Wilson and Iskratsch highlight work from the Tzima laboratory that reveals a noncanonical role for eIF6 in the regulation of focal adhesion formation and mechanotransduction.
Abstract
eIF6 is known for its role as a stimulatory translation initiation factor. In this issue, Keen et al. (2022. J. Cell Biol. https://doi.org/10.1083/jcb.202005213) identify a novel, noncanonical role, whereby eIF6 regulates focal adhesion formation, mechanosensing, and cell mechanics, independent of its translational role.
Recent developments in the field of mechanobiology have revealed crucial interactions between the cell and its microenvironment. Fundamentally, the conversion of physical forces into biochemical signals and genetic responses, termed mechanotransduction, governs cell behaviors including migration, differentiation, maturation, and organogenesis (1). Central to mechanotransduction is the cytoskeleton, comprising actin filaments, microtubules, and intermediate filaments. The cytoskeleton is sensitive to chemical and mechanical alterations of the extracellular matrix and mediates changes to cell behavior and morphology. Physical forces are propagated along tensed actin cables all the way to the nucleus, where they connect to the linker of nucleoskeleton and cytoskeleton (LINC) complex (2).
Defective mechanosensing and transduction are implicated in major diseases, where changes to adhesion composition, cytoskeleton, and downstream signaling to the nucleus ultimately interfere with appropriate gene expression. For this, several mechanisms have been uncovered, including force-dependent chromatin decondensation, leading to long-term promoter silencing (3); force-dependent changes to epigenetics (4); or nuclear shuttling of transcriptional factors. In the latter case, recent work showed that forces on the nucleus (e.g., through the cytoskeleton) allow opening of the nuclear pore complexes, enabling the nuclear shuttling of the transcriptional coactivator Yes-Associated Protein (YAP), where it regulates gene expression, proliferation, and of particular interest to the cardiovascular community, cell (i.e., cardiomyocyte) regeneration (5).
Other studies suggest an essential role for mechanical forces in regulating protein synthesis. The translational machinery—ribosomes and associated factors—are associated with the cytoskeleton and can localize to focal adhesions (6). Cytoskeletal tension, which changes over space and time, also affects which mRNAs may be recruited over others, providing an additional layer of complexity (6). Physical forces are paramount to the propagation of the amino acids within the ribosomal core complex, as well as the unwinding of mRNA structures, such as hairpin loops (7). However, these are likely only a fraction of the mechanisms in the intimate relationship between mechanical forces, gene regulation, or protein translation. It is clear that fully defining these relationships is essential to unlock the therapeutic potential required to combat pathogenic states.
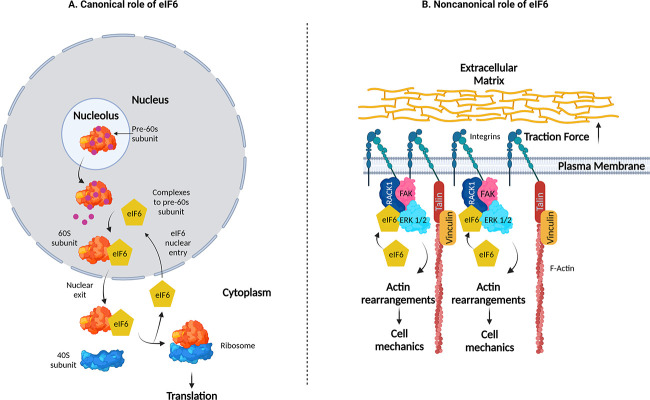
One additional layer of regulatory complexity between cellular forces and protein synthesis is outlined in the current issue by Keen et al. (8). Here, the authors present a novel, noncanonical mechanism of the eukaryotic initiation factor 6 (eIF6), a stimulatory translation initiation factor that acts downstream of insulin, or growth factors. Canonically, eIF6 binds to immature large ribosomal subunits (pre-60S) in the nucleolus and after maturation, ensures its translocation to the cytoplasm. eIF6’s release from the 60S subunit is a key step for the further formation of the 80S complex (Fig. 1). In contrast to this canonical role, the current work by Keen et al. identifies a role for eIF6 in modulating mechanical responses of endothelial cells, independent from its role in protein translation.
Figure 1.
The canonical and noncanonical activity of eIF6. (A) Canonically, eIF6 undergoes nucleocytoplasmic shuttling in order to stabilize the pre-60S subunit of the ribosome. In the cytoplasm, eIF6 release from the 60S subunit enables binding of the 40S ribosomal subunit, forming the 80S ribosome, which is paramount to translation. (B) Noncanonically, eIF6 is required to form the FAK–RACK1–ERK1/2 mechano-axis to regulate focal adhesions, the actin cytoskeleton, and cell mechanics. Created with BioRender.com.
Surprisingly, when the authors knocked down eIF6, this did not change protein synthesis, nor did it significantly affect the assembly of ribosomes. However, global mechanosensing was notably disrupted. This was evident through the observations of disrupted cell morphology, disrupted cytoskeletal organization, and decreased focal adhesion formation, as well as concomitant disruptions to traction force generation. Especially, focal adhesion protein levels did not change, but their localization was disrupted in the absence of eIF6, suggesting this protein is required for proper communication between cells and their microenvironment. Consequentially, the absence of eIF6 led to decreased elastic modulus of endothelial cells, reinforcing the finding that eIF6 is required for appropriate actin cytoskeleton formation and mechanics.
External application of tensional force further demonstrates the uncoupling of focal adhesion mechanosensing and translational functions of eIF6. Force application through paramagnetic beads, coated with an antibody against the mechanosensitive adhesion protein platelet endothelial cell adhesion molecule-1, led to increased phosphorylation of focal adhesion kinase (FAK) and focal adhesion growth, which was lost in the absence of eIF6. However, force-dependent increases to nascent protein synthesis remained unaffected by the knockdown of eIF6. The results support eIF6 as being particularly tuned to adhesion mechanosensing and as an important regulator for mechanotransduction.
Further exploring the mechanistic details, Keen et al. identify that eIF6 is necessary for the formation of mechanocomplexes comprising FAK, receptor of activated C kinase 1 (RACK1), and extracellular signal-regulated kinase 1/2 (ERK1/2). RACK1 is a scaffolding protein involved in both translation and adhesion formation, while ERK1/2 are kinases heavily involved in cardiovascular development and disease. Strikingly, eIF6 was not required for global mechano-activation of ERK1/2. Instead, it was required for the local activation of ERK1/2 at focal adhesions in response to mechanical force. Localization of ERK1/2 at focal adhesions is notably important for downstream signaling, focal adhesion remodeling, and gene regulation, and here is contingent upon eIF6 generating an eIF6–ERK–RACK1–FAK mechano-axis.
Overall, these exciting results identify a novel nontranslational role for eIF6. These results are particularly interesting as they link eIF6 directly to formation of focal adhesions, and further in regulating cell morphology, mechanosensing, and cell mechanics in endothelial cells. Thus, they ultimately outline a novel layer of involvement of eIF6 in the regulation of endothelial function, with impact on (tumor) angiogenesis (9) or cardiovascular disease. The work nicely demonstrates that translational machinery is intimately entwined with the cytoskeleton and in formation of focal adhesions, and that there may be reciprocal dialogue between these two seemingly disparate cellular constituents. It will be intriguing to see how these extratranslational roles are balanced with the canonical functions in the long term and how the balance might be shifted in cardiovascular or other diseases. Recently, eIF6 was implicated in hepatocellular carcinoma (10), and by more fully understanding its role, it is exciting to think what the future holds and how proteins involved with protein translation (or other functions) may hold novel, noncanonical roles in regulating the cell, and how we, as researchers, can harness this to our advantage.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council (BB/S001123/1) and British Heart Foundation (PG/20/6/34835).
The authors declare no competing financial interests.
References
- 1.Iskratsch, T., et al. 2014. Nat. Rev. Mol. Cell Biol. 10.1038/nrm3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang, N., et al. 2009. Nat. Rev. Mol. Cell Biol. 10.1038/nrm2594 [DOI] [PubMed] [Google Scholar]
- 3.Miroshnikova, Y.A., et al. 2017. J. Cell Sci. 10.1242/jcs.202192 [DOI] [PubMed] [Google Scholar]
- 4.Koester, J., et al. 2021. Nat. Cell Biol. 10.1038/s41556-021-00705-x [DOI] [Google Scholar]
- 5.Elosegui-Artola, A., et al. 2017. Cell. 10.1016/j.cell.2017.10.008 [DOI] [Google Scholar]
- 6.Chicurel, M.E., et al. 1998. Nature. 10.1038/33719 [DOI] [Google Scholar]
- 7.Qu, X., et al. 2011. Nature. 10.1038/nature10126 [DOI] [Google Scholar]
- 8.Keen, A.N., et al. 2022. J. Cell Biol. 10.1083/jcb.202005213 [DOI] [Google Scholar]
- 9.Pedrosa, A.R., et al. 2019. Cancer Res. 10.1158/0008-5472.CAN-18-3934 [DOI] [Google Scholar]
- 10.Sun, L., et al. 2021. J. Transl. Med. 10.1186/s12967-021-02877-4 [DOI] [Google Scholar]