Abstract
Biogenesis of the U5 small nuclear ribonucleoprotein (snRNP) is an essential and highly regulated process. In particular, PRPF8, one of U5 snRNP main components, requires HSP90 working in concert with R2TP, a cochaperone complex containing RUVBL1 and RUVBL2 AAA-ATPases, and additional factors that are still poorly characterized. Here, we use biochemistry, interaction mapping, mass spectrometry and cryoEM to study the role of ZNHIT2 in the regulation of the R2TP chaperone during the biogenesis of PRPF8. ZNHIT2 forms a complex with R2TP which depends exclusively on the direct interaction of ZNHIT2 with the RUVBL1–RUVBL2 ATPases. The cryoEM analysis of this complex reveals that ZNHIT2 alters the conformation and nucleotide state of RUVBL1–RUVBL2, affecting its ATPase activity. We characterized the interactions between R2TP, PRPF8, ZNHIT2, ECD and AAR2 proteins. Interestingly, PRPF8 makes a direct interaction with R2TP and this complex can incorporate ZNHIT2 and other proteins involved in the biogenesis of PRPF8 such as ECD and AAR2. Together, these results show that ZNHIT2 participates in the assembly of the U5 snRNP as part of a network of contacts between assembly factors required for PRPF8 biogenesis and the R2TP-HSP90 chaperone, while concomitantly regulating the structure and nucleotide state of R2TP.
INTRODUCTION
Pre-mRNA splicing factor 8 (PRPF8) is one of the largest and most conserved proteins in the spliceosome and the main component of the U5 small nuclear ribonucleoprotein particle (U5 snRNP) (1). In the nucleus, PRPF8 is forming a complex with EFTUD2 and SNRNP200 (also known as BRR2) as part of the spliceosome. The biogenesis of PRPF8 and U5 snRNP is poorly understood, but a number of factors have been implicated. In the cytoplasm, PRPF8 forms a complex with AAR2, a protein required for the assembling of U5 snRNP that is replaced by the SNRNP200 helicase after PRPF8 is transported from the cytoplasm to the nucleus (1,2). A crystal structure of the homologous yeast Prp8 in complex with Aar2 reveals that Aar2 plays a crucial role in bringing together Prp8 domains that would otherwise be only connected by disordered linkers (1).
Recent work has established that the biogenesis and assembly of U5 snRNP and PRPF8 requires the intervention of HSP90 and the R2TP complex (3,4). HSP90 is a molecular chaperone required for the maturation and activation of a number of proteins and protein complexes, including steroid hormone receptors and several signaling proteins. HSP90 is assisted by cochaperones, proteins that regulate the chaperone and contribute to bring specific clients (5). The R2TP complex functions as an HSP90 cochaperone during the biogenesis of several large macromolecular complexes including RNA polymerase II (Pol II), complexes of the PI3 kinase-like kinases (PIKKs), C/D small nucleolar RNPs (snoRNPs) and U4 small nuclear ribonucleoprotein (snRNP) (6,7). The list of complexes that require R2TP for their maturation and stability is enlarging, and PRPF8 and the U5 snRNP are recent additions (3,4).
Human R2TP is composed of four subunits, RUVBL1, RUVBL2, RPAP3 and PIH1D1 (Figure 1A) (8–10). RUVBL1 and RUVBL2 are closely related AAA-ATPases that form an hexameric RUVBL1–RUVBL2 ring of alternating subunits. Both subunits contain a domain, known as domain II (DII), that protrudes from each subunit in the RUVBL1–RUVBL2 hexamer and which has been found to interact with several proteins (8,11,12). The ATPase activity of RUVBL1 and RUVBL2, which resides in the AAA+ ring, is needed for the functions of R2TP (13), but what is the exact role of ATP hydrolysis and how this is regulated is mostly unknown. RPAP3 and PIH1D1 form a heterodimeric complex that interacts with RUVBL1–RUVBL2 (14). RPAP3 binds to HSP90 thanks to two N-terminal tetratricopeptide (TPR) domains (10,15,16).
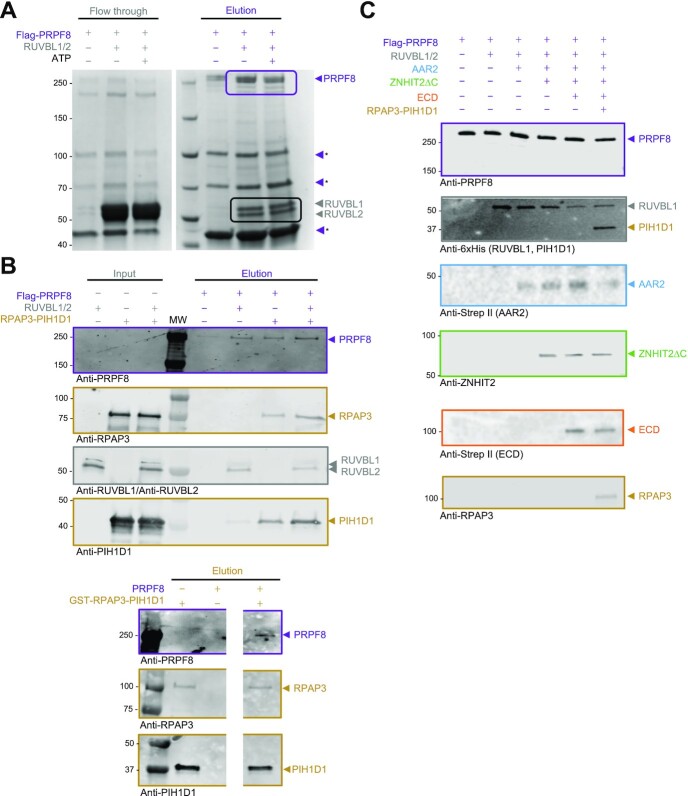
Figure 1.
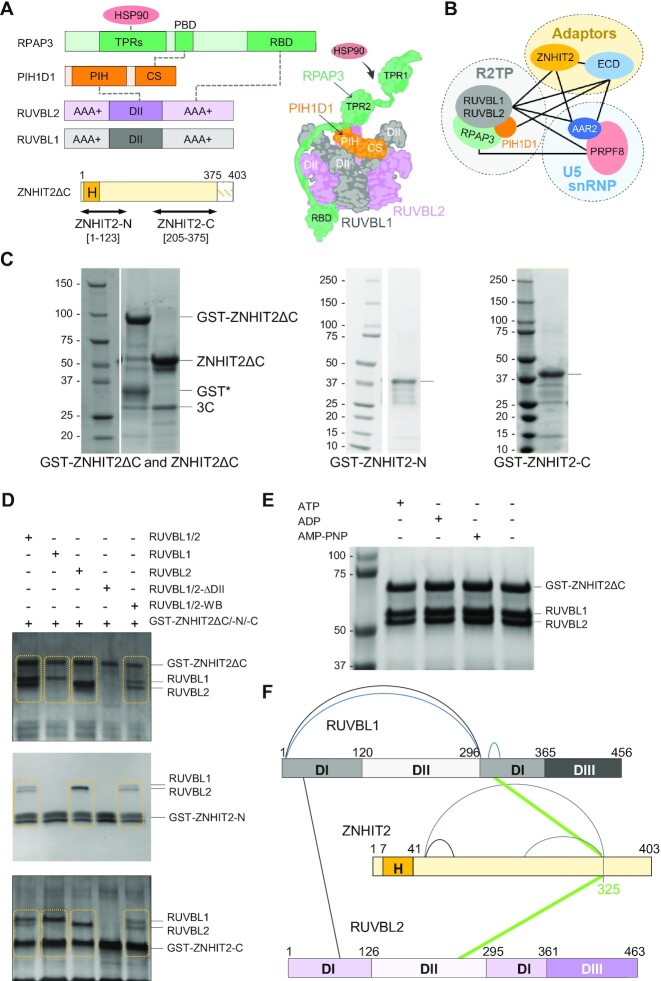
Purification and assembly of RUVBL1–RUVBL2–ZNHIT2ΔC. (A) Left top panels, domain architecture of the components of R2TP (TPRs, tetratricopeptide repeats; PBD, PIH1D1 binding domain; RBD, RUVBL2 binding domain; PIH, protein interacting with HSP90 domain; CS, CHORD-containig proteins and SGT1 domain). Domains involved in interactions within R2TP are indicated as links with dashed lines. Left bottom panel, constructs of human ZNHIT2 used in this work (H, HIT domain). Right panel, cartoon of the structure of the R2TP complex. (B) Diagram of the network of interactions described in this work for R2TP, ZNHIT2, ECD, AAR2 and PRPF8. (C) SDS-PAGE of purified ZNHIT2ΔC, GST-ZNHIT2ΔC, GST-ZNHIT2-C and GST-ZNHIT2-N. (D) GST-pull-down experiments to test the interaction of GST-ZNHIT2ΔC, GST-ZNHIT2-C and GST-ZNHIT2-N with RUVBL1, RUVBL2 and RUVBL1–RUVBL2 complexes (RUVBL1/2 for simplicity in figure legends hereafter). RUVBL1/2-ΔDII corresponds to mutants lacking part of the DII domain. RUVBL1/2-WB corresponds to mutants in the Walker B domains, able to bind ATP but affected in ATP hydrolysis. Control lanes for these experiments are shown in Supplementary Figure S1B, C. (E) GST-pull-down experiment to evaluate the effect of the presence or absence of the nucleotides ATP, ADP or AMP–PNP in the interaction between RUVBL1–RUVBL2 and ZNHIT2ΔC. Control lanes for these experiments are shown in Supplementary Figure S1D. (F) Cartoon showing the crosslinks found in RUVBL1–RUVBL2–ZNHIT2 complex detected by XL-MS.
Up to our knowledge, all the structures for human RUVBL1–RUVBL2 hetero-hexamers determined so far contain nucleotide, most frequently ADP, in the binding pocket of each AAA-subunit even if nucleotides were not added during purification (17). The crystal structures revealed that hexamerization blocks the nucleotide binding pocket, hampering the exchange from ADP to ATP and this has been used to explain the impossibility to co-crystallize nonhydrolyzable ATP analogs into these crystals (17,18). Still, some exchange of nucleotides exits since RUVBL1–RUVBL2 hydrolyzes ATP although at a low rate. Recent work has found that it is an N-terminal region of both RUVBL1 and RUVBL2 that blocks the exit route for nucleotides (8,13,17,19). DHX34, an RNA helicase that participates in nonsense-mediated mRNA decay (NMD) (12) and PIH1D1, one of the components of R2TP (8) can destabilize these N-terminal regions in RUVBL2 after binding to the DII domains. These conformational changes promote the release of the bound ADP from the nucleotide binding pocket (8,12).
Two factors, Zinc finger HIT domain-containing protein 2 (ZNHIT2) (4) and ECD (Protein ecdysoneless homolog) (20,21), have recently been found to interact with the R2TP chaperone system to regulate the biogenesis of PRPF8 and U5 snRNP. ZNHIT2 interacts with RUVBL2 and regulates the composition of U5 snRNP and splicing (4). ZNHIT2 belongs to a family of HIT-finger proteins characterized by the presence of a HIT domain, a roughly 50-residue zinc-binding domain containing conserved cysteine (Cys) and histidine (His) residues. This domain is found in several nuclear proteins involved in chromatin remodeling and the regulation of gene expression (22,23). Humans contain six HIT domain proteins and several of them interact with RUVBL1–RUVBL2 (22). ZNHIT1 and ZNHIT4 are part of the RUVBL1–RUVBL2-containing chromatin remodeling complexes SRCAP and INO80 respectively. ZNHIT2, ZNHIT3 and ZNHIT6 also interact with RUVBL1–RUVBL2. ZNHIT3 and ZNHIT6 participate in the assembly of box C/D small nucleolar ribonucleoproteins (snoRNPs) (6) and at least in the case of ZNHIT6, the interaction with RUVBL1–RUVBL2 was enhanced by the presence of ATP (24). Pull-down experiments using truncated versions of all ZNHIT proteins suggest that the HIT domain is involved in the formation of the complex with RUVBL1–RUVBL2 (4) and in yeast Ino80 complex, Ies2 (the ZNHIT4 yeast orthologue) contains a domain that resembles a HIT fold and interacts with both Rvb1 (yeast homolog of RUVBL1) and H2A in the nucleosome (25). Some experiments suggest that the interaction of some ZNHIT proteins with RUVBL1–RUVBL2 could be incompatible with the assembly of the R2TP complex (4,22). PRPF8-based purifications detect RUVBL1–RUVBL2 and ZNHIT2 but no other components of the R2TP complex, which has been interpreted as indication that ZNHIT2 competes for RPAP3 binding to RUVBL1–RUVBL2 (4,22). ECD also interacts with several components of R2TP and regulates the stability of PRPF8 and the assembly of U5 snRNP (20,21). ECD contains CK2 phosphorylation sites at consensus motifs recognized by PIH1D1, but the functional relevance of the interaction between ECD and PIH1D1 is unclear as it seems dispensable for at least some of the functions assigned to ECD. ECD also interacts directly with RUVBL1 in a phosphorylation independent manner (26).
The biogenesis and maturation of pre-mRNA splicing factor 8 (PRPF8) requires the action of HSP90, R2TP and a list of factors such as ZNHIT2 that have been poorly characterized. Here we have purified and studied ZNHIT2 and its interaction with both the R2TP chaperone machinery, PRPF8 and factors required for PRPF8 biogenesis. Several regions in ZNHIT2 are involved in the formation of a complex with RUVBL1–RUVBL2, and this interaction can occur when the ATPases are part of R2TP. Cryoelectron microscopy (CryoEM) of the RUVBL1–RUVBL2–ZNHIT2 complex revealed that ZNHIT2 regulates the structure and activity of these ATPases. In addition, ZNHIT2 interacts with several factors involved in PRPF8 biogenesis such as ECD and AAR2 (Figure 1B). Together, our results provide evidence that ZNHIT2 can be part of complexes containing factors involved in PRPF8 biogenesis and the R2TP-HSP90 chaperone, and that ZNHIT2 can regulate the nucleotide state and ATPase activity of the R2TP cochaperone.
MATERIALS AND METHODS
Cell culture
HEK293T cells (kindly provided by Dr Peinado, (CNIO, Spain)) were maintained in Dulbecco's modified Eagle's medium (DMEM) (Merck Life Sciences), supplemented with 10% (v/v) fetal bovine serum (Thermo Fisher Scientific, #10270) and Sodium Piruvate (11 mg/l, final concentration) (Merck Life Sciences). Cells were tested for mycoplasma contamination and were maintained at 37ºC in a humidified 5% CO2 atmosphere.
Cloning and mutagenesis
The cDNA of RPAP3 (NM_024604.3) was PCR amplified from the ORF clone OHu11823 (GenScript) and inserted into the pACEmam-TEV-3C-3xFlag vector (kindly provided by Dr Fernandez-Leiro (CNIO, Spain)), using the IVA cloning system (27), including homologous sequences in N- and C-terminus of the protein. The mutant M626A/F630R in RBD RPAP3 protein (comprised by residues 535–665 of RPAP3) was generated by site-directed mutagenesis of pACEmam-TEV-RPAP3-3C-3xFlag, using the IVA cloning system. Oligonucleotides used for cloning are shown in Supplementary Table S1.
Transfections and immunoprecipitations
For interaction studies in cell extracts, HEK293T cells were transiently transfected with pACEmam-TEV-3C-3xFlag, pACEmam-TEV-RPAP3-3C-3xFlag or pACEmam-TEV-RPAP3M626A/F630R-3C-3xFlag cDNA using the Lipofectamine 2000 method (Life Technologies), following manufacturer's instructions. Cells were washed 48 h later with 10 ml PBS and lysed in Triton IP Lysis Buffer (50 mM HEPES pH 7.5, 40 mM NaCl, 2 mM EDTA, 0.1 M NaF, 1% Triton X-100, 10 mM Na-Pyrophosphate, Complete Protease Inhibitor Cocktail (Roche), PhosSTOP (Roche)). Cell lysates were cleared by centrifugation and 5 mg was incubated during 2 h with 20 μl of Anti-FLAG-M2 affinity gel (Sigma Aldrich) in presence of BSA (1 mg/ml). The beads were washed ten times with PD Buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl) and bound proteins were eluted with protein sample buffer and analyzed by western blotting.
Protein purification
Human ZNHIT2 protein purification
ZNHIT2 was cloned into the pGEX-6P2 vector using the IVA cloning system (27) and the oligonucleotides described in the Supplementary Table S1. ZNHIT2 protein was expressed in BL21 (DE3) E. coli cells using the pGEX-6P2-GST-3C-ZNHIT2 construct, lacking the C-terminal 376–403 residues of ZNHIT2 (ZNHIT2ΔC) as the full-length protein tends to be proteolyzed at this region. Expression was induced for 16 h at 16°C by addition of 0.5 mM IPTG. Soluble protein extract was prepared by sonication (37% amplitude, 3 min on ice) in lysis buffer (PBS supplemented with 5 mM DTT, 1 mg/ml lysozyme, 12.5 U/ml Benzonase and EDTA-free Protease Inhibitor Cocktail (Roche) and clarified by centrifugation (Beckman rotor JA25.50 (Beckman), 22 000 rpm, 1 h). ZNHIT2ΔC was purified by affinity chromatography in a GSTprep FF 10/16 column (Cytiva) using the binding (PBS supplemented with 1 mM DTT), washing (50 mM Tris–HCl pH 8.0, 100 mM KCl, 1 mM DTT) and elution (50 mM Tris–HCl pH 8.0, 100 mM KCl, 1 mM DTT, 20 mM reduced l-glutathione) buffers, followed by several rounds of gel filtration chromatography on a Superdex 200 Increase 10/300 GL column (Cytiva) using a buffer containing 50 mM Tris–HCl pH 8.0, 100 mM KCl and 1 mM DTT. Protein sample was concentrated using 30 kDa MWCO Amicon Ultra-4 (Merck). Protein sample purity was evaluated by SDS-PAGE. When necessary, N-terminal GST tag of ZNHIT2ΔC was removed by incubating the protein complex with the GST-3C protease in the presence of Glutathione Sepharose 4B resin (Merck) for 6 h at 4°C. Untagged ZNHIT2ΔC was further purified by size-exclusion chromatography in a Superdex 75 Increase 10/300 GL column (Cytiva).
Human RUVBL1–RUVBL2 protein complex purification
The RUVBL1–RUVBL2 complex was purified as reported (28). When necessary, N-terminal His tag of RUVBL1 was removed by incubating the protein complex with the 6xHis-TEV protease in the presence of 1 mM DDT. Remaining 6xHis-RUVBL1–RUVBL2 protein complex and 6xHis-TEV was removed by incubating the sample with Ni-NTA agarose resin (Qiagen) and collecting the unbound fraction.
Human RUVBL1–RUVBL2 ΔDII as well as isolated RUVBL1, isolated RUVBL2 and RUVBL1E303Q–RUVBL2E300Q catalytic mutants were expressed and purified as reported (12,17,29).
Human ECD and AAR2 protein purification
ECD was co-expressed in BL21 (DE3) E. coli cells with the Arabidopsis thaliana α-subunit CK2 kinase to assure its phosphorylation, using the constructs pCDFduet1-Strep II-SUMO-TEV-ECD and pGEX 6P1-GST-αCK2 kindly provided by Dr V. Band and Dr. L. Pearl respectively. AAR2 cDNA (IMAGE: 2905315) was cloned into the pRSF-duet1 vector using the IVA cloning system (27) and the oligonucleotides described in Supplementary Table S1. AAR2 protein expression was induced for 16 h at 16°C, by addition of 0.5 mM ITPG to the BL21 (DE3) E. coli cells transformed with either the pCDFduet1-AAR2-3C-His or the pCDFduet1-His-3C-AAR2-Strep II construct. Cellular pellets were resuspended in lysis buffer (PBS supplemented with 1 mg/ml lysozyme, 12.5 U/ml Benzonase and EDTA-free Protease Inhibitor Cocktail (Roche)) and homogenized by sonication (37% amplitude, 3 min on ice). Protein extract was clarified by centrifugation (Beckman rotor JA25.50 (Beckman), 22 000 rpm, 45 min), filtered (0.45 μm filter pore) and loaded onto the corresponding affinity chromatography columns using the binding buffer (PBS supplemented with 1 mM DTT). Either ECD or His-AAR2-Strep II proteins were loaded onto a Strep-Tactin XT 4Flow column (IBA Lifesciences) and AAR2-His onto a HIStrap HP column (Cytiva). Unbound protein fraction was washed out with the corresponding washing buffer (ECD and His-AAR2-Strep II washing buffer is 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT; AAR2-His washing buffer is 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT, 20 mM Imidazole). Protein was recovered from the column by addition of the elution buffer (ECD and His-AAR2-Strep II elution buffer: 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT, 50 mM Biotin; AAR2-His elution buffer: 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT, 500 mM Imidazole). ECD and both tagged versions of AAR2 proteins were further purified by size-exclusion chromatography (ECD in a Superdex 200 Increase 10/300 GL column (Cytiva) and both tagged versions of AAR2 in a Superdex 75 Increase 10/300 GL (Cytiva) column), using a running buffer composed by 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT. After protein concentration, sample purity was evaluated by SDS-PAGE.
Human RPAP3–PIH1D1 protein complex purification
RPAP3 and PIH1D1 were co-expressed in BL21 (DE3) E. coli cells using the pGEX6P2-GST-3C-RPAP3 and pRSFduet1-TEV-6xHis-PIH1D1 constructs. Ready to express pGEX-RPAP3 plasmid containing human RPAP3 gene with a N-terminal GST tag and PreScission protease site was purchased from GenScript (ORF clone ID OHu11823). ORF for human PIH1D1 (ORF clone ID 2966990, Horizon Inspired Cell Cultures) was inserted into the pRSFDuet-1 vector including a N-terminal His-tag and a TEV protease site (pRSFDuet-1-His-TEV-PIH1D1) using the Gibson Assembly strategy (New England BioLabs) and the oligonucleotides described in Supplementary Table S1. RPAP3-PIH1D1 protein co-expression was induced for 16 h at 16°C by addition of 0.5 mM ITPG. Cellular pellets were homogenized in lysis buffer (25 mM HEPES–NaOH pH 7.8, 500 mM NaCl, 1 mM DTT, 0.5% Triton X-100, 1 mg/ml lysozyme, 12.5 U/ml Benzonase and EDTA-free Protease Inhibitor Cocktail (Roche)) before sonication (37% amplitude, 3 min on ice). Protein extract was clarified by centrifugation (Beckman rotor JA25.50 (Beckman), 22 000 rpm, 1 h), filtered (0.45 μm filter pore) and loaded into the GSTprep FF 10/16 affinity chromatography column (Cytiva). After washing out the unbound proteins (washing buffer is 25 mM HEPES-NaOH pH 7.8, 500 mM NaCl, 1 mM DTT), the protein complex eluted by using the washing buffer supplemented with 20 mM reduced l-glutathione. Then, the GST tag in RPAP3 was removed by incubation with GST-TEV protease during dialysis against the dialysis buffer (25 mM HEPES-NaOH pH 7.8, 100 mM NaCl, 1 mM DTT) for 16 h at 4°C. Subsequently, the sample was loaded onto a HiTrap Q HP ion exchange chromatography column pe-equilibrated in A buffer (25 mM HEPES–NaOH pH 7.8, 100 mM NaCl, 1 mM DTT). Unbound protein was removed by washing the column with A buffer. Protein eluted through a gradient from A to B buffer (25 mM HEPES–NaOH pH 7.8, 2 M NaCl, 1 mM DTT) and, after dialysis against dialysis buffer, it was concentrated. Protein sample purity was analyzed by SDS-PAGE.
Human PRPF8 protein purification
Expi 293 cells were transiently transfected with the ready to express pcDNA 3.1(+)-PRPF8-3C-Flag (DYK) construct (ORF clone ID OHu192527D, GenScript) using polyethylenimine (PEI max, Polysciences Inc.). After 72 h post-transfection, cells were harvested by centrifugation and washed with cold PBS. To purify PRPF8, cells were homogenized in buffer A (50 mM Tris–HCl pH 8.0, 200 mM NaCl, 5 mM MgCl2) supplemented with 12.5 U/ml Benzonase, EDTA-free Protease Inhibitor Cocktail (Roche) and PhosSTOP (Roche) and sonicated for 1 min (37% amplitude). The soluble protein extract clarified by centrifugation (1 h, 11 000 × g, 4°C) and filtered (0.45 μm pore size filter) was incubated with FLAG-affinity gel (Sigma Aldrich) pre-equilibrated in buffer A for 1 h at 4°C. Unbound protein fraction was removed by subsequent washing steps in A, A2 (50 mM Tris–HCl pH 8.0, 1 M NaCl, 5 mM MgCl2) and A3 (50 mM Tris–HCl pH 8.0, 200 mM NaCl, 5 mM MgCl2, 5 mM ATP) buffers. PRPF8 was finally eluted by incubation with buffer B (50 mM Tris–HCl pH 8.0, 200 mM NaCl, 5 mM MgCl2, 0.5 mg/ml of FLAG peptide) for 16 h at 4°C. In case of the PRPF8 interactions analysis, the putative PRPF8 partners (purified RUVBL1–RUVBL2, ZNHIT2ΔC, ECD, AAR2, RPAP3-PIH1D1) were incubated with the FLAG-affinity gel loaded with PRPF8 after the washing step, at a final concentration of 3 mM each. A negative control was included in the experiments by incubating the putative PRPF8 partners with the FLAG-affinity resin pre-incubated with a cell extract preparation of Expi cells without overexpressed PRPF8. Excess of the added proteins was removed by extensive washing using the A washing buffer. Finally, co-elution of PRPF8 with the potential partners was analyzed by incubation with elution buffer for 16 h at 4°C. PRPF8 protein purity and interaction analysis were visualized by SDS-PAGE and western blotting.
Design, expression and purification of ZNHIT2 truncated versions
ZNHIT2 truncated versions including the N-terminal region (ZNHIT2-N, residues 1–125) or the C-terminal region (ZNHIT2-C, residues 205–375) were design according to secondary and tertiary structure predictions using the online servers JPred4 (30) and Phyre2 (31) respectively. cDNAs were cloned into the pGEX-6P2 vector by PCR using the IVA cloning system (27) and the oligonucleotides described in Supplementary Table S1. ZNHIT2 truncated versions were expressed and purified as indicated for ZNHIT2ΔC using a Superdex 75 Increase 10/300 GL column (Cytiva) instead of a Superdex 200 Increase 10/300 GL column in the size-exclusion chromatography purification step.
In vitro pull-down experiments
In vitro pull-down assays were performed by incubation of the purified bait protein at a final concentration of 1.5 μM with the affinity chromatography resin pre-equilibrated in binding buffer. Excess of the bait protein was removed by several washing steps in washing buffer. Prey proteins were added to the resin at a final concentration of 3 μM (molar excess ration of 1:2). Prey protein excess was removed by several washing steps in washing buffer. Co-elution was carried out by incubating the resin with elution buffer for 30 min to 3 h (depending on the experiment) at 4°C. Protein interactions were analyzed by SDS-PAGE revealed with Coomassie or Silver staining. In each pull-down experiment, a negative control was included by incubating the prey proteins with the affinity resin in the absence of the bait.
GST-ZNHIT2ΔC in vitro pull-downs
GST-ZNHIT2ΔC (bait) was incubated with Glutathione Sepharose 4B resin (Merck) for 2 h at 4°C with agitation. Binding/washing buffer was 50 mM Tris–HCl pH 8.0, 100 mM KCl, 1 mM DTT; and elution buffer was 50 mM Tris–HCl pH 8.0, 100 mM KCl, 1 mM DTT, 20 mM reduced l-glutathione.
Strep II-ECD in vitro pull-downs
Strep II-ECD (bait) was incubated with Strep-Tactin XT 4Flow resin (IBA Biosciences) for 2 h at 4°C with agitation. Binding/washing buffer was 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT; and elution buffer was 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT, 50 mM Biotin.
Strep II-AAR2 in vitro pull-downs
6xHis-AAR2-Strep II (bait) was incubated with Strep-Tactin XT 4Flow resin (IBA Biosciences) for 2 h at 4°C with agitation. Binding/washing buffer was 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT; and elution buffer was 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT, 50 mM Biotin.
6xHis-AAR2 in vitro pull-downs
6xHis-AAR2 (bait) was incubated with Ni-NTA agarose resin (Qiagen) for 20 min. at 4°C with agitation. Binding buffer was 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT; washing buffer was 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT, 50 mM Imidazole; and elution buffer was 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 1 mM DTT, 500 mM Imidazole.
SDS-PAGE and immunoblot analysis
Protein concentration was measured using the Protein Assay Dye Reagent Concentrate (Bio-Rad). Protein lysates were separated on 10% SDS-PAGE gels, transferred onto nitrocellulose membrane (Amersham) and blocked for 1 h in 5% BSA–TBS. After incubation with primary antibodies (overnight) and secondary antibodies (1h, RT), the membranes were washed and analyzed using the LI-COR Odyssey Infra-red Imaging System. Immunoreactivity bands were with Image J software or by the software included in the Odyssey Infra-red Imaging System. Primary antibodies used in western blotting with dilutions were as follows: monoclonal Anti-FLAG M2 antibody (Sigma Aldrich, F1804, 1:2000), PRPF8 (Abcam, ab87433; 1:300), EFTUD2 (Abcam, ab72456, 1:200), ZNHIT2 (Abcam, ab126133; 1:500), RUVBL1 (Cell signaling #12300; 1:500), RUVBL2 (Cell signaling #8959; 1:500), PIH1D1 (Invitrogen #PA5-61482, 1:1000), RPAP3 (Invitrogen #PA5-58334D; 1:500). Secondary antibodies used were as follows: Anti-Rabbit IgG (H + L) (DyLightTM 680 Conjugate) (Cell signaling #5366; 1:15000) and Anti-Mouse IgG (H + L) (DyLightTM 800 4X PEG Conjugate) (Cell signaling #5257; 1:15000). Conjugated antibodies used were StrepMAB-Classic HPR (IBA Lifesciences #2-1509-001; 1:30000) and Anti-polyHistidine-Peroxidase (Sigma Aldrich #A7058; 1:1000).
Immunoprecipitation of RPAP3 from cells and mass spectrometry
Digestion
Immunoprecipitation of overexpressed RPAP3-WT and RPAP3M626A/F630R was performed as explained previously. Proteins were eluted twice, using 8 M urea in 100 mM Tris–HCl pH 8.0. Then, samples were digested by means of the standard FASP protocol. Briefly, proteins were reduced and alkylated (15 mM TCEP, 50 mM CAA, 30 min in the dark, RT) and sequentially digested with Lys-C (Wako) (200 ng of Lys-C per sample, o/n at RT) and trypsin (Promega) (200 ng of trypsin per sample, 6 h at 37°C). Resulting peptides were desalted using C18 stage-tips, speed-vac dried and re-dissolved in 21 μl of 0.5% formic acid.
LC–MS/MS – LC–MS/MS was done by coupling an UltiMate 3000 RSLCnano LC system to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific). Seven microliters of peptides were loaded into a trap column (Acclaim™ PepMap™ 100 C18 LC Columns 5 μm, 20 mm length) for 3 min at a flow rate of 10 μl/min in 0.1% formic acid. Then, peptides were transferred to an EASY-Spray PepMap RSLC C18 column (Thermo) (2 μm, 75 μm × 50 cm) operated at 45°C and separated using a 60 min effective gradient (buffer A: 0.1% FA; buffer B: 100% ACN, 0.1% FA) at a flow rate of 250 nl/min. The gradient used was, from 4% to 6% B in 2 min, from 6% to 33% B in 58 min, plus 10 additional minutes at 98% B. Peptides were sprayed at 1.5 kV into the mass spectrometer via the EASY-Spray source. The capillary temperature was set to 300°C. The mass spectrometer was operated in a data-dependent mode, with an automatic switch between MS and MS/MS scans using a top 15 method (Intensity threshold ≥ 4.5e4, dynamic exclusion of 10 s and excluding charges unassigned, +1 and >+6). MS spectra were acquired from 350 to 1500 m/z with a resolution of 70 000 FWHM (200 m/z). Ion peptides were isolated using a 2.0 Th window and fragmented using higher-energy collisional dissociation (HCD) with a normalized collision energy of 27. MS/MS spectra resolution was set to 35 000 (200 m/z). The ion target values were 3e6 for MS (maximum IT of 25 ms) and 1e5 for MS/MS (maximum IT of 110 ms).
Data analysis - Raw files were processed with MaxQuant (v 1.6.12.0) using the standard settings against a human protein database (UniProtKB/Swiss-Prot, 20,373 sequences) supplemented with contaminants. Carbamidomethylation of cysteines was set as a fixed modification whereas oxidation of methionine and protein N-term acetylation were set as variable modifications. Minimal peptide length was set to seven amino acids and a maximum of two tryptic missed-cleavages were allowed. Results were filtered at 0.01 FDR (peptide and protein level).
Afterwards, the ‘proteinGroup.txt’ file was loaded in Perseus (1.6.10.43) for further statistical analysis. Missing values were imputed from the observed normal distribution of intensities and two-sample Student's t-tests were performed for the RPAP3M626A/F630R versus IgG and RPAP3-WT versus IgG comparisons. Only proteins with a P-value <0.05 and a log2 ratio >3 were considered as interactors. For the RPAP3M626A/F630R versus RPAP3-WT comparison, protein intensity values were normalized using the summed intensity of all RPAP3-WT and RPAP3M626A/F630R common peptides, missing values were imputed from the observed normal distribution of intensities and a two-sample Student's T-Test was performed. Only interactors with a P-value <0.05 and a log2 ratio ←2 were considered as regulated. For every comparison, the permutation-based FDR was estimated to be below 5%.
Quantification
Immunoreactivity bands of western blot were quantified by laser densitometry with the ImageJ Software (NIH) (https://imagej.nih.gov/ij/index.html) or by the software included in the Odyssey Infra-red Imaging System.
Statistics
All data are presented as mean values ± SEM of the indicated number of independent experiments stated in the figure legend. We determined the statistical significance in instances of single comparisons by unpaired Student's t test. Statistics were obtained using GraphPad Prism 8 software (https://www.graphpad.com/). The differences were considered significant when **P < 0.01, ***P < 0.001.
Cross-linking coupled to mass spectrometry
The purified RUVBL1–RUVBL2–ZNHIT2 was cross-linked with the N-hydroxysuccinimide (NHS) esters disuccinimidyl dibutyric urea (DSBU synthesis described in (32)). The cross-linking reactions were incubated at a final excess of 200 folds that of protein concentration for 45 min at 4°C. The reactions were quenched by adding ammonium hydrogen carbonate to a final concentration of 50 mM incubating for further 15 min.
The cross-linked samples were freeze-dried and resuspended in 50 mM ammonium hydrogen carbonate, reduced with 10 mM DTT and alkylated with 50 mM iodoacetamide. Following alkylation, proteins were digested with trypsin (Promega, UK) at an enzyme-to-substrate ratio of 1:20, overnight at 37°C. The samples were acidified with formic acid to a final concentration of 2% (v/v) and fractionated by reverse phase C18 high pH chromatography on an Acquity UPLC CSH C18 1.7 μm, 1.0 × 100 mm column (Waters) over a gradient of acetonitrile 2–40% (v/v) and ammonium hydrogen bicarbonate 100 mM.
All the fractions were lyophilized and resuspended in 2% (v/v) acetonitrile and 2% (v/v) formic acid for LC–MS/MS analysis. An Ultimate U3000 HPLC (ThermoScientific Dionex, USA) was used to deliver a flow of approximately 300 nL/min. A C18 Acclaim PepMap100 5 μm, 100 μm × 20 mm nanoViper (ThermoScientific Dionex, USA), trapped the peptides before separation on a C18 Acclaim PepMap100 3 μm, 75 μm × 250 mm nanoViper (ThermoScientific Dionex, USA). Peptides were eluted with a gradient of acetonitrile. The analytical column was directly interfaced via a nano-flow electrospray ionization source, with a hybrid quadrupole orbitrap mass spectrometer (Q-Exactive HF-X, ThermoScientific, USA). MS data were acquired in data-dependent mode. High-resolution full scans (R = 120 000, m/z 350–2000) were recorded in the Orbitrap and after CID activation (stepped collision energy 30 ± 3) of the 10 most intense MS peaks, MS/MS scans (R = 45 000) were acquired. lyophilized and resuspended in 2% (v/v) acetonitrile and 2% (v/v) formic acid.
The fractions were analyzed by nano-scale capillary LC–MS/MS using an Ultimate U3000 HPLC (ThermoScientific Dionex, USA) to deliver a flow of approximately 300 nl/min. A C18 Acclaim PepMap100 5 μm, 100 μm × 20 mm nanoViper (ThermoScientific Dionex, USA), trapped the peptides before separation on a C18 Acclaim PepMap100 3 μm, 75 μm × 250 mm nanoViper (ThermoScientific Dionex, USA). Peptides were eluted with a gradient of acetonitrile. The analytical column outlet was directly interfaced via a nano-flow electrospray ionization source, with a hybrid quadrupole orbitrap mass spectrometer (Q-Exactive HF-X, ThermoScientific, USA). MS data were acquired in data-dependent mode. High-resolution full scans (R = 120 000, m/z 350–2000) were recorded in the Orbitrap and after CID activation (stepped collision energy 30 ± 3) of the five most intense MS peaks, MS/MS scans (R = 15 000) were acquired.
For data analysis, Xcalibur raw files were converted into the MGF format through MSConvert (Proteowizard) (33) and used directly as input files for MeroX (34). Searches were performed against an ad-hoc protein database containing the sequences of the complexes and a set of randomized decoy sequences generated by the software. The following parameters were set for the searches: maximum number of missed cleavages 3; targeted residues K, S, Y and T; minimum peptide length five amino acids; variable modifications: carbamidomethyl-Cys (mass shift 57.02146 Da), Met-oxidation (mass shift 15.99491 Da); BuUrBu modification fragments: 85.05276 Da and 111.03203 (precision: 5 ppm MS1 and 10 ppm MS2); false discovery rate cut-off: 5%. Finally, each fragmentation spectra were manually inspected and validated.
Cryo-electron microscopy, image processing and model building
RUVBL1–RUVBL2-ZNHIT2ΔC protein purification for cryoEM
Purified GST-ZNHIT2ΔC was incubated with the RUVBL1–RUVBL2 protein complex with a molar excess ratio 1:3 of ZNHIT2ΔC for 20 min at RT. The sample was then incubated with pre-equilibrated Glutathione Sepharose 4B resin (Merck) for 2 h at 4°C with agitation. Unbound protein fraction was subsequently removed by several washing steps (washing buffer was 50 mM Tris–HCl pH 8.0, 100 mM KCl, 1 mM DTT). RUVBL1–RUVBL2–ZNHIT2ΔC protein complex was eluted by incubation with the GST-3C protease for 30 min at 4°C that removed the GST tag that kept GST-ZNHIT2ΔC bound to the resin.
Sample vitrification
2.5 μl of purified RUVBL1–RUVBL2–ZNHIT2ΔC protein complex (0.8 mg/ml) was applied to holey carbon grids Quantifoil R1.2/1.3 Cu 300 mesh (Quantifoil) to which a glow discharge was previously applied. Buffer excess was blotted away for 3 s and plunge frozen in liquid ethane cooled at liquid nitrogen temperature, using the VitroBot Mark IV (Thermo Fisher Scientific) set to 4°C and 90% humidity. Vitrified grids were clipped and stored at liquid nitrogen.
Data collection
Data were collected over four sessions on a 300 keV Titan Krios G3 electron microscope (Thermo Fisher Scientific) using a EFTEM energy filter (Gatan) and a K2 direct electron detector operated in counting mode. Data acquisition parameters are summarized in Supplementary Table S2.
Image processing
Movies from each dataset were individually preprocessed. Beam-induced motion was initially corrected using MotionCor2 (35) and contrast transfer function parameters (CTF) were estimated using Gctf (36). Particles were automatically picked using references created from manually selected particles in Relion 3.1 (37). Selected particles were subjected to reference-free alignment and 2D classification in Relion 3.1 (37). A selection of 173 905 best quality particles were combined from each data collection session and the 2D alignment and classification repeated to extract a more homogeneous data set. Initially unmasked and then mask 3D refinements were performed in Relion 3.1 and CryoSPARC v2 (38). Several rounds of 3D classification were performed in order to both analyze data heterogeneity and recover a more homogeneous data set to perform another run of 3D refinement in Relion 3.1 (37). Per particle CTF parameters as well as micrographs aberrations were refined or corrected using the CTF refinement tool in Relion 3.1 (37). Per-particle beam induced motion was estimated and corrected using the Relion 3.1 particle polishing program. The final consensus 3D refinement was subjected to the post-processing tools in Relion 3.1 that includes modulation transfer function correction and B-factor sharpening.
In the consensus cryoEM map obtained for the RUVBL1–RUVBL2-ZNHIT2ΔC complex, the region corresponding to ZNHIT2ΔC was resolved at low resolution, suggesting flexibility and/or heterogeneity. To address this, we performed a detailed analysis of the heterogeneity in the ZNHIT2ΔC density using cryoDRGN (39). This allowed us to identify significant flexibility of ZNHIT2ΔC with respect to the RUVBL1–RUVBL2 ring. The fitting of a predicted atomic model of ZNHIT2 obtained using Alphafold (40) within the cryoEM density for ZNHIT2ΔC in the complex was performed by the automatic sequential fitting tool in Chimera (41).
Focused 3D refinements of the AAA-ring of RUVBL1–RUVBL2 in the complex was performed to improve the resolution. For this, the map was segmented using UCSF Chimera (41) and then the region of interest was masked to remove densities not corresponding to the RUVBL1–RUVBL2 ring (density subtracted particles) in Relion 3.1. Extensive 3D classifications coupled to particle selection and masked 3D refinement allowed selection of more homogeneous datasets that were used, in each case, in a final 3D refinement and post-processing jobs using both Relion 3.1 (37) and CryoSPARC v2 (38). The reconstructed AAA-ring RUVBL1–RUVBL2 map, obtained at 4.1 Å resolution, was used for model building.
Global resolution was estimated in the cryoEM maps using the gold-standard criterion and a cutoff of 0.143. Local resolution was estimated in Relion 3.1 (37) and used to local sharpen the map using the same Relion 3.1 tool.
Model building
Density subtracted cryoEM map of the AAA ring of the RUVBL1–RUVBL2 complex was used to build an atomic model of this complex region. The human RUVBL1–RUVBL2 atomic structure reported in Martino et al. (PDB ID: 6FO1) (10) was used as starting model reference. The initial model was fitted into the cryoEM map using the rigid body fitting tool in Coot (42). Real space refinement was done iteratively in Coot (42) and Refmac5 (43). Close analysis and refinement were performed at the RUVBL1 and RUVBL2 nucleotide binding pocket using Coot (42). Atoms whose density was not accounted for in the cryoEM map were removed in Coot (e.g. RUVBL2 N-terminal region). CryoEM maps and atomic models were visualized in UCSF Chimera (41) and Coot (42).
ATPase assays
ATP hydrolysis of the untagged RUVBL1–RUVBL2 protein complex was measured in a continuous spectrophotometric enzymatic assay. Pyruvate kinase (PK) and lactate dehydrogenase (LDH) enzymes couple the ATP consumption to the oxidation of NADH, which strongly absorbs to 340 nm. Thus, decrease in absorbance at 340 nm is directly correlated to ATP hydrolysis (44). The RUVBL1–RUVBL2 ATPase activity was measured at a final concentration of 3 μM (being considered monomers) in the presence or absence of 3 μM ZNHIT2ΔC. ATPase assays of ZNHIT2ΔC alone and without both RUVBL1–RUVBL2 and ZNHIT2ΔC were included as controls. Assays were performed in 100 μl reactions in a buffer containing 50 mM Tris–HCl pH 7.4, 150 mM NaCl, 20 mM MgCl2, 2 mM phosphoenolpyruvate (PEP), 0.5 mM NADH, 0.04 U/μl PK (Sigma Aldrich), 0.05 U/μl LDH (Sigma Aldrich) and 5 mM ATP. Absorbance was measured at 37°C in a Victor plate reader (Perkin Elmer) for 1h. Linear decrease in the absorbance at 340 nm was used to estimate ATP turnover and relative RUVBL1–RUVBL2 ATPase activity. Paired Student's t-tests were performed to compare the RUVBL1–RUVBL2 ATPase activity in the absence versus the presence of ZNHIT2ΔC.
RESULTS
RUVBL1–RUVBL2 binds strongly to ZNHIT2 C-terminal domain
We purified human ZNHIT2 as well as two truncated versions to analyze their interaction with RUVBL1–RUVBL2, R2TP and other proteins implicated in the biogenesis of PRPF8 (see later) (Figure 1A). ZNHIT2 generated a degradation product during our expression and purification strategy which lacked residues 376–403, as revealed by mass spectrometry, and which we were unable to remove from preparations containing a mixture of the full-length protein and the C-terminally truncated product. To facilitate subsequent analysis and interpretations, we cloned and expressed ZNHIT2 lacking this small C-terminal region (ZNHIT2ΔC from now on), which did not degrade any further. ZNHIT2ΔC was purified using a GST-tag at the N-terminus that could be removed without affecting the solubility of the protein (Figure 1C). In addition, we cloned and expressed an N-terminal fragment (ZNHIT2-N; residues 1–123) (Figure 1A–C) that included the zinc finger HIT domain (residues 12–48) whose structure has been determined by NMR (23), and a C-terminal fragment (ZNHIT2-C; residues 205–375) (Figure 1A–C) that accumulates a large number of mutations associated to several types of cancer (Supplementary Figure S1A). We also purified several versions of the RUVBL1 and RUVBL2 ATPases: the individual RUVBL1 and RUVBL2 subunits, the preassembled RUVBL1–RUVBL2 complex, RUVBL1–RUVBL2 where the OB-fold region of each DII domain in both subunits was removed (RUVBL1–RUVBL2ΔDII), and the RUVBL1E303Q–RUVBL2E318Q ATPase deficient mutant, all of these following methods described before (12).
The interaction between ZNHIT2 and RUVBL1–RUVBL2 was then studied by pull-down experiments, using the GST tag in ZNHIT2ΔC and analyzing the eluted material by SDS-PAGE (Figure 1D, top panel; Supplementary Figure S1B, C). ZNHIT2ΔC interacted with the RUVBL1–RUVBL2 complex even in the case of the ATPase deficient Walker B mutant (labeled as RUVBL1/2-WB hereafter) and independently of the nucleotide added to the sample (Figure 1E), suggesting that ATP binding or hydrolysis is not required for the formation of the RUVBL1–RUVBL2–ZNHIT2 complex. The interaction involved both RUVBL1 and RUVBL2, as each individual subunit could bind ZNHIT2ΔC on their own. Interestingly, truncation of part of the DII domain in the RUVBL1–RUVBL2 complex (residues 127–233 in RUVBL1, and 134–237 in RUVBL2, labeled as RUVBL1/2-ΔDII hereafter) abolished the interaction, indicating that these domains are necessary for the assembly of a complex between ZNHIT2ΔC and RUVBL1–RUVBL2 (Figure 1D, top panel). Although it has been proposed that the HIT domain is required for the interaction between ZNHIT2 and RUVBL1–RUVBL2 (4), our findings suggest that the C-terminal of ZNHIT2ΔC is sufficient to form a complex with the ATPases (Figure 1D, middle and bottom panels). Curiously, ZNHIT2-N seemed to interact more efficiently with RUVBL2 than RUVBL1. Whereas published experiments were performed using a mix of individually purified RUVBL1 and RUVBL2 in the presence of ATP (4), here we used pre-assembled RUVBL1–RUVBL2 complexes, and this might account for some of the differences observed.
Regardless of this, our results clearly showed that the HIT domain is dispensable for the binding of ZNHIT2 to preformed RUVBL1–RUVBL2 complexes (Figure 1D, bottom panel). The C-terminal domain of ZNHIT2ΔC binds RUVBL1 and RUVBL2 subunits and the interaction was affected by the truncation of the DII domains, thus reproducing the results obtained for ZNHIT2ΔC. Crosslinked mass spectrometry (XL-MS) performed on a purified RUVBL1–RUVBL2–ZNHIT2ΔC complex identified several internal crosslinks in ZNHIT2, RUVBL1 and RUVBL2 but also a crosslink between residue 325 at the C-terminal domain of ZNHIT2ΔC and residue 277 of RUVBL1 located in the vicinity of the DII domains (using PDB 6FO1) and a crosslink between this same residue and residue 203 within RUVBL2 DII domain (Figure 1F, Supplementary Table S3).
Together, these results show that ZNHIT2 interacts with RUVBL1 and RUVBL2 through the DII domains, and that, contrary to expectation, the HIT domain is dispensable for the interaction and the C-terminal end of the protein contributes significantly to the interaction between ZNHIT2 and preformed RUVBL1–RUVBL2 complexes.
CryoEM of the RUVBL1–RUVBL2–ZNHIT2ΔC complex
To determine how ZNHIT2 interacts with RUVBL1–RUVBL2, the complex was purified in amount and homogeneity compatible with structural studies by saturating GST-ZNHIT2ΔC with RUVBL1–RUVBL2 on the beads and eluting the complex by cleavage of the GST tag (Figure 2A). Stability of the eluted complex was probed by size-exclusion chromatography (SEC), before loading on holey carbon film for their observation using cryoEM (Supplementary Figure S2, Supplementary Table S3). 2D averaging of the cryoEM images obtained for RUVBL1–RUVBL2–ZNHIT2ΔC revealed side views corresponding to an hexameric complex with density attributed to ZNHIT2ΔC at the DII-side of the RUVBL ring, being the DII region flexible and less well defined (Figure 2B). When RUVBL1 and RUVBL2 are co-expressed and purified, a mixture of hexameric single-ring and dodecameric double-ring complexes with very distinctive side views can be observed in the electron microscope (28). Therefore, similarly to what has been described before for INO80, R2TP and other RUVBL1–RUVBL2-containing complexes (14), ZNHIT2 engages at the DII-face of hexameric rings, disrupting dodecameric complexes.
Figure 2.
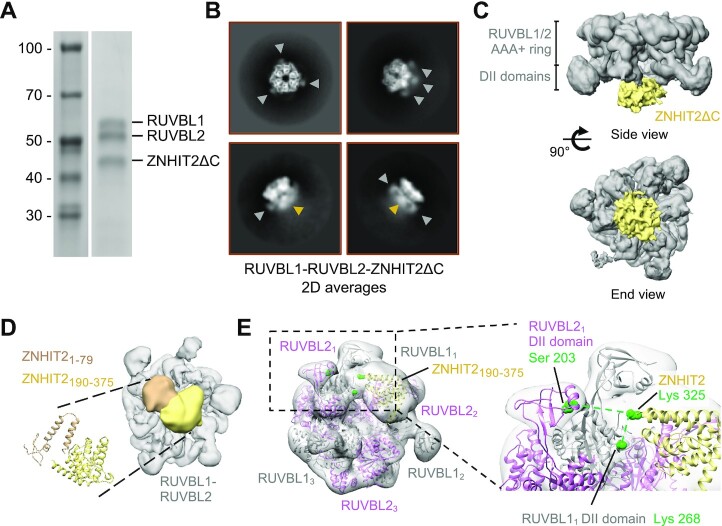
CryoEM of the RUVBL1–RUVBL2-ZNHIT2ΔC complex. (A) SDS-PAGE of the purified RUVBL1–RUVBL2–ZNHIT2ΔC complex used for structural studies. (B) Representative 2D averages obtained from the cryoEM images of RUVBL1–RUVBL2- ZNHIT2ΔC. Grey arrowheads label some of the RUVBL1–RUVBL2 DII domains, while yellow arrowheads indicate the position of ZNHIT2ΔC. (C) Two views of the consensus cryoEM map of the RUVBL1–RUVBL2-ZNHIT2ΔC complex at 4.4 Å resolution. Density for ZNHIT2ΔC (yellow color) locates at the DII-face of the ring (grey color). (D) A view of one of the cryoEM maps obtained after classification using cryoDRGN (39), showing the DII-side of the RUVBL1–RUVBL2 ring. The density for ZNHIT2ΔC complex is divided in two lobules that correlate with the two domains predicted for ZNHIT2ΔC by AlphaFold (40). (E) Left panel, a tilted view of the cryoEM map in D showing the interaction of the large ZNHIT2ΔC domain. Both the atomic model of the RUVBL1–RUVBL2 complex and the ZNHIT2ΔC C-terminal domain are fitted within the map. Linked residues, identified by XL-MS, are shown as green dots. Each RUVBL subunit is labelled from 1 to 3 (subscript). Right panel, close-up view to highlight the proximity between the cross-linked residues in ZNHIT2 and the adjacent DII domains in the RUVBL1–RUVBL2 model.
Structure of the RUVBL1–RUVBL2–ZNHIT2ΔC complex
The structure of the complex formed by RUVBL1–RUVBL2 and ZNHIT2ΔC was determined after 3D processing and classification of the cryoEM images to select homogenous subsets of particles. 173 940 particles (40% of the initial dataset) converged to a structure at 4.4 Å average resolution, with local resolutions ranging from 3.5 to 8.0 Å (Supplementary Figure S2C). As anticipated by the mapping experiments, ZNHIT2ΔC interacted with the side of the RUVBL1–RUVBL2 ring where the DII domains locate (Figure 2C).
Local resolution estimates revealed that, whereas the RUVBL1–RUVBL2 ring reached high resolution, ZNHIT2ΔC was less well defined, with an estimated resolution value around 8 Å (Supplementary Figure S2C). We hypothesized that the lack of structural features for ZNHIT2ΔC was due conformational heterogeneity. To test this, we used new methods for the reconstruction of heterogeneous cryoEM structures using neural networks (39), which revealed the flexibility of the DII domains, conformational heterogeneity in ZNHIT2ΔC, and interestingly also the flexible attachment of ZNHIT2ΔC to the RUVBL1–RUVBL2 hexameric ring (Supplementary Figure S3). Such degree of heterogeneity and flexibility hindered our attempts to improve the resolution of ZNHIT2ΔC to a level compatible with atomic modelling. However, the analysis allowed us to classify images into more homogeneous subgroups, each with improved features for ZNHIT2ΔC. These structures revealed that the density for ZNHIT2ΔC consisted of two regions, and in at least in some of the volumes a small and a large domain were clearly defined (Figure 2D, Supplementary Figure S3C).
Although we lack structural information at atomic level for most of ZNHIT2 with the exception of the HIT domain (23), we used ZNHIT2 alphafold atomic structure prediction (40) to analyze these reconstructions (Supplementary Figure S3A). According to the prediction, ZNHIT2 consists of two domains, but only the larger C-terminal domain was predicted with high confidence. We fitted the model for ZNHIT2ΔC, after removing large loops, into the two bodies of density for ZNHIT2ΔC (Figure 2D, Supplementary Figure S3). Using the automatic sequential fitting application in Chimera (41) and allowing for some flexibility between the two domains, the larger region of the ZNHIT2ΔC cryoEM density was assigned to the large C-terminal domain predicted for ZNHIT2. Interestingly, this fitting agrees with the XL-MS data (Figure 1F), since it places Lys 325 at the ZNHIT2 C-terminal domain in close proximity of the residues in RUVBL1 and RUVBL2 DII domains detected by XL-MS (Figure 2E). Taken together, our results argue in favor of a model where the C-terminal of ZNHIT2 provides important contacts between ZNHIT2 and RUVBL1–RUVBL2.
ZNHIT2 regulates the conformation and ATPase activity of RUVBL2
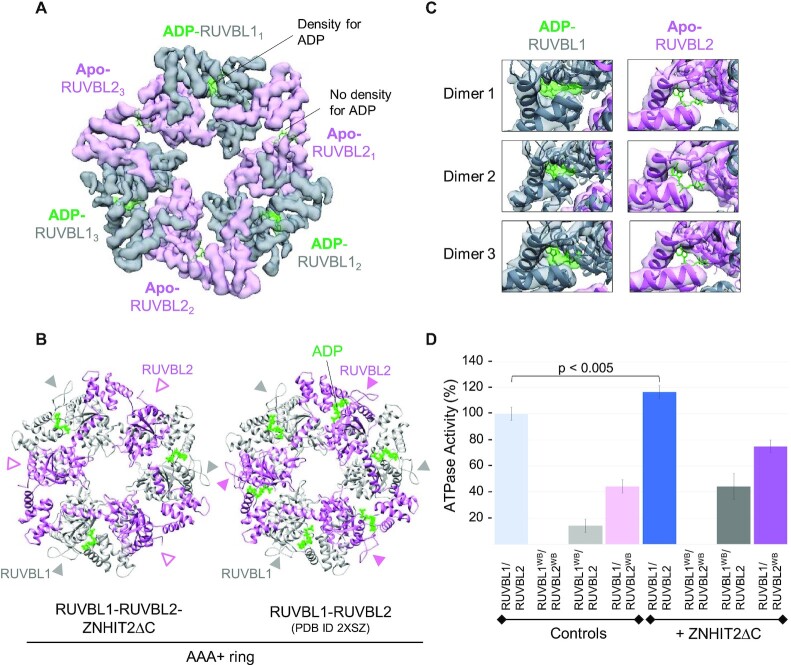
We improved the resolution of the RUVBL1–RUVBL2 ring in the complex to study the consequences of ZNHIT2ΔC binding to the ATPase ring (Figure 3A). For this, the density corresponding to the hexameric ring was subtracted from every image and processed without the influence of ZNHIT2ΔC (Supplementary Figure S2). Using this strategy, the cryoEM map reached a resolution of 4.1 Å and the structure was then modelled (Figure 3B-C). Every subunit of human RUVBL1–RUVBL2 is bound to a nucleotide in the majority, if not all, of the structures determined so far for the hetero-hexameric complex in the absence of a binding partner (PDB ID 2XSZ used in the figure), with nucleotides trapped inside their binding pockets by a short N-terminal region in RUVBL1 and RUVBL2 (8,14,19) (Figure 3B, right panel). The structure of the RUVBL1–RUVBL2 ring after binding to ZNHIT2ΔC revealed that these N-terminal regions are still visible in all RUVBL1 subunits, but it is lost, possibly disordered, in each RUVBL2 subunit (Figure 3B, left panel). In accordance, ADP was present in the nucleotide-binding pocket of every RUVBL1 subunit but absent in all RUVBL2 subunits (Figure 3C). We verified that all 3 RUVBL2 subunits in each RUVBL1–RUVBL2 hexamer were affected by these conformational changes by performing several analyzes that discarded an artefact of image processing (Supplementary Figure S4). Curiously, and as in the case previously described for DHX34 helicase in complex with RUVBL1–RUVBL2 (12), all RUVBL1 subunits were unaffected by these large changes and still bound to nucleotide, suggesting that RUVBL1 probably does not perform the same function than RUVBL2 in the complex.
Figure 3.
ZNHIT2ΔC alters the conformation and ATPase activity of RUVBL2. (A) CryoEM density of the AAA-ring in RUVBL1–RUVBL2 after being processed independently of the flexible density for ZNHIT2ΔC and the DII domains. Density for nucleotides is only present in the RUVBL1 subunits (green density with ADP fitted inside). ADP molecules, without density accounting for them, are fitted in the described nucleotide binding sites of RUVBL2 only to highlight the lack of density in the region where nucleotide is present in human RUVBL1–RUVBL2 crystal structures (PDB ID 2XSZ). (B) Right panel, crystal structure of RUVBL1–RUVBL2 (PDB ID 2XSZ). Grey and pink arrowheads indicate the N-terminal regions of RUVBL1 and RUVBL2 respectively. These were absent in the atomic structure of RUVBL1–RUVBL2–ZNHIT2ΔC (indicated as pink empty arrowheads). Nucleotides are shown in green colour. (C) Close-up of nucleotide-binding sites for every RUVBL subunit after the interaction with ZNHIT2ΔC. Nucleotide molecules are show in green and nucleotide densities are displayed in semi-transparent green density. In RUVBL2, density for nucleotide is not observed, and this is indicated by placing ADP in the binding-site. (D) ATPase activity of RUVBL1–RUVBL2, wild type and mutants (RUVBL1WB and RUVBL2WB stand for the mutants whose Walker B motif has been altered to impair ATPase activity), in the presence and absence of ZNHIT2ΔC using the value of RUVBL1–RUVBL2 alone as 100%. Significance value of the paired Student's T-Test is shown for the RUVBL1–RUVBL2–ZNHIT2ΔC complex in comparison with the RUVBL1–RUVBL2 complex alone.
We tested the effect of ZNHIT2ΔC on the intrinsically weak ATPase activity of the RUVBL1–RUVBL2 (Figure 3D, Supplementary Figure S5, Table S4). For this, we used purified RUVBL1–RUVBL2 where the His-tag initially present in RUVBL1 for purification of the complex was removed by cleavage with protease, to discard any influence of the tag in the experiments. The effect of ZNHIT2ΔC addition was analyzed in the wild type complexes, but also in complexes containing Walker B mutations in RUVBL1 and/or RUVBL2, which can bind but not hydrolyze ATP (Figure 3D). Control experiments were made to determine that the ZNHIT2ΔC preparation used had only a residual ATPase activity (Supplementary Figure S5). No ATPase activity was measured when both RUVBL1 and RUVBL2 were mutated, regardless of the addition of ZNHIT2ΔC, indicating that our measurements correspond to the activity of RUVBL1 and RUVBL2. The ATPase activity of wild type RUVBL1–RUVBL2 and of mutants with only one of the subunits mutated were stimulated after addition of ZNHIT2ΔC (Figure 3D). This suggests that the interaction of ZNHIT2ΔC with RUVBL1–RUVBL2 affects the activity of both RUVBL1 and RUVBL2 subunits.
Together, these results show that the interaction of ZNHIT2ΔC with the RUVBL1–RUVBL2 complex induces significant conformational changes which disrupt the interaction with nucleotides in all three RUVBL2 subunits and affect to the ATPase activity of the RUVBL1–RUVBL2 complex.
ZNHIT2 forms a direct complex with the R2TP-HSP90 chaperon system
The R2TP co-chaperone system is formed after the interaction between RUVBL1–RUVBL2 and RPAP3, a protein that recruits HSP90 to R2TP (Figure 1B). ZNHIT2 associates with R2TP in affinity purifications, however some data suggest that the interaction of ZNHIT2 with RUVBL1–RUVBL2 excludes the binding of the RPAP3 (4). To definitely solve this question, we used purified proteins to determine if a complex where RUVBL1–RUVBL2 interacts simultaneously with RPAP3 and ZNHIT2 can form. For this, we performed a double pull-down experiment using a His tag in RUVBL1 and a GST tag in ZNHIT2ΔC (Figure 4A). Pull-down of RUVBL1 from a mix of RUVBL1–RUVBL2, RPAP3 and ZNHIT2ΔC eluted all the components of the complex, suggesting the assembly of R2TP-ZNHIT2ΔC although we could not discard from this experiment that some RUVBL1–RUVBL2 molecules could interact with RPAP3 and others with ZNHIT2ΔC without forming a tetrameric complex. Then, the material eluted from this first pull-down was subjected to a second pull-down experiment using the tag in ZNHIT2ΔC (Figure 4A). This showed the co-purification of all four proteins, RUVBL1, RUVBL2, RPAP3 and ZNHIT2ΔC, thus confirming that ZNHIT2ΔC can form a complex with RUVBL1–RUVBL2 when these ATPases are forming part of the R2TP chaperone.
Figure 4.
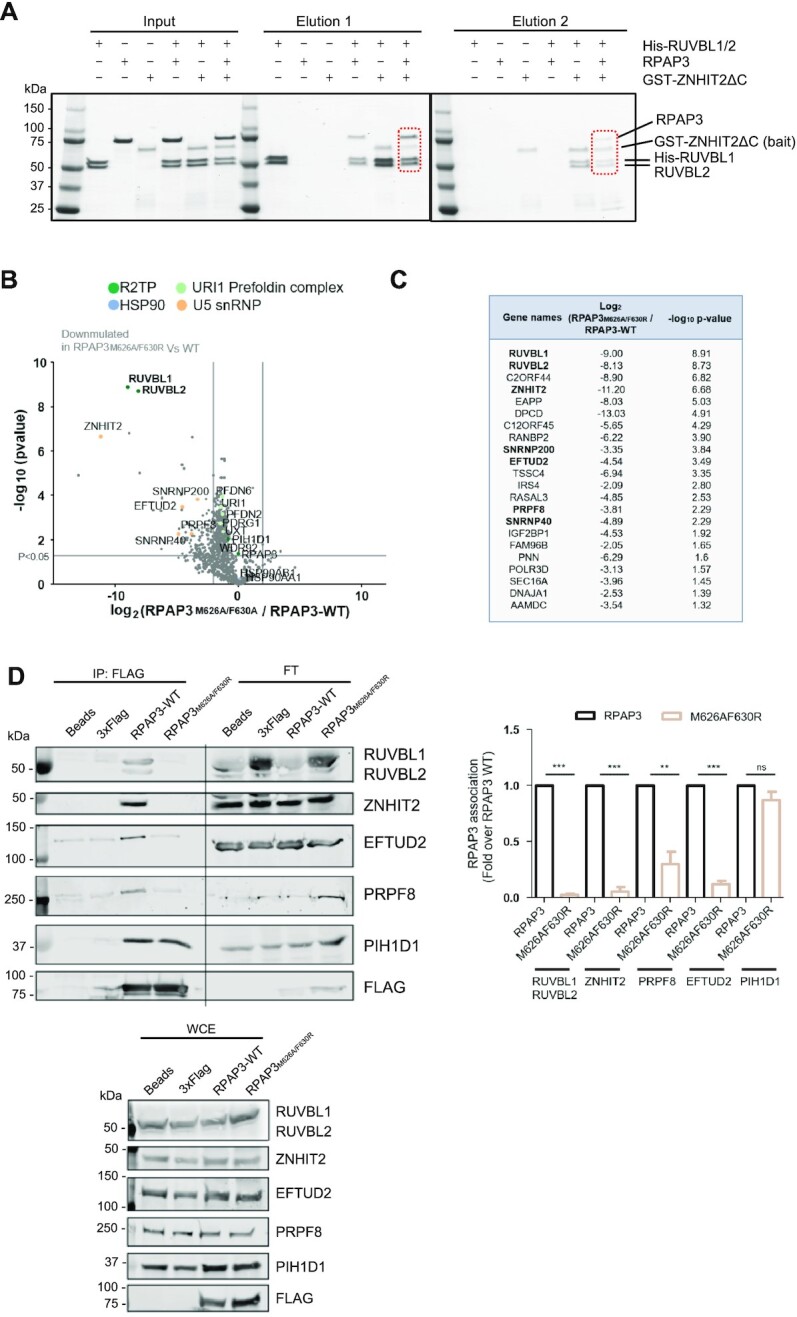
ZNHIT2 forms a complex with R2TP in vitro and in cells. (A) Consecutive pull-down experiments to analyze the formation of R2TP-ZNHIT2ΔC complexes. His-RUVBL1 was used in a first pulldown step, and then the eluted material was the input of a second pulldown experiment using the GST tag in ZNHIT2ΔC. (B) Volcano plot comparing interactors of RPAP3M626A/F630R versus RPAP3-WT is shown. Two-sample Student's t-test was performed. Only interactors with a P-value <0.05 and a log2 ratio ≤ 2 were considered downregulated in the mutant condition. The permutation-based FDR was estimated to be below 5%. (C) The 22 interactors significantly down-represented in RPAP3M626A/F630R versus RPAP3-WT are indicated in the table. Those linked with U5 snRNP or its maturation are in bold. (D) Immunoprecipitation (IP) of transiently transfected HEK293T cells with RPAP3-WT RPAP3M626A/F630R. Anti-FLAG-IPs, whole-cell extracts (WCE, 1%) and flow through (FT) were subjected to western analysis using the indicated antibodies. Blots are representative of 5 independent experiments. Data (mean ± SEM of five independent experiments) were normalized by total FLAG and expressed as fold-change of the different proteins indicated association with FLAG with respect to the RPAP3-WT condition. Statistical significance was analyzed using unpaired t-test. **P < 0.01, ***P < 0.001.
We verified that ZNHIT2 is part of R2TP in cells by making use of RPAP3, the protein that defines R2TP versus other complexes participated by RUVBL1–RUVBL2. RPAP3 interactors in HEK293T cells were analyzed by affinity purification followed by mass spectrometry (Supplementary Figure S6A). Constituents of R2TP (RUVBL1, RUVBL2 and PIH1D1) and PFDL (URI1, UXT, PDRG1, PFDN2, PFDN6) were among the most abundant interactors of RPAP3, but also ZNHIT2, indicating ZNHIT2 forms a complex with R2TP in cells. We also identified HSP90 as one the abundant interactors of RPAP3, as expected since RPAP3 contains two TPR domains that can bind HSP90 to recruit it to R2TP.
Together, these results clarify that the interaction of ZNHIT2 with RUVBL1–RUVBL2 is compatible with the binding of RUVBL1–RUVBL2 to RPAP3 and the formation of R2TP, and that this way, ZNHIT2 can associate with HSP90.
RUVBL1–RUVBL2 and ZNHIT2 associate with U5-specific proteins
Mass spectrometry revealed the association of PRPF8 and other U5-specific proteins, such as SNRNP200, SNRNP40 and EFTUD2 with RPAP3, suggesting that R2TP forms a complex not only with ZNHIT2 but also with components of the U5 snRNP (Supplementary Figure S6A). To determine the role of RUVBL1–RUVBL2 and RPAP3 in these interactions, we made use of mutations in RPAP3 (RPAP3M626A-F630R) that disrupt the interaction with RUVBL1–RUVBL2 (9). We compared the interactome of RPAP3M626A-F630R with that of RPAP3, discriminating those interactions mediated by RUVBL1–RUVBL2 from those involving RPAP3 (Figure 4B, Supplementary Figure S6B). Interestingly, this comparison revealed 22 interactors significantly down-represented in the RPAP3M626A-F630R mutant, 8 of which have been linked before to U5 snRNP and its biogenesis (Figure 4B, C). The mutation completely disrupted the interaction of RPAP3 with RUVBL1, RUVBL2 and ZNHIT2, confirming that ZNHIT2 engages R2TP thanks to the AAA-ATPases. Interestingly, the mutation also affected the interaction between RPAP3 and some of the components of U5 snRNP (PRPF8, SNRNP40, SNRNP200 and EFTUD2), but to a lesser extent. PRPF8 still showed some association after the mutation (Supplementary Figure S6B), suggesting that although RUVBL1–RUVBL2 provides the main scaffold between PRPF8 and the R2TP complex, RPAP3 or complexes formed by RPAP3 such as RPAP3-PIH1D1 could also contribute to bring PRPF8 to R2TP.
These results were confirmed by immunoprecipitating overexpressed RPAP3 or RPAP3M626A-F630R in cells (Figure 4D). Whereas RPAP3 immunoprecipitated the R2TP complex (RUVBL1–RUVBL2 and PIH1D1), ZNHIT2, EFTUD2 and PRPF8, the mutation version of RPAP3 affected the interaction with all these proteins except PIH1D1. Curiously, after quantification of the experiments, although PRPF8 was also greatly affected, it seemed to still interact with RPAP3M626A-F630R although significantly less efficiently than with RPAP3. Together, these results indicate that R2TP associates with ZNHIT2, PRPF8 and other components of U5 snRNP, and that RUVBL1–RUVBL2 provides the main scaffold for all these interactions.
ZNHIT2, ECD, AAR2 and PRPF8 can form complexes with R2TP
How R2TP deals with a diversity of clients is still not well understood, but it has been suggested that ‘adaptor’ proteins contribute to put together the chaperone with specific clients. This model is best characterized for the assembly of mTOR complexes where a TELO2-TTI1-TTI2 (TTT) complex serves to bring mTOR to R2TP (11,45). For U5 snRNP, both ZNHIT2 and ECD have been proposed to function as linker between PRPF8 and R2TP (3,4,20,26). To analyze this possibility and delineate the interactions between RUVBL1–RUVBL2, PRPF8 and several factors proposed to contribute to its biogenesis, we purified Strep II-ECD as well as a His- and/or Strep II-tagged version of AAR2, an assembly factor that forms a complex with PRPF8 in the cytoplasm (46,47). ECD was co-expressed with CK2, the kinase that mediates ECD phosphorylation at the consensus motif that is recognized by PIH1D1 (26). This phosphorylation was confirmed in the recombinant ECD protein by mass spectrometry.
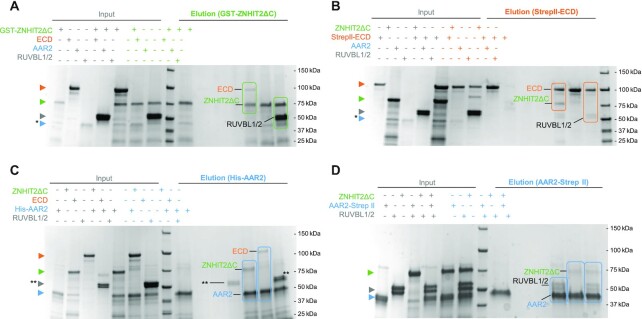
We analyzed the interactions between all these components using several pull-down strategies (Figure 5 and 6). First, we determined direct interactions using a mix of only two partners and a different protein as bait. ZNHIT2ΔC interacted with RUVBL1–RUVBL2 as expected but also with ECD (Figure 5A, B). RUVBL1–RUVBL2 interacted weakly with ECD (Figure 5B), which agrees with previous findings (26). When this experiment was repeated but RPAP3-PIH1D1 was added to the mix to allow for the assembly of R2TP, we recovered a stronger interaction of ECD with RUVBL1–RUVBL2, possibly thanks to the binding of PIH1D1 with the consensus phosphorylation-site present in ECD (Supplementary Figure S7A).
Figure 5.
RUVBL1–RUVBL2 interacts with AAR2, ECD and ZNHIT2ΔC. (A) Pull-down experiments using the GST tag in ZNHIT2ΔC showing that ZNHIT2ΔC interacts with RUVBL1–RUVBL2 and ECD. ZNHIT2ΔC contaminants (*) coelute with AAR2 so the interaction between ZNHIT2ΔC and AAR2 could not be determined in this experiment. Electrophoretic protein mobility is shown with coloured arrowheads according to the protein labels. Main results are highlighted within rectangles. (B) Pull-down experiments using the Strep II tag in ECD confirmed its interaction with ZNHIT2ΔC and RUVBL1–RUVBL2. Electrophoretic protein mobility is shown with coloured arrowheads according to the protein labels. Main results are highlighted within rectangles. (C) Pull-downs using the His-tag in AAR2 confirmed its interaction with ECD and ZNHIT2ΔC. Albeit the His-tag been removed from RUVBL1–RUVBL2 some unspecific interaction was detected in the RUVBL1–RUVBL2 control lane (**). Electrophoretic protein mobility is shown with coloured arrowheads according to the protein labels. Main results are highlighted within rectangles. (D) Pull-down experiments using the Strep II tag in AAR2 probed that AAR2 interacts with RUVBL1–RUVBL2 both in the absence and in the presence of ZNHIT2ΔC. Main results are highlighted within rectangles.
Figure 6.
Interactions between PRPF8 and RUVBL1–RUVBL2, RPAP3-PIH1D1, AAR2, ECD and ZNHIT2 (A) Flag-pull-down experiments showed a direct interaction between PRPF8 (bait) and RUVBL1–RUVBL2 (prey), which is not affected by the presence of ATP. Asterisks indicate contaminants that co-eluted with PRPF8. (B) Upper panel, pull-down experiments using Flag-PRPF8 (bait) and RPAP3-PIH1D1 in the presence or absence of RUVBL1–RUVBL2 were analyzed by western blot. Stained SDS-PAGE gels can be found in Supplementary Figure S8B. Lower panel, pull-down experiment using the GST tag in RPAP3 within the RPAP3-PIH1D1 complex, analyzed by western blot. MW, molecular weight markers. (C) In the presence of ECD, ZNHIT2ΔC, RUVBL1–RUVBL2, RPAP3-PIH1D1 and AAR2 (preys), PRPF8 (bait) interacted with RUVBL1–RUVBL2, RPAP3-PIH1D1, AAR2 and ECD as show in the Flag-pull-down assay. Control experiments using a cell extract lacking the overexpressed PRPF8, but overexpressing the Flag-tag were run in parallel SDS-PAGE for the experiments and they are shown in Supplementary Figure S8. Stained SDS-PAGE gels can be found in Supplementary Figure S8C.The presence of PRPF8, RUVBL1–RUVBL2, ECD, AAR2, RPAP3, PIH1D1 and ZNHIT2 in the Flag-pull-down assays was analyzed by western blot.
In the experiments using either GST- ZNHIT2ΔC or StrepII-ECD, we had difficulties to detect the presence of AAR2 in the elution. Thus, we addressed the analysis of direct interactions involving AAR2 using two tagged versions of this protein (Figure 5C, D). In these experiments, AAR2 formed direct interactions with ZNHIT2ΔC, ECD and RUVBL1–RUVBL2, and AAR2 was able to pull-down both ZNHIT2ΔC and RUVBL1–RUVBL2 when the proteins were mixed together (Figure 5D).
These results indicate that ECD, AAR2 and ZNHIT2ΔC can form binary complexes involving any of these proteins. RUVBL1–RUVBL2 interacts directly with all three proteins, but the interaction with ZNHIT2ΔC seems the strongest. We completed this analysis by considering the formation of complexes involving more than two proteins. For this, we mixed RUVBL1–RUVBL2, ECD, AAR2 and ZNHIT2ΔC and analyzed the complexes formed after pull-down of ZNHIT2, ECD and AAR2 (Supplementary Figure S7B). As seen before when analyzing pairwise interactions, AAR2 was only visible in experiments pulling-down from this protein. These experiments suggested the formation of several complexes or associations: RUVBL1–RUVBL2/ECD/ZNHIT2ΔC; RUVBL1–RUVBL2/ECD/AAR2; RUVBL1–RUVBL2/ECD/AAR2/ZNHIT2ΔC and ECD/ZNHIT2ΔC/AAR2. Figure 1B summarizes all these interactions.
We then explored the complexes formed by RUVBL1–RUVBL2 and PRPF8, in the absence or presence of ECD, ZNHIT2ΔC, AAR2 and RPAP3-PIH1D1. PRPF8 was produced in mammalian cells as a C-terminal Flag tagged recombinant protein and the lysates bound to affinity beads. After extensive washes, different proteins were added to the beads. Once the excess of proteins added was washed out, bound proteins were eluted using flag peptide (Figure 6). As a control to discard unspecific bindings, we performed exactly the same experiment for each of the proteins tested, but using a lysate from mammalian cells producing the C-terminal Flag but not PRPF8 (Supplementary Figure S8A). These control experiments showed that none of the proteins were eluting when PRPF8 was not present. However, when Flag-PRPF8 was present in the lysates, RUVBL1–RUVBL2 formed a direct complex with PRPF8, and this interaction was still detected when ATP was added to the reaction (Figure 6A). We then analyzed if PRPF8 could also interact with the other components of R2TP using the same methodology PRPF8 interacted directly with RPAP3-PIH1D1 in the presence and absence of RUVBL1–RUVBL2 (Figure 6B, top panels, Supplementary Figure S8B) and this was confirmed when pulling down from GST-RPAP3 pull-down (Figure 6B, bottom panels). This agrees with the results found by mass spectrometry (Figure 4) where disruption of the interaction between RPAP3 and RUVBL1–RUVBL2 affected strongly but not completely to the binding to PRPF8.
Interestingly, when RUVBL1–RUVBL2, AAR2, ECD, RPAP3-PIH1D1 and ZNHIT2ΔC were added to the Flag-affinity resin loaded with Flag-PRPF8, all these proteins eluted with PRPF8 (Figure 6C, Supplementary Figure S8C). Although we cannot discard that some of the eluted material could contain a mix of different complexes with PRPF8, the results strongly suggest that several of these proteins can be simultaneously part of the same complex.
Together, our results reveal that RUVBL1–RUVBL2, RPAP3-PIH1D1, ZNHIT2, AAR2, ECD and PRPF8 can assemble large complexes containing several of these proteins. ZNHIT2 and PRPF8 seem to make some of the stronger interactions with the RUVBL1–RUVBL2 ATPases.
DISCUSSION
The biogenesis of PRPF8 and U5 snRNP is a complex pathway requiring the intervention of several factors, including the HSP90 chaperone and the R2TP co-chaperone (3,4). R2TP contributes to the stability and assembly of several macromolecular complexes, but the mechanisms are not well understood. It is believed that one of the functions of R2TP is to facilitate the interaction between specific clients, such as PRPF8, with HSP90, acting as a scaffold between the chaperones and the clients. In addition, ATPase hydrolysis by RUVBL1–RUVBL2, the two ATPases in the complex, is required for some activities of R2TP in cells (13). This picture gets further complicated because R2TP requires additional factors to perform its functions, most of which are client dependent. All this suggests that the R2TP-HSP90 chaperone and its clients will transit though several stages containing different ancillary factors and displaying different conformations capable of recruiting and manipulating the clients.
ZNHIT2 participates in the biogenesis of PRPF8 and U5 snRNPs and interacts with RUVBL1–RUVBL2 (4), but how ZNHIT2 works and how it affects R2TP is not understood in detail. Here we have characterized the interactions of ZNHIT2 with R2TP, RUVBL1–RUVBL2, ECD, PRPF8 and AAR2. We find that ZNHIT2 performs at least two distinct functions, regulating the conformation and nucleotide state of RUVBL1–RUVBL2, and been part of a network of interactions with other factors involved in the maturation of PRPF8 and the R2TP-HSP90 chaperone system.
A nucleotide is bound in the ADP/ATP binding pocket of every RUVBL1 and RUVBL2 subunit of all the structures determined so far for human RUVBL1–RUVBL2 hetero-hexamers (17). It has been proposed that hexamerization obstructs the exchange from ADP to ATP, explaining in part the low ATPase activity of the complex (17–19). Recently, several proteins have been found to promote the release of the nucleotides trapped within the RUVBL1–RUVBL2 oligomers (8,12). DHX34, an RNA helicase involved in NMD, and PIH1D1, one of the components of the R2TP complex, interact with the DII domains of RUVBL1–RUVBL2 inducing conformational changes in a N-terminal region of RUVBL2 and the release of the bound nucleotides (8,12,19). Interestingly, we now find that ZNHIT2 interacts with the DII domains of several subunits of the RUVBL1–RUVBL2 complex, and this also induces changes in the N-terminal regions of RUVBL2, which are no longer visible in the cryoEM map of the RUVBL1–RUVBL2-ZNHIT2ΔC complex, and nucleotides are lost in all RUVBL2 subunits. ZNHIT2 modestly affects the rate of ATP hydrolysis by RUVBL1–RUVBL2 in vitro and we hypothesize that ZNHIT2 could function as a regulator of RUVBL1–RUVBL2 ATPase activity of during the biogenesis of PRPF8. What could be the functional relevance of such regulation of RUVBL1–RUVBL2 ATPase activity? Clients need to be first loaded to the R2TP-HSP90 chaperone, then manipulated and finally released once the assembly process has been completed and they are fully assembled and functional. Thus, ATP hydrolysis might be involved in the regulation of all these interactions as well as of the conformational transitions required to transform a series of components and assembly factors into a finally assembled and matured product to be released from the chaperone. Although very little is known about the conformational cycle of R2TP-HSP90 during the activation of clients, some evidence links ATP hydrolysis with the formation of interactions between R2TP and the clients. The components of the PFDL and TTT complexes as well as the PIKK clients significantly increase their association with RUVBL1–RUVBL2 when cells are incubated with an inhibitor of their ATPase activity (13).
Previous studies have shown that, at least in the presence of nucleotides and using a mix of purified RUVBL1 and RUVBL2, removal of the HIT domain greatly reduced the binding of ZNHIT2 to RUVBL1–RUVBL2 (4), providing a rational to why several members of the HIT-containing family of proteins form a complex with RUVBL1 and RUVBL2 (22). When using preassembled RUVBL1–RUVBL2 complexes, we detect binding between the HIT-containing region and RUVBL1–RUVBL2 but in contrast to the published results, the C-terminal region of ZNHIT2 was also sufficient for binding to RUVBL1–RUVBL2. This was supported by XL-MS that detects a crosslink between this C-terminal domain and the DII domain of both RUVBL1 and RUVBL2. CryoEM shows that ZNHIT2 interacts extensively with several DII domains of the RUVBL1–RUVBL2 ring in a rather flexible way, and therefore it would be expected that multiple regions of ZNHIT2 participate in the formation of the RUVBL1–RUVBL2–ZNHIT2 complex. In the cryoEM map, ZNHIT2 is resolved as a small and a large domain that can be interpreted as the N- and C-terminal domains of ZNHIT2 with the help of the predicted atomic structures. The large ZNHIT2 C-terminal domain contacts several subunits of the RUVBL1–RUVBL2 complex, and some of the contacts were identified by XL-MS. In particular, a serine residue in the OB-fold of RUVBL2 and a lysine residue in the DII domain of RUVBL1 crosslink with ZNHIT2 C-terminal domain. This interaction co-exists with other contacts between ZNHIT2 and the RUVBL1–RUVBL2 complex, which agrees with structures of the INO80 complex showing that Ies2/ZNHIT4 and Ies6 use several regions, not only the HIT domain, to interact with Rvb1/RUVBL1-Rvb2/RUVBL2 (25,48). Therefore, discrepancies between our work and that of Cloutier et al. (4) might reflect the dynamics of the R2TP chaperone system and be due in part to the use of preassembled RUVBL1–RUVBL2 complexes and no added nucleotides in our case.
In addition to its role as regulator of RUVBL1–RUVBL2 conformation, we find that ZNHIT2 forms part of a complex network of interactions with PRPF8 and with ECD and AAR2, factors involved in the biogenesis of PRPF8, and that in cells, the RUVBL1–RUVBL2–ZNHIT2 complex assembles with R2TP and HSP90. It is believed that R2TP and the RUVBL1–RUVBL2 complex assist the assembly of several unrelated complexes thanks to the help of other factors specific for each client. Some of these factors could function as adaptor proteins or scaffolds connecting specific clients to the chaperone, while others could perform other, yet unknown, regulatory functions specific to each client. For example, the TTT complex acts as a scaffold between R2TP and clients during the biogenesis of the kinases of the PIKK family, but it also regulates the ATP hydrolysis and some of the interactions taking place within the R2TP complex (11). Our results suggest that ZNHIT2 could be part of a larger complex containing other regulatory subunits together with PRPF8 and R2TP. ZNHIT2 and ECD could work as part of the same complex as they interact together and they can coexist when in complex with RUVBL1–RUVBL2. PRPF8 can interact with R2TP without the help of any additional factor, at least in vitro. Hence, ZNHIT2 and possibly ECD might be part of a list of factors that could be required to regulate the activity and conformational state of the R2TP-HSP90 chaperone during the several steps needed for the assembly of a matured PRPF8. ZNHIT2 for instance could modulate R2TP activity while PRPF8 and AAR2 are being brought together and remodeled.
Some evidence suggested that RPAP3 and ZNHIT2 could compete for the binding to RUVBL1–RUVBL2 and therefore that ZNHIT2 could not be part of R2TP (4,22). Here, we demonstrate that ZNHIT2 does assemble a complex with R2TP, using purified proteins and a double affinity purification in vitro, and analyzing the RPAP3 interactome in cells, and that the interaction between ZNHIT2 and RUVBL1–RUVBL2 can possibly coexist with the binding of PRPF8 to R2TP and HSP90. PRPF8, ZNHIT2 and RUVBL1–RUVBL2 interact with RPAP3 in cells, and also with HSP90, which binds the TPR domains in RPAP3. But when the binding of RPAP3 to RUVBL1–RUVBL2 is compromised using an RPAP3 mutant, the association of ZNHIT2 with R2TP is lost, indicating that ZNHIT2 binds to R2TP by RUVBL1–RUVBL2. PRPF8 also associates with R2TP mostly by the direct interaction with the RUVBL1–RUVBL2 complex, but the in vitro pull-down experiments and the mass spectrometry in cells suggest that RPAP3-PIH1D1 also contributes to bring PRPF8 to the R2TP complex.
Taken together, here we have characterized the interaction of ZNHIT2 with the R2TP chaperone system involved in the biogenesis of PRPF8. ZNHIT2 is part of a complex network of interactions between factors that participate in PRPF8 biogenesis while concurrently regulating the conformation of the RUVBL1–RUVBL2 ATPases, their nucleotide state, and their capacity to hydrolyze ATP. The biogenesis of PRPF8 is a complex pathway, and further work will be required to elucidate the mechanistic details of how R2TP-HSP90 contributes to U5 snRNP assembly.
DATA AVAILABILITY
Supplementary Data are available at NAR online. The cryo-EM maps for the RUVBL1–RUVBL2–ZNHIT2ΔC complex and the RUVBL1–RUVBL2 AAA+ ring subtracted from the RUVBL1–RUVBL2-ZNHIT2ΔC complex have been deposited in the EMDB with accession code EMD-13233 (access to the Electron Microscopy Data Bank: https://www.ebi.ac.uk/emdb/). The atomic model for the RUVBL1–RUVBL2 AAA+ ring subtracted from the RUVBL1–RUVBL2-ZNHIT2ΔC complex has been deposited in the Protein Data Base (PDB) with accession code 7P6X (https://doi.org/10.2210/pdb7P6X/pdb).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the EM Unit of CNIO (Jasminka Boskovic and Carmen García-Martín) for support in preparing and screening the grids, and Rafael Fernandez-Leiro at CNIO for constant help and inspiration during image processing of the cryoEM data. We also thank Carlos F. Rodriguez in Llorca′s group for his help at the initial stages of this project. We acknowledge the Diamond Light Source cryo-EM facility at the UK’s national Electron Bio-imaging Center (eBIC) under BAG proposal EM20135, for data used in this work. We also acknowledge Christos Savva from the Leicester Institute of Structural and Chemical Biology (UK) and Rebecca Thompson from the University of Leeds, for access and support during data collection. We are grateful to Vimla Band (University of Nebraska Medical Center, UNMC), Laurence Pearl (University of Sussex), Hector Peinado (CNIO) for kindly providing the ECD cDNA, the CK2 construct and HEK293T cells respectively. We acknowledge the support of the National Institute of Health Carlos III to CNIO.
Contributor Information
Marina Serna, Spanish National Cancer Research Centre (CNIO), Melchor Fernández Almagro 3, 28029 Madrid, Spain.
Ana González-Corpas, Spanish National Cancer Research Centre (CNIO), Melchor Fernández Almagro 3, 28029 Madrid, Spain.
Sofía Cabezudo, Spanish National Cancer Research Centre (CNIO), Melchor Fernández Almagro 3, 28029 Madrid, Spain.
Andrés López-Perrote, Spanish National Cancer Research Centre (CNIO), Melchor Fernández Almagro 3, 28029 Madrid, Spain.
Gianluca Degliesposti, MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge Biomedical Campus, Cambridge CB2 0QH, UK.
Eduardo Zarzuela, Spanish National Cancer Research Centre (CNIO), Melchor Fernández Almagro 3, 28029 Madrid, Spain.
J Mark Skehel, MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge Biomedical Campus, Cambridge CB2 0QH, UK.
Javier Muñoz, Spanish National Cancer Research Centre (CNIO), Melchor Fernández Almagro 3, 28029 Madrid, Spain.
Oscar Llorca, Spanish National Cancer Research Centre (CNIO), Melchor Fernández Almagro 3, 28029 Madrid, Spain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agencia Estatal de Investigación (AEI/10.13039/501100011033), Ministerio de Ciencia e Innovación and co-funded by the European Regional Development Fund (ERDF-UE) [SAF2017-82632-P and PID2020-114429RB-I00 to O.L.]; Autonomous Region of Madrid and co-funded by the European Social Fund and the European Regional Development Fund [Y2018/BIO4747 and P2018/NMT4443 to O.L., and which support the contracts of S.C. and A.G-C.]; Funding for open access charge: Agencia Estatal de Investigación (AEI/10.13039/501100011033), Ministerio de Ciencia e Innovación, co-funded by the European Regional Development Fund (ERDF-UE) [SAF2017-82632-P to O.L.]; S.C. contract is funded by the CNIO Friends Program philanthropic initiative since June 2021.
Conflict of interest statement. None declared.
REFERENCES
- 1. Galej W.P., Oubridge C., Newman A.J., Nagai K.. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature. 2013; 493:638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nguyen T.H., Li J., Galej W.P., Oshikane H., Newman A.J., Nagai K.. Structural basis of Brr2-Prp8 interactions and implications for U5 snRNP biogenesis and the spliceosome active site. Structure. 2013; 21:910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Malinova A., Cvackova Z., Mateju D., Horejsi Z., Abeza C., Vandermoere F., Bertrand E., Stanek D., Verheggen C.. Assembly of the U5 snRNP component PRPF8 is controlled by the HSP90/R2TP chaperones. J. Cell Biol. 2017; 216:1579–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cloutier P., Poitras C., Durand M., Hekmat O., Fiola-Masson E., Bouchard A., Faubert D., Chabot B., Coulombe B.. R2TP/Prefoldin-like component RUVBL1/RUVBL2 directly interacts with ZNHIT2 to regulate assembly of U5 small nuclear ribonucleoprotein. Nat. Commun. 2017; 8:15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pearl L.H. Review: The HSP90 molecular chaperone-an enigmatic ATPase. Biopolymers. 2016; 105:594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bizarro J., Charron C., Boulon S., Westman B., Pradet-Balade B., Vandermoere F., Chagot M.E., Hallais M., Ahmad Y., Leonhardt H.et al.. Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control. J. Cell Biol. 2014; 207:463–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bizarro J., Dodre M., Huttin A., Charpentier B., Schlotter F., Branlant C., Verheggen C., Massenet S., Bertrand E.. NUFIP and the HSP90/R2TP chaperone bind the SMN complex and facilitate assembly of U4-specific proteins. Nucleic Acids Res. 2015; 43:8973–8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Munoz-Hernandez H., Pal M., Rodriguez C.F., Fernandez-Leiro R., Prodromou C., Pearl L.H., Llorca O.. Structural mechanism for regulation of the AAA-ATPases RUVBL1–RUVBL2 in the R2TP co-chaperone revealed by cryo-EM. Sci. Adv. 2019; 5:eaaw1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maurizy C., Quinternet M., Abel Y., Verheggen C., Santo P.E., Bourguet M., Paiva A.C.F., Bragantini B., Chagot M.E., Robert M.C.et al.. The RPAP3-Cterminal domain identifies R2TP-like quaternary chaperones. Nat. Commun. 2018; 9:2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martino F., Pal M., Munoz-Hernandez H., Rodriguez C.F., Nunez-Ramirez R., Gil-Carton D., Degliesposti G., Skehel J.M., Roe S.M., Prodromou C.et al.. RPAP3 provides a flexible scaffold for coupling HSP90 to the human R2TP co-chaperone complex. Nat. Commun. 2018; 9:1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pal M., Munoz-Hernandez H., Bjorklund D., Zhou L., Degliesposti G., Skehel J.M., Hesketh E.L., Thompson R.F., Pearl L.H., Llorca O.et al.. Structure of the TELO2-TTI1-TTI2 complex and its function in TOR recruitment to the R2TP chaperone. Cell Rep. 2021; 36:109317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lopez-Perrote A., Hug N., Gonzalez-Corpas A., Rodriguez C.F., Serna M., Garcia-Martin C., Boskovic J., Fernandez-Leiro R., Caceres J.F., Llorca O.. Regulation of RUVBL1–RUVBL2 AAA-ATPases by the nonsense-mediated mRNA decay factor DHX34, as evidenced by Cryo-EM. eLife. 2020; 9:e63042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yenerall P., Das A.K., Wang S., Kollipara R.K., Li L.S., Villalobos P., Flaming J., Lin Y.F., Huffman K., Timmons B.C.et al.. RUVBL1/RUVBL2 ATPase activity drives PAQosome maturation, DNA replication and radioresistance in lung cancer. Cell chemical biology. 2020; 27:105–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dauden M.I., Lopez-Perrote A., Llorca O.. RUVBL1–RUVBL2 AAA-ATPase: a versatile scaffold for multiple complexes and functions. Curr. Opin. Struct. Biol. 2021; 67:78–85. [DOI] [PubMed] [Google Scholar]
- 15. Henri J., Chagot M.E., Bourguet M., Abel Y., Terral G., Maurizy C., Aigueperse C., Georgescauld F., Vandermoere F., Saint-Fort R.et al.. Deep structural analysis of RPAP3 and PIH1D1, two components of the HSP90 co-chaperone R2TP complex. Structure. 2018; 26:1196–1209. [DOI] [PubMed] [Google Scholar]
- 16. Pal M., Morgan M., Phelps S.E., Roe S.M., Parry-Morris S., Downs J.A., Polier S., Pearl L.H., Prodromou C.. Structural basis for phosphorylation-dependent recruitment of Tel2 to Hsp90 by Pih1. Structure. 2014; 22:805–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gorynia S., Bandeiras T.M., Pinho F.G., McVey C.E., Vonrhein C., Round A., Svergun D.I., Donner P., Matias P.M., Carrondo M.A.. Structural and functional insights into a dodecameric molecular machine - the RuvBL1/RuvBL2 complex. J. Struct. Biol. 2011; 176:279–291. [DOI] [PubMed] [Google Scholar]
- 18. Matias P.M., Gorynia S., Donner P., Carrondo M.A.. Crystal structure of the human AAA+ protein RuvBL1. J. Biol. Chem. 2006; 281:38918–38929. [DOI] [PubMed] [Google Scholar]
- 19. Silva S.T.N., Brito J.A., Arranz R., Sorzano C.O.S., Ebel C., Doutch J., Tully M.D., Carazo J.M., Carrascosa J.L., Matias P.M.et al.. X-ray structure of full-length human RuvB-Like 2 - mechanistic insights into coupling between ATP binding and mechanical action. Sci. Rep. 2018; 8:13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Erkelenz S., Stankovic D., Mundorf J., Bresser T., Claudius A.K., Boehm V., Gehring N.H., Uhlirova M.. Ecd promotes U5 snRNP maturation and Prp8 stability. Nucleic Acids Res. 2021; 49:1688–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Claudius A.K., Romani P., Lamkemeyer T., Jindra M., Uhlirova M.. Unexpected role of the steroid-deficiency protein ecdysoneless in pre-mRNA splicing. PLos Genet. 2014; 10:e1004287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Verheggen C., Pradet-Balade B., Bertrand E.. SnoRNPs, ZNHIT proteins and the R2TP pathway. Oncotarget. 2015; 6:41399–41400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. He F., Umehara T., Tsuda K., Inoue M., Kigawa T., Matsuda T., Yabuki T., Aoki M., Seki E., Terada T.et al.. Solution structure of the zinc finger HIT domain in protein FON. Protein Sci. 2007; 16:1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McKeegan K.S., Debieux C.M., Watkins N.J.. Evidence that the AAA+ proteins TIP48 and TIP49 bridge interactions between 15.5K and the related NOP56 and NOP58 proteins during box C/D snoRNP biogenesis. Mol. Cell. Biol. 2009; 29:4971–4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eustermann S., Schall K., Kostrewa D., Lakomek K., Strauss M., Moldt M., Hopfner K.P.. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature. 2018; 556:386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mir R.A., Bele A., Mirza S., Srivastava S., Olou A.A., Ammons S.A., Kim J.H., Gurumurthy C.B., Qiu F., Band H.et al.. A novel interaction of ecdysoneless (ECD) protein with R2TP complex component RUVBL1 is required for the functional role of ECD in cell cycle progression. Mol. Cell. Biol. 2015; 36:886–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garcia-Nafria J., Watson J.F., Greger I.H.. IVA cloning: a single-tube universal cloning system exploiting bacterial in vivo assembly. Sci. Rep. 2016; 6:27459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lopez-Perrote A., Munoz-Hernandez H., Gil D., Llorca O.. Conformational transitions regulate the exposure of a DNA-binding domain in the RuvBL1-RuvBL2 complex. Nucleic Acids Res. 2012; 40:11086–11099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lopez-Perrote A., Alatwi H.E., Torreira E., Ismail A., Ayora S., Downs J.A., Llorca O.. Structure of Yin Yang 1 oligomers that cooperate with RuvBL1-RuvBL2 ATPases. J. Biol. Chem. 2014; 289:22614–22629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Drozdetskiy A., Cole C., Procter J., Barton G.J.. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015; 43:W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hill C.H., Boreikaite V., Kumar A., Casanal A., Kubik P., Degliesposti G., Maslen S., Mariani A., von Loeffelholz O., Girbig M.et al.. Activation of the endonuclease that defines mRNA 3′ ends requires incorporation into an 8-subunit core cleavage and polyadenylation factor complex. Mol. Cell. 2019; 73:1217–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kessner D., Chambers M., Burke R., Agus D., Mallick P.. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008; 24:2534–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gotze M., Pettelkau J., Fritzsche R., Ihling C.H., Schafer M., Sinz A.. Automated assignment of MS/MS cleavable cross-links in protein 3D-structure analysis. J. Am. Soc. Mass Spectrom. 2015; 26:83–97. [DOI] [PubMed] [Google Scholar]
- 35. Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A.. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017; 14:331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016; 193:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H.. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018; 7:e42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A.. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017; 14:290–296. [DOI] [PubMed] [Google Scholar]
- 39. Zhong E.D., Bepler T., Berger B., Davis J.H.. CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Nat. Methods. 2021; 18:176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A.et al.. Highly accurate protein structure prediction with AlphaFold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 42. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A.. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Norby J.G. Coupled assay of Na+,K+-ATPase activity. Methods Enzymol. 1988; 156:116–119. [DOI] [PubMed] [Google Scholar]
- 45. Kim S.G., Hoffman G.R., Poulogiannis G., Buel G.R., Jang Y.J., Lee K.W., Kim B.Y., Erikson R.L., Cantley L.C., Choo A.Y.et al.. Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol. Cell. 2013; 49:172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Santos K., Preussner M., Heroven A.C., Weber G.. Crystallization and biochemical characterization of the human spliceosomal Aar2-Prp8(RNaseH) complex. Acta Crystallogr. F Struct. Biol. Commun. 2015; 71:1421–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weber G., Cristao V.F., Santos K.F., Jovin S.M., Heroven A.C., Holton N., Luhrmann R., Beggs J.D., Wahl M.C.. Structural basis for dual roles of Aar2p in U5 snRNP assembly. Genes Dev. 2013; 27:525–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aramayo R.J., Willhoft O., Ayala R., Bythell-Douglas R., Wigley D.B., Zhang X.. Cryo-EM structures of the human INO80 chromatin-remodeling complex. Nat. Struct. Mol. Biol. 2018; 25:37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary Data are available at NAR online. The cryo-EM maps for the RUVBL1–RUVBL2–ZNHIT2ΔC complex and the RUVBL1–RUVBL2 AAA+ ring subtracted from the RUVBL1–RUVBL2-ZNHIT2ΔC complex have been deposited in the EMDB with accession code EMD-13233 (access to the Electron Microscopy Data Bank: https://www.ebi.ac.uk/emdb/). The atomic model for the RUVBL1–RUVBL2 AAA+ ring subtracted from the RUVBL1–RUVBL2-ZNHIT2ΔC complex has been deposited in the Protein Data Base (PDB) with accession code 7P6X (https://doi.org/10.2210/pdb7P6X/pdb).