Abstract
Translational research regularly utilizes immunohistochemistry (IHC) to investigate pathological differences in the clinical or laboratory setting. However, the majority of these studies require the extensive work of a trained pathologist to analyze slides in a meaningful way. In order to explore new ways to quantitate IHC stains in a manner that is reproducible and efficient for both pathologists and research scientists, QuPath was explored as a new digital imaging tool. The hippocampal area of brains from older sleep deprived mice were stained using established IHC protocols to explore biomarker levels that would be insightful for measurable differences. Application features of QuPath are described that quantitatively show sleep deprived mice had robust differences in staining intensity for four different biomarkers compared to non-sleep deprived mice. These observations provide the rationale for QuPath as a digital imaging tool to enhance the quantitative and qualitative usefulness of IHC staining in the field of geropathology.
Keywords: Digital imaging, QuPath, immunohistochemistry, geropathology, neuropathology, machine learning
Introduction
Immunohistochemistry is a valuable tool that can visualize biomarker location in tissues. Unfortunately, there are not many free and open programs that are able to thoroughly explore quantitative results of stained slides without extensive effort or cost. In 2017, a new whole slide image analyzing software called QuPath was released by the Queen’s University Belfast [1]. This program offers bio-imaging analysis that simplifies the manual tasks performed by pathologists into machine learning that can be efficiently applied across large batches of images. The reduced labor ultimately reduces the variability and increases the reproducibility that are often hurdles in image analysis. This workflow is improved even further by a scripting function that can be saved, shared, and applied through algorithms created through the machine learning feature of QuPath. This new software holds a large range of potential that has yet to be explored for analyzing images in research.
Application
Tissues
Brain tissue from C57BL/6 mice, 16–18 months of age, was used in this study.
Immunohistochemistry staining
Immunohistochemistry (IHC) was performed on 4μm thick, paraffin embedded mouse brain tissue mounted onto slides. Slides were rehydrated with xylene, decreasing concentrations of ethanol, and deionized water. Antigen retrieval was performed by immersing the slides in a hot water bath at 98 degrees C incubated in a 1:10 Citrate Antigen Retrieval solution in autoclaved deionized water for 20 minutes. Slides were then stained using an avidin-biotin HRP kit (anti-rabbit HRP-DAB Cell & Tissue Staining Kit, R&D Systems Minneapolis, MN) with manufacturer instructions. Slides were applied with a 3% peroxidase blocking reagent for 15 minutes to quench endogenous peroxidase activity which reduces background noise on the final stain result. Slides were washed in a TBST solution for 5 minutes. To reduce non-specific hydrophobic interactions between the primary antibody and the tissue, serum blocking reagent was placed onto each section for 15 minutes. After draining the serum off the slides, avidin blocking reagent was placed onto them for 15 minutes followed by a rinse with TBST for 5 minutes. To prevent the binding of previously applied avidin, biotin blocking reagent was placed onto the slides for 15 minutes. Primary antibody in TBST was applied overnight in a humidified chamber. Slides were rinsed 3 times in TBST for 5 minutes each, then incubated with a biotinylated secondary antibody for 30 minutes and rinsed 3 times in TBST for 5 minutes each. Slides were incubated in HSS-HRP for thirty minutes and rinsed in TBST 3 time for 2 minutes each. DAB Chromogen was applied to slides and incubated in the solution for 5 minutes each before rinsing with deionized water for 5 minutes. Slides were dehydrated in an increasing concentration of ethanol and xylene then mounted with a coverslip.
Imaging
IHC slides were photographed under a Nikon Eclipse E400 microscope with a Nikon D7100 camera through a microscope camera adaptor. All photos were taken under a magnification of 4x so that the entire hippocampus could be captured within each photo field.
Using QuPath
QuPath version v0.2.0-m11 was downloaded from Github (https://QuPath.github.io/) [1]. QuPath allows the user to determine how much RAM it will take up. We used 6 GB. A new project was created on QuPath per each staining group examined. This step was done to avoid having to redo the steps of the project workflow for each image.
1. Calibration of DAB intensity.
The image type was set to H-DAB in order for QuPath to recognize the images as a DAB Chromogen stain. The RGB values for DAB were then calibrated to better represent the project by selecting the region of interest (ROI) that is representative of positive stains along the hippocampus. Smaller ROIs were used that selected only the smaller areas of positive staining to reduce any potential background noise. These ROIs were then averaged to come up with a new RGB DAB value for the project code.
2. Tissue annotation.
The hippocampus was then annotated with the polygon wand to only measure staining at the desired region of the tissue (Figure 1). However, if there were any folds or staining irregularities through a manual check, the slide was omitted from analysis.
Figure 1. Annotation of hippocampus with polygon wand tool.
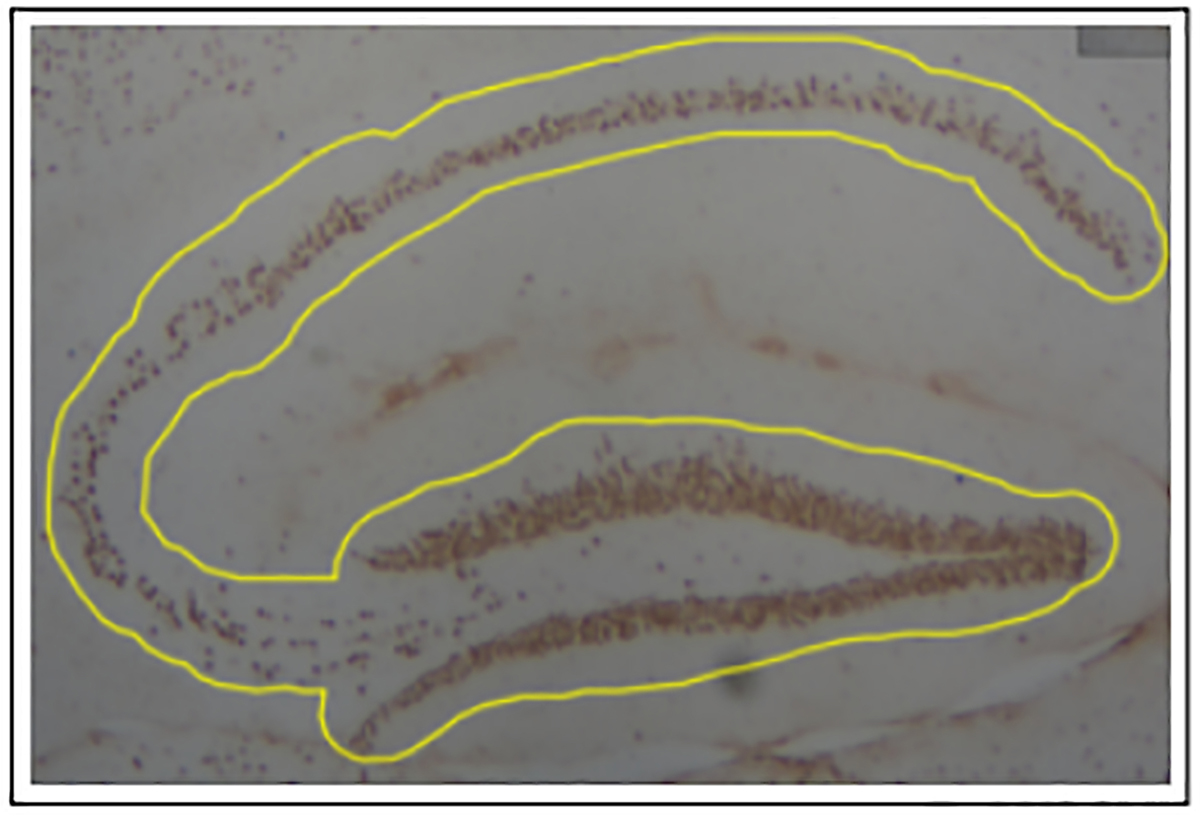
Hippocampus was outlined with a freehand tracing tool to reduce background staining. A manual check was made to ensure tissue and staining quality.
3. Superpixel analysis.
To quantify the staining, superpixels were created to analyze the hippocampus. Within the annotated hippocampus, the QuPath program groups similar pixels into a cluster called a superpixel based on the RGB values set for DAB (Figure 2). Pixel based analysis was chosen as the desired method of quantification because this study is looking at multiple stains, and pixel analysis allows us to follow an almost identical protocol between each group [2]. Superpixel size was set to 25 μm2 in order to balance capturing positively stained sections at a high resolution and processing speed. QuPath then applies a DAB intensity to each of the superpixels previously set by the initial DAB RGB calibrating. Not only does it process as positive or negative, but it also separates the staining at three levels of thresholds: 0.2, 0.4 and 0.6 or a positive at all three levels. This therefore allowed the capture of staining intensity across all positively stained cells in the annotated region.
Figure 2. Superpixel creation after tissue annotation.
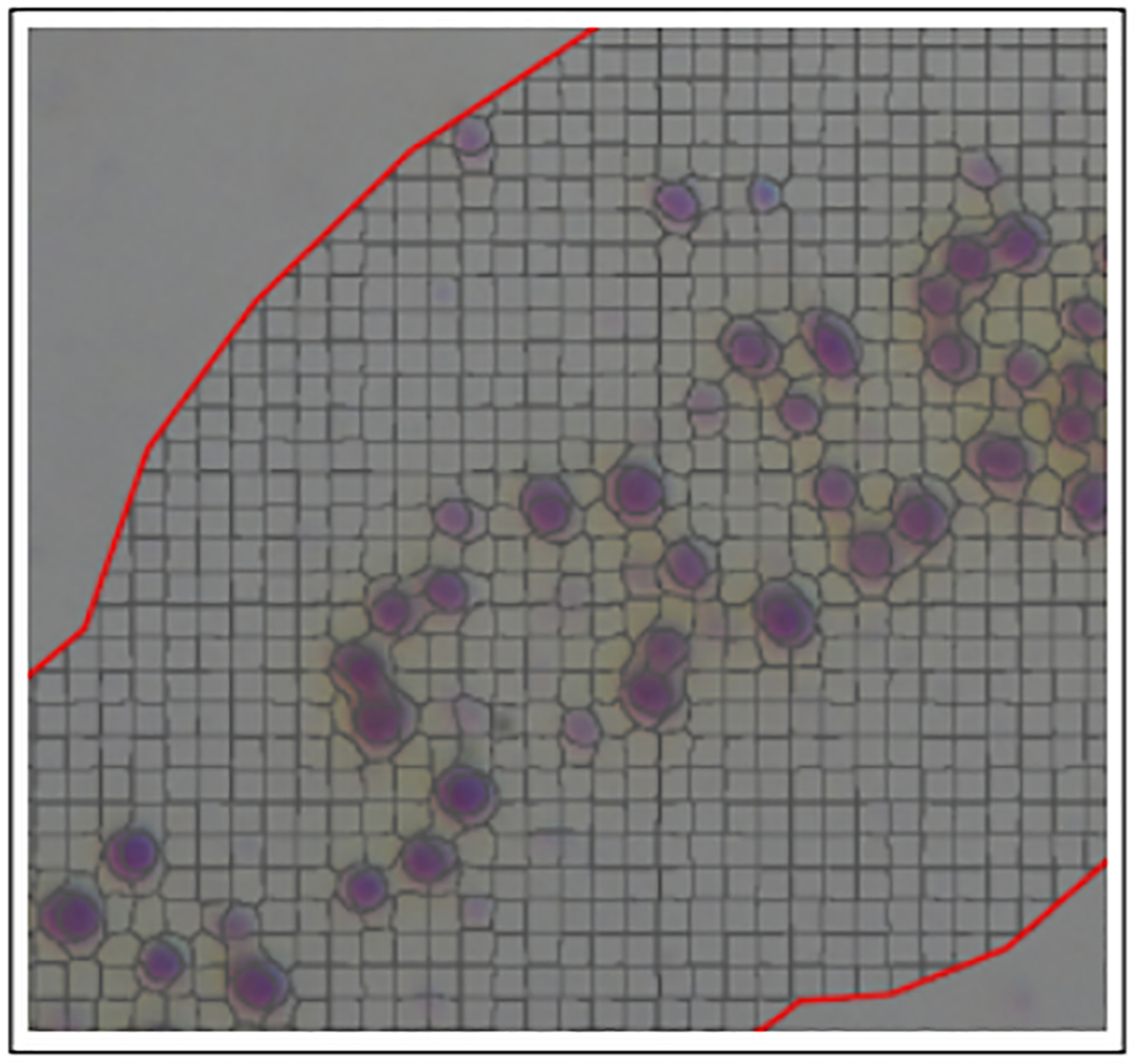
Superpixels are created through a tile feature that QuPath utilizes to capture objects. Size of tile can be adjusted to best balance resolution and processing speed. Superpixel size was set to 25 μm2.
4. Setting thresholds for heat maps.
In order to visualize the DAB staining thresholds, a “heat map” was generated for each image. QuPath allows users to apply a gradient of color according to the quantifications of DAB staining generated previously. The heat map allows a qualitative complement of the previous analyses and serves useful in identifying certain regions that have higher levels of staining. QuPath generates the heat map by assigning a color to each superpixel which indicates the different levels of DAB staining based on the previously described thresholds. However, the upper and lower limits of the color spectrum must be set by the user and made equal for each new project. This way, the results within each project can be compared to one another relative to staining distribution and intensity across the hippocampus. It should be noted, however, that the color is not indicative of positivity, but rather captures the differences in intensity of stain. In order to compare differences, four biomarkers were chosen to measure impacts of short-term sleep deprivation [3]. MCP-1 was a measure for inflammation [4]; HDAC2 was a measure for epigenetic regulation of transcriptional repression [5]; synaptophysin and BDNF were measures for synaptic function and plasticity, respectively [6–7]. As can be seen in Figure 3, there were clear differences in staining intensity in the hippocampal area of the brain from sleep deprived and non-sleep deprived mice.
Figure 3. Visual representation of positive superpixels using a heat map.
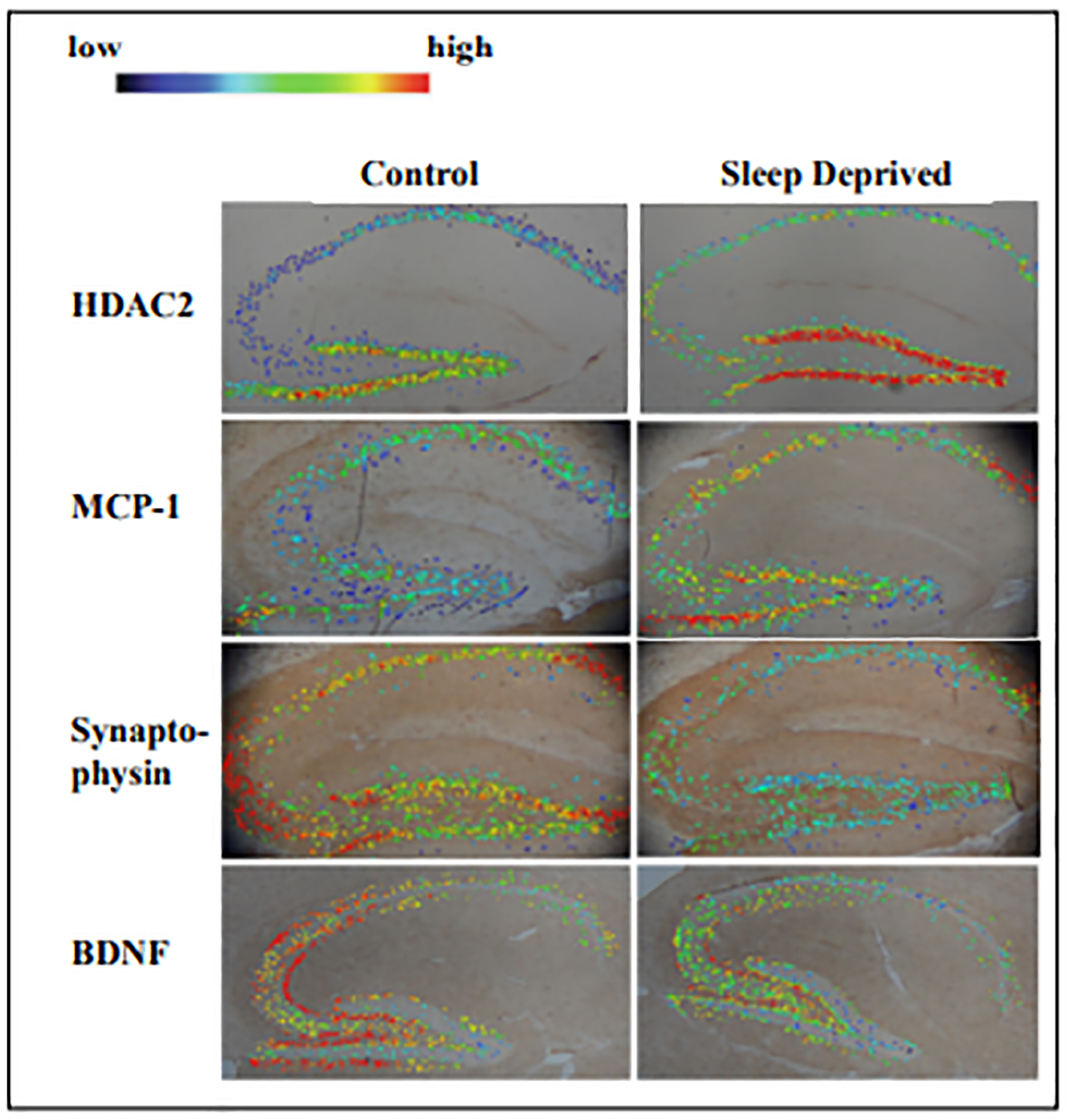
Sagittal hippocampal sections were imaged at 4x and analyzed using QuPath. Rows represent staining groups while columns show control or sleep deprived mice. Staining intensity is based on positive superpixel signal with dark blue suggesting lower levels, green being moderate, and red as high. The example slides show that sleep deprived mice had lower HDAC2 and MCP-1 but higher synaptophysin and BDNF levels than control mice.
Discussion
Immunohistochemistry results can be quickly visualized through a QuPath heat map which shows a difference in superpixel positivity among different staining groups. Although the exact mechanisms cannot be quantified, the heat map gives a clear picture of positive staining as to location and intensity based on superpixels through QuPath and provides a glimpse of the molecular pathway involved. QuPath is a relatively new software so there is still much to be explored and refined with analyzing protocols. Currently, a PubMed search only brings up a total of 22 papers in total that have used this program to analyze whole image slides. Additionally, because QuPath is only an analyzing software, it is critical that immunohistochemistry protocols be performed in a uniform and consistent manner. This would require careful planning of tissue harvesting, sectioning, and imaging that would allow for the best results. Further molecular analysis should be performed to confirm the QuPath data so that quantitative conclusions can be drawn on the molecular pathway that the study is investigating. QuPath can ultimately be used as a novel application to study correlations between positive superpixels in immunohistochemistry slides from a variety of different tissues and protocols.
Acknowledgment:
This work was supported by NIA grant R01 AG057381 (Ladiges, PI).
Footnotes
Conflict of interest: Warren Ladiges is a member of the Editorial Board of Aging Pathobiology and Therapeutics. All authors declare no conflict of interest and were not involved in the journal’s review or desicions related to this manuscript.
References
- 1.Bankhead P, Loughrey MB, Fernández J A, et al. QuPath: Open source software for digital pathology image analysis. Scientific reports, 2017, 7(1): 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morriss NJ, Conley GM, Ospina SM, et al. Automated Quantification of Immunohistochemical Staining of Large Animal Brain Tissue Using QuPath Software. Neuroscience, 2020, 429: 235–244. [DOI] [PubMed] [Google Scholar]
- 3.Lee A, Zhgu L, Ladiges W. Resilience to acute sleep deprivation is associated with attenuation of hippocampal mediated learning impairment. Geroscience, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bettcher BM, Fitch R, Wynn MJ, et al. MCP-1 and eotaxin-1 selectively and negatively associate with memory in MCI and Alzheimer’s disease dementia phenotypes. Alzheimer’s & Dementia: Diagnosis, Assessment & Disease Monitoring, 2016, 3: 91–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chouliaras L, LA van den Hove D, Kenis G, et al. Histone deacetylase 2 in the mouse hippocampus: attenuation of age-related increase by caloric restriction. Current Alzheimer Research, 2013, 10(8): 868–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calhoun ME, Kurth D, Phinney AL, et al. Hippocampal neuron and synaptophysin-positive bouton number in aging C57BL/6 mice. Neurobiology of aging, 1998, 19(6): 599–606. [DOI] [PubMed] [Google Scholar]
- 7.Quesseveur G, David DJ, Gaillard MC, et al. BDNF over-expression in mouse hippocampal astrocytes promotes local neurogenesis and elicits anxiolytic-like activities. Translational psychiatry, 2013, 3(4): e253–e253. [DOI] [PMC free article] [PubMed] [Google Scholar]


