Abstract
A cluster (14 of 18) of Streptococcus pneumoniae serotype 23F isolates that were resistant to penicillin (PEN), cephalosporin, and macrolide was found in one day care center in Kaohsiung, Taiwan. We analyzed the 18 isolates by pulsed field gel electrophoresis (PFGE). All but one serotype 23F isolate demonstrated identical PFGE patterns, which were different from the established pattern of the internationally spread Spanish 23F clone. The three strains of serotype 19F also showed a uniform pattern. These data strongly suggest that two novel clones of PEN-, cephalosporin-, and macrolide-resistant S. pneumoniae serotypes 23F and 19F are present in Taiwan.
The prevalence of penicillin (PEN) resistance among pneumococci is increasing alarmingly worldwide (3, 16, 30, 33). The international spread of a restricted number of multiresistant pneumococcal clones has significantly contributed to this increase. The most extensively studied clones are those that appear to have emerged within Spain, which include serotype 6B, 14, 23F, and 9V clones (6, 7, 8, 17, 21, 29, 34). Other clones have been reported in countries that have a high prevalence of antibiotic-resistant pneumococci, e.g., Hungary, Slovakia, South Africa, and parts of the United States (13, 15, 18, 19, 28). Apart from clonal spread, another possible mechanism of emergence of PEN resistance is in vivo selection of clonally unrelated strains with modified PEN binding proteins (PBP) (11, 24).
The prevalence of PEN-resistant Streptococcus pneumoniae in Taiwan has increased from 12 to 56.4% in the last decade (3). We have previously documented an extremely high prevalence of nasopharyngeal carriage of PEN-resistant S. pneumoniae among children attending 15 day care facilities or kindergartens in Kaohsiung, Taiwan (3). Serotypes 23F, 19F, 6B, 6A, and 14 were the most prevalent and accounted for 76% of all isolates (unpublished data). Interestingly, serotype 23 comprised 14 of 18 (78%) isolates obtained from one of the day care centers (day care center A). To further elucidate the genetic relatedness of the isolates and to investigate the possible mechanism of spread of resistance, we performed in vitro testing of susceptibilities to 10 drugs by E-test (AB Biodisk, Solua, Sweden) and pulsed field gel electrophoresis (PFGE) on the 18 isolates from day care center A.
(Presented in part at the 37th annual meeting of the Infectious Diseases Society of America, Philadelphia, Pa. 19, November 1999.)
Bacterial strains.
Nasopharyngeal-swab specimens for culture were collected by a single investigator who used a cotton swab placed 1 to 1.5 in. into the nasopharynx. The specimens were immediately placed onto 5% sheep blood (Becton Dickinson Microbiology System, Cockeysville, Md.). All plates were incubated for 24 to 48 h in 5% carbon dioxide.
S. pneumoniae isolates were identified by a typical colony appearance, alpha-hemolysis, and Gram staining. Confirmatory tests included optochin sensitivity and bile solubility tests. All strains were kept frozen at −70°C in tryptic soy broth for further analysis.
Antimicrobial susceptibility testing.
The MICs of antibiotics were determined by E-test according to the manufacturer's instructions. NCCLS breakpoints were used to interpret the E-test results (23).
Serotyping.
The serogroups of S. pneumoniae isolates were determined by Quellung reaction with 12 pools of rabbit pneumococcal antisera in the Danish checkerboard typing system (Copenhagen Serum Institute, Copenhagen, Denmark), together with pools G and I. Serotypes were further determined by the factor sera (31).
DNA preparation, restriction, enzyme, digestion, and PFGE.
PFGE of chromosomal DNA was performed as previously described (20). Interpretation of PFGE interrelationships was performed according to the criteria of Tenover et al. (32). Isolates with restriction patterns showing a one- to three-fragment difference were considered to belong to a common major PFGE restriction type.
The results of susceptibility testing and serotyping are shown in Table 1. All but one isolate were highly resistant to PEN (MIC, ≥2 μg/ml). All isolates were resistant to cefaclor, erythromycin, and tetracycline. A substantial percentage of isolates were nonsusceptible (i.e., either intermediate or highly resistant) to extended-spectrum cephalosporin (cefotaxime [83%] or ceftriaxone [83%]), imipenem (94%), chloramphenicol (44%), or trimethoprim-sulfamethoxazole (94%). The only isolate with intermediate resistance to PEN was of serotype 6B. Among the remaining isolates, 3 were serotype 19F and 14 were serotype 23F.
TABLE 1.
Antimicrobial susceptibilities and serotypes of 18 isolates from day care center A
| Isolate no. | MIC (μg/ml)
|
Serotype | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PEN | Cefaclor | Cefuroxime | Cefotaxime | Ceftriaxone | Imipenem | Erythromycin | Chloramphenicol | Tetracycline | Trim-sulfaa | ||
| 8 | 0.5 | >256 | 4 | 1 | 0.5 | 0.094 | >256 | 4 | 64 | 2 | 6B |
| 10 | 2 | >256 | 4 | 1 | 1 | 0.5 | >256 | 8 | 16 | 8 | 19F |
| 1 | 4 | >256 | 4 | 1 | 1 | 0.5 | 16 | 4 | 16 | 4 | 19F |
| 5 | 2 | >256 | 4 | 0.5 | 0.5 | 0.25 | 16 | 4 | 16 | 2 | 19F |
| 17 | 4 | >256 | 4 | 1 | 1 | 0.125 | >256 | 32 | 32 | 8 | 23F |
| 13 | 4 | >256 | 2 | 0.5 | 0.5 | 0.25 | >256 | 4 | 32 | 0.5 | 23F |
| 9 | 4 | >256 | 0.023 | 1 | 2 | 0.5 | >256 | 4 | 32 | 8 | 23F |
| 11 | 4 | >256 | 8 | 2 | 2 | 0.5 | >256 | 8 | 32 | 4 | 23F |
| 12 | 4 | >256 | 8 | 2 | 2 | 0.5 | >256 | 64 | 32 | 8 | 23F |
| 14 | 4 | >256 | 4 | 1 | 1 | 0.5 | >256 | 8 | 32 | 16 | 23F |
| 15 | 4 | >256 | 8 | 2 | 2 | 0.5 | >256 | 64 | 32 | 16 | 23F |
| 16 | 4 | >256 | 8 | 1 | 1 | 0.5 | >256 | 4 | 32 | 16 | 23F |
| 18 | 4 | >256 | 8 | 2 | 2 | 0.5 | >256 | 4 | 32 | 8 | 23F |
| 2 | 4 | >256 | 8 | 2 | 2 | 0.5 | >256 | 32 | 32 | 4 | 23F |
| 3 | 4 | >256 | 8 | 2 | 2 | 0.5 | >256 | 4 | 32 | 4 | 23F |
| 4 | 4 | >256 | 4 | 2 | 2 | 0.5 | >256 | 4 | 16 | 2 | 23F |
| 6 | 4 | >256 | 8 | 0.064 | 2 | 1 | >256 | 4 | 32 | 4 | 23F |
| 7 | 4 | >256 | 8 | 1 | 2 | 0.5 | >256 | 8 | 32 | 4 | 23F |
| MIC50 | 4 | >256 | 4 | 1 | 2 | 0.5 | >256 | 4 | 32 | 4 | |
| MIC90 | 4 | >256 | 8 | 2 | 2 | 0.5 | >256 | 32 | 32 | 16 | |
Trim-sulfa, trimethoprim-sulfamethoxazole.
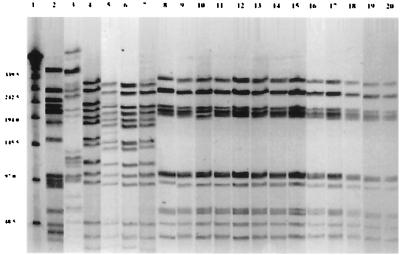
Digestion of S. pneumoniae DNA with SmaI resulted in 10 to 16 well-resolved fragments 20 to 300 kb in size for comparison between strains (Fig. 1). Three different PFGE patterns were observed: 13 strains of serotype 23F showed one pattern (designated type A), 1 serotype 23F isolate and the 3 serotype 19F isolates demonstrated another pattern (type B), and a third pattern (type C) was expressed by the 1 serotype 6B isolate. Notably, all three patterns were unique and different from those of the previously reported 6B, 19F, and 23F clones.
FIG. 1.
PFGE profiles of 18 isolates of S. pneumoniae from day care center A. Lane 1, lambda ladder; lane 2, Spanish clone 23F; lane 3, serotype 6B (strain 8); lanes 4 to 6, serotype 19F (strains 10, 1, and 5); lane 7, serotype 23F (strain 17); lanes 8 to 20, serotype 23F (strains 13, 9, 11, 12, 14, 15, 16, 18, 2, 3, 4, 6, and 7).
One serotype 23F isolate demonstrated exactly the same PFGE pattern as that of 19F isolates (Fig. 1, lane 7) (type B pattern). The possibility of capsular transformation among these isolates was high, although we did not further examine the capsular biosynthetic genes.
This study demonstrates two distinctive clones of PEN-, cephalosporin-, and macrolide-resistant S. pneumoniae in infants attending a single day care center. These clones were genetically distinct from the previously reported Spanish 23F and 19F clones in Europe and the United States but similar to clones reported from other areas of Taiwan (27). Molecular typing studies of PEN-resistant S. pneumoniae from several countries suggest that the majority of strains circulating within a geographic area are derivatives of a relatively small number of clonal lineages (22, 26). However, with the limited number of isolates tested, the precise prevalence of these two PEN-, cephalosporin-, and macrolide-resistant clones in Taiwan remains undetermined.
The fact that all but one of the 18 isolates from day care center A were highly resistant to PEN but only half of them were highly resistant to either cefotaxime or ceftriaxone is in agreement with mechanisms of resistance in extended-spectrum cephalosporins (1, 4, 5). A high level of resistance to cefotaxime and ceftriaxone is due to the production of altered low-affinity forms of only PBP1a and -2x, whereas intermediate- or high-level resistance to PEN is characterized by a reduction in the affinities of PBP1a, -2x, and -2b (1, 5). The PBP2b gene product of S. pneumoniae has very low affinity for cephalosporins, and inactivation of this PBP appears not to be involved in the killing action of cefotaxime or ceftriaxone at physiologically relevant concentrations (4, 5). As a consequence, resistance to extended-spectrum cephalosporins will not necessarily correlate with resistance to PEN (1, 4, 5).
The 13 23F isolates with identical PFGE patterns were all highly resistant to PEN, cefaclor, erythromycin, and tetracycline. On the other hand, the MICs of extended-spectrum cephalosporins, chloramphenicol, and trimethoprim-sulfamethoxazole for these isolates were different. It is notable that within this group of 13 related strains, chloramphenicol resistance varied by up to 16-fold. Identical antibiotic susceptibility patterns with different PFGE patterns have been reported before (12). Since antimicrobial susceptibility patterns are easily influenced by selective pressure exerted by several antimicrobial agents, they cannot be relied upon to predict the genetic relatedness of drug-resistant S. pneumoniae strains. From our data on PFGE and susceptibility patterns, it appears reasonable to speculate that pneumococcal resistance is a combination of the spread of resistant clones and the spread of resistance genes within those clonal lineages.
The fact that the 23F clone was clustered in one day care center raises an interesting question as to the epidemiological origin of these clones. Day care centers have been implicated as sites of augmentation and spread of drug-resistant S. pneumoniae in several recent studies (9, 10, 14, 35). The epidemic diffusion of resistant serotype 23F strains in group day care centers has been reported (2, 25). The distinctive clones we identified in a single day care center might represent the evolutionary result of a combination of antibiotic selection pressure and close contact. However, the possibility of epidemic spread in the entire population has not been excluded.
The bacterial ecological situation of the day care center, with the resident PEN-, cephalosporin-, and macrolide-resistant clones, is critical. Strategies to prevent the spread of such drug-resistant S. pneumoniae clones are increasingly important.
Acknowledgments
We thank Keith Klugman and Victor Yu for their expertise and insight in reviewing the manuscript and Lee Harrison for his assistance. We also thank Andreas Groll for his helpful comments.
This work was supported by the National Foundation for Infectious Diseases (M. C. M.) and the National Science Council in Taiwan, grant number NSC 88-2314-B057B-011.
REFERENCES
- 1.Barcus V A, Ghanekar K, Yeo M, Coffey T J, Dowson C G. Genetics of high level penicillin resistance in clinical isolates of Streptococcus pneumoniae. FEMS Microbiol Lett. 1995;126:299–303. doi: 10.1111/j.1574-6968.1995.tb07433.x. [DOI] [PubMed] [Google Scholar]
- 2.Barnes D M, Whittier S, Gilligan P H, Soares S, Tomasz A, Henderson F W. Transmission of multidrug-resistant serotype 23F Streptococcus pneumoniae in group day care: evidence suggesting capsular transformation of the resistant strain in vivo. J Infect Dis. 1995;171:890–896. doi: 10.1093/infdis/171.4.890. [DOI] [PubMed] [Google Scholar]
- 3.Chiou C C, Liu Y C, Huang T S, Hwang W K, Wang J H, Lin H H, Yen M Y, Hseih K S. Extremely high prevalence of nasopharyngeal carriage of penicillin-resistant Streptococcus pneumoniae among children in Kaohsiung, Taiwan. J Clin Microbiol. 1998;36:1933–1937. doi: 10.1128/jcm.36.7.1933-1937.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffey T J, Daniels M, McDougal L K, Dowson C G, Tenover F C, Spratt B G. Genetic analysis of clinical isolates of Streptococcus pneumoniae with high-level resistance to expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 1995;39:1306–1313. doi: 10.1128/aac.39.6.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffey T J, Dowson C G, Daniels M, Spratt B G. Genetics and molecular biology of beta-lactam-resistant pneumococci. Microb Drug Resist. 1995;1:29–34. doi: 10.1089/mdr.1995.1.29. [DOI] [PubMed] [Google Scholar]
- 6.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal gene transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 7.Coffey T J, Enright M C, Daniels M, Wilkinson P, Berrón S, Fenoll A, Spratt B G. Serotype 19A variants of the Spanish serotype 23F multiresistant clone of Streptococcus pneumoniae. Microb Drug Resist. 1998;4:51–55. doi: 10.1089/mdr.1998.4.51. [DOI] [PubMed] [Google Scholar]
- 8.Coffey T J, Berrón S, Daniels M, Garcia-Leoni E, Cercenado E, Bouza E, Fenoll A, Spratt B G. Multiply antibiotic-resistant Streptococcus pneumoniae recovered from Spanish hospitals (1988–1994): novel major clones of serotypes 14, 19F and 15F. Microbiology. 1996;142:2747–2757. doi: 10.1099/13500872-142-10-2747. [DOI] [PubMed] [Google Scholar]
- 9.Craig A S, Erwin P C, Schaffner W, Elliott J A, Moore W L, Ussery X T, Patterson L, Dake A D, Hannah S G, Butler J C. Carriage of multidrug-resistant Streptococcus pneumoniae and impact of chemoprophylaxis during an outbreak of meningitis at a day care center. Clin Infect Dis. 1999;29:1257–1264. doi: 10.1086/313451. [DOI] [PubMed] [Google Scholar]
- 10.De Lencastre H, Kristinsson K G, Brito-Avo A, Sanches I S, Sa-Leao R, Saldanha J, Sigvaldadottir E, Karlsson S, Oliveira D, Mato R, de Sousa M A, Tomasz A. Carriage of respiratory tract pathogens and molecular epidemiology of Streptococcus pneumoniae colonization in healthy children attending day care centers in Lisbon, Portugal. Microb Drug Resist. 1999;5:19–29. doi: 10.1089/mdr.1999.5.19. [DOI] [PubMed] [Google Scholar]
- 11.Doit C, Denamur E, Picard B, Geslin P, Elion J, Bingen E. Mechanisms of the spread of penicillin resistance in Streptococcus pneumoniae strains causing meningitis in children in France. J Infect Dis. 1996;174:520–528. doi: 10.1093/infdis/174.3.520. [DOI] [PubMed] [Google Scholar]
- 12.Doren G V, Brueggemann A B, Blocker M, Dunne M, Holley H P, Kehl K S, Duval J, Kugler K, Putman S, Rauch A, Pfaller M A. Clonal relationships among high-level penicillin-resistant Streptococcus pneumoniae in the United States. Clin Infect Dis. 1998;27:757–761. doi: 10.1086/514937. [DOI] [PubMed] [Google Scholar]
- 13.Figueiredo A M, Austrian R, Urbaskova P, Teixeira L A, Tomasz A. Novel penicillin-resistant clones of Streptococcus pneumoniae in the Czech Republic and Slovakia. Microb Drug Resist. 1995;1:71–78. doi: 10.1089/mdr.1995.1.71. [DOI] [PubMed] [Google Scholar]
- 14.Givon N, Fraser D, Porat N, Dagan R. Day care centers as a site for development and amplification of Streptococcus pneumoniae (Pnc) nasopharyngeal (NP) carriage. Am J Epidemiol. 1998;147:77. . (Abstract.) [Google Scholar]
- 15.Klugman K P, Coffey T J, Smith A, Wasas A, Meyers M, Spratt B G. Cluster of an erythromycin-resistant variant of the Spanish multiply resistant 23F clone of Streptococcus pneumoniae in South Africa. Eur J Clin Microbiol Infect Dis. 1994;13:171–174. doi: 10.1007/BF01982193. [DOI] [PubMed] [Google Scholar]
- 16.Klugman K P. Pneumococcal resistance to antibiotics. Clin Microbiol Rev. 1990;3:171–196. doi: 10.1128/cmr.3.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lefevre J C, Bertrand M A, Faucon G. Molecular analysis by pulsed-field gel electrophoresis of penicillin-resistant Streptococcus pneumoniae from Toulouse, France. Eur J Clin Microbiol Infect Dis. 1995;14:491–497. doi: 10.1007/BF02113426. [DOI] [PubMed] [Google Scholar]
- 18.Marton A, Meszner Z. Epidemiological studies on drug resistance in Streptococcus pneumoniae in Hungary: an update for the 1990s. Microb Drug Resist. 1999;5:201–205. doi: 10.1089/mdr.1999.5.201. [DOI] [PubMed] [Google Scholar]
- 19.McDougal L K, Rasheed J K, Biddle J W, Tenover F C. Identification of multiple clones of extended-spectrum cephalosporin-resistant Streptococcus pneumoniae isolates in the United States. Antimicrob Agents Chemother. 1995;39:2282–2288. doi: 10.1128/aac.39.10.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McEllistrem M C, Stout J E, Harrison L H. Simplified protocol for pulsed-field gel electrophoresis analysis of Streptococcus pneumoniae. J Clin Microbiol. 2000;38:351–353. doi: 10.1128/jcm.38.1.351-353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muñoz R, Coffey T C, Daniels M, Dowson C G, Laible G, Casal J, Hakenbeck R, Jacobs M, Musser J M, Spratt B G, Tomasz A. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J Infect Dis. 1991;164:302–306. doi: 10.1093/infdis/164.2.302. [DOI] [PubMed] [Google Scholar]
- 22.Muñoz R, Musser J M, Crain M, Briles D E, Marton A, Parkinson A J, Sorensen U, Tomasz A. Geographic distribution of penicillin-resistant clones of Streptococcus pneumoniae: characterization by penicillin-binding protein profile, surface protein A typing, and multilocus enzyme analysis. Clin Infect Dis. 1992;15:112–118. doi: 10.1093/clinids/15.1.112. [DOI] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M100–S10. Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 24.Nesin M, Ramirez M, Tomasz A. Capsular transformation of a multidrug-resistant Streptococcus pneumoniae in vivo. J Infect Dis. 1998;177:707–713. doi: 10.1086/514242. [DOI] [PubMed] [Google Scholar]
- 25.Reichler M R, Allphin A A, Breiman R F, Schreiber J R, Arnold J E, McDougal L K, Facklam R R, Boxerbaum B, May D, Walton R O. The spread of multiple resistant Streptococcus pneumoniae at a day care center in Ohio. J Infect Dis. 1992;166:1346–1353. doi: 10.1093/infdis/166.6.1346. [DOI] [PubMed] [Google Scholar]
- 26.Robinson D A, Turner J S, Facklam R R, Parkinson A J, Breiman R F, Gratten M, Steinhoff M C, Hollingshead S K, Briles D E, Crain M J. Molecular characterization of a globally distributed lineage of serotype 12F Streptococcus pneumoniae causing invasive disease. J Infect Dis. 1999;179:414–422. doi: 10.1086/314589. [DOI] [PubMed] [Google Scholar]
- 27.Shi Z Y, Enright M C, Wilkinson P, Griffiths D, Spratt B G. Identification of three major clones of multiply antibiotic-resistant Streptococcus pneumoniae in Taiwanese hospitals by multilocus sequence typing. J Clin Microbiol. 1998;36:3514–3519. doi: 10.1128/jcm.36.12.3514-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith A M, Klugman K P. Three predominant clones identified within penicillin-resistant South African isolates of Streptococcus pneumoniae. Microb Drug Resist. 1997;3:385–389. doi: 10.1089/mdr.1997.3.385. [DOI] [PubMed] [Google Scholar]
- 29.Soares S, Kristinsson K G, Musser J M, Tomasz A. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniae from Spain to Iceland in the late 1980's. J Infect Dis. 1993;168:158–163. doi: 10.1093/infdis/168.1.158. [DOI] [PubMed] [Google Scholar]
- 30.Song J H, Lee N Y, Ichiyama S, Yoshida R, Hirakata Y, Fu W, Chongthaleong A, Aswapokee N, Chiu C H, Lalitha M K, Thomas K, Perera J, Yee T T, Jamal F, Warsa U C, Vinh B X, Jacobs M R, Appelbaum P C, Pai C H. Spread of drug-resistant Streptococcus pneumoniae in Asian countries: Asian Network for Surveillance of Resistant Pathogens (ANSORP) Study. Clin Infect Dis. 1999;28:1206–1211. doi: 10.1086/514783. [DOI] [PubMed] [Google Scholar]
- 31.Sorensen U B. Typing of pneumococci by using 12 pooled antisera. J Clin Microbiol. 1993;31:2097–2100. doi: 10.1128/jcm.31.8.2097-2100.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomasz A. Antibiotic resistance in Streptococcus pneumoniae. Clin Infect Dis. 1997;24:S85–S88. doi: 10.1093/clinids/24.supplement_1.s85. [DOI] [PubMed] [Google Scholar]
- 34.Versalovic J, Kapur V, Mason E O, Jr, Shah U, Koeuth T, Lupski J R, Musser J M. Penicillin-resistant Streptococcus pneumoniae strains recovered in Houston: identification and molecular characterization of multiple clones. J Infect Dis. 1993;167:850–856. doi: 10.1093/infdis/167.4.850. [DOI] [PubMed] [Google Scholar]
- 35.Yagupsky P, Porat N, Fraser D, Prajgrod F, Merires M, McGee L, Klugman K P, Dagan R. Acquisition, carriage, and transmission of pneumococci with decreased antibiotic susceptibility in young children attending a day care facility in southern Israel. J Infect Dis. 1998;177:1003–1012. doi: 10.1086/515239. [DOI] [PubMed] [Google Scholar]