Abstract
Background
Enterococcus faecium is a ubiquitously distributed member of the intestinal microbiota of both humans and animals. Antibiotic resistant E. faecium are a major public health concern.
Objectives
This study aimed to detect multi‐drug resistant (MDR) E. faecium and their antibiotic resistance genes from broiler chickens in Bangladesh.
Methods
A total of 100 faecal samples of healthy broilers were screened by conventional methods and polymerase chain reaction (PCR) to detect E. faecium and their resistance genes. Disk diffusion test was employed to determine antibiotic profiles.
Results
By PCR, among 100 samples, 45% [95% confidence interval (CI): 35.62%–54.76%] were positive for E. faecium. Based on antibiogram, all the E. faecium isolates were found resistant to ampicillin, and frequently (93.33%–55.56%) resistant to ceftriaxone, cefotaxime, streptomycin, erythromycin, and imipenem; moderate to lower (26.67%–4.44%) resistance to tetracycline, ciprofloxacin, norfloxacin, chloramphenicol, gentamicin, and vancomycin. Interestingly, 80% (95% CI: 66.18%–89.10%) E. faecium isolates were MDR in nature. In addition, the indices of multiple antibiotic resistance (MAR) ranged from 0.08 to 0.83. By bivariate analysis, high positive significant correlations were observed between resistance profiles of erythromycin and imipenem, ciprofloxacin and norfloxacin, erythromycin and streptomycin, ceftriaxone and cefotaxime, tetracycline and chloramphenicol, and streptomycin and imipenem. Furthermore, the prevalence of resistance genes of E. faecium was 58.33% (tetA), 33.33% (tetB), 35.56% (blaTEM ), 60% (CITM), 13.33% (aadA1), and 12% (SHV).
Conclusions
To the best of our knowledge, this is the first study in Bangladesh to detect MDR and MAR E. faecium and their associated resistance genes. The detection of MDR and MAR E. faecium and their corresponding resistance genes from healthy broilers is of public health concern because of their potential to enter into the food chain.
Keywords: antibiotic resistance genes, Enterococcus faecium, MAR, MDR, public health
This is the first study in Bangladesh to detect multidrug‐resistant (MDR) and multiple antibiotic‐resistant (MAR) Enterococcus faecium along with their corresponding resistance genes from healthy broilers. Our findings indicated that faecal materials of broilers harbour a high level of MDR and MAR E. faecium and multiple antibiotic resistance genes in those isolates. The results obtained from our study show severe health risks to both animals and humans.
1. INTRODUCTION
Members of enterococci, under the family of Enterococcaceae, are deemed symbiotic pathogens which can develop hospital‐ and community‐acquired infections in humans and multifarious types of infections in animals (Tian et al., 2019). They are ubiquitously present in both humans’ and animals’ intestinal tracts, in addition to different environmental sources such as water and soil (Kim et al., 2019). Enterococci are used as faecal indicator organisms to track microbial sources and antibiotic resistance trends in microorganisms. Furthermore, resistance surveillance systems of humans and animals use Enterococcus spp. as important indicator organisms (Tyson et al., 2018).
All age groups of poultry can be affected by Enterococcus spp., but their devastating effects are developed in embryos and young chicks (MSD Manual Veterinary Manual, 2019). In poultry, Enterococcus spp. can colonize the intestine and cause disease conditions such as osteomyelitis, femoral head necrosis, spondylitis, skeletal disease, and arthritis in poultry. Additionally, Enterococcus faecium cause endocarditis, septicaemia, amyloid arthropathy, and spondylitis (Robbins et al., 2012). Furthermore, these organisms are directly related to musculoskeletal disease in broiler breeders and broilers (Robbins et al., 2012).
E. faecium along with E. faecalis can cause about 90% of clinical infections and more than 10% of nosocomial infections in humans (Torres et al., 2018). Importantly, E. faecium are deemed the fourth most dominant among human pathogens globally (Rehman et al., 2018). In humans, Enterococcus spp. usually develop infections in urinary and respiratory tracts, sites of surgery, skin and soft tissue, and gastrointestinal tracts (Ike, 2017). The zoonotic pathogens E. faecium can be transmitted from animals to humans and can develop bacteraemia, urinary tract infections, infective endocarditis, wound infections, sepsis, and meningitis (Hammerum, 2012).
Antimicrobial resistance (AMR) shows negative challenges to global public health (Rahman et al., 2020) and endangers all of the one‐health components (Islam, Nayeem, et al., 2021). Low‐ and middle‐developing countries are facing the ominous effects of AMR. AMR will cause a huge number of deaths in the world's human populace if effective and novel antimicrobial agents cannot be introduced in the future (Clifford et al., 2018). Antibiotic resistance has been promoted in poultry by the haphazard use of antibiotics in their production as growth promoters with treating bacterial infections (Talukder et al., 2021). These activities in poultry can promote bacteria to be resistant to multiple antibiotics. These resistant bacteria are usually developed in the microbiota of chickens and can easily be spread to the environment via faecal contamination (Hafez & Attia, 2020). Associating in the gut microbiota and environments (litter, surface, air, water, etc.) of poultry, these resistant bacteria can acquire resistance genes and keep persisting them for a long time after discontinuation of the antibiotic treatment (Obeng et al., 2013).
As Enterococcus spp. are naturally gut‐oriented pathogens, they can serve as reservoirs of resistance genes. In addition, enterococci show intrinsic resistance to multiple classes of antibiotics which assists them to acquire abilities to be highly resistance against diversified antibiotics and to be transferred horizontally to other bacteria with the help of mobile genetic determinants (Petsaris et al., 2005). Interestingly, enterococci are resistant to multiple antimicrobial drugs, for example, aminoglycosides, β–lactams, fluoroquinolones, amphenicols, macrolides, tetracyclines, and glycopeptides (Fracalanzza et al., 2007). The vast resistance characteristics of enterococci can limit therapeutic options especially antibiotic treatment in nosocomial infections in humans and in multiple kinds of diseases in poultry. Therefore, it becomes pivotal to monitor multi‐drug resistant (MDR) enterococci which have both animal and public health significance.
Globally, there are some studies that describe the detection of E. faecium from broiler chickens (Rehman et al., 2018; Robbins et al., 2012; Šeputienė et al., 2012), but to the best of our knowledge, there are no data available in Bangladesh that detect antibiotic resistance genes carrying E. faecium from broilers. In addition, the inconveniences in treating enterococci infections are connected with AMR. This study was therefore aimed to detect E. faecium from faecal materials of healthy broiler chickens using a molecular‐based approach along with detection of their antibiotic resistance phenotypes and genotypes.
2. MATERIALS AND METHODS
2.1. Sample size calculation
As there was no research on molecular detection of Enterococcus spp. in Bangladesh, the sample size was calculated with the 50% assumptive prevalence and the 95% confidence interval (CI). The previously described (Thrusfield, 1995) formula of sample size calculation was as follows: n = Z2pq/d2, where n = desired sample size, Z = the normal standard deviation (1.96 at 95% CI), p = prevalence (50% or 0.5), q = (1‐p) = (1–0.5) = 0.5, d = precision (10% or 0.1). So, n = (1.96)2 × 0.5 × 0.5/ (0.1)2 = 96.04. Therefore, we collected 100 faecal samples aseptically from broiler chickens.
2.2. Study area and sampling
From July 2019 to March 2021, this study was performed in different poultry farms within Mymensingh Sadar Upazila (24.7851° N, 90.3560° E), Mymensingh, Bangladesh. The study area is audited in Figure 1. A total of 100 freshly dropped faecal samples of broilers were collected aseptically. Each sample was collected by swirling sterile cotton buds into faecal materials and taken into a sterile zip‐lock bag having a particular tag number. After collection, samples were brought to the laboratory under cold chain maintenance and seeded immediately to 5 ml nutrient broth containing sterile test tubes. The test tubes were then incubated in aerobic condition at 37°C for 18–24 h.
FIGURE 1.
Study area map of the present study. The map was created with ArcMap 10.7 software (ESRI, Redlands, CA, USA)
2.3. Isolation of Enterococcus spp
Initial isolation of Enterococcus spp. was carried out by culturing on enterococcus agar base (EAB) (HiMedia, India) media. For this purpose, one loopful cultured broth was streaked on EAB media and subsequently incubated for optimum condition (aerobically at 37°C overnight). Oval‐shaped yellowish colonies on EAB media were initially presumed as Enterococcus spp., and further confirmed by Gram's staining and biochemical tests, for example, sugar fermentation tests, Voges–Proskauer test, indole test, and catalase test (Facklam et al., 2002).
2.4. Molecular detection of E. faecium
Isolated Enterococcus spp. were subjected to simplex polymerase chain reaction (PCR) to detect E. faecium targeting ddlE. faecium gene (Table 1).
TABLE 1.
Primers used in the present study
| Target genes | Primer Sequence (5′–3′) | Amplicon size (bp) | Annealing temperature (°C) | References |
|---|---|---|---|---|
| ddlE. faecium |
F: GCAAGGCTTCTTAGAGA R: CATCGTGTAAGCTAACTTC |
550 | 50 | Dutka‐Malen S et al., 1995 |
| tetA |
F: GGTTCACTCGAACGACGTCA R: CTGTCCGACAAGTTGCATGA |
577 | 57 | Randall et al., 2004 |
| tetB |
F: CCTCAGCTTCTCAACGCGTG R: GCACCTTGCTGATGACTCTT |
634 | 56 | Randall et al., 2004 |
| CITM |
F: TGGCCAGAACTGACAGGCAAA R: TTTCTCCTGAACGTGGCTGGC |
462 | 47 | Van et al., 2008 |
| blaTEM |
F: CATTTCCGTGTCGCCCTTAT R: TCCATAGTTGCCTGACTCCC |
793 | 56 | Randall et al., 2004 |
| ereA |
F: GCCGGTGCTCATGAACTTGAG R: CGACTCTATTCGATCAGAGGC |
419 | 52 | Van et al., 2008 |
| SHV |
F: TCGCCTGTGTATTATCTCCC R: CGCAGATAAATCACCACAATG |
768 | 52 | Van et al., 2008 |
| aadA1 |
F: TATCCAGCTAAGCGCGAACT R: ATTTGCCGACTACCTTGGTC |
447 | 58 | Van et al., 2008 |
For the PCR, the genomic DNA was extracted from isolated Enterococcus spp. by the boiling method as previously described (Ievy et al., 2020). In brief, initially, 1 ml previously enriched culture was centrifuged at 5000 rpm for 5 min; subsequently, the supernatant was discarded, followed by preparation of suspension by adding 200 μl phosphate buffer solution. The suspension was then boiled and cooled for 10 min in each step and centrifuged for 10 min at 10,000 rpm. Finally, the supernatant was collected as genomic DNA and stored at −20°C for further research.
All the PCR were done in a final volume of 20 μl reaction [nuclease free water 4 μl, master mixture (2×, Promega, Madison, WI, USA) 10 μl, forward and reverse primer 1 μl for each, and genomic DNA 4 μl. After completing amplification, the PCR products were visualized by running in 1.5% agarose gel electrophoresis, followed by staining in ethidium bromide, documenting under ultraviolet trans‐illuminator (Biometra, Göttingen, Germany). Note that 100 bp and 1 kb DNA ladder were employed to check the expected band size of the amplified PCR products (Promega).
2.5. Antibiotic susceptibility testing
Kirby–Bauer disk diffusion test (Bauer, 1966) was employed to evaluate the antibiotic susceptibility of isolated E. faecium. The test was done by spreading freshly growth cultures (equivalence to 0.5 McFarland solution) on Mueller–Hinton agar (HiMedia) plates. Here, nine classes of antibiotic comprising 12 antibiotics were employed: fluoroquinolones (ciprofloxacin 5 μg and norfloxacin 10 μg), glycopeptides (vancomycin 30 μg), tetracyclines (tetracycline‐ 30 μg), aminoglycosides (gentamicin 10 μg, streptomycin 10 μg), penicillins (ampicillin 25 μg), macrolides (erythromycin 15 μg), amphenicols (chloramphenicol 30 μg), carbapenems (imipenem 10 μg), and cephalosporins (ceftriaxone 30 μg and cefotaxime 30 μg). The results (resistant, intermediate, and sensitive) were interpreted by following the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2018), and where not possible, according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2019). Isolates showing resistance to three or more classes of antibiotics were recorded as MDR (Sweeney et al., 2018). Moreover, the multiple antibiotic resistance (MAR) index was calculated by the following formula: MAR = m/n, here ‘m’ implies the number of antibiotics resistance to a particular E. faecium isolate and ‘n’ implies the total number of antibiotics used (Krumperman, 1983).
2.6. Detection of antibiotic resistance genes
Simplex PCR was employed to detect resistance genes of E. faecium isolates associate with tetracycline (tetA and tetB), ampicillin (blaTEM and CITM), erythromycin (ereA), imipenem (SHV), and streptomycin (aadA1). The primers and targeted genes are documented in Table 1.
2.7. Statistical analysis
2.7.1. Descriptive analysis
Data were initially brought into Excel‐2013 (Microsoft Office 2013; Microsoft, Los Angeles, CA, USA) and subsequently exported to Statistical Package for Social Science (IBM SPSS 25; IBM, Chicago, IL, USA) and Graphpad Prism version 8.4.3 (GraphPad Software, Inc.) to perform descriptive and bivariate analysis. Wilson/Brown Hybrid method (Brown et al., 2001) was followed to enumerate binomial 95% CI by Graphpad Prism.
2.7.2. Bivariate analysis
Pearson correlation was performed by SPSS to observe the potential association between any of the two antibiotics that were resistant to E. faecium isolates. The statistically significant p‐value was fixed at 0.05.
3. RESULTS
3.1. Prevalence of E. faecium
Among 100 faecal samples of broiler chickens, 88 (88%; 95% CI: 80.19%–93.00%) were found positive for Enterococcus spp. based on their cultural, staining, and biochemical properties; of which 45 samples (45%, 95% CI: 35.62%–54.76%) were confirmed as E. faecium by ddl gene targeted PCR.
3.2. Antibiotic susceptibility testing
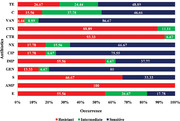
From antibiotic susceptibility test, all the E. faecium isolates (n = 45) were phenotypically resistant to ampicillin (95% CI: 92.14%–100%) and frequently resistant to ceftriaxone (93.33%; 95% CI: 82.14%–97.71%), cefotaxime (88.89%; 95% CI: 76.50%–95.16%), streptomycin (66.67%; 95% CI: 52.07%–78.64%), erythromycin (55.56%; 95% CI: 41.18%–69.06%), and imipenem (55.56%; 95% CI: 41.18%–69.06%). Moderate to lower resistance of E. faecium isolates were observed against tetracycline (26.67%; 95% CI: 15.97%–41.04%), ciprofloxacin (17.78%; 95% CI: 9.29%–31.33%), norfloxacin (17.78%; 95% CI: 9.29%–31.33%), chloramphenicol (15.56%; 95% CI: 7.75%–28.78%), gentamicin (13.33%; 95% CI: 6.26%–26.18%), and vancomycin (4.44%; 95% CI: 0.79%–14.83%). The overall antibiogram profiles are represented in Figure 2.
FIGURE 2.
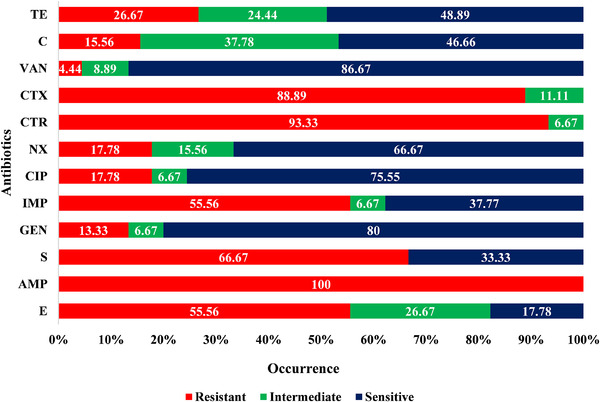
Antibiogram profiles of Enterococcus faecium isolated from faecal materials of healthy broiler chickens. Abbreviations: AMP, ampicillin; C, chloramphenicol; CIP, ciprofloxacin; CTR, ceftriaxone; CTX, cefotaxime; E, erythromycin; GEN, gentamicin; IMP, imipenem; NX, norfloxacin; S, streptomycin; TE, tetracycline; VAN, vancomycin
By bivariate analysis, high positive significant correlations were audited between resistance profiles of erythromycin and streptomycin (Pearson correlation coefficient, ρ = 0.791; p = < 0.001), erythromycin and imipenem (ρ = 0.910, p = < 0.001), streptomycin and imipenem (ρ = 0.696, p = < 0.001), ceftriaxone and cefotaxime (ρ = 0.756, p = 0.001), ciprofloxacin and norfloxacin (ρ = 0.848, p = < 0.001), and tetracycline and chloramphenicol (ρ = 0.712, p = < 0.001). In addition, moderate to lower positive significant correlations were observed between chloramphenicol and erythromycin (ρ = 0.384, p = 0.009), erythromycin and norfloxacin (ρ = 0.299, p = 0.046), norfloxacin and imipenem (ρ = 0.299, p = 0.046), erythromycin and chloramphenicol (ρ = 0.384, p = 0.009), chloramphenicol and streptomycin (ρ = 0.303, p = 0.043), chloramphenicol and imipenem (ρ = 0.384, p = 0.009), tetracycline and vancomycin (ρ = 0.358, p = 0.016), erythromycin and gentamicin (ρ = 0.351, p = 0.018), and gentamicin and imipenem (ρ = 0.351, p = 0.018). Furthermore, moderately negative significant correlation was seen between resistance profiles of gentamicin and ceftriaxone (ρ = −0.419, p = 0.004). The overall outcomes of bivariate analysis are given in Table 2.
TABLE 2.
Pearson correlation coefficients for pairs of antibiotics to assess antibiotic‐resistant Enterococcus faecium isolates from faecal samples of healthy broiler chickens
| E | AMP | S | IMP | NX | CTR | CTX | VAN | CIP | C | TE | GEN | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E | Pearson correlation | 1 | |||||||||||
| Significance (two‐tailed) | ‐ | ||||||||||||
| AMP | Pearson correlation | . a | . a | ||||||||||
| Significance (two‐tailed) | ‐ | ‐ | |||||||||||
| S | Pearson correlation | 0.791** | . a | 1 | |||||||||
| Significance (two‐tailed) | .000 | ‐ | ‐ | ||||||||||
| IMP | Pearson correlation | 0.910** | . a | 0.696** | 1 | ||||||||
| Significance (two‐tailed) | .000 | ‐ | .000 | ‐ | |||||||||
| NX | Pearson correlation | 0.299* | . a | 0.205 | 0.299* | 1 | |||||||
| Significance (two‐tailed) | .046 | ‐ | .176 | .046 | ‐ | ||||||||
| CTR | Pearson correlation | −0.060 | . a | 0.000 | −0.060 | 0.124 | 1 | ||||||
| Significance (two‐tailed) | .697 | ‐ | 1.000 | .697 | .416 | ‐ | |||||||
| CTX | Pearson correlation | −0.032 | . a | 0.050 | 0.111 | 0.164 | 0.756** | 1 | |||||
| Significance (two‐tailed) | .837 | ‐ | .744 | .469 | .281 | .000 | ‐ | ||||||
| VAN | Pearson correlation | −0.024 | . a | −0.076 | −0.024 | −0.100 | 0.058 | 0.076 | 1 | ||||
| Significance (two‐tailed) | .875 | ‐ | .619 | .875 | .512 | .707 | .619 | ‐ | |||||
| CIP | Pearson correlation | 0.182 | . a | 0.082 | 0.182 | 0.848** | 0.124 | 0.164 | −0.100 | 1 | |||
| Significance (two‐tailed) | .232 | ‐ | .591 | .232 | .000 | .416 | .281 | .512 | ‐ | ||||
| C | Pearson correlation | 0.384** | . a | 0.303* | 0.384** | −0.039 | −0.131 | −0.043 | 0.205 | −0.039 | 1 | ||
| Significance (two‐tailed) | .009 | ‐ | .043 | .009 | .798 | .391 | .777 | .177 | .798 | ‐ | |||
| TE | Pearson correlation | 0.034 | . a | 0.000 | 0.034 | −0.149 | −0.040 | 0.053 | 0.358* | −0.018 | 0.712** | 1 | |
| Significance (two‐tailed) | .826 | ‐ | 1.000 | .826 | .329 | .793 | .728 | .016 | .909 | .000 | ‐ | ||
| GEN | Pearson correlation | 0.351* | . a | 0.277 | 0.351* | −0.011 | −0.419** | −0.277 | 0.233 | −0.011 | 0.192 | 0.059 | 1 |
| Significance (two‐tailed) | .018 | ‐ | .065 | .018 | .941 | .004 | .065 | .124 | .941 | .205 | .700 | ‐ | |
Note: A p‐value <.05 was deemed as statistically significant.
Abbreviations: AMP, ampicillin; C, chloramphenicol; CIP, ciprofloxacin; CTR, ceftriaxone; CTX, cefotaxime; E, erythromycin; GEN, gentamicin; IMP, imipenem; NX, norfloxacin; S, streptomycin; TE, tetracycline; VAN, vancomycin.
Cannot be computed because at least one of the variables is constant.
Correlation is significant at the 0.05 level (two‐tailed).
Correlation is significant at the 0.01 level (two‐tailed).
3.3. Determination of MDR and MAR profiles of E. faecium
Of 45 E. faecium isolates, 36 isolates (80%; 95% CI: 66.18%–89.10%) were phenotypically MDR in nature. A total of 18 MDR patterns were observed, of which 22.22% (8/36; 95% CI: 11.72%–38.09%) isolates exhibited the resistance pattern number 9 (E‐AMP‐S‐IMP‐CTR‐CTX). Two isolates were resistant against 10 antibiotics under eight classes (Patterns 1 and 2). The ranges of MAR indices of E. faecium isolates were 0.08–0.83. Interestingly, 97.78% (44/45; 95% CI: 88.43%–99.89%) of isolates showed resistance against two or more antibiotics (Table 3).
TABLE 3.
Multi‐drug resistance and multiple antibiotic resistance profiles of Enterococcus faecium isolated from faecal materials of broiler chickens
| Pattern Number | Antibiotic resistance patterns | Number of antibiotics (classes) | Number of isolates | Overall MDR isolates (%) | MAR index |
|---|---|---|---|---|---|
| 1 | E, AMP, S, IMP, CTR, CTX, VAN, C, TE, GEN | 10 (8) | 1 |
36/45 (80) |
0.83 |
| 2 | E, AMP, S, IMP, NX, CTR, CTX, CIP, C, TE | 10 (8) | 1 | ||
| 3 | E, AMP, S, IMP, NX, CTR, CTX, CIP, GEN | 9 (6) | 1 | 0.75 | |
| 4 | E, AMP, S, IMP, CTR, CTX, C, TE | 8 (7) | 4 | 0.67 | |
| 5 | E, AMP, S, IMP, NX, CTR, CTX, CIP | 8 (6) | 4 | ||
| 6 | E, AMP, S, IMP, CTR, CTX, GEN | 7 (5) | 2 | 0.58 | |
| 7 | E, AMP, S, IMP, NX, CTR, CTX | 7 (6) | 1 | ||
| 8 | E, AMP, S, IMP, C, TE, GEN | 7 (6) | 1 | ||
| 9 | E, AMP, S, IMP, CTR, CTX | 6 (5) | 8 | 0.50 | |
| 10 | AMP, CTR, CTX, VAN, TE | 5 (4) | 1 | 0.42 | |
| 11 | AMP, S, CTR, CTX, TE | 5 (4) | 1 | ||
| 12 | E, AMP, S, IMP, GEN | 5 (4) | 1 | ||
| 13 | AMP, CTR, CTX, CIP, TE | 5 (4) | 1 | ||
| 14 | AMP, NX, CTR, CTX, CIP | 5 (3) | 1 | ||
| 15 | E, AMP, S, CTR | 4 (4) | 1 | 0.33 | |
| 16 | AMP, CTR, CTX, TE | 4 (3) | 2 | ||
| 17 | AMP, IMP, CTR, CTX | 4 (3) | 1 | ||
| 18 | AMP, S, CTR, CTX | 4 (3) | 4 | ||
| 19* | AMP, CTR, CTX | 3 (2) | 7 | – | 0.25 |
| 20* | AMP, CTR | 2 (2) | 1 | – | 0.17 |
| 21* | AMO | 1 (1) | 1 | – | 0.08 |
Abbreviations: AMP, ampicillin; C, chloramphenicol; CIP, ciprofloxacin; CTR, ceftriaxone; CTX, cefotaxime; E, erythromycin; GEN, gentamicin; IMP, imipenem; MAR, multiple antibiotic resistance; NX, norfloxacin; S, streptomycin; TE, tetracycline; VAN, vancomycin.
Non multi‐drug resistant.
3.4. Prevalence of resistance genes
By PCR, resistance gene tetA and tetB were found to be positive in 58.33% (7/12; 95% CI: 31.95%–80.67%) and 33.33% (4/12; 95% CI: 13.81%–60.94%) tetracycline resistant E. faecium isolates respectively; resistance gene blaTEM and CITM were positive in 35.56% (16/45; 95% CI: 23.22%–50.16%) and 60% (27/45; 95% CI: 45.45%–72.98%) ampicillin resistant E. faecium isolates respectively; aadA1 gene was found in 13.33% (4/30; 95% CI: 5.31%–29.68%) streptomycin resistant E. faecium isolates, and 12% (3/25; 95% CI: 4.17%–29.96%) imipenem resistant E. faecium isolates were positive for SHV resistance gene. However, all the erythromycin resistant E. faecium isolates were found negative for resistance ereA gene (Table 4).
TABLE 4.
Prevalence of antibiotic resistance genes of Enterococcus faecium isolated from faecal materials of broiler chickens
| Name of antibiotics | Name of resistance genes | Prevalence (%) | 95% CI (%) |
|---|---|---|---|
| Tetracycline (n = 12) | tetA | 7 (58.33) | 31.95–80.67 |
| tetB | 4 (33.33) | 13.81–60.94 | |
| Erythromycin (n = 25) | ereA | 0 (0) | 0.00–13.32 |
| Ampicillin (n = 45) | blaTEM | 16 (35.56) | 23.22–50.16 |
| CITM | 27 (60) | 45.45–72.98 | |
| Streptomycin (n = 30) | aadA1 | 4 (13.33) | 5.31–29.68 |
| Imipenem (carbapenem) (n = 25) | SHV | 3 (12) | 4.17–29.96 |
Abbreviation: CI, confidence interval.
4. DISCUSSION
Enterococcus spp. are considered opportunistic pathogens in both humans and animals. They represent challenges in infection control due to their rapid acquisition of antibiotic resistance capabilities. The dissemination of AMR in Enterococcus spp. associated with poultry in Bangladesh is not detailed till now. This study was therefore investigated to get better insights on the MDR and MAR observed in E. faecium isolates and their corresponding resistance genes from healthy broiler chickens in Bangladesh.
In the present investigation, E. faecium was found positive in 45% (45/100) faecal samples of broiler chickens. The high detection rate of E. faecium from the faecal materials of broilers is not unusual, as E. faecium is a ubiquitously commensal microorganism and is considered as a part of the intestinal microbiota of humans and animals (Dubin & Pamer, 2018). In addition, enterococci are present in faecal materials of different birds, mammals, reptiles, and even insects (Dubin & Pamer, 2018). Previously, Banik et al. (2018) isolated Enterococcus spp. from chicken in Bangladesh, but they used only conventional methods and did not carry any molecular approach, for example, PCR assay which was used in our present study. PCR is a robust, sensitive, and rapid method in detecting Enterococcus spp. from any kind of sample and gives higher specificity and sensitivity of the results (Maheux et al., 2011). Globally, several studies detected E. faecium from broiler chickens with variable occurrence rates (Garcia‐Migura et al., 2005; Karunarathna et al., 2017; Rehman et al., 2018; Tremblay et al., 2011). These variations might be lined with the variations in geographical and seasonal distributions, the farm management systems (hygiene, biosecurity, and sanitary), sample size and types, and methodological factors (Islam, Paul, et al., 2021).
The presence of E. faecium in faecal materials of broiler chickens reveals that the droppings of broiler chickens can shed E. faecium to other birds of the farm. In addition, contaminated faecal materials can act as vehicles to contaminate water and broiler feed. Furthermore, the enterococci contamination can be transferred into the production systems via the contaminated faeces, water, or broiler feeds. Broiler meat and its products can be contaminated by enterococci that can be transmitted to the food chain and pose a potential human health concern. Enterococci can grow under extreme temperature up to 72°C which indicates that consumption of undercooked poultry meat and products has the potential to transmit enterococci to humans (Martinez et al., 2003). Furthermore, this may reveal a high risk to human health by exposure to colonized birds or by the introduction of poultry meat contaminated with enterococci or by cross‐contaminating with ready‐to‐eat foods (Obeng et al., 2013).
Any microorganism resistant to the antibiotic is a threat to human health. Results of phenotypic antibiotic susceptibility test from our present study showed that a high number of E. faecium isolates were resistant to ampicillin, ceftriaxone, cefotaxime, streptomycin, erythromycin, imipenem, and tetracycline. High level resistance of E. faecium to streptomycin is not unusual. Streptomycin is under the aminoglycoside class of antibiotics which show intrinsic resistance to enterococci. Clinically achievable concentrations of aminoglycosides are unable to enter into the cell of enterococci especially E. faecalis, and develop enzyme‐mediated resistance in the ribosomal target site of enterococci especially E. faecium. These two factors enable enterococci to show intrinsic resistance against aminoglycoside class of antibiotics (Bertelloni et al., 2015). But in contrast, we found that another aminoglycoside‐gentamicin was highly sensitive or intermediately sensitive to E. faecium (more than 85% isolates). This variation might have a linkage with the level of concentrations of gentamicin, as enterococci show intrinsic resistance to only a low level of concentrations of aminoglycosides (Lefort et al., 2000). Previously, Tremblay et al. (2011) also found that high proportion of enterococci was sensitive to gentamicin. However, intrinsic resistance of enterococci either to streptomycin or to gentamicin has not been illustrated properly till now (Bertelloni et al., 2015).
Similarly, enterococci also are naturally resistant to cephalosporins which are lined with the higher resistance to the cephalosporin class of antibiotics – ceftriaxone and cefotaxime – obtained from our present study. Enterococci acquire intrinsic resistance to cephalosporins by a penicillin‐binding protein (Pbp5), a transduction system (CroRS), an enzyme to synthesize peptidoglycan precursors (MurAA), and a transmembrane kinase (Ser/Thr) (Kristich et al., 2014). This trait is well defined for E. faecalis, but for E. faecium, it is not well developed yet. However, gentamicin is vastly used in enterococcal infections with the combination of beta‐lactam antibiotics or glycopeptides for obtaining synergistic bactericidal effects (Emaneini et al., 2016).
Interestingly, 55.56% of E. faecium isolates were resistant to imipenem which reveals an alarming condition to both human and animal health facilities. Imipenem is under the carbapenem group of antibiotics which are only exercised for treating severe bacterial infections in humans (Lamb et al., 2002). E. faecium acquires resistance by two distinct mechanisms: one is incremented via Pbp5 which shows low‐affinity to β‐lactam antibiotics, and another one is the mutation that occurred in Pbp5 (Joste et al., 2019). Acquisition of erythromycin and tetracycline resistance in enterococci is usually developed by mobile genetic elements (Emaneini et al., 2016). E. faecium resistance to tetracycline shows importance because of its relatedness with the resistance profiles of other antibiotics (Hammerum, 2012). Erythromycin is under the macrolides class of antibiotics which have been classified as ‘critically important in human medicine’ showing importance in the treatment of different bacterial infections (WHO, 2015). We have found 4.44% vancomycin‐resistant E. faecium in this study which is also alarming to human and animal health. Vancomycin‐resistant E. faecium are WHO priority 2 high category pathogens (WHO, 2017).
By bivariate analysis, high positive significant correlations were audited between resistance profiles of erythromycin and streptomycin, erythromycin and imipenem, streptomycin and imipenem, ceftriaxone and cefotaxime, ciprofloxacin and norfloxacin, and tetracycline and chloramphenicol. The high correlation between ceftriaxone and cefotaxime, and ciprofloxacin and norfloxacin is not unusual as they are under a similar class of antibiotics (first two are under cephalosporins, and second two under fluoroquinolones). The other strong correlations might be due to the haphazard use of antibiotics in broilers. The importance of these findings is linked with developing resistance in E. faecium against other used antimicrobials (Ievy et al., 2020).
In the present study, we detected tetA (58.33%), tetB (33.33%), blaTEM (35.56%), CITM (60%), aadA1 (13.33%), and SHV (12%) genes in E. faecium isolates which were responsible for corresponding antibiotic resistance. No isolates were found positive for ereA gene. However, the presence of different resistance genes in E. faecium isolates might be due to the mobile genetic elements. The gene tetA along with tetC gene form one genetic group which shares approximately 78% of the amino acid sequences in common with each other. In addition, overlapping gene tetA encodes a classical efflux protein and a second gene tetB which codes for a protein that seems to be related to the tetracycline ribosomal protection proteins (Roberts, 1996). The results of tetA and tetB from tetracycline resistant E. faecium isolates show that the original mechanism of tetracycline resistance generated from broiler chickens is by active efflux system. Resistance gene blaTEM , CITM, and SHV are associated with β‐lactam antibiotics. These genes presenting in the organism have abilities to inactivate the antibiotics by hydrolyzing the β‐lactam ring (Livermore, 1995). The gene cassette aadA1 is associated with the resistance to streptomycin which generally presents in class 1 integrons related to transposons Tn21 (Rodríguez et al., 2006). Through conjugative plasmids, this resistance gene has the ability to be horizontally transferred from E. faecium to other bacteria (Nde & Logue, 2008). The presence of resistance genes of E. faecium in broiler chickens reveals public health significance as they can be transmitted to humans via the contaminated food supply chain and/or close contact with the animals. In addition, these resistance genes can assist in the emergence of MDR and MAR bacteria in both humans and animals.
Infections caused by MDR and MAR bacteria can have serious health repercussions for humans (Urmi et al., 2021). The abuse and overuse of antibiotics has resulted in the emergence of MDR and MAR enterococci, which has become a severe public health hazard in both humans and animals. MDR and MAR enterococci lessen the treatment option in infections developed by such strains (Obeng et al., 2013). In the present study, a high proportion of E. faecium isolates (80%) were phenotypically MDR in nature which reveals an alarming situation in poultry as well as humans. Previously, Tremblay et al. (2011) detected 100% MDR E. faecium isolates from broiler chickens in Canada. Furthermore, the index of MAR in E. faecium isolates was risen up to 0.83, which indicates that a high level of antibiotics was haphazardly used in the broiler chickens from where the enterococci were isolated. MDR and MAR in E. faecium might be developed by selective pressure triggered by the misuse, extensive use, and incorrect prescriptions of antibiotics in veterinary practices (Islam, Sobur, et al., 2021; Tawyabur et al., 2020). The MDR and MAR E. faecium obtained from our present study have the potential to contaminate the one‐health components. They can be transmitted to humans through the food chain or close contact with the broilers and to environments via contaminated water or feed sources.
5. CONCLUSION
To the best of our knowledge, we detected MDR and MAR E. faecium and their corresponding resistance genes for the first time in Bangladesh from healthy broiler chickens. A high level of E. faecium and their resistance and resistance genes detected in broiler chickens has the potential to enter into the food chain and shows a negative impact on both humans’ and animals’ health. Furthermore, the presence of MDR enterococci in broilers reveals more potential public health hazards in considering close contact with humans and animals. Though we performed our study with a limited number of broilers, this study has contributed to verify MDR enterococci and their resistance profiles in poultry. We, therefore, suggest that broiler chickens should be kept under strict biosecurity and under regular epidemiological study with a strong one‐health approach to prevent the negative effects of enterococci and to minimize the emergence of MDR and MAR E. faecium with their resistance genes in both humans and animals.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Data curation, investigation, methodology, and writing‐original draft: Krishna Roy. Data curation, formal analysis, investigation, methodology, software, visualization, writing‐original draft, and writing‐review & editing: Md. Saiful Islam. Investigation: Anamika Paul, Samina Ievy, Fatimah Muhammad Ballah, and Mithun Talukder. Formal analysis and writing‐review & editing: Md. Abdus Sobur. Supervision: Md. Shahidur Khan. Conceptualization, formal analysis, funding acquisition, methodology, supervision, validation, writing‐original draft, and writing‐review & editing: Md. Tanvir Rahman.
ETHICS STATEMENT
Ethical permissions were not required during sample collection. Samples were collected with the owners’ permission.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/vms3.669
ACKNOWLEDGEMENTS
Authors are grateful to the farmers for giving access to the samples. The authors would also like to thank the Ministry of Science and Technology, Government of the People's Republic of Bangladesh. They provided National and Science Technology (NST) fellowship to Krishna Roy.
Roy, K. , Islam, M. S. , Paul, A. , Ievy, S. , Talukder, M. , Sobur, M. A. , Ballah, F. M. , Khan, M. S. R. , & Rahman, M. T. (2022). Molecular detection and antibiotyping of multi‐drug resistant Enterococcus faecium from healthy broiler chickens in Bangladesh. Veterinary Medicine and Science, 8, 200–210. 10.1002/vms3.669
Funding information
This study was partially supported by the Ministry of Education, Government of Bangladesh (Project No. LS2018686) and the Bangladesh Agricultural University Research System (Project No. 2019/8/BAU).
REFERENCES
- Banik, A. , Mohammad, N. , Akter, T. , Fatema, K. , & Abony, M. (2018). Prevalence, identification and antibiotic susceptibility of Enterococcus Species isolated from chicken and pigeon meat in Gazipur area of Bangladesh. Open Journal of Medical Microbiology, 8(3), 74–83. 10.4236/ojmm.2018.83007 [DOI] [Google Scholar]
- Bauer, A. T. (1966). Antibiotic susceptibility testing by a standardized single disc method. American Journal of Clinical Pathology, 45, 149–158. [PubMed] [Google Scholar]
- Bertelloni, F. , Salvadori, C. , Moni, A. , Cerri, D. , Mani, P. , & Ebani, V. V. (2015). Antimicrobial resistance in Enterococcus spp. isolated from laying hens of backyard poultry flocks. Annals of Agricultural and Environmental Medicine, 22(4), 665–669. 10.5604/12321966.1185771 [DOI] [PubMed] [Google Scholar]
- Brown, L. D. , Cai, T. T. , & DasGupta, A. (2001). Interval estimation for a binomial proportion. Statistical Science, 16, 101–117. [Google Scholar]
- Clifford, K. , Desai, D. , da Costa, C. P. , Meyer, H. , Klohe, K. , Winkler, A. S. , Rahman, T. , Islam, T. , & Zaman, M. H. (2018). Antimicrobial resistance in livestock and poor quality veterinary medicines. Bulletin of the World Health Organization, 96(9), 662–664. 10.2471/BLT.18.209585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) (2018). Performance Standards for Antimicrobial Susceptibility Testing, M100‐S28. Clinical and Laboratory Standards Institute. [Google Scholar]
- Dubin, K. , & Pamer, E. G. (2018). Enterococci and their interactions with the intestinal microbiome. Microbiology Spectrum , 5, 309–330. 10.1128/microbiolspec.BAD-0014-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutka‐Malen, S. , Evers, S. , & Courvalin, P. (1995). Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. Journal of Clinical Microbiology, 33, 24–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emaneini, M. , Khoramian, B. , Jabalameli, F. , Beigverdi, R. , Asadollahi, K. , Taherikalani, M. , & Lari, A. R. (2016). Prevalence of high‐level gentamicin‐resistant Enterococcus faecalis and Enterococcus faecium in an Iranian hospital. Journal of Preventive Medicine and Hygiene, 57(4), E197–E200. [PMC free article] [PubMed] [Google Scholar]
- Facklam, R. R. , Carvalho, M. D. G. S. , & Teixeira, L. M. (2002). History, taxonomy, biochemical characteristics, and antibiotic susceptibility testing of enterococci. In Gilmore M. S., Clewell D. B., Courvalin P., Dunny G. M., Murray B. E., & Rice L. B. (Eds.), The Enterococci: Pathogenesis, molecular biology, and antibiotic resistance (pp. 1–54). ASM Press. 10.1128/9781555817923.ch1 [DOI] [Google Scholar]
- Fracalanzza, S. A. P. , Scheidegger, E. M. D. , Santos, P. F. D. , Leite, P. C. , & Teixeira, L. M. (2007). Antimicrobial resistance profiles of enterococci isolated from poultry meat and pasteurized milk in Rio de Janeiro, Brazil. Memórias do Instituto Oswaldo Cruz, 102(7), 853–859. 10.1590/S0074-02762007005000120 [DOI] [PubMed] [Google Scholar]
- Garcia‐Migura, L. , Pleydell, E. , Barnes, S. , Davies, R. H. , & Liebana, E. (2005). Characterization of vancomycin‐resistant Enterococcus faecium isolates from broiler poultry and pig farms in England and Wales. Journal of Clinical Microbiology, 43(7), 3283–3289. 10.1128/JCM.43.7.3283-3289.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafez, H. M. , & Attia, Y. A. (2020). Challenges to the poultry industry: Current perspectives and strategic future after the COVID‐19 outbreak. Frontiers in Veterinary Science, 7, 516. 10.3389/fvets.2020.00516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerum, A. M. (2012). Enterococci of animal origin and their significance for public health. Clinical Microbiology and Infection, 18(7), 619‐625. 10.1111/j.1469-0691.2012.03829.x [DOI] [PubMed] [Google Scholar]
- Ievy, S. , Islam, M. , Sobur, M. , Talukder, M. , Rahman, M. , Khan, M. F. R. , & Rahman, M. T. (2020). Molecular detection of avian pathogenic Escherichia coli (APEC) for the first time in layer farms in Bangladesh and their antibiotic resistance patterns. Microorganisms, 8(7), 1021. 10.3390/microorganisms8071021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ike, Y. (2017). Pathogenicity of enterococci. Nihon saikingaku zasshi . Japanese Journal of Bacteriology, 72(2), 189–211. 10.3412/jsb.72.189 [DOI] [PubMed] [Google Scholar]
- Islam, M. , Nayeem, M. , Hasan, M. , Sobur, M. , Ievy, S. , Rahman, S. , Kafi, M. , Ashour, H. M. , & Rahman, M. (2021). Virulence determinants and multidrug resistance of Escherichia coli isolated from migratory birds. Antibiotics, 10(2), 190. 10.3390/antibiotics10020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, M. , Paul, A. , Talukder, M. , Roy, K. , Sobur, M. A. , Ievy, S. , Nayeem, M. M. H. , Rahman, S. , Nazir, K. N. H. , Hossain, M. T. , & Rahman, M. T. (2021). Migratory birds travelling to Bangladesh are potential carriers of multi‐drug resistant Enterococcus spp., Salmonella spp., and Vibrio spp. Saudi Journal of Biological Sciences, 28(10), 5963–5970. 10.1016/j.sjbs.2021.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, M. , Sobur, M. , Rahman, S. , Ballah, F. M. , Ievy, S. , Siddique, M. P. , Rahman, M. , Kafi, M. , & Rahman, M. (2021). Detection of blaTEM , blaCTX‐M , blaCMY , and blaSHV genes among extended‐spectrum beta‐lactamase‐producing Escherichia coli isolated from migratory birds travelling to Bangladesh. Microbial Ecology, 1–9. 10.1007/s00248-021-01803-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joste, V. , Gydé, E. , Toullec, L. , Courboulès, C. , Yasmina, T. , Riverain‐Gillet, E. , Pangon, B. , & Amara, M. (2019). Enterococcus faecium and ampicillin susceptibility determination: Overestimation of resistance with disk diffusion method using ampicillin 2 μg? Journal of Clinical Microbiology, 57, e01467‐18. 10.1128/JCM.01467-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunarathna, R. , Popowich, S. , Wawryk, M. , Chow‐Lockerbie, B. , Ahmed, K. A. , Yu, C. , Liu, M. , Goonewardene, K. , Gunawardana, T. , Kurukulasuriya, S. , & Gupta, A. (2017). Increased incidence of enterococcal infection in nonviable broiler chicken embryos in Western Canadian hatcheries as detected by matrix‐assisted laser desorption/ionization‐time‐of‐flight mass spectrometry. Avian Diseases, 61(4), 472–480. 10.1637/11678-052317-Reg.1 [DOI] [PubMed] [Google Scholar]
- Kim, Y. B. , Seo, K. W. , Jeon, H. Y. , Lim, S. K. , Sung, H. W. , & Lee, Y. J. (2019). Molecular characterization of erythromycin and tetracycline‐resistant Enterococcus faecalis isolated from retail chicken meats. Poultry Science, 98(2), 977–983. 10.3382/ps/pey477 [DOI] [PubMed] [Google Scholar]
- Kristich, C. J. , Rice, L. B. , & Arias, C. A. (2014). Enterococcal infection—Treatment and antibiotic resistance. In Gilmore M. S., Clewell D. B., Ike Y., & Shankar N. (Eds.), Enterococci: From commensals to leading causes of drug resistant infection (pp. 87–134).Massachusetts Eye and Ear Infirmary. http://www.ncbi.nlm.nih.gov/books/NBK190420/ [PubMed] [Google Scholar]
- Krumperman, P. H. (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high‐risk sources of fecal contamination of foods. Applied and Environmental Microbiology, 46(1), 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb, H. M. , Ormrod, D. , Scott, L. J. , & Figgitt, D. P. (2002). Ceftriaxone. Drugs, 62(7), 1041–1089. 10.2165/00003495-200262070-00005 [DOI] [PubMed] [Google Scholar]
- Lefort, A. , Arthur, M. , Garry, L. , Carbon, C. , Courvalin, P. , & Fantin, B. (2000). Bactericidal activity of gentamicin against Enterococcus faecalis in vitro and in vivo. Antimicrobial Agents and Chemotherapy, 44(8), 2077–2080. 10.1128/aac.44.8.2077-2080.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore, D. M. (1995). beta‐Lactamases in laboratory and clinical resistance. Clinical Microbiology Reviews, 8(4), 557–584. 10.1128/CMR.8.4.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheux, A. F. , Bissonnette, L. , Boissinot, M. , Bernier, J. L. T. , Huppe, V. , Berube, E. , Boudreau, D. K. , Picard, F. J. , Huletsky, A. , & Bergeron, M. G. (2011). Method for rapid and sensitive detection of Enterococcus sp. and Enterococcus faecalis/faecium cells in potable water samples. Water Research, 45(6), 2342–2354. 10.1016/j.watres.2011.01.019 [DOI] [PubMed] [Google Scholar]
- Martinez, S. , López, M. , & Bernardo, A. (2003). Thermal inactivation of Enterococcus faecium: Effect of growth temperature and physiological state of microbial cells. Letters in Applied Microbiology, 37(6), 475–481. 10.1046/j.1472-765X.2003.01431.x [DOI] [PubMed] [Google Scholar]
- MSD Manual Veterinary Manual (2019). Enterococcosis in Poultry. https://www.msdvetmanual.com/poultry/enterococcosis/enterococcosis‐in‐poultry#:~:text=Enterococcus%20spp%20are%20part%20of,early%20stages%20of%20the%20disease
- Nde, C. W. , & Logue, C. M. (2008). Characterization of antimicrobial susceptibility and virulence genes of Salmonella serovars collected at a commercial turkey processing plant. Journal of Applied Microbiology, 104(1), 215–223. 10.1111/j.1365-2672.2007.03535.x [DOI] [PubMed] [Google Scholar]
- Obeng, A. S. , Rickard, H. , Ndi, O. , Sexton, M. , & Barton, M. (2013). Comparison of antimicrobial resistance patterns in enterococci from intensive and free range chickens in Australia. Avian Pathology, 42(1), 45–54. 10.1080/03079457.2012.757576 [DOI] [PubMed] [Google Scholar]
- Petsaris, O. , Miszczak, F. , Gicquel‐Bruneau, M. , Perrin‐Guyomard, A. , Humbert, F. , Sanders, P. , & Leclercq, R. (2005). Combined antimicrobial resistance in Enterococcus faecium isolated from chickens. Applied and Environmental Microbiology, 71(5), 2796–2799. 10.1128/AEM.71.5.2796-2799.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, M. , Sobur, M. , Islam, M. , Ievy, S. , Hossain, M. , El Zowalaty, M. E. , Rahman, A. M. M. , & Ashour, H. M. (2020). Zoonotic diseases: Etiology, impact, and control. Microorganisms, 8(9), 1405. 10.3390/microorganisms8091405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall, L. P. , Cooles, S. W. , Osborn, M. K. , Piddock, L. J. V. , & Woodward, M. J. (2004). Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty‐five serotypes of Salmonella enterica isolated from humans and animals in the UK. Journal of Antimicrobial Chemotherapy, 53(2), 208–216. 10.1093/jac/dkh070 [DOI] [PubMed] [Google Scholar]
- Rehman, M. A. , Yin, X. , Zaheer, R. , Goji, N. , Amoako, K. K. , McAllister, T. , Pritchard, J. , Topp, E. , & Diarra, M. S. (2018). Genotypes and phenotypes of Enterococci isolated from broiler chickens. Frontiers in Sustainable Food Systems, 2, 83. 10.3389/fsufs.2018.00083 [DOI] [Google Scholar]
- Robbins, K. M. , Suyemoto, M. M. , Lyman, R. L. , Martin, M. P. , Barnes, H. J. , & Borst, L. B. (2012). An outbreak and source investigation of enterococcal spondylitis in broilers caused by Enterococcus cecorum . Avian Diseases, 56(4), 768–773. 10.1637/10253-052412-Case.1 [DOI] [PubMed] [Google Scholar]
- Roberts, M. C. (1996). Tetracycline resistance determinants: Mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiology Reviews, 19(1), 1–24. 10.1111/j.1574-6976.1996.tb00251.x [DOI] [PubMed] [Google Scholar]
- Rodríguez, I. , Martín, M. C. , Mendoza, M. C. , & Rodicio, M. R. (2006). Class 1 and class 2 integrons in non‐prevalent serovars of Salmonella enterica: Structure and association with transposons and plasmids. Journal of Antimicrobial Chemotherapy, 58(6), 1124–1132. 10.1093/jac/dkl400 [DOI] [PubMed] [Google Scholar]
- Šeputienė, V. , Bogdaitė, A. , Ružauskas, M. , & Sužiedėlienė, E. (2012). Antibiotic resistance genes and virulence factors in Enterococcus faecium and Enterococcus faecalis from diseased farm animals: Pigs, cattle and poultry. Polish Journal of Veterinary Sciences, 15, 431–438. [PubMed] [Google Scholar]
- Sweeney, M. T. , Lubbers, B. V. , Schwarz, S. , & Watts, J. L. (2018). Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. Journal of Antimicrobial Chemotherapy, 73(6), 1460–1463. 10.1093/jac/dky043 [DOI] [PubMed] [Google Scholar]
- Talukder, M. , Islam, M. S. , Ievy, S. , Sobur, M. A. , Ballah, F. M. , Najibullah, M. , Rahman, M. B. , Rahman, M. T. , & Khan, M. F. R. (2021). Detection of multidrug resistant Salmonella spp. from healthy and diseased broilers having potential public health significance. Journal of Advanced Biotechnology and Experimental Therapeutics, 4(2), 248–255. 10.5455/jabet.2021.d125 [DOI] [Google Scholar]
- Tawyabur, M. , Islam, M. , Sobur, M. , Hossain, M. , Mahmud, M. , Paul, S. , Hossain, M. T. , Ashour, H. M. , & Rahman, M. (2020). Isolation and characterization of multidrug‐resistant Escherichia coli and Salmonella spp. from healthy and diseased Turkeys. Antibiotics, 9(11), 770. 10.3390/antibiotics9110770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST) , (2019). Break point tables for interpretation of MICs and zone diameters, version 9.0 Available online: http://www.eucast.org
- Thrusfield, M. (1995). Veterinary epidemiology (2nd ed.). Blackwell Science. [Google Scholar]
- Tian, Y. , Yu, H. , & Wang, Z. (2019). Distribution of acquired antibiotic resistance genes among Enterococcus spp. isolated from a hospital in Baotou, China. BMC Research Notes, 12(1), 1–5. 10.1186/s13104-019-4064-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, C. , Alonso, C. A. , Ruiz‐Ripa, L. , León‐Sampedro, R. , Del Campo, R. , & Coque, T. M. (2018). Antimicrobial resistance in Enterococcus spp. of animal origin. Antimicrobial resistance in bacteria from livestock and companion animals (pp. 185–227). ASM Press. 10.1128/9781555819804.ch9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay, C. L. , Letellier, A. , Quessy, S. , Boulianne, M. , Daignault, D. , & Archambault, M. (2011). Multiple‐antibiotic resistance of Enterococcus faecalis and Enterococcus faecium from cecal contents in broiler chicken and turkey flocks slaughtered in Canada and plasmid colocalization of tetO and ermB genes. Journal of Food Protection, 74(10), 1639–1648. 10.4315/0362-028X.JFP-10-451 [DOI] [PubMed] [Google Scholar]
- Tyson, G. H. , Nyirabahizi, E. , Crarey, E. , Kabera, C. , Lam, C. , Rice‐Trujillo, C. , McDermott, P. F. , & Tate, H. (2018). Prevalence and antimicrobial resistance of enterococci isolated from retail meats in the United States, 2002 to 2014. Applied and Environmental Microbiology, 84(1), e01902‐17. 10.1128/AEM.01902-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urmi, M. R. , Ansari, W. K. , Islam, M. S. , Sobur, M. A. , Rahman, M. , & Rahman, M. T. (2021). Antibiotic resistance patterns of Staphylococcus spp. isolated from fast foods sold in different restaurants of Mymensingh, Bangladesh. Journal of Advanced Veterinary and Animal Research, 8(2), 274–281. 10.5455/javar.2021.h512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van, T. T. H. , Chin, J. , Chapman, T. , Tran, L. T. , & Coloe, P. J. (2008). Safety of raw meat and shellfish in Vietnam: An analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. International Journal of Food Microbiology, 124(3), 217–223. 10.1016/j.ijfoodmicro.2008.03.029 [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO) (2015). 19th WHO model list of essential medicines. World health organization essential medicines and health products. Geneva. https://www.who.int/medicines/publications/essentialmedicines/EML_2015_FINAL_amended_NOV2015.pdf?ua=1
- World Health Organization (WHO) (2017). Global priority list of antibiotic‐resistant bacteria to guide research, discovery, and development of new antibiotics. https://www.who.int/medicines/publications/WHO‐PPL‐Short_Summary_25Feb‐ET_NM_WHO.pdf


