Abstract
Src homology-2 domain-containing protein tyrosine phosphatase 2 (SHP-2), encoded by the PTPN11 gene, forms a central component of multiple signalling pathways and is required for insulin-like growth factor (IGF)-induced placental growth. Altered expression of SHP-2 is associated with aberrant placental and fetal growth indicating that drugs modulating SHP-2 expression may improve adverse pregnancy outcome associated with altered placental growth. We have previously demonstrated that placental PTPN11/SHP-2 expression is controlled by miRNAs. SHP-2 regulatory miRNAs may have therapeutic potential; however, the individual miRNA(s) that regulate SHP-2 expression in the placenta remain to be established. We performed in silico analysis of 3’UTR target prediction databases to identify libraries of Hela cells transfected with individual miRNA mimetics, enriched in potential SHP-2 regulatory miRNAs. Analysis of PTPN11 levels by quantitative (q) PCR revealed that miR-758-3p increased, while miR-514a-3p reduced PTPN11 expression. The expression of miR-514a-3p and miR-758-3p within the human placenta was confirmed by qPCR; miR-514a-3p (but not miR-758-3p) levels inversely correlated with PTPN11 expression. To assess the interaction between these miRNAs and PTPN11/SHP-2, specific mimetics were transfected into first-trimester human placental explants and then cultured for up to 4 days. Overexpression of miR-514a-3p, but not miR-758-3p, significantly reduced PTPN11 and SHP-2 expression. microRNA-ribonucleoprotein complex (miRNP)-associated mRNA assays confirmed that this interaction was direct. miR-514a-3p overexpression attenuated IGF-I-induced trophoblast proliferation (BrdU incorporation). miR-758-3p did not alter trophoblast proliferation. These data demonstrate that by modulating SHP-2 expression, miR-514a-3p is a novel regulator of IGF signalling and proliferation in the human placenta and may have therapeutic potential in pregnancies complicated by altered placental growth.
Keywords: SHP-2, PTPN11, miRNA, placenta, trophoblast, proliferation, miR-514, miR-758, IGF
Introduction
The type-1 insulin-like growth factor (IGF) receptor (IGF1R) signalling pathway is key for normal human placental and fetal development (Forbes et al. 2008, Forbes & Westwood 2010). A key component of the IGF1R signalling pathway required for human placental growth is Src homology-2 (SH2) domain-containing protein tyrosine phosphatase 2 (SHP-2) (Forbes et al. 2009). SHP-2, encoded by the PTPN11 gene, is a ubiquitously expressed cytoplasmic tyrosine phosphatase (Adachi et al. 1992, Freeman et al. 1992, Ahmad et al. 1993, Feng et al. 1993, Vogel et al. 1993) that interacts with phosphorylated proteins in response to growth factor stimuli. As such, SHP-2 is also a central component of many other signalling pathways and has multiple roles in developmental processes including proliferation, differentiation and angiogenesis (Chang et al. 2002, Maile & Clemmons 2002, Mannell et al. 2008, Mannell & Krotz 2014). Studies in mice first demonstrated that defects in SHP-2 expression in the placenta lead to fetal growth restriction (FGR) and embryonic lethality (Arrandale et al. 1996, Qu et al. 1997, Chen et al. 2000, Saxton et al. 2000, Yang et al. 2006). Studies also reveal a key role for SHP-2 in the regulation of human fetal growth. Approximately, half of all cases of Noonan syndrome – a developmental disorder associated with FGR and short stature, have mutations in PTPN11resulting in defective IGF-I signalling (Limal et al. 2006, De Rocca Serra-Nedelec et al. 2012, Cessans et al. 2016). Other cases of FGR are also attributed to aberrant placental IGF-I signalling (Laviola et al. 2005, Dumolt et al. 2021); since SHP-2 is a key component of placental IGF-mediated signalling (Forbes et al. 2009), it is likely that modulation of SHP-2 levels could improve placental and subsequent fetal growth. Despite the importance of SHP-2 in normal development, the mechanism of transcriptional and post-transcriptional regulation of SHP-2 is still largely unknown. By reducing the levels of Dicer, a key enzyme in the microRNA (miRNA) biogenesis pathway, in human first-trimester villous explants, we have demonstrated that placental SHP-2 expression is controlled by miRNAs (Forbes et al. 2012).
Drugs designed to mimic or inhibit miRNAs have shown therapeutic promise in numerous diseases including hepatitis C (miR-122 inhibitor), type-2 diabetes (miR-103/107) and cancer (miR-34 mimic) (Rupaimoole & Slack 2017, Chakraborty et al. 2021), and we have previously shown the safe effective delivery of miRNA inhibitors to the placenta in mice and that targeting key placental regulatory miRNAs improves murine placental and fetal growth (Beards et al. 2017). We propose that miRNAs may be a key mechanism for the regulation of placental SHP-2 expression. While many miRNAs are predicted to target individual genes, only a few miRNAs, to date, have been shown to directly target SHP-2 in immune cells (Li et al. 2007) and in certain cancers (Patel et al. 2016, Cai et al. 2018, Long et al. 2018); none of which have reported roles in human placenta (Liang et al. 2007).
In this study, we aimed to identify miRNAs that modulate SHP-2 expression within the human placenta using high-throughput screens and to determine their potential to modulate IGF-induced cytotrophoblast growth.
Materials and methods
Human placental cells and tissue
Ethical approval
Human placentas were obtained from elective medical or surgical termination of pregnancy during the first trimester (8–12 weeks) of pregnancy. Term placentas from uncomplicated pregnancies (37–42 weeks) were collected within 30 min of vaginal or elective caesarean delivery (Supplementary Table 1, see section on supplementary materials given at the end of this article). The study had local research ethics committee approval (13/NW/0205; 08/H1010/55(+5)), and all tissue was collected from St Mary’s Hospital, Manchester, following written informed consent.
Human placental explant culture
As previously described (Farrokhnia et al. 2014), normal, late first-trimester (8–12 weeks) placental tissue was dissected under sterile conditions into 2–3 mm3 fragments comprised of small cluster of terminal villous branches. Three pieces, selected at random, were transferred into a 1:1 mixture of serum-free Dulbecco’s modified Eagle’s medium (DMEM) and Ham’s F-12 (Lonza, Cambridge, UK) containing 100 U/mL penicillin, 100 μg/mL streptomycin and 2 mM l-glutamine (Gibco). For term tissue, three random areas of each placenta were biopsied, and the chorionic, non-anchoring villi were dissected from these areas and rinsed in sterile PBS, stored in RNALater overnight at 4°C and transferred to −80°C prior to use in experiments to profile PTPN11 and miRNA expression.
Primary human cytotrophoblast and stromal cells
Primary cytotrophoblast and stromal cells were freshly isolated from first-trimester human placenta using the methods previously established (Harris et al. 2006, Ingman et al. 2010). Briefly, tissue was dissected, weighed and washed carefully in minimum essential media (MEM). Approximately, 6 g of tissue was transferred to 20 mL of MEM containing 0.125% (v/v) trypsin, 1.3 mM EDTA (Invitrogen), and 0.4 mg/mL DNase I, grade I (Sigma) and incubated for 35 min at 37°C, with occasional resuspension to remove trophoblast cells. Cells were collected, and enzymatic digestion was repeated. Remaining tissue fragments were filtered (100 μm) and used for stromal cell isolation. Trophoblast cells were loaded on a Percoll gradient and centrifuged for 30 min at 1800 g, where cytotrophoblast cells were collected from 30 to 45% range. Cells were pelleted, resuspended in DMEM/F-12 supplemented with 10% FBS (Biosera, Maidenhead, UK), penicillin (0.1 U/mL), streptomycin (100 μg/mL) and l-glutamine (2 mM) and cultured on Matrigel-coated flasks. Immunostaining for cytokeratin-7 (an epithelial marker) and vimentin (a mesenchymal marker) (data not shown) demonstrated 95% purity of the isolated cytotrophoblast cells. Stromal cells were isolated from tissue free of trophoblast cells and were washed (3 × 5 min) in DMEM containing 2.5 mg/mL collagenase and 2 mg/mL hyaluronidase and incubated for 3 h at 37°C. Settling under gravity, the suspension was then centrifuged for 20 min at 700 g, and the resulting pellet was resuspended in 3 mL of MEM. Subsequently, the cell pellet was loaded onto 25/60% Percoll and centrifuged for 30 min at 670 g. The band of stromal cells and aggregates was removed from the Percoll, added to 35 mL of MEM and centrifuged for 15 min at 100 g. The pellet was then resuspended in DMEM supplemented with 10% FBS, penicillin (0.1 U/mL), streptomycin (100 μg/mL) and l-glutamine (2 mM), and cells were seeded onto flasks coated with rat tail collagen (BD Biosciences).
Human placental cell lines
BeWo human choriocarcinoma cells (CCL-98, American Type Culture Collection (ATCC) and the immortalized human first-trimester extravillous trophoblast cell line Swan-71 (Straszewski-Chavez et al. 2009) were maintained in 1:1 DMEM:Ham’s F-12 supplemented with 10% FBS, penicillin (0.1 U/mL), streptomycin (100 μg/mL) and l-glutamine (2 mM) at 37°C, 5% CO2. JAR (ATCC. HTB-144) and JEG-3 (ATCC 92120308) choriocarcinoma cells were cultured in DMEM, containing 10% FBS and penicillin (0.1 U/mL), streptomycin (100 μg/mL)), 2 mM glutamine at 37°C, 5% CO2.
Identification of candidate SHP-2-regulatory miRNAs
To identify candidate miRNAs that regulate SHP-2 expression, we used a reverse genetics approach. Genome-wide miRNA library screening has been used successfully to identify miRNAs that regulate the expression of specific proteins (le Sage et al. 2007, Zhong et al. 2010); however, since there are currently 1917 mature miRNAs in the human genome (https://www.mirbase.org/summary.shtml?org=hsa), this can be labour intensive and time-consuming. To generate a smaller, focused miRNA library, we first performed a bioinformatics-driven approach using the miRNA:mRNA-predicted interaction programmes miRabel (Quillet et al. 2019) PITA (Kertesz et al. 2007), miRanda (John et al. 2004), SVMicrO (Liu et al. 2010) TargetScan (Lewis et al. 2005) to identify miRNAs with predicted binding sites in the 3’UTR of PTPN11, the gene encoding SHP-2. Predicted SHP-2-regulatory miRNAs present in four of the five prediction programmes were classed as potential PTPN11/SHP-2 regulatory miRNAs.
SureFind Transcriptome PCR complete miRNOME-1 mimic arrays (Qiagen) are array panels containing cDNA preparations obtained from HeLa cells that have been transfected with 90 different specific miRNA mimics to simulate the overexpression of the mature miRNAs and six experimental controls which include non-targeting miRNA mimics. We searched all commercially available SureFind transcriptome (PCR) complete miRNOME mimic array panels to identify an array panel enriched with mimics predicted to bind to PTPN11. RT2 SYBR Green quantitative (q)PCR mastermix containing PTPN11-specific primers (200 nM) (forward, 5′-CCCACAATCAAGATTCAGAACACT-3′; reverse, 5′GCCCGTGATGTTCCATGTAA-3′) or 18S rRNA-specific primers (forward, 5’GCTGGAATTACCGCGGCT-3’; reverse 5’-CGGCTACCACATCCAAGGAA-3’) were applied to the complete miRNome-3 mimic SureFIND transcriptome PCR array (product code: TCMB-303A-1; Qiagen), and qPCR was performed using a Stratagene MX3005P thermal cycler (Agilent Technologies). Data analysis was performed using Qiagen RT2 Profiler PCR Array Data Analysis Webportal which used the ΔΔCT method with normalization of the raw PTPN11 expression data to 18S rRNA. Data were expressed as fold change compared to control (non-targeting miRNA mimic). miRNA mimics found to alter PTPN11 mRNA expression by at least two-fold were defined as candidate SHP-2 regulators. Binding sites for the seed sequences of these miRNAs in the PTPN11 3’UTR were confirmed using miRANDA target prediction database (Betel et al. 2008). Validation of miRNA mimic screens was performed using qPCR and miRNA-specific mimics as described below.
Quantitative qPCR analysis of miRNA and mRNA expression in human placental cells and tissue
miRNA
Levels of candidate SHP-2 regulatory miRNAs were assessed in human first trimester and term placental tissue, primary human placental stromal or cytotrophoblast cells or human placental cells lines (BeWo, JAR, JEG, SWAN-71). Total RNA was extracted from cells or tissue using miRVANA miRNA extraction kits (Ambion). RNA quantity and purity were assessed by Nanodrop. All RNA samples had A260/A280values in the range of 1.93–2.08 and A260/A230 values in the range 2.05–2.14. Total RNA of 25 ng was reversed transcribed using miRCURY LNA™ Universal cDNA Synthesis kit (Exiqon). Individual miRNAs were detected using the miRCURY LNA™ Universal RT microRNA PCR system (Exiqon) and LNA-enhanced miRNA-specific primer sets (Exiqon) for hsa-miR-514a-3p (target sequence: AUUGACACUUCUGUGAGUAGA) and hsa-miR-758-3p (target sequence: UUUGUGACCUGGUCCACUAACC). All reactions were performed in duplicate using Stratagene MX3005P thermal cycler (Agilent Technologies) and 60°C annealing temperature. 5SrRNA was unaffected by treatment; therefore, the relative amount of miRNA was normalized to 5SrRNA. Fold changes in miRNA expression from control (mean of all ΔCt values) were calculated as 2(−ΔΔCt), where ΔΔCt = ΔCt control − ΔCt treated sample and ΔCt = CtmiRNA − Ct5SrRNA.
mRNA
Levels of PTPN11 were assessed in human first trimester and term placental tissue. Total RNA of 100ng was reverse transcribed using AffinityScript Multi-Temperature cDNA synthesis kit (Agilent). cDNA was diluted 1:10, and qPCR was performed using Brilliant III Ultra-Fast SYBR Green qPCR Master Mix (Agilent) and specific primers for PTPN11 (forward, 5′-CCCACAATCAAGATTCAGAACACT-3′; reverse, 5′GCCCGTGATGTTCCATGTAA-3′) and 18S rRNA-specific primers (forward, 5’-GCTGGAATTACCGCGGCT-3’; reverse 5’-CGGCTACCACATCCAAGGAA-3’. 18S rRNA was unaffected by treatment; therefore, the relative amount of mRNA was normalized to 18S rRNA.
Transfection of pre-miRNA mimetics and SHP-2 siRNA in first-trimester human placental explants
Pre-miR™ miRNA precursors specific for hsa-miR-514a-3p (200 nM; target sequence: AUUGACACUUCUGUGAGUAGA), hsa-miR-758-3p (200 nM; target sequence: UUUGUGACCUGGUCCACUAACC), Cy™ 3-labelled pre-miRNA precursor negative control (Pre-miR-C; 200 nM; Ambion ), short interfering RNA (siRNA) targeting PTPN11 (UAGUGUUUCAAUAUAAUGCUGGACC; 100 nM) or non-targeting siRNA (100 nM; NT; Ambion) were transfected into first-trimester human placenta explants using Nucleofector programme X-005 (Amaxa Biosystems, Germany) using basic primary mammalian epithelial cell nucleofector solution as previously described (Forbes et al. 2009). Following transfection, explants were maintained in culture for 48–72 h on 1% agarose coated 24-well plates as previously described (Farrokhnia et al. 2014). Overexpression of the miRNAs was confirmed by qPCR using specific primers as described above. The effect of the siRNA or miRNA sequences was compared to that of three controls: untreated tissue (control), tissue exposed to the transfection procedure alone (mock) or transfected with non-targeting siRNA or pre-miRNA precursor negative control. These controls formed the reference for the evaluation of the effect of the siRNA and pre-miRNA precursors on downstream protein expression and cell function.
Analysis of SHP-2 protein expression
Immunohistochemistry
First-trimester explants exposed to miRNA mimetics for up to 48 h were fixed in 4% paraformaldehyde and processed for immunohistochemistry as previously described (Forbes et al. 2008). Sections were boiled in 0.1 M sodium citrate buffer to maximize antigen retrieval. SHP-2 localization was examined using rabbit polyclonal anti-human SHP-2 antibody (1:50; Cell Signaling Technologies) followed by a biotinylated swine anti-rabbit IgG antibody (1:200; DakoCytomation Ltd.). Staining was visualized using the avidin-peroxidase method with haemotoxylin counterstain as previously described (Forbes et al. 2008).
Western blotting
To confirm the effect of the miRNA overexpression on protein expression, 72 h post-transfection first-trimester placental explant lysates were prepared in radioimmunoprecipitation assay (RIPA) buffer as previously described (Forbes et al. 2008). Protein of 50 μg from each sample was resolved by SDS-PAGE and transferred to nitrocellulose membranes for Western blotting with antiserum specific for SHP-2 (rabbit polyclonal; 1:5000, Santa Cruz Biotechnology Inc.). Immune complexes were visualized by probing with HRP-conjugated goat anti-rabbit-IgG antibody (1:2000; Dako UK ) followed by chemiluminescence reagents and exposure to x-ray film. ImageJ software was used to quantitate bands corresponding to SHP-2. Membranes were stripped (2% SDS, 100 mM β-mercaptoethanol, 50 mM Tris, pH 6.8, for 30 min at 50°C) and re-probed with an antibody specific for either SHP-1 (1:5000, Santa Cruz Biotechnology Inc.) – to confirm that SHP-2 siRNA specifically altered SHP-2 protein expression and not that of SHP-1 which has the high sequence homology to SHP-2, or for β-actin (1:1000; Sigma clone AC-15) to confirm equal protein loading.
Assessing potential interaction of candidate miRNAs with PTPN11 mRNA by analysing microRNA-ribonucleoprotein complex (miRNP)-associated mRNA
Mature miRNAs are guided to 3’UTRs of their target genes following association with ribonucleoproteins in the RNA-inducing silencing complex (RISC). The RISC complex contains a number of proteins, including members of the Agonaute (Ago) family. Antibodies to Agonaute proteins are commercially available and allow immunoprecipitation of the RISC complex (ribonucleoprotein immunoprecipitation/RIP). This method can be used to identify miRNA targets since isolated Ago immunocomplexes also contain functional RISC–miRNA–mRNA complexes (Easow et al. 2007, Wang et al. 2010). By comparing the microRNA ribonucleoprotein (miRNP)-associated mRNAs from control and miRNA-overexpressing cells or tissue, miRNAs that directly interact with specific mRNAs can be identified. First-trimester placental explants were exposed to miRNA-mimetics (200 nM) for 24 h, and miRNP–mRNA complexes were isolated using the RIP-assay kit for microRNA following the manufacturer’s instructions (MBL International Corporation, Massachusetts, USA). Briefly, explants were lysed in mi-lysis buffer and pre-cleared by incubation with protein G beads. Pre-cleared lysates of 500 mg were then applied to Ago-2 antibody (15 mg) immobilized protein-A beads (30 mL) and incubated overnight at 4°C. Beads were washed, and lysates were collected for Western blotting and for subsequent RNA extraction using miRVANA miRNA isolation kit (Ambion). Western blotting was performed to confirm successful immunoprecipitation of the RISC complex, and qPCR analysis was performed to determine the levels of PTPN11 mRNA present in the complexes.
Cell proliferation assays
As previously described, first-trimester placental explants were mock transfected, transfected with siRNA, miRNA-specific mimics or non-targeting miRNA mimics (Farrokhnia et al. 2014) and were cultured in serum-free medium for 48 h and then exposed to 5-bromo-2’-deoxyuridine (BrdU; 100 μM) and IGF-I (10 nM), or vehicle and cultured for a further 24 h. At the end of the experiment, placental tissue was fixed, embedded in wax and levels of BrdU-positive trophoblast cells were assessed by immunohistochemistry as previously described (Forbes et al. 2008).
The number of immunopositive (proliferating) cells was expressed as a percentage of total cytotrophoblast nuclei.
Statistical analysis
Data were analysed using GraphPad Prism 6 Version 6.05 (GraphPad Software Inc.). Comparisons between first trimester and term placentas were done using Mann–Whitney U test. The correlation between levels of miR-514a-3p and miR-758-3p with PTPN11 expression was assessed by Spearman rank correlation test. mRNA expression was analysed using the Wilcoxon-signed rank test, and data are presented as median and range mRNA expression relative to the control (non-transfected) sample for the corresponding experiment. Proliferation data presented as median and range were analysed using the Kruskal–Wallis ANOVA followed by Dunn’s post hoc test that was used to compare differences between groups. Results were considered significant at P < 0.05.
Results
miR-514a-3p and miR-758-3p are candidate regulators of SHP-2 expression
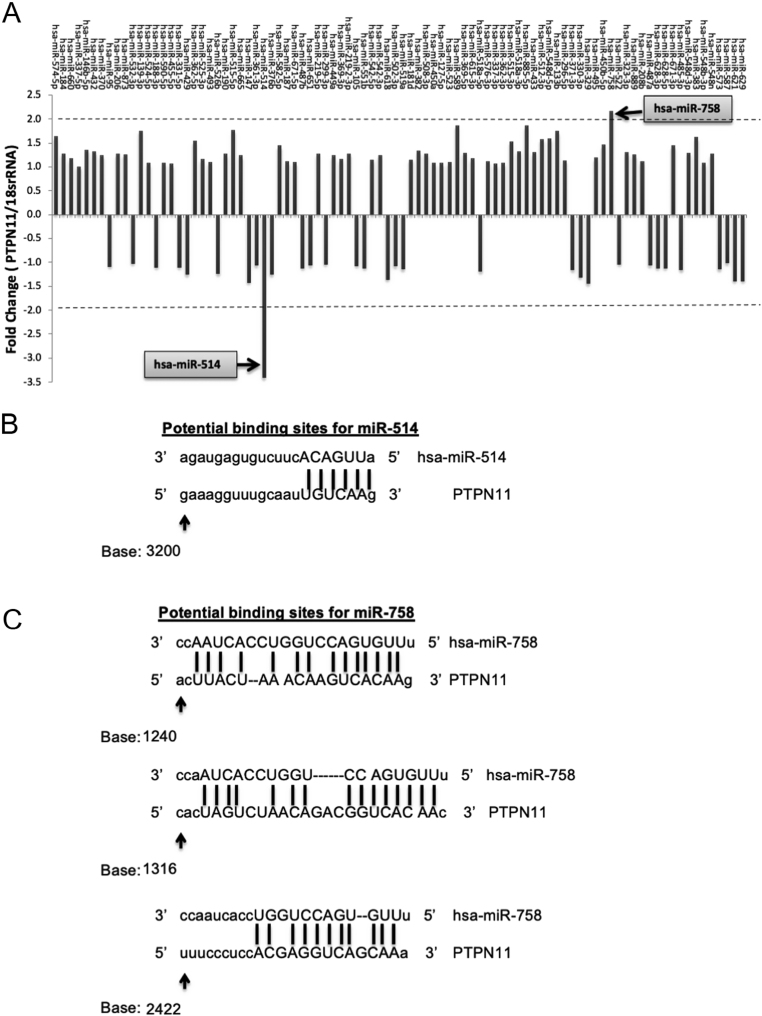
In total, 485 miRNAs were identified as potential PTPN11 regulatory miRNAs (Supplementary Table 2). Samples for 58 of these miRNAs were present in the SureFind transcriptome PCR mimic array, representing 66% of the total mimic samples present on the array (Supplementary Table 3). Despite the array library being enriched for predicted miRNAs that target SHP-2, PTPN11 expression was only altered by 2 of the 90 miRNA mimics assessed (Fig. 1A and Supplementary Table 3). PTPN11 expression was reduced by 3.40-fold in samples overexpressing miR-514a-3p and was increased (by 2.16-fold) in samples overexpressing miR-758-3p, suggesting a role for these miRNAs in the regulation of SHP-2 expression. In silico analysis using miRANDA target prediction database (Betel et al. 2008) demonstrated that there are potential binding sites for both miR-514a-3p and miR-758-3p in the PTPN11 3’UTR (Fig. 1B and C). Therefore, both miR-514a-3p and miR-758-3p were selected for subsequent analyses.
Figure 1.
Identification of SHP-2 regulator miRNAs and potential binding sites for SHP-2 mRNA. Primers specific for PTPN11 were applied to miRNA SureFind transcriptome PCR array containing cDNA from HeLa cells overexpressing 90 different miRNA mimetics and relevant controls. PTPN11 expression was expressed as fold change in expression relative to 18S rRNA. Samples with a change greater than two-fold were considered potential SHP-2 regulatory miRNAs. In silico analysis was performed to determine if the seed sequences of miRNAs that altered PTPN11mRNA expression-miR-514a-3p (B) and miR-758-3p (C) had potential binding sites in PTPN11 3’UTR. Dashed lines represent consensus binding sites between miRNAs and PTPN11. Lower case base letters represent bases with no homology between the individual miRNA and PTPN11.
miR-514a-3p negatively correlates with placental SHP-2 expression
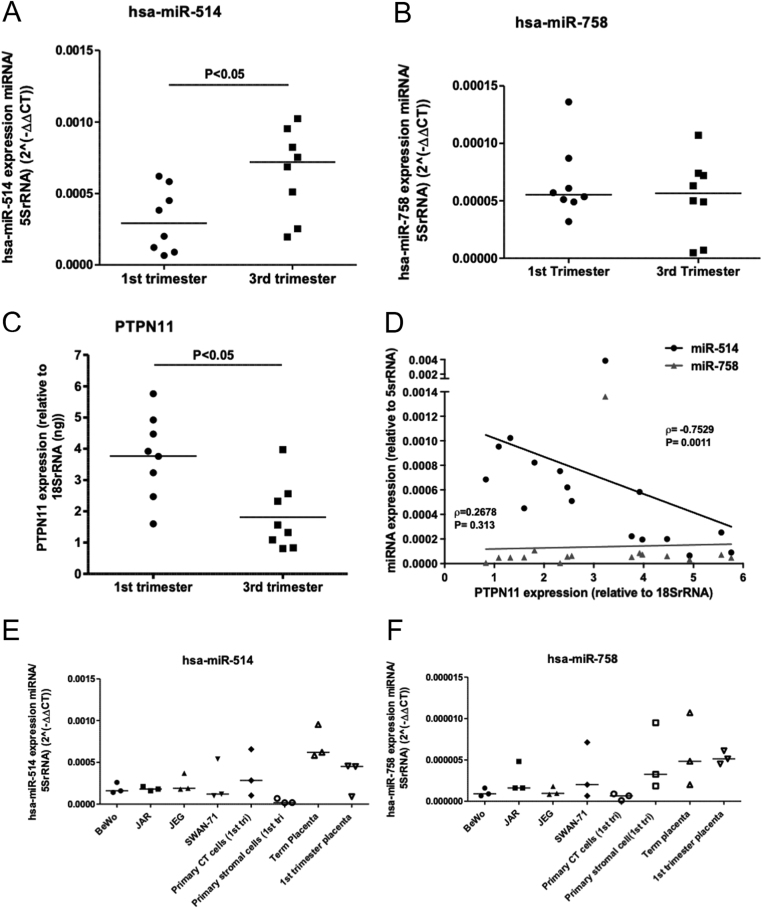
Further investigations were carried out to determine if the initial library screens in HeLa cell samples (Fig. 1A) translated into the human placenta. qPCR analysis confirmed that miR-514a-3p and miR-758-3p (Fig. 2A and B) were present in both first and third-trimester human placenta. miR-514a-3p was significantly higher in the third trimester than in the first trimester (Fig. 2A; P= 0.028; n = 8), but miR-758-3p remained at similar levels throughout pregnancy, albeit in lower levels than miR-514a-3p (Fig. 2B). Expression of PTPN11 mRNA, the transcript for SHP-2 protein, was highest in the first trimester of pregnancy (Fig. 2C; P= 0.010; n = 8). This inversely correlated with levels of miR-514a-3p, but not miR-758-3p, in the placenta (Fig. 2D; r = −0.7529, P= 0.0011). Furthermore, analysis of miRNA expression in different cell types isolated from the human placenta reveals that similar to SHP-2 (Forbes et al. 2009), miR-514a-3p is predominantly expressed in the trophoblast (Fig. 2E). In contrast, levels of miR-758-3p appeared to be higher in the placental stroma (Fig. 2F).
Figure 2.
miR-514a-3p and miR-758-3p expression in human placental tissue and cells. Total RNA was isolated from fresh first trimester (n = 8) and term (n = 8) human placental tissue, and levels of miR-514a-3p (A), miR-758-3p (B) and PTPN11 mRNA (C) were assessed by qPCR. Data were expressed relative to 5SrRNA, and lines show median relative expression levels. Differences in expression between the two gestational age periods were considered different when P< 0.05 (Mann–Whitney U-test). The correlation between the levels of miR-514a-3p and miR-758-3p with PTPN11 expression was assessed by Spearman rank correlation test (D). r and Pvalues for each miRNA are displayed. Levels of miR-514a-3p (E) and miR-758-3p (F) were assessed in a panel of placental cell lines (BeWo, JAR, JEG and SWAN-71), primary cells from first-trimester placenta (primary cytotrophoblast, CT and primary stromal cells) and fragments of whole placental tissue from term or first-trimester placenta. Levels are expressed as levels of specific miRNA relative to 5SrRNA.
miR-514a-3p directly regulates SHP-2 expression in the human placenta
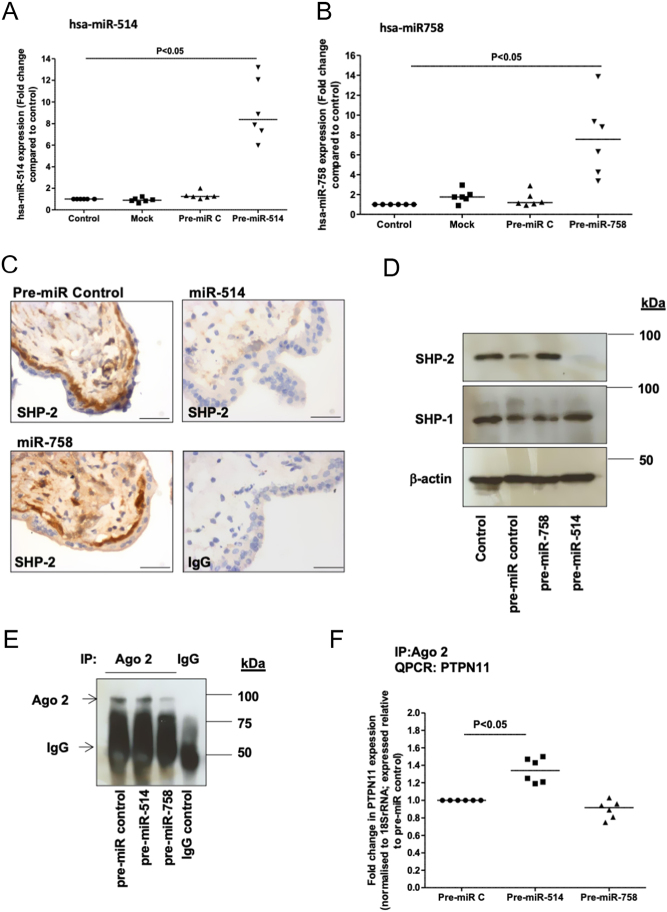
To explore if miR-514a-3p and miR-758-3p were functional in the placenta, these miRNAs were overexpressed in first-trimester placental explants using specific mimetics (compared to non-transfected, mock-transfected or non-targeting miRNA mimic controls P< 0.05; n = 6). miR-514a-3p (Fig. 3A), but not miR-758-3p (Fig. 3B) overexpression, reduced SHP-2 protein expression in the placenta (Fig. 3C and D). qPCR analysis of Ago-bound miRNA:mRNA complexes (Fig. 3E) confirmed that PTPN11 was present in immunocomplexes under control conditions (Pre-miR C; Fig. 3F). Levels of PTPN11 were significantly higher in the Ago-bound miRNA:mRNA complexes when miR-514a-3p was overexpressed (P< 0.05, n = 6). Recovery of PTPN11 from miRNA complexes was unaffected by miR-758-3p overexpression (Fig. 3F). This demonstrates direct interaction of miR-514a-3p, but not miR-758-3p, to PTPN11in the human placenta.
Figure 3.
miR-514a-3p regulates SHP-2 expression in the human placenta, but miR-758-3p does not. miR-514a-3p, miR-758-3p or negative control (pre-miR C) mimetics (200 nM) were transfected in to first-trimester human explant tissue for up to 48 h. Overexpression of miR-514a-3p (A) and miR-758-3p (B) was confirmed by qPCR, (Wilcoxon-signed rank test; n = 6; P< 0.05). SHP-2 protein expression was examined by immunohistochemistry (C) and Western blotting (D). Blots were stripped and re-probed for β-actin (as an internal control) and SHP-1 (as a control for PTP expression). For immunohistochemistry, rabbit IgG was used as a negative control. Scale bars on images represent 50 μm (C). To determine if miRNAs bound directly to PTPN11 mRNA, Ago-2-bound miRNA:mRNA complexes were immunoprecipitated (IP) from placental lysates. Recovery of Ago-2 complexes was confirmed by Western blotting (E). Assessment of Ago-2 levels in immunocomplexes recovered using mouse IgG confirmed specificity of the IP. qPCR analysis (F) revealed that levels of PTPN11 in Ago-bound miRNA:mRNA complexes were significantly increased after the overexpression of miR-514a-3p (Wilcoxon-signed rank test; n = 6; P< 0.05).
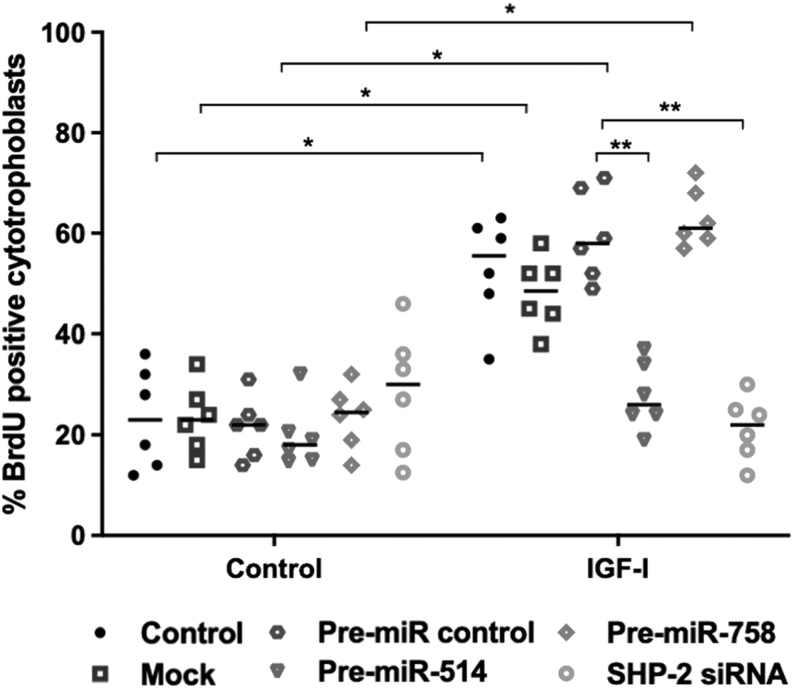
miR-514a-3p negatively regulates IGF-induced cytotrophoblast proliferation in the first-trimester human placenta
Under basal conditions, miR-514a-3p overexpression did not alter the levels of cytotrophoblast proliferation (Fig. 4). IGF-I significantly increased cytotrophoblast proliferation compared to vehicle control (Fig. 4; P< 0.05; n = 6), and miR-514a-3p overexpression prevented this response (Fig. 4; P< 0.01; n = 6). The effect of miR-514 overexpression on IGF-induced cytotrophoblast proliferation was comparable to the effect of SHP-2 siRNA on IGF-induced proliferation. miR-758-3p overexpression, mock transfection and transfection of non-targeting miRNA mimics had no effect on either basal or IGF-I-induced trophoblast proliferation.
Figure 4.
Overexpression of miR-514-a-3p attenuates IGF-induced proliferation in the human placenta. First-trimester placental explants (n = 6) were transfected with miR-514a-3p (200 nM), miR-758-3p mimics (200 nM), non-targeting miRNA-mimic (pre-miR-control; 200 nM) or SHP-2 siRNA (100 nM). IGF-I (10 nM) or vehicle was added after 48 h, then 24 h later, cytotrophoblast proliferation was assessed by immunohistochemical analysis of BrdU incorporation and expressed as the number of immunopositive cells as a percentage of total nuclei. Data are presented as median and were analysed by Kruskal–Wallis followed by Dunn’s post hoc test. n = 6; *P< 0.05; **P< 0.01.
Discussion
It has been previously shown that SHP-2 is an important component of the IGF signalling axis in the placenta (Forbes et al. 2009), and miRNAs contribute to the regulation of this signalling and placental growth (Farrokhnia et al. 2014). In this study, two miRNAs, miR-758-3p and miR-514a-3p, were identified in HeLa cells as potential SHP-2 regulatory miRNAs, and both miRNAs are expressed in first trimester and term human placenta. By enhancing the expression of miR-514a-3p and miR-758-3p in first-trimester placental explants, it was demonstrated that miR-514a-3p, but not miR-758-3p, directly influences SHP-2 expression and significantly attenuates IGF-I-induced cytotrophoblast growth.
In this study, we took the approach of combining miRNA target prediction algorithms with a miRNA mimic transcriptome library in an attempt to identify specific PTPN11 regulatory miRNAs. miRNA target prediction algorithms are useful for gaining insights into potential miRNA targets; however, they commonly provide false positives (Riolo et al. 2020). We attempted to reduce the number of false positives identified by only classing the miRNAs as potential PTPN11 regulatory miRNAs if they were predicted by four independent prediction algorithms and experimentally validating these using a miRNA mimic transcriptome array. This high throughput approach is similar to utilizing miRNA mimic libraries. These have been utilized to identify miRNAs for individual genes or for specific cellular functions (Nakano et al. 2013, Yarbrough et al. 2014, Xiao et al. 2015, Martello et al. 2018) but are expensive and time-consuming since they require transfections of multiples – sometimes hundreds – of miRNA mimics into cells/tissue prior to functional analysis. miRNA mimic transcriptome arrays overcome these limitations and have been successfully used by other researchers to identify specific gene regulatory miRNAs (Fosbrink 2011, Schaefer et al. 2011, Kumar et al. 2016, Cerro-Herreros et al. 2018). However, these too have their own limitation, for example measuring gene expression, rather than protein to assess miRNA target genes levels, does not take into account that the primary action of many mammalian miRNAs is the repression of protein translation (Ritchie et al. 2013). Furthermore, there may be cell-specific miRNA target interactions that are missed by using only HeLa cell cDNA. Indeed, these limitations may explain why, in our study, despite the mimic PCR library being enriched for other predicted miRNAs that target SHP-2, PTPN11 expression was only altered by two of the miRNA-mimetics analysed. Analysis of SHP-2 protein expression following the overexpression of miRNAs may reveal further candidate SHP-2 regulatory miRNAs identified from target prediction algorithms. Despite the limitations, our combined target prediction algorithm and miRNA mimic transcriptome array approach identified two candidate miRNAs upstream of PTPN11 which we went on to investigate in human placental tissue.
miRNAs usually function as negative regulators of their target genes, however in contrast to this, HeLa cDNA samples overexpressing miR-758-3p showed increased levels of PTPN11. It is possible that miR-758-3p is acting indirectly on PTPN11/SHP-2 via targeting of interacting partners, or that similar to some other miRNAs, miR-758-3p could be inducing target gene expression by binding to promoter regions of PTPN11 (Place et al. 2008). In silico analysis using miRanda target prediction database (Betel et al. 2008), however, demonstrated that while there are potential binding sites for miR-758-3p in the PTPN11 3’UTR, there are no binding sites for miR-758-3p in the promoter region of PTPN11; thus a positive role for miR-758-3p in regulating SHP-2 expression via this mechanism is unlikely. Nonetheless, both miR-514a-3p and miR-758-3p were selected for subsequent analyses.
Reported roles for miR-758-3p include the regulation of proliferation, migration and invasion of carcinoma cells (Wu & Liu 2020, Liu et al. 2021, Xiao et al. 2021) via modulation of c-Myc and AKT signalling and long non-coding (lincRNA) and circular RNA (circRNA) actions (Ding et al. 2021, Xiao et al. 2021, Zhang et al. 2021). While miR-758-3p do not influence proliferation in the placenta, migration and invasion are also key features of normal placentation (Knofler & Pollheimer 2012), and key roles for lincRNA and circRNA in placental and fetal development are emerging (Gong et al. 2021). Thus further studies on these aspects of placental development and function may reveal other roles for miR-758-3p in the placenta.
To our knowledge, there are limited reports in the literature of validated gene targets and functional roles for miR-514a-3p. miR-514a-3p is part of a cluster of 14 miRNAs that collectively have been reported to influence melanoma cell growth and invasion (Streicher et al. 2012, Stark et al. 2015) and is downregulated in metastatic renal cell carcinoma (Wotschofsky et al. 2013); thus, potential roles in the placenta include the regulation of cytotrophoblast proliferation and/or invasion. Arthurs et al. (2019) predicted miR-514a-3p to target components of the renin–angiotensin system, specifically downregulating angiotensinogen and angiotensin II type 1 receptor mRNA. Furthermore, miR-514 has been found to be overexpressed in the placentas of women with preeclampsia (Wang et al. 2018), commonly associated with placental insufficiency. Furthermore, in the setting of testicular germ cell tumour, loss of miR-514a-3p expression increases paternally expressed gene 3, which activates the nuclear factor kappa B pathway and protects germ cells from apoptosis (Ozata et al. 2017), thus demonstrating a pro-apoptotic function of miR-514. In this study, we have demonstrated that miR-514a-3p functions to negatively regulate IGF-mediated placental growth by modulating SHP-2 expression. This growth regulatory role for miR-514a-3p is consistent with the proposed oncogenic role for miR-514a-3p and the miR-506-514 cluster (Streicher et al. 2012, Wotschofsky et al. 2013). We have not explored the role of miR-514a-3p and/or SHP-2 in regulating other events within the placenta, but given the reported roles for both SHP-2 and miR-514a-3p in invasion and apoptosis in cancer cells, it will be interesting to investigate whether miR-514a-3p/SHP-2 regulates these events in the placenta and indeed in other tissues.
Conclusion
By utilizing a combination of in silico and molecular techniques, miR-514a-3p has been identified as a novel SHP-2 regulatory miRNA in the human placenta. Similar methodology may be useful for identifying additional miRNAs that regulate other specific proteins and/or cellular processes of interest. Ongoing studies will establish whether alterations in placental SHP-2 and/or miR-514a-3p expression are associated with fetal growth disorders. Ultimately, it is anticipated that miRNA-based approaches can be used as therapeutics for correcting abnormal placental growth and cellular proliferation seen in pathological pregnancies.
Supplementary Material
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research.
Funding
This study was funded by a University of Manchester Stepping Stones Fellowship (K F), a Society for Endocrinology Early Career Grant (awarded to K F) and a Medical Research Council New Investigator Grant (REF:MR/R023166/1). The Maternal and Fetal Health Research Centre is supported by funding from Tommy’s the Baby Charity, an Action Research Endowment Fund, and the Greater Manchester Comprehensive Local Research Network.
Author contribution statement
K F designed the study and secured funding. S L and R Q conducted the experiments and data acquisition. All authors (R Q, S L and K F) contributed to the analysis, discussion and interpretation of the data and manuscript drafting. All authors approved the final version of the manuscript.
Acknowledgements
The authors would like to thank the women that kindly donated their placental tissue to this study and the medical staff at St Mary’s Hospital, Manchester in the aid of recruiting patients.
References
- Adachi M, Sekiya M, Miyachi T, Matsuno K, Hinoda Y, Imai K, Yachi A.1992Molecular cloning of a novel protein-tyrosine phosphatase SH-PTP3 with sequence similarity to the src-homology region 2. FEBS Letters 314335–339. ( 10.1016/0014-5793(9281500-l) [DOI] [PubMed] [Google Scholar]
- Ahmad S, Banville D, Zhao Z, Fischer EH, Shen SH.1993A widely expressed human protein-tyrosine phosphatase containing Src homology 2 domains. PNAS 902197–2201. ( 10.1073/pnas.90.6.2197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrandale JM, Gore-Willse A, Rocks S, Ren JM, Zhu J, Davis A, Livingston JN, Rabin DU.1996Insulin signaling in mice expressing reduced levels of Syp. Journal of Biological Chemistry 27121353–21358. ( 10.1074/jbc.271.35.21353) [DOI] [PubMed] [Google Scholar]
- Arthurs AL, Lumbers ER, Pringle KG.2019MicroRNA mimics that target the placental renin-angiotensin system inhibit trophoblast proliferation. Molecular Human Reproduction 25218–227. ( 10.1093/molehr/gaz010) [DOI] [PubMed] [Google Scholar]
- Beards F, Jones LE, Charnock J, Forbes K, Harris LK.2017Placental homing peptide-microRNA inhibitor conjugates for targeted enhancement of intrinsic placental growth signaling. Theranostics 72940–2955. ( 10.7150/thno.18845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C.2008The microRNA.org resource: targets and expression. Nucleic Acids Research 36D149–D153. ( 10.1093/nar/gkm995) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Hao XY, Liu FX.2018MicroRNA-186 serves as a tumor suppressor in oral squamous cell carcinoma by negatively regulating the protein tyrosine phosphatase SHP2 expression. Archives of Oral Biology 8920–25. ( 10.1016/j.archoralbio.2018.01.016) [DOI] [PubMed] [Google Scholar]
- Cerro-Herreros E, Sabater-Arcis M, Fernandez-Costa JM, Moreno N, Perez-Alonso M, Llamusi B, Artero R.2018miR-23b and miR-218 silencing increase muscleblind-like expression and alleviate myotonic dystrophy phenotypes in mammalian models. Nature Communications 9 2482. ( 10.1038/s41467-018-04892-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cessans C, Ehlinger V, Arnaud C, Yart A, Capri Y, Barat P, Cammas B, Lacombe D, Coutant R, David Aet al. 2016Growth patterns of patients with Noonan syndrome: correlation with age and genotype. European Journal of Endocrinology 174641–650. ( 10.1530/EJE-15-0922) [DOI] [PubMed] [Google Scholar]
- Chakraborty C, Sharma AR, Sharma G, Lee SS.2021Therapeutic advances of miRNAs: a preclinical and clinical update. Journal of Advanced Research 28127–138. ( 10.1016/j.jare.2020.08.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Ceacareanu B, Dixit M, Sreejayan N, Hassid A.2002Nitric oxide-induced motility in aortic smooth muscle cells: role of protein tyrosine phosphatase SHP-2 and GTP-binding protein Rho. Circulation Research 91390–397. ( 10.1161/01.res.0000033524.92083.64) [DOI] [PubMed] [Google Scholar]
- Chen B, Bronson RT, Klaman LD, Hampton TG, Wang JF, Green PJ, Magnuson T, Douglas PS, Morgan JP, Neel BG.2000Mice mutant for egfr and Shp2 have defective cardiac semilunar valvulogenesis. Nature Genetics 24296–299. ( 10.1038/73528) [DOI] [PubMed] [Google Scholar]
- De Rocca Serra-Nedelec A, Edouard T, Treguer K, Tajan M, Araki T, Dance M, Mus M, Montagner A, Tauber M, Salles JPet al. 2012Noonan syndrome-causing SHP2 mutants inhibit insulin-like growth factor 1 release via growth hormone-induced ERK hyperactivation, which contributes to short stature. PNAS 1094257–4262. ( 10.1073/pnas.1119803109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Yuan X, Gu W.2021Circular RNA RBM33 contributes to cervical cancer progression via modulation of the miR-758-3p/PUM2 axis. Journal of Molecular Histology 52173–185. ( 10.1007/s10735-020-09933-1) [DOI] [PubMed] [Google Scholar]
- Dumolt JH, Powell TL, Jansson T.2021Placental function and the development of fetal overgrowth and fetal growth restriction. Obstetrics and Gynecology Clinics of North America 48247–266. ( 10.1016/j.ogc.2021.02.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easow G, Teleman AA, Cohen SM.2007Isolation of microRNA targets by miRNP immunopurification. RNA 131198–1204. ( 10.1261/rna.563707) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrokhnia F, Aplin JD, Westwood M, Forbes K.2014MicroRNA regulation of mitogenic signaling networks in the human placenta. Journal of Biological Chemistry 28930404–30416. ( 10.1074/jbc.M114.587295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng GS, Hui CC, Pawson T.1993SH2-containing phosphotyrosine phosphatase as a target of protein-tyrosine kinases. Science 2591607–1611. ( 10.1126/science.8096088) [DOI] [PubMed] [Google Scholar]
- Forbes K, Westwood M.2010Maternal growth factor regulation of human placental development and fetal growth. Journal of Endocrinology 2071–16. ( 10.1677/JOE-10-0174) [DOI] [PubMed] [Google Scholar]
- Forbes K, Westwood M, Baker PN, Aplin JD.2008Insulin-like growth factor I and II regulate the life cycle of trophoblast in the developing human placenta. American Journal of Physiology: Cell Physiology 294C1313–C1322. ( 10.1152/ajpcell.00035.2008) [DOI] [PubMed] [Google Scholar]
- Forbes K, West G, Garside R, Aplin JD, Westwood M.2009The protein-tyrosine phosphatase, SRC homology-2 domain containing protein tyrosine phosphatase-2, is a crucial mediator of exogenous insulin-like growth factor signaling to human trophoblast. Endocrinology 1504744–4754. ( 10.1210/en.2009-0166) [DOI] [PubMed] [Google Scholar]
- Forbes K, Farrokhnia F, Aplin JD, Westwood M.2012Dicer-dependent miRNAs provide an endogenous restraint on cytotrophoblast proliferation. Placenta 33581–585. ( 10.1016/j.placenta.2012.03.006) [DOI] [PubMed] [Google Scholar]
- Fosbrink M, Harris A, Devgan V.2011Identification of miRNAs that target the epithelial-mesenchymal transition regulator, Zeb2, using transcriptome PCR arrays. Cancer Research 71 (8 Suppl) abstract 1157. ( 10.1158/1538-7445.AM2011-1157) [DOI] [Google Scholar]
- Freeman Jr RM, Plutzky J, Neel BG.1992Identification of a human Src homology 2-containing protein-tyrosine-phosphatase: a putative homolog of Drosophila corkscrew. PNAS 8911239–11243. ( 10.1073/pnas.89.23.11239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Gaccioli F, Dopierala J, Sovio U, Cook E, Volders PJ, Martens L, Kirk PDW, Richardson S, Smith GCSet al. 2021The RNA landscape of the human placenta in health and disease. Nature Communications 12 2639. ( 10.1038/s41467-021-22695-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LK, Keogh RJ, Wareing M, Baker PN, Cartwright JE, Aplin JD, Whitley GS.2006Invasive trophoblasts stimulate vascular smooth muscle cell apoptosis by a Fas ligand-dependent mechanism. American Journal of Pathology 1691863–1874. ( 10.2353/ajpath.2006.060265) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman K, Cookson VJ, Jones CJ, Aplin JD.2010Characterisation of Hofbauer cells in first and second trimester placenta: incidence, phenotype, survival in vitro and motility. Placenta 31535–544. ( 10.1016/j.placenta.2010.03.003) [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS.2004Human microRNA targets. PLoS Biology 2 e363. ( 10.1371/journal.pbio.0020363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E.2007The role of site accessibility in microRNA target recognition. Nature Genetics 391278–1284. ( 10.1038/ng2135) [DOI] [PubMed] [Google Scholar]
- Knofler M, Pollheimer J.2012IFPA Award in placentology lecture: molecular regulation of human trophoblast invasion. Placenta 33 (Supplement) S55–S62. ( 10.1016/j.placenta.2011.09.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B, Khaleghzadegan S, Mears B, Hatano K, Kudrolli TA, Chowdhury WH, Yeater DB, Ewing CM, Luo J, Isaacs WBet al. 2016Identification of miR-30b-3p and miR-30d-5p as direct regulators of androgen receptor signaling in prostate cancer by complementary functional microRNA library screening. Oncotarget 772593–72607. ( 10.18632/oncotarget.12241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laviola L, Perrini S, Belsanti G, Natalicchio A, Montrone C, Leonardini A, Vimercati A, Scioscia M, Selvaggi L, Giorgino Ret al. 2005Intrauterine growth restriction in humans is associated with abnormalities in placental insulin-like growth factor signaling. Endocrinology 1461498–1505. ( 10.1210/en.2004-1332) [DOI] [PubMed] [Google Scholar]
- le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafre SAet al. 2007Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO Journal 263699–3708. ( 10.1038/sj.emboj.7601790) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP.2005Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 12015–20. ( 10.1016/j.cell.2004.12.035) [DOI] [PubMed] [Google Scholar]
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare Pet al. 2007miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell 129147–161. ( 10.1016/j.cell.2007.03.008) [DOI] [PubMed] [Google Scholar]
- Liang Y, Ridzon D, Wong L, Chen C.2007Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8 166. ( 10.1186/1471-2164-8-166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limal JM, Parfait B, Cabrol S, Bonnet D, Leheup B, Lyonnet S, Vidaud M, Le Bouc Y.2006Noonan syndrome: relationships between genotype, growth, and growth factors. Journal of Clinical Endocrinology and Metabolism 91300–306. ( 10.1210/jc.2005-0983) [DOI] [PubMed] [Google Scholar]
- Liu H, Yue D, Chen Y, Gao SJ, Huang Y.2010Improving performance of mammalian microRNA target prediction. BMC Bioinformatics 11 476. ( 10.1186/1471-2105-11-476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Song X, Li H.2021Transcription elongation factor A-like 7, regulated by miR-758-3p inhibits the progression of melanoma through decreasing the expression levels of c-Myc and AKT1. Cancer Cell International 21 43. ( 10.1186/s12935-020-01737-3) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Long J, Jiang C, Liu B, Dai Q, Hua R, Chen C, Zhang B, Li H.2018Maintenance of stemness by miR-589-5p in hepatocellular carcinoma cells promotes chemoresistance via STAT3 signaling. Cancer Letters 423113–126. ( 10.1016/j.canlet.2017.11.031) [DOI] [PubMed] [Google Scholar]
- Maile LA, Clemmons DR.2002Regulation of insulin-like growth factor I receptor dephosphorylation by SHPS-1 and the tyrosine phosphatase SHP-2. Journal of Biological Chemistry 2778955–8960. ( 10.1074/jbc.M109258200) [DOI] [PubMed] [Google Scholar]
- Mannell H, Krotz F.2014SHP-2 regulates growth factor dependent vascular signalling and function. Mini Reviews in Medicinal Chemistry 14471–483. ( 10.2174/1389557514999140506094738) [DOI] [PubMed] [Google Scholar]
- Mannell H, Hellwig N, Gloe T, Plank C, Sohn HY, Groesser L, Walzog B, Pohl U, Krotz F.2008Inhibition of the tyrosine phosphatase SHP-2 suppresses angiogenesis in vitro and in vivo. Journal of Vascular Research 45153–163. ( 10.1159/000110081) [DOI] [PubMed] [Google Scholar]
- Martello A, Mellis D, Meloni M, Howarth A, Ebner D, Caporali A, Al Haj Zen A.2018Phenotypic miRNA screen identifies miR-26b to promote the growth and survival of endothelial cells. Molecular Therapy: Nucleic Acids 1329–43. ( 10.1016/j.omtn.2018.08.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H, Yamada Y, Miyazawa T, Yoshida T.2013Gain-of-function microRNA screens identify miR-193a regulating proliferation and apoptosis in epithelial ovarian cancer cells. International Journal of Oncology 421875–1882. ( 10.3892/ijo.2013.1896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata DM, Li X, Lee L, Liu J, Warsito D, Hajeri P, Hultman I, Fotouhi O, Marklund S, Ahrlund-Richter Let al. 2017Loss of miR-514a-3p regulation of PEG3 activates the NF-kappa B pathway in human testicular germ cell tumors. Cell Death and Disease 8 e2759. ( 10.1038/cddis.2016.464) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel Y, Shah N, Lee JS, Markoutsa E, Jie C, Liu S, Botbyl R, Reisman D, Xu P, Chen H.2016A novel double-negative feedback loop between miR-489 and the HER2-SHP2-MAPK signaling axis regulates breast cancer cell proliferation and tumor growth. Oncotarget 718295–18308. ( 10.18632/oncotarget.7577) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R.2008MicroRNA-373 induces expression of genes with complementary promoter sequences. PNAS 1051608–1613. ( 10.1073/pnas.0707594105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu CK, Shi ZQ, Shen R, Tsai FY, Orkin SH, Feng GS.1997A deletion mutation in the SH2-N domain of Shp-2 severely suppresses hematopoietic cell development. Molecular and Cellular Biology 175499–5507. ( 10.1128/MCB.17.9.5499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillet A, Saad C, Ferry G, Anouar Y, Vergne N, Lecroq T, Dubessy C.2019Improving bioinformatics prediction of microRNA targets by ranks aggregation. Frontiers in Genetics 10 1330. ( 10.3389/fgene.2019.01330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riolo G, Cantara S, Marzocchi C, Ricci C.2020miRNA targets: from prediction tools to experimental validation. Methods and Protocols 41. ( 10.3390/mps4010001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie W, Rasko JE, Flamant S.2013MicroRNA target prediction and validation. Advances in Experimental Medicine and Biology 77439–53. ( 10.1007/978-94-007-5590-1_3) [DOI] [PubMed] [Google Scholar]
- Rupaimoole R, Slack FJ.2017MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nature Reviews: Drug Discovery 16203–222. ( 10.1038/nrd.2016.246) [DOI] [PubMed] [Google Scholar]
- Saxton TM, Ciruna BG, Holmyard D, Kulkarni S, Harpal K, Rossant J, Pawson T.2000The SH2 tyrosine phosphatase shp2 is required for mammalian limb development. Nature Genetics 24420–423. ( 10.1038/74279) [DOI] [PubMed] [Google Scholar]
- Schaefer JS, Montufar-Solis D, Vigneswaran N, Klein JR.2011Selective upregulation of microRNA expression in peripheral blood leukocytes in IL-10-/- mice precedes expression in the colon. Journal of Immunology 1875834–5841. ( 10.4049/jimmunol.1100922) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark MS, Bonazzi VF, Boyle GM, Palmer JM, Symmons J, Lanagan CM, Schmidt CW, Herington AC, Ballotti R, Pollock PMet al. 2015miR-514a regulates the tumour suppressor NF1 and modulates BRAFi sensitivity in melanoma. Oncotarget 617753–17763. ( 10.18632/oncotarget.3924) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straszewski-Chavez SL, Abrahams VM, Alvero AB, Aldo PB, Ma Y, Guller S, Romero R, Mor G.2009The isolation and characterization of a novel telomerase immortalized first trimester trophoblast cell line, Swan 71. Placenta 30939–948. ( 10.1016/j.placenta.2009.08.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streicher KL, Zhu W, Lehmann KP, Georgantas RW, Morehouse CA, Brohawn P, Carrasco RA, Xiao Z, Tice DA, Higgs BWet al. 2012A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene 311558–1570. ( 10.1038/onc.2011.345) [DOI] [PubMed] [Google Scholar]
- Vogel W, Lammers R, Huang J, Ullrich A.1993Activation of a phosphotyrosine phosphatase by tyrosine phosphorylation. Science 2591611–1614. ( 10.1126/science.7681217) [DOI] [PubMed] [Google Scholar]
- Wang WX, Wilfred BR, Hu Y, Stromberg AJ, Nelson PT.2010Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. RNA 16394–404. ( 10.1261/rna.1905910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lumbers ER, Arthurs AL, Corbisier De Meaultsart C, Mathe A, Avery-Kiejda KA, Roberts CT, Pipkin FB, Marques FZ, Morris BJet al. 2018Regulation of the human placental (pro)renin receptor-prorenin-angiotensin system by microRNAs. Molecular Human Reproduction 24453–464. ( 10.1093/molehr/gay031) [DOI] [PubMed] [Google Scholar]
- Wotschofsky Z, Busch J, Jung M, Kempkensteffen C, Weikert S, Schaser KD, Melcher I, Kilic E, Miller K, Kristiansen Get al. 2013Diagnostic and prognostic potential of differentially expressed miRNAs between metastatic and non-metastatic renal cell carcinoma at the time of nephrectomy. Clinica Chimica Acta: International Journal of Clinical Chemistry 4165–10. ( 10.1016/j.cca.2012.11.010) [DOI] [PubMed] [Google Scholar]
- Wu Y, Liu Y.2020miR-758-3p inhibits proliferation, migration, and invasion of clear cell renal cell carcinoma and predicts favorable prognosis. Cancer Management and Research 129285–9295. ( 10.2147/CMAR.S261882) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Chen YC, Betenbaugh MJ, Martin SE, Shiloach J.2015MiRNA mimic screen for improved expression of functional neurotensin receptor from HEK293 cells. Biotechnology and Bioengineering 1121632–1643. ( 10.1002/bit.25567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Ming X, Wu J.2021Hsa_circ_0002483 regulates miR-758-3p/MYC axis to promote acute myeloid leukemia progression. Hematological Oncology 39243–253. ( 10.1002/hon.2829) [DOI] [PubMed] [Google Scholar]
- Yang W, Klaman LD, Chen B, Araki T, Harada H, Thomas SM, George EL, Neel BG.2006An Shp2/SFK/Ras/Erk signaling pathway controls trophoblast stem cell survival. Developmental Cell 10317–327. ( 10.1016/j.devcel.2006.01.002) [DOI] [PubMed] [Google Scholar]
- Yarbrough ML, Zhang K, Sakthivel R, Forst CV, Posner BA, Barber GN, White MA, Fontoura BM.2014Primate-specific miR-576-3p sets host defense signalling threshold. Nature Communications 5 4963. ( 10.1038/ncomms5963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen F, Zhan H, Chen L, Deng Q, Xiong T, Li Y, Ye J.2021lncRNA CASC9 sponges miR7583p to promote proliferation and EMT in bladder cancer by upregulating TGFbeta2. Oncology Reports 45265–277. ( 10.3892/or.2020.7852) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Li N, Liang S, Huang Q, Coukos G, Zhang L.2010Identification of microRNAs regulating reprogramming factor LIN28 in embryonic stem cells and cancer cells. Journal of Biological Chemistry 28541961–41971. ( 10.1074/jbc.M110.169607) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 This work is licensed under a
This work is licensed under a