Abstract
Most organisms possess time-keeping devices called circadian clocks. At the molecular level, circadian clocks consist of transcription–translation feedback loops (TTFLs). Although some components of the negative TTFL are conserved across the animals, important differences exist between typical models, such as mouse and the fruit fly. In Drosophila, the key components are PERIOD (PER) and TIMELESS (TIM-d) proteins, whereas the mammalian clock relies on PER and CRYPTOCHROME (CRY-m). Importantly, how the clock has maintained functionality during evolutionary transitions between different states remains elusive. Therefore, we systematically described the circadian clock gene setup in major bilaterian lineages and identified marked lineage-specific differences in their clock constitution. Then we performed a thorough functional analysis of the linden bug Pyrrhocoris apterus, an insect species comprising features characteristic of both the Drosophila and the mammalian clocks. Unexpectedly, the knockout of timeless-d, a gene essential for the clock ticking in Drosophila, did not compromise rhythmicity in P. apterus, it only accelerated its pace. Furthermore, silencing timeless-m, the ancestral timeless type ubiquitously present across animals, resulted in a mild gradual loss of rhythmicity, supporting its possible participation in the linden bug clock, which is consistent with timeless-m role suggested by research on mammalian models. The dispensability of timeless-d in P. apterus allows drawing a scenario in which the clock has remained functional at each step of transition from an ancestral state to the TIM-d-independent PER + CRY-m system operating in extant vertebrates, including humans.
Keywords: circadian clock, reverse genetics, timeless, gene loss, Bilateria, Insecta
Introduction
From the very beginning of life on our planet, nearly every organism has been exposed to periodic changes of day and night. Often, it is a great advantage for an organism to anticipate dawn, avoid mid-day heat, or target particular activity to a specific time of day. Accordingly, organisms have evolved circadian clocks, internal genetically determined time-measuring devices that “keep ticking” with a free-running period (τ) close to 24 h, even when environmental cues are absent.
The key feature of circadian clocks is the transcription–translation feedback loop (TTFL) with positive and negative regulators. The positive regulators (transcription factors) drive the expression of mRNAs from which the negative regulators are translated, gradually accumulate, and once reach a threshold level, they suppress their own transcription by inhibiting the activity of the positive regulators. The similarities of the circadian clock shared by the fruit fly Drosophila melanogaster and mammals have facilitated elucidation of molecular mechanisms underlying rhythmicity (Allada et al. 2001; Zhang et al. 2011; Ozkaya and Rosato 2012). The core of the clock in animals is composed of the positive transcription factors, CLOCK (CLK) and BMAL/CYCLE (Darlington et al. 1998; Rutila et al. 1998; Huang et al. 2012; Menet et al. 2014), whose activity is suppressed by negative regulators. Within the negative feedback loop, we can see notable differences in the setup of the fruit fly and mammalian clocks. In the fruit fly, PERIOD and Drosophila-type TIMELESS (here referred to as TIM-d) are the key negative regulators essential for behavioral and molecular rhythmicity (Konopka and Benzer 1971; Bargiello et al. 1984; Zehring et al. 1984; Hardin et al. 1990; Sehgal et al. 1994). The chief light-cued synchronization pathway involves the interaction of TIM-d with Drosophila-type CRYPTOCHROME (CRY-d) (Stanewsky et al. 1998; Ceriani et al. 1999; Emery et al. 2000). However, mammalian clocks lack both TIM-d and CRY-d; instead, their negative feedback relies on PER and mammalian CRY (hereafter CRY-m) (Kume et al. 1999). Thus, CRY-d serves in Drosophila clock neurons as a blue light photoreceptor that triggers light-mediated degradation of the negative regulators, whereas CRY-m in mammals is itself a potent repressor of CLK-BMAL. Phylogenetic analyses indicate that CRY-m is present in numerous insect species (Yuan et al. 2007), and in some of them, CRY-m was confirmed as a component essential for circadian rhythmicity (Ikeno, Katagiri, et al. 2011; Ikeno, Numata, et al. 2011; Zhang et al. 2017). The evolution of CRY might be shaped by its additional roles, including its involvement in seasonality, as was shown in the bean bug (Ikeno, Katagiri, et al. 2011; Ikeno, Numata, et al. 2011) and in the linden bug Pyrrhocoris apterus (Urbanova et al. 2016). CRY proteins seem to participate in magnetoreception as was reported for CRY-d in the monarch butterflies (Wan et al. 2021) and Drosophila (Yoshii et al. 2009; Fedele et al. 2014), and for CRY-m in cockroaches (Bazalova et al. 2016), P. apterus (Netusil et al. 2021), and birds (Xu et al. 2021).
Mammalian-type TIM (TIM-m), first identified in mice (Zylka et al. 1998) and subsequently in Drosophila (Benna et al. 2000), was first deemed as a nonclock protein. TIM-m is essential for development, which further complicated its functional analysis (Gotter et al. 2000; Benna et al. 2010). However, conditional knockdown experiments in the rat disrupted neuronal activity rhythms in the suprachiasmatic nucleus (Barnes et al. 2003), and mutation in human TIM-m causes familial advanced sleep phase syndrome (Kurien et al. 2019) belonging to a family of circadian rhythm sleep disorders.
The above-described idiosyncrasies between the circadian clocks within Drosophila and mice prompted us to elucidate the evolution of the circadian clock set up in Bilateria, that is animals with exception of Porifera, Ctenophora, and Cnidaria. Assuming that the clock must have remained functional throughout evolution, we asked what were the transitional steps leading to the fruit fly organization (PER/TIM-d) on the one hand and the mammalian one (PER/CRY-m) on the other?
Recent progress in transcriptome sequencing of Bilateria in general and insect lineages in particular (Misof et al. 2014; Johnson et al. 2018; Kawahara et al. 2019; McKenna et al. 2019; Wipfler et al. 2019) allowed us to perform a systematic search for and subsequent evolutionary analysis of the circadian clock genes in Bilateria. Then we functionally tested all major clock components in the linden bug, P.apterus, an insect where a combination of both mammalian and Drosophila clock components were identified. Although a majority of P. apterus clock genes are functionally conserved with the fruit fly or mouse orthologs, unexpectedly, the knockout of tim-d, a gene essential for the clock ticking in Drosophila, did not compromise rhythmicity in P. apterus, but only accelerated its pace.
Results
Complex Evolution of Bilaterian Clock Setup
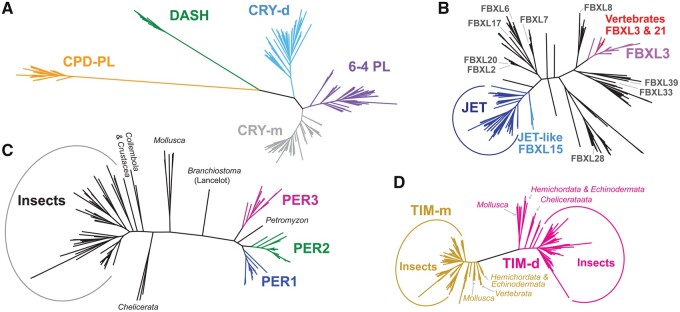
First, we retrieved genes coding for the key circadian clock negative feedback loop proteins from organisms representing major lineages of animals. To classify the CRY types (Kume et al. 1999; Yuan et al. 2007), we identified all cryptochrome/photolyase family members and reconstructed their phylogeny. The analysis revealed five well-defined clades: CRY-m, 6-4 DNA PHOTOLYASE (6-4 PL), CRY-d, CYCLOBUTANE PYRIMIDINE DIMER PHOTOLYASE (CPD-PL), and Drosophila, Arabidopsis, Synechocystis, and Human (DASH)-type CRY (fig. 1A; and supplementary fig. 1, Supplementary Material online). Although our primary interest was a reliable detection of CRY-d and CRY-m types across Bilateria, obtained phylogeny and distribution of 6-4 PL, CPD-PL, and DASH might be useful in the design of experiments (see Discussion).
Fig. 1.
Phylogeny of bilaterian clock proteins. (A) A tree illustrating relatedness among mammalian- and Drosophila-type of CRYPTOCHROME (CRY-m, grey; CRY-d, blue), 6-4 PL (deep purple), CPD-PL (orange), and DASH -type CRYPTOCHROME (green). (B) Phylogeny of JETLAG (JET, deep/ligh blue) and FBXL3/21 (purple/red) within FBXL proteins (black); (C) bilaterian PERIOD proteins with single-copy genes are shown in black and three vertebrate paralogs (PER1, PER2, and PER3) in colors. (D) TIM-m, found in all Bilateria, and TIM-d are clearly separated into two clusters. Presented trees were inferred using RAxML maximum likelihood GAMMA-based model. For detailed trees see supplementary figures 1–4, Supplementary Material online.
The stability of CRY proteins is regulated, among others, by three proteins from the FBXL subfamily: JETLAG (JET), a protein essential for the interaction between CRY-d and TIM-d in Drosophila (Koh et al. 2006; Peschel et al. 2006, 2009), and two mammalian clock components interacting with CRY-m, FBXL3, and FBXL21, known also as over-time and after-hours, respectively (Godinho et al. 2007; Siepka et al. 2007; Hirano et al. 2013). Our phylogenetic reconstruction revealed a clear separation of JET and its deuterostomian homolog JET-like (also known as FBXL15) from FBXL3 and FBXL21 (fig. 1B;supplementary fig. 2, Supplementary Material online). FBXL3 and FBXL21 share one common ancestor present in basal Deuterostomia, whereas FBXL21 is only present in Gnathostomata (fishes, amphibians, reptiles, birds, and mammals). During our search for FBXL3, FBXL 21, and JET proteins in bilaterian lineages, we retrieved all homologs, including the distantly related proteins. Therefore, the absence of target proteins reflects gene loss rather than the technical inability to identify a protein in prospected genomes and transcriptomes.
Then, we reconstructed PER and TIM phylogeny. Three paralogs (PER1–3) found in the majority of vertebrates, albeit some lineage-specific losses were identified in reptiles and birds, are separated from a single PER identified in Protostomia (fig. 1C;supplementary fig. 3, Supplementary Material online). Phylogeny of TIMELESS proteins confirmed clear separation of TIM-d from the ubiquitously present mammalian-type TIM-m (fig. 1D;supplementary fig. 4, Supplementary Material online). Furthermore, the presence of TIM-d in echinoderms and hemichordates confirms that the duplication leading to TIM-d and TIM-m arose in the ancestor of deuterostomes and protostomes.
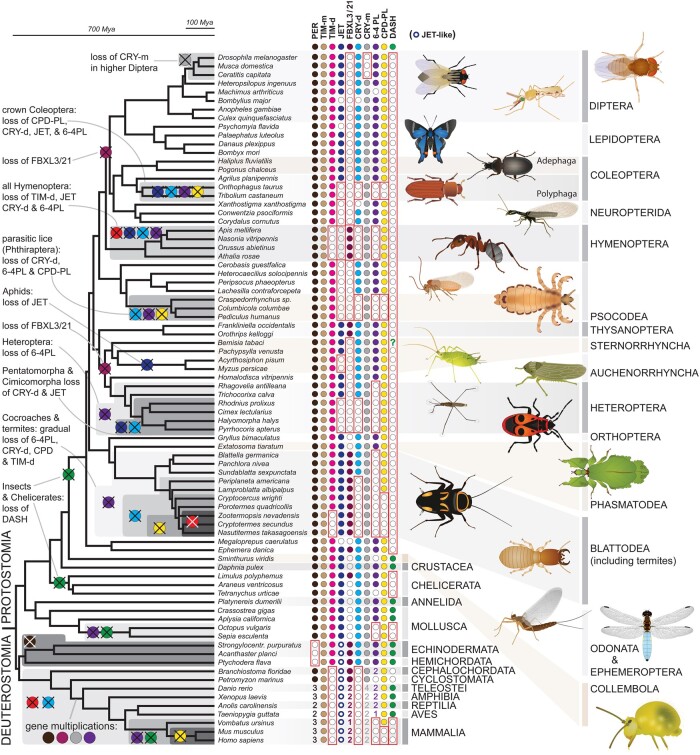
Having well-defined TIM, CRY, and PER data sets, we mapped their presence on the bilaterian phylogeny, with a particular focus on gene losses in entire lineages. Full analysis depicted in figure 2 (see supplementary tables 1 and 2, Supplementary Material online) revealed a complex evolution of the circadian clock setup. TIM-m is, consistently with its essential role in development (Gotter et al. 2000; Benna et al. 2010), the only protein identified in all organisms. Surprisingly, the canonical clock component PER has been lost in two basal deuterostomian lineages, echinoderms, and hemichordates.
Fig. 2.
Circadian clock gene losses mapped on the bilaterian phylogeny. Representative insect species are shown at the terminal nodes with indicated gene presence (full circle) or absence (empty circle) where the lineage-specific losses are highlighted with red rectangles (see details in supplementary tables 1 and 2, Supplementary Material online). The phylogenetic tree corresponds to a consensus of recent phylogenomic studies (Misof et al. 2014; Johnson et al. 2018; Kawahara et al. 2019; McKenna et al. 2019; Wipfler et al. 2019). Filled circles indicate the presence of PER (black), TIM-m (brown), TIM-d (bright magenta), JET (dark blue), FBXL3/21 (light purple), CRY-m (grey), CRY-d (blue), 6-4 PL (deep purple), CPD-PL (yellow), and DASH-type cryptochrome (green). For phylogenetic relationship see supplementary figures 1–4, Supplementary Material online. Numbers indicate the presence of multiple paralogs in one taxon. The question mark indicates a suspicious occurrence of DASH in Bemisia. Supplementary figure 16, Supplementary Material online, illustrates clear relatedness of this sequence with DASH from plants and fungi which can either be explained as contamination or as a horizontal gene transfer (HGT) from plant to insect. The latter would be consistent with a recent HGT of a plant detoxification component to Bemisia (Xia et al. 2021).
Notably, TIM-d has been lost in the entire bilaterian lineage only 3 times: in vertebrates, nearly all termites, and bees/wasps/ants (Hymenoptera; Rubin et al. 2006). In all three cases, TIM-d loss coincides with the absence of CRY-d, which seems to have been lost either simultaneously with TIM-d or even earlier, as seen in termites. Furthermore, CRY-d is absent in three additional lineages (supplementary tables 1 and 2, Supplementary Material online). In contrast, CRY-m was lost only in cyclorrhaphan Diptera, indicating the uniqueness of the Drosophila model. JET was lost in several lineages including those where either CRY-d or TIM-d is absent, such as a subset of Coleoptera, Hymenoptera, and Pentatomorpha + Cimicomorpha. FBXL3 and FBXL21 are found in Gnathostomata, their single ancestor FBXL3 was found in all analyzed basal Deuterostomia. However, FBXL3 was lost in the majority of Protostomia and has remained only in the horseshoe crab Limulus, and four insect lineages: Ephemeroptera, Blattodea, Thysanoptera, and Hymenoptera (supplementary fig. 2, Supplementary Material online). Remarkably, all three TIM-d losses observed in the entire Bilateria coincide with the presence of FBXL3/21 and CRY-m.
The Linden Bug Is a Model with Combination of Both Mammalian and Drosophila Components
Clearly, the evolution of the circadian clock architecture was more complex than previously appreciated. To understand the functionality of the clock during its evolutionary transitions, we conducted a systematic molecular-genetic analysis of the clock components in the linden bug, P.apterus, a species comprising features of both mammalian and Drosophila clock types (fig. 2;supplementary tables 1 and 3, Supplementary Material online). Paradoxically, P. apterus possesses CRY-m and TIM-d but lacks CRY-d (Bajgar et al. 2013).
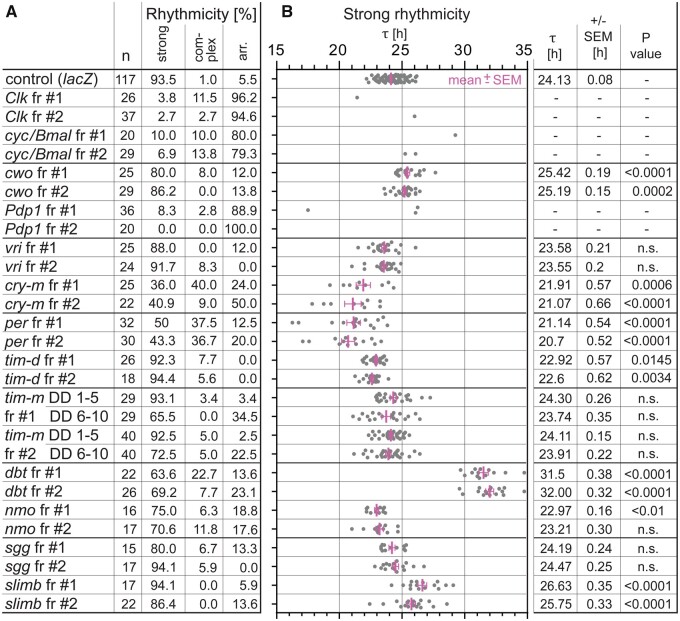
First, we employed systemic RNA interference (RNAi) to functionally test the circadian clock candidates in P. apterus. To identify possible off-target effects in RNAi experiments, each gene was separately knocked down with two nonoverlapping double-stranded RNA (dsRNA) fragments. The knockdown experiments confirmed a conserved role for the majority of P. apterus circadian clock genes (fig. 3;supplementary fig. 5, Supplementary Material online); specifically: knockdown of Clock, cycle/Bmal, or Par domain protein 1 (Pdp1) resulted in arrhythmicity, whereas clockwork orange (cwo) silencing slowed down the clock, consistent with the roles of the Drosophila homologs (Allada et al. 1998; Rutila et al. 1998; Cyran et al. 2003; Matsumoto et al. 2007). Weaker phenotypes were observed in vrille-silenced bugs, with nonsignificantly decreased τ. Consistently with the role of per and cry-m in established models, their knockdown in P. apterus resulted in severely reduced rhythmicity (fig. 3).
Fig. 3.
The role of circadian clock genes in P. apterus. (A) Summary of the gene knockdown describing its impact on the behavioral rhythmicity shown as percentage of males demonstrating strong rhythmicity, complex rhythmicity, and arrhythmicity; fr #1 and fr #2 are nonoverlapping dsRNA fragments 1 and 2, respectively. (B) Individual τ values are shown as dots for each male with strong rhythmicity; red bars represent means ± SEM (calculated if >10% individuals demonstrated rhythmicity). Columns depict the mean τ, standard error of the mean (SEM), and statistical difference from the controls (P value) (Kruskal–Wallis test with Dunn’s post hoc; calculated only if >10% individuals were rhythmic).
Silencing of posttranslational components essential for the clock, such as the casein kinase 1 doubletime (dbt; Price et al. 1998) and F-box protein slimb (Grima et al. 2002), remarkably extended τ (fig. 3;supplementary fig. 6, Supplementary Material online). In nemo (nmo)-silenced bugs, each fragment resulted in a faster clock, although only fragment’s #1 phenotype was statistically significant. Nevertheless, this observation is consistent with the role of nmo in Drosophila (Chiu et al. 2011; Yu et al. 2011).
Since RNAi is performed in adults, we were able to test the role of TIM-m on the τ without interfering with its developmental role reported for both insects and mammals (Benna et al. 2010; Kurien et al. 2019). Although tim-m silencing did not significantly impact τ, a substantial number of dsRNA-injected bugs displayed a gradual loss of rhythmicity in constant darkness. When the activity from the first 5 days of constant darkness was analyzed, only 3.4% (tim-m fr#1) and 2.5% (tim-m fr#2) of bugs were arrhythmic. However, when the rhythmicity was analyzed for Days 6–10 of constant darkness, arrhythmicity was recorded in 34.5% (tim-m fr#1) and 22.5% (tim-m fr#2) of dsRNA-injected bugs (fig. 3;supplementary fig. 7, Supplementary Material online).
TIM-d Knockdown and Knockout Linden Bugs Remain Robustly Rhythmic
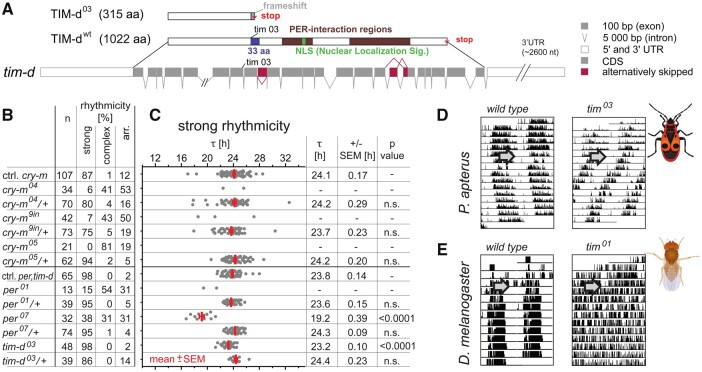
Surprisingly, tim-d-silenced linden bugs displayed robust rhythmicity (>90%) with τ shortened by >1 h (P < 0.0001). This observation strongly contrasts with the key role of tim-d in Drosophila, where its depletion results in complete arrhythmicity (Sehgal et al. 1994). Notably, silencing shaggy, a kinase essential for TIM-d-dependent phosphorylation in Drosophila (Martinek et al. 2001), had no impact on the τ in the linden bug (fig. 3). Therefore, to fully explore and clarify this unexpected tim-d phenotype, we applied the CRISPR/Cas9 technique (Kotwica-Rolinska et al. 2019) and produced well-defined genetic mutants in tim-d and two additional components of the negative feedback loop, cry-m and per, in P. apterus (fig. 4A;supplementary figs. 8–11, Supplementary Material online).
Fig. 4.
Either cry-m or per depletion completely abolishes circadian rhythmicity in P. apterus, whereas tim-d mutants demonstrate robust rhythmicity with significantly shorter τ. (A) Schematic representation of tim-d gene structure with coding regions, alternative splicing, and engineered mutation (tim03). Corresponding wt and mutant proteins are shown with major functional domains highlighted (for details, see supplementary figs. 13 and 14, Supplementary Material online). Alternative splicing of tim-d was detected in exons 9, 17, and 18. (B) Summary indicating the number and rhythmicity of the tested mutant and heterozygous animals compared with corresponding control siblings. (C) Individual τ values are plotted as a dot for each male; red bars depict means ± SEMs (calculated only if >10% individuals were rhythmic). Statistical difference from the controls is shown as P-value (Kruskal–Wallis test with Dunn’s post hoc). Double-plotted actogram of (D) wt and tim03 P. apterus compared with (E) D. melanogaster wt (Canton S) and tim01 mutant (arrow indicates the beginning of constant darkness).
Not only did the P. apterus tim-d03 homozygous mutants display robust rhythmicity (98%, fig. 4B and C), but the strength of their rhythm, plotted as PN (power of the periodogram), was comparable to values observed in the wild-type (wt; supplementary fig. 12, Supplementary Material online). The tim-d03 mutation causes a frameshift followed by a premature stop codon (fig. 4A), therefore, tim-d03 encodes only the initial one-third of the P. apterus TIM-d protein lacking the conserved PER-interaction regions, nuclear localization signal, and several key amino acid residues downstream (fig. 4A;supplementary figs. 13 and 14, Supplementary Material online). These features, essential for its proper function in Drosophila (Saez and Young 1996; Rothenfluh et al. 2000; Hara et al. 2011), are remarkably conserved in P. apterus TIM-d. Importantly, tim-d03 resulted in τ that was 0.6 h shorter than that of the wt (P < 0.0001), which corroborates the shorter τ observed after tim-d knockdown (fig. 3).
Three different mutations in cry-m were analyzed in detail (supplementary fig. 9, Supplementary Material online). The absence of CRY-m (cry-m04 and cry-m05) or an in-frame insertion (cry-m9in; supplementary fig. 10, Supplementary Material online) virtually destroyed rhythmic behavior (fig. 4B and supplementary fig. 12A–C, Supplementary Material online). However, a substantial percentage of homozygous mutants displayed aberrant complex rhythmicity, where either multiple periodic components were identified or the τ changed during the 10-day recording (fig. 4B;supplementary fig. 15, Supplementary Material online). Nevertheless, these rhythmic linden bugs, observed both in cry-m mutant homozygotes (fig. 4B) and in cry-m knockdown males (fig. 3A), displayed non-24-h rhythmicity (supplementary fig. 15B, Supplementary Material online). Similar to cry-m mutants, about one-third of P. apterus per mutants were arrhythmic, whereas the remaining bugs displayed rhythmicity with a short τ of ∼19 h (fig. 4B and C).
Discussion
Phylogenetic analyses were primarily focused on discerning clock setup and its evolution in Bilateria. However, clustering within a specific group reflects the evolutionary origin of the proteins, though, in some of them, mutations affecting biochemical properties might have occurred. It is conceivable that especially closely related groups, such as CRY-m and 6-4 PL, might relatively easily accumulate changes affecting their function. The combination and absence of particular CRY-type proteins in a given species might also be considered in experiments when antibodies recognizing conserved protein motifs are used, and vice versa, the design of either specific or universal antibodies should benefit from the available data sets.
The specific changes in the circadian clock setup, such as the absence of CRY-d and JET, pointed our attention to P. apterus. Altogether, the thorough RNAi and focused CRISPR/Cas9 analyses indicate nine P. apterus clock components (Clk, cyc/Bmal, Pdp1, vri, cwo, dbt, slimb, per, and nmo) work comparably to D. melanogaster orthologs. RNAi silencing of tim-m supports its role in the clock, which is consistent with its oscillatory role in the rat neuronal cells (Barnes et al. 2003). Nevertheless, relatively low P. apterus activity levels and their broad peaks (see Kaniewska et al. 2020) make it difficult to determine whether alterations of clock gene expression cause phase shifts in behavioral outputs. Thus, the role of TIM-m in circadian photoreception, proposed by the fruit fly experiments (Benna et al. 2010) and suggested by the altered activity phase in mammals (Kurien et al. 2019), could only be addressed in P. apterus with great difficulties.
Pyrrhocoris apterus possesses CRY-m, an ortholog of a key mammalian negative feedback loop protein, which functions consistent with its role in the monarch butterfly (Zhang et al. 2017), the bean bug (Ikeno, Katagiri, et al. 2011; Ikeno, Numata, et al. 2011), or the mouse (Kume et al. 1999). A substantial proportion of homozygous cry-m mutants (∼40–80%) displayed aberrant complex rhythmicity. Comparable aberrant residual rhythmicity has been reported for per01 Drosophila mutants, where ∼30% of flies were shown to display a non-24-h τ (Helfrich-Forster 2001). An interesting recent study on CRY-deficient mice describes their locomotor activity rhythms, which have shorter and variable τ when compared with wt controls (Putker et al. 2021).
The major difference between mammals and P. apterus is that linden bugs possess TIM-d. However, unlike in Drosophila where tim-null flies are completely arrhythmic, TIM-d only modulates the circadian period in P. apterus. This result is in agreement with earlier RNAi experiments in crickets and firebrats, which also reported shorter τ upon tim-d silencing (Danbara et al. 2010; Kamae and Tomioka 2012), dramatically contrasting with the key role of tim-d in Drosophila (Sehgal et al. 1994; fig. 4D and E).
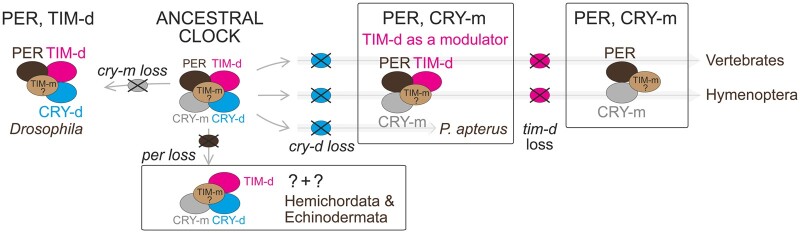
The dispensability of TIM-d in P. apterus suggests a scenario of transition between clock architectures relying on distinct components of their negative feedback loops. The proposed model implies that the clock would be functional in each step of the transition from the ancestral state to the PER + CRY-m system known today in vertebrates (fig. 5). A similar clock gene combination in Hymenoptera indicates a convergent evolution of the circadian system, although functional evidence from this insect group is not yet available. The circadian clock observed in P. apterus could then correspond to an early clock setup that facilitates tim-d loss without a complete collapse of the circadian cycling. However, the timing and causality of the proposed events might have been lineage specific, where either the loss of cry-d triggered the transition of TIM-d to its modulatory role, or alternatively, the loss of JET or change in TIM-d properties compromised its interaction with CRY-d, which in turn was subsequently lost.
Fig. 5.
Proposed scenarios of the circadian clock evolution in Bilateria. In the Drosophila ancestor, loss of cry-m gene resulted in a feedback loop relying on a PER + TIM-d dimer with CRY-d serving for light-mediated resetting of the system (Ceriani et al. 1999). A two-step process, when TIM-d first became a modulator of the τ, and only when it was lost, it allowed for a smooth transition to the PER + CRY-m system, with a functional clock in each step. In P. apterus, TIM-d is still present as a modulator of the clock, whereas CRY-d is absent. However, the timing of cry-d loss might differ between lineages. In one scenario, TIM-d could become a modulator in the presence of CRY-d, which would be lost afterward (situation observed in P. apterus). In the second scenario, the modulatory role of TIM-d led to its loss, excluding CRY-d from the loop and resulting in its subsequent loss. The involvement of TIM-m in the circadian clock is indicated both from mammalian models (Barnes et al. 2003; Kurien et al. 2019) and insect models (Benna et al. 2010; Nose et al. 2017), albeit its role is not established in detail. The third type of path observed in evolution involves the loss of the per gene. This strikingly unusual clock found in two basal deuterostomian phyla, Hemichordata, and Echinodermata, is apparently functional (Peres et al. 2014), although the mechanism remains unknown.
Materials and Methods
Data Sets and Phylogenetic Analyses
To reconstruct the evolution of circadian clock genes in animals, we applied a similar approach as in Smykal et al. (2020). First, we compared data sets from representative organisms (mouse, zebrafish, Platynereis, linden bug, pea aphid, monarch butterfly, honeybee, and fruit fly). Then we used BLAST to systematically explore all major bilaterian lineages at NCBI for CRYs, TIMs, PER, and FBXL/JET in taxon-specific searches. To ensure that all homologs were retrieved, reciprocal searches were performed for each species separately. The absence of a given gene/protein was then tested and confirmed in the corresponding lineage (order, suborder, and in some cases even family). Retrieved protein sequences were aligned using the ClustalW algorithm in Geneious Prime (www.geneious.com). Initial trees were inferred with the Fast tree, redundant sequences identified and removed from further analysis. Presented trees were constructed using RAxML maximum likelihood GAMMA-based model, although the same conclusions were obtained from trees inferred using Phyml. Pyrrhocorisapterus circadian clock genes were retrieved from in-house transcriptomic and genomic databases, verified, and completed by Sanger sequencing and PCR using primers specific to each gene. The gene models of per, tim-d, and cry-m were reconstructed in P. apterus from draft genome assemblies based on Oxford Nanopore sequencing of genomic DNA. All newly generated sequences were uploaded to GenBank (see supplementary table 3, Supplementary Material online ).
Gene Knockdowns and Gene Editing
Systemic RNAi is a well-established reverse genetic tool in P. apterus (Bajgar et al. 2013; Kotwica-Rolinska et al. 2017). Briefly, two non-overlapping cDNA fragments for each gene were designed to reduce RNAi off-targeting (see supplementary table 4, Supplementary Material online for primer’s sequences), products were cloned into plasmids from which the insert was amplified with primers introducing T7 promoter to both ends of the product (Smykal et al. 2014). Gene-specific dsRNAs were synthesized in vitro, purified by ethanol precipitation, and 8 µg injected into 2-day-old adult males. For details of gene editing in P. apterus see the Supplementary material and (Kotwica-Rolinska et al. 2019). Importantly, all mutant lines were outcrossed to the wt strain for seven to nine generations to remove possible off-target modifications.
Animal Rearing Conditions and Locomotor Activity Recordings
The majority of experiments were performed in Roana strain, whereas Oldrichovec strain was only used for cry-m gene editing. See Pivarciova et al. (2016) for details of the strain’s origin. In all experiments, adult males were used for locomotor activity analysis. Bugs were individually housed in test tubes (2.5 cm diameter, 15 cm in length) supplemented with dry linden seeds and water which the bugs were allowed to consume ad libitum and placed in the Locomotor Activity Monitors (LAM 25, TriKinetics Inc., Waltham, MA, USA). Bugs were synchronized for 5 days in LD conditions (18 h light, 6 h darkness) at 25 °C, followed by at least 10 days in constant darkness at 25 °C. To determine rhythmicity and τ in the constant darkness, the Lomb–Scargle periodogram in ActogramJ plugin of ImageJ (Schmid et al. 2011) was employed. See Pivarciova et al. (2016) and Supplementary material for details.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank V. Beneš for help with P. apterus genome assembly, Martina Hajdušková (www.BioGraphix.cz) for species’ illustration, Hana Vaněčková for technical support, and Robert Hanus, Marek Jindra, Martin Sládek, and Paul Taghert for discussions.
Funding
This work was financed by European Research Council (project InPhoTime, 726049) to D.D.
Author Contributions
Conceptualization: J.K.-R. and D.D.; investigation: J.K.-R., L.C., V.S., M.D., D.C., D.D., J.P., M.H., and B.C.-H.W.; formal analysis: J.K.-R., L.C., V.S., and D.D.; visualization: V.S. and D.D.; supervision: D.D.; and writing: D.D. with input from all coauthors.
Authors declare that they have no competing interests.
Data Availability
cDNA and genome contigs connected to the project are deposited in the GenBank (see supplenentary table 3, Supplementary Material online). All data are incorporated into the article and its online Supplementary material.
References
- Allada R, Emery P, Takahashi JS, Rosbash M.. 2001. Stopping time: the genetics of fly and mouse circadian clocks. Annu Rev Neurosci. 24:1091–1119. [DOI] [PubMed] [Google Scholar]
- Allada R, White NE, So WV, Hall JC, Rosbash M.. 1998. A mutant Drosophila homolog of mammalian clock disrupts circadian rhythms and transcription of period and timeless. Cell 93(5):791–804. [DOI] [PubMed] [Google Scholar]
- Bajgar A, Jindra M, Dolezel D.. 2013. Autonomous regulation of the insect gut by circadian genes acting downstream of juvenile hormone signaling. Proc Natl Acad Sci U S A. 110(11):4416–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargiello TA, Jackson FR, Young MW.. 1984. Restoration of circadian behavioral rhythms by gene-transfer in Drosophila. Nature 312(5996):752–754. [DOI] [PubMed] [Google Scholar]
- Barnes JW, Tischkau SA, Barnes JA, Mitchell JW, Burgoon PW, Hickok JR, Gillette MU.. 2003. Requirement of mammalian timeless for circadian rhythmicity. Science 302(5644):439–442. [DOI] [PubMed] [Google Scholar]
- Bazalova O, Kvicalova M, Valkova T, Slaby P, Bartos P, Netusil R, Tomanova K, Braeunig P, Lee HJ, Sauman I, et al. 2016. Cryptochrome 2 mediates directional magnetoreception in cockroaches. Proc Natl Acad Sci U S A. 113(6):1660–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benna C, Bonaccorsi S, Wulbeck C, Helfrich-Forster C, Gatti M, Kyriacou CP, Costa R, Sandrelli F.. 2010. Drosophila timeless2 is required for chromosome stability and circadian photoreception. Curr Biol. 20(4):346–352. [DOI] [PubMed] [Google Scholar]
- Benna C, Scannapieco P, Piccin A, Sandrelli F, Zordan M, Rosato E, Kyriacou CP, Valle G, Costa R.. 2000. A second timeless gene in Drosophila shares greater sequence similarity with mammalian tim. Curr Biol. 10(14):R512–513. [DOI] [PubMed] [Google Scholar]
- Ceriani MF, Darlington TK, Staknis D, Mas P, Petti AA, Weitz CJ, Kay SA.. 1999. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science 285(5427):553–556. [DOI] [PubMed] [Google Scholar]
- Chiu JC, Ko HW, Edery I.. 2011. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell 145(3):357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NRJ, Hardin PE, Young MW, Storti RV, Blau J.. 2003. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112(3):329–341. [DOI] [PubMed] [Google Scholar]
- Danbara Y, Sakamoto T, Uryu O, Tomioka K.. 2010. RNA interference of timeless gene does not disrupt circadian locomotor rhythms in the cricket Gryllus bimaculatus. J Insect Physiol. 56(12):1738–1745. [DOI] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TDL, Weitz CJ, Takahashi JS, Kay SA.. 1998. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280(5369):1599–1603. [DOI] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Helfrich-Forster C, Emery-Le M, Hall JC, Rosbash M.. 2000. Drosophila CRY is a deep brain circadian photoreceptor. Neuron 26(2):493–504. [DOI] [PubMed] [Google Scholar]
- Fedele G, Edwards MD, Bhutani S, Hares JM, Murbach M, Green EW, Dissel S, Hastings MH, Rosato E, Kyriacou CP.. 2014. Genetic analysis of circadian responses to low frequency electromagnetic fields in Drosophila melanogaster. PLoS Genet. 10(12):e1004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, et al. 2007. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316(5826):897–900. [DOI] [PubMed] [Google Scholar]
- Gotter AL, Manganaro T, Weaver DR, Kolakowski LF, Possidente B, Sriram S, MacLaughlin DT, Reppert SM.. 2000. A time-less function for mouse timeless. Nat Neurosci. 3(8):755–756. [DOI] [PubMed] [Google Scholar]
- Grima B, Lamouroux A, Chelot E, Papin C, Limbourg-Bouchon B, Rouyer F.. 2002. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature 420(6912):178–182. [DOI] [PubMed] [Google Scholar]
- Hara T, Koh K, Combs DJ, Sehgal A.. 2011. Post-translational regulation and nuclear entry of TIMELESS and PERIOD are affected in new timeless mutant. J Neurosci. 31(27):9982–9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M.. 1990. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343(6258):536–540. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C. 2001. The locomotor activity rhythm of Drosophila melanogaster is controlled by a dual oscillator system. J Insect Physiol. 47:877–887. [Google Scholar]
- Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, Nakagawa T, Lanjakornsiripan D, Nakayama KI, Fukada Y.. 2013. FBXL21 Regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152(5):1106–1118. [DOI] [PubMed] [Google Scholar]
- Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, Partch C, Green CB, Zhang H, Takahashi JS.. 2012. Crystal structure of the heterodimeric CLOCK: BMAL1 transcriptional activator complex. Science 337(6091):189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno T, Katagiri C, Numata H, Goto SG.. 2011. Causal involvement of mammalian-type cryptochrome in the circadian cuticle deposition rhythm in the bean bug Riptortus pedestris. Insect Mol Biol. 20(3):409–415. [DOI] [PubMed] [Google Scholar]
- Ikeno T, Numata H, Goto SG.. 2011. Photoperiodic response requires mammalian-type cryptochrome in the bean bug Riptortus pedestris. Biochem Biophys Res Commun. 410(3):394–397. [DOI] [PubMed] [Google Scholar]
- Johnson KP, Dietrich CH, Friedrich F, Beutel RG, Wipfler B, Peters RS, Allen JM, Petersen M, Donath A, Walden KKO, et al. 2018. Phylogenomics and the evolution of hemipteroid insects. Proc Natl Acad Sci U S A. 115(50):12775–12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamae Y, Tomioka K.. 2012. timeless is an essential component of the circadian clock in a primitive insect, the firebrat Thermobia domestica. J Biol Rhythms. 27(2):126–134. [DOI] [PubMed] [Google Scholar]
- Kaniewska MM, Vaněčková H, Doležel D, Kotwica-Rolinska J.. 2020. Light and temperature synchronizes locomotor activity in the Linden bug, Pyrrhocoris apterus. Front Physiol. 11:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara AY, Plotkin D, Espeland M, Meusemann K, Toussaint EFA, Donath A, Gimnich F, Frandsen PB, Zwick A, Dos Reis M, et al. 2019. Phylogenomics reveals the evolutionary timing and pattern of butterflies and moths. Proc Natl Acad Sci U S A. 116(45):22657–22663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Zheng X, Sehgal A.. 2006. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science 312(5781):1809–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S.. 1971. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 68(9):2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwica-Rolinska J, Chodakova L, Chvalova D, Kristofova L, Fenclova I, Provaznik J, Bertolutti M, Wu BC, Dolezel D.. 2019. CRISPR/Cas9 genome editing introduction and optimization in the non-model insect Pyrrhocoris apterus. Front Physiol. 10:891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwica-Rolinska J, Pivarciova L, Vaneckova H, Dolezel D.. 2017. The role of circadian clock genes in the photoperiodic timer of the linden bug Pyrrhocoris apterus during the nymphal stage. Physiol Entomol. 42(3):266–273. [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin XW, Maywood ES, Hastings MH, Reppert SM.. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98(2):193–205. [DOI] [PubMed] [Google Scholar]
- Kurien P, Hsu PK, Leon J, Wu D, McMahon T, Shi G, Xu Y, Lipzen A, Pennacchio LA, Jones CR, et al. 2019. TIMELESS mutation alters phase responsiveness and causes advanced sleep phase. Proc Natl Acad Sci U S A. 116(24):12045–12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW.. 2001. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105(6):769–779. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Ukai-Tadenuma M, Yamada RG, Houl J, Uno KD, Kasukawa T, Dauwalder B, Itoh TQ, Takahashi K, Ueda R, et al. 2007. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev. 21(13):1687–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna DD, Shin S, Ahrens D, Balke M, Beza-Beza C, Clarke DJ, Donath A, Escalona HE, Friedrich F, Letsch H, et al. 2019. The evolution and genomic basis of beetle diversity. Proc Natl Acad Sci U S A. 116(49):24729–24737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Pescatore S, Rosbash M.. 2014. CLOCK: BMAL1 is a pioneer-like transcription factor. Genes Dev. 28(1):8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, Frandsen PB, Ware J, Flouri T, Beutel RG, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346(6210):763–767. [DOI] [PubMed] [Google Scholar]
- Netusil R, Tomanova K, Chodakova L, Chvalova D, Doleze D, Ritz T, Vacha M.. 2021. Cryptochrome-dependent magnetoreception in a heteropteran insect continues even after 24 h in darkness. J Exp Biol. 224(19): jeb243000. [DOI] [PubMed] [Google Scholar]
- Nose M, Tokuoka A, Bando T, Tomioka K.. 2017. timeless2 plays an important role in reproduction and circadian rhythms in the cricket Gryllus bimaculatus. J Insect Physiol. 105:9–17. [DOI] [PubMed] [Google Scholar]
- Ozkaya O, Rosato E.. 2012. The circadian clock of the fly: a neurogenetics journey through time. Adv Genet. 77:79–123. [DOI] [PubMed] [Google Scholar]
- Peres R, Amaral FG, Marques AC, Neto JC.. 2014. Melatonin production in the sea star Echinaster brasiliensis (Echinodermata). Biol Bull. 226(2):146–151. [DOI] [PubMed] [Google Scholar]
- Peschel N, Chen KF, Szabo G, Stanewsky R.. 2009. Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, Jetlag, and Timeless. Curr Biol. 19(3):241–247. [DOI] [PubMed] [Google Scholar]
- Peschel N, Veleri S, Stanewsky R.. 2006. Veela defines a molecular link between Cryptochrome and Timeless in the light-input pathway to Drosophila's circadian clock. Proc Natl Acad Sci USA. 103(46):17313–17318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pivarciova L, Vaneckova H, Provaznik J, Wu BC, Pivarci M, Peckova O, Bazalova O, Cada S, Kment P, Kotwica-Rolinska J, et al. 2016. Unexpected geographic variability of the free running period in the Linden bug Pyrrhocoris apterus. J Biol Rhythms. 31(6):568–576. [DOI] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW.. 1998. Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94(1):83–95. [DOI] [PubMed] [Google Scholar]
- Putker M, Wong DCS, Seinkmane E, Rzechorzek NM, Zeng A, Hoyle NP, Chesham JE, Edwards MD, Feeney KA, Fischer R, et al. 2021. CRYPTOCHROMES confer robustness, not rhythmicity, to circadian timekeeping. EMBO J. 40(7):e106745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenfluh A, Abodeely M, Price JL, Young MW.. 2000. Isolation and analysis of six timeless alleles that cause short- or long-period circadian rhythms in Drosophila. Genetics 156(2):665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin EB, Shemesh Y, Cohen M, Elgavish S, Robertson HM, Bloch G.. 2006. Molecular and phylogenetic analyses reveal mammalian-like clockwork in the honey bee (Apis mellifera) and shed new light on the molecular evolution of the circadian clock. Genome Res. 16(11):1352–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC.. 1998. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93(5):805–814. [DOI] [PubMed] [Google Scholar]
- Saez L, Young MW.. 1996. Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron 17(5):911–920. [DOI] [PubMed] [Google Scholar]
- Schmid B, Helfrich-Forster C, Yoshii T.. 2011. A new ImageJ plug-in “ActogramJ” for chronobiological analyses. J Biol Rhythms. 26(5):464–467. [DOI] [PubMed] [Google Scholar]
- Sehgal A, Price JL, Man B, Young MW.. 1994. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science 263(5153):1603–1606. [DOI] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song WM, Kumar V, Hu YN, Lee C, Takahashi JS.. 2007. Circadian mutant overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129(5):1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smykal V, Bajgar A, Provaznik J, Fexova S, Buricova M, Takaki K, Hodkova M, Jindra M, Dolezel D.. 2014. Juvenile hormone signaling during reproduction and development of the linden bug, Pyrrhocoris apterus. Insect Biochem Mol Biol. 45:69–76. [DOI] [PubMed] [Google Scholar]
- Smýkal V, Pivarči M, Provazník J, Bazalová O, Jedlička P, Lukšan O, Horák A, Vaněčková H, Beneš V, Fiala I, et al. 2020. Complex rvolution of insect insulin receptors and homologous decoy receptors, and functional significance of their multiplicity. Mol Biol Evol. 37(6):1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC.. 1998. The cry(b) mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95(5):681–692. [DOI] [PubMed] [Google Scholar]
- Urbanová V, Bazalová O, Vaněčková H, Dolezel D.. 2016. Photoperiod regulates growth of male accessory glands through juvenile hormone signaling in the linden bug, Pyrrhocoris apterus. Insect Biochem Mol Biol. 70:184–190. [DOI] [PubMed] [Google Scholar]
- Wan G, Hayden AN, Iiams SE, Merlin C.. 2021. Cryptochrome 1 mediates light-dependent inclination magnetosensing in monarch butterflies. Nat Commun. 12(1):771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipfler B, Letsch H, Frandsen PB, Kapli P, Mayer C, Bartel D, Buckley TR, Donath A, Edgerly-Rooks JS, Fujita M.. 2019. Evolutionary history of Polyneoptera and its implications for our understanding of early winged insects. Proc Natl Acad Sci USA. 116:3024–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Guo Z, Yang Z, Han H, Wang S, Xu H, Yang X, Yang F, Wu Q, Xie W, et al. 2021. Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell 184(7):1693–1705 e1617. [DOI] [PubMed] [Google Scholar]
- Xu J, Jarocha LE, Zollitsch T, Konowalczyk M, Henbest KB, Richert S, Golesworthy MJ, Schmidt J, Dejean V, Sowood DJC, et al. 2021. Magnetic sensitivity of cryptochrome 4 from a migratory songbird. Nature 594(7864):535–540. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Ahmad M, Helfrich-Forster C.. 2009. Cryptochrome mediates light-dependent magnetosensitivity of drosophila's circadian clock. PLoS Biol. 7(4):e1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WJ, Houl JH, Hardin PE.. 2011. NEMO kinase contributes to core period determination by slowing the pace of the Drosophila circadian oscillator. Curr Biol. 21(9):756–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Metterville D, Briscoe AD, Reppert SM.. 2007. Insect cryptochromes: gene duplication and loss define diverse ways to construct insect circadian clocks. Mol Biol Evol. 24(4):948–955. [DOI] [PubMed] [Google Scholar]
- Zehring WA, Wheeler DA, Reddy P, Konopka RJ, Kyriacou CP, Rosbash M, Hall JC.. 1984. P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell 39(2 Pt 1):369–376. [DOI] [PubMed] [Google Scholar]
- Zhang L, Jones CR, Ptacek LJ, Fu YH.. 2011. The genetics of the human circadian clock. Adv Genet. 74:231–247. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Markert MJ, Groves SC, Hardin PE, Merlin C.. 2017. Vertebrate-like CRYPTOCHROME 2 from monarch regulates circadian transcription via independent repression of CLOCK and BMAL1 activity. Proc Natl Acad Sci USA. 114(36):E7516–E7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylka MJ, Shearman LP, Levine JD, Jin XW, Weaver DR, Reppert SM.. 1998. Molecular analysis of mammalian timeless. Neuron 21(5):1115–1122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
cDNA and genome contigs connected to the project are deposited in the GenBank (see supplenentary table 3, Supplementary Material online). All data are incorporated into the article and its online Supplementary material.