Abstract
Background and Objectives
The 2-hit model of genetic disease is well established in cancer, yet has only recently been reported to cause brain malformations associated with epilepsy. Pathogenic germline and somatic variants in genes in the mechanistic target of rapamycin (mTOR) pathway have been implicated in several malformations of cortical development. We investigated the 2-hit model by performing genetic analysis and searching for germline and somatic variants in genes in the mTOR and related pathways.
Methods
We searched for germline and somatic pathogenic variants in 2 brothers with drug-resistant focal epilepsy and surgically resected focal cortical dysplasia (FCD) type IIA. Exome sequencing was performed on blood- and brain-derived DNA to identify pathogenic variants, which were validated by droplet digital PCR. In vitro functional assays of a somatic variant were performed.
Results
Exome analysis revealed a novel, maternally inherited, germline pathogenic truncation variant (c.48delG; p.Ser17Alafs*70) in NPRL3 in both brothers. NPRL3 is a known FCD gene that encodes a negative regulator of the mTOR pathway. Somatic variant calling in brain-derived DNA from both brothers revealed a low allele fraction somatic variant (c.338C>T; p.Ala113Val) in the WNT2 gene in 1 brother, confirmed by droplet digital PCR. In vitro functional studies suggested a loss of WNT2 function as a consequence of this variant. A second somatic variant has not yet been found in the other brother.
Discussion
We identify a pathogenic germline mTOR pathway variant (NPRL3) and a somatic variant (WNT2) in the intersecting WNT signaling pathway, potentially implicating the WNT2 gene in FCD and supporting a dual-pathway 2-hit model. If confirmed in other cases, this would extend the 2-hit model to pathogenic variants in different genes in critical, intersecting pathways in a malformation of cortical development. Detection of low allele fraction somatic second hits is challenging but promises to unravel the molecular architecture of FCDs.
Focal cortical dysplasia (FCD) is a common developmental brain malformation associated with intractable epilepsy, with seizure freedom often only achievable through surgical resection.1,2 FCD is one of many malformations of cortical development, such as cortical tubers in tuberous sclerosis complex and hemimegalencephaly, associated with germline and somatic pathogenic variants of genes in the mechanistic (formerly mammalian) target of rapamycin (mTOR) pathway.1-18 Observations of germline variants with reduced penetrance suggest a 2-hit genetic model, similar to neoplasms, where inherited germline variants confer risk and interact with a second, somatic variant to produce a malformation.19 This 2-hit genetic model, where the second hit is a somatic variant present only in the brain malformation, is distinct from the 2-hit model of double germline variants observed in children with severe developmental delay.20,21
The germline and somatic 2-hit model for malformations of cortical development has been supported by recent observations showing 2 hits in the same gene for TSC2 in 2 patients with hemimegalencephaly12 and DEPDC5 in 6 patients with FCD.6,7,13-16 In all 6 patients, a germline variant was detected in blood-derived DNA, and a second-hit low allele fraction somatic variant in the same gene was found only in brain-derived DNA. Very recently, variants in different genes were reported in 2 cases: 1 had tuberous sclerosis complex with germline TSC2 and somatic DEPDC5 variants,15 and the other had hemimegalencephaly with somatic variants in MTOR and RPS6.18 Given somatic variants are often present at very low allele fraction (e.g., in <5% DNA molecules), they can be challenging to detect without high-depth, targeted sequencing, meaning such variants have been rarely detected.22
Here, we aimed to confirm the 2-hit model in a common form of FCD. In 2 brothers with focal epilepsy and FCD type IIA, we searched for germline and somatic variants, focusing on mTOR and related pathway genes. Identified variants were validated and segregated in the brothers and their parents, and functional studies were performed to assess the impact on protein function.
Methods
Standard Protocol Approvals, Registrations, and Patient Consents
The Human Research Ethics Committee of Austin Health, Melbourne, Australia, approved this study (Project No. H2007/02961). Written informed consent was obtained from the parents for themselves and their children.
Sample Collection
Whole blood was obtained from both brothers and their parents (Individuals I.1, I.2, II.2, and II.3; Figure 1A) and surgically resected fresh-frozen brain tissue from the 2 brothers with FCD (II.2 and II.3). Genomic DNA was extracted23 using the Qiagen QIAamp DNA Maxi Kit (Hilden, Germany) for blood and the Qiagen AllPrep DNA/RNA Kit for the brain.
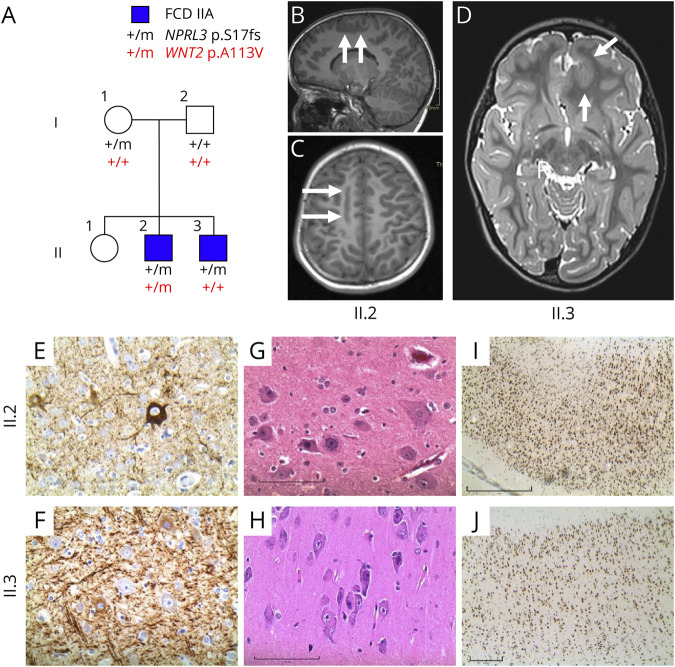
Figure 1. Genetic Analysis, Imaging and Histopathology of Family With FCDIIA.
(A) Pedigree of family and segregation of NPRL3 and WNT2 pathogenic variants. The NPRL3 mutant allele was detected in both blood and brain samples for brothers II.2 and II.3. No DNA was available from their sister (II.1). (B–D) Preoperative MRI scans (right is on the reader’s left). (B and C) MRI from II.2 showing an abnormal longitudinally oriented region of gray matter with blurred gray-white boundaries in the right posteromesial frontal region (arrows). (D) MRI from II.3 shows a globular region of abnormal gray matter in the left anteromesial frontal region (arrows). (E–J) Histopathology images showing disorganized gray matter and dysmorphic neurons for II.2 (panels E, G, and I) and II.3 (panels F, H, and J). (E and F) Phosporylated neurofilament staining of dysmorphic neurons. (G and H) Hemotoxylin and eosin staining showing dysmorphic neurons. (I and J) NeuN staining showing dyslamination. Magnification of 400× (E-H) or 40× (I and J); scale bars represent 20 µm (F), 100 µm (G and H), 1 mm (I), and 0.5 mm (J). FCDIIA = focal cortical dysplasia type IIA.
Exome Sequencing
Exome sequencing was performed using 3 μg genomic DNA sonicated to ∼200 base pair (bp) fragments and adaptor ligated to make a library for 100 bp paired-end sequencing. Following amplification and barcoding, the libraries were hybridized to biotinylated complementary RNA oligonucleotide baits from the Agilent SureSelect DNA Human All Exon V5+UTRs kit (Agilent Technologies, Santa Clara, CA) and purified using streptavidin-bound magnetic beads. Amplification was conducted before sequencing at 50-fold (blood) or 100-fold (brain) depth on the Illumina HiSeq 2500 system (San Diego, CA).
Exome Sequencing Analysis
Exome sequencing reads were aligned with Novoalign version 3.02.13 (novocraft.com/; Novocraft Technologies Sdn Bhd, Selangor, Malaysia) to the human genome assembly with ambiguous single nucleotide variations (hg19 dbSNP135-masked, UCSC Genome Browser). PCR duplicates were removed using MarkDuplicates from Picard version 2.1.1 (broadinstitute.github.io/picard/; Broad Institute, Cambridge, MA), and local realignment was performed using GATK version 3.5-0.24
Germline variant detection was performed with GATK HaplotypeCaller version 3.5-024 and variant annotation using vcfanno25 and ANNOVAR.26 Exome variants were filtered according to the following criteria: located in the splicing or coding region of a gene, appear in the gnomAD database27 less than 10 times, and the variant type is missense, nonsense, coding indel, or canonical splice site variant. Several different genetic inheritance models were considered: de novo variants shared by both affected brothers (assuming gonadal mosaicism in 1 parent); homozygous recessive, compound heterozygous recessive, X-linked recessive inheritance or autosomal dominant inheritance from an unaffected parent.
Somatic variant detection was performed using MuTect2, from GATK version 3.7-0,24 and VarScan version 2.4.3,28 taking blood and brain samples as normal and tumor, respectively. MuTect2 was run using dbSNP version 138 (excluding sites after 129) and Catalogue of Somatic Mutations in Cancer (COSMIC) v81 VCF files provided to adjust the level of evidence required to call somatic variants. Only variants that had a PASS value in the FILTER column were kept. VarScan was run in somatic mode using tumor purity of 10%, and variants of type somatic were retained. Consensus somatic variants called by both methods were retained, annotated, and filtered as described above for germline variants, with an additional requirement that variants occur in 1,384 selected genes previously associated with epilepsy or FCD or part of the mTOR and related pathways (see Supplemental Material for gene list, links.lww.com/NXG/A510).
Copy Number Variant Analysis
Copy number variant (CNV) analysis of brain-derived DNA was performed using 2 methods: chromosomal microarrays with Illumina Infinium GSA-24 v2.0 and exome sequencing data for germline and somatic CNVs using XHMM29 following a published protocol30 and cn.MOPS31 using segments defined based on the exome capture regions. Both exome analysis methods were normalized against 44 DNA samples sequenced using the same platform and exome capture kit. Identified CNVs were filtered for those which overlap exonic regions of the list of epilepsy, FCD, and mTOR and related pathway genes (Supplemental Material, links.lww.com/NXG/A510) and excluded if they had at least 80% overlap with a CNV present at least 10 times in the Structural Variant gnomAD database.27
Deeper Resequencing
A second round of deeper (200-fold target depth) exome sequencing was performed on the same brain specimen of II.3 with a separate extraction. Exome sequencing and analysis were performed as above with the following differences. Library preparation used the Illumina TruSeq Exome kit, and sequencing was performed on the Illumina HiSeq 2500/4000 platform (BGI, Hong Kong). Reads were aligned with BWA-MEM v0.7.17-r118832; then, duplicate marking and base quality score recalibration were performed using GATK v4.0.3.0.24 Somatic variants were called using MuTect2 in tumor-only mode using a panel of controls generated from 41 blood-derived DNA samples captured using the same exome kit, focusing on the list of genes associated with epilepsy, FCD, or part of the mTOR and related pathways (Supplemental Material, links.lww.com/NXG/A510).
Additional deep exome sequencing (1,000-fold target depth) was performed on newly extracted DNA from the fresh-frozen brain specimens of II.2 and II.3. Exome sequencing and analysis were performed as described above with the following differences. Library preparation used the MGIEasy Exome Capture V4, and sequencing was performed on the BGISEQ platform (BGI, Hong Kong). Reads were aligned with BWA-MEM v0.7.17-r1188,32 and duplicate marking and base quality score recalibration were performed using GATK v4.0.3.0.24 Analysis focused on searching for a second hit in NPRL3 that may have been missed on previous sequencing runs at lower depth. Somatic variants in NPRL3 were called using MuTect2 in tumor-only mode and manually inspected to filter technical artifacts. Variants that were not observed in more than 2 unique reads or were present in both brothers were removed.
Validation and Mosaicism Detection
Validation of the NPRL3 c.48delG (p.Ser17Alafs*70) exome variant was performed by PCR and Sanger sequencing. To validate low-level mosaicism, we used droplet digital PCR (ddPCR).33,34 Custom probes (WT: VIC-TTTATGCCATCTCCTCA-MGB-NFQ and A113V: FAM-TTGTTTATGTCATCTCCTCA-MGB-NFQ) and primers (FWD-TGTTTCAGGTAGTCGGGAATCTG and REV-GCCCTGGTGATGGCAAATAC) were designed to detect the WNT2 c.338C>T (p.Ala113Val) variant and wild-type allele. Droplet generation, PCR cycling, and droplet reading were performed according to the manufacturer's recommendations (Bio-Rad, Hercules, CA).35
Constructs
Active WNT2-V5 was a gift from Xi He (Addgene plasmid #43809; n2t.net/addgene:43809; RRID:Addgene_43809).36 Overlap PCR-based mutagenesis was used to produce a 987 bp product with the c.338C>T: p.Ala113Val variant (primer sequences in eTable 1, Supplemental Material, links.lww.com/NXG/A510). The mutant PCR product was digested with NotI and BstXI and cloned by ligation into the active WNT2-V5 construct cut with the same restriction enzymes. Firefly luciferase vectors, M50 Super 8x TOPFlash (Addgene plasmid # 12456; n2t.net/addgene:12456; RRID:Addgene_12456) and M51 Super 8x FOPFlash (Addgene plasmid # 12457; n2t.net/addgene:12457; RRID:Addgene_12457), were gifts from Randall Moon.37 Renilla luciferase vector, pRL-TK plasmid, was obtained from Promega (Cat #E2241).
Luciferase Reporter Assay
HEK293T cells were cultured in DMEM (Gibco, Cat #12569-010), supplemented with 10% fetal bovine serum for 24 hours before transfection. Active WNT2-V5 constructs (100 ng; wild type, mutant, or both equally combined) were cotransfected with TOPFlash or FOPFlash plasmid and pRL-TK plasmid using lipofectamine 2000 (Invitrogen, Cat #11668019). At 40 hours posttransfection, cells were harvested in passive lysis buffer. Firefly and Renilla luciferase activities were detected with dual-luciferase reporter assay system (Promega, Cat #E1910). Assay was performed in triplicate with 3 technical replicates of each condition per experiment. Firefly luciferase activity was normalized to Renilla control reporter activity to obtain relative light units (RLU) per sample. Relative luciferase activity was determined by comparing RLU of each sample to the empty vector lacking WNT2 cDNA sequence. The differences in relative luciferase activity between each construct were assessed using the Student t test.
Western Blot
Expression of WNT2 wild-type and mutant proteins was confirmed with Western blotting. Lysate from dual-luciferase reporter assay was additionally lysed in buffer containing 50 mM Tris-HCl (pH 7.4), 50 mM NaCl, 1% Triton X-100, 1 mM EDTA, 20 mM NaF, 2 mM Na3VO4, and protease inhibitor mixture (Roche, Cat#11873580001). Fifteen micrograms of extracted proteins was separated on a 7% polyacrylamide gel and then transferred to nitrocellulose membrane by electroblotting. Blots were simultaneously incubated with anti-V5 tag antibody at 1:2,000 (Thermo Fisher Scientific, Cat #R960-25) and anti–β-actin antibody AC-74 at 1:2,000 (Sigma, Cat #A2228), followed by incubation with goat anti-mouse immunoglobulin G antibody conjugated to horseradish peroxidase (Dako Cat #P0447). Exogenous WNT2 proteins and endogenous β-actin protein were detected by enhanced chemiluminescence (Bio-Rad Cat #1705061) and visualized with the ChemiDoc detection system (Bio-Rad).
Data Availability
Variants identified in this study have been submitted to ClinVar. Sequencing data are available from the corresponding authors on reasonable request.
Results
Patient Histories
Individual II.2 (Figure 1A) is a 10-year-old boy who had normal early developmental milestones. At age 2 years, he developed daily clusters of focal impaired awareness seizures characterized by staring, eyelid flickering, head deviation, and leaning to the right, sometimes associated with a fall. He was seizure free on carbamazepine until age 5 years, when treatment was weaned and seizures subsequently became refractory. He described an aura of tingling and numbness in one or both upper limbs, spreading to his forehead, with oral tingling and dysarthria. By 6 years, his aura comprised pins and needles in the left lower limb, spreading to the left side of the body. He developed sudden falls requiring a helmet. Neuropsychological evaluation showed low average to average intellect.
Ictal video-EEG captured focal seizures with bilateral tonic posturing of the upper limbs, maximal on the left, with left head and eye deviation, sometimes associated with loss of head and truncal posture to the right. The EEG showed repetitive spike-and-wave activity in the right frontocentral region, followed by attenuation and rapid spread to bilateral frontocentral regions with low-amplitude fast activity evolving to rhythmic theta activity. Neuroimaging showed abnormal gray matter in the right posteromesial frontal region, without increased T2 signal (Figures 1B and C).
Focal resection at 6 years revealed FCD type IIA with dysmorphic neurons evident at 400× (Figures 1E and G) and disorganized architecture (Figure 1I) at low power. Postoperatively, he had predominantly nocturnal focal aware emotional seizures, with fear, on 4–5 nights per week, with rare diurnal seizures. He aroused from sleep, with laughter or left upper and lower limb stiffening and head deviation. Residual dysplastic tissue was identified close to the motor cortex on MRI. By 10 years, he had focal aware seizures with giggling and paresthesia, with fine shaking or tremulousness during seizures. Behavioral concerns included inattention and distractibility. He received education support in a mainstream school.
His 6-year-old brother, II.3, had normal early development. At 6 weeks, he presented with daily series of epileptic spasms, diagnosed at 12 weeks, and controlled with phenobarbital. From 13 months, he had tonic and atonic seizures. Seizures began with a loud scream, stiffening of the whole body, associated with a fall, teeth clenching, and manual automatisms. Seizures sometimes evolved to subtle jerking of all limbs. His EEG showed a left frontal focus, consistent with a left anteromesial frontal cortical abnormality without high T2 signal on MRI (Figure 1D). Resection at age 2 years revealed a similar FCD type IIA with dysmorphic neurons (Figures 1F and H) and dyslamination (Figure 1J). He was seizure free postoperatively, and medication was withdrawn at 3 years. He is in kindergarten with age-appropriate development. Their parents (I.1 and I.2) are well without seizures; their mother had a normal MRI.
Maternally Inherited NPRL3 Germline Variant
Exome sequencing identified a novel, maternally inherited, frameshift variant in NPRL3 NM_001077350.3:c.48delG:p.Ser17Alafs*70, validated by Sanger sequencing (Figure 2A). NPRL3, encoding the nitrogen permease regulator–like 3 protein, forms a complex with NPRL2 and DEPDC5 to form the GATOR1 complex, a key negative regulator of the mTOR pathway.38
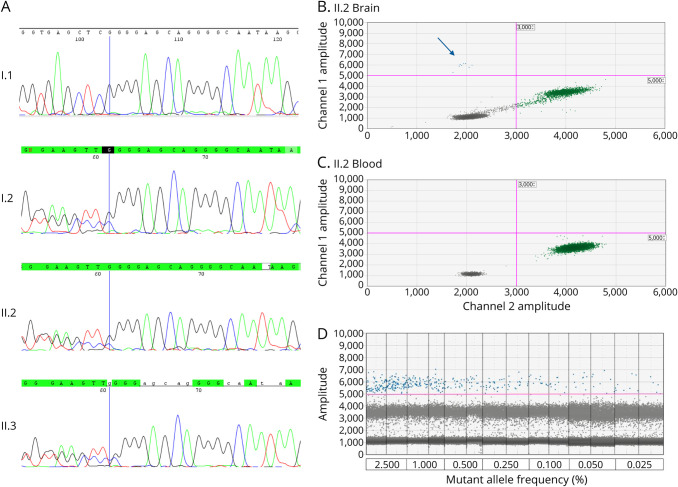
Figure 2. Validation of NPRL3 and WNT2 Variants.
(A) Sanger sequence chromatograms showing the germline NPRL3 c.48delG (p.Ser17Alafs*70) variant in individuals I.2, II.2, and II.3. (B) ddPCR readout showing WNT2 mutant template (c.338C>T, p.Ala113Val) in blue (arrow), wild-type template in green, and droplets without DNA template in gray. Individual II.2's brain sample contains low-level somatic variant with variant allele fraction of 0.3%. (C) ddPCR shows that individual II.2's blood sample contains only wild-type template (green). (D) The limit of detection of the ddPCR assay was established by serially diluting mutant gBlocks into wild-type gBlocks to obtain different mutant/(mutant + wild-type) ratios: 2.5%, 1%, 0.5%, 0.25%, 0.1%, 0.05%, and 0.025%. An amplitude of 5,000 was set as the positive mutant droplet threshold. Mutant allele at a frequency ≥0.025% could be consistently detected with at least 3 droplets.
Somatic Variant in the WNT2 Gene
We searched the paired exome sequencing data (blood and fresh-frozen brain tissue) of brothers II.2 and II.3 for second-hit somatic variants. Analysis of consensus somatic variants called by both MuTect and VarScan revealed a somatic variant in WNT2 [chr7:116955375T>G (hg19); NM_003391.3:c.338C>T; p.Ala113Val] in brother II.2. The variant was present in 5/189 sequencing reads, a variant allele fraction of 2.6%. This somatic variant is reported in renal carcinoma in the COSMIC database.39
Lists of somatic variants identified in the paired exome sequencing data for brothers II.2 and II.3 are shown in eTables 2 and 3 (links.lww.com/NXG/A510), respectively. We focused on the WNT2 variant in II.2 because of the known link between the WNT and mTOR pathways. Only 1 other somatic variant was identified in a gene previously associated with epilepsy or FCD or part of the mTOR and related pathways (see Supplemental Material for gene list, links.lww.com/NXG/A510). However, this variant, in CUX2 in II.3, was present 7 times in gnomAD and predicted to be tolerated or benign by in silico tools SIFT40 and PolyPhen2.41
The somatic WNT2 variant (c.338C>T; p.Ala113Val) was validated using ddPCR. The somatic WNT2 variant was detected at an allele fraction of ∼0.3% in II.2's brain sample (Figure 2B), but not in his blood (Figure 2C). The limit of detection of the ddPCR assay was 0.025% (Figure 2D). Blood-derived DNA from 3 family members (I.1, I.2, and II.3) and brain-derived DNA from the brother with FCD (II.3), together with brain DNA samples from 44 unrelated individuals with different malformations and epilepsy, were extracted from surgically resected fresh-frozen brain tissue of similar quality. These samples were screened for the somatic WNT2 variant (p.Ala113Val) using the same targeted ddPCR assay, and no copies were detected (data not shown). We also sought this WNT2 variant in a second cohort of 38 focal malformation of cortical development (MCD) cases with high-depth sequencing data available, but it was not found (Ann Poduri, personal communication).42
We subjected a second brain-derived DNA sample from brother II.3 to higher-depth (200-fold) exome sequencing but failed to find a plausible somatic variant in epilepsy, FCD, or mTOR and related pathway genes. We also performed additional high-depth exome sequencing (1000-fold target depth) and searched specifically for second-hit variants in NPRL3 in both brothers. A high rate of somatic variants was called in these samples, likely due to the high depth of sequencing and lack of a panel of normal samples sequenced at comparable depth on the same platform to filter technical artifacts. Exome sequencing achieved a mean coverage of 685 across the NPRL3 coding region (eFigure 1, Supplemental Material, links.lww.com/NXG/A510). No somatic variants were identified in NPRL3 in either brother. All variants called in this gene were excluded as artifacts after manual inspection because they were not observed in more than 2 unique reads or were present at low allele fraction in both brothers.
Copy Number Variant Analysis
We searched for CNVs in brain-derived DNA for second-hit somatic variants. No significant CNVs were identified from chromosomal microarrays in either brother, nor were rare CNVs identified in exome sequencing data.
Functional Characterization of the WNT2 Variant
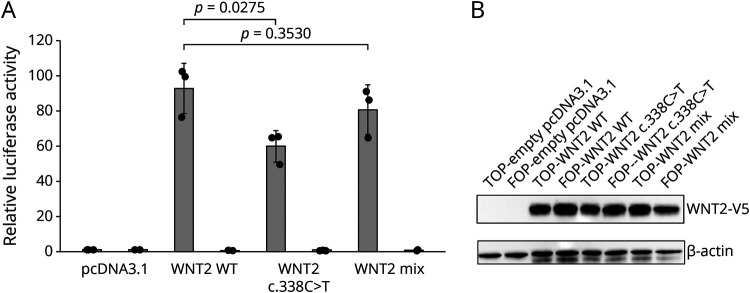
We used the TOPFlash dual-luciferase assay to determine the effects of the WNT2 p.Ala113Val variant on canonical WNT signaling (Figure 3A). The TOPflash assay is a direct read out for transcription mediated through translocation of β-catenin to the nucleus and activation of WNT target genes (or the luciferase reporter) specifically through the LEF/TCF family of transcription factors. In this context, the TOPflash assay measures the effect on WNT signaling mediated via activation through WNT2. In 3 independent experiments, we observed a consistent 35% reduction in relative luciferase activity in HEK293T cells transfected with the mutant WNT2 compared with wild-type WNT2 (p = 0.028; Student t test). This reduction in activity was unlikely due to protein instability as Western blot analysis showed equal abundance of wild-type and mutant WNT2 protein (Figure 3B). Equally mixing mutant and wild-type WNT2 constructs showed only a mild (8.5%) reduction in relative luciferase activity, suggesting that the WNT2 p.Ala113Val variant leads to partial loss of function, but not a dominant negative effect.
Figure 3. The WNT2 c.338C>T Variant Affects Canonical WNT Signaling.
(A) Relative luciferase activity measured using the TOPFlash assay in HEK293T cells transfected with expression vectors for active WNT2-V5 (WNT2 WT), mutant WNT2-V5 (WNT2 c.338C>T), or an equal mix of both (WNT2 Mix). The TOPFlash assay measures canonical WNT signaling through a reporter with functional TCF/LEF responsive promoter (gray bars; TOP) or a control FOPFlash reporter with mutated, nonfunctional TCF/LEF binding sites (black bars; FOP) by standard dual-luciferase assay. Error bars indicate SDs between 3 independent experiments. Differences assessed using the Student t test. (B) Detection of V5-tagged wild-type and mutant WNT2 proteins transfected into HEK293T cells for dual-luciferase reporter assay, and endogenous levels of β-actin to show equal loading, by Western blot.
Discussion
A number of mTOR pathway genes, such as the GATOR1 complex genes DEPDC5, NPRL2 and NPRL3, and MTOR, TSC1, and TSC2, are associated with FCD.1-3,9-13 Pathogenic variants in these genes typically activate the mTOR pathway through loss of function of negative regulators (e.g., DEPDC5, NPRL2, and NPRL3) or gain of function of mTOR activators.1,2 While we hypothesized the 2-hit model for FCD some time ago,19 germline and somatic variants causing FCD were only recently reported in patients with DEPDC513,14 and in hemimegalencephaly with TSC2.12 Recently, single patients with 2 hits in different genes were reported in hemimegalencephaly18 and tuberous sclerosis complex,15 but this has not been identified in the common forms of FCD associated with intractable focal epilepsy. In this study, we identified a novel maternally inherited germline variant in NPRL3 (c.48delG, p.Ser17Alafs*70) in 2 brothers with FCD. As both brothers have a similar phenotype, they could have inherited a second, paternally inherited, pathogenic germline variant. Our exome sequencing analysis considered a number of different inheritance models (see methods, links.lww.com/NXG/A510); however, we did not identify any additional germline variants likely to contribute to the FCD. This may be because the variant is outside the coding and splice site regions included in the exome analysis.
Pathogenic NPRL3 germline variants have been reported in families with FCD and epilepsy.8-10 NPRL3 variants are not fully penetrant; some individuals with a pathogenic variant have focal epilepsy with no detected FCD, whereas others are unaffected.9,10 Similarly, in the family we studied, the NPRL3 variant was inherited from an unaffected mother with a normal MRI. Our high-depth analysis for a second-hit somatic variant in NPRL3 in the brain-derived DNA of both brothers did not reveal any plausible variants.
A somatic variant in WNT2 (c.338C>T, p.Ala113Val) was identified in the brain-derived DNA from brother II.2 at an allele fraction estimated at 2.6% in the exome data and 0.3% by ddPCR. ddPCR assays target single DNA molecules and are highly sensitive (down to <0.1% allele fraction), making them 10-fold more sensitive than sequencing.43 Thus, the true allele fraction of the WNT2 p.Ala113Val variant is likely lower than the exome analysis suggests. The allele fraction estimated from the exome data has a wider 95% confidence interval of 0.8%–5.9% compared with the ddPCR 95% confidence interval 0.1%–0.6% (Clopper-Pearson exact method).
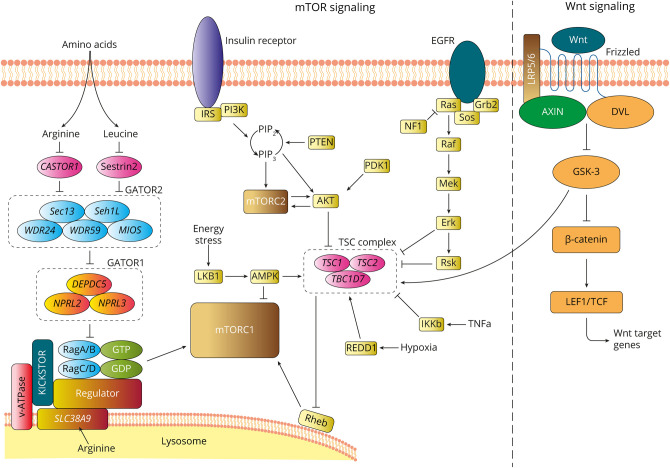
These data indicate that WNT2 is potentially associated with FCD, and of interest, it is an upstream regulator of the mTOR pathway via the TSC complex (Figure 4). TSC1 and TSC2 are negative regulators of MTOR and activating variations in both of these genes cause FCD.11,12 The canonical WNT pathway activates mTOR via inhibition of glycogen synthase kinase 3 (GSK3), which phosphorylates TSC1 and TSC2.44,45 Reduced WNT signaling reduces the inhibition of GSK3, leading to activation of the TSC complex. Furthermore, this specific WNT2 (c.338C>T, p.Ala113Val) somatic variant is not novel; it has been confirmed somatic in renal carcinoma in the COSMIC database.39 Somatic variants in TSC1 and TSC2 are also associated with renal carcinoma.46 Although loss of Wnt2 alone is correlated with decreased progenitor proliferation and decreased numbers of dopaminergic neurons in the murine ventral midbrain,47 the effect of reduced Wnt2 in the presence of a GATOR1 defect has not been modeled. It still remains mysterious whether there is physiologic coupling of these 2 signaling pathways, although a recent study intriguingly suggests that sustained mTORC1 activation impairs WNT/ß-catenin signaling to influence stem cell maintenance.48 Because our in vitro functional studies revealed the WNT2 p.Ala113Val variant itself leads to partial loss of function, this effect may be magnified in the absence of NPRL3-mediated GATOR1 regulation, further affecting growth and migration of cortical cell populations. It has been established for other FCDs caused by 2-hit germline and somatic variants in genes like TSC212 and DEPDC513 that low allele fraction somatic second-hit variants are sufficient to disrupt neuronal migration.
Figure 4. Diagram Showing Key Upstream Regulators and Activators of the mTOR and WNT Signaling Pathways.
We expect both pathogenic variants to be present in a subset of cells in the FCD. The 2-hit hypothesis predicts a cumulative effect of these variants on mTOR signaling. However, to accurately model the cumulative effect of both variants, we would need a primary or immortalized cell line derived from the dysplastic brain tissue of II:2, given the low allele fraction of the WNT2 variant. Unfortunately, we do not have such a cell line derived from this brother to study the impact in vitro.
We did not identify a second-hit somatic variant in II:3, despite interrogating NPRL3, other mTOR pathway genes, and genes in intersecting pathways at high depth. This could be because a second-hit variant was below the threshold of detection in our exome analysis, is located outside the exome, or is in another as yet unidentified pathway. Supporting the former hypothesis, analysis of pathogenic somatic variants in multiple FCD cohorts found that the average variant allele fraction was only ∼4%,12 suggesting that even lower allele fractions are likely to be pathogenic. Increasing attention to the latter hypothesis is leading to the discovery of poison exons harboring pathogenic variants49 and other variations in promoter and regulatory regions and epigenetic modifications that potentially affect gene expression; all of these are possible mechanisms leading to a potential hidden second hit.
Of interest, II.2 who has the NPRL3 and WNT2 variants has a less severe phenotype. The correlation between genotype and phenotype for somatic variants causing FCD is not well understood. In fact, it has been established that pathogenic variants associated with FCDs, such as in tuberous sclerosis complex, do not correlate with age at onset and epilepsy phenotype,50 so such a correlation would not necessarily have been expected between these brothers. This may relate to the presence of other factors such as polygenic risk haplotypes that may vary between individuals, even within the same family. Furthermore, it is difficult to draw conclusions about influences on the clinical severity of the 2 brothers without having been able to identify a second-hit variant in II.3, as discussed above.
This study had a number of limitations. We analyzed 1,384 genes selected because of a previous association with epilepsy or FCD or as part of the mTOR or related pathways. This gene panel approach introduces a potential selection bias. Exome sequencing with 100- or 200-fold coverage lacks power to confidently detect or exclude the presence of low allele fraction somatic variants (<1%–5%). We performed a third round of very-high-depth exome sequencing (1000-fold target depth) on brain-derived DNA from the 2 brothers. However, with only 2 brain DNA samples sequenced at this depth on this platform, we did not have a large pool of normal samples that could be used to efficiently remove sequencing artifacts. Thus, analysis of these samples focused specifically on searching for second-hit variants in NPRL3 as a second hit in the same gene was the most plausible alternative hypothesis.
The somatic WNT2 variant was identified at a very low allele fraction, with different fraction estimates from exome sequencing and ddPCR assay. The true somatic allele fraction likely resides within or close to the region between the 95% confidence intervals of the 2 fraction estimates (0.6%–0.8%). Such low allele fraction somatic variants have already been reported to cause focal brain lesions, including mTOR pathway variants in FCD (down to 0.25% fraction)16 and tuberous sclerosis (down to 0.7% fraction)50 and GNAQ variants in Sturge-Weber syndrome, where extremely low pathogenic allele fractions have been found (down to ∼0.09%).35,51,52 Notably, we confirmed the low-level WNT2 p.Ala113Val somatic variant in lesional tissue using an independent method with higher sensitivity than sequencing.
The data reported in this study suggest that the cumulative effect of variants in multiple genes in related pathways is worth considering in individuals with FCD or other types of MCDs. However, interpretation of low allele fraction somatic variant calls in routine high-depth exome analysis is challenging. We recommend that the functional effect of the variant and role of the gene are both considered in regard to their potential to cause a brain lesion. The WNT2 somatic variant identified in this study has been previously reported in renal carcinoma, suggesting that it may have similarly disruptive effects on brain cell growth and proliferation to cause FCD. Other genes that do not directly regulate cellular growth and proliferation may also be associated with FCD; however, additional functional studies would be required to confirm the impact of somatic variants in such genes. We hope that our findings will encourage further research to expand the genetic landscape of mosaic variants in FCD and related MCDs.
Our findings raise the intriguing possibility of a dual-pathway, 2-hit genetic model of germline and somatic variants that contribute to FCD, and we report the possible association of the WNT pathway with FCD. Our study highlights the importance of interrogating variants in multiple pathways at high depth when searching for somatic second hits.
Acknowledgment
The authors thank the family for their participation in the study. They thank Xi He and Randall Moon for providing testing reagents.
Glossary
- CNV
copy number variant
- COSMIC
Catalogue of Somatic Mutations in Cancer
- ddPCR
droplet digital PCR
- FCD
focal cortical dysplasia
- MCD
malformation of cortical development
- mTOR
mechanistic target of rapamycin
- RLU
relative light unit
Appendix. Authors
Contributor Information
Mark F. Bennett, Email: mark.bennett@wehi.edu.au.
Michael S. Hildebrand, Email: michael.hildebrand@unimelb.edu.au.
Sayaka Kayumi, Email: sayaka.kayumi@adelaide.edu.au.
Mark A. Corbett, Email: mark.corbett@adelaide.edu.au.
Sachin Gupta, Email: sachin.gupta@health.nsw.gov.au.
Zimeng Ye, Email: zimengy1@student.unimelb.edu.au.
Michael Krivanek, Email: michael.krivanek@health.nsw.gov.au.
Rosemary Burgess, Email: rosie_burgess@hotmail.com.
Olivia J. Henry, Email: henry.olivia17@gmail.com.
John A. Damiano, Email: damianojohn40@gmail.com.
Amber Boys, Email: amber.boys@vcgs.org.au.
Jozef Gécz, Email: jozef.gecz@adelaide.edu.au.
Melanie Bahlo, Email: bahlo@wehi.edu.au.
Ingrid E. Scheffer, Email: scheffer@unimelb.edu.au.
Study Funding
This study was supported by National Health and Medical Research Council (NHMRC) funding: NHMRC Program Grants 1054618 and 1091593, NHMRC Project Grants 1079058 and 1129054, NHMRC Senior Research Fellowship 1102971, NHMRC Senior Practitioner Fellowship 1104831, NHMRC Senior Principal Research Fellowship 1155224, and NHMRC R. D. Wright Career Development Fellowship 1063799. This work was also supported by a CURE Epilepsy Taking Flight Award and through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC Independent Research Institutes Infrastructure Support Scheme.
Disclosure
M.F. Bennett, M.S. Hildebrand, S. Kayumi, M.A. Corbett, S. Gupta, Z. Ye, M. Krivanek, R. Burgess, O.J. Henry, J.A. Damiano, A. Boys, J. Gecz, and M. Bahlo report no disclosures relevant to the manuscript. I.E. Scheffer has served on scientific advisory boards for UCB, Eisai, GlaxoSmithKline, BioMarin, Nutricia, and Xenon Pharmaceuticals and editorial boards of the Annals of Neurology and Neurology and Epileptic Disorders; may accrue future revenue on pending patent WO61/010176 (filed: 2008): Therapeutic Compound; a patent for SCN1A testing held by Bionomics Inc and licensed to various diagnostic companies; has received speaker honoraria from GlaxoSmithKline, Athena Diagnostics, UCB, BioMarin, Biocodex, Eisai, and Transgenomics; and has received funding for travel from Athena Diagnostics, UCB, Biocodex, GlaxoSmithKline, BioMarin, and Eisai. S.F. Berkovic discloses payments from UCB Pharma, Novartis Pharmaceuticals, Sanofi-Aventis, and Jansen Cilag for lectures and educational presentations and a patent for SCN1A testing held by Bionomics Inc and licensed to various diagnostic companies. Go to Neurology.org/NG for full disclosures.
References
- 1.Iffland PH II, Crino PB. Focal cortical dysplasia: gene mutations, cell signaling, and therapeutic implications. Annu Rev Pathol. 2017;12:547-571. [DOI] [PubMed] [Google Scholar]
- 2.Marsan E, Baulac S. Review: mechanistic target of rapamycin (mTOR) pathway, focal cortical dysplasia and epilepsy. Neuropathol Appl Neurobiol. 2018;44(1):6-17. [DOI] [PubMed] [Google Scholar]
- 3.Jansen LA, Mirzaa GM, Ishak GE, et al. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain. 2015;138(pt 6):1613-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lim JS, Kim WI, Kang HC, et al. Brain somatic mutations in MTOR cause focal cortical dysplasia type II leading to intractable epilepsy. Nat Med. 2015;21(4):395-400. [DOI] [PubMed] [Google Scholar]
- 5.Scerri T, Riseley JR, Gillies G, et al. Familial cortical dysplasia type IIA caused by a germline mutation in DEPDC5. Ann Clin Transl Neurol. 2015;2(5):575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baulac S, Ishida S, Marsan E, et al. Familial focal epilepsy with focal cortical dysplasia due to DEPDC5 mutations. Ann Neurol. 2015;77(4):675-683. [DOI] [PubMed] [Google Scholar]
- 7.Mirzaa GM, Campbell CD, Solovieff N, et al. Association of MTOR mutations with developmental brain Disorders, including megalencephaly, focal cortical dysplasia, and pigmentary mosaicism. JAMA Neurol. 2016;73(7):836-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ricos MG, Hodgson BL, Pippucci T, et al. Mutations in the mammalian target of rapamycin pathway regulators NPRL2 and NPRL3 cause focal epilepsy. Ann Neurol. 2016;79(1):120-131. [DOI] [PubMed] [Google Scholar]
- 9.Sim JC, Scerri T, Fanjul-Fernández M, et al. Familial cortical dysplasia caused by mutation in the mammalian target of rapamycin regulator NPRL3. Ann Neurol. 2016;79(1):132-137. [DOI] [PubMed] [Google Scholar]
- 10.Weckhuysen S, Marsan E, Lambrecq V, et al. Involvement of GATOR complex genes in familial focal epilepsies and focal cortical dysplasia. Epilepsia. 2016;57(6):994-1003. [DOI] [PubMed] [Google Scholar]
- 11.Lim JS, Gopalappa R, Kim SH, et al. Somatic mutations in TSC1 and TSC2 cause focal cortical dysplasia. Am J Hum Genet. 2017;100(3):454-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Gama AM, Woodworth MB, Hossain AA, et al. Somatic mutations activating the mTOR pathway in dorsal telencephalic progenitors cause a continuum of cortical dysplasias. Cell Rep. 2017;21(13):3754-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribierre T, Deleuze C, Bacq A, et al. Second-hit mosaic mutation in mTORC1 repressor DEPDC5 causes focal cortical dysplasia-associated epilepsy. J Clin Invest. 2018;128(6):2452-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee WS, Stephenson SEM, Howell KB, et al. Second-hit DEPDC5 mutation is limited to dysmorphic neurons in cortical dysplasia type IIA. Ann Clin Transl Neurol. 2019;6(7):1338-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sim NS, Ko A, Kim WK, et al. Precise detection of low-level somatic mutation in resected epilepsy brain tissue. Acta Neuropathol. 2019;138(6):901-912. [DOI] [PubMed] [Google Scholar]
- 16.Baldassari S, Ribierre T, Marsan E, et al. Dissecting the genetic basis of focal cortical dysplasia: a large cohort study. Acta Neuropathol. 2019;138(6):885-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baldassari S, Picard F, Verbeek NE, et al. The landscape of epilepsy-related GATOR1 variants. Genet Med. 2019;21(2):398-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelorosso C, Watrin F, Conti V, et al. Somatic double-hit in MTOR and RPS6 in hemimegalencephaly with intractable epilepsy. Hum Mol Genet. 2019;28(22):3755-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leventer RJ, Jansen FE, Mandelstam SA, et al. Is focal cortical dysplasia sporadic? Family evidence for genetic susceptibility. Epilepsia. 2014;55:e22-e26. [DOI] [PubMed] [Google Scholar]
- 20.Girirajan S, Rosenfeld JA, Cooper GM, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42(3):203-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veltman JA, Brunner HG. Understanding variable expressivity in microdeletion syndromes. Nat Genet. 2010;42(3):192-193. [DOI] [PubMed] [Google Scholar]
- 22.Ye Z, McQuillan L, Poduri A, et al. Somatic mutation: the hidden genetics of brain malformations and focal epilepsies. Epilepsy Res. 2019;155:106161. [DOI] [PubMed] [Google Scholar]
- 23.Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17(20):8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedersen BS, Layer RM, Quinlan AR. Vcfanno: fast, flexible annotation of genetic variants. Genome Biol. 2016;17(1):118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karczewski KJ, Francioli LC, Tiao G, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fromer M, Moran JL, Chambert K, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91(4):597-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fromer M, Purcell SM. Using XHMM software to detect copy number variation in whole-exome sequencing data. Curr Protoc Hum Genet. 2014;81:7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klambauer G, Schwarzbauer K, Mayr A, et al. Mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 2012;40:e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20(6):1698-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsao SC, Weiss J, Hudson C, et al. Monitoring response to therapy in melanoma by quantifying circulating tumour DNA with droplet digital PCR for BRAF and NRAS mutations. Sci Rep. 2015;5:11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hildebrand MS, Harvey AS, Malone S, et al. Somatic GNAQ mutation in the forme fruste of Sturge-Weber syndrome. Neurol Genet. 2018;4:e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacDonald BT, Hien A, Zhang X, et al. Disulfide bond requirements for active Wnt ligands. J Biol Chem. 2014;289(26):18122-18136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003;13(8):680-685. [DOI] [PubMed] [Google Scholar]
- 38.Bar-Peled L, Chantranupong L, Cherniack AD, et al. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forbes SA, Beare D, Boutselakis H, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777-D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40(Web Server issue):W452-W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winawer MR, Griffin NG, Samanamud J, et al. Somatic SLC35A2 variants in the brain are associated with intractable neocortical epilepsy. Ann Neurol. 2018;83(6):1133-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abyzov A, Tomasini L, Zhou B, et al. One thousand somatic SNVs per skin fibroblast cell set baseline of mosaic mutational load with patterns that suggest proliferative origin. Genome Res. 2017;27(4):512-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inoki K, Ouyang H, Zhu T, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126(5):955-968. [DOI] [PubMed] [Google Scholar]
- 45.Hermida MA, Dinesh Kumar J, Leslie NR. GSK3 and its interactions with the PI3K/AKT/mTOR signalling network. Adv Biol Regul. 2017;65:5-15. [DOI] [PubMed] [Google Scholar]
- 46.Tyburczy ME, Jozwiak S, Malinowska IA, et al. A shower of second hit events as the cause of multifocal renal cell carcinoma in tuberous sclerosis complex. Hum Mol Genet. 2015;24(7):1836-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sousa KM, Villaescusa JC, Cajanek L, et al. Wnt2 regulates progenitor proliferation in the developing ventral midbrain. J Biol Chem. 2010;285(10):7246-7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng H, Lu B, Zamponi R, et al. mTORC1 signaling suppresses Wnt/beta-catenin signaling through DVL-dependent regulation of Wnt receptor FZD level. Proc Natl Acad Sci U S A. 2018;115:E10362-E10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carvill GL, Engel KL, Ramamurthy A, et al. Aberrant inclusion of a poison exon causes dravet syndrome and related SCN1A-associated genetic epilepsies. Am J Hum Genet. 2018;103(6):1022-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogórek B, Hamieh L, Hulshof HM, et al. TSC2 pathogenic variants are predictive of severe clinical manifestations in TSC infants: results of the EPISTOP study. Genet Med. 2020;22(9):1489-1497. [DOI] [PubMed] [Google Scholar]
- 51.Shirley MD, Tang H, Gallione CJ, et al. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med. 2013;368(21):1971-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uchiyama Y, Nakashima M, Watanabe S, et al. Ultra-sensitive droplet digital PCR for detecting a low-prevalence somatic GNAQ mutation in Sturge-Weber syndrome. Sci Rep. 2016;6:22985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Variants identified in this study have been submitted to ClinVar. Sequencing data are available from the corresponding authors on reasonable request.