Abstract
Aim:
Analysis of transcriptomes from muscle tissue and cells improves our understanding of differences in gene expression and molecular function in cerebral palsy (CP) muscle.
Methods:
In this case-control study, participants were enrolled after informed consent/assent (8 CP; 11 controls) and skeletal muscle was obtained during surgery. RNA was extracted from tissue and from primary satellite cells grown to form myotubes in vitro. RNA-seq data were analyzed using validated informatics pipelines.
Results:
Analysis identified expression of 6308 genes in the tissue samples and 7459 in the cultured cells. Significant differential expression between CP and control was identified in 87 genes in the tissue and 90 genes in isolated satellite cell-derived myotube cultures.
Interpretation:
This is the first RNA-seq study of muscle and satellite cells in spastic CP. Both tissue and cell analyses identified differential expression of genes associated with muscle development and multiple pathways of interest.
Cerebral palsy (CP) is the leading cause of physical disability in childhood, occurring in 2–4 per 1000 live births.1,2 Major manifestations include disordered movement, movement control problems, and muscle dysfunction.2 Imaging studies of skeletal muscle indicate that subjects with spastic CP, which accounts for >75% of cases,1 have decreased muscle length, cross-sectional area, and volume with diminished force generation, reduced range of motion, and weakness.2 Muscles from individuals with CP exhibit increased sarcomere length,3 adipose infiltration,4 disorganized neuromuscular junctions (NMJ),5 altered gene expression,6–8 extracellular matrix (ECM) abnormalities,3 altered fiber type distributions,2,5 and loss of myogenic potential.9 CP is also associated with reduced numbers of satellite cells (SC), which are muscle-resident stem cells that have been linked to multiple aspects of the muscle phenotype seen in CP.9,10
The mechanisms contributing to the alterations in CP muscle are not well understood, but early studies of muscle gene expression using chip arrays indicated differences in pathways related to ECM expression, metabolism, and ubiquitin ligase activity.6–8 With the present study, we provide the first analyses of whole transcriptome RNA-seq data from spastic CP muscle to provide insight into genes and pathways that are dysregulated in tissue and in isolated primary satellite cell-derived myotubes (SCDMTs). These transcriptional differences may uncover molecular etiologies and potential therapeutic targets that allow therapy based on cell and tissue molecular profiles.
Method
Subject Enrollment
Nineteen subjects (8 with spastic CP and 11 controls) undergoing surgery were enrolled between 2014–2016 at the Alfred I. duPont Hospital for Children with IRB-approved consent/assent. For muscle tissue analysis, cases and controls were undergoing posterior spinal fusion (PSF) for scoliosis correction. For SC analysis, significantly more tissue was required, and additional surgeries were included for sample collection. Subjects with CP were undergoing PSF, rectus femoris resection, or varus derotation osteotomy, while controls were undergoing PSF or anterior cruciate ligament repair. Subjects with a chromosomal or genetic disorder or neurological disease other than CP were excluded. Biopsies were collected during surgery; biopsy size, which was surgery-dependent, determined if samples were used for skeletal muscle RNA-seq (n=6 CP, 7 control) and/or SC isolation with myogenic cell culture and RNA-seq (n=4 CP, 6 control) (Table 1).
Table 1:
Patient demographics.
| Sample | Diagnosis | Age | Sex | Epilepsy Diagnosis | Tissue source | Tissue RNA-seq | SCDMT RNA-seq |
|---|---|---|---|---|---|---|---|
| CN1 | Idiopathic scoliosis | 14.8 | M | N | Spinalis | * | |
| CN2 | Idiopathic scoliosis | 13.0 | M | N | Spinalis | * | |
| CN3 | Idiopathic scoliosis | 13.5 | M | N | Spinalis | * | |
| CN4 | Idiopathic scoliosis | 17.4 | M | N | Spinalis | * | |
| CN5 | Idiopathic scoliosis | 14.2 | M | N | Spinalis | * | |
| CN6 | Idiopathic scoliosis | 14.3 | F | N | Spinalis | * | * |
| CN7 | Idiopathic scoliosis | 15.1 | M | N | Spinalis | * | * |
| CN8 | Idiopathic scoliosis | 12.7 | F | N | Spinalis | * | |
| CN9 | Idiopathic scoliosis | 12.1 | F | N | Spinalis | * | |
| CN10 | Torn anterior cruciate ligament | 12.6 | M | N | Semitendinosus | * | |
| CN11 | Torn anterior cruciate ligament | 16.6 | M | N | Semitendinosus | * | |
| CP1 | Spastic quadriplegic CP | 12.2 | M | N | Spinalis | * | |
| CP2 | Spastic quadriplegic CP | 14.9 | M | Y | Spinalis | * | |
| CP3 | Spastic quadriplegic CP | 9.8 | M | Y | Spinalis | * | |
| CP4 | Spastic quadriplegic CP | 15.1 | M | Y | Spinalis | * | |
| CP5 | Spastic quadriplegic CP | 12.8 | F | Y | Spinalis | * | * |
| CP6 | Spastic quadriplegic CP | 18.0 | F | Y | Spinalis | * | * |
| CP7 | Spastic diplegic CP | 13.8 | F | N | Rectus femoris | * | |
| CP8 | Spastic quadriplegic CP | 15.6 | M | N | Vastus lateralis | * |
Sample Preparation
Muscle biopsies were snap-frozen in liquid nitrogen chilled isopentane and homogenized in TRIzol (Sigma) or underwent enzymatic digestion and immunomagnetic cell isolation to collect SCs (see Supplemental Methods). For each sample used in the skeletal muscle tissue analysis, 8–10μm sections were fixed in 10% neutral buffered formalin, stained with hematoxylin and eosin, and imaged on an EVOS FL Auto Imaging System (Thermo Fisher) to assure tissue integrity. For samples that underwent immunomagnetic isolation, SC populations that contained <90% PAX7-positive cells were excluded. Satellite cell-derived myoblasts were grown under tightly-controlled conditions (see Supplemental Methods) to allow comparison of transcriptional profiles during induced myogenesis. Briefly, cells were proliferated to confluence; then they were induced to form myotubes. After 24 hours of induction, cells were collected in TRIzol for RNA analysis.
RNA-seq library preparation, sequencing, and bioinformatics analysis
RNA-seq was carried out by the Center for Applied Genomics at the Children’s Hospital of Philadelphia. Total RNA was prepared from TRIzol, libraries were generated using TruSeq kits with ribosomal RNA depletion (Illumina); samples were sequenced using an Illumina HiSeq system with 60–80 million paired end reads. The resulting FASTQ files were processed and analyzed using established data pipelines (see Supplemental Methods).
Count data were analyzed in R Studio11 (version 1.1.383, R version 3.4.3) with the corrplot12 (version 0.84) and edgeR13 (version 3.20.7) packages using established methods to determine RNA population cross-sample correlations and to identify differentially expressed genes (DEGs). Heatmaps were generated using Heatmapper14 with Spearman’s rank correlation coefficient as the distance measure, protein association networks were generated using STRING version 11.0 (string-db.org)15 with Markov clustering, and pathway enrichment was analyzed using the reactomeFI plugin in Cytoscape (cytoscape.org)16. Benjamini-Hochberg False Discovery Rate corrected p values17 of 0.05 were used throughout to determine significance.
RNA-seq validation with qPCR
Quantitative PCR (qPCR) was performed to validate expression levels of select genes in the RNA-seq data. For samples with residual high quality RNA available, the RNA-seq samples were validated by qPCR (n=4 CP, 3 controls for tissue and n=4 CP, 6 controls for SCDMTs). RNA was reverse transcribed using High Capacity RNA-to-cDNA kits according to manufacturer’s instructions (Thermo Fisher). Taqman qPCR was performed with reactions consisting of 25ng cDNA, 1x Taqman Gene Expression Assay with FAM-MGB (Thermo Fisher), and 1x TaqMan Gene Expression Master Mix containing AmpliTaq Gold DNA Polymerase (UltraPure), Uracil-DNA glycosylase, dNTPs, and ROX passive reference (Thermo Fisher). Samples were run in triplicate by amplification on an ABI 7900 System (Applied Biosystems) under the following conditions: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of denaturing at 95°C for 15 s followed by annealing/extension at 60°C for 1 min. Results were analyzed in RQ Manager 1.2.1 (Applied Biosystems). The threshold crossing cycle value was used for relative quantification using the B2M housekeeping gene and the control samples as the calibrator group.
Results
Demographic data for the 19 unique subjects included in the study are summarized in Table 1. Spinalis tissue obtained from the concave side of the scoliotic curve during posterior spinal fusion surgery was analyzed from 7 controls with idiopathic scoliosis (6 males; 1 female) aged 14.6 ± 1.4 years and 6 CP cases (4 males; 2 females) aged 13.8 ± 2.8 years. These tissue samples were collected from the same anatomic site in each participant and were examined histologically to assure the presence of muscle fibers prior to RNA-seq analysis (Supplemental Figure 1). Interestingly, although all samples were predominantly composed of muscle fibers, a range of histological features and disruptions (e.g., variability in fiber size and morphology; endomysial/perimysial fibrosis) existed in both CP and control samples suggesting that the surgical patients enrolled in the study may have had similar muscle disruptions regardless of underlying diagnosis. The isolation of SCs required significantly more tissue, and sufficient material to allow both RNA-seq of tissue and the isolation of high quality SCs was only available from 4 subjects. To increase the number of myotube preparations that could be analyzed, additional participants were included with tissue collected from other anatomic locations: semitendinosus, rectus femoris, and vastus lateralis. The SCDMT group comprised 6 control subjects (3 males; 3 females) aged 13.9 ± 1.7 years, and 4 CP subjects (1 male; 3 females) aged 15.1 ± 2.3 years. Cells were grown to confluence; the time from seeding to confluence was 2.83 ± 0.75 days for controls and 2.75 ± 0.50 days for CP samples (p=0.852, Student’s t-test), indicating similar overall proliferation between the groups. Cells were then induced to differentiate and form myotubes for 24 hours.
In this study, we compared two different types of muscle preparation. First, we compared muscle tissue samples collected from erector spinae. Skeletal muscle is a complex tissue comprising multiple biological components and having distinctive functions depending on anatomic location and biomechanical environment. Second, we compared a subpopulation of cells that were isolated away from the multi-component, complex tissue structure and allowed to recapitulate early events in muscle formation in culture. To assess the potential impact of sample-to-sample variability in the two sets of comparisons, the level of consistency between RNA populations was assessed. RNA-seq count data showed strong technical correlation among subjects (Supplemental Figure 2) indicating that the RNA populations isolated within each arm of the study were comparable to each other overall. In particular, we found no significant global differences between the muscle types used in the SCDMT arm of the study and no differences associated with sex, age, or confounding epilepsy diagnosis.
Subsequent analyses were carried out on genes with at least 5 total counts in 50% or more of the samples. With this filtering, 6308 expressed genes were identified in muscle tissue and 7459 in SCDMTs with 4889 overlapping between the two sets. Principal component analysis (PCA) plots demonstrated the degree of separation between the two clusters in the tissue (Supplemental Figure 3A). Some overlap of the cohorts was expected as the expression patterns of the two groups were largely similar with differentially expressed genes accounting for 1.4% of the 6308 expressed genes. Although overlap remained after the dimensionality reduction, the points belonging to each cohort were distinctly grouped with the DEGs accounting for the distinct clusters. Similarly, distinctly grouped clusters with some overlap were also seen for the SCDMT data (Supplemental Figure 3B). Significant clustering was not seen related to sex, age, epilepsy diagnosis, or muscle of origin in either arm of the study.
DEG analysis of the tissue samples identified 46 down-regulated and 41 up-regulated genes in CP (Supplemental Table 1); analysis of SCDMTs identified 62 down-regulated and 28 up-regulated genes (Supplemental Table 2). To visualize patterns of expression, data were organized into heatmaps such that genes with similar expression patterns were placed near each other as rows and samples with similar patterns were near each other as columns. Heatmaps showed clear separation of samples based on diagnosis for both muscle tissue (Figure 1A) and SCDMTs (Figure 2B). Interestingly, SCDMTs from spinalis did not cluster more closely with each other than with SCDMTs from other muscle types by heatmapping or MDS algorithms. These results combined with the between-subject correlations indicated low overall variability in gene expression between SCDMTs derived from different muscles.
Figure 1: RNA-seq differential expression for muscle and satellite cells between CP and control subjects.
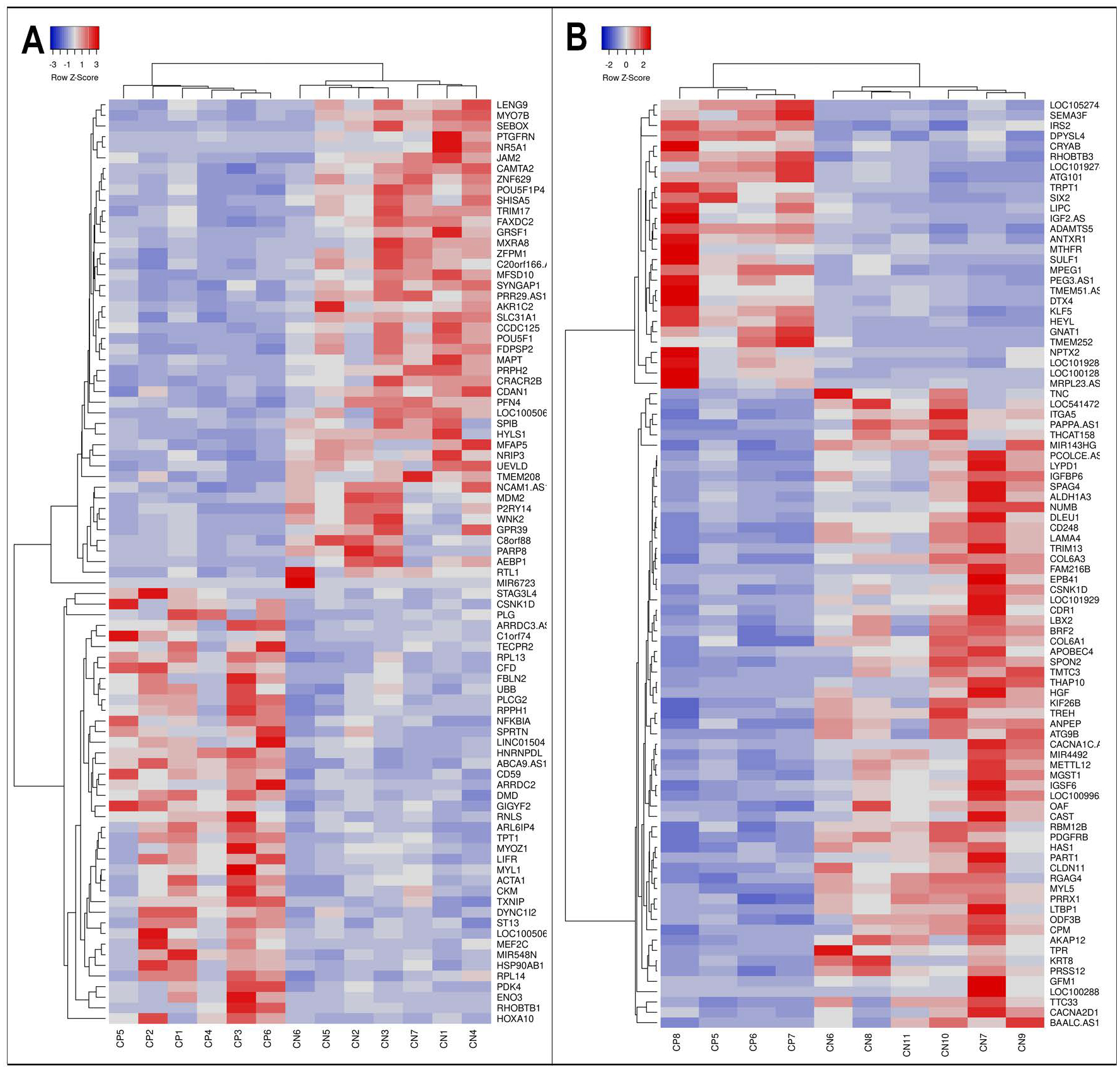
Differential expression between CP and control subjects is visualized via heatmap clustering of the genes and samples showing statistically-significant DEGs based on FDR corrected p value for skeletal muscle (A) and satellite cell-derived myotubes (B). Each column represents a single subject and each row represents a single gene with blue indicating lower expression in CP and red representing higher expression. Distinct clustering patterns were identified between the CP and control cohorts.
Figure 2: qPCR validation of RNA-seq data.
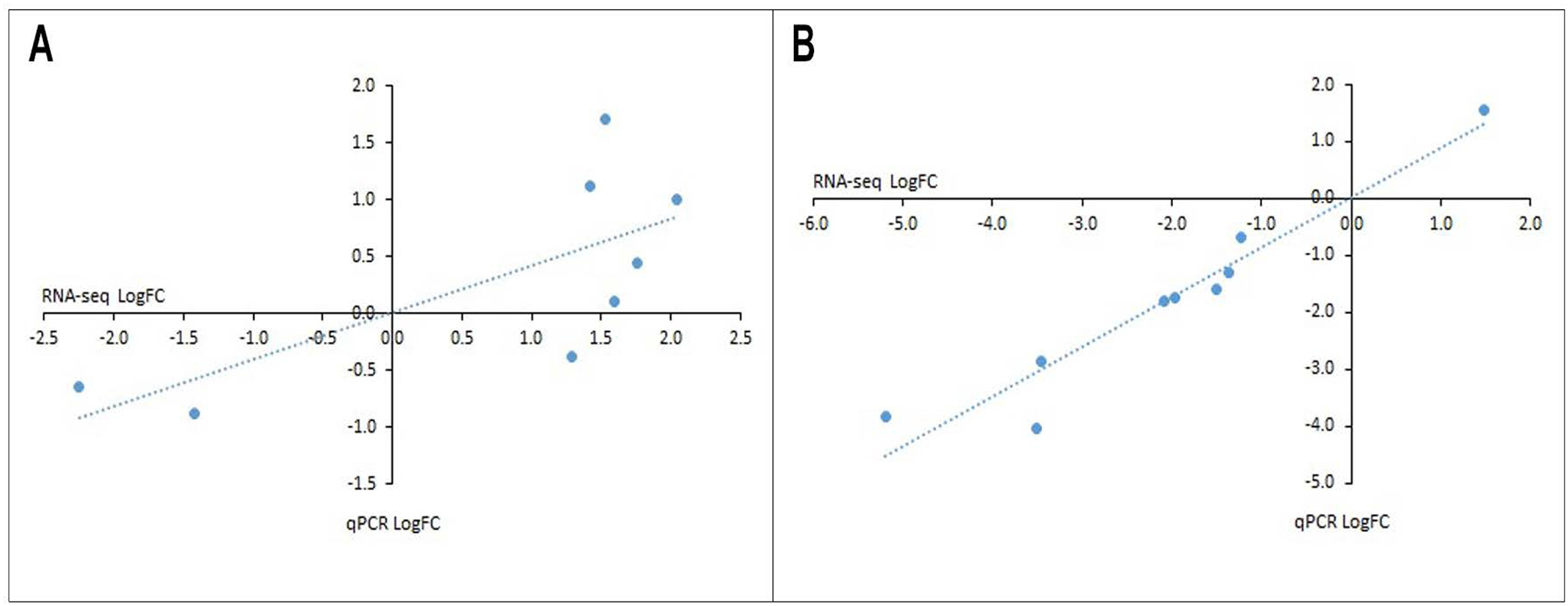
For skeletal muscle (A) and SCDMTs (B), qPCR validation was performed on the top 3 most significant genes from three different functional categories. Each point represents a single gene. Significant correlations between log2 fold change determined by RNA-seq and qPCR were observed for both tissue (Pearson’s r=0.723, p=0.043) and SCDMTs (Pearson’s r=0.960, p<0.001).
qPCR validation was performed on select genes from different functional categories (signaling, structural, and metabolic for tissue; signaling, ECM, and ECM-binding for SCDMTs). From each category, the top 3 most significant genes with RNA-seq counts > 25 in at least half the samples were chosen (for tissue, signaling = MDM2, TPT1, MXRA8, structural = DMD, DYNC1|2, ACTA1, and metabolic = PDK4, CKM, AKR1C; for SCDMTs, signaling = PDGFRB, HGF, PRRX1, ECM = LAMA4, COL6A3, COL6A1, and ECM-binding = CD248, ADAMTS5, ITGA5). Of these, only AKR1C did not amplify by qPCR. Significant correlations between RNA-seq and qPCR results were observed for both tissue (Figure 2A, Supplemental Figure 4; Pearson’s r=0.723, p=0.043, validated with permutation test as p=0.040) and SCDMTs (Figure 2B, Supplemental Figure 5; Pearson’s r=0.960, p<0.001, validated with permutation test as p<0.001).
For muscle tissue, a protein association network analysis (Figure 3A) identified several clusters, the largest contained 6 genes: UBB, MDM2, NFKB1A, SPRTN, UEVLD, and WNK2. A pathway enrichment analysis identified 11 significantly enriched pathways associated with spastic CP (Supplemental Table 3): toll-like receptor cascades, mitotic G2-G2/M phases, asparagine N-linked glycosylation, prostate cancer, C-type lectin receptor signaling pathway, striated muscle contraction, aurora A signaling, oncogene induced senescence, HSP90 chaperone cycle for steroid hormone receptors, C-type lectin receptors, and transcriptional regulation of pluripotent stem cells. In SCDMTs, the largest protein association cluster contained 3 genes (Figure 3B): ITGA5, COL6A3, and COL6A1. A pathway enrichment analysis identified 17 significantly enriched pathways (Supplemental Table 4). The most significant pathways were focal adhesion, P13K-Akt signaling pathway, beta1 integrin cell surface interaction, and ECM-receptor interactions.
Figure 3: Protein association network analyses for muscle and satellite cells.
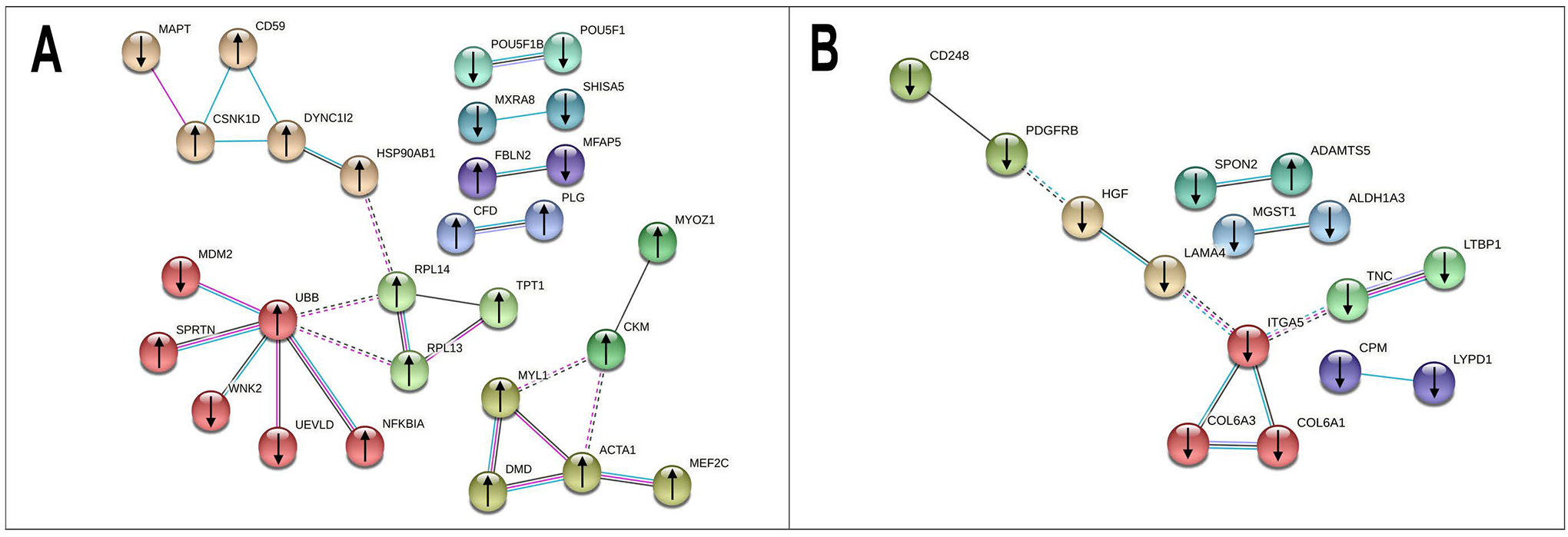
Differentially expressed genes were analyzed and rendered via STRING for muscle (A) and satellite cell-derived myotubes (B). The lines connecting the nodes represent known and predicted interactions between the proteins associated with the RNA-seq data. Nodes are colored based on cluster and edges are colored based on the type of association. Known interactions are colored cyan (from curated databases) or magenta (experimentally determined). Other interactions are colored black (co-expression) or light purple (protein homology). Inter-cluster edges are represented by dashed lines.
Discussion
Spastic CP is characterized by musculoskeletal dysfunction and disordered movement. A more complete understanding of dysregulated muscle pathways is needed to illuminate details of muscle pathophysiology and identify potential therapeutics. Previous Affymetrix U133A6,8 and PCR-based7 studies of CP muscle have reported some differences in gene expression; however, these techniques employ a limited number of probes and have technical limitations, including cross-hybridization and non-specific hybridization.18 RNA-seq provides more complete DEG detection and greater sensitivity, reproducibility, and accuracy.
We used RNA-seq to investigate DEGs in muscle of pediatric subjects with spastic CP and validated select genes with qPCR. Our results highlight fundamental differences that may be associated with the pathophysiology of spastic CP that have not been identified previously. Groups of genes were evident (Supplemental Table 5), and genes that were positively or negatively altered in CP included several associated with cytoskeletal structure, extracellular structure, metabolism, development and growth, and stress, cell death, and autophagy, suggesting complex functional alterations in CP. In addition, genes associated with antisense, non-coding RNA, or microRNA mediated pathways, receptor composition, and signaling were altered in CP. The gene that was upregulated by the largest amount was HOXA10, a transcription factor that promotes cell proliferation. HOX homeodomain transcription factors may play critical roles in adult skeletal muscle by controlling cell identity and muscle fiber type differentiation.19 Other upregulated genes in CP included CKM, ENO3, DMD, ACTA1, and MYL1. Muscle creatine kinase (CKM) is involved in energy metabolism and fatigability through the ATP regeneration from phosphocreatine and ADP.20 An increase in β-enolase (ENO3), a glycolytic enzyme prevalent in muscle, could indicate increased energy expenditure in subjects with CP21 or a difference in muscle fiber type composition.22 The upregulation of dystrophin (DMD), an important structural component of the sarcolemma, as well as the genes for contractile proteins ACTA1 and MYL1, indicates that muscle protein synthesis pathways may be upregulated in CP. Interestingly, DMD was also found to be upregulated in an Afflymetrix U133A chip study of wrist muscles from subjects with CP,6 and mutations in DMD are associated with Duchenne and Becker forms of muscular dystrophy, diseases characterized by progressive muscle weakness and atrophy. Overall, the DEGs found in spinalis tissue differed from those previously reported in upper extremity6 and lower extremity8,7 in CP, indicating that response to spasticity can be unique in different muscles.
Gene ontology analysis suggested that the most significantly enriched pathway in muscle tissue was Toll-like receptor cascades, a pathway implicated in cachexia23 and inflammation24; however, of the 141 genes in the pathway, only 47 were expressed in the tissue and only 4 were differentially expressed. We note that the lack of differences in RNA-seq signals across the set of 47 expressed genes does not diminish the significance of the finding that Toll-like receptor pathway intermediates were over-represented in our tissue data; additional work using proteomics or metabolomics may be needed to further understand the potential functional involvement of the identified pathways in CP. In addition, the DEGs identified in the Toll-like receptor pathway have important functions in other pathways that may be relevant. For example, MEF2C is a transcriptional activator which binds specifically to the MEF2 element in the regulatory regions of many muscle-specific genes and is one of the genes activated at the start of terminal differentiation. Both NF-κB inhibitor alpha (NFKB1A) and ubiquitin (UBB) are involved in ubiquitination and an upregulation of these genes suggests alterations in protein turnover in samples derived from subjects with CP.
The tissue data provided a crucial snapshot of muscle transcript levels in spastic CP, which is a chronic condition, plus any differential responses to surgical preparation. In short, differences in gene expression between the cohorts in the tissue arm of the study could arise from multiple different influences. First, tissue composition in the two cohorts may differ (e.g., CP muscle can have increased fatty infiltration). In our study, however, we found no obvious differences in muscle histology across the groups (see Supplemental Figure 1). Second, there may be differences in erector spinae function between groups; although this possibility cannot be ruled out, the samples used here were from the concave side of the scoliotic curve in patients presenting for spinal fusion surgery; thus, both cohorts had some presumed level of compromised function associated with scoliosis. Third, although not a confounding issue in the present study, the origin and lineage of different muscles may be important when comparing results from hypaxial limb muscles used in prior studies6–8 and epaxial trunk muscle like the erector spinae used here. Finally, we think the perinatal insult causing CP (e.g., hypoxia-ischemia) may have long-term sequelae in muscle and that these effects may be context dependent. More detailed studies are needed to determine whether alterations acquired at the time of CP onset may carry forward in life.
To elucidate potential gene expression differences in a model of dynamic muscle remodeling, we also investigated expression in SCDMTs that had been stimulated to differentiate in vitro. Of the 6308 genes expressed in the skeletal muscle tissue arm of the study, 4889 were also expressed in the differentiated SCDMTs; however, expression levels did not correlate (data not shown) and SCDMTs expressed an additional 2570 genes that were not present in the tissue. These results were not surprising given the differences between the mature muscle tissue samples, which represent a snapshot of the complete tissue, and satellite cells that were separated from the extra-cellular influences of the tissue and allowed to progress to an early state of muscle differentiation in culture. Not surprisingly, only one DEG was identified in both the tissue and cell analyses. CSNK1D was elevated in CP tissue but reduced in CP SCDMTs.
Groups of genes were also evident in the SCDMT analysis (Supplemental Table 5) suggesting that intrinsic alterations were retained by the cells during their isolation and culture. Published studies suggest that ECM plays key roles in muscle development,25 and several ECM markers have been reported to be dysregulated in muscle and SCs from CP subjects.26,7 Here, we found significant downregulation of critical ECM (LAMA4, COL6A1, and COL6A3) in CP, as well as the cytokine HGF. COL6A1 and COL6A3 both encode subunits of collagen VI, which is a critical component of the satellite cell niche. Collagen VI is required for satellite cell activity through modulation of muscle stiffness.27 Hepatocyte growth factor (HGF) can induce quiescent cells to enter the cell cycle, enhance their proliferation, and inhibit their differentiation.28–30 Downregulation of these genes may be related to previous findings that subjects with CP have smaller SC populations with reduced self-renewal capacity.10 In the SCDMT analysis, the most significantly enriched pathways included focal adhesion, PI3K-Akt signaling, beta1 integrin interaction, and ECM-receptor interaction. Studies suggest ECM proteins, receptors, and signaling pathways play key roles in muscle formation, growth, and maintenance, and several ECM markers have been previously reported as being dysregulated in muscle and SCs from CP subjects.
Overall, our study identified significant transcriptomic differences in spinalis muscle tissue and SCDMTs from individuals with spastic CP. The findings of our current study are compelling but limited by our reliance on patients presenting for specific surgeries, which may represent a sub-population of all individuals with spastic CP. The interpretation of results is therefore necessarily bounded by these limitations, and extrapolation to the broader CP community may require additional research. While RNA-seq has promise for diagnostic, prognostic, and therapeutic applications, additional studies are also warranted to examine gene expression changes over time in the muscle transcriptome. Additional integration with genetic, epigenetic, or gene regulatory data from subjects with CP31 could be informative. Based on the work presented here, several key genes and pathways were identified as potential targets for future investigation in spastic CP.
Supplementary Material
What This Paper Adds:
Expression differences were found in muscle tissue and in isolated muscle cells.
There was low variability in expression among cells isolated from different muscles.
Expression differences suggest complex functional alterations in spastic CP.
Genes/pathways were identified as potential targets for future investigation in spastic CP.
Acknowledgements
This work was supported by the Delaware Bioscience Center for Advanced Technology; an American Academy for Cerebral Palsy and Developmental Medicine Pedal with Pete Foundation award to RA; the Delaware CTR ACCEL Program [U54-GM104941] to EC. The authors would like to thank the Nemours Foundation, Nemours Biomedical Research, and the Department of Pediatrics for institutional support of EC, KR, SL, and RA, as well as The Swank Foundation for their support to RA allowing the establishment of the neuro-orthopedic tissue repository at Nemours that provided samples for the work. The authors thank Jeff Myers for his contributions to the RNA-seq genome alignment workflow and Adam Marsh and Jonathan Hicks for their helpful discussions and suggestions.
List of abbreviations
- CP
cerebral palsy
- DEG
differentially expressed gene
- ECM
extracellular matrix
- FDR
false discovery rate
- PCR
polymerase chain reaction
- SC
satellite cell
- SCDMT
satellite cell-derived myotube
References
- 1.Christensen D, Van Naarden Braun K, Doernberg NS, Maenner MJ, Arneson CL, Durkin MS, Benedict RE, Kirby RS, Wingate MS, Fitzgerald R, Yeargin-Allsopp M. Prevalence of cerebral palsy, co-occurring autism spectrum disorders, and motor functioning - Autism and Developmental Disabilities Monitoring Network, USA, 2008. Dev Med Child Neurol 2014; 56: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham HK, Rosenbaum P, Paneth N, Dan B, Lin JP, Damiano DL, Becher JG, Gaebler-Spira D, Colver A, Reddihough DS, Crompton KE, Lieber RL. Cerebral palsy. Nat Rev Dis Primers 2016; 2: 15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith LR, Lee KS, Ward SR, Chambers HG, Lieber RL. Hamstring contractures in children with spastic cerebral palsy result from a stiffer extracellular matrix and increased in vivo sarcomere length. J Physiol 2011; 589: 2625–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson DL, Miller F, Subramanian P, Modlesky CM. Adipose tissue infiltration of skeletal muscle in children with cerebral palsy. J Pediatr 2009; 154: 715–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson KG, Mendonca JL, Militar JL, Theroux MC, Dabney KW, Shah SA, Miller F, Akins RE. Disruption of basal lamina components in neuromotor synapses of children with spastic quadriplegic cerebral palsy. PLoS One 2013; 8: e70288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith LR, Ponten E, Hedstrom Y, Ward SR, Chambers HG, Subramaniam S, Lieber RL. Novel transcriptional profile in wrist muscles from cerebral palsy patients. BMC Med Genomics 2009; 2: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Von Walden F, Gantelius S, Liu C, Borgstrom H, Bjork L, Gremark O, Stal P, Nader GA, Ponte NE. Muscle contractures in patients with cerebral palsy and acquired brain injury are associated with extracellular matrix expansion, pro-inflammatory gene expression, and reduced rRNA synthesis. Muscle Nerve 2018; 58: 277–85. [DOI] [PubMed] [Google Scholar]
- 8.Smith LR, Chambers HG, Subramaniam S, Lieber RL. Transcriptional abnormalities of hamstring muscle contractures in children with cerebral palsy. PLoS One 2012; 7: e40686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domenighetti AA, Mathewson MA, Pichika R, Sibley LA, Zhao L, Chambers HG, Lieber RL. Loss of myogenic potential and fusion capacity of muscle stem cells isolated from contractured muscle in children with cerebral palsy. Am J Physiol Cell Physiol 2018; 315: C247–C57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dayanidhi S, Dykstra PB, Lyubasyuk V, McKay BR, Chambers HG, Lieber RL. Reduced satellite cell number in situ in muscular contractures from children with cerebral palsy. J Orthop Res 2015; 33: 1039–45. [DOI] [PubMed] [Google Scholar]
- 11.R Core Team (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/. [Google Scholar]
- 12.Wei T; Simko V R package “corrplot”: Visualization of a Correlation Matrix (Version 0.84). Available from https://github.com/taiyun/corrplot. 2017.
- 13.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 2012; 40: 4288–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res 2016; 44: W147–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019; 47: D607–D13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Croft D, O’Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, Jupe S, Kalatskaya I, Mahajan S, May B, Ndegwa N, Schmidt E, Shamovsky V, Yung C, Birney E, Hermjakob H, D’Eustachio P, Stein L. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res 2011; 39: D691–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini YHY. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological) 1995; 57: 289–300. [Google Scholar]
- 18.Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One 2014; 9: e78644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez-Prendes R, Quintanilla R, Marmol-Sanchez E, Pena RN, Ballester M, Cardoso TF, Manunza A, Casellas J, Canovas A, Diaz I, Noguera JL, Castello A, Mercade A, Amills M. Comparing the mRNA expression profile and the genetic determinism of intramuscular fat traits in the porcine gluteus medius and longissimus dorsi muscles. BMC Genomics 2019; 20: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams CJ, Williams MG, Eynon N, Ashton KJ, Little JP, Wisloff U, Coombes JS. Genes to predict VO2max trainability: a systematic review. BMC Genomics 2017; 18: 831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zogby AM, Dayanidhi S, Chambers HG, Schenk S, Lieber RL. Skeletal muscle fiber-type specific succinate dehydrogenase activity in cerebral palsy. Muscle Nerve 2017; 55: 122–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathewson MA, Lieber RL. Pathophysiology of muscle contractures in cerebral palsy. Phys Med Rehabil Clin N Am 2015; 26: 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bohnert KR, Goli P, Roy A, Sharma AK, Xiong G, Gallot YS, Kumar A. The Toll-Like Receptor/MyD88/XBP1 Signaling Axis Mediates Skeletal Muscle Wasting during Cancer Cachexia. Mol Cell Biol 2019; 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verzola D, Bonanni A, Sofia A, Montecucco F, D’Amato E, Cademartori V, Parodi EL, Viazzi F, Venturelli C, Brunori G, Garibotto G. Toll-like receptor 4 signalling mediates inflammation in skeletal muscle of patients with chronic kidney disease. J Cachexia Sarcopenia Muscle 2017; 8: 131–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goetsch SC, Hawke TJ, Gallardo TD, Richardson JA, Garry DJ. Transcriptional profiling and regulation of the extracellular matrix during muscle regeneration. Physiol Genomics 2003; 14: 261–71. [DOI] [PubMed] [Google Scholar]
- 26.Booth CM, Cortina-Borja MJ, Theologis TN. Collagen accumulation in muscles of children with cerebral palsy and correlation with severity of spasticity. Dev Med Child Neurol 2001; 43: 314–20. [DOI] [PubMed] [Google Scholar]
- 27.Urciuolo A, Quarta M, Morbidoni V, Gattazzo F, Molon S, Grumati P, Montemurro F, Tedesco FS, Blaauw B, Cossu G, Vozzi G, Rando TA, Bonaldo P. Collagen VI regulates satellite cell self-renewal and muscle regeneration. Nat Commun 2013; 4: 1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gal-Levi R, Leshem Y, Aoki S, Nakamura T, Halevy O. Hepatocyte growth factor plays a dual role in regulating skeletal muscle satellite cell proliferation and differentiation. Biochim Biophys Acta 1998; 1402: 39–51. [DOI] [PubMed] [Google Scholar]
- 29.Miller KJ, Thaloor D, Matteson S, Pavlath GK. Hepatocyte growth factor affects satellite cell activation and differentiation in regenerating skeletal muscle. Am J Physiol Cell Physiol 2000; 278: C174–81. [DOI] [PubMed] [Google Scholar]
- 30.Santos-Zas I, Cid-Diaz T, Gonzalez-Sanchez J, Gurriaran-Rodriguez U, Seoane-Mosteiro C, Porteiro B, Nogueiras R, Casabiell X, Relova JL, Gallego R, Mouly V, Pazos Y, Camina JP. Obestatin controls skeletal muscle fiber-type determination. Sci Rep 2017; 7: 2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crowgey EL, Marsh AG, Robinson KG, Yeager SK, Akins RE. Epigenetic machine learning: utilizing DNA methylation patterns to predict spastic cerebral palsy. BMC Bioinformatics 2018; 19: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


