120 ΔlchP | le2120 and lchP double-deletion mutant | This study |
| Δ3756 ΔlchP | le3756 and lchP double-deletion mutant | This study |
| Δ1158 ΔlchP | le1158 and lchP double-deletion mutant | This study |
| OH11(pBBR) | Vector control | This study |
| OH11(LchD) | Vector-based lchD overexpression strain | This study |
| OH11(LchDGGAAV) | Site-mutated lchD overexpression strain | This study |
| Escherichia coli | ||
| TOP10 | supE44 lacU169(ΔlacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA11 | 47 |
| MG1655 | Wild type | ATCC 700926a |
| XL1-Blue MRF′ | Δ(mcrA 183 83mcrCB-hsdSMR-mrr) 173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac {F[3proAB lacIqZΔM15 Tn10 (Tetr)]} | 43 |
| BL21(DE3) | Host strain for protein expression; Kmr | TaKaRa Company, Shanghai, China |
| Plasmids | ||
| pEX18GM | Suicide vector for gene in-frame deletion; Gmr | 48 |
| 0082-pEX18 | Plasmid for in-frame deletion of le0082 | This study |
| 4875-pEX18 | Plasmid for in-frame deletion of le4875 | 27 |
| 3882-pEX18 | Plasmid for in-frame deletion of le3882 | 27 |
| 0901-pEX18 | Plasmid for in-frame deletion of le0901 | This study |
| 2826-pEX18 | Plasmid for in-frame deletion of le2826 | This study |
| 0155-pEX18 | Plasmid for in-frame deletion of le0155 | 27 |
| 2120-pEX18 | Plasmid for in-frame deletion of le2120 | This study |
| 3756-pEX18 | Plasmid for in-frame deletion of le3756 | This study |
| 1158-pEX18 | Plasmid for in-frame deletion of le1158 | This study |
| pBBR1-MCS5 | Broad-host-range vector for gene overexpression; Gmr | 49 |
| lchD-pBBR | Plasmid for LchD overexpression | This study |
| lchDGGAAV-pBBR | Plasmid for site-mutated LchD overexpression | This study |
| pBAD/Myc-His B | Vector for arabinose-inducible expression; Ampr | Invitrogen |
| GGDEV-pBAD | pBAD/Myc-His B containing the cytoplasmic part of LchD | This study |
| GGAAV-pBAD | pBAD/Myc-His B containing mutated LchD | This study |
| pTRG | Plasmid for protein expression in the bacterial two-hybrid assay; Tetr | 50 |
| LchP-pTRG | pTRG containing the cytoplasmic part of LchP | 27 |
| LchD-pTRG | pTRG containing the cytoplasmic part of LchD | This study |
| PAS-pTRG | pTRG containing the PAS domain of LchP | This study |
| GGDEF-pTRG | pTRG containing the GGDEF domain of LchP | This study |
| EAL-pTRG | pTRG containing the EAL domain of LchP | This study |
| GGAAV-pTRG | pTRG containing site-mutated LchD | This study |
| 1158-pTRG | pTRG containing Le1158 | This study |
| pBT | Plasmid for protein expression in bacterial two-hybridization assays; Chlor | 50 |
| LchP-pBT | pBT containing the cytoplasmic part of LchP | 27 |
| LchD-pBT | pBT containing the cytoplasmic part of LchD | This study |
| pMAL-p2x | Vector for expression of the MBP tag fusion protein | Huayue Company, Beijing, China |
| LchP-pMAL-p2x | pMAL-p2x::LchP PAS+GGDEF+EAL | 27 |
| pET30a(+) | Vector for expression of the His tag fusion protein | TaKaRa Company |
| LchD-pET30a | pET30a::lchD GGDEV | This study |
Kmr, Gmr, Ampr, Tetr, and Chlor represent kanamycin, gentamicin, ampicillin, tetracycline, and chloramphenicol resistance, respectively.
Gene deletion and overexpression.
In-frame gene deletion in L. enzymogenes OH11 was performed as described in our previous study (27). Briefly, the up- and downstream fragments of the target gene were amplified using the corresponding primers listed in Table 2 and cloned into the suicide vector pEX18GM (Table 1). The recombinant vector was transformed into the wild-type strain by electroporation. Via double-crossover homologous recombination, the in-frame target gene deletion mutant was then picked up by PCR and further confirmed by gene sequencing (TsingKe Biological Technology Company, Beijing, China).
TABLE 2.
Primers used in this study
| Primer | Sequence (restriction enzyme)a | Purpose |
|---|---|---|
| Primers used for gene in-frame deletion | ||
| 0155F1 | CCCAAGCTTCGCAGCAGATGGGCAGCTAT (HindIII) | To amplify a 710-bp upstream homologue arm of le0155 |
| 0155R1 | GGGGTACCGCGGCGACTTCATTCCACCT (KpnI) | |
| 0155F2 | GGGGTACCCGCAGTTGCCTTGAGTCGAG (KpnI) | To amplify a 636-bp downstream homologue arm of le0155 |
| 0155R2 | GCTCTAGAGCTGTTGTTCGTCTCCACCC (XbaI) | |
| 0082F1 | CCCAAGCTTGCGGCTACTTCTGGCGGGTG (HindIII) | To amplify a 538-bp upstream homologue arm of le0082 |
| 0082R1 | CGGAATTCTAGCCCCAGTGCGAGAGGTG (EcoRI) | |
| 0082F2 | CGGAATTCCGGTTGGGGGAGGTTTGAGC (EcoRI) | To amplify a 485-bp downstream homologue arm of le0082 |
| 0082R2 | GCTCTAGAGGCTGGGGGGTGTTGAGAGG (XbaI) | |
| 4875F1 | CCCAAGCTTTCACCACCAAGGACTACCGC (HindIII) | To amplify a 692-bp upstream homologue arm of le4875 |
| 4875R1 | GGGGTACCCGCCACCAGATTGCCGTTGT (KpnI) | |
| 4875F2 | GGGGTACCGAGGATTTCGCCGTGCAGGT (KpnI) | To amplify a 421-bp downstream homologue arm of le4875 |
| 4875R2 | GCTCTAGAGATCGTGGTCTTGTCGGCGC (XbaI) | |
| 3882F1 | CCCAAGCTTACGAACAGGGCTACGAACGC (HindIII) | To amplify a 686-bp upstream homologue arm of le3882 |
| 3882R1 | GGGGTACCCGGAGGCTGTGTCGTTCGCG (KpnI) | |
| 3882F2 | GGGGTACCGTCCGCCACTGACCGCTCGC (KpnI) | To amplify a 300-bp downstream homologue arm of le3882 |
| 3882R2 | GCTCTAGACGTTGCTGGGCGAGTGCTTC (XbaI) | |
| 0901F1 | CCCAAGCTTCCTTCATCCTGCCGACCAGT (HindIII) | To amplify a 784-bp upstream homologue arm of le0901 |
| 0901R1 | GGGGTACCATGTCGTCGTAGGCGCTCTC (KpnI) | |
| 0901F2 | GGGGTACCCGCCTGGTTGCCGACGAGGG (KpnI) | To amplify a 616-bp downstream homologue arm of le0901 |
| 0901R2 | GCTCTAGACGGCAGCGGGAAACTCAGGC (XbaI) | |
| 2826F1 | CCCAAGCTTAAGGCGGTCGGCGTGGTGAT (HindIII) | To amplify a 625-bp upstream homologue arm of le2826 |
| 2826R1 | GGGGTACCCACCGCCATCACCAGCAAGC (KpnI) | |
| 2826F2 | GGGGTACCGCTTCTGATACGGCTCCTGC (KpnI) | To amplify a 616-bp downstream homologue arm of le2826 |
| 2826R2 | GCTCTAGAGATGGCGGCGAACTGGCGAT (XbaI) | |
| 2120F1 | CCCAAGCTTCGAGCGAAGGCGGATGGATG (HindIII) | To amplify a 522-bp upstream homologue arm of le2120 |
| 2120R1 | GGGGTACCACCGCCATCGCCACATCCGT (KpnI) | |
| 2120F2 | GGGGTACCAGTTCTGCCTGCTGGTCGCC (KpnI) | To amplify an 854-bp upstream homologue arm of le2120 |
| 2120R2 | GCTCTAGAATCTCCTGCGGCTCGGGCAC (XbaI) | |
| 3756F1 | CCCAAGCTTCGCCGCCCCCTGTCGTTTTT (HindIII) | To amplify a 273-bp upstream homologue arm of le3756 |
| 3756R1 | GGGGTACCTCGTCGCCACCGCCCACAAC (KpnI) | |
| 3756F2 | GGGGTACCAGCGAGAGCGAACACGAGGT (KpnI) | To amplify a 510-bp upstream homologue arm of le3756 |
| 3756R2 | GCTCTAGACCAGGCAGCGGTAGACGAAT (XbaI) | |
| 1158F1 | CCCAAGCTTATCGCCACTTCCTCCACCGC (HindIII) | To amplify a 535-bp upstream homologue arm of le1158 |
| 1158R1 | GCTCTAGAAGGACCACGACACCGCCAAG (XbaI) | |
| 1158F2 | GCTCTAGAGCCAGGACCGTTCCATTTCC (XbaI) | To amplify a 564-bp upstream homologue arm of le1158 |
| 1158R2 | GGGGTACCAGGGGGCGGGCTGAGAGGAC (KpnI) | |
| Primers used for gene overexpression | ||
| cp3756F | CCCAAGCTTCGCCGCCCCCTGTCGTTTTT (HindIII) | To amplify a 2,235-bp fragment of le3756 and its predicted promoter |
| cp3756R | GCTCTAGAGTTTCGATCAGGTAGCCCGC (XbaI) | |
| Primers used for DGC activity assay | ||
| GGDEV-F | GGGGTACCAAGCTGCAACAGCTGCG (KpnI) | To amplify a 561-bp fragment encoding the LchD cytoplasmic domain |
| GGDEV-R | CCCAAGCTTTCAAGAGACCACCTCGTGTT (HindIII) | |
| Primers used for bacterial two-hybrid assay | ||
| GGDEV-F(T) | CGGGATCCAAGCTGCAACAGCTGCGTCG (BamHI) | To amplify a 561-bp fragment encoding the LchD cytoplasmic domain |
| GGDEV-R(T) | CCGCTCGAGCCGGTCGCGGCCGGCGGACT (XhoI) | |
| PAS-F | CGGGATCCGACGATCACGAGATCCATCC (BamHI) | To amplify a 579-bp fragment encoding the PAS domain of LchP |
| PAS-R | CCGCTCGAGGTCGGTGATGTCGGTCAGCA (XhoI) | |
| GGDEF-F | CGGGATCCGCCAACTACGACACCCTCAC (BamHI) | To amplify a 477-bp fragment encoding the GGDEF domain of LchP |
| GGDEF-R | CCGCTCGAGGGTGCGCCGGCCGGCGGCCT (XhoI) | |
| EAL-F | CGGGATCCTCCGCGGCGCTGCGCAAGGT (BamHI) | To amplify a 711-bp fragment encoding the EAL domain of LchP |
| EAL-R | CCGCTCGAGCGGCTTGGCCAGCCAGTAGC (XhoI) | |
| 1158-F(PTRG) | CGGGATCCGCCAAGCTGCAGAAGCTGCG (BamHI) | To amplify a 561-bp fragment encoding the 1158 cytoplasmic domain |
| 1158-R(PTRG) | CCGCTCGAGTCAGATCGCCACTTCCTCCA (XhoI) | |
| Primers used for qRT-PCR | ||
| pksF | ACTATTTGTTGGGCGACGAC | To detect the transcript level of pks |
| pksR | GTAACCGAACAGGGTGCAAT | |
| 16sRNAF | ACGGTCGCAAGACTGAAACT | Reference gene |
| 16sRNAR | AAGGCACCAATCCATCTCTG | |
| Primers used for protein expression | ||
| LchD-His-F | GGAATTCCATATGAAGCTGCAACAGCTGCG (NdeI) | To amplify a 561-bp fragment encoding the LchD cytoplasmic domain |
| LchD-His-R | CCCAAGCTTTCAAGAGACCACCTCGTGTT (HindIII) | |
Restricted digestion enzyme sites are underlined.
Gene overexpression constructs were generated as described previously (38). The primers used in this study are listed in Table S1 in the supplemental material. In brief, the target gene and its predicted promoter were amplified by PCR and cloned into the broad-host-range vector pBBR1-MCS5 (Table 1). The construct was transformed into the wild-type strain by electroporation. The transformants were selected on LB agar plates containing gentamicin.
RNA-seq.
Strains were cultured in LB medium at 28°C overnight, and 1 mL of the culture was transformed into 50 mL of 1/10 tryptic soy broth (TSB). Cells were collected at an optical density at 600 nm (OD600) of 1.0. Next, clustering and sequencing were performed by BGI Genomics (Shenzhen, China). To analyze the differentially expressed genes between the lchP mutant and wild-type cells, the reads for all contigs were converted to reads per kilobase per million (RPKM) (39). The expression abundance of each contig in the two samples was calculated using the MARS (MA plot-based method with random sampling [“MA” indicating M values versus log-intensity averages {A values}]) model by the differentially expressed gene seq package. Differentially expressed genes between the lchP mutant strain and the wild-type strain were identified by DEGseq software (40). The FDR (false discovery rate) was used to determine the P value threshold for the analysis. An FDR of ≤0.001 and a log2 fold change of greater than 1.0 or less than −1.0 were used to indicate a significant change in the expression abundance.
qRT-PCR.
Strains were cultured in LB medium at 28°C overnight. One milliliter of the culture was transferred to 50 mL of 1/10 TSB, and cells were collected at an OD600 of 1.0. RNA extraction was performed with a bacterial RNA kit (Omega, China), and RNA concentrations were measured with a Nanodrop ND-1000 UV spectrophotometer (Thermo Fisher, USA). cDNA was generated using a PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, Japan) according to the manufacturer’s protocol. qRT-PCR was performed using an Applied Biosystems 7500 system, and the 16S rRNA gene was used as an internal control, as described previously (41). The primers used in this study are listed in Table 2.
HSAF extraction and detection.
HSAF extraction and quantification were performed as described previously (42). In brief, HSAF was extracted from L. enzymogenes cultures in 1/10 TSB with an equal volume of ethyl acetate. HSAF was detected by HPLC (high-performance liquid chromatography) and quantified per OD600 unit. Biological experiments for each treatment were performed three times and assayed in triplicate.
DGC activity assay.
The DGC activity assay was based on an E. coli motility reporter system as described previously (27). Briefly, the gene encoding the DGC under analysis was cloned into the pBAD/Myc-His B vector (Table 1). The construct was transformed into the E. coli MG1655 strain. Next, the swimming zone of the MG1655 strain expressing the construct or the original vector was observed on LB soft agar plates (0.25% agar) supplemented with 0.1% arabinose. Three biological replicates were tested.
Bacterial two-hybrid assay.
The bacterial two-hybrid (B2H) assay was performed according to the protocol of the BacterioMatch II two-hybrid system (Agilent Technologies, USA). Genes encoding the target proteins were cloned into the pBT and pTGR plasmids and transformed into E. coli XL1-Blue MRF′ Kan. E. coli XL1-Blue MRF′ Kan containing the original pBT and pTGR plasmids served as a negative control, and the strain containing pBT-GacS and pTRG-GacS served as a positive control (43). All cotransformants were spotted onto selective plates and grown at 28°C for 48 h. According to the protocol from the kit manufacturer, selective plates contained 3-amino-1,2,4-triazole (3-AT) and Str to enable the growth of the positive colonies and disable the growth of the negative colonies. Three biological replicates were performed.
Protein expression and purification.
The gene encoding cytoplasmic LchD was amplified and cloned into pET-30a(+) (Table 1). The construct was transformed into E. coli strain BL21(DE3) (Table S1) for protein expression and purification. Two milliliters of the culture grown overnight was transferred to 200 mL of fresh LB medium at 37°C and grown with shaking at 200 rpm to an OD600 of 0.6. Subsequently, isopropyl β-d-1-thiogalactopyranoside (IPTG; Sigma) was added to the culture to a final concentration of 0.5 mM, followed by growth at 28°C for 4 h. The cells were collected by centrifugation (13,000 rpm) at 4°C and sonicated using a 250 sonifier (Branson digital sonifier). After centrifugation, the soluble proteins were mixed with preequilibrated Ni2+ resin (GE Healthcare, Shanghai, China), and the target protein was eluted with 250 mM imidazole. Protein purity was assessed by SDS-PAGE, and the protein concentration was determined using a bicinchoninic acid (BCA) assay kit (TransGen Biotech, China).
Pulldown assay.
Purified His-fused LchD-GGDEV and MBP-fused LchP-PGE (27) were subjected to a pulldown assay. The reaction system contained 800 μL of phosphate-buffered saline (PBS) with 5 μM LchD-GGDEV-His and LchP-PGE-MBP and 50 μL of amylose resin. Samples were incubated at 4°C overnight. The resins were collected by centrifugation and washed 10 times with PBS containing 1% Triton X-100 to remove unbound proteins. The immunoprecipitated proteins were eluted by boiling in the presence of 6× SDS loading dye for 10 min. Western blotting was performed using specific anti-MBP (catalogue number ab49923) and anti-His (catalogue number ab18184) antibodies from Abcam.
c-di-GMP extraction and quantification.
c-di-GMP extraction and quantification were performed as described previously (27). Cultures were grown in 1/10 TSB at 28°C to an OD600 of 1.5. c-di-GMP was extracted with 0.6 M HClO4 and 2.5 M K2CO3 as described previously (44, 45). Two milliliters of the cells was harvested for protein quantification by the BCA assay (TransGen Biotech, China). The samples were analyzed by liquid chromatography-mass spectrometry (LC-MS) on an Agilent 6460 triple-quadrupole LC-MS instrument.
Statistical analysis.
Some experiments included in this study were performed three times with three replicates each. The means from three replicates are shown, with error bars indicating standard errors. Statistical comparisons were performed with GraphPad software (GraphPad, La Jolla, CA) using one-way analysis of variance (ANOVA). A P value of <0.05 was used to indicate significance.
Data availability.
RNA-seq data have been uploaded to the Sequence Read Archive in the NCBI database with accession number PRJNA716139.
ACKNOWLEDGMENTS
We thank Jialei Wang and Ning Wang from the Central Laboratory at the Jiangsu Academy of Agricultural Sciences for their technical support with the LC-MS (HPLC) analysis.
This study was funded by the National Natural Science Foundation of China (numbers 31901931 and 31872018), the Natural Science Foundation of Jiangsu Province (number BK20190266), and the China Agriculture Research System of MOF and MARA.
G.X. contributed to conceptualization, investigation, writing and editing the manuscript, and funding acquisition. G.X. carried out the experiments and analyzed data. L.Z. contributed to RNA-seq data analysis. G.Q. modified the main body of the manuscript. F.L. contributed to reviewing the manuscript, supervision, and funding acquisition. All authors contributed to the article and approved the submitted version.
Footnotes
Supplemental material is available online only.
Contributor Information
Fengquan Liu, Email: fqliu20011@sina.com.
Gladys Alexandre, University of Tennessee at Knoxville.
REFERENCES
- 1.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281. 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 2.Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet 40:385–407. 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- 3.Romling U, Amikam D. 2006. Cyclic di-GMP as a second messenger. Curr Opin Microbiol 9:218–228. 10.1016/j.mib.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Hengge R. 2009. Principles of cyclic-di-GMP signaling in bacteria. Nat Rev Microbiol 7:263–273. 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 5.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan C, Paul R, Samoray D, Amiot N, Giese B, Jenal U, Schirmer T. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc Natl Acad Sci USA 101:17084–17089. 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev 18:715–727. 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J Biol Chem 280:30829–30837. 10.1074/jbc.M504429200. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol 187:4774–4781. 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamayo R, Tischler AD, Camilli A. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J Biol Chem 280:33324–33330. 10.1074/jbc.M506500200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellini D, Caly DL, McCarthy Y, Bumann M, An SQ, Dow JM, Ryan RP, Walsh MA. 2014. Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre. Mol Microbiol 91:26–38. 10.1111/mmi.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang ZX. 2015. The expanding roles of c-di-GMP in the biosynthesis of exopolysaccharides and secondary metabolites. Nat Prod Rep 32:663–683. 10.1039/c4np00086b. [DOI] [PubMed] [Google Scholar]
- 13.Ryjenkov DA, Simm R, Romling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310–30314. 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- 14.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev 23:93–104. 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang X, Ahmad I, Blanka A, Schottkowski M, Cimdins A, Galperin MY, Römling U, Gomelsky M. 2014. GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol Microbiol 93:439–452. 10.1111/mmi.12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang YC, Chin KH, Tu ZL, He J, Jones CJ, Sanchez DZ, Yildiz FH, Galperin MY, Chou SH. 2016. Nucleotide binding by the widespread high-affinity cyclic di-GMP receptor MshEN domain. Nat Commun 7:12481. 10.1038/ncomms12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ha D-G, Richman ME, O’Toole GA. 2014. Deletion mutant library for investigation of functional outputs of cyclic diguanylate metabolism in Pseudomonas aeruginosa PA14. Appl Environ Microbiol 80:3384–3393. 10.1128/AEM.00299-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newell PD, Yoshioka S, Hvorecny KL, Monds RD, O’Toole GA. 2011. Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0-1. J Bacteriol 193:4685–4698. 10.1128/JB.05483-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindenberg S, Klauck G, Pesavento C, Klauck E, Hengge R. 2013. The EAL domain protein YciR acts as a trigger enzyme in a c-di-GMP signaling cascade in E. coli biofilm control. EMBO J 32:2001–2014. 10.1038/emboj.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahlstrom KM, Giglio KM, Collins AJ, Sondermann H, O’Toole GA. 2015. Contribution of physical interactions to signaling specificity between a diguanylate cyclase and its effector. mBio 6:e01978-15. 10.1128/mBio.01978-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valentini M, Laventie BJ, Moscoso J, Jenal U, Filloux A. 2016. The diguanylate cyclase HsbD intersects with the HptB regulatory cascade to control Pseudomonas aeruginosa biofilm and motility. PLoS Genet 12:e1006354. 10.1371/journal.pgen.1006354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Römling U, Gomelsky M, Galperin MY. 2005. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol 57:629–639. 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- 23.Li S, Du L, Yuen G, Harris SD. 2006. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol Biol Cell 17:1218–1227. 10.1091/mbc.e05-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu F, Zaleta-Rivera K, Zhu X, Huffman J, Millet JC, Harris SD, Yuen G, Li XC, Du L. 2007. Structure and biosynthesis of heat-stable antifungal factor (HSAF), a broad-spectrum antimycotic with a novel mode of action. Antimicrob Agents Chemother 51:64–72. 10.1128/AAC.00931-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lou L, Qian G, Xie Y, Hang J, Chen H, Zaleta-Rivera K, Li Y, Shen Y, Dussault PH, Liu F, Du L. 2011. Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J Am Chem Soc 133:643–645. 10.1021/ja105732c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Xia J, Su Z, Xu G, Gomelsky M, Qian G, Liu F. 2017. Lysobacter PilR, the regulator of type IV pilus synthesis, controls antifungal antibiotic production via a cyclic di-GMP pathway. Appl Environ Microbiol 83:e03397-16. 10.1128/AEM.03397-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu G, Han S, Huo C, Chin KH, Chou SH, Gomelsky M, Qian G, Liu F. 2018. Signaling specificity in the c-di-GMP-dependent network regulating antibiotic synthesis in Lysobacter. Nucleic Acids Res 46:9276–9288. 10.1093/nar/gky803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Zhao Y, Zhang J, Zhao Y, Shen Y, Su Z, Xu G, Du L, Huffman JM, Venturi V, Qian G, Liu F. 2014. Transcriptomic analysis reveals new regulatory roles of Clp signaling in secondary metabolite biosynthesis and surface motility in Lysobacter enzymogenes OH11. Appl Microbiol Biotechnol 98:9009–9020. 10.1007/s00253-014-6072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galperin MY, Nikolskaya AN, Koonin EV. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett 203:11–21. 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 30.Ren X, Ren S, Xu G, Dou W, Chou SH, Chen Y, Qian G. 2020. Knockout of diguanylate cyclase genes in Lysobacter enzymogenes to improve production of antifungal factor and increase its application in seed coating. Curr Microbiol 77:1006–1015. 10.1007/s00284-020-01902-x. [DOI] [PubMed] [Google Scholar]
- 31.Anantharaman V, Aravind L. 2003. Application of comparative genomics in the identification and analysis of novel families of membrane-associated receptors in bacteria. BMC Genomics 4:34. 10.1186/1471-2164-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joung JK, Ramm EI, Pabo CO. 2000. A bacterial two-hybrid selection system for studying protein-DNA and protein-protein interactions. Proc Natl Acad Sci USA 97:7382–7387. 10.1073/pnas.110149297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richter AM, Possling A, Malysheva N, Yousef KP, Herbst S, von Kleist M, Hengge R. 2020. Local c-di-GMP signaling in the control of synthesis of the E. coli biofilm exopolysaccharide pEtN-cellulose. J Mol Biol 432:4576–4595. 10.1016/j.jmb.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cruz DP, Huertas MG, Lozano M, Zárate L, Zambrano MM. 2012. Comparative analysis of diguanylate cyclase and phosphodiesterase genes in Klebsiella pneumoniae. BMC Microbiol 12:139. 10.1186/1471-2180-12-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dahlstrom KM, O’Toole GA. 2017. A symphony of cyclases: specificity in diguanylate cyclase signaling. Annu Rev Microbiol 71:179–195. 10.1146/annurev-micro-090816-093325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tuckerman JR, Gonzalez G, Sousa EH, Wan X, Saito JA, Alam M, Gilles-Gonzalez MA. 2009. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 48:9764–9774. 10.1021/bi901409g. [DOI] [PubMed] [Google Scholar]
- 37.Bock A, Annibale P, Konrad C, Hannawacker A, Anton SE, Maiellaro I, Zabel U, Sivaramakrishnan S, Falcke M, Lohse MJ. 2020. Optical mapping of cAMP signaling at the nanometer scale. Cell 182:1519–1530. 10.1016/j.cell.2020.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu G, Shi X, Wang R, Xu H, Du L, Chou SH, Liu H, Liu Y, Qian G, Liu F. 2018. Insights into the distinct cooperation between the transcription factor Clp and LeDSF signaling in the regulation of antifungal factors in Lysobacter enzymogenes OH11. Biol Control 120:52–58. 10.1016/j.biocontrol.2016.08.006. [DOI] [Google Scholar]
- 39.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 40.Wang L, Feng Z, Wang X, Wang X, Zhang X. 2010. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138. 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 41.Qian G, Wang Y, Liu Y, Xu F, He Y, Du L, Venturi V, Fan J, Hu B, Liu F. 2013. Lysobacter enzymogenes uses two distinct cell-cell signaling systems for differential regulation of secondary-metabolite biosynthesis and colony morphology. Appl Environ Microbiol 79:6604–6616. 10.1128/AEM.01841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qian G, Xu F, Venturi V, Du L, Liu F. 2014. Roles of a solo LuxR in the biological control agent Lysobacter enzymogenes strain OH11. Phytopathology 104:224–231. 10.1094/PHYTO-07-13-0188-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo M, Feng H, Zhang J, Wang W, Wang Y, Li Y, Gao C, Chen H, Feng Y, He Z. 2009. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res 19:1301–1308. 10.1101/gr.086595.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu B, Liu C, Liu S, Cong H, Chen Y, Gu L, Ma LZ. 2016. Membrane association of SadC enhances its diguanylate cyclase activity to control exopolysaccharides synthesis and biofilm formation in Pseudomonas aeruginosa. Environ Microbiol 18:3440–3452. 10.1111/1462-2920.13263. [DOI] [PubMed] [Google Scholar]
- 46.Qian G, Hu B, Jiang Y, Liu F. 2009. Identification and characterization of Lysobacter enzymogenes as a biological control agent against some fungal pathogens. Agr Sci China 8:68–75. 10.1016/S1671-2927(09)60010-9. [DOI] [Google Scholar]
- 47.Han Y, Wang Y, Tombosa S, Wright S, Huffman J, Yuen G, Qian G, Liu F, Shen Y, Du L. 2015. Identification of a small molecule signaling factor that regulates the biosynthesis of the antifungal polycyclic tetramate macrolactam HSAF in Lysobacter enzymogenes. Appl Microbiol Biotechnol 99:801–811. 10.1007/s00253-014-6120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 49.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 50.Fang X, Gomelsky M. 2010. A post-translational, c-di-GMP-dependent mechanism regulating flagella motility. Mol Microbiol 76:1295–1305. 10.1111/j.1365-2958.2010.07179.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, Fig. S1 to S3. Download AEM.01895-21-s0001.pdf, PDF file, 0.7 MB (672.1KB, pdf)
Data Availability Statement
RNA-seq data have been uploaded to the Sequence Read Archive in the NCBI database with accession number PRJNA716139.