Abstract
A quantitative-competitive PCR for the quantification of porcine cytomegalovirus (PCMV) was developed. The virus was detected in a variety of pig organs (including potential xenotransplant donations), with viral loads ranging from <10 to 97 genome copies/μg of DNA. This assay will have significant utility for studying the activation and replication of PCMV and in swine models for allo- and xenotransplantation.
Porcine cytomegalovirus (PCMV) is a betaherpesvirus which causes generalized infection in newborn piglets. It has a worldwide distribution and is present in 90% of herds in the United Kingdom, many of which are from well-managed, high-health-status farms (3). Epidemiological features of the virus include evidence of latency and crossplacental infection. Xenotransplantation has the potential to relieve the current donor organ shortage associated with human allotransplantation, and for both practical and ethical reasons, the pig has become the most likely source of such tissues. As clinical trials of xenotransplantation are beginning, concerns have been raised about the potential transmission of porcine organisms to immunocompromised human recipients of porcine tissues (1, 4, 5). As with allotransplantation, there are potential risks associated with the transmission of porcine viruses through xenograft tissues. Although transmission to humans has not been demonstrated in vivo, porcine endogenous retroviruses have been shown to replicate in human cells in vitro (11, 15). Human cytomegalovirus (HCMV) has a variety of direct and indirect effects in the allograft recipient (6). Given the high frequency of seropositivity of swine for PCMV and the efficiency of transmission of HCMV from donor to transplant recipient (6), PCMV may represent another potential risk to humans.
At present the laboratory detection of PCMV is carried out by serology or qualitative PCR (8, 13, 14). We now describe the development of a quantitative-competitive PCR (QC-PCR) assay and its application to the measurement of viral load in porcine tissues. The QC-PCR is based on the coamplification of the test sample with an internal competitor, which differs from the wild-type sequence by the presence of a restriction endonuclease site in the middle of the amplicon. To our knowledge this is the first quantitative PCR assay for the quantification of PCMV to be reported.
The qualitative and QC-PCR assays amplify a region of the polymerase gene (14). Primers PCMVF1 (5′ CCTATGTTGGCACTGATACTTGAC 3′) and PCMVR1 (5′ CCCTGAAAATCACCGTCTGAGAGA 3′) were initially used to amplify a 236-bp region of the gene from tissue culture supernatant of porcine alveolar macrophages infected with PCMV strain B6. The amplicon was then cloned into the pGEM-T Easy vector (Promega). PCR-mediated site-directed mutagenesis, similar to the method described by Fox et al. (7), was used to create a SmaI restriction endonuclease site in the middle of the PCMV amplicon sequence by altering two nucleotides. Two complementary mutagenic primers were used in two separate PCRs; primers PCMVF1 and PCMVFM (5′ TAAGCATGTCCCGGGCTATGCTGG 3′) were used in one PCR, and primers PCMVR1 and PCMVRM (5′ CCAGCATAGCCCGGGACATGCTTA 3′) were used in the other. The products of these PCR experiments were purified by Wizard PCR Prep (Promega), mixed in equimolar amounts, heated to 95°C for 12 min, and then cooled to room temperature over a 30-min period. The annealed products were 3′ extended using Klenow polymerase and deoxynucleoside triphosphates (dNTPs) (both from Promega), and then used as the template for amplification with primers PCMVF1 and PCMVR1. The 236-bp PCR product was cloned into the pGEM-T Easy vector, and the mutation was confirmed by restriction endonuclease mapping and DNA sequencing.
For the qualitative nested PCR, the first-round primers PCMVF2 (5′ AAGCAGCAGCTTGCCCTCAAGGTG 3′) and PCMVR1 amplify a 212-bp amplicon and the nested primers PCMVFB (5′ ACGTGCAATGCGTTTTACGGCTTC 3′) and PCMVR2 (5′ ACTTCTCTGACACGTATTCTCTAG 3′) amplify a 160-bp product. Three of the primers show a degree of overlap with those described previously (8, 14). The four primers show 100% nucleotide homology to the DNA polymerase nucleotide sequences of three PCMV strains (55b, B6, and OF-1) deposited in GenBank, suggesting their ability to amplify different isolates. Each PCR mixture contained 100 ng of each primer, 1.5 mM MgCl2 (Bioline), 200 μM each dNTP, 1 U of Taq polymerase (Bioline), and the target DNA, in (NH4)2SO4 PCR buffer (Bioline) to a final volume of 50 μl. The cycle parameters used for the first round were initial denaturation at 95°C for 6 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. The final extension was increased to 10 min. In the nested PCR the annealing temperature was reduced to 55°C. Using plasmid dilution experiments the sensitivity of the qualitative PCR assay was <10 genome copies/reaction. The specificity of the PCR assay was tested by amplifying DNA from other betaherpesviruses: human herpesvirus 6 (HHV-6), HHV-7, and HCMV. None of these samples yielded detectable amplification products (data not shown).
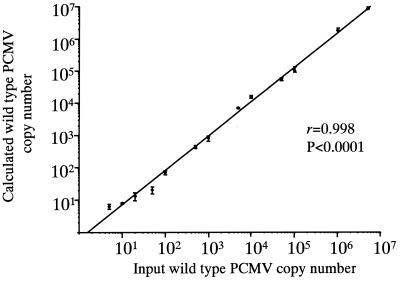
To assess the utility of the control sequence in the QC-PCR, a range of known copy numbers of the wild-type and control plasmids were coamplified. A single round of PCR was performed when 106 to 103 input copies were used, while a nested PCR of 15 second-round cycles was used for 102 to 101 input copies. PCR conditions and cycle parameters were the same as those for the qualitative assay. Following amplification, 10 μl of the amplicons was digested with 10 U of SmaI (Boehringer Mannheim) and separated by polyacrylamide gel electrophoresis (PAGE) on a 10% (first round) or 12% (nested round) gel. The gels were stained with ethidium bromide and photographed. The intensities of the bands for wild-type and control amplicons were analyzed as previously described (10), and the input copy number of wild-type sequence was calculated. The range of quantification for each control sequence copy number was ±0.5 logunit, i.e., with a control plasmid copy number of 100, between 50 and 500 copies of wild-type plasmid were tested. The mean result from three experiments was plotted against the known input copy numbers of wild-type sequence. Figure 1 shows that the calculated and actual copy numbers of PCMV were highly correlated (r = 0.998; P < 0.0001 for a linear fit curve), suggesting that the QC-PCR method was highly reproducible and could be used to accurately quantify PCMV.
FIG. 1.
Characterization of the PCMV QC-PCR assay. Known copy numbers of the plasmid containing the wild-type target, ranging from 101 to 5 × 106, were coamplified with known copy numbers of the plasmid containing the control sequence (ranging from 101 to 106). Calculated wild-type copy numbers were plotted against actual input wild-type copy numbers. Each point represents the mean of triplicate determinations. Error bars, standard errors. The line of best fit was plotted through the points.
To test the applicability of the qualitative and QC-PCR assays, five pig tissues (lung, liver, salivary gland, kidney, and gut) derived from multiple animals from a well-characterized herd of inbred miniature swine being sacrificed for other ongoing studies (12) were tested for the presence of PCMV. All animal care and study protocols were approved by the Animal Studies Committee of the Massachusetts General Hospital. DNA was extracted by overnight incubation of tissue samples with proteinase K at 65°C, followed by phenol-chloroform extraction and ethanol precipitation. Qualitative analysis tested 1 μg of extracted DNA, and four of the five tissues were PCR positive (lung, liver, salivary gland, and kidney). The gut tissue sample was consistently PCR negative. QC-PCR was then applied to determine viral loads, utilizing 10 and 100 input copy numbers of control plasmid. When viral loads were obtained for a test sample using both input copy numbers of control sequence, the value used was that where the wild-type-to-control-amplicon signal ratio was closest to equivalence. The viral loads in the liver and salivary gland were below the threshold of quantification and were given an arbitrary value of <10 PCMV genome copies/μg of DNA. The kidney contained 38 genome copies/μg of DNA, and the lung contained 97 genome copies/μg of DNA.
In conclusion, this report describes the development and application of a QC-PCR assay for the quantification of PCMV, similar to assays currently used in our laboratory for the quantification of human herpesviruses including HHV-6, HHV-7, and HCMV (2, 7, 9). This assay will be useful in further studying the pathogenesis of PCMV and will enable the detection and accurate quantification of PCMV in porcine organs in pigs being bred for use in xenotransplantation.
Acknowledgments
We thank David H. Sachs, Harvard Medical School, Boston, Mass., for providing the porcine tissues and Malcolm Banks, Veterinary Laboratories Agency, Addlestone, United Kingdom, for providing PCMV strain B6.
This work was funded by the Stanley Thomas Johnson Foundation.
REFERENCES
- 1.Auchincloss H, Jr, Sachs D H. Xenogeneic transplantation. Annu Rev Immunol. 1998;16:433–470. doi: 10.1146/annurev.immunol.16.1.433. [DOI] [PubMed] [Google Scholar]
- 2.Clark D A, Ait-Khaled M, Wheeler A C, Kidd I M, McLaughlin J E, Johnson M A, Griffiths P D, Emery V C. Quantification of human herpesvirus 6 in immunocompetent persons and post-mortem tissues from AIDS patients by PCR. J Gen Virol. 1996;77:2271–2275. doi: 10.1099/0022-1317-77-9-2271. [DOI] [PubMed] [Google Scholar]
- 3.Edington N. Porcine cytomegalovirus. In: Leman A D, Straw B, Glock R D, Mengeling W L, Penny R H C, Scholl E, editors. Diseases of swine. 6th ed. Ames: Iowa State University Press; 1986. pp. 330–336. [Google Scholar]
- 4.Fishman J A. Infection and xenotransplantation. Developing strategies to minimize risk. Ann N Y Acad Sci. 1998;862:52–66. doi: 10.1111/j.1749-6632.1998.tb09117.x. [DOI] [PubMed] [Google Scholar]
- 5.Fishman J A. Xenosis and xenotransplantation: addressing the infectious risks posed by an emerging technology. Kidney Int. 1997;51(Suppl. 58):S41–S45. [PubMed] [Google Scholar]
- 6.Fishman J A, Rubin R H. Infection in organ-transplant recipients. N Engl J Med. 1998;338:1741–1751. doi: 10.1056/NEJM199806113382407. [DOI] [PubMed] [Google Scholar]
- 7.Fox J C, Griffiths P D, Emery V C. Quantification of human cytomegalovirus DNA using the polymerase chain reaction. J Gen Virol. 1992;73:2405–2408. doi: 10.1099/0022-1317-73-9-2405. [DOI] [PubMed] [Google Scholar]
- 8.Hamel A L, Lin L, Sachvie C, Grudeski E, Nayar G P. PCR assay for detecting porcine cytomegalovirus. J Clin Microbiol. 1999;37:3767–3768. doi: 10.1128/jcm.37.11.3767-3768.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kidd I M, Clark D A, Ait-Khaled M, Griffiths P D, Emery V C. Measurement of human herpesvirus 7 load in peripheral blood and saliva of healthy subjects by quantitative polymerase chain reaction. J Infect Dis. 1996;174:396–401. doi: 10.1093/infdis/174.2.396. [DOI] [PubMed] [Google Scholar]
- 10.Kidd I M, Clark D A, Emery V C. A non-radioisotopic quantitative competitive polymerase chain method: application in measurement of human herpesvirus 7 load. J Virol Methods. 2000;87:177–181. doi: 10.1016/s0166-0934(00)00164-6. [DOI] [PubMed] [Google Scholar]
- 11.Patience C, Takeuchi Y, Weiss R A. Infection of human cells by an endogenous retrovirus of pigs. Nat Med. 1997;3:282–286. doi: 10.1038/nm0397-282. [DOI] [PubMed] [Google Scholar]
- 12.Sachs D H. MHC homozygous miniature swine. In: Swindle M M, Moody D C, Phillips L D, editors. Swine as models in biomedical research. 1st ed. Ames: Iowa State University Press; 1992. p. 3. [Google Scholar]
- 13.Tajima T, Hironao T, Kajikawa T, Kawamura H. Application of enzyme-linked immunosorbent assay for the seroepizootiological survey of antibodies against porcine cytomegalovirus. J Vet Med Sci. 1993;55:421–424. doi: 10.1292/jvms.55.421. [DOI] [PubMed] [Google Scholar]
- 14.Widen B F, Lowings J P, Belak S, Banks M. Development of a PCR system for porcine cytomegalovirus detection and determination of the putative partial sequence of its DNA polymerase gene. Epidemiol Infect. 1999;123:177–180. doi: 10.1017/s0950268899002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson C A, Wong S, Muller J, Davidson C E, Rose T M, Burd P. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J Virol. 1998;72:3082–3087. doi: 10.1128/jvi.72.4.3082-3087.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]