Abstract
MicroRNAs (miRNAs) are short endogenously expressed RNAs that have the potential to regulate the expression of any RNA. This potential has led to the publication of several thousand papers each year connecting miRNAs to many different genes and human diseases. By contrast, relatively few papers appear that investigate the molecular mechanism used by miRNAs. There is a disconnect between rigorous understanding of mechanism and the extraordinary diversity of reported roles for miRNAs. Consequences of this disconnect include confusion about the assumptions underlying the basic science of human miRNAs and slow development of therapeutics that target miRNAs. Here, we present an overview of investigations into miRNAs and their impact on gene expression. Progress in our understanding of miRNAs would be aided by a greater focus on the mechanism of miRNAs and a higher burden of evidence on researchers who seek to link expression of a particular miRNA to a biological phenotype.
INTRODUCTION
microRNAs (miRNAs) are endogenously expressed short nucleic acids that function through RNA interference (RNAi) (1,2). miRNAs have the potential to bind to many different sequences within RNA, endowing them with the potential to be sequence-specific regulators of any gene.
Many thousands of publications appear each year describing the function of miRNAs in normal physiology and disease. These studies have been encouraged by: (i) the conceptual ease of imagining complementary recognition between a miRNA and an interesting RNA target; (ii) the potential of miRNAs to bind to almost any gene; (iii) conservation of miRNAs across species (3,4); (iv) examples of miRNA function in model organisms (5–9) and (v) the routine success of fully complementary duplex RNAs as experimental tools for regulating gene expression in human cells (10).
While the case for the potential impact of miRNA-mediated gene regulation in human cells is compelling, after decades of research there has been little success developing approved drugs that take advantage of miRNAs as targets (11,12) (Table 1). This lack of success is especially striking when compared to the recent clinical progress of duplex RNA and antisense oligonucleotide therapeutics (13–15). For investigators examining prior literature and attempting to form new hypotheses, it can be difficult to discern underlying lessons, make accurate predictions, or draw general conclusions about the scope of recognition by miRNAs.
Table 1.
The clinical trials of anti-miR miRNA inhibitors
| Clinical trial | |||||
|---|---|---|---|---|---|
| Drug name | miRNA | Diseases/disorders | Number | Phase | Status (Year) |
| Miravirsen (SPC3649) | miR-122 | Hepatitis C | NCT00688012 | I | Completed (2009) |
| NCT00979927 | I | Completed (2011) | |||
| NCT01646489 | I | Completed (2012) | |||
| NCT01200420 | II | Completed (2012) | |||
| NCT01727934 | II | Unknown (2014) | |||
| NCT01872936 | II | Unknown (2014) | |||
| pSil-miR200c/PMIS miR200a | miR-200a/c | Tooth Extraction Status Nos | NCT02579187 | I | Withdrawn (2019) |
| RG-125 (AZD4076) | miR-103/107 | Type 2 diabetes mellitus with non-alcoholic fatty liver disease | NCT02826525 | I | Completed (2019) |
| Non-alcoholic Steatohepatitis | NCT02612662 | I | Active | ||
| MRG-110 (S95010) | miR-92 | Wound healing | NCT03603431 | I | Completed (2019) |
| Cardiovascular Diseases | NCT03494712 | I | Completed (2020) | ||
| CDR132L | miR-132 | Heart Failure | NCT04045405 | I | Completed (2020) |
| Cobomarsen (MRG-106) | miR-155 | Lymphoma; Mycosis Fungoides; Leukemia | NCT02580552 | I | Completed (2020) |
| Cutaneous T-Cell Lymphoma/Mycosis Fungoides | NCT03837457 | II | Terminated (2020) | ||
| NCT03713320 | II | Terminated (2020) | |||
| Lademirsen (RG-012) | miR-21 | Alport Syndrome | NCT03373786 | I | Completed (2019) |
| NCT02855268 | II | Recruiting | |||
| RGLS4326 | miR-17 | Polycystic Kidney Disease, Autosomal Dominant | NCT04536688 | I | Completed (2021) |
| LNA-i-miR-221 | miR-221 | Multiple Myeloma, Refractory; Hepatocarcinoma; Advanced Solid Tumor | NCT04811898 | I | Recruiting |
Notes. Information taken from https://clinicaltrials.gov
Our purpose here is to describe the foundation of knowledge about miRNAs, how that foundation has been used to gain insights into mammalian physiology, and why many studies fall short of providing conclusive insights. We end by advising how researchers interested in this field should prioritize their evaluation of the thousands of papers that describe miRNAs.
BIOGENESIS OF miRNAs
miRNAs are chromosomally-encoded short ∼22 nucleotide (nt) duplex RNAs (1,5–8). The first step in the biogenesis of miRNAs is the synthesis of a relatively long structured primary transcript (pri-miRNA) (Figure 1A) (16,17). This transcript is processed to an intermediate length hairpin precursor miRNA (pre-miRNA) by the nuclear microprocessor complex consisting of the proteins Drosha (18) and DGCR8 (19–21) and exported into cytoplasm (Figure 1B). In the cytoplasm, Dicer (22–24) cleaves the precursor to produce the mature miRNA (Figure 1C). miRNA biogenesis has been the subject of several recent primary reports and reviews (2,25–28).
Figure 1.
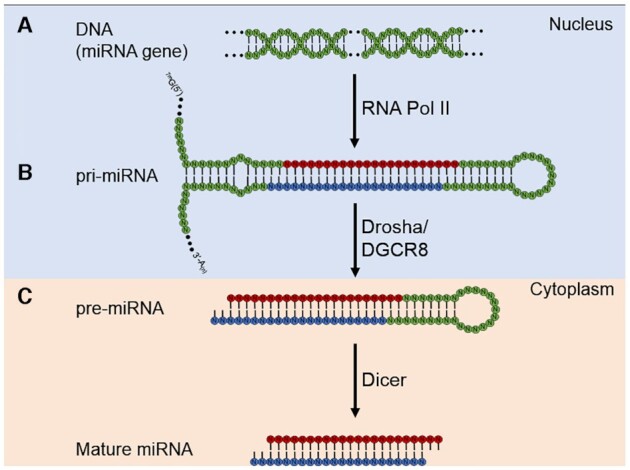
Biogenesis of miRNAs. (A) A miRNA is transcribed into pri-miRNA by Pol II. (B) Drosha/DCGR8 microprocessor complex cleaves the pri-miRNA to pre-miRNA, which enters the cytoplasm. (C) The mature duplex miRNA is generated by Dicer.
SEQUENCE SPECIFIC RECOGNITION
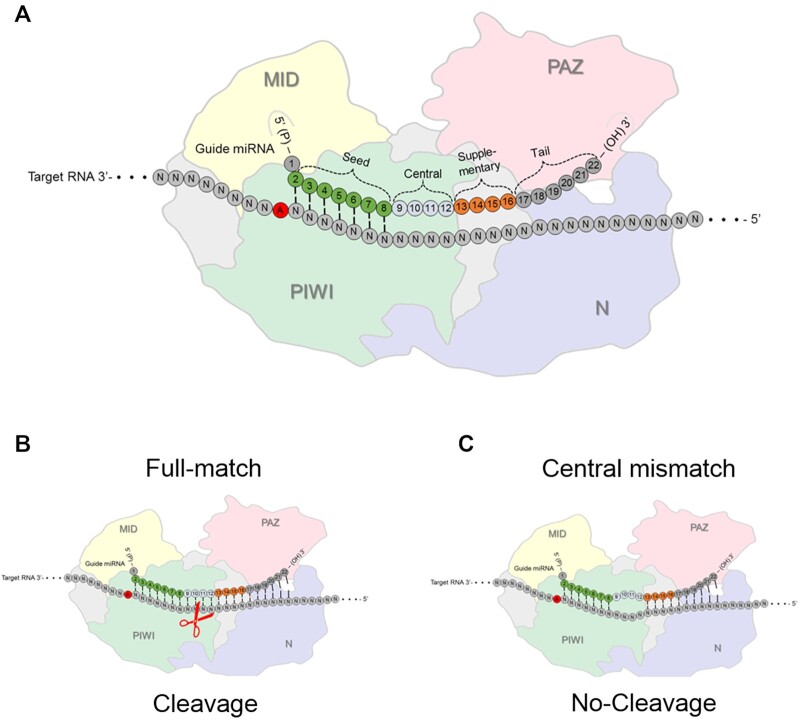
Mature miRNAs are loaded into cytoplasmic argonaute (AGO) protein to form a programmable ribonucleoprotein complexes (29–31) (Figure 2A). The miRNA ‘programs’ the complex with the complementary sequence information necessary to recognize RNA target sequences within cells. The role of AGO protein is to protect RNA from being degraded, prioritize one strand of an miRNA over the other, spatially organize the miRNA for binding complementary sequences, and facilitate the search for and recognition of RNA targets inside cells (32–34).
Figure 2.
Interactions between AGO protein, an miRNA guide strand, and an RNA target. AGO proteins are composed of four domains: The N-terminal domain supports miRNA loading, the PAZ domain anchors the 3′ end while the MID domain binds the 5′ end of the miRNA. (A) Argonaute loaded miRNA seed region and other regions. (B) PIWI is the protein domain responsible for cleaving RNA substrates by AGO2 when there is full complementarity. (C) Centrally mismatched bases will block cleavage while permitting binding of a miRNA to a target RNA. PAZ – PIWI-Argonaute-Zwille, N – N-terminal (amino-terminal), PIWI – P-element-induced whimpy testes and MID – middle domains.
There are four AGO variants in mammalian cells, AGO1-AGO4 (30,31). In cell lines where quantities of the AGO variants have been measured, AGO1, AGO2, and AGO3 are the prevalent variants with AGO4 being much less detectable (29,35–37). AGO2 (29,30), and to a lesser extent AGO3 (38), have the ability to promote cleavage of target RNA when the match between short RNA is fully complementary.
Efficient cleavage of fully complementary target sequences by RNA:AGO2 complexes facilitated the widespread application of duplex RNAs as laboratory tools, either as expressed hairpin RNAs (shRNAs) (39) or synthetic RNA duplexes (10). Potent control of gene expression had led to the development of synthetic duplex RNAs as successful drugs (14) (Figure 2B). The introduction of base mismatches into the central region of the small RNA relative to its RNA target does not block sequence-specific binding but does prevent cleavage by AGO2 (Figure 2C).
Most miRNAs are only partially complementary to their target sequences – full or even majority complementarity is not necessary (40,41). It is generally assumed that ‘seed’ base-pairing at position 2–7 or 8 of the miRNA is the primary factor governing recognition (Figure 2A). It is also often assumed that binding occurs within the 3′-untranslated region (3′-UTR) to inhibit gene translation in the cytoplasm through RNA interference (RNAi) (1). Other reports describe non-canonical binding beyond 3′-UTR and seed regions, making the rules governing recognition even more complex (42–46). Repression can occur through destabilization of target transcripts or inhibition of gene translation (47–49).
Binding by a miRNA to cellular RNA can be ‘on-target’ or ‘off-target’ (50,51). On-target binding produces a beneficial effect on gene expression through recognition of a predictable complementary target site. Off-target interactions do not produce a beneficial effect on cell fitness. They can have two detrimental consequences by (i) reducing the amount of the miRNA available for on-target action and (ii) perturbing the expression of genes in ways that reduce fitness.
A given six or seven base sequence will occur many times within the transcriptome. Therefore, the seed sequences of miRNAs have the potential to bind to the 3′-UTRs of many different genes (3,40,52,53) as well as to sequences within coding regions and noncoding RNAs (43,46,54,55). Because of this inherent potential for association with many targets, the binding of miRNAs within the transcriptome will always lead to a mixture of on- and off-target effects. The mechanisms for balancing these effects should be understood.
It is important to acknowledge that the concept of ‘on-target’ and ‘off-target interactions may be an oversimplification when it is applied to physiologic regulation. Physiologic control may be the additive outcome of many interactions between miRNAs and complementary miRNAs, including some that may appear to bind with low affinity. Perhaps a better strategy is to consider ‘primary’ target sites where high affinity interactions take place and ‘secondary’ lower affinity sites, all functioning together to regulate gene expression. This complexity adds to the difficult of understanding the action of miRNAs.
The potential for an individual miRNA to recognize many different sequences raises important questions: How does a miRNA evolve selectivity for the control of a physiologically advantageous subset of potential target genes? What distinguishes the magnitude of effects miRNAs have on a given gene? How is the finite pool of a particular miRNA within a cell partitioned between the relatively large number of binding sites that are not points for physiological regulation and the likely smaller number of binding sites that are candidates for regulation? Are biological impacts due to regulation of just one or perhaps a handful of genes, or do outcomes arise from modest changes to many genes?
DIVERSITY OF miRNAs AND miRNA FAMILIES
RNA sequencing (RNAseq) has identified thousands of potential miRNAs (56–58). RNAseq is a sensitive technique and not every miRNA detected will be expressed at levels that permit biologically relevant gene regulation. It is important, therefore, to understand the level of expression of individual miRNAs and miRNA ‘families’.
Many miRNAs have the same or similar seed sequences, allowing them to be classified into families (59), with the impact on gene expression being the sum of the individual effects of each family member. Some families are expressed at higher levels than others (57). For example, in HCT116 cells, just six families account for ∼50% of all miRNAs detected after immunoprecipitation with an anti-AGO2 antibody (60) (Figure 3A). The Let-7-5p/98-5p family in HCT116 cells has eight different members that contribute to overall expression, each expressed at a different locus. By contrast, the miR-96-5p/1271-5p family has only one member (Figure 3B). While seed sequences among family members are mostly identical, there can be significant variation outside the seed sequence (Figure 3C).
Figure 3.
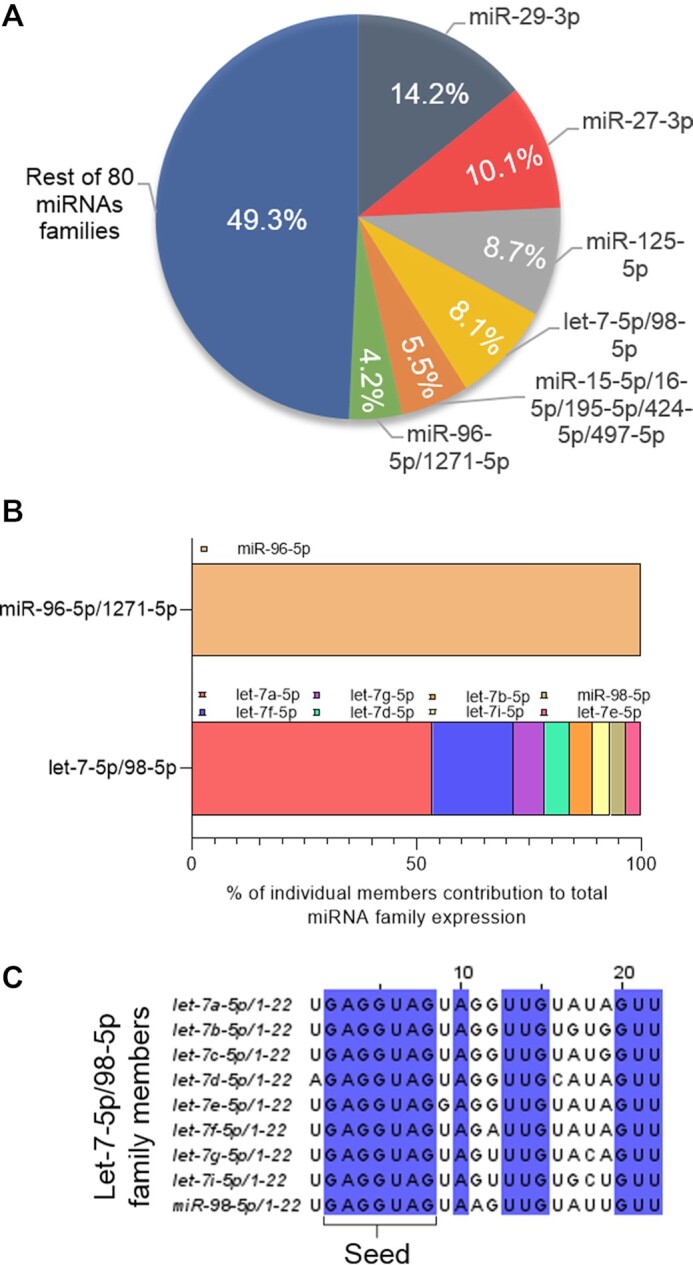
miRNA families. (A) Six miRNA families account for ∼50% of top 100 miRNAs loaded on AGO2 in HCT116 cells (60). (B) miR-96 is the only member of its family while Let-7 has several family members expressed. (C) Sequence variation among Let-7–5p/98–5p family members.
NOT ALL ‘miRNAs’ ARE EQUAL
Thousands of genes are annotated as miRNAs in miRBase (61). Too often, these annotations are not treated with adequate critical evaluation. Recent studies have shown that only a fraction of annotated miRNAs are Drosha substrates in vitro (28,58,62). It is possible that many studies investigate ‘miRNAs’ that are more likely to be degradation products and thus biological noise. Simply assuming that a miRNA is biologically relevant because it appears on a published list is not an adequate foundation for research.
The observations that many annotated ‘miRNAs’ might not be relevant to biological function emphasizes the need to carefully consider the biochemical basis for choosing a miRNA. There should be justification for the belief that the RNAs in question are processed by the proteins responsible for production of miRNAs and are loaded to AGO proteins. As will be discussed at more length below, the quantity of a candidate miRNA per cell and the stoichiometry of miRNA relative to target RNA should be considered. All papers should make a clear, transparent case that there is a plausible physical basis for believing that a miRNA might have a biological function.
COOPERATIVE BINDING
As noted above, the primary determinant of recognition for miRNAs, the seed sequence, is just six or seven bases long. These short sequences have the potential to bind to many places within the transcriptome—approximately once per 4100 bases for six base recognition and once per 16 000 bases for seven base complementarity. If binding were distributed over all potential binding sites two complications become apparent: (i) significant gene repression due to off-target interactions and (ii) less miRNA available to bind to target RNA sequences and produce a biologically significant reduction in gene expression. Mechanisms that prioritize ‘on-target’ recognition over ‘off target’ recognition would enhance the likelihood of achieving gene repression at levels adequate to have a real effect on cell biology.
One solution to the problem of achieving selective gene regulation by miRNAs is cooperative binding between two or more AGO:miRNA complexes to nearby target sequences. The potential for cooperativity was first suggested by the observation that regulation of a luciferase reporter system became more efficient as the number of miRNA binding sites was increased within the reporter gene's 3′-UTR (63). Subsequent studies supported the conclusion that adjacent binding sites and allow cooperative interactions were important for the action of miRNAs (42,64–66).
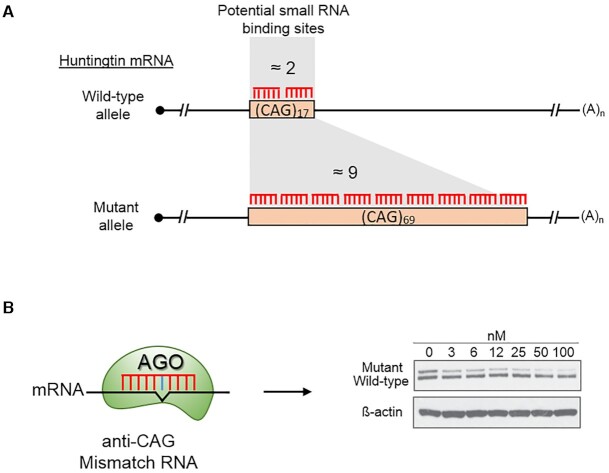
One example of the power of adjacent target sites to control gene expression is provided by the allele selective inhibition of mutant huntingtin protein (67). Mutant huntingtin is the cause of Huntington's disease and contains an expanded CAG repeat sequence within its coding region. The wild-type gene has less than twenty CAG repeats, whereas the mutant gene usually contains >40 repeats (Figure 4A).
Figure 4.
An example of the impact of cooperative binding on repression of gene expression by duplex RNAs. In this example an anti-CAG repeat duplex RNA is introduced into patient-derived Huntington's disease cells. (A) The wild-type huntingtin (HTT) allele has 17 repeats, while the mutant allele has 69 repeats. 1–2 anti-CAG small RNAs can bind to the wild-type allele while as many as nine anti-CAG RNAs can bind to the mutant allele. (B) Allele selective inhibition by a small RNA with a central mismatch relative to the RNA target.
As noted above (Figure 2C), the introduction of one or more mismatched bases relative to the mRNA target sequences eliminates the potential for cleaving the target while retaining the ability to bind target RNA (68). When duplex anti-CAG RNAs that contain central mismatched bases (Figure 4B) are introduced into mutant cells, they yield robust inhibition of mutant (greater than six potential binding sites) gene expression while leaving wild-type (one or two potential binding sites for the anti-CAG RNA) expression largely unaffected.
Cooperative binding provides a foundation for understanding how miRNAs might discriminate between binding to a site that has a single seed sequence match (Figure 5A) and a site with multiple matches (Figure 5BC). The simplest ‘multiple match’ scenario would be multiple seed matches for particular miRNA or members of the same family of miRNAs (Figure 5B). It is also possible to have a more complex scenario when members of different miRNA families have the potential to bind near to one another (Figure 5C). This more complex scenario has obvious implications for the challenges face by investigators seeking to define the action of a particular gene by an individual miRNA.
Figure 5.
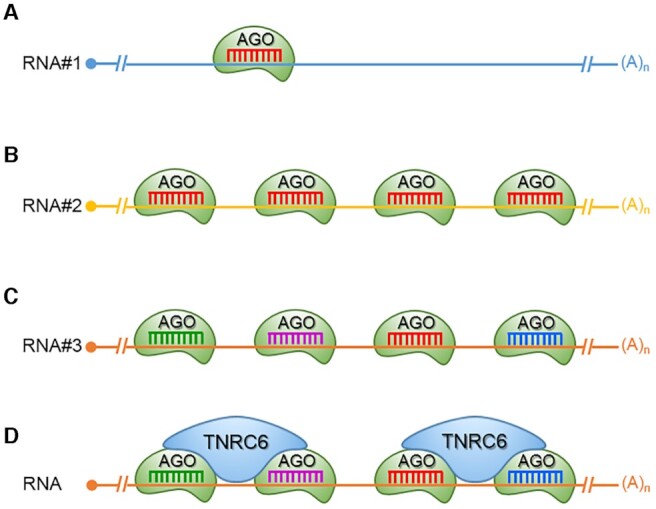
Single versus multiple binding of miRNAs and cooperativity. (A) Binding of a single miRNA to a target, no potential for cooperativity. Multiple miRNAs of the (B) same or (C) different miRNA families. (D) Bridging of AGO:miRNA complexes by TNRC6 scaffolding protein provides a structural basis for cooperativity.
STRUCTURAL BASIS OF COOPERATIVITY
What is the structural basis for cooperativity? All AGO variants bind trinucleotide repeat binding containing six protein A (TNRC6A), also known as human GW182, and its two paralogs, TNRC6B and TNRC6C (69–71). The TNRC6 paralogs are multidomain proteins that act as protein scaffolds. Part of the scaffolding function involves the recruitment of proteins that can facilitate the repression of translation.
The TNRC6 paralogs also possess an N-terminal AGO-binding domain containing Glycine/Tryptophan (GW)-repeats that has the capacity to recognize up to three AGO proteins simultaneously (72,73). A TNRC6 protein can bind up to three AGO proteins while each AGO protein can bind one TNRC6 protein (71). The capacity of TNRC6 proteins to bridge more than one AGO protein gives it the capacity to facilitate cooperative binding by miRNAs that recognize adjacent sequences (Figure 5D). The consequence of the AGO:TNRC6 partnership is that a single miRNA may bind relatively weakly, but two or more miRNAs binding to adjacent sites can form multi-valent interactions bridged by TNRC6. Briskin et al. (73) have recently confirmed that TNRC6 increases affinity of adjacent bound miRNAs, independently of type of miRNA and loaded AGO protein. This was achieved by slowing dissociation of the miRNA-AGO complex from the target.
PRIORITIZING CANDIDATE miRNAs FOR STUDY
Thousands of miRNAs have been annotated, each miRNA has seed sequence complementarity to many different genes, and the potential for gene regulation by a particular miRNA will likely differ depending on cell type or environmental conditions. As a result, connecting a miRNA to a known set of genes to affect a demonstrated cellular function can be like finding a needle in a haystack. The first question for a productive investigation into miRNA function is to identify the most promising candidate miRNAs for in depth investigation.
One obvious starting point is to quantify the expression of miRNAs relative to one another. As noted above, miRNAs can form both ‘on-target’ and ‘off-target’ interactions. The ‘off-target’ interactions may have no biologically significant impact on cell physiology, but when a miRNA is bound to the ‘off-target’ site it is not available to contribute to the regulation at biologically relevant control points. A miRNA with a relatively high concentration within a cell will be more likely to have a biological impact than miRNAs that are present at lower concentrations (32,74–76).
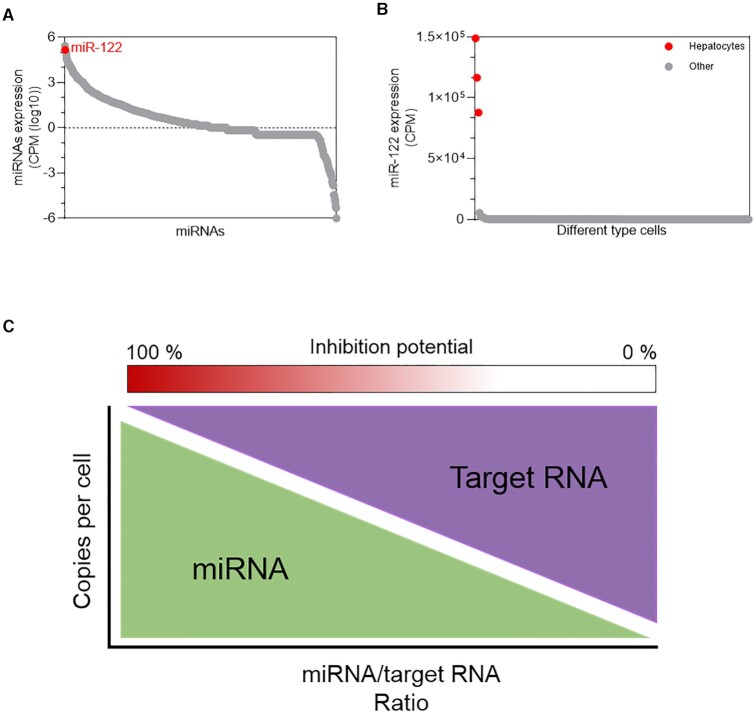
A second factor is whether expression of a miRNA is increased in a cell type or tissue relative to other tissues (77). A large increase in expression of a miRNA might signal that the miRNA is assuming a regulatory role unique to that cell type or tissue. Similarly, miRNA expression might increase when environmental conditions change, suggesting a potential role in responding to that change. miR-122, which will be discussed below, has stood out as a candidate for robust studies of miRNA action because it is highly expressed relative to most other miRNAs in hepatocytes and its expression in hepatocytes is much higher than in other cell types (Figure 6A, B).
Figure 6.
Interplay of miRNA and target concentration. miR-122 was prioritized for study because of (A) high expression in hepatocyte cells relative to most other miRNAs and (B) other cell types. (C) Model showing the correlation between miRNA concentration, target mRNA concentration, and potential for biologically significant inhibition of gene expression. CPM – counts per million. Data obtained from FANTOM5 database (77).
Relative concentrations are useful to initially rank candidate miRNAs but, at a molecular level, the absolute number of miRNAs per cell determines biological impact. The number of miRNA molecules per cell can be calculated by parallel comparisons with standards of known concentration. Quantitation in tissue confronts the difficulty separating and counting intact cells can be expressed as copy/number per ng of total RNA relative to reference samples. Quantification can extend to RNA targets. A highly expressed mRNA target will require a higher concentration of a miRNA to achieve a given outcome (78–80) (Figure 6C). The stoichiometry between an mRNA and a potential regulatory RNA is a critical consideration governing activity. This readily quantitated value should be reported and is essential for any evaluation of potential physiologic activity.
anti-miRs, TOOLS FOR BLOCKING miRNA ACTION
Synthetic antisense oligonucleotides (ASOs) that bind RNA sequences and control gene expression are reliable experimental tools and form a growing class of successful drugs (13,15,81). In the clinic, some ASOs induce RNase H-mediated cleavage of target genes and function through lowering the levels of disease-causing mRNA. Alternatively, ASOs can block splice sites to affect alternative splicing act through defined mechanisms that yield predictable outcomes.
ASOs that affect alternative splicing have been used to change the splicing of dystrophin to created drugs to treat muscular dystrophy (82,83). Spinraza, an ASO that affects splicing of the survival motor neuron 2 (SMN2) RNA has proven to be a remarkably effective treatment for spinal muscular atrophy (84). An outstanding question is whether the clinical success of ASOs when used for steric blocking of splice sites can be recreated in drug development programs that target miRNAs.
Studies have shown that ASOs can bind miRNAs to block their activities (85–87) (Figure 7). These compounds are known as antagomirs or anti-miRs. The combination of a well-known technology (ASOs) and an emerging biological target of obvious broad potential importance (miRNAs) led to creation of several companies aiming to target miRNAs with synthetic oligonucleotides, modulate gene expression, and treat disease (88).
Figure 7.
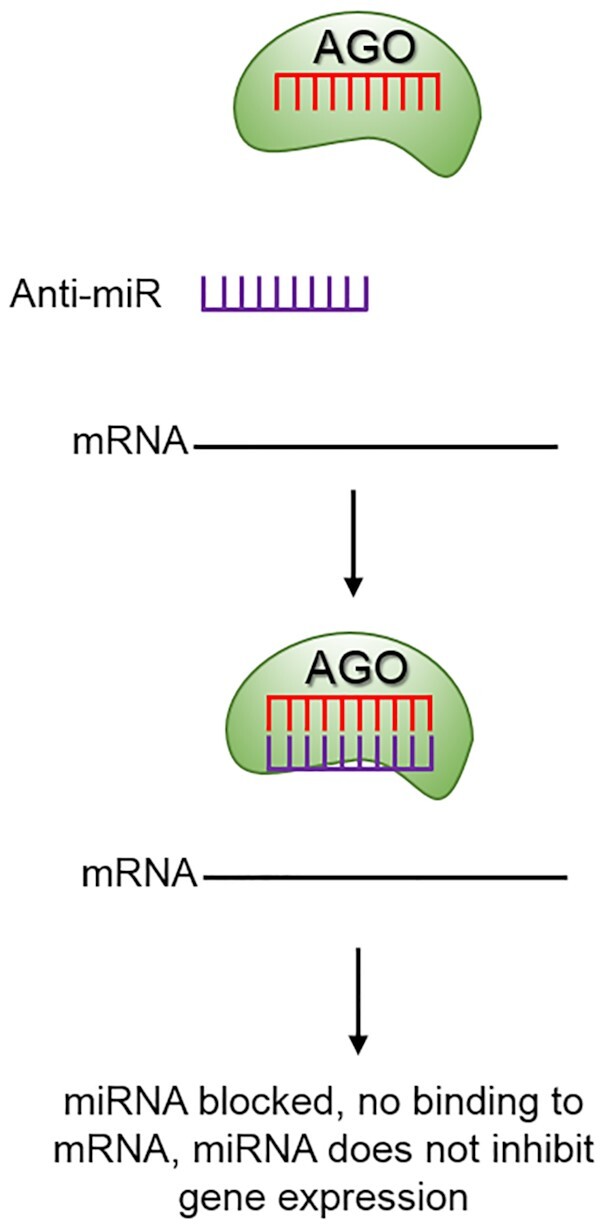
Anti-miRs, tools for investigating the function of miRNAs. When an anti-miR is present it has the potential to block the miRNA and prevent miRNA-mediated repression of the target RNA.
These companies benefited from two decades of practical experience developing ASOs that target mRNA. Previous efforts with ASOs provided a strong understanding of the chemistry needed to create oligonucleotide drugs and their pharmacological properties. This foundation, in combination with the simplicity of the steric blocking mechanism for interfering with miRNA action, created optimism that targeting miRNAs might become an important modality for therapeutic development. In spite of these advantages and substantial investment, the main companies in this area have either ceased operation, rebranded to pursue other modalities, or continue at a reduced level.
We note that the successful application of ASOs in the clinic required many years of research and over two decades of work in industry before there were substantial benefits to patients. It is not surprising, therefore, that the pace of miRNA drug development has been slow. One lesson from ASO development is that a steady focus on mechanism, robust basic science, and shrewd selection of development targets makes it more likely that programs will succeed. Rigor is the cornerstone of progress.
miR-122: CASE STUDIES FOR CLINICAL USE OF miRNA INHIBITORS
miR-122 is a case study for targeting miRNAs for therapeutic development (89). miRNA-122 first attracted attention because of its high expression levels in adult liver, where it is composes ∼70% of the total RNA pool (9) with an abundance of 50 000 (90) to 150 000 copies per cell. High expression levels made efficient target recognition more likely and miR-122 was a target for all initial studies using antagomirs (85,86). These studies revealed that blocking miR-122 with a synthetic oligonucleotide altered lipid metabolism and reduced cholesterol levels.
Several genes involved in cholesterol metabolism had potential target sites for miR-122. These genes were up-regulated by addition of anti-miR, as would be expected for the anti-miR acting by reversing miRNA-mediated inhibition of translation. Control oligonucleotides with altered sequences were not active. While these studies did not directly implicate miRNA binding at the target sites, they did show that miRNAs could orchestrate the control of multiple genes and produce a physiologically relevant outcome.
miRNA-122 also has a second important physiologic role. It was found that the Hepatitis C virus had incorporated recognition of miR-122 into its life cycle with an interaction between miR-122 and the 5′-UTR enhancing viral replication (91,92). While not a typical miRNA:mRNA target interaction, these studies are additional evidence that miRNAs can regulate gene expression through binding to an RNA target.
Two different companies, Santaris Pharma and Regulus Therapeutics initiated clinical trials using anti-miRs that target miR-122 for the treatment of HCV infection (Table 1). Neither advanced past Phase II clinical trials because adverse events outweighed the potential for clinical benefit. The failure of drug development of anti-miRs that target miRNAs that have well-demonstrated biological relevance highlights the challenges of targeting other miRNAs whose biological roles and disease relevance are less understood.
A CAUTION ABOUT THE USE OF ANTI-miRs
Anti-miRs can be powerful tools for investigating the mechanism and biological function of miRNAs. Many papers, however, are based on the assumption that because anti-miR ‘A’ is complementary to miRNA ‘B’, recognition will automatically occur and block miRNA ‘B’ from binding to mRNA ‘C’. The authors further assume that by blocking miRNA:mRNA recognition, expression of target gene ‘C’ will increase. The observation of an increase in expression is often taken as conclusive evidence that their initial hypothesis about miRNA ‘A’ function is correct.
All too often, these assumptions and the circular reasoning underlying them are inaccurate. It is possible that an anti-miR can act through the hypothesized direct effect. However, it is also possible that the anti-miR is changing gene expression through many different indirect mechanisms that have no relationship to engagement at the intended target. Demonstrating a direct ‘on-target’ mechanism is not trivial. Indeed, the effort necessary to build a strong (but probably not definitive) case for an on-target effect is likely to require a substantial effort.
Some of the experimental strategies available to acquire enough evidence to make a plausible case for on-target action are outlined below.
THEORY TO FUNCTION: STRUGGLING WITH ASSIGNING PHYSIOLOGIC ROLES FOR miRNAs
In contrast to the scaling back of drug development efforts, a literature search from 2000 to 2021 of the term miRNA reveals continued growth of publications related to miRNAs. There have been over 90 000 citations with over 10 000 new citations appearing every year (Figure 8). 56 000 papers (7000 in 2020) appear on a PubMed search of ‘cancer’ and ‘miRNA’. These numbers suggest that the science of miRNA action is well settled and that there should be many opportunities to gain insights into basic biology and begin well-reasoned drug development.
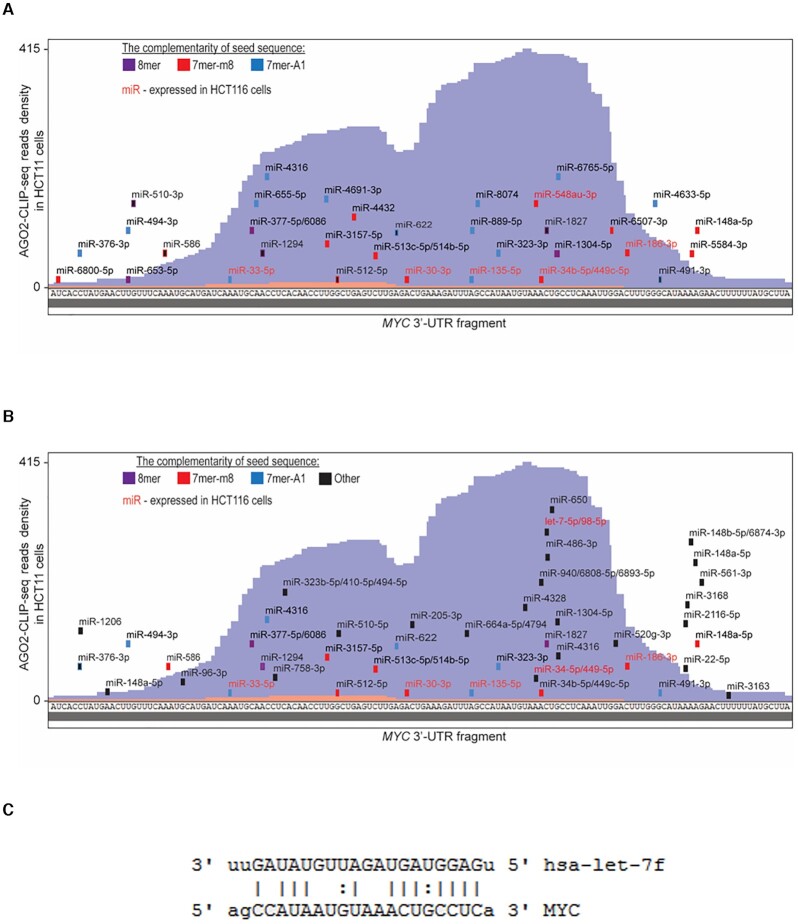
Figure 8.
Challenges to identifying candidate miRNAs from CLIP data and predictions programs. (A, B) Show AGO2 eCLIP data (60) in HCT116 cells for the MYC 3′-UTR. miRNAs were predicted using (A) TargetScan or (B) miRANDA. (C) Let-7f, can example of a highly expressed miRNA that was not predicted by TargetScan but was predicted by miRANDA. Red labeled miRNAs significantly loaded on AGO2 over AGO2 KO cell line.
The proliferation of published work stands in contrast to the slow progress encountered when developing drugs that target miRNAs (Table 1). Slow progress towards important goals presents a paradox–miRNA activity in human cells seems ubiquitous when viewed as a body of thousands of peer-reviewed publications, but the connection of a miRNA to a function can be impossible to reproduce or rationalize when miRNAs are examined closely. Since almost every 3′-UTR has binding sites for many different miRNAs, it is tempting to form hypotheses that relate almost any miRNA to almost any human disease.
Many of these papers describe a change in miRNA expression in response to some alteration in physiology. Gene expression changes are observed and a gene that has a seed sequence match with a miRNA with changed expression is identified. Often, manipulation of the miRNA:target gene interaction is observed to change cell proliferation or some other physiologic readout. Closer examination, however, reveals that many papers lack the minimum controls and experimentation necessary to make convincing conclusions linking complementary recognition by a miRNA to a functional effect on gene expression (93).
High quality data, not just quantities of data
Many papers characterizing the action of miRNAs span a broad range of experiments, from initial identification of a candidate miRNA, to characterization in cultured cells and demonstration of phenotypes in an animal model. In many cases, superficial evaluation suggests that the experiments are appropriate and the data appear to support the overall conclusion. Closer inspection, however, often reveals that conclusions are inadequately supported and controls lacking. In many cases, individual figures involve complex biological systems that are better suited as the focus of an entire paper rather than one part of an expansive manuscript. Because the individual experiments are unconvincing, the grand conclusions are unsupported.
A more constructive approach might be to focus initial reports on well-controlled studies that demonstrate that miRNA ‘A’ has the potential to regulate mRNA ‘B’ through an RNAi mechanism. Because experiments in animals are often difficult or expensive, it would be reasonable to expect that these mechanistic experiments focus on cell culture. Once the link between miRNA and an mRNA has a strong mechanistic foundation, further follow-up investigations in more complex cell culture systems or animals would be justified.
Lessons from a retracted paper
A paper describing the impact of miR-34a on osteoporosis and bone metastasis was recently retracted from Nature (94). It is instructive to retrospectively examine this report. The format of the paper is similar to many others, emphasizing extensive characterization of physiologic change over investigation into the mechanism of action of the miRNA. The initial reason for choosing to focus on miR-34a is only briefly described. Little mechanistic data is shown and miRNA numbers are not quantitated. Synthetic tool compounds are used to evaluate mechanism but controls are insufficient. In vivo experiments assume delivery of anti-miR oligonucleotides to target tissues where there is little precedent for expecting successful uptake. There was no consideration that multiple genes and multiple miRNAs might be acting in concert or how that might affect analysis.
In tandem with the retraction, penalties were applied by the National Institutes of Health and the author's home institution (https://ori.hhs.gov/content/case-summary-wan-yihong). Scientific misconduct is often assumed to involve plagiarism or doctored images. That was not the case here - superficial examination of the data does not reveal misconduct and the manuscript is indistinguishable from many hundreds of similar manuscripts. Instead, meticulous study of the spreadsheets containing the numerical data underpinning the phenotypic imaging was performed. This difficult detective work raised questions about the veracity of the analysis.
The hallmark potential shortcomings of this manuscript might have been less apparent to referee with primary expertise in cancer biology or bone physiology because such referees could not be expected to have a deep understanding of the limitations or need for controls related to the study of nucleic acids (just as a nucleic acid expert could not be expected to understand scholarly standards related to bone physiology). This case study emphasizes the need to prioritize transparent mechanistic data as a foundation before focusing on extensive descriptive studies that examine physiologic change. While editors may be attracted to a manuscript for its biological conclusions, they should include reviewers with a demonstrated understanding of nucleic acid mechanisms. Editors should also have a basic understanding of the controls necessary when evaluating miRNAs.
Paper mills: Adding to the challenge of discriminating signal from noise
It has become apparent that hundreds, perhaps thousands, of published papers are the products of ‘paper mills’ that construct scientific manuscripts. While many of these papers re-use images making them relatively susceptible to detection, others are less obvious. General advice for how to detect the products of paper mills is available (95,96).
Unfortunately, a substantial proportion of retracted papers involve miRNAs. The practical consequence of the proliferation of such papers is their sheer numbers complicate attempts to use literature searches to discern research trends and identify papers containing legitimate findings. The poor scientific practice exemplified by the output of paper mills reinforces the need for research to prioritize focused adherence to transparent and convincing experiments over broad superficial explorations.
THE GLASS IS HALF FULL: RECENT MECHANISTIC INSIGHTS INTO MAMMALIAN RNAi
Paper mills, scientific misconduct, and inadequate rigor can create a dispiriting impression of the current state of RNAi/miRNA exploration. In reality, the importance of RNAi as a powerful mechanism for controlling mammalian gene expression has remained clear. Important questions are unanswered and increasingly refined techniques are now available to address them. While these gaps in knowledge can be frustrating, filling them will provide opportunities for discovery.
One recent example of laying the foundation to understanding complex issues is work from MacRae et al. investigating the structural basis for PIWI-interacting RNA (piRNA) function (97). piRNAs are molecular cousins of miRNAs that guard the germline from transposable elements and their study confronts challenges that parallel those confronting the study of miRNAs. This study used cryo-EM structural analysis in combination with quantitative binding assays to address the fundamental question of how piRNA and miRNA recognition differ. They found that PIWI protein, the AGO analog in the piRNA system, forms weaker seed sequence interactions with piRNAs relative to typical AGO/miRNA interactions. These interactions are compensated by stronger interactions outside the sequence. One possibility is that PIWI proteins have evolved to minimize off-target interactions, a provocative basis for future studies that aim to understand how mRNAs control different targets.
Another recent example from Kim and colleagues (28) asked the basic question: How many annotated miRNAs are processed by DROSHA protein? This comprehensive study examined processing of a full set of 1881 human pri-miRNAs. Of these 1881 candidate pri-miRNAs, only 758 were clearly processed by DROSHA. This study better defined the mechanism of DROSHA-mediated processing and re-emphasized the caution that the existence of a functional miRNA cannot be assumed solely on its appearance in a database. An understanding of what is or is not a miRNA is the foundation for understanding biological function.
A third example that highlights aspects of basic miRNA functions that are largely unexplored involves recent insights into a mechanism termed target-directed miRNA degradation (TDMD) (98,99). In TDMD, RNA target sequences with high degree of complementarity to a miRNA induce the degradation of the miRNA. The two recent reports discovered that this process involves a specific E3 ubiquitin ligase complex inducing proteolysis of the bound AGO protein. The data are transparent and prioritize mechanism over broad physiologic or disease-related descriptions. While important aspects of this process remain to be explored, the findings demonstrate the continued potential to discover new perspectives on the fundamental components of gene regulation by RNAi.
HCT116, A MODEL CELL LINE FOR TESTING THE CONNECTION BETWEEN miRNA, TARGET AND ACTIVITY
The function of miRNAs is complex and varies from one cell type to the next. As a result, assumptions about universal rules governing function that are based on experiments in one cell should be made with caution. Nevertheless, as is the case for any complex system, well controlled studies of defined model cell types can be instructive.
HCT116 is a widely used colorectal cancer-derived cell line. A recent study of over 1000 cell lines (100) reported that HCT116 cells expressed miRNAs at levels that are typical. Because miRNA expression in HCT116 cells falls within the norm, it is reasonable to believe that the relationship of miRNAs and gene expression in HCT116 cells will broadly reflect trends found in many other cell lines.
HCT116 is also a good model because it has been used extensively in miRNA research. A search of ‘HCT116’, ‘cancer’, and ‘miRNA’ on PubMed reveal over 900 publications. We examined the most recent 29 publications from early 2020 to March 2021. These papers implicate dozens of miRNAs in the repression of dozens of different genes (Table 2). These data report that miRNAs play a major role directly controlling the expression of a remarkably large set of different genes involved in cancer cell proliferation. Close examination of miRNA-mediated regulation in HCT116 cells would facilitate understanding the value of these data.
Table 2.
The summary of recent publications reporting functions of miRNAs in HCT116 cells
| miRNA | Rank abundance | Target | Citation |
|---|---|---|---|
| miR-142-3p | >50 | beta-Catenin | Front Oncol. 2021 Feb 10;10:552944. |
| miR-128-3p | >50 | FOXO4 | Front Cell Dev Biol. 2021 Feb 9;9:568738. |
| miR-206 | >50 | c-Met | Oncol Lett. 2021 Feb;21(2):147. |
| miR-424-5p | >50 | PLSCR4 | Eur Rev Med Pharmacol Sci. 2021,25:749-757. |
| miR-106a | >50 | ULK1 | Genes 2021 Feb 9;12(2):245. |
| miR-133b | >50 | LUCAT-1 | Future Oncol. 2021 Mar;17(9):1013-1023. |
| miR-1254 | >50 | MEGF6 | Am J Transl Res. 2021 Jan 15;13(1):183-196. |
| miR-423-5p | >50 | BCL-2 | Front Oncol. 2021 Jan 11;10:582239. |
| Let-7b, miR-203a | >50 | Survivin | Cancer Res. 2021 Jan 20:canres.3157.2020. |
| miR-34a | >50 | n/a | Mol Biol Rep. 2021 Jan;48(1):203-218. |
| miR-145 | >50 | MYC, FSCN1 | J Biomed Nanotechnol. 2020;16(8):1183-1195. |
| miR-143,145 | >50 | ADAM17, K-Ras, XPO5, SET | Epigenetics. 2020 Dec 28:1-18. |
| miR-144 | >50 | KLF4 | J Biomed Nanotechnol. 2020;16(7):1102-1109. |
| miR149,150 | 27, >50 | linc00460 | Mol Ther Nucleic Acids. 2020;22:1004-1015. |
| miRNA-140 | >50 | TRAF6 | Onco Targets Ther. 2020;13:11991-12001. |
| miRNA-486-5p | >50 | n/a | Cancers. 2020 Nov 19;12(11):3432. |
| miR-141 | 29 | PHLPP2 | Cancer Manag Res. 2020;12:11341-11350. |
| mR-608 | >50 | MRPL43 | DNA Cell Biol. 2020 Nov;39(11):2017-2027. |
| miR-942 | >50 | DLG2 | Mol Carcinog. 2020 Dec;59(12):1323-1342. |
| miR-103a-5p | >50 | PDHB | Neoplasma. 2020 Oct 30:200813N858. |
| miR-20b-5p | >50 | CCND1 | Cell Cycle. 2020 Nov;19(21):2939-2954. |
| miR-708 | >50 | ZNF549 | Sci Rep. 2020 Oct 7;10(1):16729. |
| miR-30a-5p | >50 | HSPA5 | Int J Mol Sci. 2020 Oct 3;21(19):7315. |
| miR-409-3p | >50 | ERCC1 | Evid Based Comp Alternat Med. 2020:8394574. |
| miR-488 | >50 | PFKB3 | J Clin Lab Anal. 2021 Jan:e23578. |
| miR-421 | >50 | CASP3 | Cancer Manag Res. 2020 Aug;12:7579-7587. |
| miR-548a-3p | >50 | TPX2 | Cancer Biother Radiopharm. 2020. doi:10.1089/cbr.2020.3767 |
| miR-34a-5p | >50 | lncASPR | Cancer Sci. 2020 Oct;111(10):3938-3952. |
| miR-195-5p | >50 | CEP55 | J Envir. Pathol Toxicol Oncol. 2020;39:101-111. |
Notes. n/a – not available.
To test the impact of miRNAs on HCT116 cells, the Corey laboratory used enhanced crosslinking immunoprecipitation (eCLIP) to identify the locations for AGO2 binding within 3′-UTRs (60). Sequences within RNA that bind AGO2 are strong candidates for interactions with miRNAs (45). CLIP protocols combine RNA sequencing and immunoprecipitation to provide an important tool for identifying potential binding sites between RNA and proteins. eCLIP is a modified protocol to enhance discrimination between promising sites for biologically relevant interactions and background. To further enhance the focus on bona fide AGO2:RNA interactions, we compared wild-type and AGO2 knockout cells to reduce the likelihood of false positive identification of AGO2:RNA interactions.
We examined the effect of the expression of genes that were associated with AGO2 in AGO1, AGO2, AGO1/2 and AGO1/2/3 knock out cell lines. The standard expectation for the action of miRNAs suggests that knocking out AGO gene expression should reverse the action of miRNAs and increase the expression of genes with significant engagement between 3′-UTRs and AGO protein. Contrary to that expectation, we observed little correlation between binding of AGO2 and change in gene expression. Genes with strong evidence of association were as likely to show decreased expression as increased expression.
For example, the strongest eCLIP/RNAseq signal within 3′-untranslated regions was within the Myc gene (Figure 8A). However, upon knocking out AGO expression, expression of Myc RNA and protein were decreased rather than increased (60) as would be expected from standard assumptions about miRNA action. Complicating experimental exploration, many miRNAs possessed seed sequence complementary to sites defined by the RNAseq read cluster, and the identity of these miRNAs varied depending on which miRNA prediction program was used (Figure 8B, C). Variation of outcomes depending on prediction programs has been noted previously (101,102). The uncertainty of prediction in combination with the many possible combinations of miRNAs that might be involved repressing a particular gene complicate experimental validation.
Our data demonstrated that an experimentally verified association between AGO2 and a 3′-UTR cannot be assumed to lead to a predictable effect on gene expression. By extension, a predicted seed sequence match—a primary criteria for forming hypothesis in many papers—also is not a reliable predictor. Our experience with HCT116 cells suggests that demonstrating on-target miRNA control at a particular gene requires extensive experimentation to support conclusion about mechanism. Other laboratories have reported similar unpredictable relationships between miRNA recognition and gene expression (101,103) and differences in outcomes depending on which algorithm was used for predicting potential sites for miRNA association. Structure within mRNA can also complicate the potential for binding to miRNAs (104). These same uncertainties make predictions based solely on seed sequence complementarity even less predictable, raising the bar for experimental validation.
BENCHMARKS FOR EVALUATING CLAIMS FOR CELLULAR REGULATION BY miRNAs
Evaluating the scientific value of papers that describe the action of miRNAs is an important task for editors, reviewers, and researchers. With tens of thousands of papers in the literature (Figure 9), designing new projects requires separating those papers that provide a firm foundation for future research from those that do not. Authors have a responsibility to make a strong and transparent case for their conclusions. Benchmarks for judging studies that involve human miRNAs include:
Figure 9.
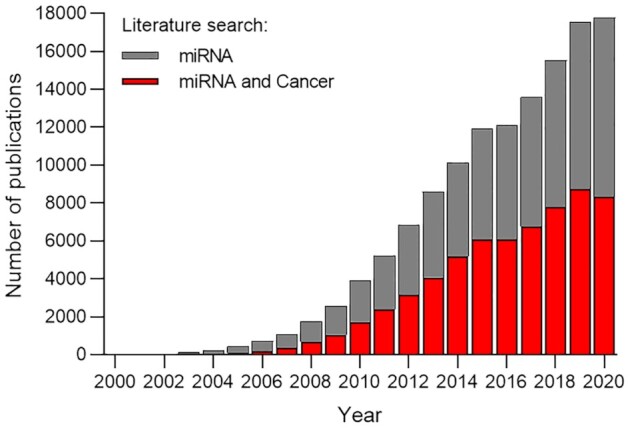
The miRNA literature. Publications identified by PubMed searches for the terms ‘miRNA’ and ’miRNA and cancer’, 2000–2020.
Strong justification for focusing on a miRNA
Many different miRNAs are expressed in cells and miRNAs have seed sequence complementary to many different genes. It is easy to form a hypothesis that miRNA ‘A’ recognizes mRNA ‘B’ to cause phenotype ‘C’. It is essential that persuasive reasons be supplied for focusing on a particular miRNA for in-depth experimental investigation. This evidence is necessary to build a case that the miRNA being brought into focus has not been ‘cherry picked’ to fit into a particular endogenous or disease-related biological pathway.
Quantitate the level of miRNA expression to verify it has the potential to be compatible with biologically relevant regulation
Highly expressed miRNAs in a cell or tissue will probably be the most promising candidates for investigation. It is essential, therefore, to have data on the number of a particular miRNA per cell. As shown above (Figure 3) a handful of miRNA families are typically the prevalent miRNAs inside cells. Quantitation and insights into stoichiometry are especially important for projects that implicate miRNAs outside these families.
Quantitative PCR (qPCR) alone or newer approaches like droplet digital PCR are inadequate for determining ‘per cell’ numbers for miRNAs because the data provides relative amounts of a give RNA under different conditions. Even these relative measurements are not reliable when different primer sets are used, preventing comparisons of the quantity of different miRNAs.
The experimental solution is to standardize measurements of mRNA targets or miRNAs using known quantities of the RNA species, providing reliable benchmarks for evaluation. These measurements of miRNA and mRNA quantities are straightforward. For miRNAs, they allow rapid decisions to be made about physiologic relevance – low amounts are probably not relevant, while levels of miRNAs that are high are better candidates for in-depth experimental investigation.
Of the twenty-nine papers from 2020–2021 describing the action of miRNAs in HCT116 cells (Table 2), not a single RNA from the top seven families was implicated in the control of gene expression. Only a handful appeared among the top ranked fifty miRNAs determined by our eCLIP and none were estimated to have concentrations of more than one thousand per cell. It is not easy, therefore, to understand the physical basis for the interactions and physiologic effects being described in these manuscripts.
Quantitate variation of miRNA expression in different cell types, tissues, or environmental conditions
One common hypothesis is that an miRNA will have increased expression in one cellular or environmental context and that this increased expression will allow the miRNA to exert its biological effect. The most reliable starting point for these studies is an unbiased screen to identify a particular miRNA that is highly up-regulated in a cell-type of interest.
Fold-change from one cell type or tissue relative to another is an imperfect measurement. A 10-fold increase in expression of an miRNA that is lowly expressed may leave it expressed at level that remains too low to have an impact, whereas a 10-fold increase in expression of a highly expressed miRNA may push the equilibrium of recognition into a range where biological impacts begin to be important. Fold-change should be accompanied by absolute quantitation, as noted above. While qPCR is a useful technique for comparing the relative amounts of RNA expression in different cell lines or under different conditions, it is important to understand that the method should not be viewed as a ‘black box’, its limitations be understood, and that appropriate guidelines be followed (105,106).
Prioritize building a foundation of molecular mechanism over physiologic function
As noted, many papers hypothesize that miRNA ‘A’ recognizes mRNA ‘B’ to cause phenotype ‘C’ and then supply data to match that formula. In many cases, data (often in vivo data) about phenotype dominates the paper. While understanding the physiologic impact of miRNAs is an important goal, that goal cannot be achieved without first building the foundation for understanding mechanism.
Editors and readers should understand that building a reliable foundation supporting the link between a miRNA and a function is difficult. They should not demand expansive publications that begin with identification of a candidate miRNA and end with demonstration of a function in animals. In many cases, rather than superficial explorations of mechanism, better outcomes would be achieved by having multiple papers, each of which demonstrates one important finding in a persuasive and transparent manner.
Transparent data
Transparent data helps build a case that conclusions are trustworthy and, therefore, its presence or absence is a critical factor affecting confidence in papers that examine miRNAs and lncRNAs. Experimenters may differ on how they define transparent data, but some guidelines include: (i) primary data that can be directly inspected for quality (i.e. western blots for protein expression); These complement secondary data (i.e. bar graphs of RNA expression) by providing a direct window on data quality; (ii) microscopy is not useful when it is supplied as anecdotal pictures. A sufficient number of images should be obtained and evaluated through unbiased means before claiming an effect; (iii) replicate experiments should be performed and replicate data shown as Supplemental information; (iv) it is routine to observe small variations from day to day or experimenter to experimenter. When effects are small, it is necessary that authors persuasively justify why the effects have biological significance. Proper use of control conditions because especially important when variation is small and (v) methods for obtaining large datasets from RNAseq or mass spectrometry should be described in detail, justifying the reliability of the data and precautions taken against cherry picking an experimental focus.
The RNAseq and mass spectrometry data should be presented in a transparent fashion where thought has been given to reducing the inherent complexity of the data to visual representations that can be easily interpreted by readers outside the laboratory acquiring the data. Proper execution of RNAseq and mass spectrometry experiments is not trivial and data should not be trusted absent clear support from the researchers responsible for generating it.
Controls
miRNAs are nucleic acids that control gene expression by recognizing target nucleic acids. Many experiments use synthetic anti-miRs or miRNA mimics to control gene expression and test hypotheses. The potential for synthetic oligonucleotides and duplex RNAs to cause confounding off-target effects is well known (15,107–111). It is essential that experiments be carefully controlled and adhere to commonly accepted guidelines (93). Failure to adhere to the use of proper controls should be easy to spot. For example, a paper that uses an anti-miR with a single control oligonucleotide (or, even worse, a buffer only control) to show a change in gene expression or cell proliferation in a novel system is not likely to be persuasive. Failure to persuasively address the use of controls provides a simple means for editors or readers to determine their confidence in a paper's results. The advent of CRISPR has made gene editing widely accessible and provides another approach to directly address the importance of potential target sequences.
SUMMARY
Thousands of papers have appeared describing the actions of miRNAs in human cells and potential impacts on normal physiology and disease. It can be difficult for researchers to discriminate between papers that offer convincing results and those that do not. Lack of confidence in published results, in combination with an incomplete understanding of miRNA mechanism, is an obstacle to miRNA-directed therapeutics achieving the same high level of success as fully complementary synthetic duplex RNAs.
We note that similar calls for renewed emphasis on rigorous and quantitative experiments have been for studies investigating phase separation (112–114), RNA:protein binding interactions (115), and miRNA sponges (75,116,117). miRNAs, phase separation, circular RNAs, and specific RNA:protein interactions share the potential to reshape views of how gene expression is regulated. All these fields, however, have been weighted towards descriptive research rather than the detailed biochemical investigations necessary to build the strong framework for making testable predictions.
Journals, especially journals that set publishing trends, should prioritize studies that focus on mechanism rather than superficial investigations spanning a broad (and often unrealistic) swath of science ranging from identification of a miRNA to an in vivo physiologic impact. A focus on mechanism is necessary to discriminate sound from unsound science among the thousands of papers appearing every year. Such papers may sometimes appear ‘incremental’ at first glance, but robust data that clarifies critical issues and contributes to the foundation necessary for progress should always be welcome.
RNAi is a powerful mechanism for controlling mammalian gene expression. Studies of detailed molecular mechanisms for individual miRNAs will build a better understanding of how RNAi proteins, miRNAs, and cellular RNA targets act in concert to regulate gene expression. This foundation of rigorous research, coupled with an unbiased view about the boundaries of RNAi, will unlock discoveries and likely point the field in new directions both unexpected and exciting.
ACKNOWLEDGEMENTS
This study was supported by R35GM118103 (D.R.C.) from the National Institutes of Health and the Robert A. Welch Foundation I-1244 (D.R.C.).
Contributor Information
Audrius Kilikevicius, Department of Pharmacology and Biochemistry, UT Southwestern Medical Center, 6001 Forest Park Road, Dallas, TX, USA.
Gunter Meister, Regensburg Center for Biochemistry (RCB), Laboratory for RNA Biology, University of Regensburg, Regensburg, Germany.
David R Corey, Department of Pharmacology and Biochemistry, UT Southwestern Medical Center, 6001 Forest Park Road, Dallas, TX, USA.
FUNDING
National Institutes of Heatlh [GM118103]. Funding for open access charge: NIH [GM118103].
Conflict of interest statement. D.R.C. is Executive Editor of NAR.
REFERENCES
- 1. Bartel D.P. Metazoan microRNAs. Cell. 2018; 173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Treiber T., Treiber N., Meister G.. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019; 20:5–20. [DOI] [PubMed] [Google Scholar]
- 3. Banerjee D., Slack F.. Control of developmental timing by small temporal RNAs: a paradigm for RNA-mediated regulation of gene expression. Bioessays. 2002; 24:119–129. [DOI] [PubMed] [Google Scholar]
- 4. Pasquinelli A.E., Ruvkun G.. Control of developmental timing by micrornas and their targets. Annu. Rev. Cell Dev. Biol. 2002; 18:495–513. [DOI] [PubMed] [Google Scholar]
- 5. Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75:843–854. [DOI] [PubMed] [Google Scholar]
- 6. Wightman B., Ha I., Ruvkun G.. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993; 75:855–862. [DOI] [PubMed] [Google Scholar]
- 7. Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P.. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001; 294:858–862. [DOI] [PubMed] [Google Scholar]
- 8. Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T.. Identification of novel genes coding for small expressed RNAs. Science. 2001; 294:853–858. [DOI] [PubMed] [Google Scholar]
- 9. Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T.. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002; 12:735–739. [DOI] [PubMed] [Google Scholar]
- 10. Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T.. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001; 411:494–498. [DOI] [PubMed] [Google Scholar]
- 11. Zhang S., Cheng Z., Wang Y., Han T.. The Risks of miRNA therapeutics: in a drug target perspective. Drug Des. Dev. Ther. 2021; 15:721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Winkle M., El-Daly S.M., Fabbri M., Calin G.A.. Noncoding RNA therapeutics - challenges and potential solutions. Nat. Rev. Drug Discov. 2021; 20:629–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shen X., Corey D.R.. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018; 46:1584–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Setten R.L., Rossi J.J., Han S.P.. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019; 18:421–446. [DOI] [PubMed] [Google Scholar]
- 15. Crooke S.T., Baker B.F., Crooke R.M., Liang X.H.. Antisense technology: an overview and prospectus. Nat. Rev. Drug Discov. 2021; 20:427–453. [DOI] [PubMed] [Google Scholar]
- 16. Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N.. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002; 21:4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N.. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004; 23:4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S.et al.. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003; 425:415–419. [DOI] [PubMed] [Google Scholar]
- 19. Landthaler M., Yalcin A., Tuschl T.. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004; 14:2162–2167. [DOI] [PubMed] [Google Scholar]
- 20. Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N.. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004; 18:3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R.. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004; 432:235–240. [DOI] [PubMed] [Google Scholar]
- 22. Grishok A., Pasquinelli A.E., Conte D., Li N., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C.. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001; 106:23–34. [DOI] [PubMed] [Google Scholar]
- 23. Ketting R.F., Fischer S.E., Bernstein E., Sijen T., Hannon G.J., Plasterk R.H.. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001; 15:2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hutvagner G., McLachlan J., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D.. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001; 293:834–838. [DOI] [PubMed] [Google Scholar]
- 25. Ha M., Kim V.N.. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014; 15:509–524. [DOI] [PubMed] [Google Scholar]
- 26. Kim B., Jeong K., Kim V.N.. Genome-wide mapping of DROSHA cleavage sites on primary microRNAs and noncanonical substrates. Mol. Cell. 2017; 66:258–269. [DOI] [PubMed] [Google Scholar]
- 27. Jin W., Wang J., Liu C.P., Wang H.W., Xu R.M.. Structural basis for pri-miRNA recognition by Drosha. Mol. Cell. 2020; 78:423–433. [DOI] [PubMed] [Google Scholar]
- 28. Kim K., Baek S.C., Lee Y.Y., Bastiaanssen C., Kim J., Kim H., Kim V.N.. A quantitative map of human primary microRNA processing sites. Mol. Cell. 2021; 81:3422–3439. [DOI] [PubMed] [Google Scholar]
- 29. Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J.. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004; 305:1437–1441. [DOI] [PubMed] [Google Scholar]
- 30. Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T.. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004; 15:185–197. [DOI] [PubMed] [Google Scholar]
- 31. Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013; 14:447–459. [DOI] [PubMed] [Google Scholar]
- 32. Wee L.M., Flores-Jasso C.F., Salomon W.E., Zamore P.D.. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012; 151:1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chandradoss S.D., Schirle N.T., Szczepaniak M., MacRae I.J., Joo C.. A dynamic search process underlies microRNA targeting. Cell. 2015; 162:96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Salomon W.E., Jolly S.M., Moore M.J., Zamore P.D., Serebrov V.. Single-molecule imaging reveals that Argonaute reshapes the binding properties of its nucleic acid guides. Cell. 2015; 162:84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Petri S., Dueck A., Lehmann G., Putz N., Rudel S., Kremmer E., Meister G.. Increased siRNA duplex stability correlates with reduced off-target and elevated on-target effects. RNA. 2011; 17:737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Valdmanis P.N., Gu S., Schuermann N., Sethupathy P., Grimm D., Kay M.A.. Expression determinants of mammalian argonaute proteins in mediating gene silencing. Nucleic Acids Res. 2012; 40:3704–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Voller D., Linck L., Bruckmann A., Hauptmann J., Deutzmann R., Meister G., Bosserhoff A.K.. Argonaute family protein expression in normal tissue and cancer entities. PLoS One. 2016; 11:e0161165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park M.S., Sim G., Kehling A.C., Nakanishi K.. Human Argonaute2 and Argonaute3 are catalytically activated by different lengths of guide RNA. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:28576–28578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Paddison P.J., Caudy A.A., Bernstein E., Hannon G.J., Conklin D.S.. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002; 16:948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B.. Prediction of mammalian microRNA targets. Cell. 2003; 115:787–798. [DOI] [PubMed] [Google Scholar]
- 41. Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 42. Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P.. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007; 27:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Loeb G.B., Khan A.A., Canner D., Hiatt J.B., Shendure J., Darnell R.B., Leslie C.S., Rudensky A.Y.. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell. 2012; 48:760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Helwak A., Kudla G., Dudnakova T., Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013; 153:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moore M.J., Scheel T.K., Luna J.M., Park C.Y., Fak J.J., Nishiuchi E., Rice C.M., Darnell R.B.. miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 2015; 6:8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McGeary S.E., Lin K.S., Shi C.Y., Pham T.M., Bisaria N., Kelley G.M., Bartel D.P.. The biochemical basis of microRNA targeting efficacy. Science. 2019; 366:eaav1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Behm-Ansmant I., Rehwinkel J., Izaurralde E.. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb. Symp. Quant. Biol. 2006; 71:523–530. [DOI] [PubMed] [Google Scholar]
- 48. Subtelny A.O., Eichhorn S.W., Chen G.R., Sive H., Bartel D.P.. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014; 508:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Eichhorn S.W., Guo H., McGeary S.E., Rodriguez-Mias R.A., Shin C., Baek D., Hsu S.H., Ghoshal K., Villen J., Bartel D.P.. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell. 2014; 56:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S.. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003; 21:635–637. [DOI] [PubMed] [Google Scholar]
- 51. Jackson A.L., Burchard J., Schelter J., Chau B.N., Cleary M., Lim L., Linsley P.S.. Widespread siRNA ‘off-target’ transcript silencing mediated by seed region sequence complementarity. RNA. 2006; 12:1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lai E.C. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002; 30:363–364. [DOI] [PubMed] [Google Scholar]
- 53. Saxena S., Jonsson Z.O., Dutta A.. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J. Biol. Chem. 2003; 278:44312–44319. [DOI] [PubMed] [Google Scholar]
- 54. Imig J., Brunschweiger A., Brummer A., Guennewig B., Mittal N., Kishore S., Tsikrika P., Gerber A.P., Zavolan M., Hall J.. miR-CLIP capture of a miRNA targetome uncovers a lincRNA H19-miR-106a interaction. Nat. Chem. Biol. 2015; 11:107–114. [DOI] [PubMed] [Google Scholar]
- 55. Zhang K., Zhang X., Cai Z., Zhou J., Cao R., Zhao Y., Chen Z., Wang D., Ruan W., Zhao Q.et al.. A novel class of microRNA-recognition elements that function only within open reading frames. Nat. Struct. Mol. Biol. 2018; 25:1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ludwig N., Leidinger P., Becker K., Backes C., Fehlmann T., Pallasch C., Rheinheimer S., Meder B., Stahler C., Meese E.et al.. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016; 44:3865–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. de Rie D., Abugessaisa I., Alam T., Arner E., Arner P., Ashoor H., Astrom G., Babina M., Bertin N., Burroughs A.M.et al.. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017; 35:872–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Alles J., Fehlmann T., Fischer U., Backes C., Galata V., Minet M., Hart M., Abu-Halima M., Grasser F.A., Lenhof H.P.et al.. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019; 47:3353–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Friedman R.C., Farh K.K., Burge C.B., Bartel D.P.. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009; 19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chu Y., Kilikevicius A., Liu J., Johnson K.C., Yokota S., Corey D.R.. Argonaute binding within 3′-untranslated regions poorly predicts gene repression. Nucleic Acids Res. 2020; 48:7439–7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kozomara A., Birgaoanu M., Griffiths-Jones S.. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019; 47:D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rice G.M., Shivashankar V., Ma E.J., Baryza J.L., Nutiu R.. Functional atlas of primary miRNA maturation by the microprocessor. Mol. Cell. 2020; 80:892–902. [DOI] [PubMed] [Google Scholar]
- 63. Doench J.G., Petersen C.P., Sharp P.A.. siRNAs can function as miRNAs. Genes Dev. 2003; 17:438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Saetrom P., Heale B.S., Snove O. Jr, Aagaard L., Alluin J., Rossi J.J. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007; 35:2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Broderick J.A., Salomon W.E., Ryder S.P., Aronin N., Zamore P.D.. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA. 2011; 17:1858–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Flamand M.N., Gan H.H., Mayya V.K., Gunsalus K.C., Duchaine T.F.. A non-canonical site reveals the cooperative mechanisms of microRNA-mediated silencing. Nucleic Acids Res. 2017; 45:7212–7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hu J., Liu J., Corey D.R.. Allele-selective inhibition of huntingtin expression by switching to an miRNA-like RNAi mechanism. Chem. Biol. 2010; 17:1183–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J.. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008; 456:921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Baillat D., Shiekhattar R.. Functional dissection of the human TNRC6 (GW182-related) family of proteins. Mol. Cell. Biol. 2009; 29:4144–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Takimoto K., Wakiyama M., Yokoyama S.. Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. RNA. 2009; 15:1078–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Elkayam E., Faehnle C.R., Morales M., Sun J., Li H., Joshua-Tor L.. Multivalent recruitment of human Argonaute by GW182. Mol. Cell. 2017; 67:646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sheu-Gruttadauria J., MacRae I.J.. Phase transitions in the assembly and function of human miRISC. Cell. 2018; 173:946–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Briskin D., Wang P.Y., Bartel D.P.. The biochemical basis for the cooperative action of microRNAs. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:17764–17774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mukherji S., Ebert M.S., Zheng G.X., Tsang J.S., Sharp P.A., van Oudenaarden A.. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 2011; 43:854–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Denzler R., Agarwal V., Stefano J., Bartel D.P., Stoffel M.. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell. 2014; 54:766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kataruka S., Modrak M., Kinterova V., Malik R., Zeitler D.M., Horvat F., Kanka J., Meister G., Svoboda P.. MicroRNA dilution during oocyte growth disables the microRNA pathway in mammalian oocytes. Nucleic Acids Res. 2020; 48:8050–8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Abugessaisa I., Ramilowski J.A., Lizio M., Severin J., Hasegawa A., Harshbarger J., Kondo A., Noguchi S., Yip C.W., Ooi J.L.C.et al.. FANTOM enters 20th year: expansion of transcriptomic atlases and functional annotation of non-coding RNAs. Nucleic Acids Res. 2021; 49:D892–D898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Arvey A., Larsson E., Sander C., Leslie C.S., Marks D.S.. Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol. 2010; 6:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Garcia D.M., Baek D., Shin C., Bell G.W., Grimson A., Bartel D.P.. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 2011; 18:1139–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Aoki Y., Wood M.J.A.. Emerging oligonucleotide therapeutics for rare neuromuscular diseases. J. Neuromuscul. Dis. 2021; 8:869–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Aartsma-Rus A., Corey D.R.. The 10th oligonucleotide therapy approved: golodirsen for duchenne muscular dystrophy. Nucleic Acid Ther. 2020; 30:67–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mendell J.R., Khan N., Sha N., Eliopoulos H., McDonald C.M., Goemans N., Mercuri E., Lowes L.P., Alfano L.N.Eteplirsen Study, G . Comparison of long-term ambulatory function in patients with Duchenne muscular dystrophy treated with eteplirsen and matched natural history controls. J. Neuromuscul. Dis. 2021; 8:469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bennett C.F., Kordasiewicz H.B., Cleveland D.W.. Antisense drugs make sense for neurological diseases. Annu. Rev. Pharmacol. Toxicol. 2021; 61:831–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M.. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005; 438:685–689. [DOI] [PubMed] [Google Scholar]
- 86. Esau C., Davis S., Murray S.F., Yu X.X., Pandey S.K., Pear M., Watts L., Booten S.L., Graham M., McKay R.et al.. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006; 3:87–98. [DOI] [PubMed] [Google Scholar]
- 87. Hogan D.J., Vincent T.M., Fish S., Marcusson E.G., Bhat B., Chau B.N., Zisoulis D.G.. Anti-miRs competitively inhibit microRNAs in Argonaute complexes. PLoS One. 2014; 9:e100951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Li Z., Rana T.M.. Therapeutic targeting of microRNAs: current status and future challenges. Nat. Rev. Drug Discov. 2014; 13:622–638. [DOI] [PubMed] [Google Scholar]
- 89. Girard M., Jacquemin E., Munnich A., Lyonnet S., Henrion-Caude A.. miR-122, a paradigm for the role of microRNAs in the liver. J. Hepatol. 2008; 48:648–656. [DOI] [PubMed] [Google Scholar]
- 90. Chang J., Nicolas E., Marks D., Sander C., Lerro A., Buendia M.A., Xu C., Mason W.S., Moloshok T., Bort R.et al.. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004; 1:106–113. [DOI] [PubMed] [Google Scholar]
- 91. Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P.. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005; 309:1577–1581. [DOI] [PubMed] [Google Scholar]
- 92. Sarnow P., Sagan S.M.. Unraveling the mysterious interactions between hepatitis C virus RNA and liver-specific microRNA-122. Annu Rev Virol. 2016; 3:309–332. [DOI] [PubMed] [Google Scholar]
- 93. Gagnon K.T., Corey D.R.. Guidelines for experiments using antisense oligonucleotides and double-stranded RNAs. Nucleic Acid Ther. 2019; 29:116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Krzeszinski J.Y., Wei W., Huynh H., Jin Z., Wang X., Chang T.C., Xie X.J., He L., Mangala L.S., Lopez-Berestein G.et al.. Retraction note: miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2020; 582:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Byrne J.A., Christopher J.. Digital magic, or the dark arts of the 21(st) century-how can journals and peer reviewers detect manuscripts and publications from paper mills?. FEBS Lett. 2020; 594:583–589. [DOI] [PubMed] [Google Scholar]
- 96. Seifert R. How Naunyn-Schmiedeberg's Archives of Pharmacology deals with fraudulent papers from paper mills. Naunyn Schmiedebergs Arch. Pharmacol. 2021; 394:431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Anzelon T.A., Chowdhury S., Hughes S.M., Xiao Y., Lander G.C., MacRae I.J.. Structural basis for piRNA targeting. Nature. 2021; 597:285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Han J., LaVigne C.A., Jones B.T., Zhang H., Gillett F., Mendell J.T.. A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. Science. 2020; 370:eabc9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Shi C.Y., Kingston E.R., Kleaveland B., Lin D.H., Stubna M.W., Bartel D.P.. The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science. 2020; 370:eabc9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. 3rd, Barretina J., Gelfand E.T., Bielski C.M., Li H.et al.. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 2019; 569:503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Agarwal V., Bell G.W., Nam J.W., Bartel D.P.. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015; 4:e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kern F., Krammes L., Danz K., Diener C., Kehl T., Kuchler O., Fehlmann T., Kahraman M., Rheinheimer S., Aparicio-Puerta E.et al.. Validation of human microRNA target pathways enables evaluation of target prediction tools. Nucleic Acids Res. 2021; 49:127–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Xu P., Palmer L.E., Lechauve C., Zhao G., Yao Y., Luan J., Vourekas A., Tan H., Peng J., Schuetz J.D.et al.. Regulation of gene expression by miR-144/451 during mouse erythropoiesis. Blood. 2019; 133:2518–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ruijtenberg S., Sonneveld S., Cui T.J., Logister I., de Steenwinkel D., Xiao Y., MacRae I.J., Joo C., Tanenbaum M.E.. mRNA structural dynamics shape Argonaute-target interactions. Nat. Struct. Mol. Biol. 2020; 27:790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Huggett J.F., Foy C.A., Benes V., Emslie K., Garson J.A., Haynes R., Hellemans J., Kubista M., Mueller R.D., Nolan T.et al.. The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin. Chem. 2013; 59:892–902. [DOI] [PubMed] [Google Scholar]
- 106. Chapman J.R., Waldenstrom J.. With reference to reference genes: a systematic review of endogenous controls in gene expression studies. PLoS One. 2015; 10:e0141853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Stein C.A., Krieg A.M.. Problems in interpretation of data derived from in vitro and in vivo use of antisense oligodeoxynucleotides. Antisense Res. Dev. 1994; 4:67–69. [DOI] [PubMed] [Google Scholar]
- 108. Stein C.A. Keeping the biotechnology of antisense in context. Nat. Biotechnol. 1999; 17:209. [DOI] [PubMed] [Google Scholar]
- 109. Myers K.J., Dean N.M.. Sensible use of antisense: how to use oligonucleotides as research tools. Trends Pharmacol. Sci. 2000; 21:19–23. [DOI] [PubMed] [Google Scholar]
- 110. Stein C.A. The experimental use of antisense oligonucleotides: a guide for the perplexed. J. Clin. Invest. 2001; 108:641–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Editorial Whither RNAi?. Nat. Cell Biol. 2003; 5:489–490. [DOI] [PubMed] [Google Scholar]
- 112. McSwiggen D.T., Mir M., Darzacq X., Tjian R.. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev. 2019; 33:1619–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Alberti S., Gladfelter A., Mittag T.. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019; 176:419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Narlikar G.J., Myong S., Larson D., Maeshima K., Francis N., Rippe K., Sabari B., Strader L., Tjian R.. Is transcriptional regulation just going through a phase?. Mol. Cell. 2021; 81:1579–1585. [DOI] [PubMed] [Google Scholar]
- 115. Davidovich C., Wang X., Cifuentes-Rojas C., Goodrich K.J., Gooding A.R., Lee J.T., Cech T.R.. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol. Cell. 2015; 57:552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Bosson A.D., Zamudio J.R., Sharp P.A.. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell. 2014; 56:347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Broderick J.A., Zamore P.D.. Competitive endogenous RNAs cannot alter microRNA function in vivo. Mol. Cell. 2014; 54:711–713. [DOI] [PubMed] [Google Scholar]