Abstract
Island biogeography is one of the most powerful subdisciplines of ecology: its mathematical predictions that island size and distance to mainland determine diversity have withstood the test of time. A key question is whether these predictions follow at a population-genomic level. Using rigorous ancient-DNA protocols, we retrieved approximately 1,000 genomic markers from approximately 100 historic specimens of two Southeast Asian songbird complexes from across the Sunda Shelf archipelago collected 1893–1957. We show that the genetic affinities of populations on small shelf islands defy the predictions of geographic distance and appear governed by Earth-historic factors including the position of terrestrial barriers (paleo-rivers) and persistence of corridors (Quaternary land bridges). Our analyses suggest that classic island-biogeographic predictors may not hold well for population-genomic dynamics on the thousands of shelf islands across the globe, which are exposed to dynamic changes in land distribution during Quaternary climate change.
Keywords: Sundaland, Quaternary glacial cycles, babblers, ancient DNA, paleorivers
Introduction
Islands contribute disproportionately toward our understanding of the evolutionary process (MacArthur and Wilson 1967; Whittaker et al. 2017). They provide a window into the workings of the evolutionary forces of isolation, migration, speciation and extinction, as well as the interactions among them (MacArthur and Wilson 1967; Borregaard et al. 2016; Whittaker et al. 2017). MacArthur and Wilson’s (1967) seminal work on island biogeography suggested that the two main factors determining the species diversity of a given island depended upon its area and distance from the mainland. However, island size and distance to mainland may not provide a complete picture (Borregaard et al. 2016; Fernández‐Palacios et al. 2016; Weigelt et al. 2016; Whittaker et al. 2017), with the role of Quaternary glaciations and concomitant sea-level fluctuations also pertinent to island biogeography (Fernández‐Palacios et al. 2016; Weigelt et al. 2016). Cyclical periods of global cooling and warming over the past approximately 2.6 My have not only been an engine of diversification at higher latitudes and altitudes, but also affected gene flow and speciation patterns in shelf-island regions (i.e., islands on continental shelves), especially in Southeast Asia (Heaney 1986; Hewitt 2000, 2004; Moyle et al. 2009; Lim and Sheldon 2011; Brown et al. 2013; Chattopadhyay et al. 2017; Ng et al. 2017; Garg et al. 2018, 2020; Cros, Chattopadhyay, et al. 2020; Rheindt et al. 2020). Many present-day shelf-islands have been connected by land bridges during cooler Quaternary periods with lower sea levels, allowing for possible dispersal of terrestrial organisms and gene flow (Lim and Sheldon 2011; Lim et al. 2011, 2017; Leonard et al. 2015; Ng et al. 2017; Garg et al. 2018; Cros, Chattopadhyay, et al. 2020).
A key question is understanding the mechanisms by which Quaternary land bridges provide a conduit of gene flow for terrestrial organisms. Such land bridges can be highly species-specific conduits of gene flow, depending upon ecological requirements, and may be semipermeable by allowing more gene flow in one direction than in the other (Garg et al. 2018; Cros, Chattopadhyay, et al. 2020). However, a precise quantification of the amount of gene flow that is facilitated by Quaternary land bridges remains problematic, as many organismic groups are capable of active or passive overwater dispersal (Mayr et al. 2001), obviating the need to evoke land bridges as the sole explanatory variable for dispersal events among present-day shelf-islands (Ng et al. 2017).
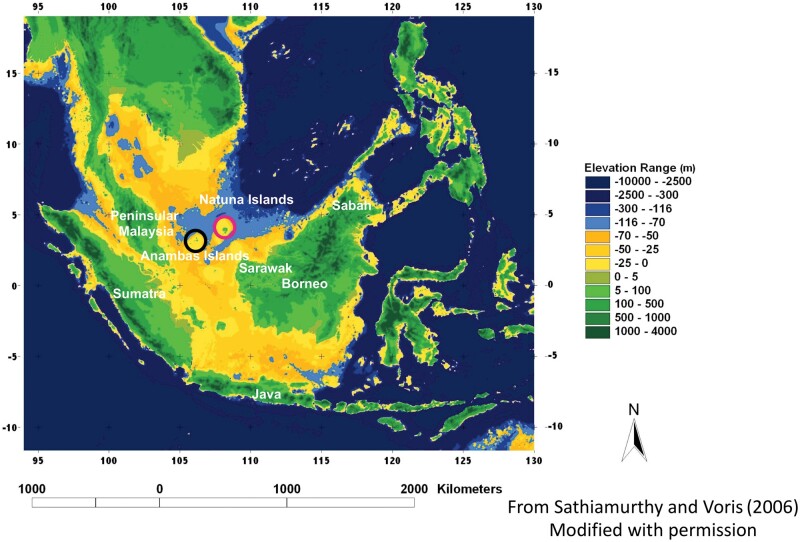
In this study, we directly tested the importance of Quaternary land bridges in facilitating gene flow through population-genomic analysis of two terrestrial rainforest songbird species complexes across the Sundaic Region (Sundaland) in Southeast Asia. Sundaland constitutes the most complex shelf-island archipelago in the world, encompassing the present-day landmasses of Sumatra, Java, Borneo, Peninsular Malaysia (henceforth: the Peninsula), plus numerous satellite islands (fig. 1), and has undergone the most pronounced changes in land distribution globally across the Quaternary cooling cycles of the past 2.6 My. During Quaternary cooling cycles, sea levels drop as water is locked up in the form of ice at higher latitudes, leading to the emergence of land bridges connecting Sumatra, Java, Borneo, and numerous smaller islands with the mainland, forming Sundaland (fig. 1). The widely disjunct historic distribution of orangutans and Asian rhinoceroses on different islands, separated by approximately 500 km of open sea, bears testament to the frequent occurrence of connecting land bridges in the recent past (Nater et al. 2017; Mays et al. 2018). Consequently, Sundaland has become one of the main model regions to test impacts of Quaternary sea level change on evolutionary processes (Lim and Sheldon 2011; Lim et al. 2011, 2017; Leonard et al. 2015; Cros, Chattopadhyay, et al. 2020).
Fig. 1.
Map showing Sundaland and sea depths across the Sunda Shelf. Note that land would have extended approximately to the −116 m isobath (border between light-blue and midblue) at the peak of glacial episodes.
We leveraged historic museum specimens collected during the 1800s and early 1900s to include samples from remote islands that are difficult to sample in modern times. By determining the genomic affinity of populations on key strategic island groups, we disentangle overwater dispersal from terrestrial dispersal across past land bridges. The study of underwater depth—bathymetry—is important in determining the level of island isolation: for instance, whereas the small Natuna islands are geographically close to Borneo, bathymetric data indicate that their land connection to Borneo severed far earlier than did their connection to Sumatra and the Peninsula (fig. 1). A genomic affinity of Natuna populations with those on Sumatra or the Peninsula, despite Natuna’s proximity to Borneo, would be a powerful indication that gene flow among these birds is purely governed by the distribution of Quaternary land bridges. Equally, a closer affinity with Borneo would argue for considerable levels of overwater dispersal, especially during periods when intervening water distances were smaller.
We focus on two widespread and characteristic Sundaic bird species complexes, the black-capped babbler (Pellorneum capistratum) and short-tailed babbler (P. malaccense), both sedentary rainforest denizens foraging in the understory (Eaton et al. 2016). Both these songbirds are known for their ubiquitous occurrence across a wide range of Sundaic lowland and hill rainforests and their general inability to cross even narrow nonforest gaps, especially open water (Zakaria et al. 2014; Sadanandan and Rheindt 2015; Cros, Ng, et al. 2020). We characterized and compared variation in plumage patterns using a series of museum specimens to assess whether morphological differentiation is congruent with the population-genomic signal. Museums are a treasure trove of important phenotypic and genomic information from remote areas. However, DNA from century-old museum specimens, especially from the tropics, is often heavily degraded and only minuscule amounts can be salvaged (Dabney et al. 2013; Chattopadhyay et al. 2019). Hence, we used a target capture approach to ensure high quality genome-wide data would be obtained from multiple samples. This approach has been successfully utilized previously for similarly degraded samples (Chattopadhyay et al. 2019). At the same time, we implemented multiple cleanup steps and analytical approaches to account for excess DNA damage and increased levels of C to T and G to A substitutions in the ancient DNA of our samples (Dabney et al. 2013; Chattopadhyay et al. 2019).
We harvested approximately 1,000 genome-wide sequence loci targeting approximately 1% of the genomes and employed modern approaches of admixture analysis, allowing us to shed light on patterns of isolation and divergence across Sundaland and the role of Quaternary land bridges in dictating dispersal. We tested whether distance to the mainland is the main determinant of population genomic patterns, or whether bathymetric data—reflective of Quaternary land bridges and the course of paleo-rivers—define the population genomic structure of our target species. Based on the first hypothesis, the population genomic affinity of small-island bird populations, for example, on Natuna, will be similar to that of birds from nearby island of Borneo (fig. 1). However, if the history of land connections is more important, then the affinity of these island populations should be with populations from the Peninsula and Sumatra (fig. 1).
Results
Plumage Analysis
We compared the plumages of museum specimens of P. capistratum and P. malaccense (see supplementary table 1, Supplementary Material online) deposited at the Lee Kong Chian Natural History Museum (Singapore). Within the P. capistratum complex, we observed three groups based on plumage. The most striking plumage difference among populations was the color of the supercilium, which divided specimens into: 1) a cluster in western Sundaland (subspecies P. c. nigrocapitatum; Peninsula, Sumatra, and Natuna) characterized by a gray supercilium with thin white shaft streaks, 2) a Bornean cluster (subspecies P. c. capistratoides at least from Sarawak and P. c. morrelli at least from Sabah) with a wholly white supercilium, and 3) a Javan cluster (subspecies P. c. capistratum) with an orange anterior and white posterior supercilium (supplementary fig. 1A–C, Supplementary Material online). Similarly, lores were white or pale gray in Javan specimens, midgray in western Sundaic specimens, and dark grey in Bornean specimens (supplementary fig. 1A, B, and D, Supplementary Material online). All western Sundaic specimens had a black moustachial stripe (supplementary fig. 1C, Supplementary Material online) that was absent in Javan and Bornean specimens. Additionally, specimens from Java had a paler tail and back than all other populations (supplementary fig. 1D, Supplementary Material online), and a uniquely orange lateral suffusion on the white throat.
In western Sundaland, we observed consistent differences across specimens from the Thai-Malay Peninsula, Sumatra, and Natuna. Specimens from the Peninsula had a brownish black crown, whereas Natuna and Sumatran specimens usually had a more saturated black crown. Similarly, on Borneo, specimens from Sabah had a paler orange belly and breast than specimens from Sarawak. Interestingly, Sarawak specimens’ belly and breast coloration resembled that of specimens from western Sundaland. We did not compare the coloration of legs, bills, and iris due to postmortem color change and incomplete label data.
We observed a high variability in plumage within the P. malaccense complex, much of it seemingly independent of location (supplementary fig. 1E–G, Supplementary Material online). Nevertheless, a few geographically consistent differences did emerge: Bornean populations (subspecies P. m. poliogene and P. m. saturatum) differed from those in western Sundaland (subspecies P. m. malaccense) in having a more olive rather than chestnut back (mantle and wings) (supplementary fig. 1E–G, Supplementary Material online). Within Borneo, specimens from Sabah (subspecies P. m. poliogene) exhibited a more rufous tail, whereas specimens from Sarawak showed a less warm-colored brown tail (supplementary fig. 1E and G, Supplementary Material online). Interestingly, all the other differences between Sarawak and Sabah also differentiated Sarawak populations from western Sundaland. Specimens from Sarawak appeared the most distinct population based on plumage alone: they had a darker crown, darker ear coverts, and darker moustachial stripe than all other taxa. They also had more intensely orange flanks and breast whereas other populations had paler, more apricot-colored flanks and breast (supplementary fig. 1E, Supplementary Material online). Again, comparisons of the coloration of legs, bills, and iris were impossible because of postmortem color change and a lack of sufficient label information.
Sampling, DNA Extraction, and Raw Data Filtering
We obtained toepads of specimens of both P. capistratum (n = 50) and P. malaccense (n = 46) from the Lee Kong Chian Natural History Museum (Singapore) and Yale Peabody Museum of Natural History (New Haven, Connecticut) (supplementary table 1, Supplementary Material online). These specimens were collected over a period of approximately 70 years from 1893 to 1957. All historical samples were processed in a separate dedicated facility and with fresh gloves, forceps, and scalpels for each sample to avoid cross contamination.
We successfully isolated DNA from approximately 80% of historic samples (n = 77). Presence of DNA was confirmed using Qubit and an AATI Fragment Analyzer. No detectable DNA was observed in negative controls. All 77 samples along with negative controls were further processed for library preparation and target enrichment. Target enrichment protocols have been shown to be highly effective for ancient DNA samples (Chattopadhyay et al. 2019). We designed target loci (960 loci) for sequence capture protocols that are useful for both population genomic and phylogenomic studies targeting both conserved exons and variable intronic regions (see Materials and Methods for details). We supplemented historical DNA with fresh samples from Sabah (n = 12) (supplementary table 1, Supplementary Material online) following the fieldwork protocols of Cros, Chattopadhyay, et al. (2020). All enriched libraries were sequenced on multiple lanes of HiSeq 4000 (150-bp paired-end runs). The fresh samples from Sabah were processed separately and sequenced in a dedicated lane. We retained approximately 0.97 billion reads after cleanup steps (supplementary table 1, Supplementary Material online). The average number of reads per sample was approximately 11 million (SD=∼5 million).
Data Matrix
For DNA sequence-based analysis, we obtained sequence data for 944 out of 960 target loci designed for both species complexes using the HybPiper pipeline. We removed ten historic P. malaccense samples from downstream processing as their missing data exceeded 85%. After multiple sequence alignments, we performed stringent filtering of alignments using Gblocks and removed 117 loci from the P. capistratum data set and 385 loci from the P. malaccense data set due to high missing data. After removal of loci <200 bp and Z-chromosomal loci, we retained 652 loci for P. capistratum and 314 loci for P. malaccense. The total sequence matrix length for P. capistratum was 454,712 bp (average locus length = 697 bp; minimum = 201 bp; maximum = 4,723 bp) and 145,877 bp for P. malaccense (average locus length = 465 bp; minimum = 201 bp; maximum = 2,607 bp).
For SNP-based analysis, we generated four different data sets for each species complex (data set I: all SNPs obtained after mapping to Mixornis gularis genome; data set II: only transversions obtained after mapping to M. gularis genome; data set III: single random SNP per target locus; data set IV: only transversions obtained after mapping to Parus major genome). We retrieved between 960 and 208,186 SNPs for P. capistratum and between 958 and 198,711 SNPs for P. malaccense across data sets before filtering (table 1). For data set IV, after filtering for linkage, deviations from Hardy–Weinberg equilibrium, neutrality, and DNA damage, we retained 40,611 transversions for P. capistratum and 34,809 transversions for P. malaccense. As admixture graph analysis can accommodate SNPs located on the Z chromosome, we included these in our analysis of gene flow. In contrast, for analyzing population structure, we removed the outgroup and any resulting monomorphic loci along with SNPs located on the Z chromosome and retained 38,463 transversions for P. capistratum and 32,735 transversions for P. malaccense (table 1). The overall level of missing data for data set IV was less than 10% for both species after all cleanup steps. For the other data sets, the number of SNPs after cleanup is summarized in table 1.
Table 1.
Number of SNPs Identified and Filtered for Different Data Sets.
| Data Set | Number of Unfiltered SNPs | Number of SNPs Removed due to Linkage | Number of SNPs Removed That Were Not in HWE | Number of Nonneutral SNPs Removed | Number of SNPs Removed Mapping to Z Chromosome | Number of Transitions Removed | Number of SNPs Retained | Number of SNPs with No Missing Data |
|---|---|---|---|---|---|---|---|---|
| Pellorneum capistratum | ||||||||
| Data set I: all SNPs obtained after mapping to Mixornis gularis genome | 82,468 | 24,025 | 1,904 | 82 | 1,246 | NA | 54,906 | 54,906 |
| Data set II: only transversions obtained after mapping to M. gularis genome | 82,468 | 24,025 | 1,904 | 82 | 1,246 | 37,690 | 17,216 | 17,216 |
| Data set III: single random SNP per target locus | 960 | NA | 30 | 12 | 35 | NA | 883 | 883 |
| Data set IV: only transversions obtained after mapping to Parus major genome | 208,186 | 67,451 | 3,357 | 1,130 | 1,470 | 96,315 | 38,463 | 10,848 |
| Pellorneum malaccense | ||||||||
| Data set I: all SNPs obtained after mapping to M. gularis genome | 57,300 | 21,595 | 440 | 240 | 1,317 | NA | 33,708 | 33,708 |
| Data set II: only transversions obtained after mapping to M. gularis genome | 57,300 | 21,595 | 440 | 240 | 1,317 | 22,558 | 11,150 | 11,150 |
| Data set III: single random SNP per target locus | 958 | NA | 17 | 2 | 44 | NA | 895 | 895 |
| Data set IV: only transversions obtained after mapping to Parus major genome | 198,711 | 83,460 | 749 | 1,490 | 1,491 | 78,786 | 32,735 | 6,091 |
Note.—HWE, Hardy–Weinberg equilibrium; NA, not applicable.
Phylogenomic Reconstruction
We used both concatenation approaches and species tree reconstruction for phylogenomic analysis (see Materials and Methods). Sumatran and Natuna populations were embedded within the peninsular population based on the concatenated maximum likelihood trees in both species complexes (supplementary fig. 2A and C, Supplementary Material online). Our sole P. malaccense individual from the Anambas archipelago (fig. 1) also formed part of the large peninsular clade (supplementary fig. 2C, Supplementary Material online). In P. capistratum, the Javan population was distinct from both peninsular/Sumatran and Bornean populations, emerging as sister to the latter (supplementary fig. 2A and B, Supplementary Material online). In the case of P. malaccense, the Sabah population formed a clade distinct from Sarawak and basal to all members of the complex (supplementary fig. 2C and D, Supplementary Material online).
Population Structure
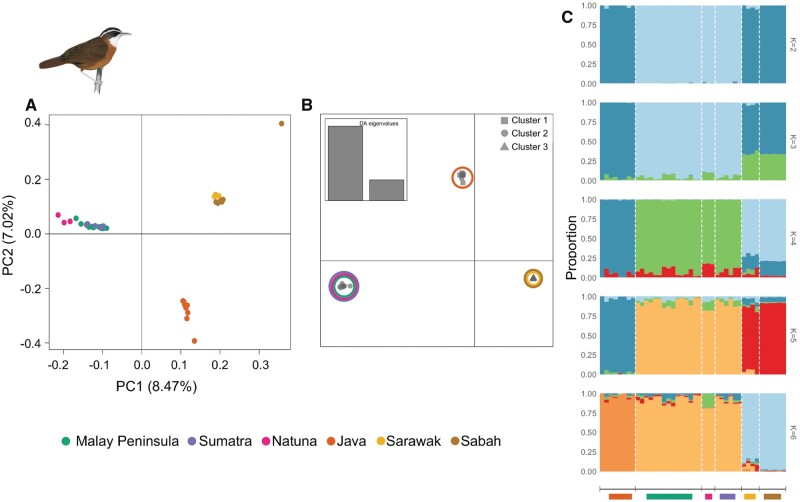
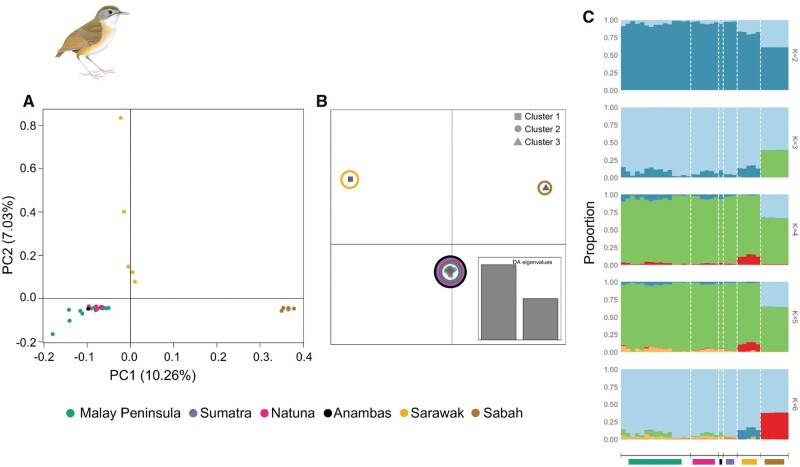
We employed multiple approaches to understand population structure within each babbler species complex and observed similar trends of subdivision across all four SNP data sets generated in this study (figs. 2 and 3; supplementary figs. 3–8, Supplementary Material online). Principal component analysis (PCA) and discriminant analysis of principal components (DAPC) suggested three distinct population groupings in each complex in agreement with phylogenomic results (figs. 2A and Band 3A and B; supplementary fig. 2, Supplementary Material online). In P. capistratum, the division entailed: 1) a western Sundaic group comprising populations from the Peninsula, Sumatra, and Natuna, 2) a Javan group, and 3) a Bornean group (fig. 2 and supplementary figs. 3–5, Supplementary Material online). In P. malaccense, Bornean populations separated into two deeply divergent groups, one from Sarawak and the other from Sabah (fig. 3 and supplementary figs. 6–8, Supplementary Material online) in agreement with previously published studies based on mitochondrial DNA (Lim and Sheldon 2011; Sadanandan and Rheindt 2015) and the phylogenomic analysis presented in this study (supplementary fig. 2C and D, Supplementary Material online). The Sarawak cluster emerged as more closely related to the third group consisting of individuals from the Peninsula, Sumatra, Natuna, and Anambas (fig. 3 and supplementary figs. 6–8, Supplementary Material online).
Fig. 2.
Population subdivision observed in Pellorneum capistratum based on transversions isolated after mapping to the Parus major genome. Three population clusters observed based on (A) principal component analysis (number of transversions=10,848), (B) discriminant analysis (DA) of principal components, in which clusters are color coded based on population affinity (number of transversions=38,463), (C) genetic assignment of P. capistratum individuals using STRUCTURE for K = 2 to K = 6 (number of transversions=38,463).
Fig. 3.
Population subdivision observed in Pellorneum malaccense based on transversions isolated after mapping to the Parus major genome. Three population clusters observed based on (A) principal component analysis (number of transversions=6,091), (B) discriminant analysis (DA) of principal components, in which clusters are color coded based on population affinity (number of transversions=32,735), (C) genetic assignment of P. malaccense individuals using STRUCTURE for K = 2 to K = 6 (number of transversions=32,735).
The results based on the Bayesian clustering program STRUCTURE were congruent with other analyses for P. capistratum, in which the western Sundaic cluster (Peninsula, Sumatra, Natuna) separated from the other clusters (Borneo and Java) at K = 2 (fig. 2C), with the latter two separating at K = 3 (fig. 2C and supplementary figs. 3C, 4C, and 5C, Supplementary Material online). STRUCTURE results were less clean for P. malaccense, in which only Sabah emerged as clearly distinct for K = 3 (and sometimes K = 2), whereas the Sarawak cluster did not emerge as visually distinct before K = 4 (fig. 3C and supplementary figs. 6C, 7C, and 8C, Supplementary Material online).
Gene Flow Dynamics
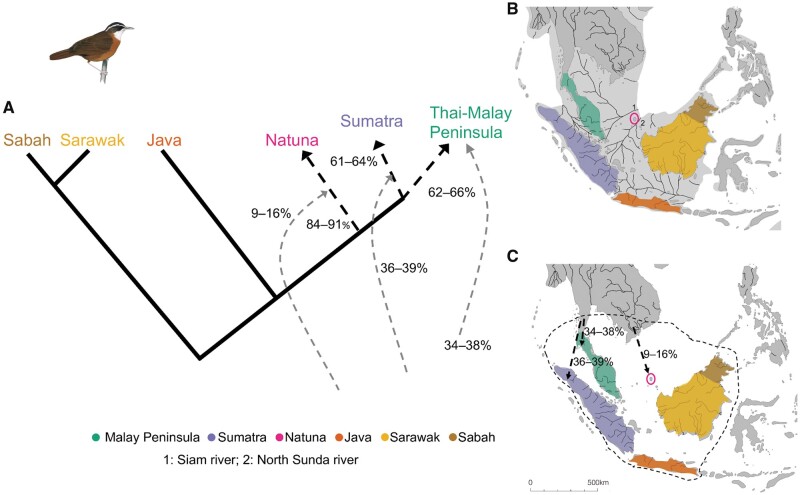
The application of D-statistics and admixture graph analysis allowed us to infer the complicated nature of gene flow events and dynamics among populations of all the major Sundaic landmasses investigated (figs. 4 and 5; supplementary table 2, Supplementary Material online). For P. capistratum, 781 out of 63,725 possible graphs tested by qpbrute exhibited a fit with our data when considering all populations. As this number of graphs was computationally intractable for Bayes factor estimation, we performed subsequent qpbrute analysis in two steps. Initially, we included all populations other than Natuna in our analysis and obtained 41 possible solutions. After Bayes factor estimation, we selected ten graphs as the most likely starting models. For the selected ten graphs, we then included the Natuna population, resulting in 12 unique graphs. Following another round of Bayes factor estimation, five of these admixture graphs displayed a good fit with the data (fig. 4A). These five admixture graphs had identical topologies and only differed slightly in estimates of admixture proportions for the western Sundaic populations (fig. 4). They suggested a lack of substantial gene flow between lineages from Java, Borneo, and western Sundaland, but pronounced allelic contributions into the three western Sundaic populations from unsampled sources, likely now-extinct populations from the north.
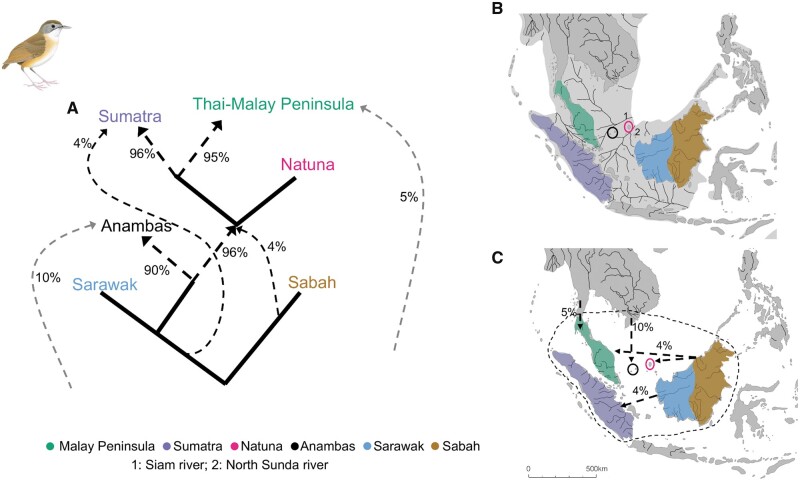
Fig. 4.
Admixture graph of gene flow dynamics observed in Pellorneum capistratum. (A) Schematic of genetic ancestry and admixture proportions among populations of P. capistratum based on a combination of five best-fit admixture graphs, all of which exhibited an identical topology and only differed slightly in estimates of the extent of admixture from an unknown source for the western Sundaic population; (B) map during sea level recession of approximately 120 m at glacial maximum around 20,000 years before present, showing major paleo-rivers and present-day taxon distributions; (C) current map of South East Asia with taxon distributions, major rivers, and arrows indicating potential gene flow from unsampled or extinct populations (see A). The black-stippled line indicates potential habitat during sea level lows. Panels (B) and (C) modified with permission from Voris (2000).
Fig. 5.
Admixture graph of gene flow dynamics observed in Pellorneum malaccense. (A) Schematic of genetic ancestry and admixture proportions among populations of P. malaccense based on single best-fit admixture graph; (B) map during sea level recession of approximately 120 m at glacial maximum around 20,000 years before present, showing major paleo-rivers and present-day taxon distributions; (C) current map of South East Asia with taxon distributions, major rivers, and arrows indicating potential gene flow among populations or from unsampled or extinct populations (see A). The black-stippled line indicates potential habitat during sea level lows. Panels (B) and (C) modified with permission from Voris (2000).
We obtained a single possible admixture graph for P. malaccense out of 9,083 unique graphs tested by qpbrute (fig. 5). This graph supported postdivergence gene flow from Sabah into western Sundaic populations but not into the adjacent Sarawak population, indicating a potentially strong reproductive barrier between the two Bornean lineages. It also suggested various streams of ancestral allelic contributions into most western Sundaic populations from unsampled, possibly extinct sources (fig. 5).
Discussion
Bathymetric Topography Predicts Genomic Affinity of Island Populations
For both songbird species complexes under study, all phylogenomic and population-genomic approaches unanimously confirmed that Natuna and Anambas island populations are firmly embedded with the peninsular—not Bornean—population cluster (figs. 2 and 3; supplementary figs. 2–8, Supplementary Material online). These conclusions were further corroborated by plumage analysis (supplementary fig. 1, Supplementary Material online). These insights defy the fact that Natuna is only approximately 220 km from the nearest populations on Borneo, less than half (∼46%) the distance to the nearest population in Peninsular Malaysia (∼480 km) (fig. 1). Our results unequivocally support the hypothesis that the history of land connections, as dictated by sea level changes and bathymetry, has determined the genetic affinity of shelf-island populations, and that overwater dispersal-related processes have been of much less importance.
The distance of an island to the nearest mainland has long served as one of the central tenets of classical island biogeography in making inferences about island biota (MacArthur and Wilson 1967). Geographic distance to the mainland has been accepted as the natural criterion explaining why many island biotas are exclusively recruited from one landmass versus the other. Well-documented examples include the American origin of most species inhabiting Bermuda (Sterrer et al. 2004), the European provenance of most species on the Azores (Wallace 1872), and the affinities of British animal populations with those from nearby France, rather than with Scandinavian or Central European populations that share more similarities in climatic regimes they are adapted to (Taberlet et al. 1998; Hewitt 2000, 2004; Teacher et al. 2009).
An improvement in our understanding of paleo-climate has led to a realization of the importance of Quaternary sea level change in defining the evolutionary history of terrestrial biota (Heaney et al. 2005; Lim et al. 2011; Ng et al. 2017; Garg et al. 2018; Cros, Chattopadhyay, et al. 2020), and has resulted in a new appreciation of bathymetry as a crucial indicator of the extent of land bridges which existed only a few thousand years ago (Garg et al. 2018; Rheindt et al. 2020). Inspection of the bathymetric profile of Sundaland suggests that the land connection between the Peninsula and Natuna/Anambas persisted approximately 1,000 years longer than between Borneo and Natuna/Anambas (Sathiamurthy and Voris 2006) (fig. 1), supporting the importance of bathymetry and paleo-island distribution in determining the genomic composition of smaller island populations. At the macroevolutionary level, a reanalysis of species diversity patterns across the planet has shown that Quaternary land connections are an important but overlooked parameter in defining island species diversity (Weigelt et al. 2016) and faunal turnover (Lohman et al. 2011). For instance, one of the steepest and most renowned faunal transition zones runs across Wallace’s line (Wallace 1860; Huxley 1868; Lohman et al. 2011), separating Sundaland from the Australo-Papuan faunal region. Although only separated by narrow straits, land masses to the east and west of Wallace’s line harbor biota of extremely different affinities, attesting to the importance of the deep sea trenches that have precluded the formation of land bridges across this narrow gap (Wallace 1860).
At the population-genetic level, there has so far been a distinct lack of understanding whether the genomic composition of terrestrial populations on present-day shelf islands is largely defined by geographic distance to the nearest large landmass, or by bathymetric topography and consequently by the duration of land connections during the Pleistocene. Genome-wide markers have been instrumental in answering whether shelf island populations are mostly the product of overwater dispersal or of dispersal across historic land connections, which would be difficult to ascertain based on phenotypic data alone (Ng et al. 2017). As Quaternary glacial cycles leave a track record within the genome, patterns of genomic diversity can be used to reconstruct evolutionary history (Lim et al. 2017; Ng et al. 2017; Papadopoulou and Knowles 2017; Garg et al. 2018; Cros, Chattopadhyay, et al. 2020; Rheindt et al. 2020).
Paleo-Rivers: A Long-Overlooked Determinant of Population-Genetic Structure
The role of big rivers in shaping population structure in large areas of tropical rainforest has received much interest. In South America, the Amazon River and its tributaries are known as important barriers between neighboring subspecies and closely related species, many of which have recently been taxonomically upgraded with an improved biological understanding (Isler et al. 2001; Rheindt et al. 2008, 2009; Burney and Brumfield 2009; Isler and Maldonado-Coelho 2017). Likewise, in Africa, the Congo River is an important divide between young, recently separated species (Prüfer et al. 2012). In Southeast Asia, on the other hand, most rainforest is fragmented archipelagically, and open sea dividing different islands is considered the main shaping force of population structure, with rivers afforded a minor role (Lim et al. 2011, 2017; Lohman et al. 2011; Mason et al. 2019; Cros, Chattopadhyay, et al. 2020). At the same time, the present division of Sundaland into a handful of large and numerous smaller islands only represents a snapshot in time, as all larger landmasses in Sundaland have been connected by land bridges for a cumulative total of approximately 90% of the last 1 My (Cannon et al. 2009; Mason et al. 2019; Sarr et al. 2019). Therefore, rivers may have played a much larger role in shaping population structure here than previously appreciated.
Our historic museum samples did not encompass a sufficient number of sites to systematically test the divergence effects of large paleo-rivers in Sundaland. However, it did allow us to inspect whether population-genetic divisions are consistent with the river barrier effect specifically in the case of the two small shelf-island archipelagos of Natuna and Anambas. During the global sea-level lows, Natuna has been connected to the Peninsula further west by a hilly watershed with adjacent flat valleys, covered by evergreen tropical rainforest ideal for the babblers under study (Bird et al. 2005; Cannon et al. 2009). On the eastern side, a large paleo-river, the North Sunda River, originating in the Central Sumatran mountains and debouching in the South China Sea, separated Natuna from Bornean land extensions further south (Voris 2000; Bird et al. 2005) (figs. 4 and 5). Natuna is situated close to the former delta of the North Sunda River, which is the longest exclusively Sundaic river during times of land emergence (Voris 2000). With its length of almost 2,000km forming a vast tropical rainforest watershed, it would have featured a wide lower course and delta, and would have been equivalent in impact to some Amazonian tributaries of similar length, for example, the Xingu River, that are also known to constitute important population-genetic barriers (da Costa et al. 2016; Isler and Maldonado-Coelho 2017). Paleo-rivers, which have hitherto been afforded little importance in accounting for population subdivisions in Sundaland, offer a compelling explanation for Natuna’s deeper genetic rift from Borneo: Natuna’s placement just north-west of the delta of the North Sunda Paleo-River, the largest tropical Asian river at the time, would have prevented small and poorly dispersive forest inhabitants from easily crossing over toward Borneo even at times when extensive land connections existed.
Our data indicate that Sundaic paleo-rivers other than the North Sunda River may also have had an important imprint on population structure. North of present-day Natuna and Anambas, the Siam Paleo-River system constituted an extension of the present-day Chao Phraya, the most dominant river in the Thai plains (figs. 4 and 5), extending the length of the latter 2- to 3-fold during periods of land emergence (Voris 2000). North of the Siam river, there would have been extensive areas of lowland rainforest that are largely submerged now (Cannon et al. 2009), but have historically survived at the southernmost tip of Vietnam, where a number of Sundaic vertebrates have their northernmost isolated outposts (e.g., see Robson [2005]). Pellorneum malaccense and P. capistratum no longer occur in this Sundaic outpost but may have survived in this area into the present interglacial and gone extinct as a result of the historic destruction of all rainforests here. An ancestral allelic contribution of 10% in the Anambas population of P. malaccense from an unsampled, possibly extinct population may well relate to gene flow across the Siam Paleo-River from a diverged northern population that still existed there at the time (figs. 4 and 5). By the same token, a similar ancestral allelic contribution of 9–16% into the Natuna population of P. capistratum may reflect gene flow from a northern, now-extinct stronghold across the Siam Paleo-River into populations that are now stranded on Natuna (fig. 4).
Importance of Museum Collections
Our study demonstrates the timeless importance of historic specimen collections for evolutionary research. In modern times when DNA collection is becoming ever more restrictive, one of the best paths to comprehensive genomic sampling is through historic museum collections. Ancient DNA is prone to degradation and damage through an excess in C to T and G to A substitutions, and the implementation of multiple safeguards is necessary to avoid bias in DNA sequence generation from historic specimens. Our study highlights the utility of target enrichment methods to isolate homologous genomic regions across multiple degraded historic museum samples, and to harvest the DNA signal of hundreds of genomic markers which can capture both phylogenomic and population genomic information. Our study was based on specimens containing highly degraded DNA from the Lee Kong Chian Museum (formerly the Raffles Museum) in Singapore, held at tropical temperatures for many decades before air-conditioning was introduced in the approximately 1980s. Our successful retrieval of thousands of genome-wide SNPs through the rigorous application of ancient DNA protocols to reduce artifacts due to excess damage demonstrates that even degraded historic museum material can serve an important purpose in molecular research. We hope this approach will be applied to numerous additional organismic groups across the tropics to solve evolutionary problems.
Materials and Methods
Plumage Analysis
We examined the plumages of 41 museum specimens of P. capistratum and 45 museum specimens of P. malaccense (supplementary table 1, Supplementary Material online) deposited at the Lee Kong Chian Natural History Museum (Singapore). Specifically, we laid out specimen series arranged by geographic area and compared plumage hues for wings, tail, upperparts, underparts, and head. We checked label information for the color of beak, legs, and iris on live birds.
DNA Extraction
DNA extractions of historical samples were performed in a separate dedicated ancient DNA facility within a biosafety cabinet. We used DNeasy Blood and Tissue Kits (QIAGEN, Germany) with modifications to extract highly degraded DNA (see Chattopadhyay et al. [2019] for details). We used one or two toepads per sample for DNA extraction. Prior to DNA extraction the toepads were washed two to three times with molecular grade water to remove any PCR inhibitors. We used 360 µl of ATL buffer and approximately 100 µl of Proteinase K per sample to digest the tissue. The toepads generally required approximately 3–5 days to completely digest. The volumes of AL buffer and ethanol were adjusted according to the total volume of ATL buffer and Proteinase K. We used Minelute columns (QIAGEN, Germany) for DNA extraction instead of the regular columns provided by the manufacturer as these can help elute single stranded DNA as well as small DNA fragments. As historical samples are highly degraded, these columns were helpful in isolating poor quality DNA. DNA extracted from historical samples was then quantified using an AATI Fragment Analyzer as well as high sensitivity Qubit Assay kits (Invitrogen). For all DNA extractions, we carried through a negative control to detect possible contamination. For fresh samples, we extracted DNA from blood using DNeasy Blood and Tissue Kits following the manufacturer’s instructions and quantified DNA using high sensitivity Qubit Assay kits.
Library Preparation
We prepared whole-genome libraries using NEB II Ultrakits (New England BioLabs). For ancient samples, we included an additional step of DNA repair using the FFPE DNA repair kit (New England BioLabs) prior to library preparation. Historical samples are prone to DNA damage and this step reduced the levels of damage observed in later steps (Chattopadhyay et al. 2019). We carried through a dedicated negative control for each batch of library preparation. For fresh samples, we used a bioruptor pico (Diagenode, Belgium) to fragment the DNA, performing 13 cycles of 30 s ON and 30 s OFF to obtain DNA fragments of approximately 250 bp. Samples were then subjected to library preparation using the NEB Ultra II kit. All samples were dual indexed using 8-bp barcodes.
Design of Target Loci
Target enrichment protocols have been shown to be highly effective for ancient DNA samples (Chattopadhyay et al. 2019). We designed target loci for sequence capture protocols that are useful for both population genomic and phylogenomic studies targeting both conserved exons and variable intronic regions. We used EvolMarkers (Lim et al. 2011) to identify conserved single copy coding sequences in the striped tit-babbler genome (M. gularis, QVAJ00000000.1), collared flycatcher genome (Ficedula albicollis, GCA_000247815.1), and zebra finch genome (Taeniopygia guttata, GCF_003957565.1). The striped tit-babbler genome is the phylogenetically closest genome available for both target species. To identify conserved exons, EvolMarkers performs a BLAST search, for which we set a minimum of 55% identity and e-value of less than 10E-15. Only exons longer than 500 bp were used for downstream analysis. We identified a total of 1,161 exons. Then we isolated 500 bp upstream and downstream of these conserved exons from the striped tit-babbler genome to recover variable intronic regions using bedtools 2.28.0 (Quinlan and Hall 2010). We further checked for overlapping targets and merged all overlapping loci in bedtools. We then removed any loci with a GC content <40% or >60%. Loci which contained repeat elements were identified using RepeatMasker 4.0.7 (Smit et al. 2015) and removed. We finally retained 960 loci (1.99 Mb), which were used by MYcroarry to design RNA baits. We used 73,928 100-bp baits with 4X tiling density for in-solution target enrichment.
Target Enrichment
We performed in-solution hybrid capture for whole-genome libraries to enrich target loci. A modified version of the myBaits protocol was used for hybridization (myBaits manual version 3). For ancient samples, we diluted the baits and used them at 50% strength, carrying out independent hybridization reactions at 60 ºC for 40 h. For fresh samples, we pooled three uniquely barcoded samples at equimolar concentrations and carried out hybridization at 65 ºC for 20 h. We used a lower temperature and longer duration for the hybridization of ancient samples as suggested by the MYBaits manual. Following hybridization, the samples were cleaned as suggested in the MYBaits manual and we performed PCR for the enriched libraries using IS5 and IS6 primers (Kircher et al. 2012). The final libraries were cleaned using Ampure beads and pooled at equimolar concentrations. We sequenced the enriched libraries on multiple lanes of HiSeq 4000 (150-bp paired-end runs). Fresh and ancient samples were run on separate lanes. All negative controls were also sequenced to rule out contamination.
Data Filtering and Cleanup
We obtained 1.4 billion 150-bp paired-end reads. Reads with a PHREAD score below ten were removed by the service provider. We ran FASTQC 0.11.7 (Andrews 2010) to check for adapter contamination. We used cutadapter 1.12 (Martin 2011) to trim adapters from the reads and performed another quality check in FastQC. Subsequently, we used Trimmomatic 0.38 (Bolger et al. 2014) to remove any remaining adapter sequences. Trimmomatic also removes any low-quality reads and reads less than 36 bp in length. Finally, we performed a third FastQC run to check for sequence quality and confirm complete adapter removal. Next, we removed PCR duplicates using the dedupe program within bbmap 36.84 (Bushnell 2014). Furthermore, for the ancient samples, we used mapDamage 2.0 (Jónsson et al. 2013) to remove preservation-related postmortem substitutions. We observed high rates of transitions (G to A and C to T) at the ends of reads. Hence, we first rescaled our bam files using mapDamage. Then the rescaled bam files were converted to fastq reads and we trimmed 10 bp from both the 5′- and 3′-ends in the historic samples using Seqtk 1.2-r94 (https://github.com/lh3/seqtk) to reduce bias due to DNA degradation. The cleaned reads were used for downstream processing.
Data Matrix Generation
We used two different approaches to generate data matrices for each of the two babbler species complexes. In the first approach, we generated sequence data matrices for target loci. In the second approach, we generated genome-wide SNPs.
Sequence Data Generation
To generate a sequence data matrix of target loci, we used the HybPiper 1.2 pipeline (Johnson et al. 2016), which is specifically designed for sequence capture protocols. We generated sequence data with an outgroup individual for phylogenomic analysis. The resultant sequences were then aligned using MAFFT v7.130b (Katoh and Standley 2016). We used auto settings within MAFFT to identify the best approach for sequence alignment. The alignments were further cleaned using strict settings (default settings) within Gblocks 0.91b (Castresana 2000) to remove poorly aligned regions. We further removed loci with a sequence length less than 200 bp and those located on the Z chromosome for phylogenomic analysis.
We obtained sequence data for 944 out of 960 target loci designed for both species complexes using the HybPiper pipeline. We removed ten historic P. malaccense samples from downstream processing as their missing data exceeded 85%. After multiple sequence alignments, we performed stringent filtering of alignments using Gblocks and removed 117 loci from the P. capistratum data set and 385 loci from the P. malaccense data set due to high missing data. After removal of loci <200 bp and Z-chromosomal loci, we retained 652 loci for P. capistratum and 314 loci for P. malaccense. The total sequence matrix length for P. capistratum was 454,712 bp (average locus length = 697 bp; minimum = 201 bp; maximum = 4,723 bp) and 145,877 bp for P. malaccense (average locus length = 465 bp; minimum = 201 bp; maximum = 2,607 bp).
SNP Data Generation
We generated four different SNP sets for each of the two babbler species complexes. For the first three SNP sets, we mapped the clean reads to the striped tit-babbler genome using BWA-MEM 0.7.17-r1188 (Li and Durbin 2009). The mapped reads were sorted and converted to a bam format using SAMTOOLS 1.9 (Li et al. 2009; Li 2011). We used the GATK SNP caller within ANGSD 0.923-3-ga8ed56f (Korneliussen et al. 2014) to identify SNPs for each species complex separately. For both species complexes, we used a P value cut-off of 1E-6. Further, only SNPs with a PHREAD score ≥30 (99.9% accuracy) were retained. We allowed for no missing data during SNP calling. The SNPs were further filtered using VCFtools 0.1.16 (Danecek et al. 2011), and any locus with a read depth less than ten per sample was removed. We then removed linked loci using PLINK 1.9 (Purcell et al. 2007) by applying the indep-pairwise algorithm with a sliding window size of 50 SNPs, a step size of ten and an r2 correlation coefficient cut-off of 0.9. We also removed any loci not in Hardy–Weinberg equilibrium using PLINK while correcting the P value for multiple comparisons. We further tested for selection in BAYESCAN 2.1 (Foll and Gaggiotti 2008) using default settings and removed any locus under positive selection. The striped tit-babbler genome was also blasted to the chromosomal assembly of the great tit genome (Parus major, GCA_001522545.3) using BlastN to identify scaffolds which map to the Z chromosome. The great tit genome is the phylogenetically closest genome available that has been assembled to the chromosome level. SNPs which mapped to the Z chromosome of the great tit genome were removed from downstream processing. The cleaned, unlinked, neutral autosomal SNPs constituted data set I. C to T and G to A substitutions are the most common postmortem substitutions observed in historic samples and hence can bias results (Chattopadhyay et al. 2019). To account for DNA damage due to historic sampling, we therefore generated a second data set in which only transversions were included in analyses (data set II). We also generated a third data set which included only a single random SNP from each target locus. We did not perform any test for linkage for this data set (data set III).
For the final data set (data set IV), we mapped clean reads to the great tit genome and carried out SNP calling in ANGSD as mentioned above. We included an outgroup for this SNP set and allowed for 20% missing data. We further pruned the data set for linkage disequilibrium and deviations from neutrality and Hardy–Weinberg equilibrium using the same approaches as mentioned above. We only retained transversions for data set IV. This latter data set had the advantage of chromosomal information required for admixture graph analyses (see ABBA-BABA Tests and Admixture Graph Analysis). For the population structure analysis using data set IV, we further pruned the loci located on the Z chromosome.
In the end, we retrieved between 960 and 208,186 SNPs for P. capistratum and retained 883 to 54,906 SNPs after filtering (table 1). For P. malaccense between 958 and 198,711 SNPs were obtained across data sets before filtering and we retained 895 to 33,708 SNPs for further analysis (table 1).
Phylogenomic Analysis
We used both concatenation approaches and species tree reconstruction for phylogenomic analysis. For concatenation-based analyses, we used RAxML 8.2 (Stamatakis 2014) to reconstruct a maximum likelihood tree using the GTR+GAMMA model of substitution. We ran the AMAS pipeline (Borowiec 2016) to concatenate loci. For species tree reconstruction, we used the phyluce pipeline 1.6.6 (Faircloth 2016) to process the sequence data. We generated individual maximum likelihood gene trees using RAxML within the phyluce pipeline for downstream use in MP-EST 1.6 (Liu et al. 2010) to estimate the species tree. We followed Liu et al. (2017) for species tree estimation and generated 100 bootstrap trees per locus. We further generated 100 bootstrap files containing one bootstrap replicate per locus. These files were then used for MP-EST species tree estimation. The 100 bootstrap species trees were further used to estimate nodal support and to generate a majority-rule consensus tree using Phylip v3.69 (Felsenstein 2005). Both the concatenated tree and species tree were viewed in FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Population Genomic Analysis
We employed multiple approaches to understand population subdivision within each babbler species complex. First, we performed PCA using the SNPrelate package in R (Zheng et al. 2012). PCA is a multivariate approach to identify structure within data sets, but is at the same time independent of population genetic assumptions. For data set IV, we removed the loci with missing data for PCA analysis. Second, we performed DAPC in the adegenet 2.1.1 R package (Jombart 2008). One major feature that sets DAPC apart from PCA is that it tends to maximize between-group differences while placing less emphasis on within-group variability (Jombart 2008). Finally, we adopted a Bayesian clustering approach using STRUCTURE (Pritchard 2000) to identify subdivision within each species complex. We used the structure threader program for STRUCTURE analysis and performed ten runs each for K = 1–10. For each K, we performed 100,000 burn-ins and 500,000 Markov chain Monte Carlo steps. Structure runs were performed without any a priori assumptions of population assignment. We ran the pophelper R package (Francis 2017) to visualize STRUCTURE results and compared across multiple K values to assess genomic assignments of individuals. We used R version 3.5.3 (R Core Team 2020) for all analyses.
ABBA-BABA Tests and Admixture Graph Analysis
We used the four-taxon ABBA-BABA test to understand gene flow among various island populations of babblers (Green et al. 2010; Durand et al. 2011). The ABBA-BABA test is a powerful method to differentiate between secondary admixture and incomplete lineage sorting (Green et al. 2010; Durand et al. 2011). We performed ABBA-BABA tests using ANGSD considering only transversions to account for DNA damage. Only sites with a mapping quality and PHRED score ≥30 were considered. To test for significance, we performed jackknifing of 20-kb blocks. Only test scenarios congruent with the phylogenomic analysis were considered. Significant secondary admixture was inferred when Z scores exceeded −3 or +3. For ABBA-BABA analyses, we used the bam files obtained by mapping clean reads to the chromosomal assembly of the great tit genome.
In addition to ABBA-BABA tests, we performed admixture graph analysis with qpbrute (Leathlobhair et al. 2018; Liu et al. 2019), which implements a heuristic algorithm to iteratively fit complex admixture models using qpGraph (part of ADMIXTOOLS 5.1; Patterson et al. 2012) as well as estimate Bayes factors to determine the best admixture graph: qpGraph reconstructs relationships among populations using a phylogenetic approach allowing for admixture events. For a given topology, f2, f3, and f4 statistics are estimated for all taxa and the observed and expected allele frequencies are calculated for the observed data and model. Within qpbrute, for a given outgroup, taxa are added iteratively. If a node cannot initially be included and outliers are observed for the f4 statistic, then all possible admixture events are attempted. If a node ends up being excluded, its subgraph is discarded, but if a node is successfully included, remaining nodes are added iteratively to the subgraph. All possible combinations of taxa are tested within qpbrute to ensure a complete coverage of graph space. We used data set IV along with an outgroup and also included SNPs mapping to the Z chromosome for this analysis.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was supported by a South East Asian Biodiversity Genomics (SEABIG) Grant (WBS R-154-000-648-646 and WBS R-154-000-648-733) and a Singapore Ministry of Education Tier II Grant (WBS R-154-000-C41-112 to F.E.R.). B.C. acknowledges the startup funding from Trivedi School of Biosciences, Ashoka University, India and K.M.G. acknowledges the support from the DBT-Ramalingaswami Fellowship (No. BT/HRD/35/02/2006). The authors thank Kelvin Lim at the Lee Kong Chian Natural History Museum and Kristof Zyskowski at the Yale Peabody Museum of Natural History for providing toepad samples and Lau On Sun for help with the bioruptor. The authors thank Chyi Yin Gwee, Pratibha Baveja, Tang Qian, Arina Adom, Ling Lih Chua, and Lu Wee Tan for logistical and lab assistance and Evan K. Irving-Pease for help with qpBrute.
Author Contributions
KMG and FER designed the research. KMG performed laboratory work with input from BC. KMG and BC designed the target enrichment loci with input from FER. EC performed toepad sampling and plumage analysis. EC conducted fieldwork with the help of SB, ST and DPE. KMG and FER wrote the paper with input from all co‐authors.
Ethics Statement
This study complied with all ethical regulations. Protocols were approved by the National University of Singapore Institutional Animal Care and Use Committee (IACUC, Protocol No.: L2017–00459). Permits for sampling in Sabah were approved by the Danum Valley Management Committee, the Sabah Forestry Department, and the Sabah Biodiversity Council (Permit Nos.: JKM/MBS.1000-2/2 JLD.3 [118]; JHL 100.7/27) and the export of samples was under permit (Permit Number: JKM/MBS.1000-2/3 JLD.2 [65]).
Data Availability
The data underlying this article are available in the article and in its Supplementary Material online. Raw data generated in this study have been submitted to the NCBI SRA database (BioProject ID: PRJNA701111).
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Available from: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bird MI, Taylor D, Hunt C.. 2005. Palaeoenvironments of insular Southeast Asia during the Last Glacial Period: a savanna corridor in Sundaland? Quat Sci Rev. 24(20–21):2228–2242. [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec ML. 2016. AMAS: a fast tool for alignment manipulation and computing of summary statistics. PeerJ 4:e1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borregaard MK, Matthews TJ, Whittaker RJ, Field R.. 2016. The general dynamic model: towards a unified theory of island biogeography? Glob Ecol Biogeogr. 25(7):805–816. [Google Scholar]
- Brown RM, Siler CD, Oliveros CH, Esselstyn JA, Diesmos AC, Hosner PA, Linkem CW, Barley AJ, Oaks JR, Sanguila MB, et al. 2013. Evolutionary processes of diversification in a model island archipelago. Annu Rev Ecol Evol Syst. 44(1):411–435. [Google Scholar]
- Burney CW, Brumfield RT.. 2009. Ecology predicts levels of genetic differentiation in Neotropical birds. Am Nat. 174(3):358–368. [DOI] [PubMed] [Google Scholar]
- Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner. Berkeley (CA: ): Lawrence Berkeley National Lab (LBNL; ). [Google Scholar]
- Cannon CH, Morley RJ, Bush AB.. 2009. The current refugial rainforests of Sundaland are unrepresentative of their biogeographic past and highly vulnerable to disturbance. Proc Natl Acad Sci U S A. 106(27):11188–11193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay B, Garg KM, Gwee CY, Edwards SV, Rheindt FE.. 2017. Gene flow during glacial habitat shifts facilitates character displacement in a Neotropical flycatcher radiation. BMC Evol Biol. 17(1):210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay B, Garg KM, Mendenhall IH, Rheindt FE.. 2019. Historic DNA reveals Anthropocene threat to a tropical urban fruit bat. Curr Biol. 29(24):R1299–R1300. [DOI] [PubMed] [Google Scholar]
- Cros E, Chattopadhyay B, Garg KM, Ng NS, Tomassi S, Benedick S, Edwards DP, Rheindt FE.. 2020. Quaternary land bridges have not been universal conduits of gene flow. Mol Ecol. 29(14):2692–2706. [DOI] [PubMed] [Google Scholar]
- Cros E, Ng EY, Oh RR, Tang Q, Benedick S, Edwards DP, Tomassi S, Irestedt M, Ericson PG, Rheindt FE.. 2020. Fine‐scale barriers to connectivity across a fragmented South‐East Asian landscape in six songbird species. Evol Appl. 13(5):1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa MJ, do Amaral PJ, Pieczarka JC, Sampaio MI, Rossi RV, Mendes-Oliveira AC, Noronha RC, Nagamachi CY.. 2016. Cryptic species in Proechimys goeldii (Rodentia, Echimyidae)? A case of molecular and chromosomal differentiation in allopatric populations. Cytogenet Genome Res. 148(2–3):199–210. [DOI] [PubMed] [Google Scholar]
- Dabney J, Meyer M, Pääbo S.. 2013. Ancient DNA damage. Cold Spring Harb Perspect Biol. 5(7):a012567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M.. 2011. Testing for ancient admixture between closely related populations. Mol Biol Evol. 28(8):2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton JA, van Balen S, Brickle NW, Rheindt FE.. 2016. Birds of the Indonesian Archipelago: Greater Sundas and Wallacea. Barcelona: Lynx Edicions. [Google Scholar]
- Faircloth BC. 2016. PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics 32(5):786–788. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 2005. PHYLIP version 3.6, Software package distrbuted by the author. Seattle (WA: ): Department of Genetics, University of Washington. [Google Scholar]
- Fernández‐Palacios JM, Rijsdijk KF, Norder SJ, Otto R, Nascimento L, Fernández‐Lugo S, Tjørve E, Whittaker RJ, Santos A.. 2016. Towards a glacial‐sensitive model of island biogeography. Glob Ecol Biogeogr. 25(7):817–830. [Google Scholar]
- Foll M, Gaggiotti O.. 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180(2):977–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis RM. 2017. pophelper: an R package and web app to analyse and visualize population structure. Mol Ecol Resour. 17(1):27–32. [DOI] [PubMed] [Google Scholar]
- Garg KM, Chattopadhyay B, Koane B, Sam K, Rheindt FE.. 2020. Last Glacial Maximum led to community-wide population expansion in a montane songbird radiation in highland Papua New Guinea. BMC Evol Biol. 20(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg KM, Chattopadhyay B, Wilton PR, Prawiradilaga DM, Rheindt FE.. 2018. Pleistocene land bridges act as semipermeable agents of avian gene flow in Wallacea. Mol Phylogenet Evol. 125:196–203. [DOI] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH-Y, et al. 2010. A draft sequence of the Neandertal genome. Science 328(5979):710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaney LR. 1986. Biogeography of mammals in SE Asia: estimates of rates of colonization, extinction and speciation. Biol J Linn Soc. 28(1–2):127–165. [Google Scholar]
- Heaney LR, Walsh JS Jr, Townsend Peterson A.. 2005. The roles of geological history and colonization abilities in genetic differentiation between mammalian populations in the Philippine archipelago. J Biogeogr. 32(2):229–247. [Google Scholar]
- Hewitt GM. 2000. The genetic legacy of the Quaternary ice ages. Nature 405(6789):907–913. [DOI] [PubMed] [Google Scholar]
- Hewitt GM. 2004. Genetic consequences of climatic oscillations in the Quaternary. Philos Trans R Soc Lond B Biol Sci. 359(1442):183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley TH. 1868. On the classification and distribution of the Alectoromorphae and Heteromorphae. Proc Zool Soc Lond. 294:319. [Google Scholar]
- Isler ML, Alonso JA, Isler PR, Whitney BM.. 2001. A new species of Percnostola antbird (Passeriformes: Thamnophilidae) from Amazonian Peru, and an analysis of species limits within Percnostola rufifrons. Wilson J Ornithol. 113:164–176. [Google Scholar]
- Isler ML, Maldonado-Coelho M.. 2017. Calls distinguish species of Antbirds (Aves: Passeriformes: Thamnophilidae) in the genus Pyriglena. Zootaxa 4291(2):275–294. [Google Scholar]
- Johnson MG, Gardner EM, Liu Y, Medina R, Goffinet B, Shaw AJ, Zerega NJ, Wickett NJ.. 2016. HybPiper: extracting coding sequence and introns for phylogenetics from high‐throughput sequencing reads using target enrichment. Appl Plant Sci. 4(7):1600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24(11):1403–1405. [DOI] [PubMed] [Google Scholar]
- Jónsson H, Ginolhac A, Schubert M, Johnson PL, Orlando L.. 2013. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29(13):1682–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32(13):1933–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Sawyer S, Meyer M.. 2012. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40(1):e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen TS, Albrechtsen A, Nielsen R.. 2014. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leathlobhair MN, Perri AR, Irving-Pease EK, Witt KE, Linderholm A, Haile J, Lebrasseur O, Ameen C, Blick J, Boyko AR, et al. 2018. The evolutionary history of dogs in the Americas. Science 361(6397):81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JA, den Tex RJ, Hawkins MT, Muñoz‐Fuentes V, Thorington R, Maldonado JE.. 2015. Phylogeography of vertebrates on the Sunda Shelf: a multi‐species comparison. J Biogeogr. 42(5):871–879. [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21):2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HC, Gawin DF, Shakya SB, Harvey MG, Rahman MA, Sheldon FH.. 2017. Sundaland’s east–west rain forest population structure: variable manifestations in four polytypic bird species examined using RAD‐Seq and plumage analyses. J Biogeogr. 44(10):2259–2271. [Google Scholar]
- Lim HC, Rahman MA, Lim SL, Moyle RG, Sheldon FH.. 2011. Revisiting Wallace’s haunt: coalescent simulations and comparative niche modeling reveal historical mechanisms that promoted avian population divergence in the Malay Archipelago. Evolution 65(2):321–334. [DOI] [PubMed] [Google Scholar]
- Lim HC, Sheldon FH.. 2011. Multilocus analysis of the evolutionary dynamics of rainforest bird populations in Southeast Asia. Mol Ecol. 20(16):3414–3438. [DOI] [PubMed] [Google Scholar]
- Liu L, Bosse M, Megens HJ, Frantz LA, Lee YL, Irving-Pease EK, Narayan G, Groenen MA, Madsen O.. 2019. Genomic analysis on pygmy hog reveals extensive interbreeding during wild boar expansion. Nat Commun. 10(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Yu L, Edwards SV.. 2010. A maximum pseudo-likelihood approach for estimating species trees under the coalescent model. BMC Evol Biol. 10:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang J, Rheindt FE, Lei F, Qu Y, Wang Y, Zhang Y, Sullivan C, Nie W, Wang J, et al. 2017. Genomic evidence reveals a radiation of placental mammals uninterrupted by the KPg boundary. Proc Natl Acad Sci U S A. 114(35):E7282–E7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman DJ, de Bruyn M, Page T, von Rintelen K, Hall R, Ng PK, Shih HT, Carvalho GR, Von Rintelen T.. 2011. Biogeography of the Indo-Australian archipelago. Annu Rev Ecol Evol Syst. 42(1):205–226. [Google Scholar]
- MacArthur RH, Wilson EO.. 1967. The theory of island biogeography. New York: : Princeton University Press. [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–12. [Google Scholar]
- Mason VC, Helgen KM, Murphy WJ.. 2019. Comparative phylogeography of forest-dependent mammals reveals Paleo-forest corridors throughout Sundaland. J Hered. 110(2):158–172. [DOI] [PubMed] [Google Scholar]
- Mayr E, Diamond JM, Diamond J.. 2001. The birds of Northern Melanesia: speciation, ecology & biogeography. Oxford: Oxford University Press. [Google Scholar]
- Mays HL Jr, Hung CM, Shaner PJ, Denvir J, Justice M, Yang SF, Roth TL, Oehler DA, Fan J, Rekulapally S, et al. 2018. Genomic analysis of demographic history and ecological niche modeling in the endangered Sumatran rhinoceros Dicerorhinus sumatrensis. Curr Biol. 28(1):70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle RG, Filardi CE, Smith CE, Diamond J.. 2009. Explosive Pleistocene diversification and hemispheric expansion of a “great speciator”. Proc Natl Acad Sci U S A. 106(6):1863–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nater A, Mattle-Greminger MP, Nurcahyo A, Nowak MG, de Manuel M, Desai T, Groves C, Pybus M, Sonay TB, Roos C, et al. 2017. Morphometric, behavioral, and genomic evidence for a new orangutan species. Curr Biol. 27(22):3487–3498.e3410. [DOI] [PubMed] [Google Scholar]
- Ng NS, Wilton PR, Prawiradilaga DM, Tay YC, Indrawan M, Garg KM, Rheindt FE.. 2017. The effects of Pleistocene climate change on biotic differentiation in a montane songbird clade from Wallacea. Mol Phylogenet Evol. 114:353–366. [DOI] [PubMed] [Google Scholar]
- Papadopoulou A, Knowles LL.. 2017. Linking micro‐and macroevolutionary perspectives to evaluate the role of Quaternary sea‐level oscillations in island diversification. Evolution 71(12):2901–2917. [DOI] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D.. 2012. Ancient admixture in human history. Genetics 192(3):1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P.. 2000. Inference of population structure using multilocus genotype data. Genetics 155(2):945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, Munch K, Hellmann I, Akagi K, Miller JR, Walenz B, Koren S, Sutton G, Kodira C, Winer R, et al. 2012. The bonobo genome compared with the chimpanzee and human genomes. Nature 486(7404):527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81(3):559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rheindt FE, Christidis L, Cabanne GS, Miyaki C, Norman JA.. 2009. The timing of neotropical speciation dynamics: a reconstruction of Myiopagis flycatcher diversification using phylogenetic and paleogeographic data. Mol Phylogenet Evol. 53(3):961–971. [DOI] [PubMed] [Google Scholar]
- Rheindt FE, Christidis L, Norman JA.. 2008. Habitat shifts in the evolutionary history of a Neotropical flycatcher lineage from forest and open landscapes. BMC Evol Biol. 8:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheindt FE, Prawiradilaga DM, Ashari H, Gwee CY, Lee GW, Wu MY, Ng NS.. 2020. A lost world in Wallacea: description of a montane archipelagic avifauna. Science 367:167–170. [DOI] [PubMed] [Google Scholar]
- Robson C. 2005. Birds of Southeast Asia. New York: : Princeton University Press. [Google Scholar]
- Sadanandan KR, Rheindt FE.. 2015. Genetic diversity of a tropical rainforest understory bird in an urban fragmented landscape. Condor 117(3):447–459. [Google Scholar]
- Sarr AC, Husson L, Sepulchre P, Pastier AM, Pedoja K, Elliot M, Arias-Ruiz C, Solihuddin T, Aribowo S.. 2019. Subsiding Sundaland. Geology 47:119–122. [Google Scholar]
- Sathiamurthy E, Voris HK.. 2006. Maps of Holocene sea level transgression and submerged lakes on the Sunda Shelf. Nat Hist J Chulalongkorn Univ. Suppl 2:1–43. [Google Scholar]
- Smit A, Hubley R, Green P.. 2015. RepeatMasker Open-4.0. [Internet]. 2013–2015. Available from: http://www.repeatmasker.org/. [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterrer W, Glasspool A, De Silva H, Furbert J.. 2004. Bermuda—an island biodiversity transported. In: Davenport J, Davenport J, editors. The effects of human transport on ecosystems: cars and planes, boats and trains. Dublin (Ireland: ): Royal Irish Academy. p. 118–170. [Google Scholar]
- Taberlet P, Fumagalli L, Wust‐Saucy AG, Cosson JF.. 1998. Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol. 7(4):453–464. [DOI] [PubMed] [Google Scholar]
- Teacher A, Garner T, Nichols R.. 2009. European phylogeography of the common frog (Rana temporaria): routes of postglacial colonization into the British Isles, and evidence for an Irish glacial refugium. Heredity (Edinb). 102(5):490–496. [DOI] [PubMed] [Google Scholar]
- Voris HK. 2000. Maps of Pleistocene sea levels in Southeast Asia: shorelines, river systems and time durations. J Biogeogr. 27(5):1153–1167. [Google Scholar]
- Wallace A. 1872. Flora and fauna of the Azores. Am Nat. 6:176–177. [Google Scholar]
- Wallace AR. 1860. On the zoological geography of the Malay Archipelago. J Proc Linn Soc. 4(16):172–184. [Google Scholar]
- Weigelt P, Steinbauer MJ, Cabral JS, Kreft H.. 2016. Late Quaternary climate change shapes island biodiversity. Nature 532(7597):99–102. [DOI] [PubMed] [Google Scholar]
- Whittaker RJ, Fernández-Palacios JM, Matthews TJ, Borregaard MK, Triantis KA.. 2017. Island biogeography: taking the long view of nature’s laboratories. Science 357(6354):885–892. [DOI] [PubMed] [Google Scholar]
- Zakaria M, Rajpar MN, Moradi HV, Rosli Z.. 2014. Comparison of understorey bird species in relation to edge–interior gradient in an isolated tropical rainforest of Malaysia. Environ Dev Sustain. 16(2):375–392. [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS.. 2012. High-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28(24):3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its Supplementary Material online. Raw data generated in this study have been submitted to the NCBI SRA database (BioProject ID: PRJNA701111).