Abstract
PURPOSE
Human epidermal growth factor receptor 2 (HER2) overexpression or amplification (ERBB2amp) are biomarkers for approved anti-HER2 therapies. ERBB2amp may better predict response compared with immunohistochemistry or in situ hybridization, and quantitative copy number (CN) may further stratify patients. We characterized ERBB2amp in advanced gastroesophageal adenocarcinomas (GEA) and hypothesized that increased CN was associated with better outcome to trastuzumab.
METHODS
Comprehensive genomic profiling, including assessment of ERBB2amp, was performed for 12,905 GEA tissue cases. Clinical outcomes were assessed using a clinicogenomic database linking deidentified electronic health record–derived clinical data to genomic data. Multivariable Cox proportional hazard models were used for real-world progression-free survival (rwPFS) comparisons.
RESULTS
ERBB2amp (CN ≥ 5) was detected in 15% (1,934 of 12,905) of GEA; median CN 22 (interquartile range 9-73). Median ERBB2 amplicon size was 0.27 megabase (interquartile range 0.13-0.95), and smaller amplicons were associated with higher CN (P < .001). In the clinicogenomic database, of 101 evaluable first-line trastuzumab-treated patients, ERBB2 CN was a significant predictor of rwPFS as a continuous variable (adjusted hazard ratio = 0.73; 95% CI, 0.60 to 0.89; P = .002), whereas ERBB2 CN was not predictive of rwPFS on chemotherapy (adjusted hazard ratio = 0.93; 95% CI, 0.73 to 1.20; P = .59). Among trastuzumab-treated patients, no significant associations with ERBB2 CN were observed for disease site, age, stage at advanced diagnosis, or most selected coalterations.
CONCLUSION
ERBB2amp was detected in 15% of GEA tissue samples, with significant diversity in ERBB2 CN and amplicon focality. ERBB2 CN was predictive of rwPFS as a continuous variable for patients treated with trastuzumab. Further studies exploring the clinical utility of quantitative ERBB2 CN, particularly in the setting of the evolving anti-HER2 landscape and combination therapies, are warranted.
INTRODUCTION
The ERBB2 gene encodes the human epidermal growth factor receptor 2 (HER2) receptor tyrosine kinase (RTK), a member of the epidermal growth factor receptor (EGFR)-related family of RTKs, and ligand-independent activation of HER2 can result from HER2 overexpression or ERBB2 amplification (ERBB2amp).1-4 HER2-targeted therapy revolutionized cancer care by providing one of the first demonstrations of clinical utility of biomarker-selected approaches and led to the approval of anti-HER2 therapies in breast cancers with HER2 overexpression.5-7 The anti-HER2 therapy trastuzumab is also approved in advanced gastroesophageal adenocarcinoma (advGEA) in combination with first-line chemotherapy on the basis of results from the Trastuzumab for Gastric Cancer trial,8 and investigational use of anti-HER2 therapies is being explored in other tumor types including colorectal cancer.9-11
CONTEXT
Key Objective
Human epidermal growth factor receptor 2 (HER2) overexpression or ERBB2 amplification are established biomarkers in gastroesophageal adenocarcinoma (GEA), and studies have suggested that degree of expression or amplification may further inform outcomes to anti-HER2 therapies. We sought to determine whether quantitative ERBB2 copy number (CN) as determined by next-generation sequencing was directly associated with better outcomes to trastuzumab in a real-world data set of patients with GEA.
Knowledge Generated
ERBB2 amplification is present in multiple tumor types including 15% of GEA, but the degree of CN gain and amplicon focality is variable. We show that ERBB2 CN is a significant predictor of improved real-world overall survival and real-world progression-free survival as a continuous variable in advanced GEA patients treated with first-line trastuzumab.
Relevance
Genomic profiling to quantitatively assess ERBB2 amplification should be performed to inform treatment selection and trial enrollment in GEA, particularly as new anti-HER2 therapies and combinations continue to be developed.
High concordance for HER2 expression by immunohistochemistry (IHC) or in situ hybridization (ISH) and other modalities, among each other and with and ERBB2amp by next-generation sequencing (NGS), has been demonstrated in GEA12-15; however, in one study evaluating discordant cases, those positive for both expression and amplification had better efficacy outcomes to anti-HER2 therapy compared with those positive for HER2 expression but negative for ERBB2amp.15 Stein et al and others have also presented data suggesting that higher ERBB2 copy number (CN) gain as quantified by NGS is associated with better efficacy outcomes to anti-HER2 therapy compared with lower levels of ERBB2amp in GEA.15,16 Earlier studies have also presented data showing that higher ERBB2 CN by ISH is predictive of better response to anti-HER2 therapy.17 The NCCN guidelines for esophageal and esophagogastric junction cancer and for gastric cancer (version 2.2021) recommend IHC or ISH for adenocarcinomas as the gold standard for selection of anti-HER2 therapy and NGS as an alternative subsequent option; however, these studies suggest a role for ERBB2amp detection by NGS, including CN quantification, as a further predictive marker of HER2 therapy benefit. Thus, further assessment of the utility of ERBB2amp as assessed by NGS, as well as the potential additive benefit of quantitative ERBB2 CN as a biomarker for anti-HER2 therapy, is needed.
We evaluated the frequency and distribution of CN gain and amplicon size in primary disease site subtypes of ERBB2amp GEA using a large genomic database of > 14,000 GEA tissue samples. We further analyzed treatment patterns for patients with ERBB2amp GEA captured in the Foundation Medicine-Flatiron Health clinicogenomic database (CGDB) and assessed the predictive impact of quantitative ERBB2 CN on real-world progression-free survival (rwPFS) to anti-HER2 therapy.
METHODS
Foundation Medicine Comprehensive Genomic Profiling
Hybrid capture–based comprehensive genomic profiling (CGP) was performed on formalin-fixed paraffin-embedded tumor tissue samples collected from 12,905 patients with primarily advanced GEA, as well as a comparison cohort of 34,629 primarily advanced breast cancer patients, during routine clinical care (January 2011-September 2020). Testing was performed in a Clinical Laboratory Improvement Amendments–certified, College of American Pathologists–accredited, New York State–regulated reference laboratory (Foundation Medicine, Inc, Cambridge, MA). Approval for this study, including a waiver of informed consent and a Health Insurance Portability and Accountability Act waiver of authorization, was obtained from the WIRB-Copernicus Group Institutional Review Board (IRB; protocol no. 20152817).
As previously described, DNA (> 50 ng) was extracted from formalin-fixed paraffin-embedded specimens, and NGS was performed by hybridization-captured, adaptor ligation–based libraries to high, uniform coverage (> 500×) for all coding exons of up to 324 cancer-related genes (FoundationOne or FoundationOne CDx) plus selected introns.18 The results were analyzed for base substitutions (subs), short insertions and deletions (indels), CN gains or losses, and rearrangements. ERBB2amp was defined as gene CN ≥ +3 of the tumor base ploidy (ie, CN = 5 in diploid, CN = 6 in triploid, CN = 7 in tetraploid, etc) and gene CN of ≥ 5 with 80% of baited targets amplified.19 For additional methods for tumor mutational burden (TMB) and genomic ancestry determination, see the Data Supplement.
Flatiron Health-Foundation Medicine CGDB
This study used the nationwide deidentified Flatiron Health-Foundation Medicine CGDB (FH-FMI CGDB). Retrospective longitudinal clinical data were derived from electronic health records (EHRs), comprising patient-level structured and unstructured data, curated via technology-enabled abstraction, and were linked to genomic data derived from FMI CGP tests by deidentified, deterministic matching.20,21 During the study period, the deidentified data originated from approximately 280 US cancer clinics (approximately 800 sites of care). Genomic alterations were identified via CGP as described above. To date, more than 400,000 samples have been sequenced from patients across the United States. The study included 2,270 patients who had a diagnosis of advGEA, received care within the FH network between January 2011 and September 2020, and underwent tissue CGP (FoundationOne or FoundationOne CDx). IRB approval with waiver of informed consent was obtained before study conduct from WIRB-Copernicus Group IRB.
Patients who were diagnosed with advanced GEA > 90 days before their first structured activity within the FH network or who received their FMI report > 60 days after their last FH structured activity date were excluded to ensure all therapies received before CGP were captured and to exclude patients who left the FH network before CGP. This left 270 unique patients eligible for this study. Clinical characteristics and treatment information were obtained via technology-enabled abstraction of clinical notes and radiology and pathology reports and linked to CGP data. Real-world progression (rwP) events were captured as episodes in which the treating clinician documented in the EHR that there had been growth or worsening of disease.22,23
The Fisher exact test or chi-squared test was used to assess significance of categorical relationships, and the Kruskal-Wallis test was used to assess significance of continuous variables. False discovery rate correction was performed by the Benjamin-Hochberg procedure to correct P values for multiple tests.
Outcome Assessment
The primary study outcome measurement was rwPFS, which was defined as the time from therapy of interest initiation to the first rwP date > 14 days after therapy initiation or to death, and patients were censored at their last clinic note date if no progression or death was observed. Real-world overall survival (rwOS), which was defined as the time from therapy of interest initiation to death, was also measured as secondary outcome. rwPFS and rwOS were compared across different ERBB2 CN subgroups using the Kaplan-Meier method and the log-rank test. Median rwPFS and rwOS values were estimated in months with 95% CIs. Multivariable Cox proportional hazard models were fitted on rwPFS or rwOS to estimate the adjusted hazard ratio and its significance level of ERBB2 CN. Log-transformed CN was used in Cox models. Other items included in the Cox model were age at advanced diagnosis, practice type (academic or community), sex, origin of disease site (esophageal, gastroesophageal junction [GEJ], or gastric), Eastern Cooperative Oncology Group (ECOG) performance status, and therapy line number for rwPFS analysis of chemotherapy only.
RESULTS
Foundation Medicine Genomic Database
Analysis of the Foundation Medicine genomic database of 12,905 GEA tumor samples (Data Supplement) identified ERBB2amp in 15.0% (n = 1,934) cases including 19.8% (1,384 of 6,975) of esophageal or GEJ adenocarcinomas and 9.3% (550 of 5,930) of gastric adenocarcinomas (Fig 1A). The clinocogenomic characteristics of 1,934 cases of ERBB2amp GEA compared with GEA without ERBB2 alterations (ERBB2wt) are shown in the Data Supplement. For ERBB2-amplified cases, median patient age was 63 years, 82.5% of patients were male, and for 71.6% of cases the primary tumor location was esophageal or GEJ. Both male sex and esophageal tumor location were further enriched relative to ERBB2wt GEA patients. The most frequent coaltered genes with ERBB2amp were TP53 (89.2%), CDKN2A% (27.3%), and CCNE1 (19.2%; Data Supplement). Coalteration frequencies were similar for ERBB2amp versus ERBB2wt GEA, although TP53 alterations were significantly more common in ERBB2amp cases (89.2% v 71.2%; P < .001), whereas KRAS alterations were significantly less common in ERBB2amp cases (9.2% v 21.9%; P < .001) and overall gene amplifications were more frequent in the ERBB2amp subset (Data Supplement). High microsatellite instability was rare in ERBB2amp GEA samples (0.3%) compared with ERBB2wt GEA (0.3% v 3.2%; P < .001), whereas TMB distribution was similar regardless of ERBB2 amplification status (median 4.3 v 3.8 mutations per megabase (Mb) Data Supplement).
FIG 1.
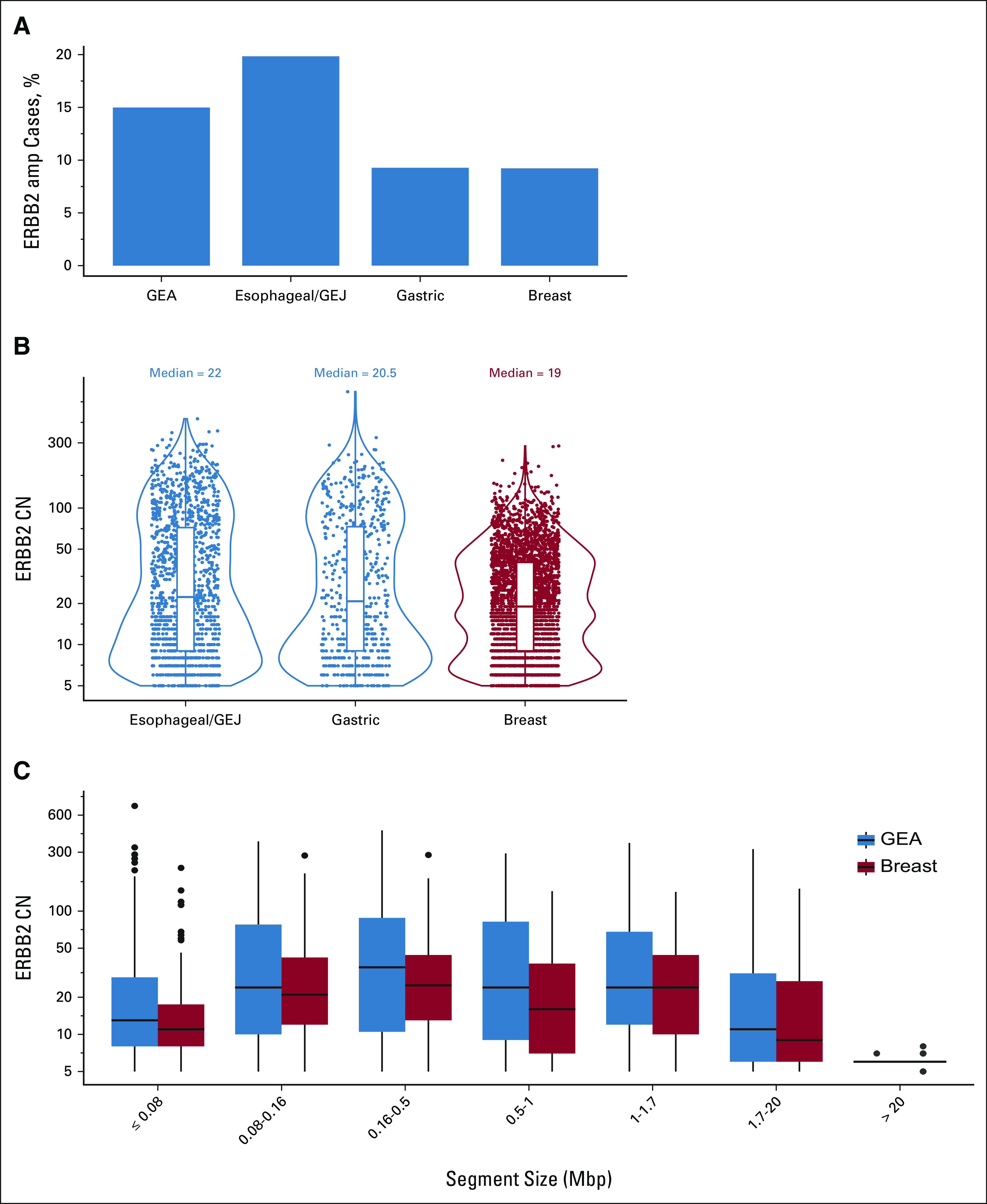
ERBB2 amplification frequencies, CN distribution, and amplicon size across GEA samples. (A) Frequency of ERBB2 amplification detected in all GEA tissue samples as well as in esophageal/GEJ and gastric subsets and breast cancer samples in the Foundation Medicine Genomic Database. (B) Similar ERBB2 CN distribution was observed in ERBB2-amplified esophageal/GEJ, gastric, and breast subsets. (C) ERBB2 CN distribution in ERBB2-amplified GEA and breast cancer samples bucketed by ERBB2 amplicon size. For amplicons > 0.16 Mbp, increased focality significantly correlated with higher ERBB2 CN (P < .001). CN, copy number; GEA, gastroesophageal adenocarcinoma; GEJ, gastroesophageal junction; Mbp, megabase pairs.
In ERBB2amp GEA samples, median ERBB2 CN was 22 (interquartile range [IQR] 9-73) and was similar across esophageal or GEJ (median CN = 22, IQR = 9-72) and gastric (median CN = 20.5, IQR = 9-73) subtypes (Fig 1B). We further assessed the size of the ERBB2 amplicon in ERBB2amp GEA cases. The median ERBB2 amplicon size was 0.27 Mb (IQR 0.13-0.95) for all GEA cases and was also similar for esophageal or GEJ (median 0.28 Mb, IQR 0.13-1.26) and gastric (median 0.26 Mb, IQR 0.13-0.85) subtypes. We also examined the correlation between ERBB2 CN and amplicon size in GEA samples and found that more focal amplification significantly correlated with higher CN (P < .001) for amplicons larger than 0.16 Mb (Fig 1C). In an orthogonal comparison, ERBB2 was amplified in 9.2% (3,193 of 34,629) of breast carcinoma cases analyzed, and CN (median 19, IQR 9-40), amplicon size (median 0.32 Mb, IQR 0.13-1.37), and correlation between CN and amplicon focality were similar (Fig 1). In both GEA and breast cases, for amplicons ≤ 0.08 Mb, partial ERBB2 gene amplification was more common and cases with < 100% of baited ERBB2 targets amplified were associated with lower ERBB2 CN gains (Data Supplement).
CGDB Outcomes Analysis
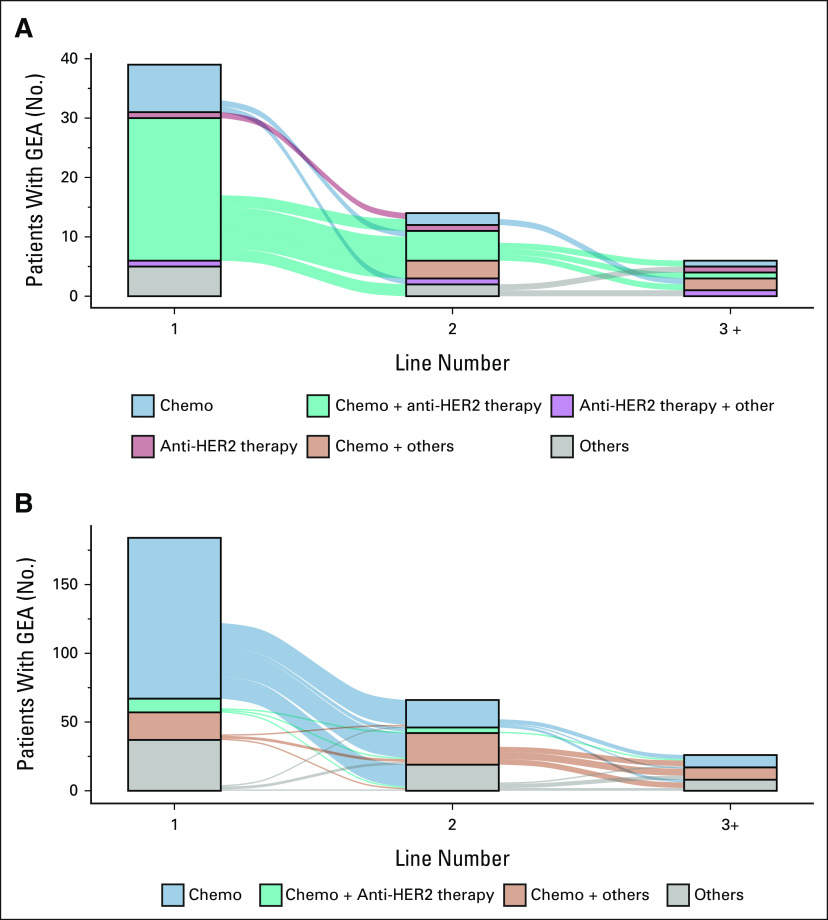
Of 2,270 patients with advGEA in the CGDB, ERBB2amp was detected in 15% (342 of 2,164) of cases with CGP performed on tissue biopsies (Data Supplement). Of 39 evaluable patients with ERBB2amp whose CGP report was obtained before first-line, 66.7% of patients received a first-line treatment regimen containing anti-HER2 therapy (most commonly chemotherapy plus trastuzumab [61.5%]; Fig 2A). No anti-HER2 therapy was included in the first-line treatment regimen in 25.6% of cases including 20.5% of patients who received chemotherapy alone. The remaining 7.7% of cases received an unspecified clinical study drug. In the subset of patients who did not receive anti-HER2 therapy in the first-line post-CGP report, the median ERBB2 CN was 7 (IQR 6-7.5) and a prior negative HER2 IHC or ISH test was recorded in 20% (2 of 10) of cases; of the remaining patients, four had a prior positive HER2 IHC and/or ISH test and four had no documentation of prior HER2 IHC or ISH testing. Furthermore, of the 10 patients with ERBB2amp who did not receive anti-HER2 therapy in the first-line post-CGP, only two had documentation of second-line therapy at the time of data cutoff and in both cases that second-line regimen included an anti-HER2 treatment. By contrast, for ERBB2wt patients, the most common first-line therapies post-CGP report were chemotherapy alone (63.6%) or other therapies (20.1%) including immunotherapy, non–HER2-targeted therapies, or unspecified clinical study drugs (Fig 2B). Of 10 patients with ERBB2wt tumors on CGP who received an anti-HER2 containing regimen in the first-line post-CGP, seven had prior HER2 and/or IHC test results and 7 of 7 results were positive.
FIG 2.
Treatment patterns for patients with GEA with ERBB2 amplification or ERBB2 wild-type tumors. Sankey diagrams showing treatment patterns for patients with GEA receiving first-line therapy post-CGP report. (A) Patients with GEA with ERBB2 amplification detected by CGP commonly received anti–HER2-containing regimens. (B) Patients with GEA without ERBB2 amplification or other ERBB2 alterations detected by CGP rarely received anti-HER2 therapy. Therapies grouped as others include immunotherapy, non–HER2-targeted therapies, and unknown clinical study drugs. CGP, comprehensive genomic profiling; chemo, chemotherapy; GEA, gastroesophageal adenocarcinoma; HER2, human epidermal growth factor receptor 2.
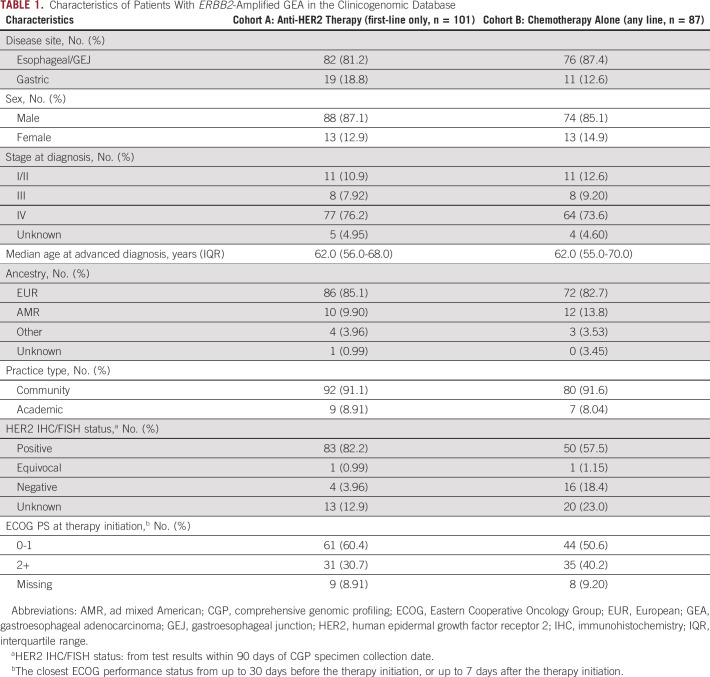
Among 270 patients with ERBB2amp advGEA meeting criteria for assessment, 101 received anti-HER2 therapy in the first-line setting regardless of CGP report timing (cohort A). A control set (cohort B) of 87 ERBB2amp patients treated with chemotherapy alone was also assessed; these patients received chemotherapy in any line because of prohibitively small numbers available for assessment when limited to the first-line setting. Clinical characteristics, genomic characteristics, and therapeutic regimens comparing cohorts A and B are shown in Table 1. Patients with ERBB2 amplified GEA by CGP who received first-line anti-HER2 therapy (cohort A) were significantly more likely to have had a documented positive HER2 IHC or FISH result than those who received chemotherapy alone in the first-line (82.2% v 57.9%; P < .001) or in any line (cohort B, 82.2% v 57.5%; P < .001).
TABLE 1.
Characteristics of Patients With ERBB2-Amplified GEA in the Clinicogenomic Database
In patients with ERBB2amp advGEA treated with first-line trastuzumab (cohort A), higher ERBB2 CN was a significant predictor as a continuous variable of longer rwPFS (hazard ratio [HR], 0.73; 95% CI, 0.60 to 0.89; P < .01). In contrast to the anti-HER2 cohort, higher ERBB2 CN was not significantly associated with longer rwPFS in patients receiving chemotherapy (cohort B, HR, 0.93; 95% CI, 0.73 to 1.20; P = .59). Age at advanced diagnosis, practice type, sex, and disease site did not show a statistically significant association with rwPFS in cohort A. Other patient characteristics assessed were also not significantly associated with rwPFS in the chemotherapy cohort (Data Supplement). ECOG performance status at the start of therapy was marginally lower for patients receiving HER2-targeted therapy versus chemotherapy (Table 1), and higher ECOG significantly correlated with worse outcome in trastuzumab-treated patients (Data Supplement). Also, as expected, increasing line number was significantly associated with an increased risk of progression or death in patients receiving chemotherapy.
No single ERBB2 CN cutoff was identified as an optimal predictor of rwPFS on trastuzumab-based therapy (Data Supplement). To visualize the relationship between ERBB2 CN and rwPFS, patients in cohort A were divided into five subgroups by quintiles of quantitative ERBB2 CN. For each increasing CN quartile, median rwPFS increased from 4.5 months for patients with CN 5-9 to 11.0 months for patients with CN > 107 (Fig 3). To assess whether longer rwPFS might have been the result of ERBB2 CN association with other factor(s), we analyzed the relationship between ERBB2 CN and multiple covariates in cohort A and saw no significant differences in ERBB2 CN for disease site, age and stage at advanced diagnosis, sex, ancestry, practice type, co-KRAS mutation, co-EGFR amplification, or co-FGFR1 or FGFR2 amplification (Data Supplement). We did see a suggestive association between PIK3CA mutation and lower ERBB2 CN (unadjusted P = .03). In patients with ERBB2amp advGEA treated with first-line trastuzumab, higher ERBB2 CN was a significant predictor as a continuous variable of longer rwOS (HR, 0.79; 95% CI, 0.64 to 0.97; P = .02), although the increasing trend for rwOS by each ERBB2 CN quintile was not as consistent as that for rwPFS (Data Supplement). Patients with CN 34 approximately 107 had longest median rwOS (20.4 months), whereas patients with CN > 107 had similar median rwOS to other subgroups. We did observe longer rwOS for patients with advGEA positive for HER2 by IHC or FISH treated with first-line trastuzumab with ERBB2 amplification detected by NGS versus those negative for amplification by NGS (Data Supplement), consistent with existing literature.15
FIG 3.
ERBB2 CN as a predictor of rwPFS in patients with advanced GEA treated with anti-HER2 therapy in the first-line setting. Association of ERBB2 CN with rwPFS in patients with GEA treated with first-line trastuzumab in the CGDB. ERBB2 CN was predictive as a continuous variable, and no single CN cutoff was identified as an optimal predictor of rwPFS. To visualize Kaplan-Meier rwPFS curves, the cohort was split into ERBB2 CN quintiles. Curves are truncated at 30 months as the number at risk was reduced to one in the entire ERBB2amp cohort. Median and IQR rwPFS for each CN quintile is shown. CGDB, clinicogenomic database; CN, copy number; GEA, gastroesophageal adenocarcinoma; HER2, human epidermal growth factor receptor 2; IQR, interquartile range; rwPFS, real-world progression-free survival.
DISCUSSION
In a large genomic data set of 12,905 GEA cases, ERBB2amp was detected in 15% of tissue samples tested using NGS-based CGP, with enrichment in esophageal or GEJ samples relative to gastric samples consistent with published studies.24 The ERBB2amp CN distribution and amplicon size were similar across esophageal or GEJ and gastric subsets, and more focal amplification generally correlated with higher copy gains. We compared a cohort of ERBB2amp breast carcinoma samples and observed a similar distribution of ERBB2 CN gain and amplicon size, as well as correlation between focality and CN. However, more comprehensive pan-tumor analysis is needed to further elucidate the genomics of ERBB2 amplification across disease subtypes.
In a real-world CGDB, we observed that most patients with ERBB2amp detected by CGP received trastuzumab in combination with chemotherapy postreceipt of their genomic test results, whereas those without ERBB2amp detected received chemotherapy alone, consistent with current guidelines. However, a smaller subset of ERBB2amp cases received first-line treatment regimens absent of anti-HER2 therapy, which was likely because of relatively low-level ERBB2amp (median CN 7), and in two cases, a prior negative HER2 IHC and/or ISH result. Furthermore, for two of these patients with documented second-line therapy at the time of data cutoff, both received anti-HER2 therapy at that time. We also observed that a small subset of patients without ERBB2amp detected on CGP received chemotherapy in combination with anti-HER2 therapy following CGP report, and in the majority of these cases, the patient had a prior positive HER2 IHC and/or ISH result, despite the generally high reported concordance between IHC and ISH testing for HER2 expression and the CGP methodology used for ERBB2 amplification detection in this study (88%-98%).14,15
Historically, IHC testing for HER2 overexpression has been considered the gold standard and remains so according to published guidelines. However, many testing modalities including NGS for ERBB2amp are now clinically available as validated US Food and Drug Administration–approved assays and studies correlating testing results with clinical outcomes to anti-HER2 therapies are needed. Published data in GEA as well as in colorectal cancer show that borderline positivity for HER2 expression and lower associated levels of ERBB2 amplification are associated with reduced benefit from anti-HER2 therapies17,25,26 Recent data from our group suggest that patients with GEA with positive IHC and/or ISH results for HER2 expression but negative results for ERBB2amp by NGS have worse outcomes on anti-HER2 therapy compared with patients testing positive by both or multiple methodologies.15 Further exploratory analysis using median ERBB2 CN as a cutoff suggested that ERBB2 CN can be used effectively to further stratify best responders to anti-HER2 therapy, suggesting additional utility for quantitative NGS testing.15,16
In the current study, we expanded on these analyses and showed that ERBB2 CN is predictive as a continuous variable for efficacy outcome to anti–HER2-targeted therapy in advGEA. Although no single CN cutoff appeared to be clinically optimal, increased rwPFS was observed for each increase in ERBB2 CN quintile, with a median rwPFS of 5.4 months for patients with ERBB2 CN of 5-9 compared with 11.0 months for patients with ERBB2 CN > 107. We further assessed whether covariates associated with higher ERBB2 CN could actually be driving anti-HER2 therapy outcome; however, none of the covariates assessed were significantly associated with ERBB2 CN in our cohort of patients treated with first-line trastuzumab with the exception of PIK3CA mutations, which were suggestively associated with lower ERBB2 CN (P = .03) and could contribute to anti-HER2 resistance. In our genomic analysis, we also found that higher ERBB2 CN was associated with more focal gene amplification; however, ERBB2 amplicon size itself was not an independent predictor of outcome to anti-HER2 therapy (HR, 1.12; 95% CI, 0.94 to 1.35; P = .21).
Additional RTKs have been shown to be oncogenic drivers, and therapeutic strategies have been implemented targeting overexpression or amplification of EGFR, MET, and FGFR2.27 Historically, IHC and/or ISH methodologies, similar to those approved for HER2, have been used in trials investigating EGFR and MET expression as biomarkers with limited success.28 However, more recently, promising results for MET amplification as a biomarker in NSCLC have been reported, and within the METamp cohort, increased MET CN was further predictive of improved outcome to MET-targeted therapy.29 Promising studies assessing EGFR amplification as a predictive biomarker in GEA have also been presented.30 Finally, innovative trial designs have been implemented in which personalized antibody therapy was selected and prioritized on the basis of presence and degree of gene amplification.31 These data suggest that the observations made herein for ERBB2 in GEA may potentially be applicable to other RTKs and disease subtypes.
Limitations of this study include absence of clinical information and treatment history for most patients in the FMI genomic database. For the CGDB cohort, clinical data were derived from EHR and data not documented in the EHR may be incomplete or missing, particularly for events occurring outside of the FH network. rwPFS was defined as time from therapy start to progression or death, where rwP events are abstracted from EHR and are limited by clinician interpretation and documentation. Selection bias is likely also present due to all patients in this study having received CGP. Additional retrospective and prospective analyses to assess quantitative ERBB2 CN as a more sensitive predictive biomarker in GEA, and potentially in other tumor types, are needed.
In conclusion, in patients with advGEA treated with first-line trastuzumab, quantitative ERBB2 CN was a significant predictor of efficacy outcome to anti-HER2 therapy, where higher CN was associated with longer rwPFS. These data along with results of prior studies suggest that although traditional IHC and ISH methodologies for detection of HER2 expression are generally concordant, NGS-based methodologies to detect and quantify ERBB2 copy gains may allow for more robust stratification of HER2-positive patients and better prediction of efficacy benefit from approved and investigational HER2-targeted therapies. Ultimately, in caring for patients, we seek to gather maximal data to inform prognostic and predictive clinical abilities and feel our data suggest that quantitative ERBB2 CN information may help extend these goals in ERBB2amp GEA.
Liangliang Zhang
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Omar Hamdani
Employment: Foundation Medicine
Ole Gjoerup
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Cheryl Cho-Phan
Employment: Syapse, Roche (I), Flatiron Health, Gilead Sciences (I)
Leadership: Gilead Sciences (I)
Stock and Other Ownership Interests: Roche (I), Gilead Sciences (I)
Jeremy Snider
Employment: Flatiron Health
Stock and Other Ownership Interests: Flatiron Health
Emily Castellanos
Employment: Flatiron Health
Stock and Other Ownership Interests: Flatiron Health, Roche
Halla Nimeiri
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Garrett Frampton
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Jeffrey M. Venstrom
Employment: Foundation Medicine, Genentech/Roche
Leadership: Foundation Medicine
Stock and Other Ownership Interests: Roche
Consulting or Advisory Role: Synkrino Biotherapeutics
Travel, Accommodations, Expenses: Foundation Medicine, Genentech/Roche
Geoffrey Oxnard
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Samuel J. Klempner
Stock and Other Ownership Interests: TP Therapeutics
Honoraria: Natera
Consulting or Advisory Role: Lilly, Astellas Pharma, Bristol Myers Squibb, Pieris Pharmaceuticals, Merck, Daiichi Sankyo/UCB Japan, Sanofi/Aventis
Research Funding: Leap Therapeutics (Inst), BeiGene (Inst)
Other Relationship: NCCN
Alexa B. Schrock
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine, Roche
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented in part as a poster at the ASCO 2021 Annual Meeting.
SUPPORT
Supported by Foundation Medicine, Inc.
DATA SHARING STATEMENT
All data relevant to the study are included in the article or uploaded in the Data Supplement.
AUTHOR CONTRIBUTIONS
Conception and design: Liangliang Zhang, Omar Hamdani, Ole Gjoerup, Cheryl Cho-Phan, Jeremy Snider, Garrett Frampton, Jeffrey M. Venstrom, Alexa B. Schrock
Collection and assembly of data: Jeremy Snider, Emily Castellanos, Garrett Frampton
Data analysis and interpretation: Liangliang Zhang, Ole Gjoerup, Cheryl Cho-Phan, Jeremy Snider, Emily Castellanos, Halla Nimeiri, Garrett Frampton, Jeffrey M. Venstrom, Geoffrey Oxnard, Samuel J. Klempner, Alexa B. Schrock
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Liangliang Zhang
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Omar Hamdani
Employment: Foundation Medicine
Ole Gjoerup
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Cheryl Cho-Phan
Employment: Syapse, Roche (I), Flatiron Health, Gilead Sciences (I)
Leadership: Gilead Sciences (I)
Stock and Other Ownership Interests: Roche (I), Gilead Sciences (I)
Jeremy Snider
Employment: Flatiron Health
Stock and Other Ownership Interests: Flatiron Health
Emily Castellanos
Employment: Flatiron Health
Stock and Other Ownership Interests: Flatiron Health, Roche
Halla Nimeiri
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Garrett Frampton
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Jeffrey M. Venstrom
Employment: Foundation Medicine, Genentech/Roche
Leadership: Foundation Medicine
Stock and Other Ownership Interests: Roche
Consulting or Advisory Role: Synkrino Biotherapeutics
Travel, Accommodations, Expenses: Foundation Medicine, Genentech/Roche
Geoffrey Oxnard
Employment: Foundation Medicine
Stock and Other Ownership Interests: Roche
Samuel J. Klempner
Stock and Other Ownership Interests: TP Therapeutics
Honoraria: Natera
Consulting or Advisory Role: Lilly, Astellas Pharma, Bristol Myers Squibb, Pieris Pharmaceuticals, Merck, Daiichi Sankyo/UCB Japan, Sanofi/Aventis
Research Funding: Leap Therapeutics (Inst), BeiGene (Inst)
Other Relationship: NCCN
Alexa B. Schrock
Employment: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine, Roche
No other potential conflicts of interest were reported.
REFERENCES
- 1. Indini A, Rijavec E, Grossi F. Trastuzumab deruxtecan: Changing the destiny of HER2 expressing solid tumors. Int J Mol Sci. 2021;22:4774. doi: 10.3390/ijms22094774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meric-Bernstam F, Johnson AM, Dumbrava EEI, et al. Advances in HER2-targeted therapy: Novel agents and opportunities beyond breast and gastric cancer Clin Cancer Res 252033–20412019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moasser MM.The oncogene HER2: Its signaling and transforming functions and its role in human cancer pathogenesis Oncogene 266469–64872007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shitara K, Baba E, Fujitani K, et al. Discovery and development of trastuzumab deruxtecan and safety management for patients with HER2-positive gastric cancer Gastric Cancer 24780–7892021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pegram MD, Lipton A, Hayes DF, et al. Phase II study of receptor-enhanced chemosensitivity using recombinant humanized anti-p185HER2/neu monoclonal antibody plus cisplatin in patients with HER2/neu-overexpressing metastatic breast cancer refractory to chemotherapy treatment J Clin Oncol 162659–26711998 [DOI] [PubMed] [Google Scholar]
- 6.Robert N, Leyland-Jones B, Asmar L, et al. Randomized phase III study of trastuzumab, paclitaxel, and carboplatin compared with trastuzumab and paclitaxel in women with HER-2-overexpressing metastatic breast cancer J Clin Oncol 242786–27922006 [DOI] [PubMed] [Google Scholar]
- 7.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2 N Engl J Med 344783–7922001 [DOI] [PubMed] [Google Scholar]
- 8.Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial Lancet 376687–6972010 [DOI] [PubMed] [Google Scholar]
- 9.Dumbrava EEI, Balaji K, Raghav K, et al. Targeting ERBB2 (HER2) amplification identified by next-generation sequencing in patients with advanced or metastatic solid tumors beyond conventional indications JCO Precis Oncol 31–122019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Javle M, Borad MJ, Azad NS, et al. Pertuzumab and trastuzumab for HER2-positive, metastatic biliary tract cancer (MyPathway): A multicentre, open-label, phase 2a, multiple basket study Lancet Oncol 221290–13002021 [DOI] [PubMed] [Google Scholar]
- 11.Nowak JA.HER2 in colorectal carcinoma: Are we there yet? Surg Pathol Clin 13485–5022020 [DOI] [PubMed] [Google Scholar]
- 12.Catenacci DVT, Liao WL, Zhao L, et al. Mass-spectrometry-based quantitation of Her2 in gastroesophageal tumor tissue: Comparison to IHC and FISH Gastric Cancer 191066–10792016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janjigian YY, Sanchez-Vega F, Jonsson P, et al. Genetic predictors of response to systemic therapy in esophagogastric cancer Cancer Discov 849–582018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross DS, Zehir A, Cheng DT, et al. Next-generation assessment of human epidermal growth factor receptor 2 (ERBB2) amplification status: Clinical validation in the context of a hybrid capture-based, comprehensive solid tumor genomic profiling assay J Mol Diagn 19244–2542017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stein SM, Snider J, Ali SM, et al. Real-world association of HER2/ERBB2 concordance with trastuzumab clinical benefit in advanced esophagogastric cancer Future Oncol 174101–41142021 [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Shi M, Li X, et al. HER2 copy number as predictor of disease-free survival in HER2-positive resectable gastric adenocarcinoma J Cancer Res Clin Oncol 1471315–13242021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez-Martin C, Plaza JC, Pazo-Cid R, et al. Level of HER2 gene amplification predicts response and overall survival in HER2-positive advanced gastric cancer treated with trastuzumab J Clin Oncol 314445–44522013 [DOI] [PubMed] [Google Scholar]
- 18.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing Nat Biotechnol 311023–10312013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.US Food and Drug Administration PMA P170019/S006: FDA summary of safety and effectiveness data. www.accessdata.fda.gov/cdrh_docs/pdf17/P170019S006B.pdf
- 20.Singal G, Miller PG, Agarwala V, et al. Association of patient characteristics and tumor genomics with clinical outcomes among patients with non-small cell lung cancer using a clinicogenomic database JAMA 3211391–13992019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Birnbaum B, Nussbaum N, Seidl-Rathkopf K, et al. Model-assisted cohort selection with bias analysis for generating large-scale cohorts from the EHR for oncology research. arXiv. 2020 2001.09765. [Google Scholar]
- 22.Griffith SD, Miksad RA, Calkins G, et al. Characterizing the feasibility and performance of real-world tumor progression end points and their association with overall survival in a large advanced non-small-cell lung cancer data set JCO Clin Cancer Inform 31–132019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffith SD, Tucker M, Bowser B, et al. Generating real-world tumor burden endpoints from electronic health record data: Comparison of RECIST, radiology-anchored, and clinician-anchored approaches for abstracting real-world progression in non-small cell lung cancer Adv Ther 362122–21362019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Cutsem E, Bang YJ, Feng-Yi F, et al. HER2 screening data from ToGA: Targeting HER2 in gastric and gastroesophageal junction cancer Gastric Cancer 18476–4842015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haffner I, Schierle K, Raimundez E, et al. HER2 expression, test deviations, and their impact on survival in metastatic gastric cancer: Results from the prospective multicenter VARIANZ study J Clin Oncol 391468–14782021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sartore-Bianchi A, Trusolino L, Martino C, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): A proof-of-concept, multicentre, open-label, phase 2 trial Lancet Oncol 17738–7462016 [DOI] [PubMed] [Google Scholar]
- 27. Wainberg ZA, Enzinger PC, Kang Y-K, et al. Randomized double-blind placebo-controlled phase 2 study of bemarituzumab combined with modified FOLFOX6 (mFOLFOX6) in first-line (1L) treatment of advanced gastric/gastroesophageal junction adenocarcinoma (FIGHT) J Clin Oncol. 2021;39 suppl 3; abstr 160. [Google Scholar]
- 28.Drilon A, Cappuzzo F, Ou SI, et al. Targeting MET in lung cancer: Will expectations finally be MET? J Thorac Oncol 1215–262017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolf J, Seto T, Han JY, et al. Capmatinib in MET exon 14-mutated or MET-amplified non-small-cell lung cancer N Engl J Med 383944–9572020 [DOI] [PubMed] [Google Scholar]
- 30.Maron SB, Alpert L, Kwak HA, et al. Targeted therapies for targeted populations: Anti-EGFR treatment for EGFR-amplified gastroesophageal adenocarcinoma Cancer Discov 8696–7132018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Catenacci DVT, Moya S, Lomnicki S, et al. Personalized antibodies for gastroesophageal adenocarcinoma (PANGEA): A phase II study evaluating an individualized treatment strategy for metastatic disease Cancer Discov 11308–3252021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data relevant to the study are included in the article or uploaded in the Data Supplement.