Abstract
In vivo, left-handed DNA duplex (usually refers to Z-DNA) is mainly formed in the region of DNA with alternating purine pyrimidine (APP) sequence and plays significant biological roles. It is well known that d(CG)n sequence can form Z-DNA most easily under negative supercoil conditions, but its essence has not been well clarified. The study on sequence dependence of Z-DNA stability is very difficult without modification or inducers. Here, by the strong topological constraint caused by hybridization of two complementary short circular ssDNAs, left-handed duplex part was generated for various sequences, and their characteristics were investigated by using gel-shift after binding to specific proteins, CD and Tm analysis, and restriction enzyme cleavage. Under the strong topological constraint, non-APP sequences can also form left-handed DNA duplex as stable as that of APP sequences. As compared with non-APP sequences, the thermal stability difference for APP sequences between Z-form and B-form is smaller, which may be the reason that Z-DNA forms preferentially for APP ones. This result can help us to understand why nature selected APP sequences to regulate gene expression by transient Z-DNA formation, as well as why polymer with chirality can usually form both duplexes with left- or right-handed helix.
Graphical Abstract
Graphical Abstract.
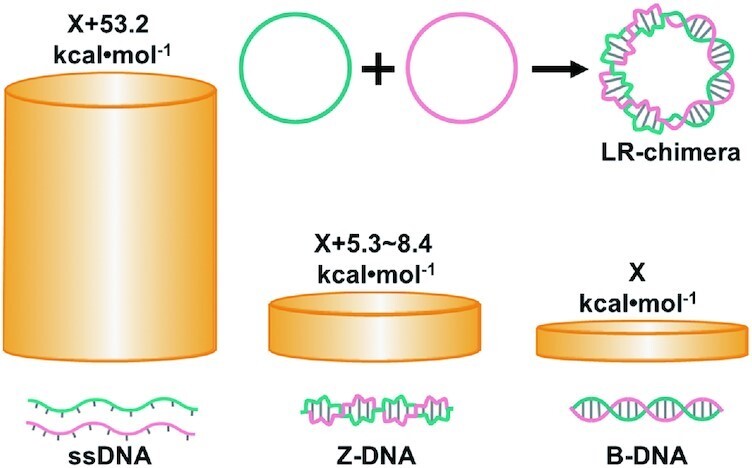
By formation of the LR chimera from two complementary ssDNA circles, we found that non-APP sequences can form left-handed DNA duplex, and APP sequences prefer to form Z-DNA due to its smaller difference in free energy between Z-form and B-form.
INTRODUCTION
The binding of Z-DNA (the left-handed DNA duplex) specific antibody to Drosophila polytene chromosome first proved that Z-DNA may exist in organisms (1). Subsequently, scientists found a variety of proteins that can specifically bind Z-DNA in organisms, such as vertebrate adenosine deaminase acting on RNA 1 (ADAR1) (2), mammalian Z-DNA binding protein (3,4), fish Z-DNA binding protein kinase PKZ (5,6), and poxvirus E3L protein (7). It is believed that these proteins are closely related to immune response and virus resistance (8–11). In addition, a variety of biological functions of Z-DNA have been revealed: participating in gene regulation (12,13), inducing genetic instability (2,14), relating to several diseases (15–20). On the other hand, clear conclusions are hard drawn and controversies are constant because Z-DNA can be present only under abnormal conditions such as chemical modification (21–26), high ionic strength (27–35), induced by Z-DNA-binding molecules (36–40), and strong negative supercoiling (41,42). Normally, it is hard to form under physiological conditions so that its study is difficult on basic molecular properties, biological functions, and practical applications. For example, the essence of sequence dependence of Z-DNA stability has confused scientists for many years.
Since most detected Z-DNA has alternating purine pyrimidine (APP) sequences, and each two base pairs of 5′-d(PyPu)-3′/5′-d(PyPu)-3′ (e.g. 5′-d(CA)-3′/5′-d(TG)-3′) can be considered as one structural unit of Z-DNA, many scientists believe that nonalternating purine pyrimidine (non-APP) sequences cannot form Z-DNA. Here, we tried to study whether non-APP sequence (containing no longer than 6 bp of continuous alternating purine pyrimidine) cannot form left-handed DNA duplex (designated as lh-DNA) absolutely or other factors inhibit its formation. In other words, what kind of condition can allow a non-APP sequence to form lh-DNA? Here, the name of lh-DNA (all left-handed DNA duplex forms including Z-DNA) is used, because even a left-handed duplex is formed for non-APP sequences, it does not necessarily exist in Z-form. Wang et al. showed that a sequence of 5′-d(CGATCG)-3′ with the cytosines brominated or methylated on C-5 can also form Z-DNA, indicating that modified non-APP base pairs (5′-d(GA)-3′/5′-d(TC)-3′) are allowed to be present in a Z-DNA structure (43). Wu et al. also showed that chiral ruthenium complexes may induce random sequences to form Z-DNA (37).
Interestingly, as early as 1979, Stettler et al. showed that after hybridization of two long ssDNA circles (4361 nt) with complementary sequences, CD spectra different from that of B-conformation were observed, and they called it V DNA (44). Later, Pohl et al. found that antibodies to Z-DNA could bind to V DNA, indicating that V DNA might contain left-handed duplex of Z-type (45). We suppose that some non-APP sequences in V DNA they used may form lh-DNA. However, it is not clear that all lh-DNA sequences in V DNA take Z-conformation or not. In their case, it is also unknown that the Z-DNA antibody binds to non-APP sequences or not, because more than 30 APP sequences are present of 6 bp or longer. They used the CD spectra of poly(d(GC)n) under 2.3 M NaCl (about 50% of B-from and 50% of Z-form) as the control. It is well known that CD spectra of B-form changes greatly with some repetitive sequences such as poly(dG)/poly(dC) and poly(dA)/poly(dT), and higher ion concentrations (e.g. comparison between 10 mM or 3.5 M Na+) also caused big changes (46). Obviously, the CD spectra of V DNA (containing about 50% B-DNA) cannot represent those of lh-DNA with non-APP sequences. In addition, supercoil can also form for this long plasmid DNA, and it is hard to analyze the exact ratio of lh-DNA because the density of supercoils affects it. Furthermore, the number of B–Z-junctions is also not clear for V DNA. More controllable molecular design is required to study lh-DNA of non-APP sequences.
Recently, we successfully prepared stable natural Z-DNA using two complementary circular ssDNAs of 74–111 bp in a buffer of 10 mM MgCl2 (47). For these short circular dsDNA, no supercoil can form. More interestingly, we showed that a 15 bp long non-APP sequence may also form lh-DNA (47). All above researches enlighten us to answer the question that non-APP sequences can really form stable lh-DNA or not. If formed, does the lh-DNA take Z-formation or a new one? This can help us to understand more helix structures of biopolymers. In this study, we designed several sequences with various content of APP and prepared topologically constrained circular dsDNAs in a buffer (10 mM HEPES, pH 7.5, 10 mM MgCl2) with ionic strength close to physiological conditions. Its linking number is zero, containing two related topological domains, one has linking number contribution of + n (left-handed) and the other has -n (right-handed) (Figure 1). Similarly, as we reported, the Z–B-chimeras (also designated as LR-chimeras, LR means left and right) are formed involving the lh-DNA part and the B-form one. The results also showed that non-APP sequences can form lh-DNA with similar thermal stability as APP sequences. Furthermore, we confirmed our suppose that APP sequences prefer to form lh-DNA with Z-conformation because its thermal stability is more close to its isomer of B-DNA, as compared with non-APP ones.
Figure 1.
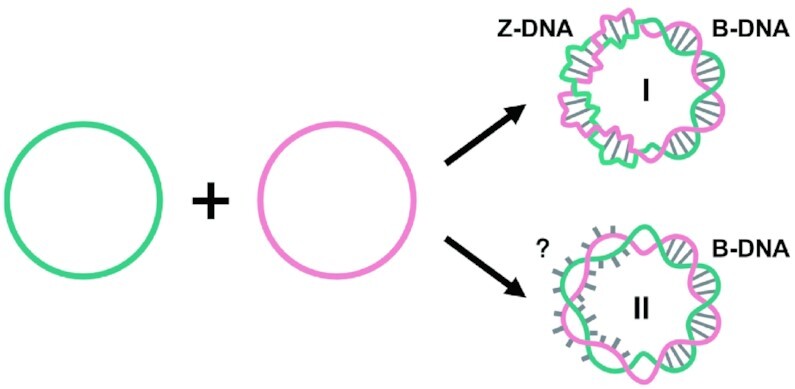
Schematic diagram of duplex formation by hybridization of two entirely complementary circular ssDNA under topological constraints. The two possible cases of Z-conformation (I) and non-Z-formation (II) are shown. For structure I, the left-handed part takes the Z-conformation. For II, base pairs are not formed in the left-handed part. If mismatched sequences are put at the left-handed part, structure of form II prefers to form (another possibility is to form a mismatched duplex with Z-conformation).
MATERIALS AND METHODS
Materials
T4 DNA ligase and Exonuclease I were obtained from Thermo Scientific (Pittsburgh, PA, USA). EcoRI, MboI, Hpych4Iv and SphI were purchased from New England Biolabs Inc. (Ipswich, MA, USA). The fluorescent dye of EvaGreen was from Biotium (Fremont, CA, USA), and Ultra GelRed (a dye staining both dsDNA and ssDNA) was purchased from Vazyme (Nanjing, China). Z-DNA-specific antibody (Z22) was from Absolute Antibody Ltd. (Oxford, UK), and ZBP1 (recombinant Z-DNA-binding protein) was from Wuhan USCN Business Co., Ltd. (Wuhan, China). All other chemicals were from Sigma-Aldrich (St. Louis, MO, USA).
All oligonucleotides used in this study (see Supplemental Table S1 for sequences) were purchased from GENEWIZ (Suzhou, China). Sequence were designated as follows. ls: linear ssDNA strand; cs: circular ssDNA strand; Sp: direct the head-tail ligation of a linear oligonucleotide to a circular one; lc: hybridization of a linear strand (α) and a circular one (β); cl: hybridization of a circular strand (α) and a linear one (β); cc: hybridization of two circular strands. 740: the duplex containing a continuous APP sequence; 741 and 743 are duplexes containing non-APP sequences; 741–20 and 741–35 are duplexes containing 20 bp mismatches and 35 bp mismatches, respectively. cc-742 is a duplex formed with one strand of poly(purine) and the other strand of poly(pyrimidine).
Preparation of circular DNA
For a typical reaction, to 3.5 μl of pure water, 1.0 μl of 1 × T4 DNA ligase buffer, ssDNA oligonucleotides ls74α1 (10 μM, 2.0 μl) and Spα1 (20 μM, 3.0 μl) were added (see Supplemental Table S1 for the sequences). After being heated to 90°C for 3 min, the solution was cooled gradually (0.1°C/s) to 37°C and kept for 20 min. Then, 0.5 μl of T4 DNA ligase (2.5 U) was added. The ligation reaction (10 μl in total) was carried out at 37°C for 2 h. The ligase was deactivated by incubation at 65°C for 10 min. The cyclization of other sequences was the same as above. To the above reaction containing cs74 solution (2.0 μM, 10 μl), 1.0 μl of 10 × Exonuclease I reaction buffer (pH9.5 at 25°C, 670 mM glycine–KOH, 67 mM MgCl2, 10 mM DTT) and 0.5 μl of Exonuclease I (10 U) were added, and the digestion reaction was carried out at 37°C for 10 h. The Exonuclease I was deactivated at 80°C for 15 min. After gel cutting and recovery, DNA was precipitated with ethanol to remove proteins and ions (46). Finally, the obtained circular DNA samples were stored in the buffer of 0.1× TE (1.0 mM Tris–HCl and 0.1 mM EDTA, pH8.0) and analyzed by Nanodrop 2000 spectrophotometer (Thermo Scientific, USA).
Preparation of circular double-stranded DNA (dsDNA)
First, 1.0 μl of 10× HEPES buffer (100 mM HEPES, pH7.5, 100 mM MgCl2) was added to 5.0 μl pure water, and then 2.0 μl cs74α1 (20 μM) and 2.0 μl cs74β1 (20 μM) were added (10 μl in total). After being heated to 90°C for 3 min, the solution was cooled gradually (0.1°C/s) to 25°C and remained for 20 min, and finally cc-741 (LR-chimera) was obtained. The same treatment was used to obtain ll hybrid, lc hybrid, cl hybrid, and cc hybrid (l means linear, and c means circular). To obtain B-form cc, 2.0 μl cs74β1 (20 μM), 2.0 μl ls74α1 (20 μM) and 1.0 μl of 10× HEPES buffer were added to 4.5 μl pure water. After being heated to 90°C for 3 min, the solution was cooled gradually (0.1°C/s) to 37°C and kept for 20 min. Then, 0.5 μl of T4 DNA ligase (2.5 U) was added. The ligation reaction (10 μl in total) was carried out at 37°C for 12 h. The obtained 740 and 741 hybrids were analyzed on an 8% native polyacrylamide gel in 1× TBE running buffer, and the temperature of gel was kept at 10°C by a water-bath (cooling water circulating machine, Zhixin, China). 741–20, 741–35, 742 and 743 hybrids were analyzed on 8% native polyacrylamide gel with 10 mM MgCl2 in 1× TBE running buffer containing 1.0 mM MgCl2, and the temperature of gel was also kept at 10°C.
Measurement of Tm by HRM
The Tms of the hybrids were determined with the high resolution melting (HRM) method (48). The 2.5 μl hybrid (4.0 μM) and 1.0 μl 10× EvaGreen fluorescent dye (specifically stain dsDNA part) were dissolved in the 6.5 μl 1× HEPES buffer (10 mM HEPES, pH7.5, 10 mM MgCl2). The fluorescence data were collected with 0.1°C increments (holding 2 s) from 37°C to 90°C by a PikoReal Real-Time PCR instrument (Thermo Scientific, Finland). At least three parallel tests were made for one plate, and Tms were calculated by the first derivatives of melting curves.
Mfold calculation
The Tm and Gibbs free energy (ΔG) of the 39 bp dsDNA in this study were simulated by “The mfold Web Server” using “Hybridization of Two Differernt Strands of DNA or RNA” (http://www.unafold.org/mfold.php) (49). The temperature range is from 25°C by the intervals of 1°C to 100°C, [dsDNA] = 1.0 μM, [Na+] = 10 mM, [Mg2+] = 10 mM. The ΔG values at 25°C were used.
Gel shift assay with antibodies and ZBP1
Z-DNA-specific antibodies (Z22) and ZBP1 (a protein containing the Zα domain) were used. For 10 μl reaction system, the 5.0 μl cc-741 (0.5 μM) and 0.5 μl 10 × HEPES buffer (100 mM HEPES, pH7.5, 100 mM MgCl2) were added to 2.0 μl pure water, and then 2.5 μl Z22 (5.0 μM) or 2.5 μl ZBP1 (10 μM) was added and incubated at 25°C for 2 h. The gel shift assay of other sequences with Z22 and ZBP1 was carried out as above. The products of hybrids were subjected to 6% native polyacrylamide gel electrophoresis in 1× TBE running buffer. The products of 741–20, 741–35 and 742 hybrids were also subjected to 6% native polyacrylamide gel electrophoresis with 10 mM MgCl2 in 1× TBE running buffer containing 1.0 mM MgCl2.
Circular dichroism (CD) spectroscopy
Spectra were recorded from 210 to 320 nm at 100 nm/min by a J-815 spectropolarimeter. Quartz cuvette with a path length of 1 mm was used. Each CD spectrum presented here was an average of at least 3 scans. Samples were equilibrated at room temperature (20 ± 2°C) for at least 3 min before the measurement. The typical solution contained the hybrid (4.0 μM) in 1× HEPES buffer (10 mM HEPES, pH7.5, 10 mM MgCl2). The baseline was calibrated with the corresponding buffer. Difference spectra were obtained by subtracting 0.47 × [the spectrum of the corresponding cl hybrid] from [the original spectrum of the cc hybrid], where the length of the B-DNA portion in the hybrids was evaluated by using the factor 0.47. This value was calculated by 10.5/(10.5 + 12) since the twisting number of the Z-DNA portions (∼12 bp/turn) in the hybrids must be equal to the number of the B-DNA portions (∼10.5 bp/turn).
Cleavage of the dsDNA by restriction enzymes
The circular DNA duplexes with or without a nick were cleaved by restriction enzyme SphI (5′-GCATG↓C-3′), EcoRI (5′-G↓AATTC-3′), MboI (5′-↓GATC-3′), or Hpych4Iv (5′-A↓CGT-3′). To 3.0 μl 1 × HEPES buffer (10 mM HEPES, pH7.5, 10 mM MgCl2), 5.0 μl LR-chimera (0.5 μM), 1.0 μl restriction enzyme (5.0 U) and 1.0 μl 10× NEB cutsmart buffer (50 mM KAc, 20 mM Tris-acetate, 10 mM Mg(Ac)2 and 100 μg/ml BSA) were added. After incubating at 37°C for 5 min, the reaction was terminated by adding 1.0 μl of EDTA (0.5 M, pH8.0). The products were subjected to an 8% native polyacrylamide gel in 1× TBE running buffer, and the temperature of gel was kept at 10°C.
Cleavage of LR-chimeras by restriction enzymes after Z22 binding
For a 16 μl reaction system, after mixing the 10 μl LR-chimera (0.5 μM), 1.0 μl 10× HEPES buffer (100 mM HEPES, pH7.5, 100 mM MgCl2), and 5.0 μl Z22 (5.0 μM), the solution was incubated at 25°C for 2 h. Then 2.0 μl restriction enzyme (5.0 U) and 2.0 μl 10× NEB cutsmart buffer (50 mM KAc, 20 mM Tris-acetate, 10 mM Mg(Ac)2 and 100 μg/ml BSA) were added. After incubating at 37°C for 30 min, the reaction was terminated by adding 2.0 μl of EDTA (0.5 M, pH 8.0). The proteins were extracted by phenol/chloroform and then subjected to an 8% native polyacrylamide gel in 1× TBE running buffer, and the temperature of gel was kept at 10°C.
RESULTS
The strategy used in this study is illustrated in Figure 1. By using the approach we reported (47), the left-handed duplex (lh-DNA) is forced to form by hybridization of two mutually complementary circular ssDNAs under a ionic strength condition close to that in the physiological environment. The hybrid of two complementary ssDNA circles is designated as LR-chimera, involving left-handed part and right-handed part. These two parts have the same twist number in the contrary direction so that the total linking number is zero. APP or non-APP sequences are introduced in the LR-chimera, and the thermal stability and structure of the left-handed part can be investigated by CD, Tm measurement, and interaction with Z-DNA binding proteins. The left-handed part can take Z-form (I in Figure 1) or another conformation (II in Figure 1, e.g. two strands twist around each other with bases extruded to outside of the duplex).
Sequence design
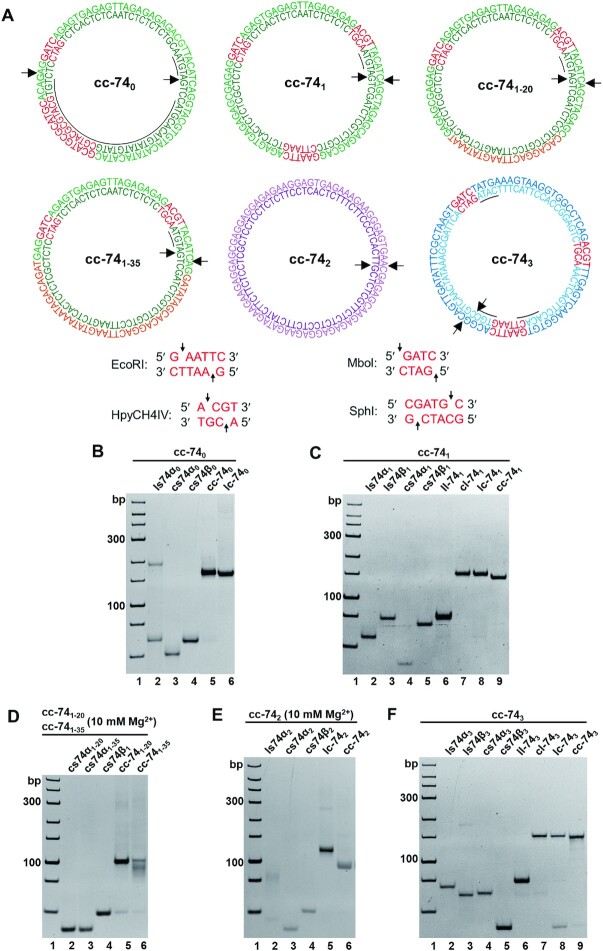
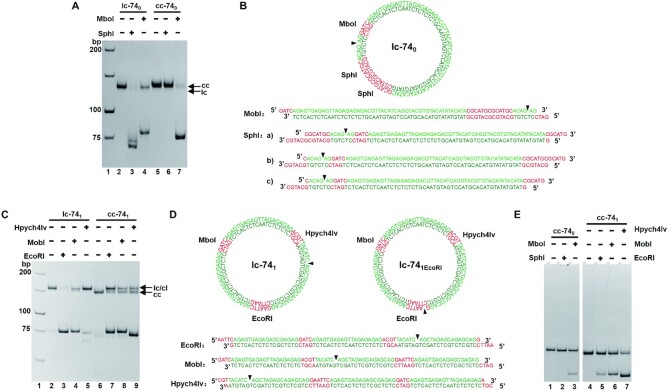
For sequences in genome DNA, short APP sequences (<6 bp) are usually scattered, and complete non-APP sequences (i.e. poly(purine) or poly(pyrimidine)) are very rare. Actually, short APP sequences (<6 bp) are believed to hardly form Z-DNA. We designed six groups of LR-chimeras (Figure 2A): cc-740 contains a continuous APP sequence (34 bp) with only very short d(CG)n (n ≤ 2); cc-741 is a non-APP sequence and there is only one 4 bp continuous APP sequence (5′-TACA-3′); cc-743 contains only three 4 bp continuous APP sequences (5′-CATA-3′, 5′-CGTG-3′, 5′-CACA-3′). We also designed cc-742, a non-APP sequence basically with one strand of poly(purine) and the other strand of poly(pyrimidine). In addition, we designed cc-741–20 with 20 bp base mismatches and cc-741–35 with 35 bp base mismatches. Four hybrids (cc-740, cc-741, cc-741–20, cc-741–35) have the same consecutive 39 bp of non-APP sequence. In order to analyze the location of left-handed part, recognition sites for restriction enzymes were introduced to above sequences. For cc-740, recognition sites for restriction enzymes SphI (5′-GCATG↓C-3′) and MboI (5′-↓GATC-3′) were designed in APP and non-APP sequences, respectively. For cc-741 and cc-743, we used three restriction sites (EcoRI (5′-G↓AATTC-3′), MboI (5′-↓GATC-3′) and Hpych4Iv (5′-A↓CGT-3′)), whose positions are separated to ensure at least one restriction site locates in the left-handed region.
Figure 2.
Designed sequences and hybridization results for LR-chimeras with various sequences. (A) Sequences of six LR-chimeras of APP (cc-740), non-APP (cc-741, cc-742 and cc-743), and two mismatched sequences (cc-741–20 and cc-741–35, containing 20- and 35-bp mismatches (orange), respectively). The red parts show restriction enzyme recognition sites, and the underlines show continuous APP sequences. Arrows indicate the terminals before circularization. (B–F) PAGE analysis of hybridization for cc-740, cc-741, cc-741–20, cc-741–35, cc-742 and cc-743 respectively. Name of samples are shown on top of the gel. The abbreviations are shown as follows. ls: linear ssDNA strand; cs: circular ssDNA strand; lc: hybridization of liner α strand and circular β strand; cl: hybridization of circular α strand and linear β strand; cc: hybridization of two circular strands. For (D) and (E), 10 mM MgCl2 was present in the gel, and 1.0 mM MgCl2 was added in the running buffer. All the gels were kept at 10°C during electrophoresis.
Stability analysis of LR-chimera formed from two mutually complementary circular ssDNAs
At first, circular ssDNAs of designed sequences were prepared and purified (Supplemental Figure S1). After two complementary strands of purified circular ssDNAs were hybridized at ionic strength close to physiological conditions (10 mM HEPES, pH7.5, 10 mM MgCl2), the formation of LR-chimera was analyzed by 8% PAGE (Figure 2B-F). As expected, new bands assigned as dsDNA of cc-740 (lane 5, Figure 2B), cc-741 (lane 9, Figure 2C) and cc-743 (lane 9, Figure 2F) were observed, which have similar mobility as the dsDNA formed from the circular ssDNA and its complementary linear one (lane 6, Figure 2B; lanes 7 and 8, Figure 2C; lanes 7 and 8, Figure 2F). It should be noted that during PAGE analysis, only 89 mM Tris-H3BO4 and no MgCl2 were present, indicating that the formed cc-740, cc-741 and cc-743 were even stable under these low ionic strength conditions. Interestingly, for cc-741–20 and cc-741–35, in which 20 or 35 mismatches are present (Figure 2A), no hybridization was observed under the same conditions as above (data not shown). When 10 mM of MgCl2 was present in the gel, efficient hybridization was observed for cc-741–20, and partial hybridization was observed for cc-741–35 (lanes 5 and 6, Figure 2D). These results showed that cc-741–20 and cc-741–35 are not as stable as cc-741, indicating that non-APP in cc-741 did not form a similar structure as cc-741–20 or cc-741–35 (e.g. form II, Figure 1). For poly(purine)/poly(pyrimidine) sequence of cc-742, only trace of hybridization was observed if no MgCl2 was present in the gel (data not shown). Complete hybridization of cc-742 was observed when 10 mM MgCl2 was present in the gel (lane 6, Figure 2E).
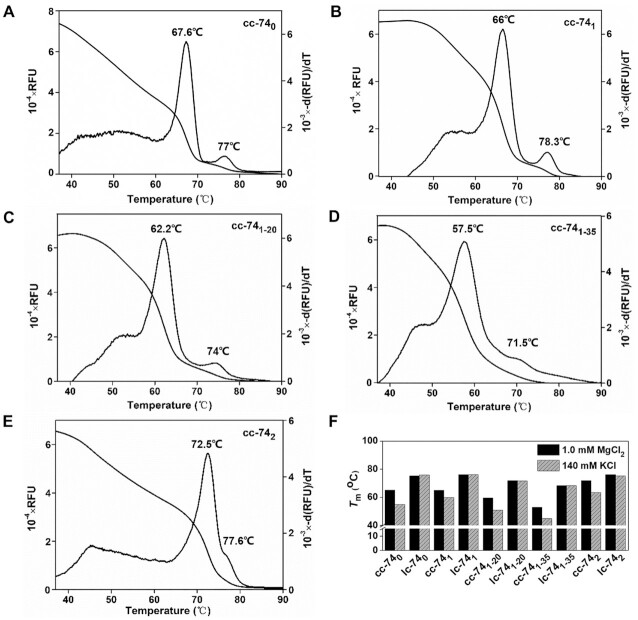
We have developed an approach to measure Tm of the short duplex by HRM, based on the fluorescence change of the dyes binding specifically to DNA duplex (48,50), and it can be used to evaluate the stability of the LR-chimera (47). Here we used this approach to analyze the stability of LR-chimeras and lc duplexes (1.0 μM) in detail (Figure 3). For each measurement, besides the main peak (57∼73°C), another weak peak appeared at higher temperatures (71∼79°C). By measuring the Tm of a hybrid (cl) between a ring and its complementary strand, the peak at higher temperature was determined to be cl's Tm. This should be caused by the breakage of a small amount of circular ssDNA during the recovery process. As a result, the peak at lower temperature was assigned to be the Tm of cc. Interestingly, the Tm values of cc-740 and cc-741 were very close, which are 67.6°C and 66°C, respectively (Figure 3A, 3B). The corresponding Tms of 39 bp B-form duplexes (the part designed to form left-handed helix) of 740 and 741 were almost the same, which were determined by mfold (49) to be 78.3°C and 78.7°C, respectively. These results show that the stability of left-handed part formed by non-APP sequence may be comparable to that by APP sequence. It demonstrates that left-handed part in cc-741 formed lh-DNA (base-paring left-handed part), but not mismatched one like structure II (Figure 1).
Figure 3.
Thermal stability (Tm) of circular ssDNA hybrids measured by HRM. (A–E) Melting curves and differential curves of cc-740, cc-741, cc-741–20, cc-741–35 and cc-742, respectively. [dsDNA] = 1.0 μM, [HEPES] = 10 mM, [MgCl2] = 10 mM, pH7.5. The small peaks of differential curves at higher temperatures are assigned to be the Tms of lc- or cl- duplexes formed by a linear ssDNA and a circular ssDNA. (F) Tms of cc and lc measured by HRM in 1.0 mM MgCl2 or 140 mM KCl. [dsDNA] = 0.25 μM, [HEPES] = 10 mM, pH7.5.
To further prove that cc-740 and cc-741 did not form structure II, Tms of cc-741–20 and cc-741–35 were measured, which are 62.2°C and 57.5°C, respectively (Figure 3C, D). It should be noted that for cc-741–20 and cc-741–35, if a 54 bp B-form duplex (10.4 bp/turn) forms, the left 20 bp mismatched duplex should twist at least 5 turns of left-handed, which is very difficult. The relatively lower Tms may be caused by the constraint that only 3 or 4 turns of B-form duplex (31–42 bp) can form, and the left 30–40 bp duplex has to take a left-handed conformation of 3 or 4 turns. Thus, at least 10 to 20 bp of complementary part form left-handed part for cc-741–20. For cc-741–35, basically, the 35 bp mismatches should form structure II (Figure 1) or mismatched lh-DNA. Accordingly, from the fact that Tm for cc-741 was much higher than that of cc-741–35, it can be deduced that base pairs of left-handed duplex should form in cc-741. Obviously, for cc-740, Z-DNA should form in the APP sequence region (47). Unexpectedly and interestingly, the Tm of cc-742 was determined to be 72.5°C, which is quite higher as compared with other sequences. This result indicates that poly(purine)/poly(pyrimidine) could also form stable lh-DNA in 10 mM MgCl2 (Figure 3E) once the topological constraint is strong enough, which is completely different as we knew previously (51,52). However, this does not necessarily mean that these sequences form lh-DNA more easily than APP ones (as discussed later).
More interestingly, we noticed that the difference in Tm between cl and cc of the same sequence for the APP sequence was 9.4°C (Figure 3A), and for the non-APP sequence was 12.3°C (Figure 3B). As mentioned previously, the difference in Tm of 39 bp B-form duplexes (the part designed to form left-handed helix) of 740 (Tm = 78.3°C) and 741 (Tm = 78.7°C) was only 0.4°C. This shows that the difference in Tm between 741 and 740 is really caused by the stability difference of left-handed part. Although we do not know the difference in Tm between B-DNA and its corresponding lh-DNA part with the same sequence for cc-740 (ΔTmAPP) or cc-741 (ΔTmnon-APP), we can estimate that the ΔΔTm (ΔTmnon-APP – ΔTmAPP) is about 2°C.
We further measured the Tms at various concentrations of MgCl2 and 140 mM KCl (Supplemental Table S2, Supplemental Figure S2). Tms of cc-740, cc-741, and cc-742 in buffers of various concentrations of MgCl2 (1.0–10 mM) are almost unchanged, which means that they are stable at 1.0 mM MgCl2 (Supplemental Table S2), indicating that the LR-chimeras by non-APP sequences are stable even at 1.0 mM MgCl2 under topological constraints. Very interestingly, we found that Tms for all the LR-chimeras (cc) decreased obviously (5.0–10.6°C) if 140 mM KCl was used instead of 1.0 mM MgCl2, but for their lc isomers almost no change of Tm was observed (Figure 3F). This result is coincident with the fact that divalent cations can stabilize Z-DNA much more than monovalent cations, as compared with B-DNA (53). Interestingly, for cc-741 (non-APP), Tm in 140 mM KCl is only 5.0°C lower than that in 1.0 mM MgCl2, but for cc-740 (APP) this value is as large as 10.6°C. The lh-DNA of APP sequences (Z-DNA) seems more sensitive to cations. In other words, cation of Mg2+ stabilizes lh-DNA for APP sequences more than that for non-APP sequences.
Binding of specific proteins to LR-chimeras
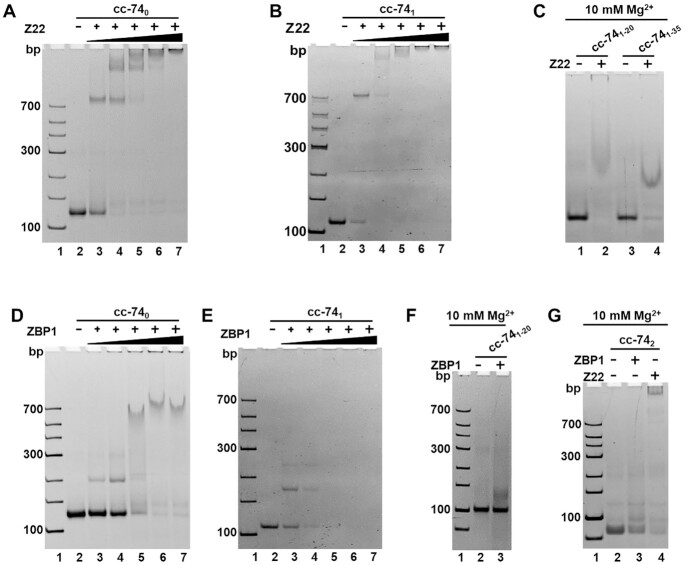
In 1981, Lafer et al. obtained a high-affinity antibody to Z-DNA with brominated modification (54). This antibody can specifically recognize and bind to the skeleton of Z-DNA without sequence specificity (55,56). Combined with modern biotechnology, the recombinant Z-DNA antibody (Z22) was prepared for Z-DNA detection. Here, we used Z22 to verify whether Z-DNA conformation was present in the LR-chimera we obtained. The results of Z22 (0.25, 0.5, 0.75, 1.0, 1.25 μM) binding to LR-chimeras (0.25 μM) in 1
× HEPES buffer (10 mM HEPES, pH7.5, 10 mM MgCl2) are shown in Figure 4A–C, G. The band for cc-740 (Figure 4A) and cc-741 (Figure 4B) disappeared gradually with the increase of Z22 concentration, and new bands were observed, demonstrating that Z22 binds strongly. For cl-740, cc-740B, cl-741 and cc-741B (0.25 μM) without topological constraint, almost no binding was observed even for 1.25 μM of Z22 (Supplemental Figure S6A, S6C, S6E). Very interestingly, for cc-741–20 and cc-741–35 with mismatched base pairs, the binding of Z22 was also observed when 10 mM of MgCl2 was present in the Gel (Figure 4C), although no binding was observed if MgCl2 was absent in the Gel (data not shown). This showed that Z-DNA-like conformation may form even with mismatched base pairs to some extent.
Figure 4.
Gel shift assay for binding of Z22 (Z-DNA-specific antibody) and ZBP1 to LR-chimera of various sequences. (A) Z22 to cc-740; (B) Z22 to cc-741; (C) Z22 to cc-741–20 and cc-741–35; (D) ZBP1 to cc-740; (E) ZBP1 to cc-741; (F) ZBP1 to cc-741–20; (G) Z22 or ZBP1 to cc-742. 10 mM MgCl2 was present in the gel for cc-741–20, cc-741–35 and cc-742. The gel was kept at 10°C during running the gel. 0.25 μM of LR-chimera was used. For (A, B) 0.25, 0.5, 0.75, 1.0 and 1.25 μM of Z22 were used. For (D, E) 0.25, 0.625, 1.25, 1.875 and 2.5 μM of ZBP1 were used; For (C, F and G) 2.5 μM ZBP1 or 1.25 μM of Z22 was used.
When another Z-DNA binding protein ZBP1 was used for cc-740 and cc-741 (57), similar results were obtained but with weaker binding activities (Figure 4D–F). Another difference was that the cc-741 band becomes weaker with the increase of ZBP1 concentration, but there are no new clear bands observed. Again, for cl-740, cc-740B, cl-741 and cc-741B, no binding to ZBP1 (2.5 μM) was observed (Supplemental Figure S6B, D, E). Not like Z22, ZBP1 did not bind to cc-741–20 protein at all (lane 3, Figure 4F).
Binding results of ZBP1 and Z22 to cc-742 are shown in Figure 4G. Interestingly, even for this poly(purine)/poly(pyrimidine)-rich sequence, which is most different from the APP sequences, binding of these two Z-DNA specific binding proteins was also observed (lanes 3 and 4, Figure 4G). For this PAGE analysis, 10 mM MgCl2 was present in the gel. When normal PAGE without MgCl2 was carried out, no binding was observed (data not shown), showing that ZBP1 and Z22 binding to cc-742 requires relatively strong ionic strength. As compared with Z22, ZBP1 bound to cc-742 much more difficult (see lane 3 with lane 4). The dsDNA cc-743, cc-92 and cc-109 formed by other non-APP sequences (containing only very small content of APP sequences) also showed strong binding with Z22 and ZBP1 (Supplemental Figure S3-S5), which confirmed that any random sequence may form Z-DNA-like lh-DNA by strong topological constraints.
Circular dichroism (CD) analysis of LR-chimera with APP and non-APP sequences
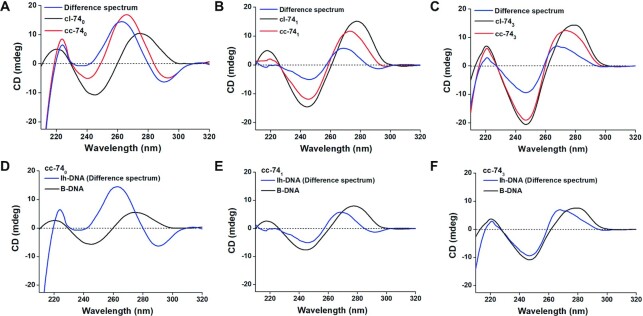
Circular dichroism (CD) spectroscopy brings us the information about helix structures. For DNA structure, CD spectra at 210–320 nm can reflect the interaction between base pairs in a helix (46,58). Figure 5 shows the CD results for LR-chimeras of cc-740, cc-741 and cc-743. Circular dsDNAs with a nick of cl-740, cl-741 and cl-743 with B form structures were used for comparison. Obviously, cc-740 showed a quite different CD spectrum compared with cl-740 (Figure 5A). After subtraction of CD spectrum of B-form, a typical Z-DNA spectrum was obtained (Blue line, Figure 5A). For cc-741 and cc-743, however, relatively weak difference spectra were obtained (Blue line, Figure 5B and C). Obviously, this result shows that left-handed duplex structures of APP and non-APP sequences are quite different. Very interestingly, for APP sequences (cc-740), CD intensity of lh-DNA (difference spectrum) is much stronger than that of B-DNA of the same length (Figure 5D). It should be noted that we used continuous APP sequence involving both G–C and A–T base pairs (about 43% GC content). However, the CD intensity of lh-DNA for non-APP sequences is weaker to some extent than that of B-DNA (Figure 5E and F).
Figure 5.
CD spectra of the cc-740, cc-741 and cc-743. (A) Spectrum of cc-740 with APP sequence; (B, C) spectra of cc-741 and cc-743 with non-APP sequence. Red lines show the original spectra; Black lines show the spectra cl-740 and cl-741 (typical B-DNA); blue lines show the difference spectra of cc-74–0.47 × cl-74. (D–F) Spectra of lh-DNA and B-DNA. Spectra for lh-DNAs here are the same ones as difference spectra in A-C. Spectra for B-DNA are 0.53 (39/74) times of those for cl-74 DNAs. [dsDNA] = 4.0 μM, [MgCl2] = 10 mM, [HEPES] = 10 mM, pH 7.5, 20 ± 2°C.
Determination of the position of left-handed DNA sequence in the LR-chimeras using RE cleavage
It has been shown that restriction enzymes (REs) cannot cleave the left-handed duplex with their recognition sites (59–62). Usually, they are very sensitive to the duplex structure, and B-DNA with only one different base pair at the recognition site cannot be cleaved. Here, based on the difference in cleavage efficiency of several restriction enzymes (recognition sites are shown in Figure 2A), we tried to determine the position of left-handed part in cc-740 and cc-741 (Figure 6, Supplemental Figure S7). As shown in Figure 6A, for cc-740, MboI cleaved more than 90% into linear dsDNA (lane 7), but no cleavage was observed for SphI (lane 6). As expected, SphI cleaved almost 100% of B-form duplex with the same sequence (lc-740, lane 3). MobI did not completely cleave lc-740 (lane 4). It should be noted that the nick is very close to its recognition site (Figure 6B). When the B-form circular duplex (cc-740B) was used, almost all DNA was cut (Supplemental Figure S7C).
Figure 6.
Analysis of the positions of left-handed DNA in the LR-chimeras (8% PAGE). (A) Cleavage (5 min) of lc-740 and cc-740. Lane 2: lc-740; lanes 3 and 4: lc-740 cleaved by SphI and MobI, respectively; lane 5: cc-740; lanes 6,7: cc-740 cleaved by SphI and MobI, respectively. (B) Schematic illustration of lc-740 and linear products of cleavage by SphI and MobI, respectively. (C) Cleavage (5 min) of lc-741 and cc-741. Lane 2: lc-741; lanes 3–5: lc-741 cleaved by EcoRI, MobI, and Hpych4Iv, respectively; lane 6: cc-741; lanes 7–9: cc-741 cleaved by EcoRI, MobI, and Hpych4Iv, respectively. (D) Schematic illustration of lc-741, lc-741EcoRI and linear products of cleavage by EcoRI, MobI, and Hpych4Iv, respectively. (E) Cleavage (30 min) of cc-740 and cc-741 after binding to Z22. Lane 1: Z22 bind to cc-740; lanes 2, 3: Cleavage of cc-740 by SphI and MboI after binding to Z22, respectively; lane 4: Z22 bind to cc-741; lanes 5–7: Cleavage of cc-741 by EcoRI, MboI, and Hpych4Iv after binding to Z22, respectively.
Cleavage of cc-740 was further carried out by SphI and MboI after Z22 binding (Figure S6E). Interestingly, MboI cleaved only 25% of cc-740, showing that binding of Z22 affected the cleavage by Mbol, whose recognition site is located in B-DNA near the B-Z junction. It should be noted that the APP sequence is only 4 bp away from this recognition site. Again, no cleavage of cc-740 was observed for Sphl after Z22 binding (lane 2, Figure 6E), confirming that the APP region where the SphI recognition site is located formed lh-DNA in cc-740. As expected, when Z22 was present in the case of lc-740 cleavage by SphI and MboI, no effect was observed (data not show). These results showed that the APP sequence in cc-740 took Z-conformation, which is consistent with our previous results for other APP sequences (47).
Cleavage results of lc-741 and cc-741 (almost no APP sequences) by EcoRI, MboI, and Hpych4Iv are shown in Figure 6C. For EcoRI, almost all lc-741 disappeared, and a new band with much higher mobility was observed (lane 3). This new band can be assigned as a linear dsDNA with a nick (formed by two terminals of linear strand ls74α1) and two 4-bp-long sticky ends (Figure 6D). For cc-741, about 53% of LR-chimera was cleaved to a linear duplex by EcoRI (lane 7, Figure 6C). A new band with the same mobility as lc-741 was observed, showing that the cc-741 was nicked for 5 min by EcoRI. After 30 min of reaction these nicked products were further cleaved to linear dsDNA (Supplemental Figure S7F). It can be seen that EcoRI cleaved cc-741 more slowly as compared with lc-741. The nicking can be explained as follows. Once EcoRI cleaves one strand of cc-741, the topological constraint is released, and a duplex of lc-741EcoRI forms (Figure 6D). As the dsDNA is only 74 bp long, several base pairs prefer to dissociate due to the rigidity of dsDNA, so that it is not so easy to be cleaved again by EcoRI. When the B-form circular dsDNA (cc-741B) was cleaved, which was obtained by circularization after hybridization of a linear ssDNA and the circular complementary strand, all cc-741B was almost cleaved (Supplemental Figure S7A). Because cc-741B has only B-form duplex, EcoRI can bind strongly and cut both strands simultaneously.
For Mbol, similar cleaving efficiency was observed for cc-741, lc-741 and cc-741B (Figure 6C, Supplemental Figure S7A). For Hpych4Iv, cleavage of cc-741 was even more efficient than lc-741 (comparing lane 9 with lane 5). Hpych4Iv cannot cleave lc-741 efficiently due to the fact that the nick on lc-741 is only 6 bp away from the recognition site. When we used another sequence on which the nick is far from the recognition site of Hpych4Iv, the cleaving efficiency was greatly improved under the same conditions (Supplemental Figure S7B). Hpych4Iv cleavage of cc-741B was also carried out, and similar cleavage activity for cc-741 and cc-741B was observed (Supplemental Figure S7A).
From above results it is hard to determine exactly the location of lh-DNA. To clarify this, effect of Z22 binding on cleavage of cc-741 by REs was further investigated (Figure 6E). The results showed that, even after 30 mins, EcoRI, MboI and Hpych4Iv cleaved about 25%, 40% and 70% of cc-741, respectively. Obviously, Z22 binding affects greatly the activity of cc-741 cleavage by all these three REs. Accordingly, the recognition site of EcoRI may prefer to form lh-DNA. However, the location of lh-DNA part is not so distinct. Similar results were also obtained for cc-743 after Z22 binding, with cleavage by EcoRI being affected more than that by MboI and Hpych4Iv, demonstrating that the recognition site for EcoRI prefers more to form lh-DNA (Supplemental Figure S7G-I).
DISCUSSION
In this study, we tried to find the clues to answer the questions of whether non-APP sequences can form stable Z-DNA and why it was believed that only APP sequences can form Z-DNA in vivo. Usually, scientists consider that polymer with chiral backbone prefer to form only helix of one direction, because DNA duplex with D-ribose usually takes right-handed direction. The α-helix of peptide (involving only L-amino acid) usually takes right-handed direction. However, Yashima et al. found that many artificial polymers with chirality can form either left-handed or right-handed helix with similar probability (63). Actually, for DNA sequence d(CG)n, it takes about 50% of left-handed structure and 50% of right-handed structure when about 2.5 M NaCl is present (53). Obviously, under this condition, Z-DNA shows the same stability as B-DNA. By analyzing these facts, we consider that linear dsDNA takes right-handed structure (B-DNA) under normal conditions only because B-DNA is more stable than Z-DNA to a small extent (Figure 7). It does not mean that Z-DNA is so unstable, especially in the case that B-DNA cannot form with some constraint. For complementary sequences of non-APP, they can form left-handed duplex structure, which is much more stable as compared with the single-stranded state (Figure 7). We believe that the free energy between lh-DNA and B-DNA may be very close even for non-APP sequences, although the free energy of B-DNA is usually lower. Here, we estimated the difference in free energy at 25°C between lh-DNA and B-DNA (including APP sequences) for a 39 bp duplex is 5.3–8.4 kcal/mol (Figure 7). However, it should be pointed out that this value may be even lower if the B–Z junction is taken into account.
Figure 7.
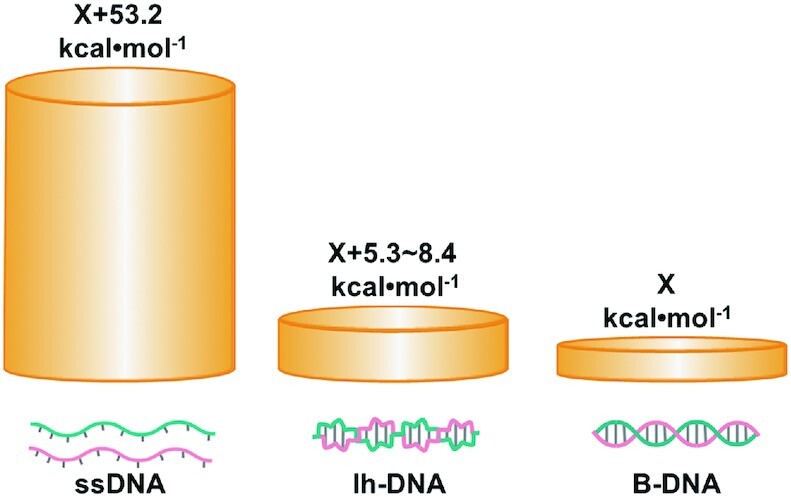
Comparison of free energy levels for linear ssDNA, lh-DNA (including Z-DNA), and B-DNA with the same sequence. Here, the sequence of 5′-d(AGCTAGAGCAGAGCAGGAATTCAGAGTGAGAGAGCGAGA)-3′ is used as an example. Suppose the absolute free energy for B-DNA is X kcal/mol, the corresponding free energy (at 25°C) of ssDNA is calculated by mfold to be X + 53.2 kcal/mol under the following conditions. ([dsDNA] = 1.0 μM, [Na+] = 10 mM, [Mg2+] = 10 mM). Suppose Tm for lh-DNA (Tm = 73–75.4°C) is 3∼6°C lower than B-DNA (Tm = 78.7°C), the free energy of lh-DNA should be X + 5.3–8.4 kcal/mol).
Basically, the above speculation was consistent with the results of hybridization, Tm measurement, and Z-DNA specific binding analysis of the LR-chimeras we designed. For cc-741 (non-APP), for example, it has the similar Tm and binding affinity to Z22 and ZBP1 as cc-740 (APP). The base pairs at the left-handed part should also form for cc-741, otherwise, it should have the similar Tm as the mismatched ones of cc-741–35 (Figure 3). Similar results were obtained for other non-APP sequences of cc-743, cc-92 and cc-109, indicating that this is the common nature of DNA (Supplemental Figure S3–S5). Similar CD spectrum as that of cc-741 was also obtained for another non-APP sequence of cc-743 (Figure 5). However, CD spectra for cc-741 and cc-743 (non-APP) are quite different from that for cc-740 (Figure 5). This result shows us that left-handed duplex structure formed by non-APP sequences may be different from normal Z-DNA one (with the zig-zag shape of backbone). Here we describe it as Z-DNA-like structure (another form of lh-DNA), because both Z22 and ZBP1 bind to it. Further analysis is required to determine its detailed structure. The different CD spectrum for non-APP sequences from that for APP sequences can be explained as follows. The unit of normal Z-DNA is formed by two base pairs of 5′-d(PyPu)-3′/5′-d(PyPu)-3′ with alternating anti-syn glycosidic bonding (64), but non-APP sequences cannot have this kind of very regular structural unit. The strong CD of Z-DNA for APP sequences (Figure 5D) may be attributed the strong interaction of two base pairs within one structure unit (58). For non-APP sequences, some of the two base pairs within the same unit may take a right-handed step to a small extent so that its lh-DNA has CD patterns similar as B-DNA (Figure 5E and F). Another possibility is that the random sequence may average the CD signal so that it is weaker and not so typical and obvious. The weak CD spectra of lh-DNA for non-APP sequences, whose pattern is also similar as B-DNA, probably made it hard be discovered.
We believe that the difference in Tm between cc-741 and cc-740 (1.6°C) is caused by the stability difference of left-handed part, because they have the same 35-nt-long sequence which forms B-form in cc-740 and tends to form B-form in cc-741. In addition, the left 39-nt-long different sequence (between cc-741 and cc-740) forms B-DNA with very close Tm. Accordingly, the thermal stability of lh-DNA of non-APP sequence in cc-741 is quite high, and even close to that of APP sequence in cc-740. On the other hand, these results also indicated that the difference in Tm between B-DNA and its corresponding lh-form with the same sequence for cc-741 (ΔTmnon-APP) is about 2°C bigger than that for cc-740 (ΔTmAPP). As a result, when topological constraint, such as negative supercoil, is present, we speculate that APP sequences are more preferable to form lh-DNA than non-APP ones, although the ΔΔTm (ΔTmnon-APP – ΔTmAPP) is not so big. In other words, as compared with non-APP sequences, the thermal stability difference for APP sequences between Z-form and B-form is smaller, which may be the reason that Z-DNA forms preferentially for APP sequences.
The unexpected high Tm (72.5°C,) for cc-742 (containing poly(purine)/poly(pyrimidine) basically) is a surprise. The migration rate of cc-742 is significantly faster than that of cl-742 (Figure 2E), which may be due to the easier bending of poly(purine)/poly(pyrimidine) (51,52). The Tm for lc-742 was determined to be 77.6°C, which is even a little bit lower than that of lc-740 and lc-741. One possible reason of this high Tm for cc-742 may be due to the regular structure, which is favorable for forming a regular left-handed duplex, as compared with random sequences. Another possible reason is that the duplex of this special sequence may be easier to bend like a duplex of polydA/polydT, so that free energy due to bending is less. We failed to obtain its CD spectra because the yield for the circularization of poly(purine) strand was extremely low, and not enough circular DNA was obtained.
For most of the sequences used, binding of ZBP1 and Z22 to LR-chimera was observed, although the mismatched ones and cc-742 showed very weak binding to ZBP1. Basically, we can deduce that ZBP1 and other Z-DNA binding proteins in organisms can only bind to left-handed DNA after it is formed by some constraint. Their function is to stabilize the formed Z-DNA but not inducing the transformation of B-DNA to Z-DNA. They can prevent Z-DNA from quickly changing back to B-conformation or other structures such as a cruciform or an open state. It has been reported that Z22 prefers to bind to Z-DNA part close to two B-Z junctions (65). This kind of binding is helpful for stabilizing the formed Z-DNA. Probably for the similar reason, clear bands were observed when the first Z22 molecule bound to LR-chimera (Figure 4). With the increase of Z22 concentration, the second Z22 molecule could bind to another Z–B junction. When concentration of Z22 increased further, Z22 started to bind to the middle part of lh-DNA with a relative smaller binding affinity. Multiple and diffuse binding bands were observed, probably because Z22 molecules bound to the middle region were easier to dissociate compared with those bound to the B-Z junctions. Another possibility is that the holding of two Z-B junctions helped fix the lh-DNA so that Z22 became easier to bind. As a result, the band for two Z22 binding to the LR-chimera was not so clear-cut. It should be noted that more alternating B-Z regions (>2) are hard to form in our LR-chimera, because B–Z junctions increase greatly the free energy (5.0 kcal/mol for each junction at 25°C) (66,67). However, binding of ZBP1 to lh-DNA seems quite different from that of Z22 (Figure 4). As compared with Z22, ZBP1 gel shift assay gave more smear bands for from cc-741, probably because ZBP1 molecules bind and dissociate from cc-741 quickly. In addition, the binding affinity of ZBP1 to various LR-chimeras (cc-740, cc-741 and cc-742) changed greatly as compared with Z22, indicating ZBP1 has more stringent structural requirements for binding to lh-DNA. Because these Z-DNA binding proteins can bind to lh-DNA for non-APP sequences, we speculate that it takes a Z-DNA-like structure. Considering that Z22 has been shown to mainly bind to the phosphate backbone, lh-DNA for non-APP sequences may have zig-zag backbone with alternating anti and syn nucleotides, but not so regular one as normal Z-DNA with APP sequences.
Our RE cleavage results also showed that there was no distinct location for lh-DNA in cc-741 (Figure 6). It is reasonable because the sequence is non-APP. This may be the reason that the activity of EcoRI cannot be completely inhibited. On the other hand, the results using the combination of RE and Z22 binding (Figure 6E) showed that lh-DNA preferred to form around the recognition site of EcoRI (lower half of cc-741 in Figure 2A). We found that the numbers of 3-bp-APP (3-bp-long continuous APP) sequence in the upper half (GTG, ACG (CGT), TAC(ACA)) and lower half (GCA, GCA, CAG, GTG, GCG) are 3 and 5, respectively. More (or high density of) short alternating purine pyrimidine sequences may be energy favorable to form lh-DNA. For cleavage of cc-741, the fact can be also explained that EcoRI gave more lc products compared with MboI and Hpych4Iv (Figure 6C). Only the rigidity factor of short dsDNA is not enough to edge off EcoRI after it forms a nick. However, if the recognition site of EcoRI prefers to form lh-DNA, the topological factor plus the rigidity one can edge off EcoRI after it cleaves one strand to form a nick. Accordingly, cl products (with a nick) were observed to a smaller extent for MboI and Hpych4Iv (lanes 8, 9, Figure 6C). Due to this preference to form lh-DNA at EcoRI recognition site, only EcoRI showed less efficient cleavage for cc-741 compared with lc-741 (lanes 3 and 7, Figure 6C). For LR-chimeras without APP sequences longer than 6 bp, a dynamic equilibrium may be present so that the two B–Z junctions can move (change locations) (60), causing not complete inhibition of EcoRI cleavage of cc-741, as well as of cc-743 (Supplemental Figure S7F and S7H). After strong binding of Z22, the structure should be fixed so that RE cleavage can give us more correct structural information.
Obviously, left-handed duplex prefers to form at the sequence where the free energy difference between these two conformations (left and right) is small enough. The relatively stable left-handed duplex can form in the case that either the local density of supercoil is high enough or some proteins can produce constraint by specific binding to left-handed conformation. From the crystal structure reported up to now, most Z-conformations have been found to be APP sequences, and the non-APP sequences are less than 7% (68). On the other hand, we may find some other sequences can form relative stable left-handed duplex, which has different structure to normal Z-DNA more or less. It should be noted that the chirality that causes DNA to prefer B-conformation comes only from the deoxyribose. There should be not a huge free energy difference between left-handed and right-handed duplex structure. In vivo, however, regulation of gene expression can smartly use this subtle difference between APP and non-APP sequences. Our results also show that, by forming left-handed duplex, non-APP sequences may also temporarily alleviate the abnormal stress of negative supercoil caused by transcription. Without protein binding, relatively long ssDNA (>20 bp) can only be present in a very short time scale, because it has higher free energy than left-handed duplex.
In summary, we found that non-APP sequences can form Z-DNA-like structure with close thermal stability as APP sequences under ionic strength close to physiological conditions. The skeleton of left-handed duplex formed by non-APP sequence seems similar as that of the traditional ‘Z-shape’ but not so perfect. The CD spectra of lh-DNA with non-APP sequences are not so distinct and much weaker as compared with those of APP ones. In addition, we provided some evidence that APP prefers to form Z-DNA because its thermal stability is closer to its isomer of B-DNA than non-APP ones (its B-form is much more stable). Our discussion provides a theoretical basis for special sequence such as APP to participate in gene regulation by forming left-handed duplexes in vivo. Further detailed studies are required to clarify the characteristics of this interesting structure. This study is promising to help understanding more about left-handed DNA, as well as left-handed RNA, and promote researches on thermal stability, dynamics, sequence dependence, and physiological characteristics of this significant duplex structure. The small free energy difference between B-DNA and lh-DNA may be the motive force for nature to select only D-ribose (eliminating L-ribose) during the molecular evolution on earth. The quantitative analysis of protein binding to lh-DNA of non-APP sequence is carrying out underway in our lab.
Supplementary Material
Notes
Present address: Yaping Zhang, State Key Laboratory of Chemical Oncogenomics, The School of Chemical Biology and Biotechnology, Peking University, Shenzhen Graduate School, Shenzhen 518055, China.
Contributor Information
Lin Li, College of Food Science and Engineering, Ocean University of China, Qingdao 266003, China.
Yaping Zhang, College of Food Science and Engineering, Ocean University of China, Qingdao 266003, China.
Wanzhi Ma, College of Food Science and Engineering, Ocean University of China, Qingdao 266003, China.
Hui Chen, College of Food Science and Engineering, Ocean University of China, Qingdao 266003, China.
Mengqin Liu, College of Food Science and Engineering, Ocean University of China, Qingdao 266003, China.
Ran An, College of Food Science and Engineering, Ocean University of China, Qingdao 266003, China; Laboratory for Marine Drugs and Bioproducts, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266235, China.
Bingxiao Cheng, College of Food Science and Engineering, Ocean University of China, Qingdao 266003, China.
Xingguo Liang, College of Food Science and Engineering, Ocean University of China, Qingdao 266003, China; Laboratory for Marine Drugs and Bioproducts, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266235, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fundamental Research Funds for Co-construction of Universities in Qingdao [to X.L.]; Shandong Provincial Natural Science Foundation, China [ZR2019BC096 to R.A.]; National Natural Science Foundation of China [31571937 to X.L.]. Funding for open access charge: Shandong Provincial Natural Science Foundation, China [ZR2019BC096 to R.A.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Nordheim A., Pardue M.L., Lafer E.M., Möller A., Stollar B.D., Rich A. Antibodies to left-handed Z-DNA bind to interband regions of drosophila polytene chromosomes. Nature. 1981; 294:417–422. [DOI] [PubMed] [Google Scholar]
- 2. Ng S.K., Weissbach R., Ronson G.E., Scadden A.D.J. Proteins that contain a functional Z-DNA-binding domain localize to cytoplasmic stress granules. Nucleic Acids Res. 2013; 41:9786–9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang Y., Ramelot T.A., Lee H.W., Xiao R., Everett J.K., Montelione G.T., Prestegard J.H., Kennedy M.A. Solution structure of the free zα domain of human DLM-1 (ZBP1/DAI), a Z-DNA binding domain. J. Biomol. NMR. 2014; 60:189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim K., Khayrutdinov B.I., Lee C.K., Cheong H.K., Kang S.W., Park H., Lee S., Kim Y.G., Jee J., Rich A. et al. Solution structure of the zβ domain of human DNA-dependent activator of IFN-regulatory factors and its binding modes to B- and Z-DNAs. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:6921–6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu C.X., Wang S.J., Lin G., Hu C.Y. The zα domain of PKZ from carassius auratus can bind to d(GC)n in negative supercoils. Fish Shellfish Immunol. 2010; 28:783–788. [DOI] [PubMed] [Google Scholar]
- 6. Lee A.R., Seo Y.J., Choi S.R., Ryu K.S., Cheong H.K., Lee S.S., Katahira M., Park C.J., Lee J.H. NMR elucidation of reduced B-Z transition activity of PKZ protein kinase at high NaCl concentration. Biochem. Biophys. Res. Commun. 2017; 482:335–340. [DOI] [PubMed] [Google Scholar]
- 7. Cao H., Dai P., Wang W., Li H., Yuan J., Wang F., Fang C.M., Pitha P.M., Liu J., Condit R.C. et al. Innate immune response of human plasmacytoid dendritic cells to poxvirus infection is subverted by vaccinia E3 via its Z-DNA/RNA binding domain. PLoS One. 2012; 7:e36823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin J., Kumari S., Kim C., Van T.M., Wachsmuth L., Polykratis A., Pasparakis M. RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation. Nature. 2016; 540:124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dondelinger Y., Hulpiau P., Saeys Y., Bertrand M.J.M., Vandenabeele P. An evolutionary perspective on the necroptotic pathway. Trends Cell Biol. 2016; 26:721–732. [DOI] [PubMed] [Google Scholar]
- 10. Thapa R.J., Ingram J.P., Ragan K.B., Nogusa S., Boyd D.F., Benitez A.A., Sridharan H., Kosoff R., Shubina M., Landsteiner V.J. et al. DAI senses influenza a virus genomic RNA and activates RIPK3-Dependent cell death. Cell Host Microbe. 2016; 20:674–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang T., Yin C., Boyd D.F., Quarato G., Ingram J.P., Shubina M., Ragan K.B., Ishizuka T., Crawford J.C., Tummers B. et al. Influenza virus Z-RNAs induce ZBP1-mediated necroptosis. Cell. 2020; 180:1115–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu R., Liu H., Chen X., Kirby M., Brown P.O., Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001; 106:309–318. [DOI] [PubMed] [Google Scholar]
- 13. Ditlevson J.V., Tornaletti S., Belotserkovskii B.P., Teijeiro V., Wang G., Vasquez K.M., Hanawalt P.C. Inhibitory effect of a short Z-DNA forming sequence on transcription elongation by T7 RNA polymerase. Nucleic Acids Res. 2008; 36:3163–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kha D.T., Wang G., Natrajan N., Harrison L., Vasquez K.M. Pathways for double-strand break repair in genetically unstable Z-DNA-forming sequences. J. Mol. Biol. 2010; 398:471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Geng J., Zhao C., Ren J., Qu X. Alzheimer's disease amyloid beta converting left-handed Z-DNA back to right-handed B-form. Chem. Commun. 2010; 46:7187–7189. [DOI] [PubMed] [Google Scholar]
- 16. Ray B.K., Dhar S., Henry C., Rich A., Ray A. Epigenetic regulation by Z-DNA silencer function controls cancer-associated ADAM-12 expression in breast cancer: cross-talk between mecp2 and NF1 transcription factor family. Cancer Res. 2013; 73:736–744. [DOI] [PubMed] [Google Scholar]
- 17. Khan N., Kolimi N., Rathinavelan T. Twisting right to left: A…A mismatch in a CAG trinucleotide repeat overexpansion provokes left-handed Z-DNA conformation. PLoS Comput. Biol. 2015; 11:e1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suram A., Rao K.S., Latha K.S., Viswamitra M.A. First evidence to show the topological change of DNA from B-DNA to Z-DNA conformation in the hippocampus of alzheimer's brain. Neuromol. Med. 2002; 2:289–297. [DOI] [PubMed] [Google Scholar]
- 19. Nishino M., Ikegami H., Fujisawa T., Kawaguchi Y., Kawabata Y., Shintani M., Ono M., Ogihara T. Functional polymorphism in Z-DNA-forming motif of promoter of SLC11A1 gene and type 1 diabetes in Japanese subjects: association study and meta-analysis. Metabolism. 2005; 54:628–633. [DOI] [PubMed] [Google Scholar]
- 20. Xie K.T., Wang G., Thompson A.C., Wucherpfennig J.I., Reimchen T.E., MacColl A.D.C., Schluter D., Bell M.A., Vasquez K.M., Kingsley D.M. DNA fragility in the parallel evolution of pelvic reduction in stickleback fish. Science. 2019; 363:81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang J., Wang S., Zhong C., Tian T., Zhou X. Novel insights into a major DNA oxidative lesion: its effects on Z-DNA stabilization. Org. Biomol. Chem. 2015; 13:8996–8999. [DOI] [PubMed] [Google Scholar]
- 22. Möller A., Nordheim A., Kozlowski S.A., Patel D.J., Rich A. Bromination stabilizes poly(dG-dC) in the Z-DNA form under low-salt conditions. Biochemistry. 1984; 23:54–62. [DOI] [PubMed] [Google Scholar]
- 23. Temiz N.A., Donohue D.E., Bacolla A., Luke B.T., Collins J.R. The role of methylation in the intrinsic dynamics of B- and Z-DNA. PLoS One. 2012; 7:e35558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gannett P.M., Heavner S., Daft J.R., Shaughnessy K.H., Epperson J.D., Greenbaum N.L. Synthesis, properties, and NMR studies of a C8-phenylguanine modified oligonucleotide that preferentially adopts the Z DNA conformation. Chem. Res. Toxicol. 2003; 16:1385–1394. [DOI] [PubMed] [Google Scholar]
- 25. Train B.C., Bilgesü S.A., Despeaux E.C., Vongsutilers V., Gannett P.M. Single C8-arylguanine modifications render oligonucleotides in the Z-DNA conformation under physiological conditions. Chem. Res. Toxicol. 2014; 27:1176–1186. [DOI] [PubMed] [Google Scholar]
- 26. Vongsutilers V., Phillips D.J., Train B.C., McKelvey G.R., Thomsen N.M., Shaughnessy K.H., Lewis J.P., Gannett P.M. The conformational effect of para-substituted C8-arylguanine adducts on the B/Z-DNA equilibrium. Biophys. Chem. 2011; 154:41–48. [DOI] [PubMed] [Google Scholar]
- 27. Pohl F.M., Jovin T.M., Baehr W., Holbrook J.J. Ethidium bromide as a cooperative effector of a DNA structure. Proc. Natl. Acad. Sci. U.S.A. 1972; 69:3805–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Owczarzy R., You Y., Moreira B.G., Manthey J.A., Huang L., Behlke M.A., Walder J.A. Effects of sodium ions on DNA duplex oligomers: improved predictions of melting temperatures. Biochemistry. 2004; 43:3537–3554. [DOI] [PubMed] [Google Scholar]
- 29. Maity A., Singh A., Singh N. Differential stability of DNA based on salt concentration. Eur. Biophys. J. 2017; 46:33–40. [DOI] [PubMed] [Google Scholar]
- 30. Pan F., Roland C., Sagui C. Ion distributions around left- and right-handed DNA and RNA duplexes: a comparative study. Nucleic Acids Res. 2014; 42:13981–13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chatake T., Sunami T. Direct interactions between Z-DNA and alkaline earth cations, discovered in the presence of high concentrations of MgCl2 and CaCl2. J. Inorg. Biochem. 2013; 124:15–25. [DOI] [PubMed] [Google Scholar]
- 32. Santangelo M.G., Antoni P.M., Spingler B., Jeschke G. Can copper(II) mediate hoogsteen base-pairing in a left-handed DNA duplex? a pulse EPR study. ChemPhysChem. 2010; 11:599–606. [DOI] [PubMed] [Google Scholar]
- 33. Drozdzal P., Gilski M., Kierzek R., Lomozik L., Jaskolski M. High-resolution crystal structure of Z-DNA in complex with Cr3+ cations. J. Biol. Inorg. Chem. 2015; 20:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nayak A.K., Mishra A., Jena B.S., Mishra B.K., Subudhi U. Lanthanum induced B-to-Z transition in self-assembled Y-shaped branched DNA structure. Sci. Rep. 2016; 6:26855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bhanjadeo M.M., Nayak A.K., Subudhi U. Cerium chloride stimulated controlled conversion of B-to-Z DNA in self-assembled nanostructures. Biochem. Biophys. Res. Commun. 2017; 482:916–921. [DOI] [PubMed] [Google Scholar]
- 36. D’Urso A., Holmes A.E., Berova N., Balaz M., Purrello R. Z-DNA recognition in B-Z-B sequences by a cationic zinc porphyrin. Chem. Asian. J. 2011; 6:3104–3109. [DOI] [PubMed] [Google Scholar]
- 37. Wu Z., Tian T., Yu J., Weng X., Liu Y., Zhou X. Formation of sequence-independent Z-DNA induced by a ruthenium complex at low salt concentrations. Angew. Chem. Int. Ed. Engl. 2011; 50:11962–11967. [DOI] [PubMed] [Google Scholar]
- 38. Choi J.K., Reed A., Balaz M. Chiroptical properties, binding affinity, and photostability of a conjugated zinc porphyrin dimer complexed with left-handed Z-DNA and right-handed B-DNA. Dalton Trans. 2014; 43:563–567. [DOI] [PubMed] [Google Scholar]
- 39. Holmes A.E., Choi J.K., Francis J., D’Urso A., Balaz M. Sulfonated Ni(II)porphyrin improves the detection of Z-DNA in condensed and non-condensed BZB DNA sequences. J. Inorg. Biochem. 2012; 110:18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choi J.K., D’Urso A., Balaz M. Chiroptical properties of anionic and cationic porphyrins and metalloporphyrins in complex with left-handed Z-DNA and right-handed B-DNA. J. Inorg. Biochem. 2013; 127:1–6. [DOI] [PubMed] [Google Scholar]
- 41. Rahmouni A.R., Wells R.D. Stabilization of Z DNA in vivo by localized supercoiling. Science. 1989; 246:358–363. [DOI] [PubMed] [Google Scholar]
- 42. Wittig B., Dorbic T., Rich A. The level of Z-DNA in metabolically active, permeabilized mammalian cell nuclei is regulated by torsional strain. J. Cell Biol. 1989; 108:755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang A.H., Gessner R.V., van der Marel G.A., van Boom J.H., Rich A. Crystal structure of Z-DNA without an alternating purine-pyrimidine sequence. Proc. Natl. Acad. Sci. U.S.A. 1985; 82:3611–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stettler U.H., Weber H., Koller T., Weissmann C. Preparation and characterization of form V DNA, the duplex DNA resulting from association of complementary, circular single-stranded DNA. J. Mol. Biol. 1979; 131:21–40. [DOI] [PubMed] [Google Scholar]
- 45. Pohl F.M., Thomae R., DiCapua E. Antibodies to Z-DNA interact with form V DNA. Nature. 1982; 300:545–546. [DOI] [PubMed] [Google Scholar]
- 46. Kypr J., Kejnovská I., Renciuk D., Vorlícková M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009; 37:1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y., Cui Y., An R., Liang X., Li Q., Wang H., Wang H., Fan Y., Dong P., Li J. et al. Topologically constrained formation of stable Z-DNA from normal sequence under physiological conditions. J. Am. Chem. Soc. 2019; 141:7758–7764. [DOI] [PubMed] [Google Scholar]
- 48. Wang J., Pan X., Liang X. Assessment for melting temperature measurement of nucleic acid by HRM. J. Anal. Methods Chem. 2016; 2016:5318935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang J., Dong P., Wu W., Pan X., Liang X. High-throughput thermal stability assessment of DNA hairpins based on high resolution melting. J. Biomol. Struct. Dyn. 2018; 36:1–13. [DOI] [PubMed] [Google Scholar]
- 51. Hizver J., Rozenberg H., Frolow F., Rabinovich D., Shakked Z. DNA bending by an adenine-thymine tract and its role in gene regulation. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8490–8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Barbic A., Zimmer D.P., Crothers D.M. Structural origins of adenine-tract bending. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:2369–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B-Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc. Natl. Acad. Sci. U.S.A. 1981; 78:1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lafer E.M., Möller A., Nordheim A., Stollar B.D., Rich A. Antibodies specific for left-handed Z-DNA. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:3546–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Möller A., Gabriels J.E., Lafer E.M., Nordheim A., Rich A., Stollar B.D. Monoclonal antibodies recognize different parts of Z-DNA. J. Biol. Chem. 1982; 257:12081–12085. [PubMed] [Google Scholar]
- 56. Lafer E.M., Sousa R., Rich A. Anti-Z-DNA antibody binding can stabilize Z-DNA in relaxed and linear plasmids under physiological conditions. EMBO J. 1985; 4:3655–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang Y., Ramelot T.A., Lee H.W., Xiao R., Everrtt J.K., Montelione G.T., Prestegard J.H., Kennedy M.A. Solution structure of the free Zα domain of human DLM-1 (ZBP1/DAI), a Z-DNA binding domain. J. Biomol. NMR. 2014; 60:189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kawara K., Tsuji G., Taniguchi Y., Sasaki S. Synchronized chiral induction between [5]helicene-spermine ligand and B-Z DNA transition. Chemistry. 2017; 23:1763–1769. [DOI] [PubMed] [Google Scholar]
- 59. Lesnik E.A., Khalimullina Zh.A. The effect of Z-conformation on enzymatic activity of restriction endonucleases. Mol. Biol. 1989; 23:1638–1644. [PubMed] [Google Scholar]
- 60. Singleton C.K., Klysik J., Wells R.D. Conformational flexibility of junctions between contiguous B- and Z-DNAs in supercoiled plasmids. Proc. Natl. Acad. Sci. U.S.A. 1983; 80:2447–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Vardimon L., Rich A. In Z-DNA the sequence G-C-G-C is neither methylated by Hha I methyltransferase nor cleaved by Hha I restriction endonuclease. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:3268–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Azorin F., Hahn R., Rich A. Restriction endonucleases can be used to study B-Z junctions in supercoiled DNA. Proc. Natl. Acad. Sci. U.S.A. 1984; 81:5714–5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yashima E., Maeda K., Iida H., Furusho Y., Nagai K. Helical polymers: synthesis, structures, and functions. Chem. Rev. 2009; 109:6102–6211. [DOI] [PubMed] [Google Scholar]
- 64. Harvey S.C. DNA structural dynamics: longitudinal breathing as a possible mechanism for the B in equilibrium Z transition. Nucleic Acids Res. 1983; 11:4867–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Runkel L., Nordheim A. Chemical footprinting of the interaction between left-handed Z-DNA and anti-Z-DNA antibodies by diethylpyrocarbonate carbethoxylation. J. Mol. Biol. 1986; 189:487–501. [DOI] [PubMed] [Google Scholar]
- 66. Peck L.J., Wang J.C. Energetics of B-to-Z transition in DNA. Proc. Natl. Acad. Sci. U.S.A. 1983; 80:6206–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lee M., Kim S.H., Hong S. Minute negative superhelicity is sufficient to induce the B-Z transition in the presence of low tension. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. D’Ascenzo L., Leonarski F., Vicens Q., Auffinger P. Z-DNA like’ fragments in RNA: a recurring structural motif with implications for folding, RNA/protein recognition and immune response. Nucleic Acids Res. 2016; 44:5944–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.