Rottner and Stradal discuss new work from the Weiner laboratory unveiling that WASP integrates substrate topology, cell polarity, and migration in immune cells.
Abstract
WASP is a remodeler of the actin cytoskeleton, but its mechanistic contribution to neutrophil migration is unclear. In this issue, Brunetti et al. (2021. J. Cell Biol. https://doi.org/10.1083/jcb.202104046) show that WASP is recruited to substrate-induced membrane deformations near the cell front, where it induces Arp2/3 complex–mediated local actin assembly to direct migration.
Neutrophils are the most abundant type of white blood cells in humans and constitute an important first line of defense of the innate immune system. Neutrophils exit the blood stream in response to chemoattractants that signal danger in the form of damage, infection, or inflammation, ultimately removing dangerous particles by phagocytosis. Efficient migration is crucial to properly execute such functions, but how precisely the coordination of polarity establishment and complex shape changes needed for migratory processes occurs in these cells is just beginning to be elucidated. The dynamic remodeling of actin filaments, which can differentially assemble and disassemble at both ends, is key to the development of pulling and pushing forces below the plasma membrane of most eukaryotic cells or on the surfaces of their intracellular organelles (1).
Actin filaments are organized into either bundles or networks, with networks most commonly generated and maintained by the continuous filament branching activity of the actin-related protein 2/3 (Arp2/3) complex (2). Arp2/3 complex–driven branching in actin networks has often been associated with the development of pushing forces, even in a mechano-responsive fashion (3, 4), and less so linked with pulling or traction forces as developed, for instance, by adhesions. The heteropentameric Arp2/3 complex, which catalyzes the formation of daughter filaments off the sides of mother filaments, is intrinsically inactive, but can be activated by nucleation promoting factors (NPFs). The canonical NPFs include the Wiskott-Aldrich syndrome protein (WASP), its ubiquitous orthologue neural WASP (N-WASP), three WASP family verprolin-homologous protein (WAVE) isoforms as well as WASH, WHAMM, and JMY (5). The C-termini of all these factors physically interact with the Arp2/3 complex, driving it into an active conformation, and aid daughter filament polymerization by adding actin monomers onto Arp-2 and -3, forming the base of the branch. Coordinating the branching of daughter filaments by Arp2/3 complex and their elongation appears to be a major function of NPFs such as WASP. The role of WASP in stimulating actin assembly and the observation that patients with Wiskott-Aldrich syndrome harboring mutations in this gene have white blood cells that are unable to reach the sites of infections (6) points to WASP as a regulator of guided migration of immune cells. In this issue, Brunetti et al. establish the precise cellular function by which WASP controls neutrophil migration: WASP is recruited to sites of inward membrane deformations to stimulate the formation of spot-like actin structures that aid in the adhesion to or in the grabbing onto extracellular structures of high diversity and flexibility such as extracellular matrix fibrils, thus linking substrate topology, cell polarity, and migration (7).
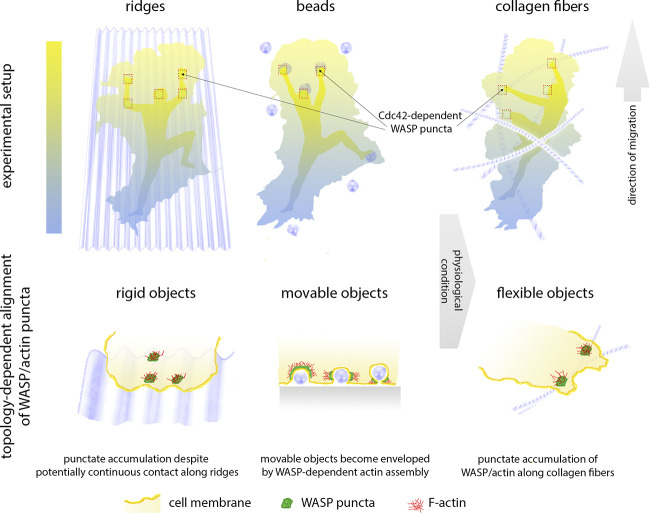
The authors first determined the spatiotemporal dynamics of EGFP-tagged, endogenous WASP in the human neutrophil–like cell line HL-60 using total internal reflection fluorescence microscopy. WASP formed puncta that were largely devoid of clathrin accumulation, thus likely not embodying endocytic pits, and exogenously triggered by submicron-sized beads, microfabricated ridges, or collagen fibers. Substratum-associated WASP puncta exhibited adhesive functions, as integrin inactivation by ion chelation largely dissociated them, stimulating their sliding and centripetal displacement. Surprisingly, the prominent WASP accumulation to beads below the plasma membrane was biased to the front half of the cell, and coincided with a gradient of activity of its major activator, the Rho GTPase Cdc42 from front to rear. This observation led Brunetti et al. to postulate WASP as a factor linking substrate topology to cell polarization and effective migration.
A closer look at the recruitment of WASP to inward plasma membrane invaginations (positive membrane curvature) stimulated by submicron-sized beads revealed two patterns of association: either following the bead bodies as U-shaped accumulations or to the necks of invaginations reminiscent of endocytic pits, which could reflect later stages of bead–plasma membrane interactions. Although the mechanistic significance of these distinct association patterns remains elusive, it was clear that WASP favored associations at the plasma membrane with sites of comparably high, positive curvature (small, 100-nm-diameter beads more attractive than larger beads, for instance) and in a punctate fashion (even along ridges). In our view, this much diverges from WAVE isoforms exhibiting a prominent, more linear association with the rapidly protruding membranes of negative curvature in lamellipodia (8). Finally, the authors showed that Cdc42 loss-of-function diminished WASP puncta at the cell front, and that lack of WASP reduced actin puncta formation and the frequency of Arp2/3 complex accumulation at beads. WASP-null cells migrated less in the direction of ridges, although their perpendicular movement across the nanopatterns was not affected—indeed, it was increased. These observations connect the role of WASP in reading substrate topology to Arp2/3-dependent actin filament branching and network formation. The findings of Brunetti et al. appear to be of broader impact and relevance than just being a human neutrophil-specific phenomenon—a separate study has recently shown that sliding, WASP-dependent, punctate adhesion sites in murine dendritic cells and T cells, which are triggered in response to mechanical load, polymerize actin networks orthogonally to the plasma membrane, and aid in squeezing and dragging these cells through dense tissues (9).
Taking all this together, the study by Brunetti et al. highlights that at least immune cells primarily respond to mechanical impact and indentations of their plasma membranes with rapid accumulation of WASP and Arp2/3 complex–dependent actin assembly, thereby pushing back or grabbing extracellular material to push and pull themselves forward during migration through the complex, dense, and three-dimensional environments on their way through tissues (Fig. 1).
Figure 1.
Neutrophils use WASP puncta to crawl like mountain climbers. Punctate accumulation of WASP and consequently F-actin brings about friction points that support pushing and pulling during cell translocation. In the front half of the cell, formation of these points strictly depends on Cdc42 mediating WASP activation and focal actin assembly. Even on substrates that would allow continuous adhesion (ridges or collagen fibers), WASP is recruited in a punctate fashion and preferentially to the front half of the cell that is dominated by Cdc42 activity signaling to cell polarity. Upon contact, movable objects like beads lead to a strong WASP/actin response potentially culminating in object envelopment.
Interestingly, according to Brunetti et al., mechanisms of WASP/N-WASP recruitment to inward membrane deformations might even be conserved during early stages of formation of podosomes and invadopodia, which are WASP- and N-WASP–dependent, both protrusive and adhesive structures operating in matrix degradation of hematopoietic and cancer cells, respectively (10). This hypothesis, however, remains to be experimentally validated. Furthermore, additional questions remain unsolved, including the precise signals, aside from Cdc42, contributing to WASP accumulation and turnover at the plasma membrane. Where does the bias for focal, punctate WASP accumulation come from? WASP family NPFs are unlikely to themselves harbor curvature-sensing activities, so which are the decisive factors for interaction with positively curved membranes? How is specificity brought about? The F-BAR domain-containing TOCA family proteins would have been potential candidates, but surprisingly, disruption of two of their most prominent members, FBP17 and CIP4, did not cause severe defects, as Brunetti et al. showed in this study. Future work will surely improve our insights into the differential sorting and subcellular functions of distinct NPFs in both immune and other cells.
Acknowledgments
The authors declare no competing financial interests.
References
- 1.Rottner, K., et al. 2017. J. Cell Sci. 10.1242/jcs.206433 [DOI] [PubMed] [Google Scholar]
- 2.Gautreau, A.M., et al. 2021. Trends Cell Biol. 10.1016/j.tcb.2021.10.006 [DOI] [Google Scholar]
- 3.Bieling, P., et al. 2016. Cell. 10.1016/j.cell.2015.11.057 [DOI] [Google Scholar]
- 4.Papalazarou, V., and Machesky L.M.. 2021. Curr. Opin. Cell Biol. 10.1016/j.ceb.2020.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alekhina, O., et al. 2017. J. Cell Sci. 10.1242/jcs.199570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivers, E., and Thrasher A.J.. 2017. Eur. J. Immunol. 10.1002/eji.201646715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunetti, R.M., et al. 2022. J. Cell Biol. 10.1083/jcb.202104046 [DOI] [Google Scholar]
- 8.Pipathsouk, A., et al. 2021. J. Cell Biol. 10.1083/jcb.202003086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaertner, F., et al. 2022. Dev. Cell. 10.1016/j.devcel.2021.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paterson, E.K., and Courtneidge S.A.. 2018. FEBS J. 10.1111/febs.14123 [DOI] [PMC free article] [PubMed] [Google Scholar]