Abstract
Proteins are the building blocks for almost all the functions in cells. Understanding the molecular evolution of proteins and the forces that shape protein evolution is essential in understanding the basis of function and evolution. Previous studies have shown that adaptation frequently occurs at the protein surface, such as in genes involved in host–pathogen interactions. However, it remains unclear whether adaptive sites are distributed randomly or at regions associated with particular structural or functional characteristics across the genome, since many proteins lack structural or functional annotations. Here, we seek to tackle this question by combining large-scale bioinformatic prediction, structural analysis, phylogenetic inference, and population genomic analysis of Drosophila protein-coding genes. We found that protein sequence adaptation is more relevant to function-related rather than structure-related properties. Interestingly, intermolecular interactions contribute significantly to protein adaptation. We further showed that intermolecular interactions, such as physical interactions, may play a role in the coadaptation of fast-adaptive proteins. We found that strongly differentiated amino acids across geographic regions in protein-coding genes are mostly adaptive, which may contribute to the long-term adaptive evolution. This strongly indicates that a number of adaptive sites tend to be repeatedly mutated and selected throughout evolution in the past, present, and maybe future. Our results highlight the important roles of intermolecular interactions and coadaptation in the adaptive evolution of proteins both at the species and population levels.
Keywords: intermolecular interaction, coadaptation, Drosophila, population, physical interaction, adaptive and nonadaptive changes
Introduction
Natural selection plays an important role in the molecular evolution of protein sequences. Recent advances in genome sequencing and reliable inference methods at both phylogenetic and population levels have enabled fast and robust estimation of evolutionary rates and adaptation driven by natural selection. In addition, the increased availabilities of structural and functional data of proteins have made it possible to study how structural and functional constraints affect protein sequence evolution and adaptation. Different proteins and different sites within a protein have varying rates of evolution and adaptation due to both structural and functional constraints (Kosiol et al. 2008; Lindblad-Toh et al. 2011; Zhang and Yang 2015; Echave et al. 2016). For example, genes that are highly expressed or perform essential functions are often under strong purifying selection and tend to evolve slowly (Pál et al. 2001; Drummond et al. 2005; Zhang and He 2005; Zhang and Yang 2015; Moutinho et al. 2019); genes involved in host–pathogen interactions, for example, immune responses and antivirus responses, show exceptionally high rates of adaptive changes (Nielsen et al. 2005; Sackton et al. 2007; Obbard et al. 2009; Sironi et al. 2015; Enard et al. 2016; Palmer et al. 2018; Uricchio et al. 2019); and residues that are intrinsically disordered or at the protein surface are fast evolving and proved to be hotspots of adaptive evolution (Goldman et al. 1998; Lin et al. 2007; Ramsey et al., 2011; Afanasyeva et al. 2018; Moutinho et al. 2019). More recently, Slodkowicz and Goldman (2020) employed genomic-scale integrated structural and evolutionary phylogenetic analysis in mammals and showed that positively selected residues are clustered near ligand binding sites, especially in proteins that are associated with immune responses and xenobiotic metabolism. However, it remains unclear how adaptive sites are distributed in the genome and how adaptation is related to functions and structures. Moreover, most of the existing literature is focused on protein differences between species, and it remains unclear how much within-species selective processes like spatially varying selection may contribute to long-term evolution.
Although evidence has shown that adaptation is more likely to occur at intrinsically disordered regions (IDRs; Afanasyeva et al. 2018) and clustered at the surface of proteins (Dasmeh et al. 2013; Moutinho et al. 2019; Slodkowicz and Goldman 2020), it remains unclear how functional and structural properties of proteins shape adaptation at the species and population scale. Moreover, due to the lack of structural and functional information of many proteins in the genome, the underlying evolutionary mechanism derived from current studies might be incomplete. Here, we systematically investigated the evolution and adaptation of protein-coding genes in Drosophila melanogaster by comparing it to its closely related species and their own populations, to distinguish the main factors that impact evolution and adaptation at the protein-coding level. We applied large-scale bioinformatic and structural analysis to obtain the structural and functional properties of proteins. We then classified residues into different structural and functional sites. By comparing rates of sequence evolution and adaptation between different proteins and sites, we were able to locate hotspots of adaptation at the genome scale. We found that functional properties are better predictors of protein adaptation rates than structural properties. Interestingly, we found that adaptation rates of a protein positively correlate with the fraction of residues that are involved in intermolecular interactions inside the protein. In agreement with this finding, we found that putative binding regions including allosteric sites at protein surface show higher rates of adaptive evolution than other sites. For proteins under fast-adaptive evolution, defined as proteins with high rates of adaptive evolution, we showed that they tend to interact with each other more frequently than random expectations, suggesting fast-adaptive genes might undergo coadaptive evolution. We further discovered that coadaptation might be universal for many interacting proteins in D. melanogaster.
Our results suggest that intermolecular interactions in D. melanogaster are an important driver of protein adaptive evolution. We further hypothesized and provided evidence that intermolecular interactions, such as physical interactions, might be an important mechanism that contributes to the coadaptive evolution of interacting proteins in D. melanogaster genome. One intriguing question is that how the adaptive signals between-populations (short term) and between-species (long term) are correlated. We then asked if those patterns hold for the selective processes occurring within-species. Despite the abundance of literature studying geographic variation in Drosophila species (Kolaczkowski et al. 2011; Fabian et al. 2012; Langley et al. 2012; Pitchers et al. 2013; Bergland et al. 2014; Reinhardt et al. 2014; Lack et al. 2015; Svetec et al. 2016), very little is known for a systematic evaluation of the protein properties affected by spatially varying selection. We thus investigated protein adaptation signals of strongly differentiated amino acids across geographic regions, which were often associated with within-species local adaptations (Matthey-Doret and Whitlock 2019). We showed that most of the patterns found between-species in fact hold at the within-species levels. This may partly be because most sites contributing to within-species local adaptation tend to also contribute to long-term adaptive evolution in D. melanogaster, suggesting that a subset of protein-coding loci are constantly or repeatedly utilized for the adaptive purposes.
Results
Putative Molecular Interaction Sites Are Hotspots for Protein Adaptive Evolution
To uncover the main factors that impact the evolutionary rates of genes, we analyzed 13,528 protein-coding genes in D. melanogaster using genomic data from melanogaster subgroup species and D. melanogaster population genomics data from 205 inbred lines from Drosophila Genetic Reference Panel, Freeze 2.0 (DGRP2; Huang et al. 2014). We applied a maximum likelihood method (Yang 2007) to compute the dN/dS ratio (ω) using the protein-coding sequences of five closely related melanogaster subgroup species (D. melanogaster, D. simulans, D. sechellia, D. yakuba, and D. erecta). We estimated the proportions of adaptive changes (α) in each gene by applying an extension of McDonald–Kreitman (MK) test named asymptotic MK (Messer and Petrov 2013; Uricchio et al. 2019) using D. yakuba as outgroup. We then calculated the rate of adaptive changes (ωa) of each gene by multiplying ω to α (ωa = αω) (Moutinho et al. 2019; see Material and Methods). The rate of nonadaptive changes can be further calculated by ωna = ω − ωa. Finally, we successfully assigned ω to 12,118 protein-coding genes and ωa and ωna to 7,192 genes. For each of D. melanogaster genes subjecting the same analysis pipeline, we further obtained 17 different structural or functional properties (see Material and Methods, supplementary file S1 and tables S1 and S2, Supplementary Material online). We calculated Pearson’s correlations of ω, ωa, and ωna with all these properties (supplementary table S1, Supplementary Material online). Many of these genome-wide correlations were expected (for details, see supplementary tables S1, S2, and figs. S1–S5, Supplementary Material online). Interestingly, we found that some previously unexplored properties, fractions of molecular-interaction sites (including protein–protein interaction (PPI)-site ratio, ratio of residues involved in PPIs, and DNA-site ratio, ratio of residues involved in protein–DNA interactions) were strongly positively correlated with ω, ωa, and ωna (supplementary section Molecular interactions contribute to the variations of protein sequence evolution and adaptation, table S1, figs. S1 and S2, Supplementary Material online). The results indicate that molecular interactions might act as an important factor that drives protein adaptive evolution in the Drosophila genome.
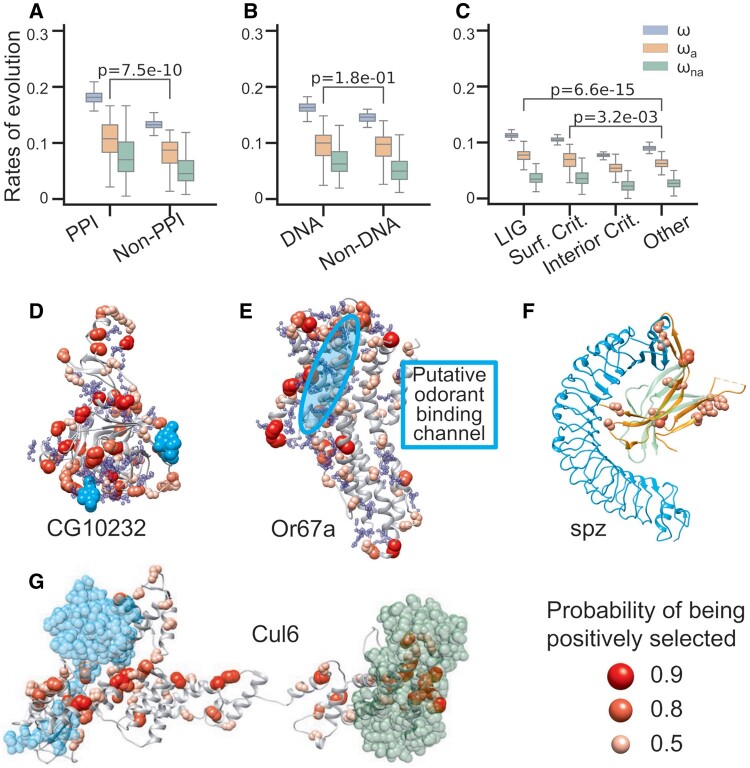
We then investigated whether residues involved in molecular interactions are targets for adaptive evolution. To tackle this question, we predicted PPI-sites and DNA binding sites (DNA-sites) for each of D. melanogaster protein sequences (see Materials and Methods). In addition, we characterized allosteric residues as surface and interior critical residues with STRESS model (Clarke et al. 2016) for all the structural models. We also extracted putative binding sites from STRESS Monte Carlo simulations. We calculated ω, ωa, and ωna for residues in each of the putative molecular interaction categories. Strikingly, we observed that residues involved in PPIs, DNA binding, and ligand binding exhibited higher rates of adaptive evolution compared with their corresponding null sites (t-test, P = 7e-10, 0.18, and 7e-15, respectively; fig. 1A–C). In addition, allosteric residues at protein surface showed higher adaptation rates than allosteric residues at protein interior (t-test, P = 3e-10) or residues that are not involved in ligand binding (t-test, P = 0.003; fig. 1C and supplementary fig. S6, Supplementary Material online).
Fig. 1.
Adaptive evolution in molecular interaction sites. PPI sites (A), DNA binding sites (B), and putative ligand binding sites (C) show higher adaptation rates than none binding sites. The t-test P-values between the adaptation rates of binding sites and nonbinding sites were highlighted in (A–C). t-test P-values between other evolutionary rates of binding sites and nonbinding sites were shown in supplementary figure S2, Supplementary Material online. Examples of positive selection around molecular interaction sites in high-quality structural models of CG10232 (D), Or67a (E), spz (F), and Cul6 (G). Except for spz (PDB code 3e07), the other proteins are obtained from SWISS model repository. Putative ligand binding pockets of CG10232 (D) and Or67a (E) are shown in blue spheres. Ligands including interacting proteins are shown in cyan or green: NAG of CG10232 in cyan (D), Toll receptor of spz in cyan (F), Rbx protein in cyan and F-box protein in green for Cul6 (G). The putative odorant binding channel of Or67a is highlighted in cyan circle (E). The ligand poses in (D, F, and G) are obtained by superimposition from structures 2XXL, 4BV4, and 1LDK, respectively.
To gain a better understanding of adaptation in molecular interaction sites, we further visualized positive selections that are associated with molecular interactions. We first investigated whether adaptive evolution is associated with particular protein structures or protein families. To do this, we looked into fast-adaptive proteins with the largest ∼15% rates of adaptation (ωa > 0.15) that are linked to high-quality structural models. Interestingly, among these proteins, we found 45 enriched as trypsin-like cysteine/serine peptidase domain and 17 7TM chemoreceptors, suggesting widespread adaptive evolution acting on these protein families or protein domains in D. melanogaster (supplementary table S3, Supplementary Material online). Many of the 7TM chemoreceptors are olfactory and gustatory genes and show adaptive evolution in various species such as Drosophila and mosquito (Hill et al. 2002; Lawniczak and Begun 2007; McBride 2007; Wu et al. 2009). In addition to these two protein families, previous studies identified recurrent positive selections acting on some other fast-adaptive proteins in Drosophila and mammals, and the possible adaptive evolution mechanisms have been linked to exogenous ligand binding, for example, serine protease inhibitors (serpin), Toll-like receptor 4 (TLR-4), and cytochrome P450 (Jiggins and Kim 2007; Slodkowicz and Goldman 2020).
We used the two representative cases of fast-adaptive protein evolution of CG10232 and Or67a—a trypsin-like cysteine/serine peptidase domain and a 7TM chemoreceptor, respectively—to illustrate the link between adaptive evolution and molecular interactions in the two protein families with frequent adaptive evolution. We observed that in both cases, positively selected sites were significantly closer to predicted or inferred binding sites in the protein 3D structure (t-test, P = 8e-133 for CG10232 and 3e-169 for Or67a, fig. 1D and E) and were overlapped with predicted or inferred binding pockets for CG10232 (Fisher’s exact test, P-values 0.02, fig. 1D). There might be an overlap with predicted or inferred binding pockets for Or67a, but it is not statistically significant (Fisher’s exact test, P = 0.19, fig. 1E). Specifically, in CG10232, we found clusters of positively selected sites around NAG binding sites that are inferred from a crystal structure of serine protease (PDB code: 2XXL; fig. 1D), whereas in Or67a, positively selected sites expand around the putative odorant-binding channel formed by helices S1–S6 in extracellular regions (Butterwick et al. 2018; fig. 1E).
Besides the examples that are associated with exogenous ligand or exogenous peptide binding, we also identified two previously undescribed examples where adaptive evolution might be linked to endogenous protein binding: Spaztle (spz, fig. 1F) and Cul6 (fig. 1G). Spaztle can bind to TLRs and trigger a humoral innate immune response. We built the missing loop in Spaztle in the crystal structure of Toll/Spaztle complex (PDB code 4BV4) according to the dimeric crystal structure of Spaztle (PDB code 3E07). In this complex structural model, we observed several positively selected sites in Toll-4/Spaztle interfaces (fig. 1F). Cul6, another example, is a protein in the cullins family in D. melanogaster. The cullins protein family is known as scaffold proteins that assemble multisubunit Cullin-RING E3 ubiquitin ligase by forming SCF complex with F box and RING-box (Rbx) proteins (Zheng et al. 2002). We constructed the putative Cul6 contained SCF complex by superimposition to the crystal structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex (Zheng et al. 2002). In the structural model, we observed positively selected sites in Cul6 clustered around the binding sites of Rbx protein, Rbx1, and F-box protein, Skp1 (fig. 1G). The examples above suggest that intermolecular interactions, including both exogenous and endogenous binding, could contribute to protein adaptive evolution.
Frequent Adaptive Evolution and Coadaptative Evolution in Genes Involved in Reproduction, Immune System, and Environmental Information Processing
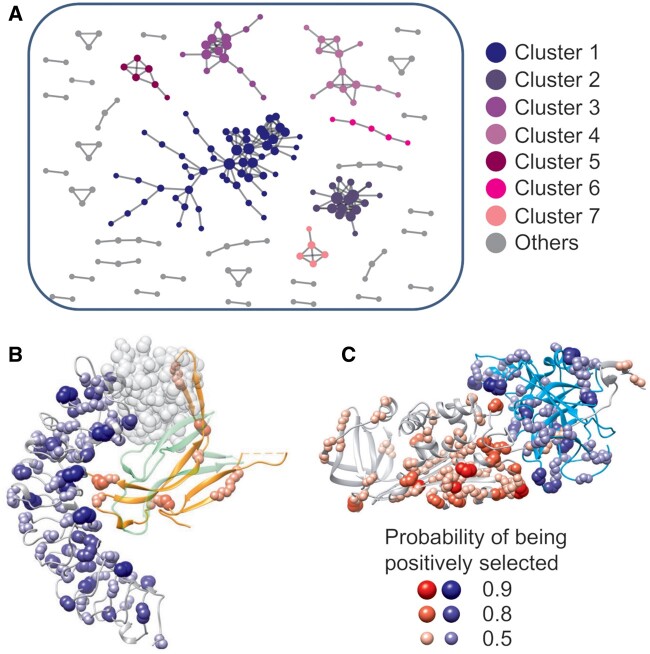
To find out whether specific biological functions were associated with fast-adaptive genes, we applied Gene Ontology (GO) analysis using DAVID tool to the genes with the largest rates of adaptation (ωa > 0.15, top ∼15%). The significant GO terms are frequently linked to serine-type endopeptidase activity, reproduction, protein lysis, chemosensory, and other related biological functions (supplementary table S4, Supplementary Material online). As these fast-adaptive genes tend to be enriched in similar biological functions, we asked whether these genes evolved coadaptively that is, whether these proteins are interacting with each other frequently. To test this possibility, we obtained PPI of D. melanogaster from STRING database (Szklarczyk et al. 2019) and analyzed PPIs among fast-adaptive proteins. We found that fast-adaptive proteins tend to interact with each other more frequently than expected (PPI enrichment P-value < 1.0e-16). In the PPI network of fast-adaptive proteins, we observed seven strongly connected subclusters with at least five members (fig. 2A and supplementary table S5, Supplementary Material online, e.g., fig. 2B and C). Proteins in these subclusters are enriched in biological processes such as reproduction, immune response, defense response to bacterium and virus, RNA interference, chitin metabolic, etc. (supplementary tables S5–S11, Supplementary Material online), which are in line with the GO analysis of fast-adaptive genes (supplementary table S4, Supplementary Material online) and previous enrichment analysis of positively selected genes identified from genome-wide studies (Nielsen et al. 2005; Begun et al. 2007; Enard et al. 2016).
Fig. 2.
Coadaptation of fast-adaptive proteins. (A) Subclusters of PPI networks of fast-adaptive proteins. Only proteins with at least one partner were shown. Examples of molecular interactions that might regulate coadaptation in fast-adaptive proteins: (B) Toll-4 (gray) and spz (orange, with green representing the other spz monomer), (C) Spn28Db (gray, serine protease inhibitor 28Db) and CG18563 (cyan, with Go term “serine-type endopeptidase activity”). A putative N-terminus (transparent beads) of Toll-4 was built by superimposition from 4LXR, since the N-terminus was missing in the structural model. Complex structural models of Spn28Db and CG18563 were inferred from 1EZX.
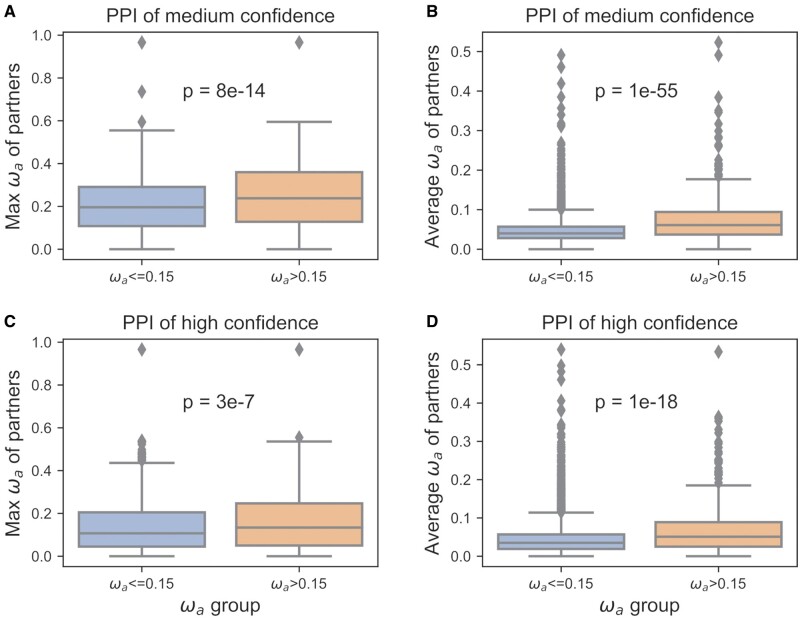
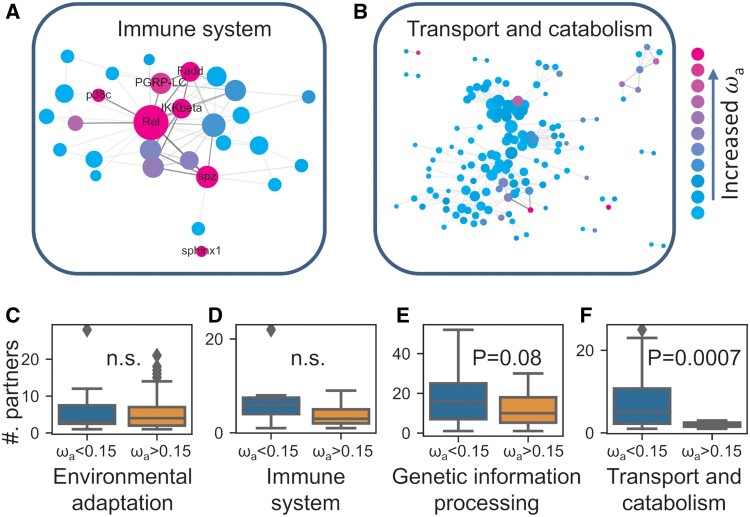
We next asked whether coadaptation plays a role in the adaptive evolution of interacting proteins to a broader extend, including both fast- and slow-adaptive proteins. To address this question, we analyzed and compared adaptation rates of all D. melanogaster PPIs available in STRING database with high confidence, and we found that protein partners of fast-adaptive proteins (ωa > 0.15) have significantly larger maximum/average ωa compared with slow-adaptive proteins (fig. 3). We further analyzed and visualized adaptive evolutionary rates of proteins in PPI networks of nine different biological pathways extracted from Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, including the immune system, xenobiotics biodegradation, response to the environment, aging and development, genetic information processing, sensory system, transport and catabolism, cell growth and death, and metabolism. We observed that, in these PPI networks, proteins with relatively large ωa tend to interact with each other (fig. 4A and B). We also noticed that, for pathways that are previously known as adaptation-hotspots (e.g., immune system), fast-adaptive proteins can act as central nodes and are coadaptively evolving with other fast-adaptive proteins (fig. 4A and C). Although in pathways such as transport and catabolism, fast-adaptive proteins are mainly at PPI periphery. Specifically, we observed that in pathways related to the immune system and environmental adaptation, where adaptive evolution often occurs, fast-adaptive proteins have a comparable number of interactors as other genes (fig. 4C and D), whereas in conserved pathways such as transport and catabolism, fast-adaptive genes are often at network peripheries and have significantly fewer interactors (fig. 4E and F). In line with these findings, we found that ωa are larger in pathways that harbor fast-adaptive proteins as central nodes than other pathways (supplementary fig. S7, Supplementary Material online).
Fig. 3.
Coadaptation of PPIs in D. melanogaster. For fast-adaptive proteins, adaptation rates of their partners (orange box plot) are significantly larger compared with slow adaptive proteins (blue box plot). Max ωa of protein partners are shown in (A and C) and averaged ωa, of protein partners are shown in (B and D). PPI from STRING with median confidence (combined score larger than 0.4) are shown in (A and B), and PPI with high confidence (combined score larger than 0.7) are shown in (C and D).
Fig. 4.
Rates of protein sequence adaptive evolution in the PPI network of different functional pathways. The PPI networks showed the adaptive evolution in the immune system (A) and transport and catabolism (B). In pathways that are hotspots of adaptive evolution, for example, environmental adaptation (C) and immune system (D), fast-adaptive proteins can act as central nodes. Although in conserved pathways, for example, genetic information processing (E) and transport and catabolism (F), fast-adaptive proteins are often at the periphery of the PPI network.
Physical Interactions Contribute to Coadaptation of Fast-Adaptive Genes
Having established that molecular interactions contribute to the adaptive evolution of protein sequence, we then investigated whether these physical molecular interactions could drive protein–protein coadaptation. To do this, we looked into interacting fast-adaptive protein pairs that are associated with known or inferred complex structural models. For inferred complex structural models, we superimposed the structural models of the pair of proteins onto their high-resolution homologous complex structures. Here we illustrated coadaptation at the PPI interface in two examples: Toll-4/Spatzle and Spn28Db/CG18563 (fig. 2B and C).
Toll-4/Spatzle
Toll-4 is a member of TLRs. Previous studies have shown strong evidence of adaptive evolution of Toll-4 in Drosophila and mammals (Levin and Malik 2017; Slodkowicz and Goldman 2020), and implied the important roles of ligand binding in the positive selection and coevolution of Toll receptors and Spatzle proteins (Lima et al. 2021). Toll-4 can bind to Spatzle and trigger further innate immune responses with high confidence (inferred from STRING database). In the previous section, we showed that several positively selected sites in Spatzle overlap with Toll-Spatzle interfaces (fig. 1F). Here, we further showed that, in Toll-4, many significantly positively selected sites were located at the interface for Spatzle (fig. 2B), which is in line with a previous study of Toll-4 in Drosophilawillistoni (Levin and Malik 2017).
Spn28Db/CG18563
Spn28Db is one of the serine protease inhibitors in D. melanogaster expressed in male accessory glands, whereas CG18563 belongs to the protein family of trypsin-like cysteine/serine peptidase domain. The interactions between the two proteins were predicted with high confidence from the STRING database, and the molecular interactions can be inferred from the existing crystal structure of serpin and bacteria protease complex (PDB code 1EZX). We observed many positive selected sites at the molecular interface between the two proteins (fig. 2C), suggesting that physical interactions might play a role in the coadaptation of the two proteins.
Most Geographically Differentiated Nonsynonymous Single Nucleotide Polymorphisms in Protein-Coding Genes Are Adaptive
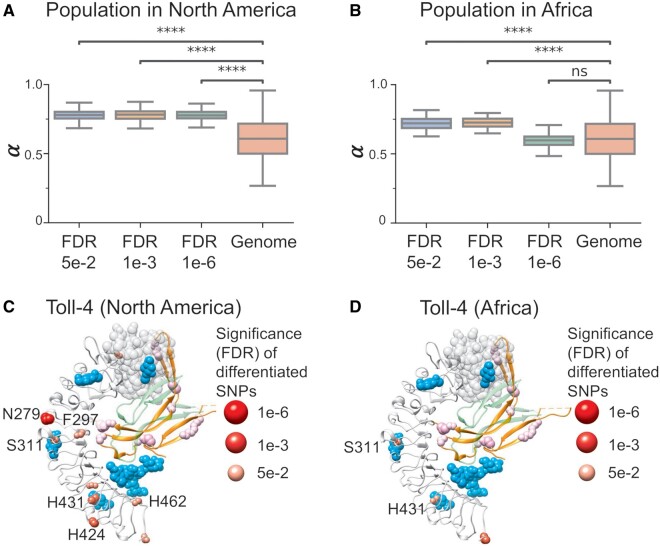
To learn more about the relationship between short-term adaptation to local environments and long-term adaptive evolution, we extracted residues with significant allele frequency differentiation across latitudes in North America (Svetec et al. 2016) and Africa (Lack et al. 2015). For the DPGP3 African population data, we followed the same protocol as Svetec et al. (2016) to identify significantly differentiated single nucleotide polymorphism (SNPs; see Population Genetics of DPGP3 African Population). We then computed evolutionary rates (ω), adaptation rates (ωa), nonadaptation rates (ωna), and proportions of adaptive changes (α) of these residues as in the previous section (fig. 5A and B and supplementary fig. S8, Supplementary Material online). We observed that, in both the North American population and the African population, these sites have significantly higher proportions of adaptive changes (fig. 5A and B) than other SNPs, suggesting that they can be hotspots for adaptive evolution. To find out whether these SNPs are related to even longer-term adaptive evolution, we inferred positive selection sites of each protein-coding gene from phylogenic data (see Material and Methods). We found that geographically differentiated nonsynonymous SNPs are significantly enriched for long-term positive selection (supplementary fig. S9, Supplementary Material online). To further characterize structural and functional properties of short-term genetic variations, we mapped geographically differentiated nonsynonymous residues to different structural and functional characteristics, such as intrinsic structural disorder (ISD), relative solvent accessibility (RSA), PPI-sites, DNA-sites, and ligand-binding sites. We found that these nonsynonymous SNPs were significantly enriched in disordered regions and protein surfaces, as well as in PPIs and ligand binding (supplementary fig. S9, Supplementary Material online). To better visualize the characteristics of these SNPs, we used Toll-4 as an example. We mapped its geographically differentiated nonsynonymous sites onto its structural model. We found that these sites are either positively selected or are located very close to positively selected sites (fig. 5C and D). For example, highly differentiated sites in the North American population, N279 (false discovery rate [FDR] 3e-7) and H431 (FDR 3e-6) were predicted to be positively selected both at a probability of P = 0.9. Although another highly differentiated site, D424 was close to three positively selected sites S401 (P = 0.8), H431 (P = 0.95), and V448 (P = 0.8). We also noticed some differentiated sites that may be located within ligand binding sites, including F297 (FDR 3e-3), S311 (FDR 3e-3), H431 (FDR 3e-6), and H462 (FDR 1e-2). In the structure of Toll4, we also observed three highly differentiated sites in African populations, which are S311 (FDR 4e-2), H431 (FDR 2e-2), and S490 (FDR 2e-4). We noticed that all the three sites overlapped with differentiated SNPs in North America and two of them (S311 and H431) localized within ligand binding sites (fig. 5D), further supporting our observation that ligand binding sites in some genes may undergo recurrent adaptive evolution.
Fig. 5.
Adaptive evolution in significantly differentiated SNPs. The significantly differentiated SNPs at different FDR cutoffs all show much higher proportions of adaptative changes (α) than genome-wide expectation in the North American population (A) and African population (B) (t-test, ****, P < 1e-4). (C) Significantly differentiated nonsynonymous SNPs of North American populations in Toll-4. Ligands are shown in cyan by superimposing crystal structure of Toll-Spatzle (PDB code 4BV4) on to Toll-4 structural model. Residues N279 and H431 are both highly differentiated (FDR 3e-7 and 3e-6) and positively selected (both at a probability of P = 0.9). Other highly differentiated sites, F297, S311, H424, H431, and H462 are located near ligand binding sites or positively selected sites. (D) Significantly differentiated nonsynonymous SNPs of African populations in Toll-4. Two highly differentiated SNPs, S311 (FDR 4e-2), and H431 (FDR 2e-2), exist in the North American population and are located near ligand-binding sites.
Discussion
In this study, we systematically studied the impact of structure- and function-related gene properties on protein sequence evolution and adaptation in D. melanogaster genome. We found that molecular interactions in proteins contribute to the variation of protein sequence adaptive evolution. A novel discovery of this work is that molecular interaction sites including PPI sites and protein–DNA interaction sites are hotspots for adaptative evolution. We revealed that fast-adaptive proteins tend to interact with each other frequently and protein partners of these fast-adaptive proteins tend to have higher adaptation rates, suggesting that coadaptive evolution might be common in D. melanogaster. By visualizing examples of interacting fast-adaptive proteins, we further demonstrated that physical interactions may contribute to the coadaptation of fast-adaptive proteins.
Protein surface and intrinsic disorder regions are frequent targets for adaptive evolution and contribute to the variations of protein sequence adaptive evolution (Afanasyeva et al. 2018; Moutinho et al. 2019). However, the detailed mechanisms underlying these observations remain unclear. One possible explanation would be that these regions are frequently linked to intermolecular interactions (Afanasyeva et al. 2018; Moutinho et al. 2019). For example, Moutinho et al. (2019) hypothesized that molecular interactions involved in host–pathogen coevolution were the major driver of protein adaptation. Here, we further identified that proportions of possible molecular interaction sites inside proteins contribute to the variations of protein sequence adaptive evolution. These molecular interaction sites or regulatory sites at protein surfaces can be hotspots of protein adaptation. Indeed, some specific molecular interactions have been linked to adaptive evolution in several case studies (Hughes and Nei 1988; Bachtrog 2008; Schott et al. 2014; Levin and Malik 2017), a recent study on sweet taste receptors in the songbird radiation (Toda et al. 2021), and large-scale studies based on proteins with high-quality structural models (Slodkowicz and Goldman 2020). In the latter study, the authors showed that amino acids under positive selection in mammals tend to cluster closer to binding sites of exogenous ligands than expected by chance (Slodkowicz and Goldman 2020), suggesting an important role of functionally important regions in adaptive evolution. Here, we extend the conclusion to D. melanogaster genome, including proteins with or without high-resolution structural models. We also showed that in addition to exogenous ligands, endogenous ligands might also contribute to adaptive evolution, while the latter might explain why interacting proteins tend to evolve coadaptively.
Notably, previous studies showed that multi-interface proteins tend to evolve more slowly than single-interface proteins (Kim et al. 2006) and that proteins with many interactors tend to evolve slowly (Jordan et al. 2003), which seems to be contradictory to our results that proteins with more interaction sites evolve faster and have faster adaptation rates. Here, we argue that, in our study, we used sequence profiles to predict molecular interaction sites in proteins at a genomic scale, rather than only looking into proteins with high-resolution structures. In this way, we may capture many weak or transient interactions, which are evolving faster than obligate and conserved interactions (Mintseris and Weng 2005). Meanwhile, we did not exclude IDRs or intrinsically disordered proteins (IDPs) in our study, which are widespread in D. melanogaster genome. It has been suggested that IDR/IDP tend to evolve fast due to the lack of structural restraints (Echave et al. 2016). In the functional aspect, IDR/IDP are thought to be promiscuous binders through many multiple binding mechanisms, including forming static, semistatic, and fuzzy or dynamic complexes (Uversky 2019), suggesting that the evolution of IDR/IDP cannot be explained merely by the lack of structural restraints. Indeed, IDP and IDR in the human genome were found to be undergoing extensive adaptive evolution (Afanasyeva et al. 2018). At last, it has been recognized that, except for allosteric regulations, encounter complexes (Gabdoulline and Wade 1999) might also play an important role in mediating intermolecular interactions, such as protein–protein association (Tang et al. 2006) and protein-ligand binding (Re et al. 2019). Since encounter residues that are responsible for encounter complexes do not reside in conserved binding interfaces, these residues could be under relaxed purifying selection or even positive selection, which could be another yet-to-identify mechanism that contributes to protein sequence adaptive evolution.
We showed that fast-adaptive proteins are enriched in molecular functions such as reproduction, immunity, and environmental information processing (Begun and Whitley 2000; Lazzaro et al. 2004; Begun and Lindfors 2005). We further demonstrated that fast-adaptive proteins tend to interact with each other more frequently than random expectations, suggesting coadaptation might be common among fast-adaptive proteins. Mechanisms contributing to the coadaptation could be: 1) interacting fast-adaptive proteins are often enriched in similar molecular functions and under similar selective pressure; and 2) interacting fast-adaptive undergo coevolution through physical interactions. In this study, we showed two examples that adaptive evolution could occur at PPI, which suggest that physical interactions could contribute to the coadaptation of fast-adaptive proteins in D. melanogaster. Moreover, we showed that coadaptation might exist to a broader extend rather than only among fast-adaptive proteins. Specifically, proteins that interact with fast-adaptive proteins tend to have higher adaptation rates. Since molecular interactions contribute to adaptive evolution, it is reasonable to hypothesize that coadaptation at a broader extend could be regulated by these interactions. Actually, it has been suggested that interacting proteins tend to have similar evolutionary rates and the possible mechanism would be the coevolution of physical interactions (Pazos and Valencia 2008).
In this study, we found that amino acids showing great geographical differences often overlap with sites that show adaptive signals between species. These loci follow similar patterns as adaptive changes, that is, they are enriched in disordered regions, protein surfaces, and functionally important regions. These results suggest that population differentiation of protein-coding genes can be an important basis for long-term adaptive evolution. In other words, many SNPs are repeatedly selected for the adaptive processes in evolution. Importantly, our results indicate that most of the strongly differentiated clinal amino-acid changes are adaptive, suggesting that nonselective forces play a less essential role in the SNPs that show great geographical differences. Our results also support a large effect of spatially varying selection on protein sequence and structures (Storz and Kelly 2008). Interestingly, our previous work showed that geographically differentiated SNPs often occur on the same orthologous genes between species but rarely the same SNPs (Zhao et al. 2015). Thus, it would be interesting to extend this work to D. simulans to study parallel protein evolution at the structural level.
It should be noted that studies at the genomic scale that aim to uncover the function- or structure-related constraints imposed on protein sequence evolution and adaptation share similar limitations that for most of the proteins or residues, structural or functional information would be incomplete or even missing. To overcome this, in this study, we used highly accurate neural network-based tools to predict molecular interactions, secondary structures, ISD, RSA for each of the proteins. In this way, we were able to identify key factors that impact protein sequence evolution and adaptation in a less accurate but rather systematic fashion. Notably, it has been reported that false positives are non-negligible in methods to the estimation of adaptive evolution (Markova-Raina and Petrov 2011), and other mechanisms including translational selection were acting on the evolution of protein-coding genes (Larracuente et al. 2008), which together hinder our understanding toward protein evolution and protein adaptation. Another limitation of our study is that we did not include indels, especially nonframeshift indels, as it is very difficult to address the adaptive effects of indels. However, with method development and increased knowledge of protein structures, this would be an important question to investigate in the future. In recent years, deep learning-based predictors and estimators have contributed to our knowledge of protein structure (Jumper et al. 2021), protein function (Kulmanov and Hoehndorf 2020), or even protein evolution (Schrider and Kern 2018). We hope that with the availability of more and more curated structural, functional information, and complex structural models of proteins in the near future, we will be able to uncover the precise role of molecular interactions in protein sequence adaptive evolution.
Materials and Methods
dN/dS Ratio (ω)
We used a maximum likelihood method to infer dN/dS ratio (ω) of D. melanogaster protein-coding genes using the genome sequences of five species in melanogaster subgroup (D. melanogaster, D. simulans, D. sechellia, D. yakuba, and D. erecta). The protein-coding sequences were extracted from the alignments of 26 insects, which were obtained from UCSC Genome Browser (http://hgdownload.soe.ucsc.edu/downloads.html). The sequences were further processed by GeneWise (Birney et al. 2004) to remove possible insertions and deletions using the longest isoforms of the corresponding D. melanogaster protein sequences as references (FlyBase version r6.15) (Thurmond et al. 2019). The processed sequences were then realigned by PRANK -codon function (Löytynoja 2014). The conservation scores and coverages of the alignments can be found in supplementary fig. S10, Supplementary Material online. We used codeml in PAML (Yang 2007) to compute gene-specific ω using M0 model. We removed sequences that have more than 15% of their nucleotides not aligned (gaps) to D. melanogaster genes in more than two species. To further avoid numeric errors and ensure reasonable estimations, we only retained relatively divergent sequences that are: 1) divergent with dS larger than 0.3 and (2) less divergent with dS larger than 0.1 and dN smaller than 0.001 (dS≫dN). At last, there were 12,118 genes in total that passed all the criteria and were assigned gene-specific ω, containing 6,538,872 amino acids. We also calculated site-specific ω by using likelihood ratio tests (LRT) comparing M7 model against M8 model, and M8fix model against M8 model (Yang et al. 2005), respectively. For genes that passed LRT, we extracted potentially positively selected sites with the probability being positively selected (P) greater than 0.5. In the examples of positive selection around molecular interaction sites in high-quality structural models, we list sites with P > 0.5, P > 0.8, and P > 0.9. In cases where we did not specify the probabilities being positively selected, we used the more stringent probability cutoff of 0.9.
Rate of Adaptive and Nonadaptive Changes
We recalled all SNPs of 205 inbred lines from the DGRP, Freeze 2.0 (Huang et al. 2014; http://dgrp2.gnets.ncsu.edu). We then generated 410 alternative genomes using all monoallelic and bi-allelic SNP data sets. We extracted the coding sequences of D. melanogaster genes from the generated alternative genomes, removed all possible insertions and deletions using GeneWise (Birney et al. 2004) as described earlier. We then align all the coding sequences to their corresponding aligned coding sequences using PRANK -codon function (Löytynoja 2014). We removed polymorphisms segregating at frequencies smaller than 5% to reduce possible slightly deleterious mutations (Charlesworth and Eyre-Walker 2008). In order to avoid possible effects of low divergence between D. simulans and D. melanogaster (Keightley and Eyre-Walker 2012), we used D. yakuba as outgroup to estimate nonsynonymous polymorphisms (Pn), synonymous polymorphisms (Ps), nonsynonymous substitutions (Dn), and synonymous substitutions (Ds) by MK.pl (Begun et al. 2007; Langley et al. 2012). Similar to Begun et al. (2007), we only analyzed genes with at least six variants for each of substitutions, polymorphisms, nonsynonymous changes, and synonymous changes. We used an extension of MK test, asymptotic MK (Messer and Petrov 2013; Uricchio et al. 2019), to estimate the proportions of adaptive changes (α). The rate of adaptive changes (ωa) was then calculated as ωa = ωα and the rate of non-adaptive changes as ωna = ω − ωa. Details of the asymptotic MK test were as follows:
(1) Classical MK test. According to Smith and Eyre-Walker (2002), the proportions of adaptive changes for protein-coding genes can be calculated as follows:
According to this equation, we could estimate the proportion of adaptive changes and carry out classical MK test by applying Fisher’s exact test.
(2) Asymptotic estimation of α. A known problem of the classical estimation of α above is the accumulation of slightly deleterious mutations at low frequencies. We therefore used an extension of MK test, asymptotic MK test approach (Messer and Petrov 2013) to estimate the proportions of adaptive changes. As in original aMK, we defined α(x) as a function of derived allele frequency (x):
where Pn(x) and Ps(x) are number of nonsynonymous and synonymous polymorphisms at frequency x, respectively. However, the original approach may suffer from numeric errors when there were very few polymorphic sites, which is quite common in many of D. melanogaster genes. To make the estimations more robust while preserving the same asymptote, we further defined Pn(x) and Ps(x) as total number of Pn and Ps above frequency x as described in Uricchio et al. (2019). We fitted α(x) to an exponential curve of α(x) ≈ exp(−bx)+c using lmfit (Newville et al. 2014) and determined the asymptotic value of α at the limit of x, 1.0. We then estimate the rate of adaptive changes (ωa) as
where Na is the number of adaptive changes and dNa = Na/LN is the number of adaptive changes per nonsynonymous site. Finally, we calculated the rate of nonadaptive changes (ωna) as ωna = ω − ωa. The final data set contains 7,192 protein-coding genes, with the smallest ωa being 0.00 and largest being 1.29.
Structure-/Function-Related Properties of D. melanogaster Proteins
We obtained function-related properties mentioned in the main text as following. We derived D. melanogaster gene ages (Zhang et al. 2010; Kondo et al. 2017) for genes that are specific to Drosophila, and from GenTree (Shao et al. 2019) for genes that are beyond Drosophila clade. We then assigned a pseudo-age to each of the genes. Specifically, there are 11 age groups from “cellular organisms,” assigning to a pseudo age value of 0, to “melanogaster,” assigning a pseudo age value of 10. We downloaded D. melanogaster PPI from STRING database (Szklarczyk et al. 2019). A cutoff of a combined score larger than 0.7 was used to retain high confident PPI for further analysis. We then used BSpred (Mukherjee and Zhang 2011) to predict PPI sites and DRNApred (Yan and Kurgan, 2017) to predict DNA binding sites. For each protein, we calculated ratios of protein interaction residues (PPI-site ratio) and ratios of DNA binding residues (DNA-site ratio) by dividing total predicted protein interaction sites and DNA binding sites over protein length, respectively. For structure-related properties, we used DeepCNF (Wang et al. 2016) to predict these properties for each gene, including three-state secondary structures (helix, sheet, and coil), structural disorder, RSA. Further, we calculated the ratios of helix, sheet, helix+sheet, and coil residues of each gene from predicted secondary structures. DeepCNF (Wang et al. 2016) is a deep learning method to capture complex sequence–structure relationships and was proved to have high accuracy in the prediction of protein secondary structures (Yang et al. 2018), structural disorder (Necci et al. 2021), and RSA (Wang et al. 2016). For each gene, we computed ISD and RSA, as protein-length normalized summations of the probabilities of each residue being disorder and exposed, respectively.
Gene Expression Patterns
We downloaded the gene expression profile from FlyAtlas2 (Leader et al. 2018). We converted Fragments Per Kilobase of transcript per Million mapped reads (FPKM) to Transcript per Million mapped reads (TPM) by normalizing FPKM against the summation of all FPKMs as follows:
After TPM conversion, we only retained genes with expression level larger than 0.1 TPM for further analysis. We treated male and female whole-body TPM as male and female expression levels. We calculated the mean expression level by averaging male and female TPM. We used the following Z-score to describe male specificities of D. melanogaster genes:
We calculated tissue specificities of genes using tau values (Yanai et al. 2005) based on the expression profiles of 27 different tissues.
High-Quality 3D Structures of D. melanogaster Proteins
We downloaded high-quality structures or structural models of D. melanogaster proteins from protein data bank (PDB; Burley et al. 2019), SWISS-MODEL Repository (Bienert et al. 2017), and MODBASE (Pieper et al. 2011), with descending priorities. For example, if there were 3D structures of the same protein or protein region in multiple databases, we first considered high-resolution structures from PDB; if no structures were found in PDB, we then considered SWISS-MODEL Repository; and at last from MODBASE. In addition, we used blastp (Camacho et al. 2009) to search homologs of each D. melanogaster protein against all PDB sequences with an E-value threshold of 0.001. We further carried out comparative structural modeling using RosettaCM (Song et al. 2013) to model high-quality structural models of proteins or protein regions that were not available in PDB, SWISS-MODEL Repository and MODBASE. For each RosettaCM simulation, we used no more than five most significant hits from blastp search. For proteins that are in complex forms, we only extracted monomers for further analysis. Finally, we obtained 14,543 high-quality structural models, corresponding to 11,284 genes. These structural models contain 2,691,913 unique amino acids, 41.2% of all the residues in genes that were assigned ω.
Evolutionary Rates of Different Structural/Functional Sites
We classified amino acids into different classes of structural/functional properties. Specifically, we classified three classes for both ISD and RSA according to the probability of residues being disordered or exposed: ordered or buried (0.00–0.33), medium (0.33–0.67), disordered or exposed (0.67–1.00). For both PPI and DNA binding, we classified two classes: PPI- or DNA-site (binding sites), None-PPI or None-DNA (corresponding null sites for PPI or DNA binding). For residues that have 3D structures, we used STRESS (Clarke et al. 2016) to predict putative ligand binding sites and allosteric sites from all the high-quality structures or structural models. We chose STRESS over other programs because it takes both geometry and protein dynamics into account and has been used in genome-wide studies and explained many poorly understood diseases associated variants in humans (Clarke et al. 2016). The allosteric sites were further classified as surface critical or interior critical according to their locations. We then classified these residues into four groups: LIG (ligand binding sites), Surf. Crit. (surface critical sites), Interior Crit. (interior critical sites), and Others (other sites). For each of the site classes, we randomly sampled 100 sequences, each containing 10,000 amino acids. We computed ω, ωa, and ωna for the randomly sampled sequences similar to the steps described in the above sections.
Population Genetics of DPGP3 African Population
We analyzed 20 high-quality genomes in high latitude South Africa and 30 high-quality genomes in low latitude Ethiopia (Lack et al. 2015). We called all biallelic SNPs and removed SNPs segregating at frequencies smaller than 5% to reduce possible slightly deleterious mutations. Similar to Svetec et al. (2016), for each SNP in both populations, we calculated the fixation index, FST, and used ormidp.test from epitools package in R to perform the odds ratio test for independence. For SNPs at each chromosome arm, we calculated the FDR using the Bioconductor q-value package (https://github.com/jdstorey/qvalue, last accessed December 1, 2021).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank members of the Zhao Lab for helpful discussions, especially Evan Witt for gene expression analysis and Christopher Langer for critically reading an early version of the article. The work was supported by NIH MIRA R35GM133780, the Robertson Foundation, a Monique Weill-Caulier Career Scientist Award, an Alfred P. Sloan Research Fellowship (FG-2018-10627), a Rita Allen Foundation Scholar Program, and a Vallee Scholar Program (VS-2020-35) to L.Z. J.P. is supported by a C. H. Li Memorial Scholar Fund Award at the Rockefeller University.
Author Contributions
J.P. and L.Z. conceived the study. J.P. performed all the analysis with the input from L.Z., N.S. conceptualized the population genetic analysis and helped with the interpretation, J.P. and L.Z. wrote the manuscript.
Data Availability
Codes and scripts for this study are available on GitHub: https://github.com/LiZhaoLab/DrosophilaProteinEvolution (last accessed December 1, 2021).
References
- Afanasyeva A, Bockwoldt M, Cooney CR, Heiland I, Gossmann TI.. 2018. Human long intrinsically disordered protein regions are frequent targets of positive selection. Genome Res. 28(7):975–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. 2008. Positive selection at the binding sites of the male-specific lethal complex involved in dosage compensation in Drosophila. Genetics 180(2):1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Lindfors HA.. 2005. Rapid evolution of genomic Acp complement in the melanogaster subgroup of Drosophila. Mol Biol Evol. 22(10):2010–2021. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Whitley P.. 2000. Adaptive evolution of relish, a Drosophila NF-kappaB/IkappaB protein. Genetics 154(3):1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, Hillier LW, Poh Y-P, Hahn MW, Nista PM, Jones CD, Kern AD, Dewey CN, et al. 2007. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 5(11):e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland AO, Behrman EL, O'Brien KR, Schmidt PS, Petrov DA.. 2014. Genomic evidence of rapid and stable adaptive oscillations over seasonal time scales in Drosophila. PLoS Genet. 10(11):e1004775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert S, Waterhouse A, De Beer TAP, Tauriello G, Studer G, Bordoli L, Schwede T.. 2017. The SWISS-MODEL repository-new features and functionality. Nucleic Acids Res. 45(D1):D313–D319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R.. 2004. GeneWise and genomewise. Genome Res. 14(5):988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Berman HM, Bhikadiya C, Bi C, Chen L, Di Costanzo L, Christie C, Dalenberg K, Duarte JM, Dutta S, et al. 2019. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 47(D1):D464–D474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterwick JA, del Mármol J, Kim KH, Kahlson MA, Rogow JA, Walz T, Ruta V.. 2018. Cryo-EM structure of the insect olfactory receptor Orco. Nature 560(7719):447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth J, Eyre-Walker A.. 2008. The McDonald-Kreitman test and slightly deleterious mutations. Mol Biol Evol. 25(6):1007–1015. [DOI] [PubMed] [Google Scholar]
- Clarke D, Sethi A, Li S, Kumar S, Chang RWF, Chen J, Gerstein M.. 2016. Identifying allosteric hotspots with dynamics: application to inter- and intra-species conservation. Structure 24(5):826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasmeh P, Serohijos AWR, Kepp KP, Shakhnovich EI.. 2013. Positively selected sites in cetacean myoglobins contribute to protein stability. PLoS Comput Biol. 9:e1002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH.. 2005. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci USA. 102(40):14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echave J, Spielman SJ, Wilke CO.. 2016. Causes of evolutionary rate variation among protein sites. Nat Rev Genet. 17(2):109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard D, Cai L, Gwennap C, Petrov DA.. 2016. Viruses are a dominant driver of protein adaptation in mammals. Elife 5:e12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian DK, Kapun M, Nolte V, Kofler R, Schmidt PS, Schlötterer C, Flatt T.. 2012. Genome-wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol Ecol. 21(19):4748–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabdoulline RR, Wade RC.. 1999. On the protein-protein diffusional encounter complex. J Mol Recognit. 12(4):226–234. [DOI] [PubMed] [Google Scholar]
- Goldman N, Thorne JL, Jones DT.. 1998. Assessing the impact of secondary structure and solvent accessibility on protein evolution. Genetics 149(1):445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CA, Fox AN, Pitts RJ, Kent LB, Tan PL, Chrystal MA, Cravchik A, Collins FH, Robertson HM, Zwiebel LJ.. 2002. G protein-coupled receptors in Anopheles gambiae. Science 298(5591):176–178. [DOI] [PubMed] [Google Scholar]
- Huang W, Massouras A, Inoue Y, Peiffer J, Ràmia M, Tarone AM, Turlapati L, Zichner T, Zhu D, Lyman RF, et al. 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24(7):1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Nei M.. 1988. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335(6186):167–170. [DOI] [PubMed] [Google Scholar]
- Jiggins FM, Kim KW.. 2007. A screen for immunity genes evolving under positive selection in Drosophila. J Evol Biol. 20(3):965–970. [DOI] [PubMed] [Google Scholar]
- Jordan IK, Wolf YI, Koonin EV.. 2003. No simple dependence between protein evolution rate and the number of protein-protein interactions: only the most prolific interactors tend to evolve slowly. BMC Evol Biol. 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596(7873):583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A.. 2012. Estimating the rate of adaptive molecular evolution when the evolutionary divergence between species is small. J Mol Evol. 74(1-2):61–68. [DOI] [PubMed] [Google Scholar]
- Kim PM, Lu LJ, Xia Y, Gerstein MB.. 2006. Relating three-dimensional structures to protein networks provides evolutionary insights. Science 314(5807):1938–1941. [DOI] [PubMed] [Google Scholar]
- Kolaczkowski B, Kern AD, Holloway AK, Begun DJ.. 2011. Genomic differentiation between temperate and tropical australian populations of Drosophila melanogaster. Genetics 187(1):245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Vedanayagam J, Mohammed J, Eizadshenass S, Kan L, Pang N, Aradhya R, Siepel A, Steinhauer J, Lai EC.. 2017. New genes often acquire male- specific functions but rarely become essential in Drosophila. Genes Dev. 31:1841–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosiol C, Vinař T, da Fonseca RR, Hubisz MJ, Bustamante CD, Nielsen R, Siepel A.. 2008. patterns of positive selection in six mammalian genomes. PLoS Genet. 4(8):e1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulmanov M, Hoehndorf R.. 2020. DeepGOPlus: improved protein function prediction from sequence. Bioinformatics 36(2):422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack JB, Cardeno CM, Crepeau MW, Taylor W, Corbett-Detig RB, Stevens KA, Langley CH, Pool JE.. 2015. The Drosophila genome nexus: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics 199(4):1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley CH, Stevens K, Cardeno C, Lee YCG, Schrider DR, Pool JE, Langley SA, Suarez C, Corbett-Detig RB, Kolaczkowski B, et al. 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192(2):533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente AM, Sackton TB, Greenberg AJ, Wong A, Singh ND, Sturgill D, Zhang Y, Oliver B, Clark AG.. 2008. Evolution of protein-coding genes in Drosophila. Trends Genet. 24(3):114–123. [DOI] [PubMed] [Google Scholar]
- Lawniczak MKN, Begun DJ.. 2007. Molecular population genetics of female-expressed mating-induced serine proteases in Drosophila melanogaster. Mol Biol Evol. 24(9):1944–1951. [DOI] [PubMed] [Google Scholar]
- Lazzaro BP, Sceurman BK, Clark AG.. 2004. Genetic basis of natural variation in D. melanogaster antibacterial immunity. Science 303(5665):1873–1876. [DOI] [PubMed] [Google Scholar]
- Leader DP, Krause SA, Pandit A, Davies SA, Dow JAT.. 2018. FlyAtlas 2: a new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucleic Acids Res. 46(D1):D809–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin TC, Malik HS.. 2017. Rapidly evolving Toll-3/4 genes encode male-specific Toll-like receptors in Drosophila. Mol Biol Evol. 34(9):2307–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima LF, Torres AQ, Jardim R, Mesquita RD, Schama R.. 2021. Evolution of Toll, Spatzle and MyD88 in insects: the problem of the Diptera bias. BMC Genomics 22(1):562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YS, Hsu WL, Hwang JK, Li WH.. 2007. Proportion of solvent-exposed amino acids in a protein and rate of protein evolution. Mol Biol Evol. 24(4):1005–1011. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. ; Genome Institute at Washington University. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478(7370):476–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A. 2014. Phylogeny-aware alignment with PRANK. Methods Mol Biol. 1079:155–170. [DOI] [PubMed] [Google Scholar]
- Markova-Raina P, Petrov D.. 2011. High sensitivity to aligner and high rate of false positives in the estimates of positive selection in the 12 Drosophila genomes. Genome Res. 21(6):863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthey-Doret R, Whitlock MC.. 2019. Background selection and FST: consequences for detecting local adaptation. Mol Ecol. 28(17):3902–3914. [DOI] [PubMed] [Google Scholar]
- McBride CS. 2007. Rapid evolution of smell and taste receptor genes during host specialization in Drosophila sechellia. Proc Natl Acad Sci USA. 104:4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer PW, Petrov DA.. 2013. Frequent adaptation and the McDonald-Kreitman test. Proc Natl Acad Sci USA. 110(21):8615–8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintseris J, Weng Z.. 2005. Structure, function, and evolution of transient and obligate protein-protein interactions. Proc Natl Acad Sci USA. 102(31):10930–10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho AF, Trancoso FF, Dutheil JY, Zhang J.. 2019. The impact of protein architecture on adaptive evolution. Mol Biol Evol. 36(9):2013–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Zhang Y.. 2011. Protein-protein complex structure predictions by multimeric threading and template recombination. Structure 19(7):955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necci M, Piovesan D, Hoque MT, Walsh I, Iqbal S, Vendruscolo M, Sormanni P, Wang C, Raimondi D, Sharma R, et al. ; DisProt Curators. 2021. Critical assessment of protein intrinsic disorder prediction. Nat Methods 18(5):472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newville M, Stensitzki T, Allen DB, Ingargiola A.. 2014. Non-Linear Least-Squares Minimization and Curve-Fitting for Python. Zenodo. Available from: 10.5281/zenodo.11813. [DOI]
- Nielsen R, Bustamante C, Clark AG, Glanowski S, Sackton TB, Hubisz MJ, Fledel-Alon A, Tanenbaum DM, Civello D, White TJ, et al. 2005. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 3(6):e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Welch JJ, Kim K-W, Jiggins FM.. 2009. Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 5(10):e1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C, Papp B, Hurst LD.. 2001. Highly expressed genes in yeast evolve slowly. Genetics 158(2):927–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer WH, Hadfield JD, Obbard DJ.. 2018. RNA-interference pathways display high rates of adaptive protein evolution in multiple invertebrates. Genetics 208(4):1585–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos F, Valencia A.. 2008. Protein co-evolution, co-adaptation and interactions. EMBO J. 27(20):2648–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper U, Webb BM, Barkan DT, Schneidman-Duhovny D, Schlessinger A, Braberg H, Yang Z, Meng EC, Pettersen EF, Huang CC, et al. 2011. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 39(Database issue):D465–D474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitchers W, Pool JE, Dworkin I.. 2013. Altitudinal clinal variation in wing size and shape in African Drosophila melanogaster: one cline or many? Evolution 67:438–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey DC, Scherrer MP, Zhou T, Wilke CO.. 2011. The relationship between relative solvent accessibility and evolutionary rate in protein evolution. Genetics 188(2):479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Re S, Oshima H, Kasahara K, Kamiya M, Sugita Y.. 2019. Encounter complexes and hidden poses of kinaseinhibitor binding on the free-energy landscape. Proc Natl Acad Sci USA. 116(37):18404–18409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt JA, Kolaczkowski B, Jones CD, Begun DJ, Kern AD.. 2014. Parallel geographic variation in Drosophila melanogaster. Genetics 197(1):361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG.. 2007. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 39(12):1461–1468. [DOI] [PubMed] [Google Scholar]
- Schott RK, Refvik SP, Hauser FE, López-Fernández H, Chang BSW.. 2014. Divergent positive selection in rhodopsin from lake and riverine cichlid fishes. Mol Biol Evol. 31(5):1149–1165. [DOI] [PubMed] [Google Scholar]
- Schrider DR, Kern AD.. 2018. Supervised machine learning for population genetics: a new paradigm. Trends Genet. 34(4):301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Chen C, Shen H, He BZ, Yu D, Jiang S, Zhao S, Gao Z, Zhu Z, Chen X, et al. 2019. GenTree, an integrated resource for analyzing the evolution and function of primate-specific coding genes. Genome Res. 29(4):682–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi M, Cagliani R, Forni D, Clerici M.. 2015. Evolutionary insights into host-pathogen interactions from mammalian sequence data. Nat Rev Genet. 16(4):224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slodkowicz G, Goldman N.. 2020. Integrated structural and evolutionary analysis reveals common mechanisms underlying adaptive evolution in mammals. Proc Natl Acad Sci USA. 117(11):5977–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith NGC, Eyre-Walker A.. 2002. Adaptive protein evolution in Drosophila. Nature 415(6875):1022–1024. [DOI] [PubMed] [Google Scholar]
- Song Y, Dimaio F, Wang RYR, Kim D, Miles C, Brunette T, Thompson J, Baker D.. 2013. High-resolution comparative modeling with RosettaCM. Structure 21(10):1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz JF, Kelly JK.. 2008. Effects of spatially varying selection on nucleotide diversity and linkage disequilibrium: insights from deer mouse globin genes. Genetics 180(1):367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetec N, Cridland JM, Zhao L, Begun DJ.. 2016. The adaptive significance of natural genetic variation in the DNA damage response of Drosophila melanogaster. PLoS Genet. 12(3):e1005869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47(D1):D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Iwahara J, Clore GM.. 2006. Visualization of transient encounter complexes in protein-protein association. Nature 444(7117):383–386. [DOI] [PubMed] [Google Scholar]
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, et al. ; FlyBase Consortium 2019. FlyBase 2.0: the next generation. Nucleic Acids Res. 47(D1):D759–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda Y, Ko MC, Liang Q, Miller ET, Rico-Guevara A, Nakagita T, Sakakibara A, Uemura K, Sackton T, Hayakawa T, et al. 2021. Early origin of sweet perception in the songbird radiation. Science 373(6551):226–231. [DOI] [PubMed] [Google Scholar]
- Uricchio LH, Petrov DA, Enard D.. 2019. Exploiting selection at linked sites to infer the rate and strength of adaptation. Nat Ecol Evol. 3(6):977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. 2019. Intrinsically disordered proteins and their “Mysterious” (meta)physics. Front Phys. 7:10. Available from: 10.3389/fphy.2019.00010. [DOI] [Google Scholar]
- Wang S, Li W, Liu S, Xu J.. 2016. RaptorX-Property: a web server for protein structure property prediction. Nucleic Acids Res. 44(W1):W430–W435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DD, Wang GD, Irwin DM, Zhang YP.. 2009. A profound role for the expansion of trypsin-like serine protease family in the evolution of hematophagy in mosquito. Mol Biol Evol. 26(10):2333–2341. [DOI] [PubMed] [Google Scholar]
- Yan J, Kurgan L.. 2017. DRNApred, fast sequence-based method that accurately predicts and discriminates DNA- and RNA-binding residues. Nucleic Acids Res. 45:gkx059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Benjamin H, Shmoish M, Chalifa-Caspi V, Shklar M, Ophir R, Bar-Even A, Horn-Saban S, Safran M, Domany E, et al. 2005. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics 21(5):650–659. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Y, Gao J, Wang J, Heffernan R, Hanson J, Paliwal K, Zhou Y.. 2018. Sixty-five years of the long march in protein secondary structure prediction: the final stretch? Brief Bioinform. 19:482–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R.. 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 22(4):1107–1118. [DOI] [PubMed] [Google Scholar]
- Zhang J, He X.. 2005. Significant impact of protein dispensability on the instantaneous rate of protein evolution. Mol Biol Evol. 22(4):1147–1155. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yang J-R.. 2015. Determinants of the rate of protein sequence evolution. Nat Rev Genet. 16(7):409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Krinsky BH, Long M.. 2010. Age-dependent chromosomal distribution of male-biased genes in Drosophila. Genome Res. 20(11):1526–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Wit J, Svetec N, Begun DJ.. 2015. Parallel gene expression differences between low and high latitude populations of Drosophila melanogaster and D. simulans. PLoS Genet. 11(5):e1005184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416(6882):703–709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Codes and scripts for this study are available on GitHub: https://github.com/LiZhaoLab/DrosophilaProteinEvolution (last accessed December 1, 2021).