Abstract
Wireless electroceutical dressing (WED) fabric kills bacteria and disrupts bacterial biofilm. This work tested, comparing with standard of care topical antibiotic ketoconazole, whether the weak electric field generated by WED is effective to manage infection caused by ketoconazole-resistant yeast Candida albicans. WED inhibited Candida albicans biofilm formation and planktonic growth. Unlike ketoconazole, WED inhibited yeast to hyphal transition and downregulated EAP1 curbing cell attachment. In response to WED-dependent down-regulation of biofilm-forming BRG1 and ROB1, BCR1 expression was markedly induced in what seems to be a futile compensatory response. WED induced NRG1 and TUP1, negative regulators of filamentation; it down-regulated EFG1, a positive regulator of hyphal pathway. Consistent with the anti-hyphal properties of WED, the expression of ALS3 and HWP1 were diminished. Ketoconazole failed to reproduce the effects of WED on NRG1, TUP1 and EFG1. WED blunted efflux pump activity; this effect was in direct contrast to that of ketoconazole. WED exposure compromised cellular metabolism. In the presence of ketoconazole, the effect was synergistic. Unlike ketoconazole, WED caused membrane depolarization, changes in cell wall composition and loss of membrane integrity. This work presents first evidence that weak electric field is useful in managing pathogens which are otherwise known to be antibiotic resistant.
Keywords: antibiotic textile, wound, antibiotic resistance, electric field, yeast infection, biofilm
1. Introduction
Textile is in contact with the exfoliating, warm and moist human skin, making it well suited to harbor and propagate microbial pathogens. The threat is substantially heightened in the setting of healthcare delivery [1]. Thus, specialized textiles have been developed and studied for antibiotic properties [2]. Broadly, such biomaterials may be classified into two categories: those with inherent antibiotic property of its components, and those where in antibiotic properties are added. Plant-derived natural fibers contain many active compounds such as phenolics, polyphenols, essential oils, alkaloids, lectins, polypeptides, tannins, coumarins, and flavonoids which present broad spectrum antimicrobial activity [3]. Microbicidal constituents present in hemp have exhibit efficacy against Bacillus subtilis, Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, Aspergillus niger and Candida albicans. Other fiber plants such as jute, bamboo, flax, sisal, kenaf, banana and pineapple contain biologically active microbicidal compounds like soluble phenols, triterpenoids, sterols, lignans, alpha-linoleic acid. Organic extracts from barks, leaves or fibers of these plants have exhibited antimicrobial activity against commonly tested pathogens [3]. In commercial textile, quaternary ammonium compounds (QAC) bestow antibiotic properties. For example, in non-woven fabric surface modification of poly(D,L-lactide) with QAC inhibit E coli and S aureus [4]. The halogenated phenol Triclosan has been used for six decades as antibiotic in textile [5]. Chitosan, a natural antimicrobial copolymer, extracted from exoskeleton of crabs and shrimps has been used in fabrics as a complex with bivalent metal ions. These chitosan-metal complexes have successfully demonstrated microbicidal activity against a wide range of bacteria and fungi [2]. Other examples of chemical additives that are used to add antibiotic properties to textile include poly(hexamethylene biguanide) [6]. In addition to natural as well as synthetic chemicals, fortification of textiles with metals and metal-based compounds (Ag, ZnO, Cu, TiO2, and other minor metals based such as iron based, CeO2, Au, SiO2, Ni, and Pt) represents a common antibiotic strategy [7]. The use of chemical additives as well as metal-complexed fabric has given rise to antimicrobial resistant pathogens raising concerns. For example, triclosan is associated with a high risk of developing resistance and cross-resistance in S aureus and E coli [8]. Similarly, evidence of widespread silver resistance in clinical isolates of microbial pathogens is alarming [9]. Our work studying the placement of electrochemically coupled metals in a specific geometric pattern on fabric has resulted in the generation of weak electric field upon contact of the fabric with electrolyte containing liquid including body fluids [10–13]. Such strategic placement of Ag and Zn dots on fabric kills pathogens that cannot be killed Ag alone [11]. The fabric, wireless electroceutical dressing (WED), has been FDA cleared for use as wound care dressing [14]. Developing work show that coronavirus infectivity can be rapidly neutralized upon contact with WED fabric proposing the benefit of using such fabric for the development of personal protective equipment during the ongoing pandemic [10]. Following a series of studies demonstrating anti-biofilm properties of the dressing, a clinical trial is in progress to testing WED in a setting of burn wound infection (NCT04079998). In this work, we ask whether the weak electric field generated by WED is effective to instill antibiotic sensitivity to an antibiotic resistant pathogen. Candida albicans poses major threat to skin and wound infection [15, 16]. Ketoconazole is the azole drug of choice to manage such skin infection topically [17]. Fluconazole and itraconazole are used to manage systemic infection [18, 19]. The objective of this work is to test whether WED can kill ketoconazole-resistant Candida albicans. The potential synergy of WED and ketoconazole in the management of ketoconazole-resistant Candida albicans was also tested.
2. Materials and methods
2.1. Candida albicans culture and maintenance
Ketoconazole-resistant Candida albicans (Robin) Berkhout (ATCC® 64124™) was procured from American Type Culture Collection (ATCC). This strain was maintained at 4°C on Yeast extract Peptone Dextrose (YPD, Cat no: DF0427–17-6; BD™ Difco™, Franklin Lakes, NJ) agar plates. Planktonic yeast cultures (seed inoculums) were initiated in 5 ml of YPD broth (Cat no: DF0428–17-5; BD™ Difco™) from a single colony of the yeast strain. Cells were incubated at 27°C, under shaker conditions (200 rpm) (Excella E24, New Brunswick Scientific, Enfield, CT) for 24h, followed by initiation of master inoculums for respective experiments in 5 ml YPD broth (OD600nm ~0.15). Yeast cells were incubated at 27°C for all assays, except for hyphal transition experiments. For all assays, ketoconazole (Cat no: PHR1385–1G, Sigma Aldrich, St. Louis, MO) was used at a final concentration of 100 µg/ml (dissolved in 100% Methanol; stored at −20°C).
2.2. Wireless electroceutical dressing as a source of electric field
In this work, a FDA cleared Wireless Electroceutical Dressing (WED) was used as a source of weak electric field. This dressing, co-developed by our laboratory [10–13], has been commercialized by Vomaris Inc. (Phoenix, AZ). It is made of polyester fabric printed with alternating circular dots of silver (Ag) and zinc (Zn) metals (ø2 mm and 1 mm, respectively), generating electric potential in the range of 0.6 – 0.9 V [10–13]. A polyester fabric without any metal deposition (hence unable to generate electric field) was used as an experimental control and is referred to as unprinted fabric. Fabric with only silver dots was used as ‘Ag-only fabric’ control in all experiments, to rule out the effects of any microbicidal activity of silver alone. All dressings were provided to us by the manufacturer.
2.3. Contact potential mapping for dressings
Two-dimensional maps of the surface potential of the electroceutical dressing were collected by the modified version of a method reported previously [20]. Briefly, a gold-coated probe tip (100 µm diameter) was mounted on a microprobing system consisting of miniature linear actuators (Zaber Technologies Inc., Vancouver, Canada) with independent control in the XYZ dimensions. The dressings were moistened by YPD broth and mounted on the sample stage. With the microprobing system, the probe tip raster scanned to contact the sample surface for potential measurement at each pixel. The scan area was 5 × 5 mm in 20 × 20 pixels covering 2 Ag dots, 2 Zn dots and empty dressing. The potential at the probe tip was referenced to the empty dressing, as measured by a high precision and high input impedance voltmeter (EMF 16, Lawson Lab Inc., Malvern, PA, USA) at each pixel. The collected data were plotted and processed by Gwyddion [21].
2.4. Voltage measurements for dressings
Since electroceutical dressing was used as the source of weak electric fields, voltage generated by dressings was measured immediately after wetting with YPD broth and after incubation in YPD broth for 24h, 48h or 72h at 27°C under static conditions [11]. For measurements, probes of a multimeter (Amprobe 34XR-A, Everett, WA) were placed on adjacent silver spots of Ag-only fabric or adjacent silver and zinc dots in WED. Voltage was first measured by placing the probes on dry dressings. This was followed by wetting the dressings by addition of 200 µl of YPD broth and continuous DC voltage measurement for the next 5 mins. For 24h, 48h or 72h voltage readings, dressings incubated in YPD broth (for respective time intervals) were placed in separate petri plates and voltage was measured as earlier. All voltage measurements were performed at room temperature. Four independent measurements (each lasting for 5 mins) were taken from WED and Ag-only fabric for each time point.
2.5. Candida albicans in vitro biofilm formation in response to fabric
Log phase Candida albicans (1 × 104 cells) were allowed to form biofilms, in vitro, in two sets of 24-well polystyrene microtiter plates. One set had one sterilized round cover glass (1.2 cm diameter, sterilized by treatment 100% ethanol for 10 mins) in each well while in the second set, direct attachment of C albicans biofilms on polystyrene surface was assayed using Microtiter plate crystal violet assay [22]. Untreated cells were provided with only YPD broth and incubated at static condition. Treatment groups were exposed to moistened fabrics (discs of ~1.5 cm diameter, with their conducting side facing biofilms) or ketoconazole or a combination of both with same media and incubation conditions. After every 24h, old media was discarded, and fresh YPD broth was added in all wells to replenish nutrition for growing biofilms. After 72h, biofilms formed on cover glasses were stained with modifications in previously described method [23]. Biofilms were washed thrice thoroughly with 500 µl of 1X PBS to remove loosely adhered planktonic cells and fixed with 100 µl of 4% paraformaldehyde for 1h, at room temperature. Fixed biofilms were washed once with 500 µl of 1X PBS and stained with 200 µl of Film Tracer™ SYPRO™ Ruby (1X) (biofilm matrix stain) (Cat no: F10318, Invitrogen) for 30 mins followed by counterstaining with Calcofluor white (cell wall chitin binding dye; 25 µg/ml) for 10 mins, to visualize C albicans cells in biofilms. All staining procedures were done in dark (covered with aluminum foil) at 27°C. After staining, biofilms were washed once again with 200 µl of 1XPBS and mounted between glass slides and cover glasses with 5 µl of Vectashield® Antifade mounting medium (Cat no: P36930, Vector Laboratories, Burlingame, CA). Biofilms were observed on a CLSM at 63X to study their spatial arrangement and thickness.
The second set of biofilms on polystyrene plates (without cover glasses) were washed thrice with 1 ml of with double distilled water thrice followed by air drying and staining with 1 ml of crystal violet (0.1%; Cat no: S25275, Fisher Scientific, Waltham MA) per well for 15 mins. After staining, plates were washed thrice with 1 ml of 1X PBS, air dried and crystal violet was extracted from wells using 1 ml of 90% ethanol per well. Plates were incubated with ethanol for 30 mins followed by absorbance measurements at 590 nm. For Scanning Electron Microscopy (SEM), 10 µl of C albicans cell suspension (1 × 104 cells/ml) was spotted on 0.22 µm polycarbonate membrane discs (cut as 1 cm diameter; Cat no: 1215309, GVS Life Sciences, Sanford, ME) placed on YPD agar plates (with or without ketoconazole). Once the culture spots dried, they were overlaid with moistened fabrics (~ 1.2 cm diameter; to cover the discs). After every 24h, these discs were carefully transferred to fresh YPD agar plates (with or without ketoconazole). At respective time points, discs with biofilms were placed in 24 well polystyrene plates and processed for SEM as previously described [11]. Biofilms were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer with 50 mM sucrose (300 µl/well) at 4°C for overnight followed by washing twice in phosphate buffer with sucrose. Biofilms were then dehydrated in graded ethanol series (50% to 100%) for 15 mins each. After ethanol dehydrations, discs were processed with a series of ethanol: hexamethyldisilazane (HMDS) (Cat no: 16710, Electron Microscopy Services, Hatfield, PA) gradients (3:1, 2:2, 1:3) for 20 mins each and finally biofilms were treated with HMDS alone for 20 mins and left for complete drying. Discs were then mounted on aluminum stubs with the help of a double-sided carbon tape (Cat no: 77816, Electron Microscopy Services) and sputter coated with gold for 60s followed by microscopic observations on SEM (JEOL 7800F, JEOL USA Inc., Peabody, MA) operating at 5 kV in the secondary electron mode.
2.6. Planktonic growth assessment
The effect of electroceutical fabrics on planktonic growth was assessed by culturing C albicans in sterile 14 ml round bottom polypropylene tubes with 5 ml YPD broth [11]. For fabric treatments, fabrics were cut (5 cm x 5 cm) and added to the culture tubes, by rolling them with their conducting side rolled up inside. With this arrangement, cells were in contact with the conducting side of the fabric. Cells were then incubated for 24h, 48h or 72h at 27°C under shaker conditions. Culture tubes for untreated cells and only ketoconazole treated cells did not have any fabrics. After incubation for respective time intervals, 900 µl aliquots were taken from all groups and absorbance was measured using spectrophotometer (Nanodrop 2000c, ThermoFisher Scientific, Waltham, MA) at OD600 nm.
2.7. Transition from yeast to hyphal morphology
The morphological transition of C albicans from yeast form to hyphal form was studied with hyphae inducing conditions (YPD broth with 10 % Fetal Bovine Serum and incubation at 37°C, static condition) [24]. Cells were cultured in 5 ml of hyphae inducing medium, with or without fabrics or ketoconazole. An aliquot of cells (1 ml) was taken from all treatment groups at 6h, 24h, 48h and 72h. Hyphal induction and elongation were monitored microscopically (63X) and images were captured at 6h, 24h, 48h and 72h. For hyphal length measurements, aliquots were taken after 2h in hyphae inducing medium and images were captured at 20X magnification. Hyphal lengths were measured from these images using Accuview software (50 hyphae were measured per experimental group; four biological replicates were assayed in total).
2.8. Assessment of efflux pump activity by Nile Red accumulation assay
The efflux pump activity in C albicans cells, in response to fabrics, was assayed with Nile red accumulation assay [25]. Candida albicans cells were cultured in liquid media (YPD broth), with or without fabrics or ketoconazole. At respective time intervals, an aliquot of cells (1 × 108) was taken and washed with 1 ml of 1X PBS followed by treatment with an assay mixture (200 µl) of 1X PBS with 2% glucose and 7µM Nile red (Cat no: 19123–100MG, Sigma Aldrich). One set of cells was not stained and used as an unstained control. Heat-killed cells (1 × 108 cells, treated at 100°C for 30 mins) were used as a positive control with 100% impaired efflux pump activity and showing maximum Nile red accumulation. After 30 mins of dark incubation at room temperature, cells were washed with 1 ml of 1X PBS followed by flow cytometry (with FL2-PE filter) (10,000 events per sample) to assess Nile red positive cells in control and test samples. Data generated by flow cytometry were analyzed using FlowJo software and graphically represented. Scatter plots with ratio of fluorescence intensity and forward scatter were plotted to show the shift in cell populations with respect to Nile red accumulation. Samples remaining after flow cytometry were also observed at 63X magnification as a microscopic confirmation. Two technical replicates were assayed for each experimental group and in total six biological replicates were assayed.
2.9. Metabolic state assessment by FUN1 staining
The metabolic state of C albicans cells, in response to fabrics alone or with ketoconazole, was measured using a two-color fluorescent stain, FUN®1 (Cat no: F7030, Invitrogen, Carlsbad, CA) [26]. After treatments with electroceutical fabrics, an aliquot of C albicans cells (1 X 108) was washed with 1 ml of 1X PBS followed by treatment with 100 µl of FUN®1 (1:1000; freshly diluted in 1X PBS) for 40 mins under dark condition. Cells were then washed once with 1 ml of 1X PBS followed by counter staining with 100 µl of Calcofluor white (25 µg /ml; Cat no: 18909–100ML-F, Sigma Aldrich) for 10 mins under dark conditions. After counter staining, cells were again washed with 1 ml of 1X PBS; an aliquot of cells was trapped between a cover glass and glass slide and observed at 63X on a Confocal Laser Scanning Microscope (CLSM; LSM 880, Carl Zeiss, Oberkochen, Germany). Images captured at this magnification were analyzed using ZEN blue software (Carl Zeiss Microscopy LLC, White Plains, NY) to calculate the ratio of red and green fluorescence intensities. Two technical replicates were assayed for each experimental group and in total six biological replicates were assayed.
2.10. Assessment of cell membrane potential changes by DiBAC4(3) staining
Weak electric field responsive change in C albicans cell membrane potential was assayed using slow response membrane potential sensitive dye, DiBAC4(3) [27]. Candida albicans cells were cultured in liquid media (YPD broth), with or without fabrics or ketoconazole (as specified in respective figure legends). At respective time intervals, an aliquot of cells (1 × 108) was taken and washed with 1 ml of 1X PBS followed by treatment with either 200 µl of 10 µM DiBAC4(3) [Bis-(1,3 – dibutylbarbituric acid) trimethine oxonol; Cat no: AS-84700, Anaspec, Fremont, CA] [for DiBAC4(3) single stained controls] or 10 µM propidium iodide (Cat no: P4170–100MG, Sigma Aldrich) [for PI single stained controls] or a combination of both stains. Heat-killed cells (1 × 108; treated at 100°C for 30 mins) and cells treated with Amphotericin B (3 µg /ml; Cat no: A2411–250MG, Sigma Aldrich) for 6h were used as controls for dual stain positive and DiBAC4(3) only positive cells, respectively. After staining, cells were washed with 1XPBS followed by flow cytometry (BD Accuri C6, BD Biosciences, San Jose, CA). Cells were assessed with FL1-FITC filter for DiBAC4(3) and FL2-PE filter for PI fluorescence (10,000 events per sample). Cells from all experimental groups stained only with single fluorescent stain were used for setting compensation of the flow cytometry analysis. Data generated by flow cytometry were analyzed using FlowJo software and graphically represented. Scatter plots with ratio of fluorescence intensities through FL1-FITC filter and FL2-PE filter were plotted. Samples remaining after flow cytometry were also observed at 63X as a microscopic confirmation. Two technical replicates were assayed for each experimental group and in total six biological replicates were assayed.
2.11. Ultrastructure analysis
C albicans cells were processed for ultrastructure studies, with modifications in previously described protocol [28]. After 24h of respective treatments in YPD broth, C albicans cells were initially fixed for overnight with 500 µl of 3% glutaraldehyde/0.1 M phosphate buffer followed by fixation with same fixative with 0.15% tannic acid for 1h. After three rinses in 0.1 M phosphate buffer, the specimens were subjected to secondary fixation with 1% osmium tetroxide/0.1 M phosphate buffer for 1h. Samples were then rinsed for five times with distilled water followed by Enbloc staining with 1% uranyl acetate in distilled water for 1h. Samples were thoroughly rinsed in distilled water and then dehydrated through a range of 70–100% ethanol. After dehydration, samples were infiltrated with acetone and embedding resin (50:50) for 48 h. Specimens were then embedded with 100% resin (Embed 812, Electron Microscopy Sciences, Hatfield, PA) and polymerized overnight. Sections were cut, 80–85 nm and placed on copper mesh grids and viewed on Transmission Electron Microscope (TEM) (ThermoFisher, Tecnai Spirit, Hillsoboro, OR) and images were captured with CCD camera (Advanced Microscopy Techniques Danvers, MA). Analysis of cell wall thickness was done with TEM images using ImageJ software (n = 20 cells).
2.12. Semi-quantitative analysis for cell wall carbohydrates – chitin, beta-glucan and mannan
After 24h treatment, with fabrics alone or in combination with ketoconazole, 500 µl of C albicans cells were washed once with 1 ml of 1X PBS followed by chitin staining with Calcofluor white (Sigma Aldrich) (25 µg /ml) for 10 mins [29]. For beta-glucan staining, cells were incubated with Aniline Blue (0.05%; Cat no: AN105–10GM, Spectrum Chemical, Tucson, AZ) for 15 mins [29]. Cell wall mannans were stained with Concanavalin A – Texas Red conjugate (25 µg /ml, Cat no: C825, Life Technologies, Carlsbad, CA) for 40 mins [30]. All staining procedures were performed under dark conditions. After staining, cells were washed once with 1 ml of 1X PBS and observed under 63X magnification. Microscopic images (10 per group per biological replicate) were used for measuring fluorescence intensities using ZEN blue software.
2.13. Secondary cell wall stress assay
After exposure to weak electric fields, C albicans were subjected to secondary cell wall stress assay as previously reported [31, 32]. Following abiotic stress agents were used for this assay: heat stress (42°C), osmotic stress (1M KCl and 1M NaC) and cell wall perturbing agents (Calcofluor white - 50 µg /ml and SDS – 0.01%). Briefly, cells were cultured in liquid media (YPD broth), with or without fabrics or ketoconazole. At respective time intervals, an aliquot was taken, and cells were washed with 1 ml of 1X PBS followed by adjusting the cell count to 1×108 cells/ml with 1X PBS. These cells were then serially diluted (10−1 to 10−6) with 1X PBS, followed by spotting 10 µl each of two selected dilutions for that time point along with the neat (undiluted) culture on YPD agar with aforementioned stressors (osmotic and cell wall perturbing agents). These plates along with control group plates (cells spotted on YPD agar plates without any stress agents) were incubated at 27°C. For high temperature stress, cells were spotted on YPD agar plates and incubated at 42°C. After 48–72h incubation, plates were observed for C albicans growth followed by imaging the plates. Two technical replicates were assayed for each experimental group and in total four biological replicates were assayed.
2.14. Quantitative Real time PCR
For real time PCR analysis, C albicans cells were cultured in three different culture conditions. One set of cells were cultured in YPD broth, either with fabrics or with ketoconazole or a combination of both. This set was used for gene expression analysis for cell wall related and efflux pump activity related genes. A second set of cells were cultured in hyphae inducing medium (as mentioned earlier) and cells were harvested after 24h incubation. Third set of cells were allowed to form biofilms on 6-well polystyrene plates (as described earlier) and after 72h of biofilm formation, supernatant was discarded, and wells were washed thrice with 2 ml of 1XPBS (each wash) to remove planktonic and loosely attached cells. After washing, 1 ml of 1X PBS was added to the wells and wells were scrapped with sterile cell scrapers to detach cells adhered to polystyrene bottom followed by collection in 1.7 ml tubes. Cells were centrifuged at 10,000 x g for 5 mins at room temperature, supernatant discarded and processed for RNA extraction and cDNA preparation as follows. RNA was extracted using RiboPure™ Yeast RNA purification kit (Cat no: AM1926, Invitrogen) as per the manufacturer’s protocol. Briefly, ~3 × 108 C albicans cells (for each experimental group) were first re-suspended in Lysis buffer + 10 % SDS + Phenol:Chloroform:IAA mixture (480 µl + 48 µl + 480 µl) and then lysed by vigorous vortexing (10 min) in presence of ~750 of ice-cold Zirconia bead. After lysis, cells were centrifuged at 16,100 x g for 5 mins at room temperature to separate the aqueous, phase containing the RNA, from the organic phase. This aqueous phase (~ 500 µl) was then transferred to a 15 ml polypropylene tube, followed by addition of 1750 µl of Binding buffer and 1175 µl of 100 % Ethanol. This sample mixture was then allowed to bind to the Filter Cartridge through a series of centrifugation steps, each at 16,100 x g for 5 mins at room temperature. After complete binding, the Filter Cartridges were first washed with 700 µl of Wash Solution 1 followed by two washes with 500 µl each of Wash Solution 2. After sufficient washing, RNA from the Filter Cartridges was eluted in 50 µl of Ultra-Pure Water (UPW) followed by DNAse 1 treatment with 1/10th volume of 10X DNase 1 buffer and 4 µl of DNase (8 U) for 30 mins at 37°C. DNase inactivation was carried out by incubating with 0.1 volume of DNase 1 Inactivation Reagent for 5 mins at room temperature. RNA supernatant was transferred to fresh 1.7 ml tubes after centrifugation at 16,100 x g for 3 mins at room temperature. RNA concentration and purity (260/280 nm ratio) was checked on Nanodrop followed by cDNA preparation using Superscript VILO cDNA synthesis kit (Cat no: 11754–050, Invitrogen) [33]. Briefly, 4 µl of 5X Vilo reaction mix and 2 µl of 10X Superscript enzyme mix were added to 2 µg RNA and the volume was made up to 20 µl with UPW. The contents were mixed thoroughly and incubated at 25°C for 10 mins followed by incubation at 42°C for 60 mins. The reaction was terminated at 85°C (5 mins). RNA thus obtained was used for real time PCR analysis or stored at −80°C till further use.
Primers used for real time PCR in the current study were procured from Integrated DNA Technologies (Coralville, IA) and their sequences are given in Table 1. Sequences for some primer pairs were obtained from previous studies while remaining were designed using Primer 3 software [34] and the specificity of these primers was checked by comparing their sequences to the C albicans database http://www.candidagenome.org/ using BLAST [35]. All primers were used at a final concentration of 100 nM (per reaction). Real-time PCR was performed in 96-well plates on a QuantStudio™ 3 Real-Time PCR system (A28131, Applied Biosystems, Foster City, CA). Five microliters of 1:10 diluted cDNA samples (prepared as above) and 20 μl of mastermix (containing 5 µl of PowerUp™ SYBR™ Green Master Mix [Cat no: A25743, Applied Biosystems] along with UPW and primers) were added to the plates. Real-time PCR reactions were performed by holding the reaction at 50°C for 5 min followed by denaturation at 95°C for 20 s and 40 cycles of 0.01 s at 95°C and 20 s at 55°C. Following PCR, samples were subjected to incubations of increasing temperature starting from 60°C to 95°C to obtain melt curves. Real-time PCR data were normalized to one reference gene - ACT1. Expression levels were quantified employing the 2 (−ΔΔct) relative quantification method [11, 33].
2.15. Agar lawn assay for fungicidal activity
Fabrics (5 cm x 5 cm) were moistened with sterile YPD broth and placed in empty petri plates with the conducting side of the electroceutical fabric facing up [11]. One hundred microliter of log phase C albicans cells (1 × 108 cells) was thoroughly mixed in sterile molten YPD agar (10 ml) and poured evenly in plates with fabrics. For untreated and ketoconazole treatment, plates did not have any fabric. After incubation for 48–72h, plates were observed for any zone of growth inhibition around fabrics. Images of these culture plates were captured, and the area of growth inhibition was calculated with ImageJ software.
2.16. Statistical Analysis
GraphPad Prism (GraphPad Software) v8.0 was used for statistical analyses. Statistical analysis between multiple groups was performed using one-way or two-way analysis of variance with the post-hoc Sidak multiple comparison test. P <0.05 was considered statistically significant. Significance levels and exact P values are indicated in all relevant figures. Data for independent experiments were presented as means ± SD. Individual data points are plotted reflecting ‘n’ for each experiment.
3. Results
3.1. Wireless electroceutical dressing
The electroceutical dressing tested in the present study is composed of alternating metal deposition spots of silver and zinc metals printed on a polyester fabric (Fig. 1a–b). The silver dots have a diameter of 2mm while the zinc dots are of 1mm diameter. These dots are placed in close proximity (~ 1mm) to each other. In the presence of a electrolyte containing liquid medium, the Ag/Zn redox couple generates weak electric field the strength of which depend on the composition of the wetting medium. Unprinted fabric without any metal deposition was used in this study as a control for the dressing fabric (Figure S1a). Polyester fabric with only silver metal spots was used in the present study as a control for the microbicidal activity of silver ions (Figure S1a). Under dry conditions, the electroceutical dressing did not generate measurable electric potentials. After being moistened by YPD broth, the Ag spots displayed positive potentials while the Zn spots were highly negative. As shown in the two-dimensional contact surface potential map (Fig. 1c), both Ag and Zn spots can be clearly visualized and their sizes matches well with the factual diameter, indicating that electrical potential can be generated stably at the region of printed metal patterns. The averaged potential of Ag spots and Zn spots of the measured region with reference to unprinted dressing were recorded as +40 mV and −680 mV, respectively. The voltage measurements for two adjacent Ag and Zn dots in the dressing were also performed with different incubation times. Finite element simulations showed that electric fields project to a significant degree above the WED fabric, with field strength dropping to 5 V/m around 3 mm above the fabric surface (Fig. 1d). Upon addition of YPD broth to the dry dressing, WED was activated within 12–15s generating peak DC voltage ~0.2 V for two adjacent Ag and Zn dots (electrodes; Fig. 1e). Voltage generation in WED increased in a range between 0.1–0.5V after 24h incubation in YPD broth (Fig. 1f–h). The unprinted fabric without any electrode did not provide any such reading. Testing the two adjacent Ag dots in the Ag-only control fabric showed negligible DC voltage generation upon wetting with YPD broth or after incubation in YPD broth for 24h or 72h (Figure S1b–e).
Figure 1: Electroceutical fabric as source of weak electric fields.
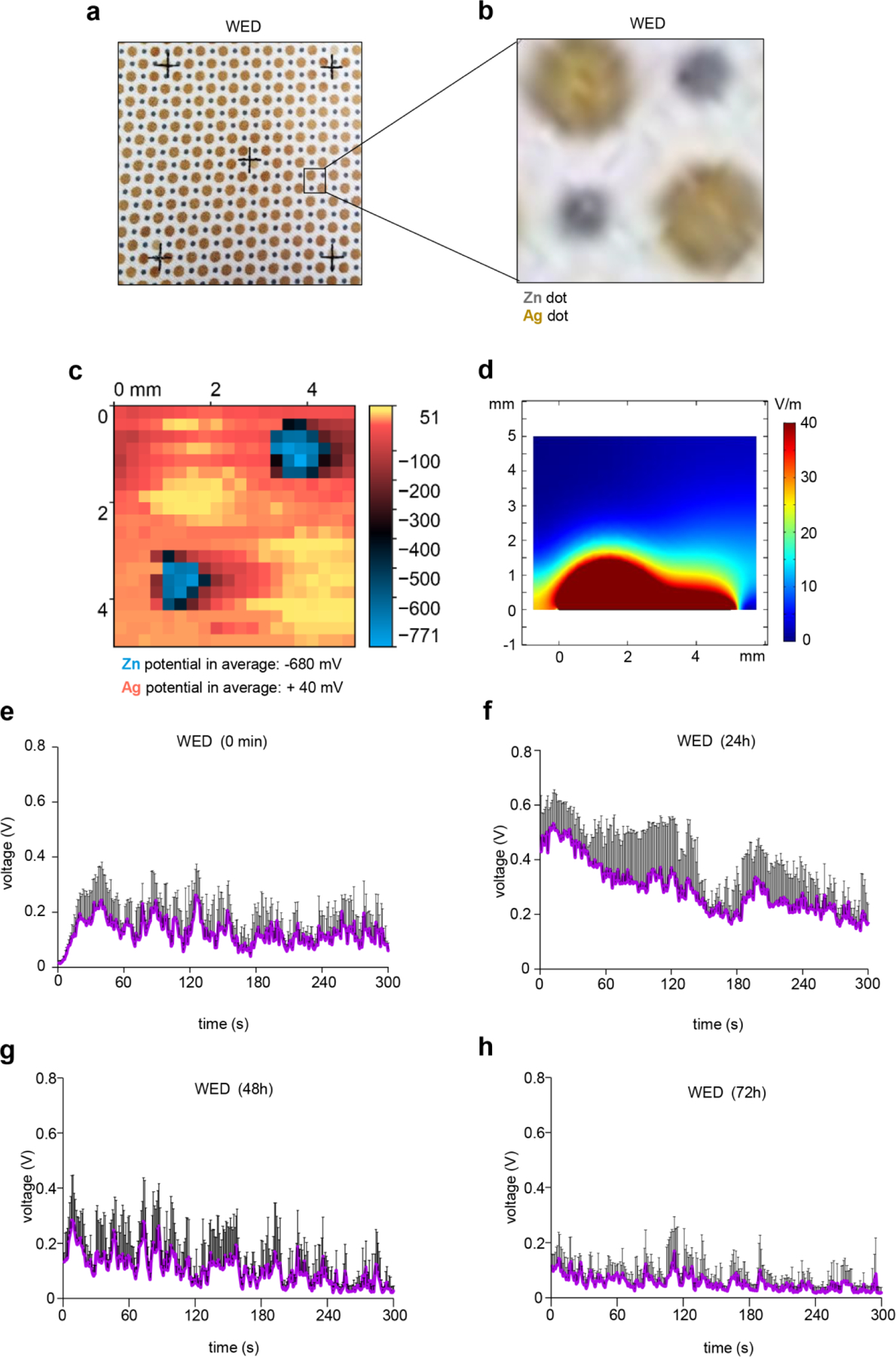
(a) Photomicrograph of WED. (B) Zoomed in image of a region of interest on WED corresponding to arrangement of silver and zinc dots. (c) Two-dimensional surface potential map of a region similar to (b) of the dressing in presence of YPD broth. The averaged potentials of Zn and Ag were measured as −680 mV and +40 mV, respectively. (d) Calculated E-field map from FEM simulations in perpendicular direction from WED fabric surface showing 3 mm above the fabric has E-field of 5 V/m. A slice of the field values was selected that crossed both a Ag and Zn electrode. (e) – (h) Voltage measurements for WED. Voltage generated by WED was measured using the Amprobe multimeter in presence of YPD broth. The multimeter probes were placed on adjacent Ag and Zn dots. At 0s, 200 microliters of YPD broth was added to the respective fabric and DC voltage was measured. Further, voltage measurements were recorded for these fabrics after incubation in YPD broth for 24h, 48h and 72h. n = 4. Data are represented as the mean ± SD.
3.2. Inhibition of Candida albicans biofilm formation and planktonic growth
CLSM analyses of biofilm formation demonstrated no change of biofilm formation in response to ketoconazole, control fabric or Ag-only fabric. Significant reduction in biofilm thickness was noted when cells were exposed to WED alone or in combination with ketoconazole (Fig. 2a, Figure S3a). Akin to the outcome of ketoconazole alone, addition of the drug to WED did not cause any further difference in biofilm outcome (Fig. 2a, Figure S3a). Visualization of biofilm aggregate architecture using SEM revealed biofilms hyphal morphologies within 24h (Fig. 2b, Figure S2). Such structures were abundant in untreated cells, cells treated with unprinted fabric or Ag-only fabric. In biofilm-forming cells treated with ketoconazole, the filamentation appeared after 48h. In WED treated cells, hyphal morphology was absent (Fig. 2b, Figure S2, Figure S3b). Attachment to polystyrene plates, a common biofilm assay, was studied using crystal violet assay. All groups studied, except those involving WED, tested positive demonstrating biofilm forming characteristic. WED, alone or in combination with ketoconazole, markedly abrogated the attachment properties of the cell (Fig. 2c). The presence of ketoconazole with WED did not make any further difference. Expression levels of EAP1, a gene responsible for enhanced adherence to polystyrene [36, 37] was down-regulated in response to WED, alone or comparably in combination with ketoconazole (Figure S3f). While ketoconazole did lower the expression of EAP1 (Figure S3f) no difference in attachment was observed in the functional assay (Fig. 2c).
Figure 2: WED inhibited biofilm formation and planktonic growth in Candida albicans.
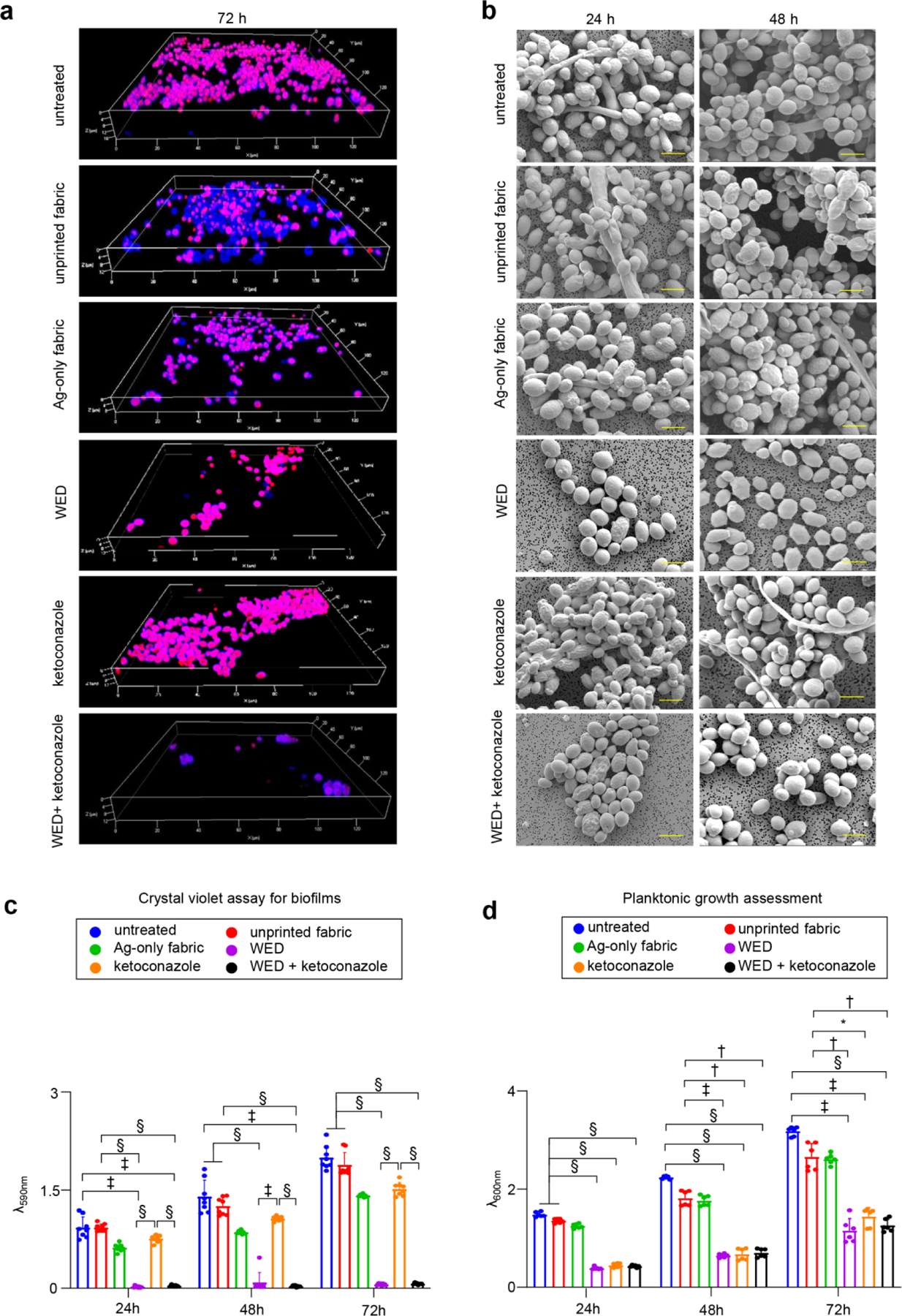
(a) Representative CLSM images for C albicans biofilm thickness. Biofilms were allowed to form in presence or absence of fabrics (alone or in combination with ketoconazole). After 72h, biofilms were stained with Calcofluor white and Sypro Ruby (biofilm matrix stain) and observed at 63X magnification. Display settings for all images were kept same. (b) Representative SEM images for C albicans biofilm architecture. In vitro biofilms (24h and 48h old) formed on polycarbonate membrane discs were processed for SEM and images were captured at 4000X magnification. Scale bar represents 5 µm. (c) Effect on in vitro biofilm attachment. C albicans cells were allowed to form biofilms on six well polystyrene plates, in presence of fabrics or ketoconazole. At respective time intervals, planktonic cells were washed and biofilms attached to the wells were stained with crystal violet followed by ethanol extraction and spectrophotometric analysis at 590 nm. n = 8. §P < 0.0001, ‡P < 0.0005 and *P < 0.05 (Two-way ANOVA followed by post-hoc Sidak multiple comparison test). Data are represented as the mean ± SD. (d) Effect on planktonic growth measured through absorbance. C albicans cells (untreated or with respective treatments) were cultured in YPD broth. At respective time intervals, absorbance was measured at 600 nm to assess the rate of planktonic growth. n = 6. §P < 0.0001, ‡P < 0.0005 and *P < 0.05 (Two-way ANOVA followed by post-hoc Sidak multiple comparison test). Data are represented as the mean ± SD.
To further address the mechanisms responsible for the anti-biofilm properties of WED, BCR1, BRG1 and ROB1 representing three key positive transcriptional regulators of biofilm formation were studied [38, 39]. Compared to that of other groups, WED treatment markedly down-regulated the expression of BRG1 and ROB1. Comparable effects on these two genes were noted with ketoconazole treatment alone (Figure S3c–d). Interestingly, in response to WED-dependent down regulation of BRG1 and ROB1, BCR1 expression was markedly induced in what seems to be a futile compensatory response. Of further interest is the observation that while ketoconazole alone did not display such compensatory effect, it acted synergistically with WED to cause a surge in BCR1 expression (Figure S3e).
Exposure to WED not only resisted biofilm formation but it suppressed planktonic growth as well. Cells treated with the dressing showed two-fold reduction in planktonic growth (Fig. 2d). This reduction was comparable to the tested MIC of ketoconazole (Fig. 2d). When grown under conditions of Agar lawn assay, WED showed a prominent zone of inhibition. WED was embedded 5mm deep in agar. The zone of inhibition was larger than the size of the fabric (Figure S4a–b). Ketoconazole did not further increase the zone of inhibition when treated in conjunction with WED. Under these conditions, Candida albicans growth was not inhibited in presence of unprinted or Ag-only fabric (Figure S4a–b).
3.3. Inhibition of Candida albicans morphological switch
Transition of yeast cells to hyphal form was studied with hyphae inducing conditions i.e., YPD broth with 10% FBS and incubation at 37°C [24]. Hyphal length measurements were performed after 2h incubation in hyphae inducing medium (Fig. 3a). Hyphal induction was observed in untreated cells or cells treated with unprinted fabric or Ag-only fabric. However, in the presence WED hyphae were not induced. Transient pseudohyphal forms were observed (2h) instead under such conditions of weak electric field (Fig. 3a, Figure S5a). Over a longer time period (72h) pseudohyphae failed to convert to hyphae and disappeared. Thus, the effect of WED against hyphal induction was persistent. On the other hand, ketoconazole had a transient effect against hyphal induction which held until 24h. In 48–72h, filamentation was evident in the presence of ketoconazole. When used in combination, the effects of WED+ketoconazole were comparable to that of WED alone (Fig. 3b). These results demonstrate that weak electric field is effective in perturbing yeast to hyphal switch in C albicans cells.
Figure 3: WED inhibited inducible hyphal transition in Candida albicans.
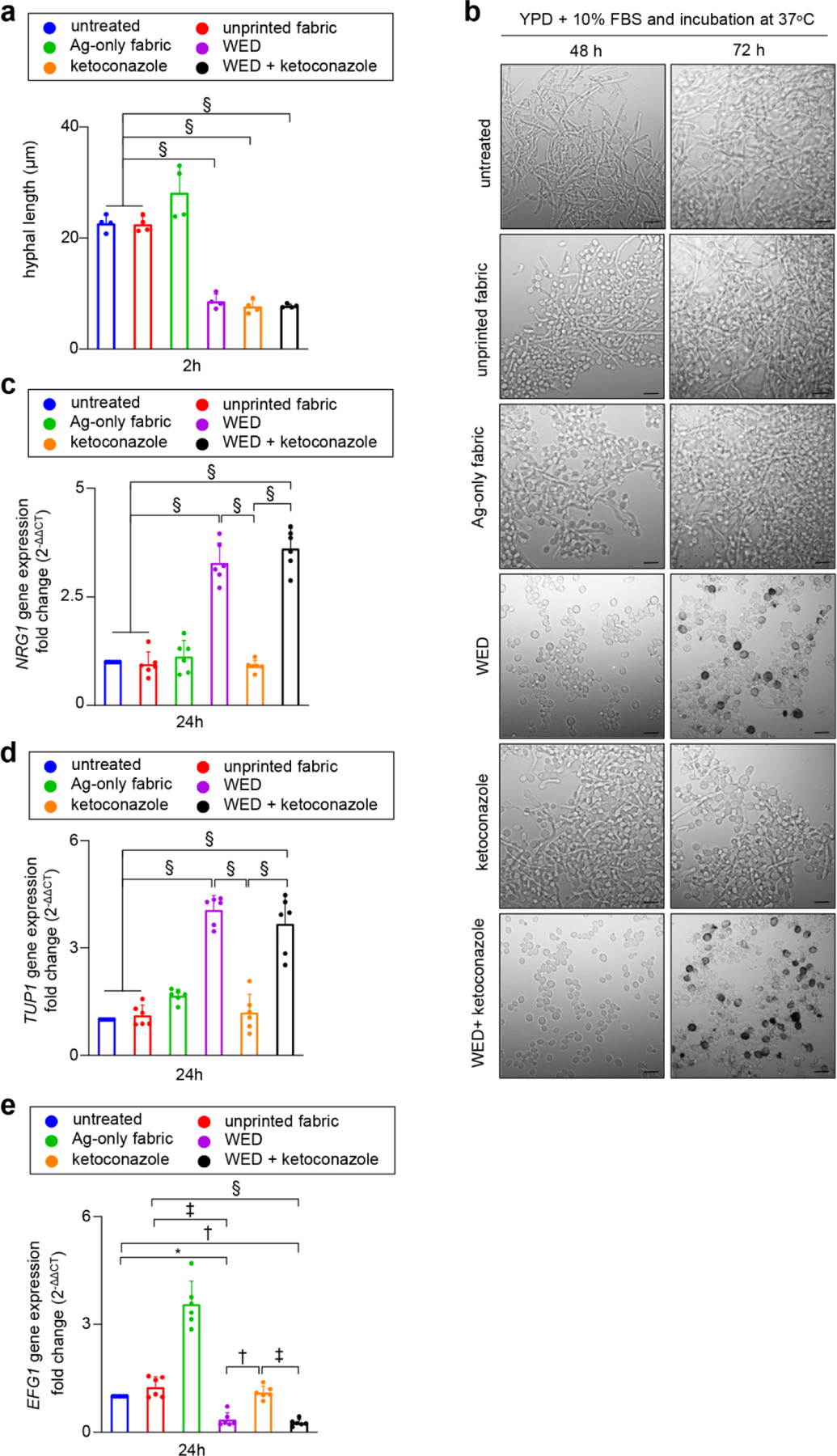
(a) Hyphal length assessment. Candida albicans cells (untreated or with respective treatments) were cultured in hyphae inducing condition (YPD broth with 10% FBS and incubated at 37°C) for 2h, followed by microscopic observation and image capture at 20X. These images were used to measure hyphal lengths with AccuView software. n = 4 biological replicates (40–70 hyphae measured per experimental group per replicate). §P ≤ 0.0001 (One-way ANOVA followed by post-hoc Sidak multiple comparison test). (b) Representative images for inhibitory effect of WED on inducible hyphal transition. Candida albicans were cultured in in hyphae inducing condition. After 48 and 72h an aliquot from each sample was observed at 63X magnification. Scale bar represents 5 µm. (c) – (e) Quantitative real-time PCR analysis for yeast to hyphal transition pathway related genes – NRG1, TUP1 and EFG1. C albicans cells were cultured in hyphae inducing medium (all six experimental groups) for 24h at 37ᴼC. RNA was extracted from these cells and cDNA was prepared. Real time PCR analysis was performed for transcripts: NRG1, TUP1 and EFG1 with aforementioned cDNA. Candida albicans actin gene (ACT1) was used as a housekeeping control. Expression levels were quantified employing the 2 (−ΔΔct) relative quantification method. n = 6. §P < 0.0001, ‡P < 0.0005 and *P < 0.05 (One-way ANOVA followed by post-hoc Sidak multiple comparison test). Data are represented as the mean ± SD.
The molecular basis of the effect of WED on yeast to hyphal transition was examined by studying EFG1, NRG1, TUP1, RFG1, key regulators of yeast to hyphae transition pathway; and two important markers of hyphal cell wall as represented by ALS3 and HWP1 [40]. Exposure to WED alone induced NRG1 and TUP1, negative regulators of filamentation (Fig. 3c–d). In addition, WED down-regulated EFG1, a positive regulator of hyphal pathway (Fig. 3e). With these effects that are consistent with the hyphal induction outcomes data (Fig. 3b), we observed yet another effect which appear to be compensatory but functionally futile. WED downregulated the expression of RFG1, another negative regulator of filamentation (Figure S5b). Consistent with the anti-hyphal properties of WED, the expression of ALS3 and HWP1 were also significantly diminished in cells exposed to weak electric field (Figure S5c–d). Albeit with a lower magnitude, ketoconazole showed effects on the expression of RFG1, ALS3 and HWP1 expressions that were comparable to that of WED (Figure S5b–d). However, ketoconazole failed to reproduce the effects of WED on NRG1, TUP1 and EFG1 (Fig. 3c–e). In contrast to the effect of WED, the fabric with Ag-only markedly induced the pro-hyphal EFG1 (Fig. 3e). Expression levels of all aforementioned genes in C albicans cells treated with unprinted fabric were equivalent to that of untreated cells (Fig. 3c–e, Figure S5b–d).
3.4. Impaired efflux pump activity
Efflux pump activity was studied by Nile red accumulation assay [25]. Nile red, a fluorescent lipophilic dye, upon entry into the cells, binds to lipids and shows increased fluorescence intensity. Healthy active cells with fully functional efflux activity will efflux out Nile red when provided with glucose as a competitor in the assay conditions. Cells with impaired efflux activity retain Nile red. Untreated cells, cells treated with unprinted fabric or Ag-only fabric displayed fully functional efflux pump activity (Fig. 4a–c, Figure S6). Exposure of cells to WED resulted in Nile red accumulation (Fig. 4a–c). In contrast, consistent with the reported literature [25], treatment of cells with ketoconazole boosted efflux pump activity (Fig. 4a–b). Results from microscopic as well as flow cytometric analyses were consistent demonstrating impairment of efflux pump activity in response to WED (Fig. 4a–c, Figure S6). When ketoconazole was added with WED, the results were identical to what was seen with WED alone. In such case, the pump boosting effect of ketoconazole was overridden by WED.
Figure 4: WED impaired efflux pump activity in Candida albicans.
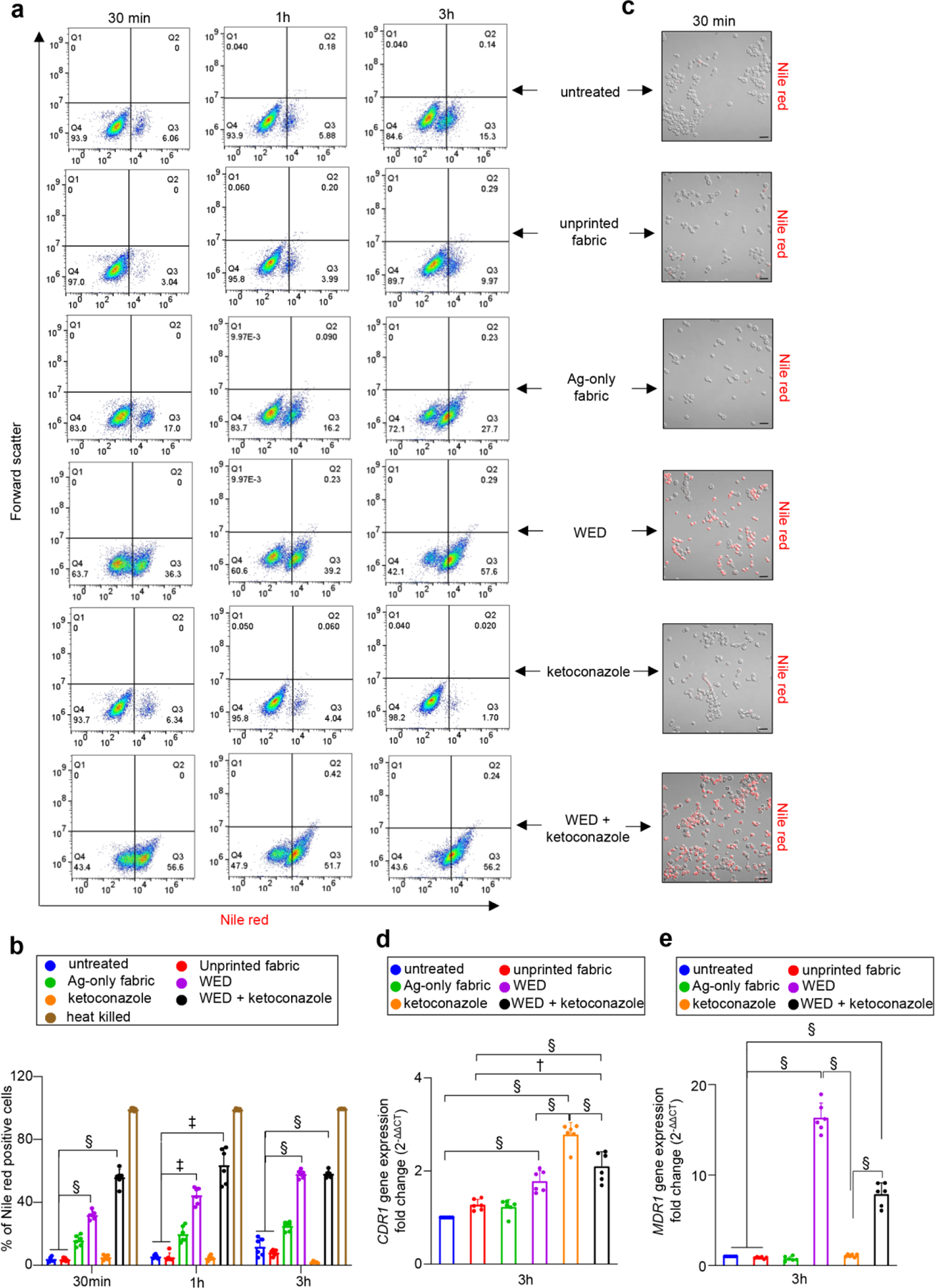
(a) Representative scatter plots for flow cytometry analysis of Nile red positive population. C albicans cells (untreated or with respective treatments) were cultured in YPD broth. At respective time intervals, an aliquot was taken and treated with 7 µM Nile red in presence of 2% glucose. After 30 mins of Nile red treatment, cells were washed and processed for flow cytometry analysis. Scatter plots with fluorescence intensity (for Nile red) versus forward scatter were plotted. (b) Graphical representation of flow cytometry analysis. Nile red positive populations, analyzed using FlowJo software, were plotted in this graph. n = 6. §P < 0.0001, ‡P < 0.0005 (Two-way ANOVA followed by post-hoc Sidak multiple comparison test). Data are represented as the mean ± SD. (c) Representative images of C albicans cells after Nile red accumulation assay. Aforementioned samples (after 30 mins of fabric treatment) were observed at 63X magnification. Scale bar represents 10 µm. Display settings for all images were kept same. (d) and (e) Quantitative real-time PCR analysis of C albicans efflux pump activity related genes – CDR1 and MDR1, respectively. After 3h exposure to fabrics or ketoconazole, RNA was extracted from C albicans cells and cDNA was prepared. Real time PCR analysis was performed for transcripts: CDR1 and MDR1 with aforementioned cDNA. Candida albicans actin gene (ACT1) was used as a housekeeping control. Expression levels were quantified employing the 2 (−ΔΔct) relative quantification method. n = 6. §P < 0.0001 and †P ≤ 0.001 (One-way ANOVA followed by post-hoc Sidak multiple comparison test). Data are represented as the mean ± SD.
The efflux pump is encoded CDR1 and MDR1. Under conditions of impaired efflux pump activity is response to WED as reported above, there seems to be a futile induction of both CDR1 and MDR1 (Fig. 4d–e). Increased efflux pump activity in response to ketoconazole can be explained by induction of CDR1 (Fig. 4d). MDR1 remained unchanged (Fig. 4e). The effects of WED+ketoconazole on CDR1 were comparable to that of ketoconazole alone (Fig. 4d). For MDR1, the effect was comparable to that of WED alone (Fig. 4e).
3.5. Impaired cellular metabolism
The metabolic state of C albicans cells was measured using a two-color fluorescent stain, FUN1™ [26]. Metabolically active cells with intact plasma membrane convert FUN1 into orange-red or yellow-orange fluorescent intravacuolar cylindrical structures; cells with intact plasma membrane but little or no metabolic activity exhibit diffused green cytoplasmic fluorescence with no intravaculoar bodies. Dead cells exhibit bright, diffuse, orange-red fluorescence. FUN1™ staining showed distinct intravacuolar structures in untreated cells and cells treated with unprinted fabric or ketoconazole (Fig. 5a) indicating healthy state of metabolism and intact plasma membrane in these cells. WED exposure sharply compromised cellular metabolic status. In the presence of ketoconazole, the effect was synergistic. In response to Ag-only fabric, cell metabolism was slightly dampened (Fig. 5a–b).
Figure 5: WED impaired metabolism in Ketoconazole resistant Candida albicans.
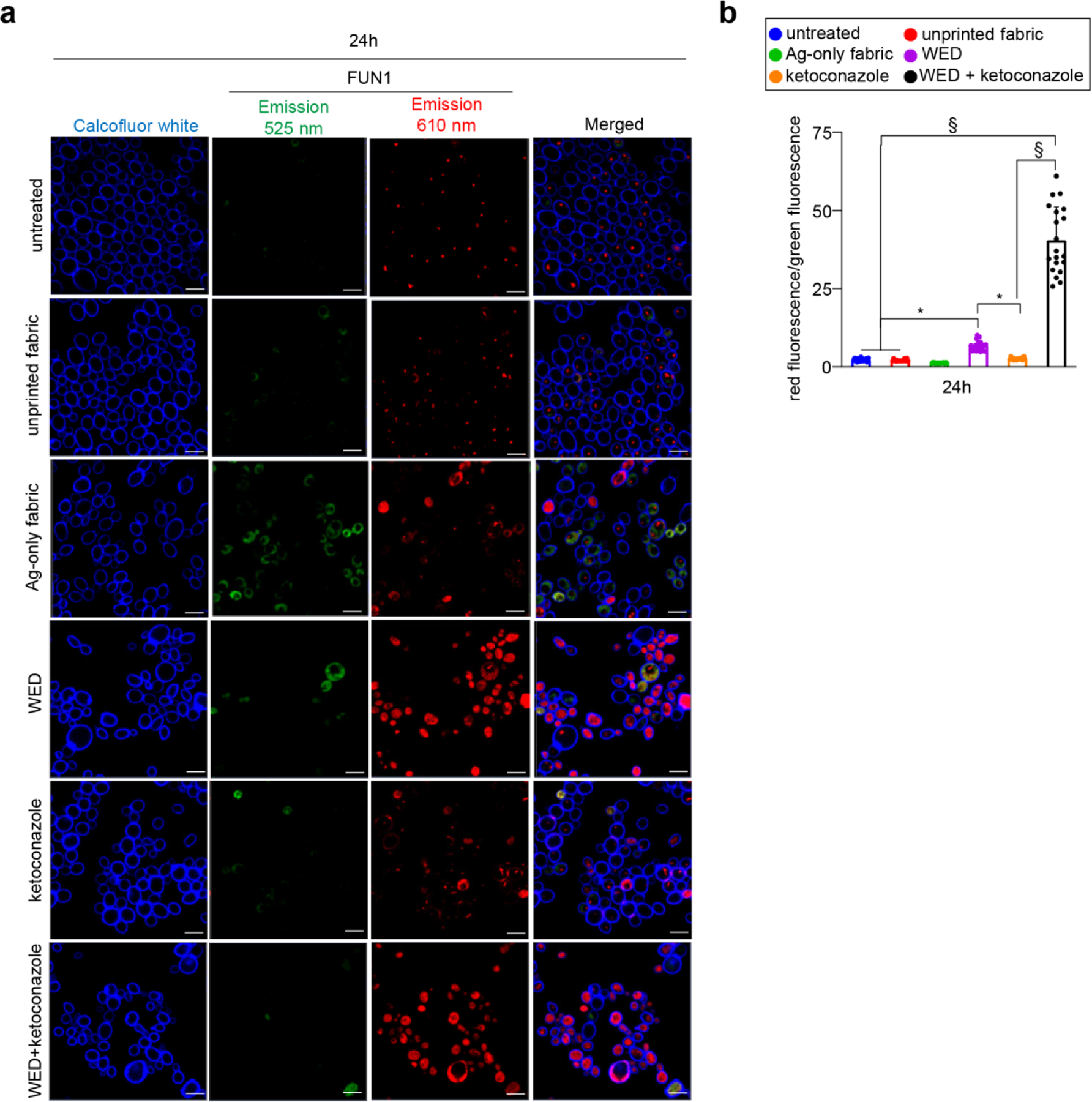
(a) Representative images showing effect of fabrics or ketoconazole on C albicans metabolism. After 24h growth in YPD broth, C albicans were stained with FUN1™ and Calcofluor white. After staining, cells were imaged at 63X magnification for differential staining patterns of FUN1™. Scale bar represents 5 µm. Display settings for all images were kept same. (b) Quantitative assessment of metabolic impairment in C albicans cells. Aforementioned microscopic images were analyzed for red and green fluorescence intensities using Zen Blue software and a ratio of red fluorescence intensity over green fluorescence intensity was plotted for all groups. This ratio is an indirect representation of metabolism impairment and viability loss in C albicans cells. n = 20 images. §P ≤ 0.0001 and *P ≤ 0.05 (One-way ANOVA followed by post-hoc Sidak multiple comparison test). Data are represented as the mean ± SD.
3.6. Cell membrane depolarization
DiBAC4(3) and PI staining were performed to assess membrane depolarization and integrity. C albicans cells treated with Amphotericin B were used as a positive control for DiBAC4(3) only staining while membrane damage associated membrane depolarization in heat killed cells acted as a positive control for dual staining in this assay (Figure S7a–b). Untreated cells or treatment with unprinted fabric, Ag-only fabric and ketoconazole alone resulted in stable membrane potential. WED caused membrane depolarization with considerable loss of membrane integrity over 24h. These effects of WED were in contrast with the effects of ketoconazole which did cause any membrane depolarization. WED+ketoconazole had effects comparable to that of WED alone (Fig. 6a–c, Figure S7c–d, Figure S8a–b).
Figure 6: WED depolarized Candida albicans cell membrane.
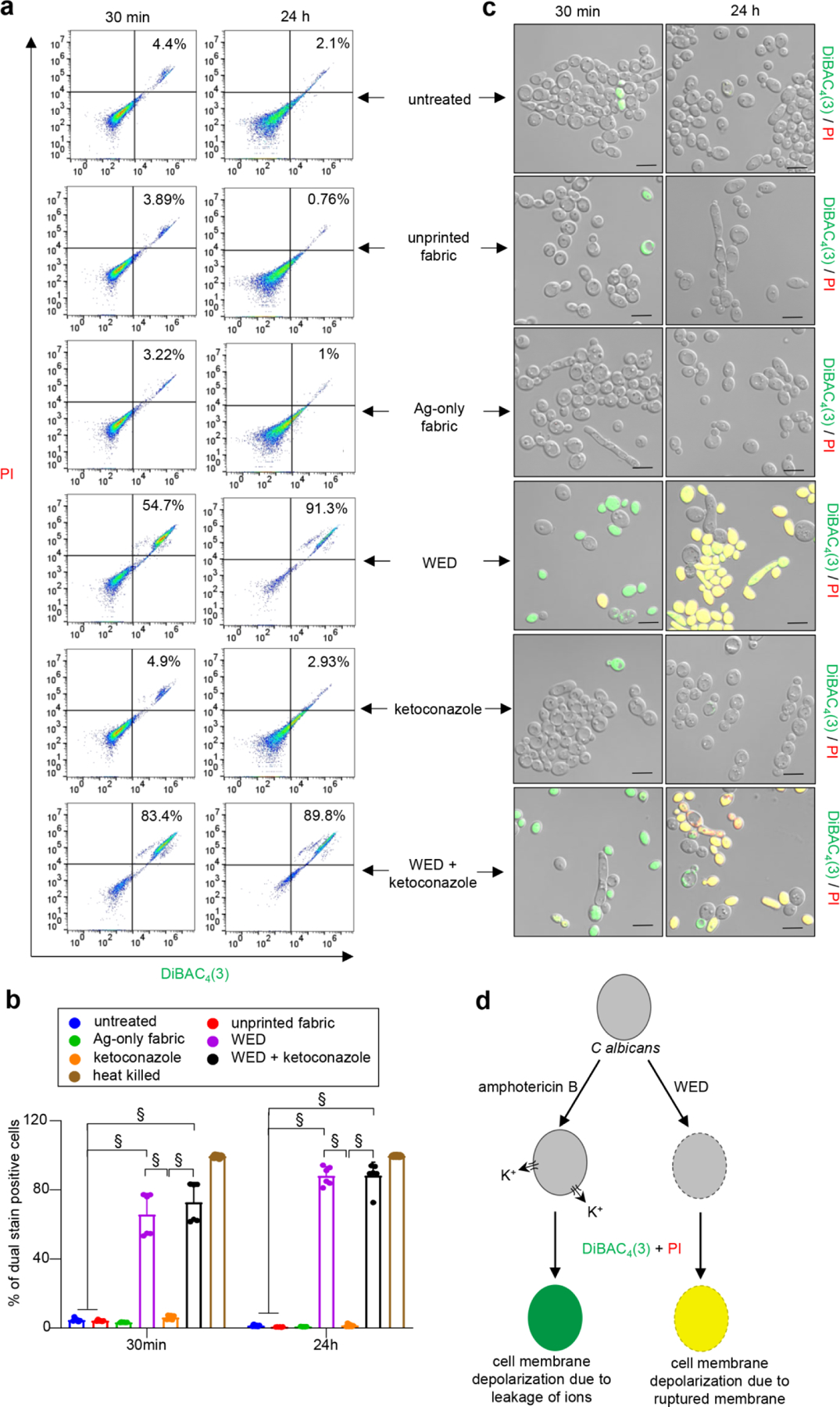
(a) Representative scatter plots for flow cytometry analysis of DiBAC4(3)+ and PI+ population. C albicans cells (untreated or with respective treatments) were cultured in YPD broth. At respective time intervals, an aliquot was taken and stained with DiBAC4(3) and PI. After 30 mins of dual staining, cells were washed and processed for flow cytometry analysis. Scatter plots with DiBAC4(3)+ and PI+ population were plotted. (b) Graphical representation of flow cytometry analysis. Dual stain positive populations, analyzed using FlowJo software, were plotted in this graph. n = 6. §P < 0.0001 (Two-way ANOVA followed by post-hoc Sidak multiple comparison test). Data are represented as the mean ± SD. (c) Representative images of C albicans cells confirming membrane depolarization after treatment with WED, alone or in combination with ketoconazole. Aforementioned samples remaining after flow cytometry were observed at 63X magnification. Scale bar represents 5 µm. Display settings for all images were kept same. (d) Schematic representation of possible mechanism for WED mediated cell membrane depolarization in C albicans cells. Black border on cell represents cell membrane. Dotted border represents damaged cell membrane. Amphotericin B causes cell membrane depolarization by forming pores causing rapid leakage of ions, especially potassium ions (K+), without damaging the cell membrane. Hence cells treated with amphotericin B get stained with DiBAC4(3) only and give a green fluorescence signal. On the other hand, WED treated cells are observed to take up both fluorescent stains [DiBAC4(3) and PI] as witnessed by a yellow fluorescence signal. This indicates WED causes cell membrane depolarization by cell membrane damage.
3.7. Cell wall remodeling
In response to azole drugs, C albicans is known to increase cell wall thickness as defense response [41]. Consistently, exposure of cells to ketoconazole resulted in cell wall thickening. Such response was not observed in response to WED. However, in combination with ketoconazole, WED acted synergistically to augment cell wall thickening (Fig. 7a–b). Cells treated with unprinted fabric or Ag-only fabric did not cause any change in cell wall thickness compared to reference untreated cells. The cell wall is made up of three principal carbohydrate components: chitin, glucans and mannans [29, 30]. Efforts to elucidate changes in cell wall composition led to the observation that cells treated with unprinted fabric or Ag-only fabric did not cause any change compared to reference untreated cells. WED alone, although did not alter cell wall thickness, resulted in significant increase in mannan at the expense of reduction in β-glucan; chitin remained unchanged. In contrast, ketoconazole exposure increased chitin levels; β-glucan and mannan remain unchanged. Exposure to a combination of WED+ketoconazole resulted in an additive response such that mannan and chitin were elevated; β-glucan levels were lower (Fig. 7c–f, Figure S9a–b).
Figure 7: Candida albicans cell wall thickening as observed in response to WED.
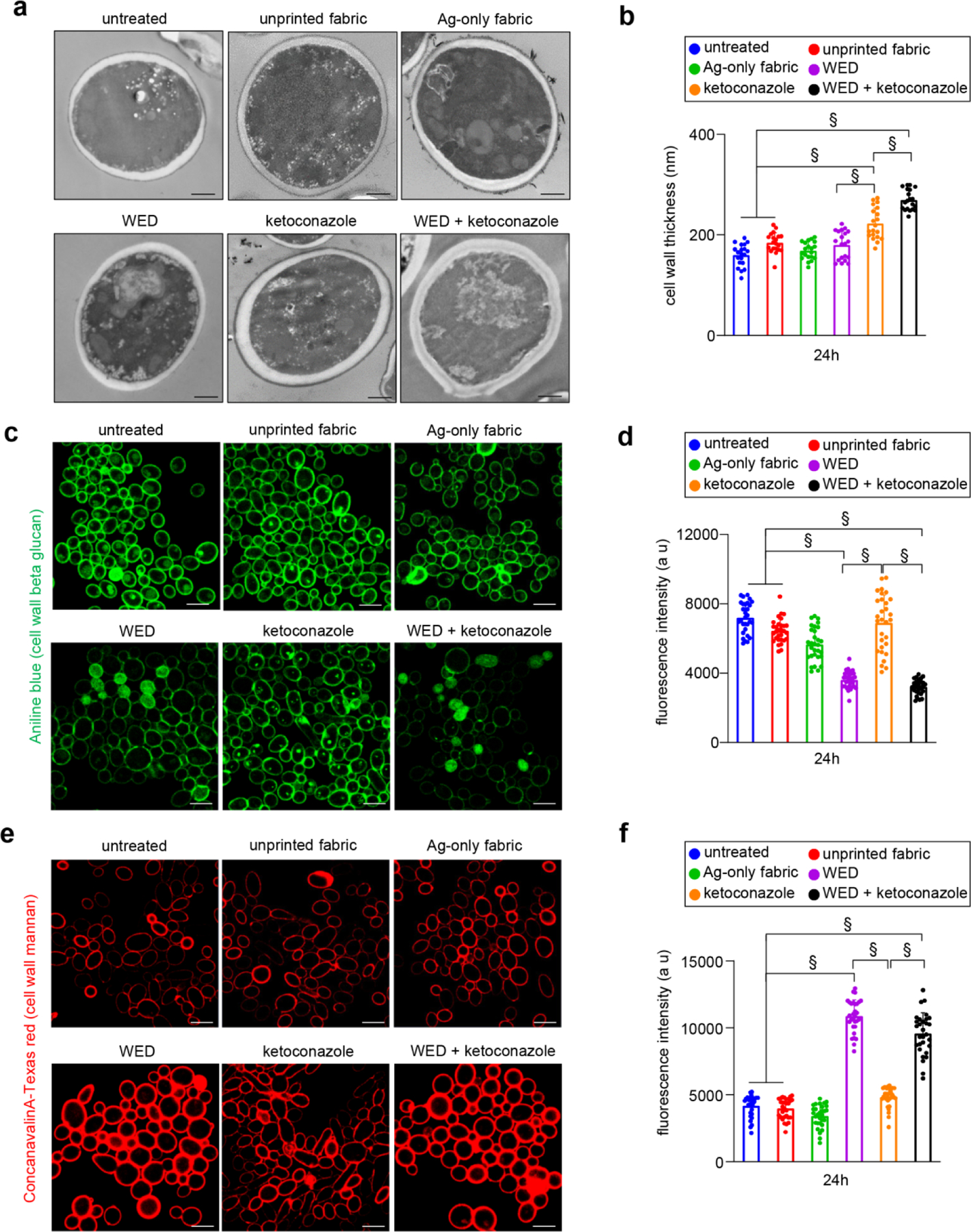
(a) Representative images for ultrastructure analysis to assess C albicans cell wall thickness. C albicans cells (untreated or with respective treatments) were cultured in YPD broth. After 24h, cells were pelleted and processed for TEM. Scale bar represents 600 nm. (b) Graphical representation of C albicans cell wall thickness. Cell wall thickness was measured using ImageJ software. n = 20 cells. §P ≤ 0.0001 (One-way ANOVA followed by post-hoc Sidak multiple comparison test). (c) and (e) Representative images for qualitative assessment of cell wall β-glucan and mannan content, respectively. C albicans cells, after 24h of planktonic growth (untreated or with respective treatments) were stained with Aniline blue (for β-glucan) or Conacavalin A conjugated with Texas red (for mannan) and observed at 63X magnification. Scale bar represents 10 µm. Display settings for all images were kept same. (d) and (f) Semi-quantitative assessment of cell wall β-glucan and mannan content, respectively. Microscopic images captured for aforementioned samples were analyzed for fluorescence intensities using Zen Blue software. These values are graphically represented in terms of arbitrary units (a u). n = 31 images. §P ≤ 0.0001 (One-way ANOVA followed by post-hoc Sidak multiple comparison test). Data are represented as the mean ± SD.
In pursuit of the mechanistic underpinnings of the observed cell wall thickness, the following three genes of functional significance in the context of this study were investigated: CHS3 (major chitin synthase), ERG11 (critical for ergosterol biosynthesis, a major azole target) and GSL1 (encodes beta-1,3-glucan synthase subunit) [42–44]. Cells treated with unprinted fabric did not cause any change in cell wall thickness compared to reference untreated cells. While the Ag-only fabric did not have any effect of cell wall thickness or composition as described above, ERG11 was marginally yet significantly lower; CHS3 and GSL1 remained unchanged. Consistent with its effect on cell wall composition, WED repressed GSL1 and CHS3 remained unchanged; like azole drugs WED alone induced ERG11. Ketoconazole induced ERG11, albeit more strongly than WED. Consistent with its effect in increasing chitin composition in the cell wall, CHS3 was induced by the azole drug. Although ketoconazole had no effect on the beta-glucan composition of the cell wall, it down-regulated GSL1. When ketoconazole was added to WED, it did not cause any further change compared to the effects of WED alone as described above (Figure S9c–e).
Thickening of cell wall is typically viewed as a defense response. Yet WED+ketoconazole, which in other experiments were observed to cause loss of cell integrity, increased cell wall thickness. Thus, a secondary stressor assay was performed to test the significance of cell wall thickness observed in our studies [45]. Secondary stressors high temperature (42°C), osmotic agents (KCl and NaCl), and cell wall stress agents (Calcofluor white and SDS) were independently tested. Indeed, the cell wall thickening induced by ketoconazole fortified the cells with enhanced defense against all three classes of stressors tested. This undesirable effect of ketoconazole was not only neutralized but reversed by the addition of WED. Exposed to this combination, cell wall thickens with clear changes in composition. The net result is that cells become more vulnerable to secondary stressors. This is a diametrically opposite to the effect of ketoconazole alone. The desirable effects of this combination were exactly replicated by WED making it a potent therapeutic candidate. (Fig. 8a–g, Figure S10a–f, Figure S11a–f).
Figure 8: WED mediated Candida albicans cell wall alterations increased cell susceptibility towards secondary stressors.
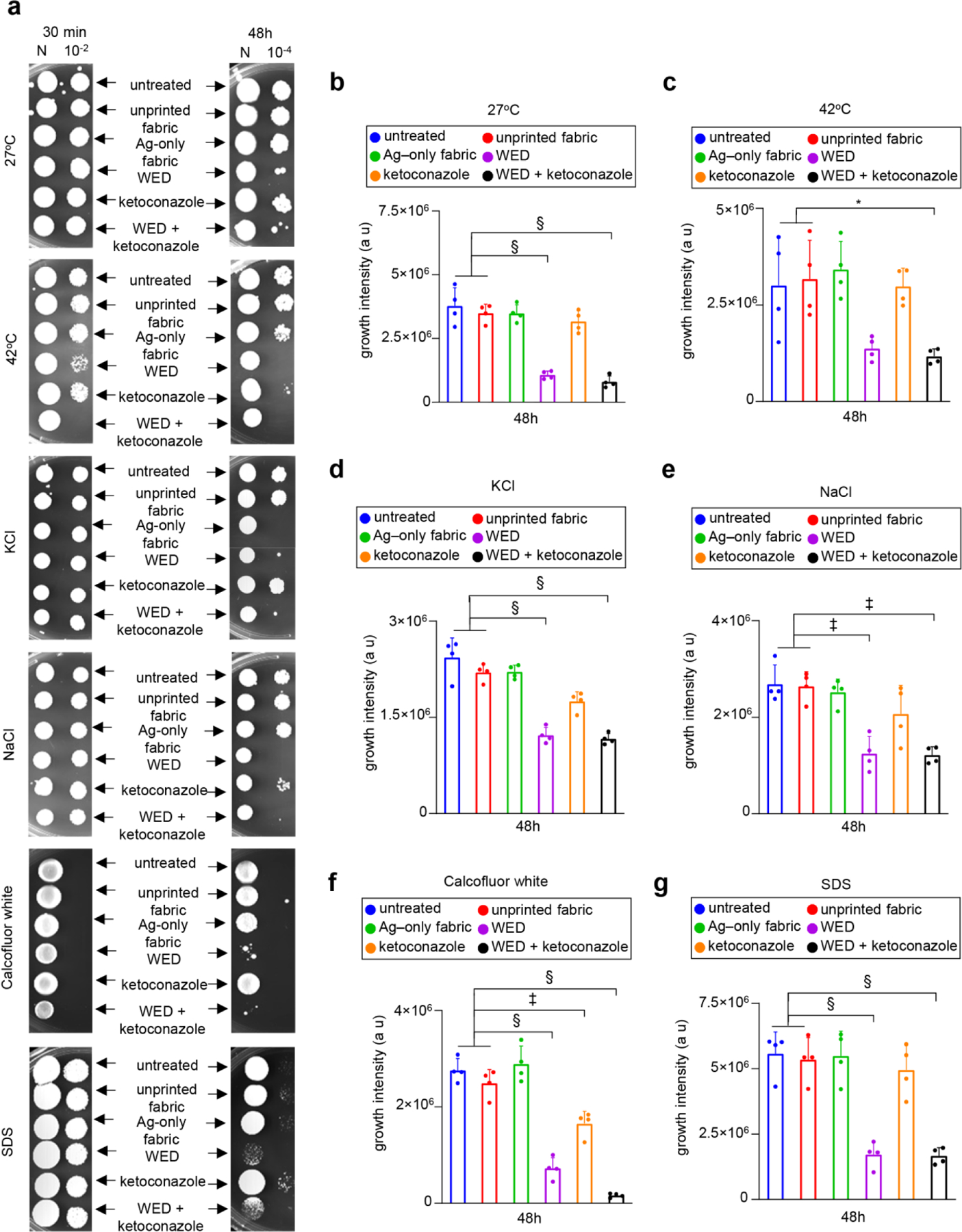
(a) Representative plate images for spot viability assay with secondary cell wall stressors. Candida albicans cells were first cultured in YPD broth with fabrics or ketoconazole or a combination of both. After respective time intervals (30 min and 48h), these stressed cells were washed and spotted on to YPD agar plates with a secondary cell wall stress agent such as 1M KCl, 1M NaCl, 50 µg /ml Calcofluor white or 0.01% SDS. One set of cells were also incubated at 42°C (heat stress). All plates except heat stress plates were incubated at 27°C. After 48–72h incubation, plates were observed for growth. (b) – (g) Growth intensity measurement plots. An area of interest was selected around the growth spots on secondary cell wall stress assay plate images (after 48h) and the intensity of growth was calculated using ImageJ software. This data was graphically represented for 27°C, 42°C, KCl, NaCl, Calcofluor white and SDS respectively. n = 4 plates. §P < 0.0001, ‡P < 0.0005 and *P < 0.05 (One-way ANOVA followed by post-hoc Sidak multiple comparison test). Data are represented as the mean ± SD.
4. Discussion
Rampant clinical use of antibacterial and immunosuppressive agents selectively facilitates fungi to establish infections. Fungal invasion into deep tissues is a major contributor of delayed wound healing in diabetic and burn patients, resulting in prolonged hospitalization, amputation and morbidity [16, 46]. Among yeast infections in such patients, Candida albicans is one of the most prevalent pathogens [15, 16]. This organism is a part of the normal human microbiome found on skin, genital and gastrointestinal tracts. However, being an opportunistic pathogen, it can cause cutaneous candidiasis, oral thrush or candidemia in immunocompromised, elderly or hospitalized patients [18, 19]. Candida albicans has also been implicated in polymicrobial infections wherein it exists in a symbiotic association with vicinal bacterial counterparts [47]. Depending on type and severity of C albicans infection as well as the anatomical site where the infection has occurred, four classes of antifungal drugs are currently used for treating C albicans infections: azoles (target ergosterol biosynthesis), echinocandins (target cell wall glucan synthesis), polyenes (bind to ergosterol leading to pore formation and rapid leakage of monovalent ions) or nucleoside analogues (inhibitors of DNA/RNA synthesis) [18, 19]. Of these, azoles are most widely preferred for managing C albicans infections. For systemic infection, fluconazole and itraconazole represent common choices [18, 19]. Among the azoles, ketoconazole is the primary choice for topical infections [17]. Because the current work involves the testing of an electroceutical skin wound care dressing, ketoconazole was studied as the drug of choice.
Azole drug resistance is an emerging threat in the management of skin infection [48, 49]. A recent report claims that the resistance rates for all C albicans isolates from vulvovaginal and oral infections (n=206) was highest for fluconazole, itraconazole and ketoconazole [50]. All these reports provide an insight into the emergence of multidrug resistance in C albicans thereby warranting the need for alternative ways for C albicans treatment. During the last six years, our laboratory has published a series of publications on the development of WED for the management of bacterial biofilm infection [11, 13]. Reported findings include improved epithelial cell migration, necessary for wound closure [12]. On the other hand, pre-clinical studies provided mechanistic insight into the protective effects of WED against polymicrobial bacterial biofilm infection [13]. Emergent studies in our laboratory have also demonstrated that coronavirus loses infectivity upon contact with WED [10]. These findings warrant considering WED for use in personal protective equipment. Taken together, WED has established efficacy in benefiting host response and disrupting pathogen action. With this backdrop, our current study is the first to test whether an antibiotic-resistant pathogenic yeast can be managed by WED. To test WED against C albicans, the WED textile had to be wetted with broth medium used for culture of the yeast. Because the chemical composition of the wetting medium determines the strength of the electric field generated by WED, results of the current work show that WED in YPD broth generated an electric field weaker than other examples published using other biological wetting media. Interestingly, even such weak electric field produced striking beneficial outcomes as evident in this work.
The ability of microbes to transport ions against large ionic gradients drive numerous biochemical processes central to the survival of microbes. Such transport mechanisms, along with electron transport necessary for microbial bioenergetics, establish a biophysical milieu critical for their survival. The application of biophysical modalities in the treatment of Candida infections has included direct electric current (DC) [51–54] and pulsed electric field [55–57]. Limitations of the published work are: a. in vitro studies, b. bulky power supply and invasive procedures not consistent with long term application. c. testing not carried out with anti-microbial resistant strains, and d. mechanistic underpinnings not established. WED is an FDA-cleared wound care dressing that generates current using electrolytes of body fluids. The proposed work is based on a new approach that has high translational value because of its presentation as simple textile-based wound care dressing and FDA clearance for clinical application.
In candidemia patients, biofilm-forming ability of the isolated Candida strains is associated with azole drug resistance and high mortality rates [58, 59]. In high biofilm forming C albicans hyphal morphologies are dominant [60]. Transition from yeast to hyphal morphology is an important event for C albicans invasive biofilms. Hyphae produce hydrolytic enzymes needed for successful host niche damage and colonization [61]. Hyphal growth positively correlates with eDNA release and antifungal resistance in C albicans [62]. Additionally, a hyphae specific fungal toxin, candidalysin has been implicated in epithelial damage [63]. In polymicrobial biofilms, agglutinin-like sequence proteins on C albicans hyphae facilitate adherence of bacterial counterparts [41]. Resistance towards azole and echinocandin drugs is a common trait among C albicans high biofilm forming isolates [59]. Hence studies involving non-drug approaches for managing yeast biofilms warrant attention. This work presents first evidence demonstrating the feasibility that weak electric field may blunt hyphal induction and biofilm formation. While WED subdued BRG1, a positive regulator of biofilm; BCR1, another inducer of biofilm formation was upregulated. Under conditions of low BRG1, elevated BCR1 is expected to support biofilm formation [64]. However, in the presence of WED biofilm formation is disrupted. The induction of BCR1 may thus be viewed as a microbial response intended to be compensatory in bolstering its biofilm but it turns out to be futile in the presence of WED.
In non-biofilm planktonic configuration, C albicans exists in dimorphic states [24]. Depending on the environmental cues presented, the transition from yeast to hyphal morphology follows complex pathways involving transcriptional regulators and their downstream effectors [65]. Hyphal initiation requires removal of transcriptional repression of hypha-specific genes (HSG) by Nrg1 and Tup1 proteins [40]. Rfg1 protein, another negative regulator of filamentation, promotes pseudohyphal morphology when over-expressed under yeast growth conditions [66]. Although RFG1 represses filamentation in C albicans, the effects of its deletion are more subtle than NRG1 and TUP1 [67]. Transcriptional repression of two key HSG, HWP1 and ALS3, is under direct control of these repressors [65]. On the contrary, Efg1 is a downstream component of protein kinase A (PKA) pathway and a major activator of filamentation [65]. Efg1 upregulates HWP1 and ALS3 expression. Hyphal induction and maintenance, in presence of serum and at 37°C, requires down-regulation of NRG1 transcripts. However, this requirement is defied in strain deleted for EFG1 suggesting the possible negative regulation of NRG1 by Efg1 [67]. Thus, an interplay of these proteins, along with others, decides the fate of yeast to hypha morphological switch. This work observes that weak electric fields inhibit hyphal transition; induction of NRG1 and TUP1 transcripts coupled with decrease in EFG1 transcripts explain such inhibition. Downstream of transcription, the effects of WED were evident as down-regulation of hyphal cell wall proteins encoded by ALS3 and HWP1. Interestingly, WED appeared to acutely induce morphological characteristics reminiscent of pseudohyphae. However, such pseudohyphae failed to convert to true hyphae. This futility is in line with repression of RFG1 which would be expected to bolster filamentation [66]. Thus, WED clearly suppressed pathogenic hyphae formation and in the process the yeast countered with responses to restore the deficit but in vain. Considering that invasive C albicans infections require yeast to hyphal transition in the host niche, these effects of WED are of consequential value [24]. Hyphal morphogenesis has been targeted by several antifungal agents for prophylactic therapy [68–71]. However, these agents posed limitations either with concentration for active dose or restricted activity to only a few hyphae inducing conditions in vitro or inactivity in presence of serum or lack of scope for further development. This work presents weak electric field as a novel therapeutic platform.
C albicans efflux pumps help the pathogen remove unwanted substrates such as drugs [18]. Elevated efflux pump function and related up-regulation of efflux pump encoding genes (CDR1/CDR2 and/or MDR1), helps the pathogen develop antifungal drug resistance, especially towards azole drugs [19]. Disabling this pathogen defense response therefore represents a sought-after therapeutic strategy. Beauvericin, a funicone related compound extracted from fungus Beauveria species, reversed the multi-drug resistant phenotype of C albicans by specifically blocking Cdr1p and Cdr2p efflux pump activity [72]. This efflux pump inhibitor showed synergistic antifungal effect, with ketoconazole or itraconazole, against diverse multi drug resistant Candida species. In another example, synthetic organotellurium compounds blunted efflux pump activity but relevant genes were induced possibly as a retaliatory response from the pathogen which ended up being functionally futile [73]. Weak electric field acted in a similar manner. Efflux pump was functionally impaired while CDR1 and MDR1 were induced in what may be viewed as a futile compensatory response by the pathogen.
Metabolism provides the energy for a successful C albicans infection in a niche-specific fashion. During an infection, metabolic adaptations synchronize viability and virulence in response to the host microenvironments [74]. The most dramatic effects for stress or antifungal drug resistance responses are based on the available carbon source in the vicinity. These effects include cell wall remodeling via changes in cell wall proteome and differences in cell wall chitin, β-glucan and mannan proportions [75]. Furthermore, metabolism influences yeast to hyphal transition [74]. Compounds interrupting glucose metabolism, by targeting key enzymes of this pathway, present encouraging observations for exploring these alternatives as antifungal agents [76]. Severe impairment of cellular metabolism was observed in response to weak electric field administered by WED. This harm to the metabolic framework of the cells could be viewed as a cornerstone that enables the other beneficial effects of WED such as hyphal/biofilm inhibition and cell wall changes.
Cell wall acts as a significant defense barrier for protecting yeast cells from external stressors and antifungal drugs [77]. In C albicans, cell wall is made up of chitin (for cell wall shape and strength), β-glucan (structural scaffold) and mannan (role in immune recognition). Widely used antifungal drugs for managing C albicans infections are target various components of the yeast cell wall [18]. Increased chitin content in response to a decrease in β-glucan content is a widely reported response in fungi that aids in antifungal resistance or compensates for maintenance of cell wall integrity [78]. However, in these studies the cell wall mannan content was either decreased or no appreciable change in mannans was reported. A cell wall mutant deficient in CWT1 (cell wall transcription factor) decreased β-glucan and increased mannan while the chitin content remained unchanged [79]. This mutant also shows hypersensitivity to calcofluor white and minor sensitivity to SDS. WED treatment profoundly changed cell wall composition. Significant increases in mannan and decrease in β-glucan was accompanied with no change in chitin. These changes in the cell wall weakened the ability of the pathogen to cope with secondary stressors. On the contrary, an increase in chitin content and cell wall thickening were observed in cells treated with ketoconazole alone. Unfortunately, in this case, changes in the cell wall helped C albicans defend against secondary stressors. This adverse effect of ketoconazole was neutralized and reversed in the presence of WED.
Of the four chitin synthase genes in C albicans, CHS3 is responsible for the synthesis of majority cell wall chitin. Disruption of CHS3 causes 80% reduction in cell wall chitin content in yeast cell as well as hyphal cell stage [80]. This emphasizes the importance of this gene in context of cell wall chitin. Azole drugs induce CHS3 which explains the increase in chitin content of azole drug treated cells [81]. Another important target for azole drugs is ERG11, a gene encoding a key enzyme in ergosterol biosynthesis pathway [81]. Azole drug resistance commonly results by mutation in this gene leading to an altered protein product that can no longer be targeted by these drugs. β-Glucan constitutes 30–40% of C albicans cell wall. Its biosynthesis is co-ordinated by glucan synthase enzymes. One major component of this pathway is GSL1 [82]. C albicans resistance to echinocandins, antifungal drugs that target cell wall β-glucan, is a result of mutations in genes encoding for β-glucan synthases including GSL1 [83]. CHS3 expression was insensitive to WED. However, exposure to weak electric field induced ERG11. Thus, WED can target azole drug targets in an azole drug independent manner. Down-regulation of GSL1 in these cells explains the reduction in β-glucan content. A biophysical signal in the form of a weak electric field can either mimic the pharmacological effect or act independently to achieve antifungal benefits.
Metabolism plays an important role in maintaining cellular ion homeostasis and vice versa. The activity of Pma1 (plasma membrane H+-ATPase) increases by ten-fold in presence of an external glucose stimulus [84]. Similarly, glucose and other fermentable sugars activate potassium uptake pathways in yeast [85]. Furthermore, a link between ion transport, metabolism and plasma membrane potential (ΔΨ) can be explained with the fact that yeast cells in a suspension (devoid of glucose) exhibit depolarized plasma membrane due to efflux of K+ ions through Tok1p channel [86]. In yeasts, the maintenance of ΔΨ is highly regulated by cation fluxes (H+, K+, Ca+2 and Na+) [87]. Additionally, these fluxes regulate other physiological outcomes such as hyphal transition, vesicle trafficking, host association and invasion [87]. Because dysregulation of ion homeostasis can trigger cell death in yeasts, antifungal agents are being developed to target ion-signaling pathways resulting in an undesired change in ΔΨ as well [87]. The main polyene drug Amphotericin B, which is used to treat systemic fungal infections, primarily acts on fungal cell membrane ergosterol inducing pore formation and rapid leakage of monovalent cations (especially K+ ions) [88]. This results in plasma membrane depolarization followed by loss of membrane integrity and eventual cell death. Plant based membrane active phenolic compounds such as carvacol, thymol, eugenol and amiodarone display a similar activity against yeasts [87]. Although AmB remains a drug of choice for treating azole and echinocandin non-responsive fungi, resistance towards AmB is spreading at an alarming rate. This is achieved by decrease in ergosterol content, modification in ergosterol synthesis pathway, cell wall alterations and up-regulation of oxidative stress responsive machinery [88]. Treatment of C albicans cells with nanosecond pulsed electric fields (100 pulses, 100 ns in duration, intensities of 20, 40 kV/cm) resulted in decreased planktonic growth accompanied with increase in intracellular calcium ion concentration [89]. An atypical increase in influx or efflux of ions does have a detrimental effect on ion homeostasis coupled with an adverse change in ΔΨ. This could be a possible reason for WED mediated plasma membrane depolarization and hence studies examining involvement of voltage gated ion channels are warranted. This depolarization in response to weak electric fields was accompanied with membrane damage and loss of viability.
5. Conclusion
Pharmacological approaches to manage surgical infection are limited by the emergence of antibiotic resistant pathogens and the rising threat of hospital acquired infection. Cell membrane depolarization in response to WED was accompanied with significant loss of membrane integrity. While treatment with ketoconazole alone did not display any such effect, its addition with WED too failed to reverse this cellular damage. WED acted on its own, and in several cases in co-operation with ketoconazole, to neutralize ketoconazole-resistant C albicans. WED inhibited planktonic growth, biofilm formation and filamentation. This was achieved by impairing cellular metabolism and efflux pump activity. Cellular metabolism in C albicans was severely compromised in presence of WED. This effect was significantly enhanced by addition of ketoconazole with WED. While ketoconazole alone did not cause such response, it acted in synergism with WED resulting in an added surge in BCR1 expression as compared to WED alone. WED inhibited inducible yeast to hyphae morphological transition. On the contrary, ketoconazole showed this effect transiently. This inhibition was a result of up-regulation of canonical negative regulators of this pathway (NRG1 and TUP1) and down-regulation of positive regulator, EFG1. A detrimental downstream effect of this differential expression was evident by blunted expression of hyphal cell wall protein encoding genes, ALS3 and HWP1. Electric field emitting WED caused cell membrane depolarization and changed composition of the cell wall. WED alone caused an increase in cell wall mannan and decrease in β–glucan content. Akin to azole and echinocandins, WED resulted in differential expression of genes encoding C albicans cell wall components. The cell was thus more vulnerable to secondary stress. This work reports on how weak electric field, generated by a textile in contact with bodily fluids, functions in a multi-pronged manner to treat azole-resistant C albicans infection. This biophysical, as opposed to pharmacological, therapeutic principle may be leveraged to develop novel intervention strategies to prevent and manage yeast infection.
Supplementary Material
Schematic 1:
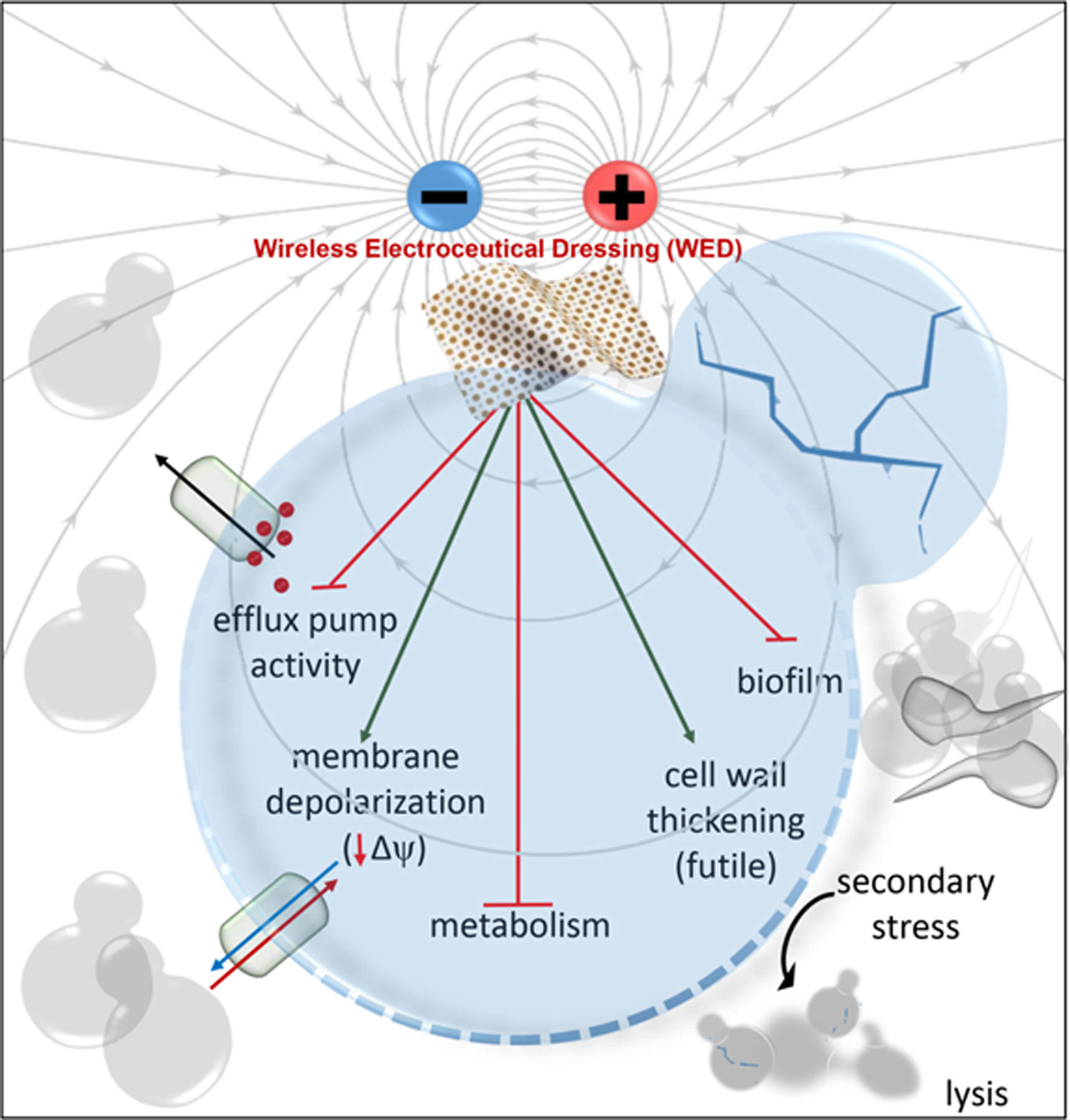
Mechanisms by which Ketoconazole Resistant Candida albicans is Sensitive to a Wireless Electroceutical Wound Care Dressing
Wireless electroceutical dressing (WED) inhibited Candida albicans biofilm formation.
WED inhibited hyphal transition, compromised efflux pump and cellular metabolism.
In the presence of ketoconazole, the effect of WED was synergistic.
First evidence of weak electric field in managing antibiotic-resistant pathogens.
Funding source
This work was supported in part by NIH grants NR015676 and DK125835 to C.K.S and by W81XWH-15–9–0001–0005 (MT17002.18) from the Department of Defense to S.R.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
CKS, in his capacity as a co-developer of the technology, has minor commercial interest in Vomaris Inc. He is also a paid consultant for the company.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Datasets are available from the corresponding author upon reasonable request. All Correspondence and requests for materials should be addressed to C.K.S.
References
- [1].Bockmuhl DP, Schages J, Rehberg L, Laundry and textile hygiene in healthcare and beyond, Microb Cell, 6 (2019) 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Morais DS, Guedes RM, Lopes MA, Antimicrobial Approaches for Textiles: From Research to Market, Materials (Basel), 9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Khan BA, Warner P, Wang H, Antibacterial Properties of Hemp and Other Natural Fibre Plants: A Review, Bioresources, 9 (2014) 3642–3659. [Google Scholar]
- [4].Yao C, Li XS, Neoh KG, Shi ZL, Kang ET, Antibacterial Poly(D,L-Lactide) (Pdlla) Fibrous Membranes Modified with Quaternary Ammonium Moieties, Chinese J Polym Sci, 28 (2010) 581–588. [Google Scholar]
- [5].McArthur JV, Tuckfield RC, Baker-Austin C, Antimicrobial textiles, Handb Exp Pharmacol, (2012) 135–152. [DOI] [PubMed]
- [6].Dilamian M, Montazer M, Masoumi J, Antimicrobial electrospun membranes of chitosan/poly(ethylene oxide) incorporating poly(hexamethylene biguanide) hydrochloride, Carbohydr Polym, 94 (2013) 364–371. [DOI] [PubMed] [Google Scholar]
- [7].Tan LY, Sin LT, Bee ST, Ratnam CT, Woo KK, Tee TT, Rahmat AR, A review of antimicrobial fabric containing nanostructures metal-based compound, J Vinyl Addit Techn, 25 (2019) E3–E27. [Google Scholar]
- [8].Westfall C, Flores-Mireles AL, Robinson JI, Lynch AJL, Hultgren S, Henderson JP, Levin PA, The Widely Used Antimicrobial Triclosan Induces High Levels of Antibiotic Tolerance In Vitro and Reduces Antibiotic Efficacy up to 100-Fold In Vivo, Antimicrob Agents Chemother, 63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hosny AEM, Rasmy SA, Aboul-Magd DS, Kashef MT, El-Bazza ZE, The increasing threat of silver-resistance in clinical isolates from wounds and burns, Infect Drug Resist, 12 (2019) 1985–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Abhishek S, Dolly K, Subhadip G, Vinoj G, Kenneth C, Sashwati R, Savita K, Chandan S, Electroceutical Fabric Lowers Zeta Potential and Eradicates Coronavirus Infectivity upon Contact, 2020.
- [11].Banerjee J, Das Ghatak P, Roy S, Khanna S, Hemann C, Deng B, Das A, Zweier JL, Wozniak D, Sen CK, Silver-zinc redox-coupled electroceutical wound dressing disrupts bacterial biofilm, PLoS One, 10 (2015) e0119531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Banerjee J, Das Ghatak P, Roy S, Khanna S, Sequin EK, Bellman K, Dickinson BC, Suri P, Subramaniam VV, Chang CJ, Sen CK, Improvement of human keratinocyte migration by a redox active bioelectric dressing, PLoS One, 9 (2014) e89239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Barki KG, Das A, Dixith S, Ghatak PD, Mathew-Steiner S, Schwab E, Khanna S, Wozniak DJ, Roy S, Sen CK, Electric Field Based Dressing Disrupts Mixed-Species Bacterial Biofilm Infection and Restores Functional Wound Healing, Ann Surg, 269 (2019) 756–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].FDA, Procellera 510(k) Summary of Safety and Effectiveness, Available at https://www.accessdata.fda.gov/cdrh_docs/pdf8/K081977.pdf., Accessed August 23, 2020. (2008).
- [15].Dowd SE, Delton Hanson J, Rees E, Wolcott RD, Zischau AM, Sun Y, White J, Smith DM, Kennedy J, Jones CE, Survey of fungi and yeast in polymicrobial infections in chronic wounds, J Wound Care, 20 (2011) 40–47. [DOI] [PubMed] [Google Scholar]
- [16].Maurel V, Denis B, Camby M, Jeanne M, Cornesse A, Glavnik B, Alanio A, Rousseau AF, Lefloch R, Lagrange-Xelot M, Textoris J, Wiramus S, de Tymowski C, Legrand M, Outcome and characteristics of invasive fungal infections in critically ill burn patients: A multicenter retrospective study, Mycoses, 63 (2020) 535–542. [DOI] [PubMed] [Google Scholar]
- [17].Kalra MG, Higgins KE, Kinney BS, Intertrigo and secondary skin infections, Am Fam Physician, 89 (2014) 569–573. [PubMed] [Google Scholar]
- [18].Bhattacharya S, Sae-Tia S, Fries BC, Candidiasis and Mechanisms of Antifungal Resistance, Antibiotics (Basel), 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Spampinato C, Leonardi D, Candida infections, causes, targets, and resistance mechanisms: traditional and alternative antifungal agents, Biomed Res Int, 2013 (2013) 204237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Park SS, Kim H, Makin IR, Skiba JB, Izadjoo MJ, Measurement of microelectric potentials in a bioelectrically-active wound care device in the presence of bacteria, J Wound Care, 24 (2015) 23–33. [DOI] [PubMed] [Google Scholar]
- [21].Necas D, Klapetek P, Gwyddion: an open-source software for SPM data analysis, Cent Eur J Phys, 10 (2012) 181–188. [Google Scholar]
- [22].Deng B, Ghatak S, Sarkar S, Singh K, Das Ghatak P, Mathew-Steiner SS, Roy S, Khanna S, Wozniak DJ, McComb DW, Sen CK, Novel Bacterial Diversity and Fragmented eDNA Identified in Hyperbiofilm-Forming Pseudomonas aeruginosa Rugose Small Colony Variant, iScience, 23 (2020) 100827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Montelongo-Jauregui D, Srinivasan A, Ramasubramanian AK, Lopez-Ribot JL, An In Vitro Model for Candida albicans(−)Streptococcus gordonii Biofilms on Titanium Surfaces, J Fungi (Basel), 4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Boyce KJ, Andrianopoulos A, Fungal dimorphism: the switch from hyphae to yeast is a specialized morphogenetic adaptation allowing colonization of a host, FEMS Microbiol Rev, 39 (2015) 797–811. [DOI] [PubMed] [Google Scholar]
- [25].Ivnitski-Steele I, Holmes AR, Lamping E, Monk BC, Cannon RD, Sklar LA, Identification of Nile red as a fluorescent substrate of the Candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p, Anal Biochem, 394 (2009) 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Essary BD, Marshall PA, Assessment of FUN-1 vital dye staining: Yeast with a block in the vacuolar sorting pathway have impaired ability to form CIVS when stained with FUN-1 fluorescent dye, J Microbiol Methods, 78 (2009) 208–212. [DOI] [PubMed] [Google Scholar]
- [27].Trunk K, Peltier J, Liu YC, Dill BD, Walker L, Gow NAR, Stark MJR, Quinn J, Strahl H, Trost M, Coulthurst SJ, The type VI secretion system deploys antifungal effectors against microbial competitors, Nat Microbiol, 3 (2018) 920–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mesa-Arango AC, Rueda C, Roman E, Quintin J, Terron MC, Luque D, Netea MG, Pla J, Zaragoza O, Cell Wall Changes in Amphotericin B-Resistant Strains from Candida tropicalis and Relationship with the Immune Responses Elicited by the Host, Antimicrob Agents Chemother, 60 (2016) 2326–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sherrington SL, Sorsby E, Mahtey N, Kumwenda P, Lenardon MD, Brown I, Ballou ER, MacCallum DM, Hall RA, Adaptation of Candida albicans to environmental pH induces cell wall remodelling and enhances innate immune recognition, PLoS Pathog, 13 (2017) e1006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tkacz JS, Cybulska EB, Lampen JO, Specific staining of wall mannan in yeast cells with fluorescein-conjugated concanavalin A, J Bacteriol, 105 (1971) 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gil-Bona A, Reales-Calderon JA, Parra-Giraldo CM, Martinez-Lopez R, Monteoliva L, Gil C, The Cell Wall Protein Ecm33 of Candida albicans is Involved in Chronological Life Span, Morphogenesis, Cell Wall Regeneration, Stress Tolerance, and Host-Cell Interaction, Front Microbiol, 7 (2016) 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Heilmann CJ, Sorgo AG, Mohammadi S, Sosinska GJ, de Koster CG, Brul S, de Koning LJ, Klis FM, Surface stress induces a conserved cell wall stress response in the pathogenic fungus Candida albicans, Eukaryot Cell, 12 (2013) 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li J, Ghatak S, El Masry MS, Das A, Liu Y, Roy S, Lee RJ, Sen CK, Topical Lyophilized Targeted Lipid Nanoparticles in the Restoration of Skin Barrier Function following Burn Wound, Mol Ther, 26 (2018) 2178–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG, Primer3--new capabilities and interfaces, Nucleic Acids Res, 40 (2012) e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, Basic local alignment search tool, J Mol Biol, 215 (1990) 403–410. [DOI] [PubMed] [Google Scholar]
- [36].Desai JV, Mitchell AP, Candida albicans Biofilm Development and Its Genetic Control, Microbiol Spectr, 3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Li F, Palecek SP, EAP1, a Candida albicans gene involved in binding human epithelial cells, Eukaryot Cell, 2 (2003) 1266–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Glazier VE, Murante T, Murante D, Koselny K, Liu Y, Kim D, Koo H, Krysan DJ, Genetic analysis of the Candida albicans biofilm transcription factor network using simple and complex haploinsufficiency, PLoS Genet, 13 (2017) e1006948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Nobile CJ, Johnson AD, Candida albicans Biofilms and Human Disease, Annu Rev Microbiol, 69 (2015) 71–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lu Y, Su C, Liu H, Candida albicans hyphal initiation and elongation, Trends Microbiol, 22 (2014) 707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nishiyama Y, Itoyama T, Yamaguchi H, Ultrastructural alterations of Candida albicans induced by a new imidazole antimycotic omoconazole nitrate, Microbiol Immunol, 41 (1997) 395–402. [DOI] [PubMed] [Google Scholar]
- [42].Bulawa CE, Miller DW, Henry LK, Becker JM, Attenuated virulence of chitin-deficient mutants of Candida albicans, Proc Natl Acad Sci U S A, 92 (1995) 10570–10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ruiz-Herrera J, Elorza MV, Valentin E, Sentandreu R, Molecular organization of the cell wall of Candida albicans and its relation to pathogenicity, FEMS Yeast Res, 6 (2006) 14–29. [DOI] [PubMed] [Google Scholar]
- [44].Whaley SG, Berkow EL, Rybak JM, Nishimoto AT, Barker KS, Rogers PD, Azole Antifungal Resistance in Candida albicans and Emerging Non-albicans Candida Species, Front Microbiol, 7 (2016) 2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Jung WH, Son YE, Oh SH, Fu C, Kim HS, Kwak JH, Cardenas ME, Heitman J, Park HS, Had1 Is Required for Cell Wall Integrity and Fungal Virulence in Cryptococcus neoformans, G3 (Bethesda), 8 (2018) 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Chellan G, Shivaprakash S, Karimassery Ramaiyar S, Varma AK, Varma N, Thekkeparambil Sukumaran M, Rohinivilasam Vasukutty J, Bal A, Kumar H, Spectrum and prevalence of fungi infecting deep tissues of lower-limb wounds in patients with type 2 diabetes, J Clin Microbiol, 48 (2010) 2097–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Allison DL, Willems HME, Jayatilake J, Bruno VM, Peters BM, Shirtliff ME, Candida-Bacteria Interactions: Their Impact on Human Disease, Microbiol Spectr, 4 (2016). [DOI] [PubMed] [Google Scholar]
- [48].Cen H, Wu Z, Wang F, Han C, Pathogen distribution and drug resistance in a burn ward: a three-year retrospective analysis of a single center in China, Int J Clin Exp Med, 8 (2015) 19188–19199. [PMC free article] [PubMed] [Google Scholar]
- [49].Zhou J, Tan J, Gong Y, Li N, Luo G, Candidemia in major burn patients and its possible risk factors: A 6-year period retrospective study at a burn ICU, Burns, 45 (2019) 1164–1171. [DOI] [PubMed] [Google Scholar]
- [50].Zida A, Yacouba A, Bamba S, Sangare I, Sawadogo M, Guiguemde T, Kone S, Traore LK, Ouedraogo-Traore R, Guiguemde RT, In vitro susceptibility of Candida albicans clinical isolates to eight antifungal agents in Ouagadougou (Burkina Faso), J Mycol Med, 27 (2017) 469–475. [DOI] [PubMed] [Google Scholar]
- [51].Barbosa GM, Dos Santos EG, Capella FN, Homsani F, de Pointis Marçal C, Dos Santos Valle R, de Araújo Abi-Chacra É, Braga-Silva LA, de Oliveira Sales MH, da Silva Neto ID, da Veiga VF, Dos Santos AL, Holandino C, Direct electric current modifies important cellular aspects and ultrastructure features of Candida albicans yeasts: Influence of doses and polarities, Bioelectromagnetics, 38 (2017) 95–108. [DOI] [PubMed] [Google Scholar]
- [52].Berger TJ, Spadaro JA, Bierman R, Chapin SE, Becker RO, Antifungal properties of electrically generated metallic ions, Antimicrob Agents Chemother, 10 (1976) 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Parry-Nweye E, Onukwugha NE, Balmuri SR, Shane JL, Kim D, Koo H, Niepa THR, Electrochemical Strategy for Eradicating Fluconazole-Tolerant Candida albicans Using Implantable Titanium, ACS Appl Mater Interfaces, 11 (2019) 40997–41008. [DOI] [PubMed] [Google Scholar]
- [54].Ruiz-Ruigomez M, Badiola J, Schmidt-Malan SM, Greenwood-Quaintance K, Karau MJ, Brinkman CL, Mandrekar JN, Patel R, Direct Electrical Current Reduces Bacterial and Yeast Biofilm Formation, Int J Bacteriol, 2016 (2016) 9727810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Arendrup MC, Patterson TF, Multidrug-Resistant Candida: Epidemiology, Molecular Mechanisms, and Treatment, J Infect Dis, 216 (2017) S445–s451. [DOI] [PubMed] [Google Scholar]
- [56].Novickij V, Staigvila G, Gudiukaitė R, Zinkevičienė A, Girkontaitė I, Paškevičius A, Švedienė J, Markovskaja S, Novickij J, Lastauskienė E, Nanosecond duration pulsed electric field together with formic acid triggers caspase-dependent apoptosis in pathogenic yeasts, Bioelectrochemistry, 128 (2019) 148–154. [DOI] [PubMed] [Google Scholar]
- [57].Novickij V, Švedienė J, Paškevičius A, Markovskaja S, Girkontaitė I, Zinkevičienė A, Lastauskienė E, Novickij J, Pulsed electric field-assisted sensitization of multidrug-resistant Candida albicans to antifungal drugs, Future Microbiol, 13 (2018) 535–546. [DOI] [PubMed] [Google Scholar]
- [58].Tascini C, Sozio E, Corte L, Sbrana F, Scarparo C, Ripoli A, Bertolino G, Merelli M, Tagliaferri E, Corcione A, Bassetti M, Cardinali G, Menichetti F, The role of biofilm forming on mortality in patients with candidemia: a study derived from real world data, Infect Dis (Lond), 50 (2018) 214–219. [DOI] [PubMed] [Google Scholar]
- [59].Tumbarello M, Fiori B, Trecarichi EM, Posteraro P, Losito AR, De Luca A, Sanguinetti M, Fadda G, Cauda R, Posteraro B, Risk factors and outcomes of candidemia caused by biofilm-forming isolates in a tertiary care hospital, PLoS One, 7 (2012) e33705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sherry L, Rajendran R, Lappin DF, Borghi E, Perdoni F, Falleni M, Tosi D, Smith K, Williams C, Jones B, Nile CJ, Ramage G, Biofilms formed by Candida albicans bloodstream isolates display phenotypic and transcriptional heterogeneity that are associated with resistance and pathogenicity, BMC Microbiol, 14 (2014) 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Naglik JR, Moyes D, Makwana J, Kanzaria P, Tsichlaki E, Weindl G, Tappuni AR, Rodgers CA, Woodman AJ, Challacombe SJ, Schaller M, Hube B, Quantitative expression of the Candida albicans secreted aspartyl proteinase gene family in human oral and vaginal candidiasis, Microbiology (Reading), 154 (2008) 3266–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Rajendran R, Sherry L, Lappin DF, Nile CJ, Smith K, Williams C, Munro CA, Ramage G, Extracellular DNA release confers heterogeneity in Candida albicans biofilm formation, BMC Microbiol, 14 (2014) 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Hofs S, Gratacap RL, Robbins J, Runglall M, Murciano C, Blagojevic M, Thavaraj S, Forster TM, Hebecker B, Kasper L, Vizcay G, Iancu SI, Kichik N, Hader A, Kurzai O, Luo T, Kruger T, Kniemeyer O, Cota E, Bader O, Wheeler RT, Gutsmann T, Hube B, Naglik JR, Candidalysin is a fungal peptide toxin critical for mucosal infection, Nature, 532 (2016) 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Huang MY, Woolford CA, May G, McManus CJ, Mitchell AP, Circuit diversification in a biofilm regulatory network, PLoS Pathog, 15 (2019) e1007787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Biswas S, Van Dijck P, Datta A, Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans, Microbiol Mol Biol Rev, 71 (2007) 348–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Cleary IA, Mulabagal P, Reinhard SM, Yadev NP, Murdoch C, Thornhill MH, Lazzell AL, Monteagudo C, Thomas DP, Saville SP, Pseudohyphal regulation by the transcription factor Rfg1p in Candida albicans, Eukaryot Cell, 9 (2010) 1363–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Braun BR, Kadosh D, Johnson AD, NRG1, a repressor of filamentous growth in C.albicans, is down-regulated during filament induction, EMBO J, 20 (2001) 4753–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Bar-Yosef H, Vivanco Gonzalez N, Ben-Aroya S, Kron SJ, Kornitzer D, Chemical inhibitors of Candida albicans hyphal morphogenesis target endocytosis, Sci Rep, 7 (2017) 5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Fazly A, Jain C, Dehner AC, Issi L, Lilly EA, Ali A, Cao H, Fidel PL Jr., Rao RP, Kaufman PD, Chemical screening identifies filastatin, a small molecule inhibitor of Candida albicans adhesion, morphogenesis, and pathogenesis, Proc Natl Acad Sci U S A, 110 (2013) 13594–13599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Shareck J, Belhumeur P, Modulation of morphogenesis in Candida albicans by various small molecules, Eukaryot Cell, 10 (2011) 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Toenjes KA, Stark BC, Brooks KM, Johnson DI, Inhibitors of cellular signalling are cytotoxic or block the budded-to-hyphal transition in the pathogenic yeast Candida albicans, J Med Microbiol, 58 (2009) 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Tong Y, Liu M, Zhang Y, Liu X, Huang R, Song F, Dai H, Ren B, Sun N, Pei G, Bian J, Jia XM, Huang G, Zhou X, Li S, Zhang B, Fukuda T, Tomoda H, Omura S, Cannon RD, Calderone R, Zhang L, Beauvericin counteracted multi-drug resistant Candida albicans by blocking ABC transporters, Synth Syst Biotechnol, 1 (2016) 158–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Reis de Sa LF, Toledo FT, Goncalves AC, Sousa BA, Dos Santos AA, Brasil PF, Duarte da Silva VA, Tessis AC, Ramos JA, Carvalho MA, Lamping E, Ferreira-Pereira A, Synthetic Organotellurium Compounds Sensitize Drug-Resistant Candida albicans Clinical Isolates to Fluconazole, Antimicrob Agents Chemother, 61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Brown AJP, Brown GD, Netea MG, Gow NAR, Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels, Trends in Microbiology, 22 (2014) 614–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Ene IV, Adya AK, Wehmeier S, Brand AC, MacCallum DM, Gow NAR, Brown AJP, Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen, Cell Microbiol, 14 (2012) 1319–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Chen XQ, Zhang ZW, Chen ZZ, Li YM, Su S, Sun SJ, Potential Antifungal Targets Based on Glucose Metabolism Pathways of Candida albicans, Frontiers in Microbiology, 11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ene IV, Walker LA, Schiavone M, Lee KK, Martin-Yken H, Dague E, Gow NA, Munro CA, Brown AJ, Cell Wall Remodeling Enzymes Modulate Fungal Cell Wall Elasticity and Osmotic Stress Resistance, mBio, 6 (2015) e00986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Garcia-Rubio R, de Oliveira HC, Rivera J, Trevijano-Contador N, The Fungal Cell Wall: Candida, Cryptococcus, and Aspergillus Species, Front Microbiol, 10 (2019) 2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Moreno I, Martinez-Esparza M, Laforet LC, Sentandreu R, Ernst JF, Valentin E, Dosage-dependent roles of the Cwt1 transcription factor for cell wall architecture, morphogenesis, drug sensitivity and virulence in Candida albicans, Yeast, 27 (2010) 77–87. [DOI] [PubMed] [Google Scholar]
- [80].Mio T, Yabe T, Sudoh M, Satoh Y, Nakajima T, Arisawa M, YamadaOkabe H, Role of three chitin synthase genes in the growth of Candida albicans, Journal of Bacteriology, 178 (1996) 2416–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].De Backer MD, Ilyina T, Ma XJ, Vandoninck S, Luyten WHML, Vanden Bossche H, Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray, Antimicrob Agents Ch, 45 (2001) 1660–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Murciano C, Moyes DL, Runglall M, Islam A, Mille C, Fradin C, Poulain D, Gow NAR, Naglik JR, Candida albicans Cell Wall Glycosylation May Be Indirectly Required for Activation of Epithelial Cell Proinflammatory Responses, Infect Immun, 79 (2011) 4902–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Suwunnakorn S, Wakabayashi H, Kordalewska M, Perlin DS, Rustchenko E, FKS2 and FKS3 Genes of Opportunistic Human Pathogen Candida albicans Influence Echinocandin Susceptibility, Antimicrob Agents Ch, 62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Goossens A, de la Fuente N, Forment J, Serrano R, Portillo F, Regulation of yeast H+-ATPase by protein kinases belonging to a family dedicated to activation of plasma membrane transporters, Mol Cell Biol, 20 (2000) 7654–7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Alijo R, Ramos J, Several Routes of Activation of the Potassium Uptake System of Yeast, Biochim Biophys Acta, 1179 (1993) 224–228. [DOI] [PubMed] [Google Scholar]
- [86].Zahumensky J, Jancikova I, Drietomska A, Svenkrtova A, Hlavacek O, Hendrych T, Plasek J, Sigler K, Gaskova D, Yeast Tok1p channel is a major contributor to membrane potential maintenance under chemical stress, Bba-Biomembranes, 1859 (2017) 1974–1985. [DOI] [PubMed] [Google Scholar]
- [87].Zhang YQ, Muend S, Rao R, Dysregulation of ion homeostasis by antifungal agents, Frontiers in Microbiology, 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Mesa-Arango AC, Scorzoni L, Zaragoza O, It only takes one to do many jobs: Amphotericin B as antifungal and immunomodulatory drug, Front Microbiol, 3 (2012) 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Guo JS, Dang J, Wang KL, Zhang J, Fang J, Effects of nanosecond pulsed electric fields (nsPEFs) on the human fungal pathogen Candida albicans: an in vitro study, J Phys D Appl Phys, 51 (2018). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets are available from the corresponding author upon reasonable request. All Correspondence and requests for materials should be addressed to C.K.S.


