Abstract
An integrated microfluidic platform (IMP) utilizing real-time reverse-transcription loop-mediated isothermal amplification (RT-LAMP) was developed here for detection and quantification of three genes of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; i.e., coronavirus diseases 2019 (COVID-19)): RNA-dependent RNA polymerase, the envelope gene, and the nucleocapsid gene for molecular diagnosis. The IMP comprised a microfluidic chip, a temperature control module, a fluidic control module that collectively carried out viral lysis, RNA extraction, RT-LAMP, and the real-time detection within 90 min in an automatic format. A limit of detection of 5 × 103 copies/reaction for each gene was determined with three samples including synthesized RNAs, inactive viruses, and RNAs extracted from clinical samples; this compact platform could be a useful tool for COVID-19 diagnostics.
Keywords: COVID-19, Loop-mediated isothermal amplification, Microfluidics, Molecular diagnostics, SARS-CoV-2
Nomenclature and abbreviations
- BIP
Backward internal primer
- COVID-19
Coronavirus diseases 2019
- ddH2O
Double-distilled water
- EMV
Electromagnetic valve
- E
Envelope protein
- FIP
Forward internal primer
- InFA & InFB, respectively
Influenza A and B
- IMP
Integrated microfluidic platform
- Kp
Klebsiella pneumoniae
- LOD
Limit of detection
- LAMP
Loop-mediated isothermal amplification
- BCG
Mycobacterium bovis
- NCKUH
National Cheng Kung University Hospital
- NEB
New England Biolabs
- ORFs
Open reading frames
- PMT
Photomultiplier tube
- PDMS
Polydimethylsiloxane
- PMMA
Polymethylmethacrylate
- RT
Reverse transcription
- RT-PCR
Reverse-transcription polymerase chain reaction
- RdRp
RNA-dependent RNA polymerase
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- Sp
Streptococcus pneumoniae
- TE
Thermoelectric
- WHO
World Health Organization
1. Introduction
The coronavirus diseases 2019 (COVID-19) pandemic, which began in China in late 2019, was caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; an RNA virus). The outbreak of SARS-CoV-2 soon became a global pandemic recognized by World Health Organization (WHO) [1]. Total deaths reached four million people worldwide by September 2021, in part due to high rates of transmission between asymptomatic individuals [2]. Developing fast and accurate diagnostic methods could effectively prevent the spread of infections, and SARS-CoV-2 detection devices have indeed been critical. This is normally undertaken via reverse-transcription polymerase chain reaction (RT-PCR), which is highly sensitive and specific [3] although it may require a lengthy thermal-cycling process on a costly apparatus. Coronavirus genomes consist of 8–10 open reading frames (ORFs) [4]; For instance, ORF1 is translated into 16 non-structural proteins that include the RNA-dependent RNA polymerase enzyme (RdRp). Envelope (E), nucleocapsid (N), S spikes, and membrane proteins are also essential for completion of the viral replication cycle [5], and RdRp, E, and N, genes have been widely used as SARS-CoV-2 biomarkers [6]. In addition to PCR [7], virus antigen and antibody testing have been used [8], [9]; such procedures are relatively fast and simple although they lack the specificity and sensitivity of nucleic acids-based approaches and may generate false-positive or false-negative results. In addition, antibodies are only observed in blood in the middle and later stages of the illness and thus are not suitable for early detection [9], [10].
RT-PCR is relatively time-consuming (~2–4 hr) and labor-intensive; furthermore, viral lysis and RNA extraction must be undertaken by well-trained personnel. In contrast, loop-mediated isothermal amplification (LAMP) can amplify nucleic acids without thermocycling [11]; this is associated with reductions in detection time compared to RT-PCR [12]. Efforts have been made to miniaturize and automate LAMP [13], [14], [15], and LAMP technology has been integrated with microfluidic technologies [16], [17] for rapid and sensitive detection of pathogens [18]. Compared with traditional LAMP, the main advantages of these microfluidics-based systems are 1) lower sample and reagent volumes, 2) compact size for on-site testing, and 3) capacity for automation via integration of microfluidic devices (e.g., micropumps, micromixer, & microreactors). For instance, we demonstrated that sample purification, RNA extraction, and LAMP could be performed on an integrated microfluidic platform without human intervention for influenza diagnosis [19]. However, this device was used for end-point testing only (i.e., only qualitative, not quantitative diagnosis), which may not be satisfactory for SARS-CoV-2 diagnostics since quantitative diagnosis of viral load is required.
Several devices/systems for detecting SARS-CoV-2 have been reported [20], [21], [22]. For example, a portable platform for rapid virus detection was characterized by a limit of detection (LOD) of 5 × 104 copies/mL of the N gene in 30 min though RNA extraction was carried out off-chip [20]. Similarly, an LOD of only 1 copy/mL was reported for a device featuring an integrated microfluidic platform (IMP) controlled by a smartphone [21]. Another smartphone-controlled device, a centrifugal microfluidic platform, was characterized by an LOD of 104 copies/mL of the Orf1ab gene, and the detection time was only 60 min [22]. However, none of these approaches are fully automated (i.e., requiring human intervention during detection). They also feature only 1–2 genes while a 3-gene approach has been shown to reduce chances of false-negative or false-positive diagnoses [23]. Given these limitations, we devised an IMP for rapid detection and quantification of three viral genes (E, N, and RdRp) of SARS-CoV-2. The entire process including rival lysis, RNA extraction, RT-LAMP and quantification could be automated within 90 min with a limit of detection as low as 5 × 103 copies/reaction (i.e., 2 × 102 copies/μL). Therefore, this new platform could provide useful and accurate detection for SARS-CoV-2.
2. Materials and methods
2.1. Experimental process for viral lysis, RNA extraction, RT- LAMP, and real-time quantification
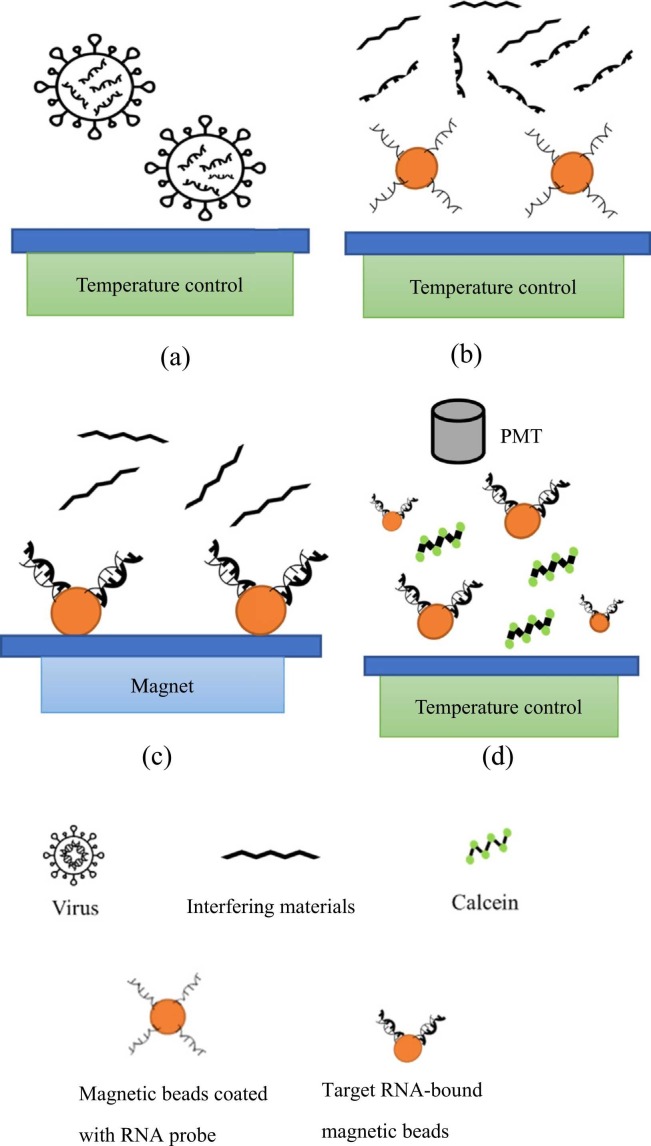
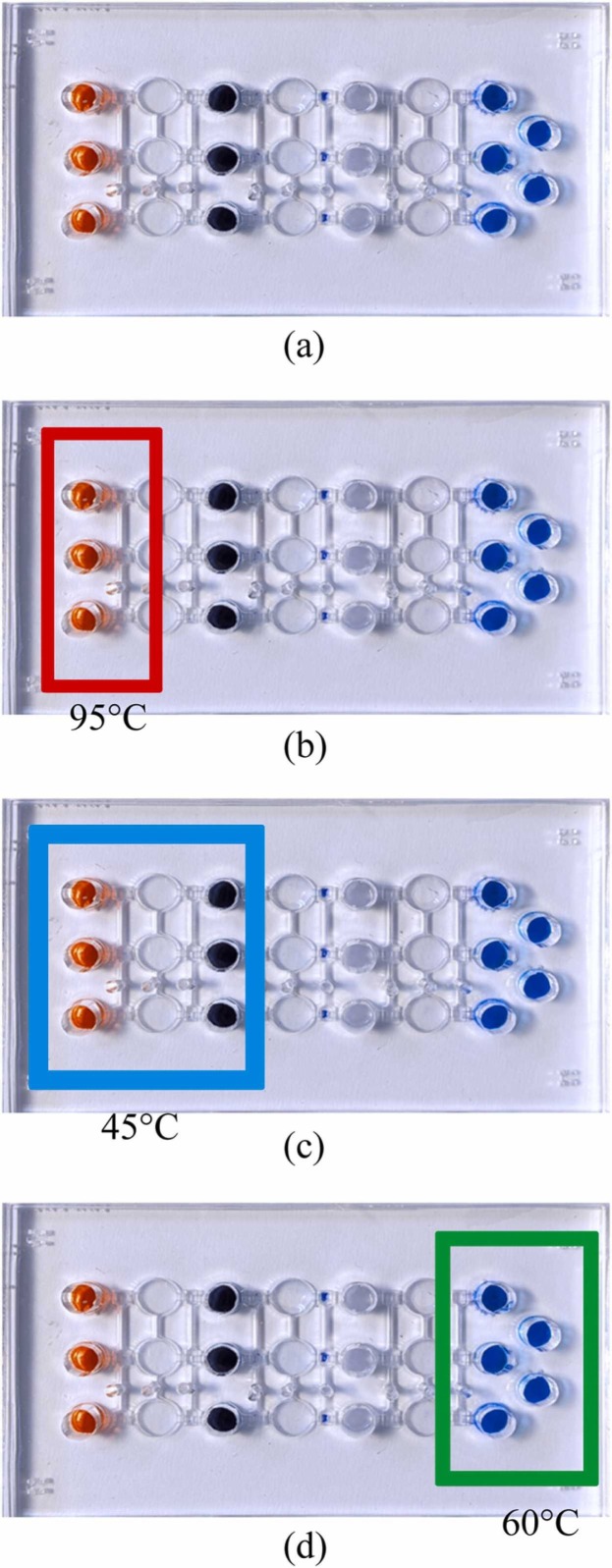
Fig. 1 shows a schematic of the entire experimental procedure, including viral lysis, RNA extraction, RT-LAMP, and real-time quantification ( Fig. 3 for on-chip details.). First, 35 μL of test sample (inactive SARS-CoV-2 viruses (Qnostics, UK) with different concentrations was firstly lysed thermally for 5 min at 95 °C or chemically (lysis buffer from ABP biosciences, USA; catalog# R145) for 20 min at room temperature in the corresponding chambers (Fig. 3a-b and Table S3). In other experiments, synthesized RNA (35 μL) with different concentrations for the three target genes (Antech Diagnostics, USA) or previously extracted viral RNA (35 μL) with different concentrations were instead used; the latter were obtained from National Cheng Kung University Hospital (NCKUH), Taiwan (with a stock concentration of 108 copies/μL). In order to test primer specificity, Streptococcus pneumoniae (Sp), Pseudoemonas aeruginosa (PA), Mycobacterium bovis (BCG), Influenza A and B (InfA & InfB, respectively), and Klebsiella pneumonia (Kp) were also prepared and tested (10 ¹¹ copies/reaction). Note that they are common acute upper respiratory viruses/bacteria. Released RNAs were transported to chambers (described below) to be mixed with 5 μL of 1.05 µm-diameter Dynabeads MyOne™ (carboxylic acid, Invitrogen, USA; 106 beads/μL) that had been conjugated with three amine-modified RNA-specific probes (Table S1) via carboxylation with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide.
Fig. 1.
Schematic illustration of the experimental procedure for detection and quantification of SARS-CoV-2 in the microfluidic device. (a) Sample loading. (b) Viral lysis and addition of magnetic beads surface-coated with RNA probes. (c) RNA extraction under an external magnetic field. (d) RT-LAMP of the extracted RNAs and real-time optical detection of the products.
Fig. 3.
A schematic illustration showing where various procedures occurred on the microfluidic chip. (a) Reagents were loaded into corresponding chambers and (b) viral lysis (at 95 °C or chemical lysis at room temperature) was executed to release RNA. (c) Then magnetic beads surface-coated with RNA probes captured the target RNA for three genes at 45 °C. (d) Next, RT-LAMP was conducted at 60 °C for 60 min after washing away wastes. Finally, signals were optically detected during real-time RT-LAMP.
After RNA extraction with beads for 10 min at 45 °C, an external magnet (2000 Gauss) was placed under the chambers to collect the magnetic bead-RNA complexes for 1 min. Then, unbound RNAs and interfering materials were washed out, and the complexes were concentrated by the magnet for 5 min. Next, unbound RNAs were again washed away with 35 μL of double-distilled water (ddH2O) for 5 min. Next, RT-LAMP reagents (24 μL/reaction) were mixed with magnetic bead-RNA complexes and then transported to the corresponding chambers for RT-LAMP using calcein for fluorescent detection as in [19]. In addition to the LAMP primers F3 (5 μL of 10 μM), B3 (0.5 μL of 10 μM), FIP (1.5 μL of 20 μM), and BIP (1.5 μL of 20 μM; Table S2; all from Protech, Taiwan), RT-LAMP reactions comprised 2.5 μL of 10X isothermal amplification buffer (New England Biolabs [NEB], USA), 3 μL of 10 mM dNTP mix (Protech), 2 μL of 100 mM MgSO4 (NEB), 0.8 μL of 5 M betaine (0.8 M final concentration, Sigma-Aldrich, USA), 1 μL of Bst DNA polymerase (8000 U/mL, NEB), 0.5 μL of reverse transcriptase (Promega, USA), 1 μL of 0.5 mM calcein (C0875, Sigma-Aldrich), and 9.2 μL of ddH2O. For negative/positive control, ddH2O/target gene mixture (1 μL/reaction) was added with RT-LAMP reagents (24 μL/reaction). In a 60-min amplification at 60 °C (Fig. 3d), calcein fluorescence was observed with a photomultiplier tube (PMT), and data were analyzed by OriginLab (USA).
2.2. Design, microfabrication, and operation of the microfluidic chip
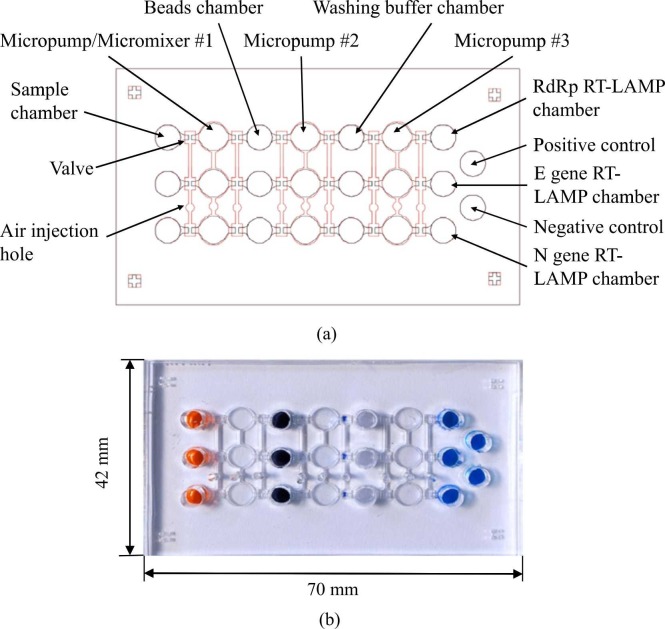
The microfluidic chip (Figs. S2 and 2a), which consisted of an air channel layer, a liquid control layer, 9 micropumps/mixers, 18 microvalves, 14 microchambers, and 18 air injection holes, was made from polydimethylsiloxane (PDMS, Sylgard 184 A/B, Sil-More Industrial, USA) that had been poured into polymethymethacrylate (PMMA, Dateh Industrial, Taiwan) molds [24]. The PDMS was mechanically demolded from PMMA, cured at 80 °C for 5 hr, treated with oxygen plasma (Cute MP/R, Atlas Technology, Taiwan) for 3 min, and bound to a 0.7 mm-thick glass slide (Taiwan Glass, Taiwan). The 70 × 42 × 10 mm (L x W x H) chip (Fig. 2b) was equipped with five parallel microchambers for simultaneous detection of three genes, a positive control, and a negative control (water; Fig. 2a). Both samples and reagents were transported by pneumatically driven membrane-type micropumps (Fig. S1), which also functioned as micromixers. For liquid transport, a positive gauge pressure was provided by closing the left microvalves and activating the micropumps (Fig. S1a); the liquid was drawn into the micropump with a negative gauge pressure provided (Fig. S1b). The left microvalve was closed via a positive gauge pressure, and the right microvalve was opened via a negative gauge pressure for liquid transport (Fig. S1c); in other words, liquid transport was achieved by providing a positive gauge pressure for activating the micropump and then providing a positive gauge pressure for closing the right microvalve (Fig. S1d). For mixing, liquids were drawn into the micropump while providing a negative gauge pressure for opening the left microvalve and micropump. The micropump then transported the liquid from chamber to chamber in turn by providing positive and negative gauge pressures.
Fig. 2.
(a) Schematic of the main components of the microfluidic chip: 9 micropumps/mixers, 18 microvalves, 14 microchambers, and 18 air injection holes. (b) A photograph of the chip, which included 5 RT-LAMP chambers (3 for target genes & 1 each for the positive & negative controls).
2.3. Experimental setup
The microfluidic platform included a fluidic control module, a temperature control module, and an optical detection module, all controlled by a microcontroller (Arduino UNO, Italy; Fig. S3). The fluidic control module, which was adapted from [24], consisted of an air compressor (TC-10, Centenary Material, Taiwan), a vacuum pump (DC-18 V-12, Uni-Crown, Taiwan), two regulators (IR1000–01-a & IRV10-C06B [SMC Pneumatics, Japan] for the compressor & vacuum, respectively), and electromagnetic valves (EMVs, S070B-SBG-05, SMC Pneumatics, Taiwan). The compressor provided compressed air for actuating the chip’s micropumps/micromixers and microvalves, and air passage was controlled by the microcontroller (which regulated the EMVs). The vacuum pump provided a negative gauge pressure (i.e., suction) to the microvalves, micropumps and micromixers that lifted the PDMS membranes, and the mentioned regulators established the supplied air pressures. The microcontroller (Fig. S6a) was also used to keep the two thermoelectric (TE) coolers (TEC1.127.10, Tande, Taiwan) at different temperatures, as confirmed by temperature sensors (Max6675, Centenary Materials, Taiwan) inserted between two, 10-mm-thick copper plates (NTHU Scientific Instrument Center, Taiwan) that were placed on top of the TE coolers to maintain heating uniformity. Since the platform consisted of three modules, the casing featured two units. The first was for the fluidic control and temperature control modules (19 cm × 19 cm x 11 cm; Fig. S6a), which was designed to be compact for on-site applications. The second was for the optical detection module (45 cm × 30 cm x 30 cm; Fig. S6(b), as described below). Both units were fabricated by a 3D printer (i3 Mega, Anycubic, China).
2.4. Statistical analysis and real-time detection of optical detection module
The optical detection module consisted of a PMT (C6271 +R928, Hamamatsu, Japan), an optical shutter, a laser, a set of optical filters and mirrors, and a microcontroller (Fig. S4). The light source used for the optical detection module was a diode-pumped, solid-state laser (MBL-473, CNI, China; 473 ± 1 nm, [for calcein]). A shutter controlled by a stepper motor (MG995, TowerPro, Taiwan) was used to block the laser when not needed (Fig. S5). The optical detection module was mounted with band-pass filters (500–550 nm) for fluorescence detection, and signals were captured by the PMT under microcontroller regulation. The fluorescence intensities (i.e. optical detection signals) of serially diluted test samples were evaluated with the negative control among triple experiments in this study. Thus, the two-tailed student t test analysis was used to calculate whether there was a significant difference (p < 0.05) for LOD determination.
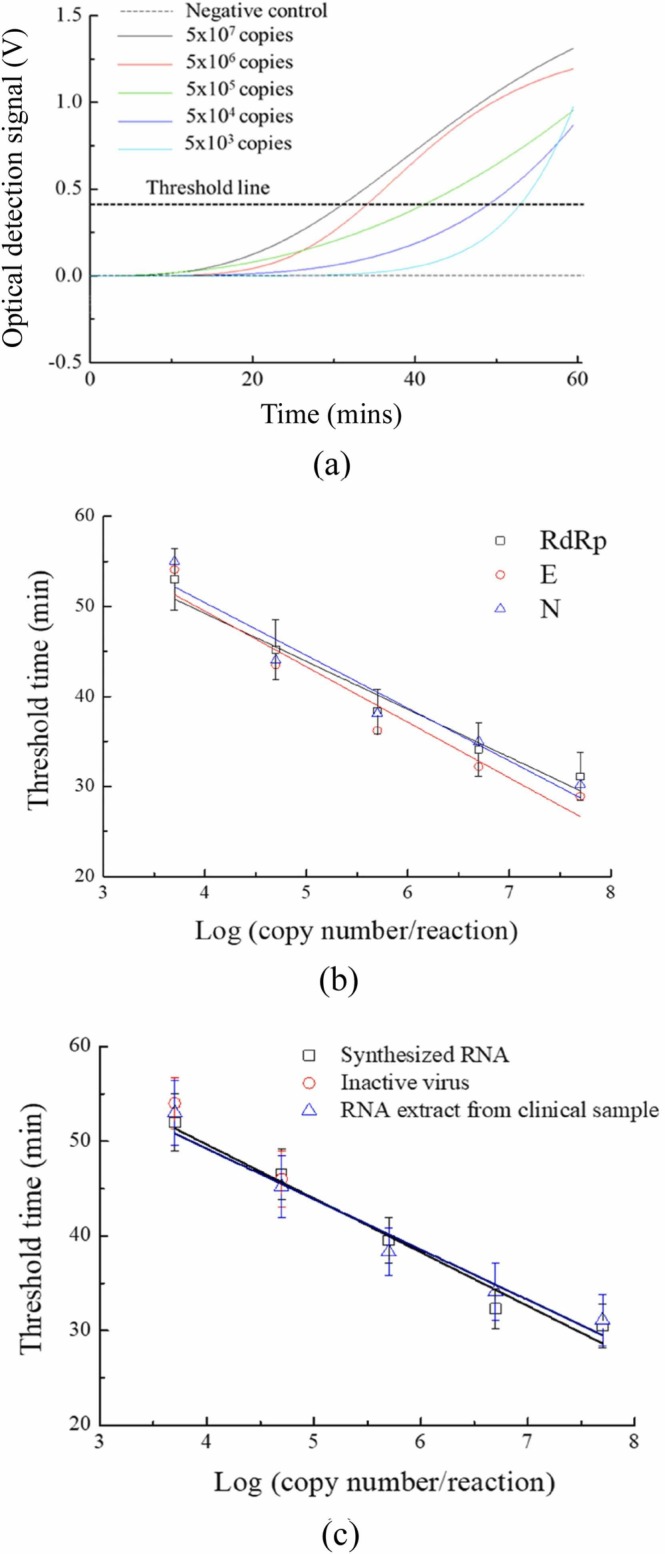
During the 60-min RT-LAMP, signals were acquired every minute, and a threshold time could be defined as the time required for the fluorescence signal to cross over a threshold, which was set as in [18], [25], [26]; since calcein was used, the threshold time was inversely proportional to the amount of initial target in the sample. With this approach, real-time fluorescence signal could be used to quantify the initial concentration of viruses. The data were fitted to a sigmoidal curve, and the threshold time was calculated as the sum of the mean fluorescence signal during the initial 5 min and 5 times of the standard deviation values during the same time as in the previous study [26].
| (1) |
Therefore, the threshold time, which was inversely proportional to the initial amount of the target, could be determined by the point of intersection of the threshold value and the fitted curve.
3. Results and discussion
3.1. Characterization of the temperature and fluidic control modules
Temperature variations within the two chambers were both < 0.5 °C (Fig. S7), and the target values of 95, 45, and 60 °C were maintained for viral lysis, RNA extraction, and RT-LAMP, respectively. When compared with a previous work featuring a different packing size (55 cm × 40 cm x 50 cm) [25], our far smaller (19 cm × 19 cm x 11 cm) temperature control module achieved comparable performance. Furthermore, it could maintain required temperature profiles automatically without human intervention.
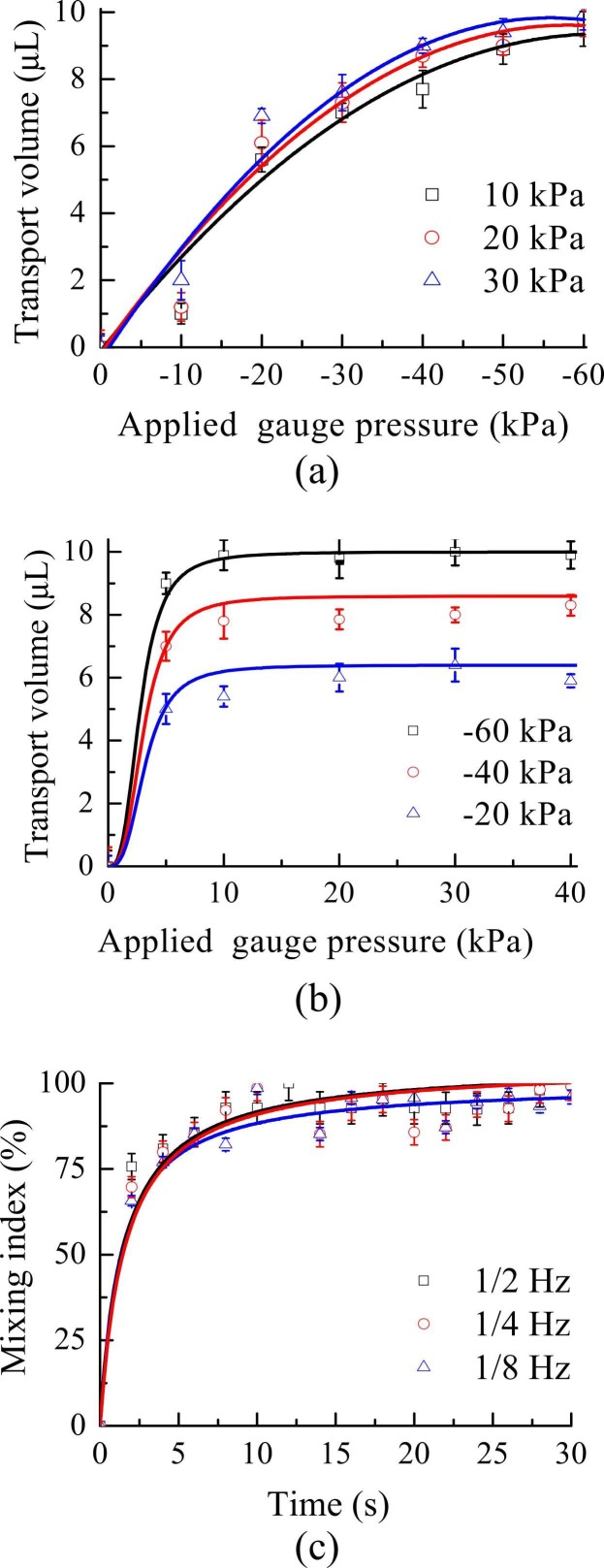
Since the microfluidic chip was equipped with four liquid chambers connected by micropumps and liquid channels within the liquid channel layer, the relationship between the pumping volume and the applied negative gauge pressure (when operated at positive gauge pressures of 10, 20 and 30 kPa) was explored ( Fig. 4a); performance was similar while applying the same negative gauge pressure, and transport volume plateaued at − 40 kPa. The relationship between the pumping volume and the applied positive gauge pressure (when operated at negative gauge pressures of −20, −40 and −60 kPa) was next explored, and the transport volume was saturated after 10 kPa in all cases. Thus, when the micropump was activated at positive/negative gauge pressures of 10 kPa/− 40 kPa in the subsequent experiments, precise transport of virus samples and magnetic beads was achieved. However, when mixing released RNA with magnetic beads coated with RNA probes, the applied pressure could affect bead capture [27]. Therefore, the gauge pressures for the liquid mixing/washing steps were decreased to positive/negative gauge pressures of 10/−15 kPa, respectively. According to previous studies [13], [27], a gentle mixing should be applied to achieve an optimal capture. If the mixing was too strong such that the resulting shear force was much higher than the binding force between released RNA and probes on the beads, then the capture rate could be deteriorated.
Fig. 4.
(a) Relationship between pumping volume and the applied negative gauge pressure at 10, 20, or 30 kPa (n = 3). (b) Relationship between pumping volume and the applied positive gauge pressure at − 20, − 40, or − 60 kPa (n = 3). (c) The mixing index of the micromixer at different driving frequencies while operated at positive/negative gauge pressures of 10/− 15 kPa. Error bars represent standard error of the mean (n = 3).
The mixing performance of the micromixer was further explored. The mixing index of the micromixer was similar (Fig. 4c) at 1/2, 1/4, and 1/8 Hz, with the former used for subsequent 10-min incubations during bead-based RNA extraction. Since the micropumps/mixers were activated by the fluidic control module (regulating pressure range from 0 to 1.01 × 102 kPa, with a resolution of 0.1 kPa), the liquid transportation and mixing could be controlled precisely and automatically without human intervention. Furthermore, the fluidic and temperature control modules were packaged in an integrated, compact casing (19 cm × 19 cm x 11 cm), that could provide more convenient than prior designs [20], [21].
3.2. Specificity and sensitivity analysis
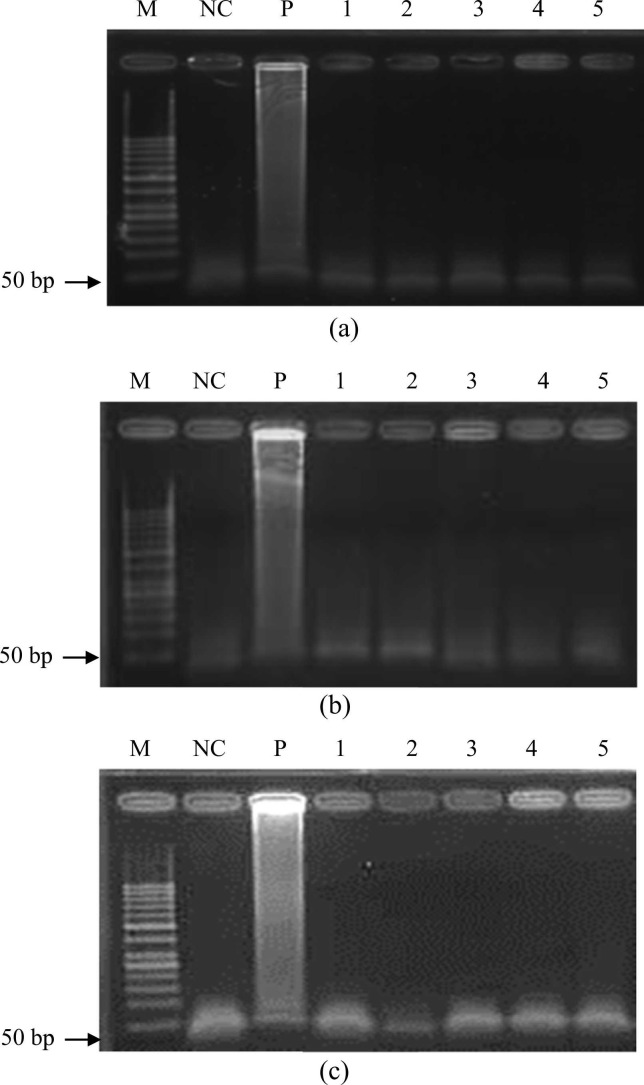
The specificity of the RT-LAMP assay was explored with synthesized RNA samples, as well as common acute upper respiratory viruses/bacteria (Sp, PA, BCG, InfA, InfB, & Kp). Agarose gel electrophoresis ( Fig. 5) showed that only the genes of the SARS-CoV-2 virus could be successfully amplified, indicating that the designed primers exhibited satisfactory specificity.
Fig. 5.
Specificity tests of on-chip RT-LAMP for the (a) E, (b) N, and (c) RdRp genes. M: 50-bp DNA ladder; NC: Negative control (ddH2O mixed with RT-LAMP reagents); P: LAMP products of the target RNA; 1: LAMP products of Streptococcus pneumoniae; 2: LAMP products of Mycobacterium bovis; 3: LAMP products of Influenza A virus; 4: LAMP products of Influenza B virus; 5: LAMP products of Klebsiella pneumoniae. The concentrations of the target genes were 103 copies/reaction (1011 copies/reaction for the off-target types).
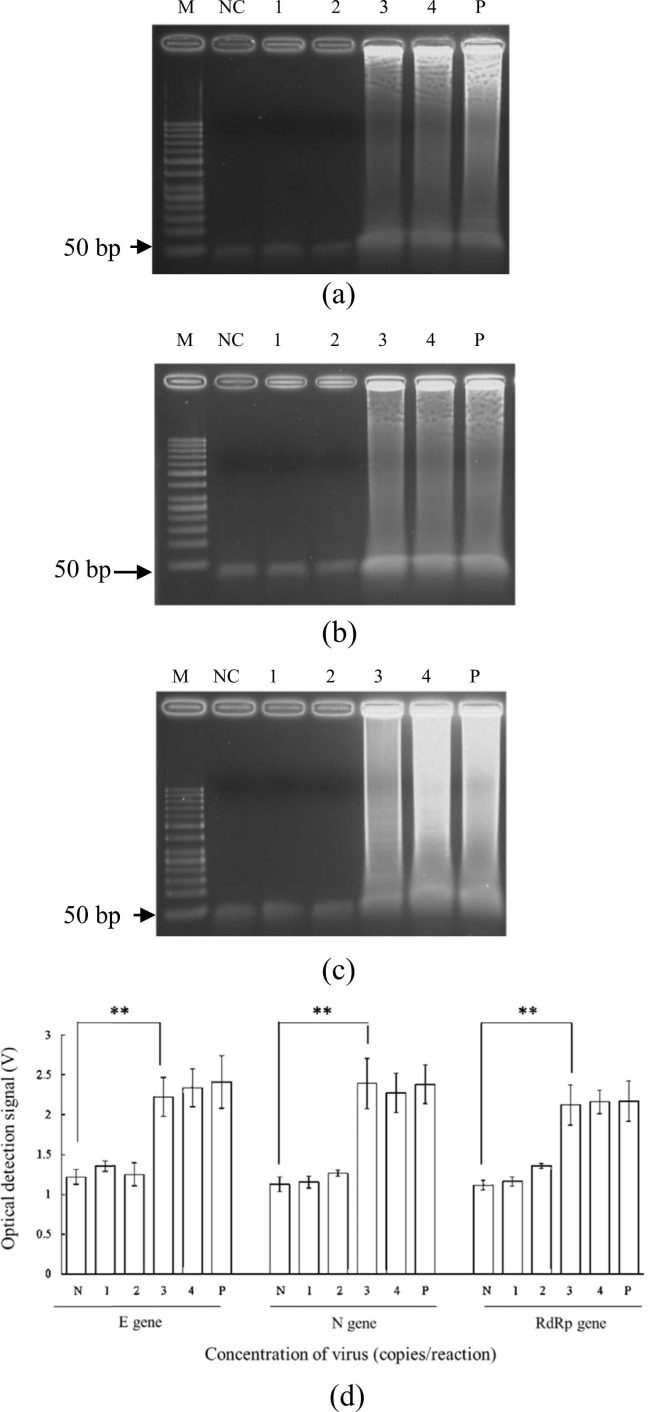
Next, E, N, and RdRp gene constructs were serially diluted from 5 × 104 to 5 × 101 copies/reaction for exploring the sensitivity of RNA extraction+RT-LAMP on-chip. After RNA extraction and RT-LAMP, each gene was successfully amplified ( Fig. 6). The LODs were found to be 5 × 103 copies/reaction for E (Fig. 6a), N (Fig. 6b), and RdRp (Fig. 6c) genes when using synthesized RNA. To evaluate the fluorescence intensities in real-time, the optical detection module was used to detect fluorescence every minute, and, after a 60-min reaction (Fig. 6d), fluorescent signals were evident and varied in intensity across differing starting concentrations. The two-tailed student t test showed that there was a significant difference (p < 0.05) for LOD determination for 3 genes with a concentration higher than 5 × 103 copies/reaction, which indicated that the optical detection was reliable.
Fig. 6.
Gel electrophoresis results of RNA extraction+RT-LAMP with synthesized RNA on chip for the (a) E, (b) N, and (c) RdRp genes. (d) Relationship between the voltage of the optical detection signal and the sample concentration. M: 50-bp DNA ladder; NC: Negative control (ddH2O mixed with RT-LAMP reagents); 1: 5 × 101 copies/reaction; 2: 5 × 102 copies/reaction; 3: 5 × 103 copies/reaction; 4: 5 × 104 copies/reaction; P: Positive control (5 × 105 copies/reaction). (d) Plot of RT-LAMP optical detection signal versus starting concentrations for the three target genes, negative control, and positive control. Error bars represent standard error of the mean (n = 3). * * indicated as p < 0.05 by the two-tailed student t test assay. The p values for E, N, and RdRp genes were calculated to be 0.0026, 0.0026, and 0.0026, respectively.
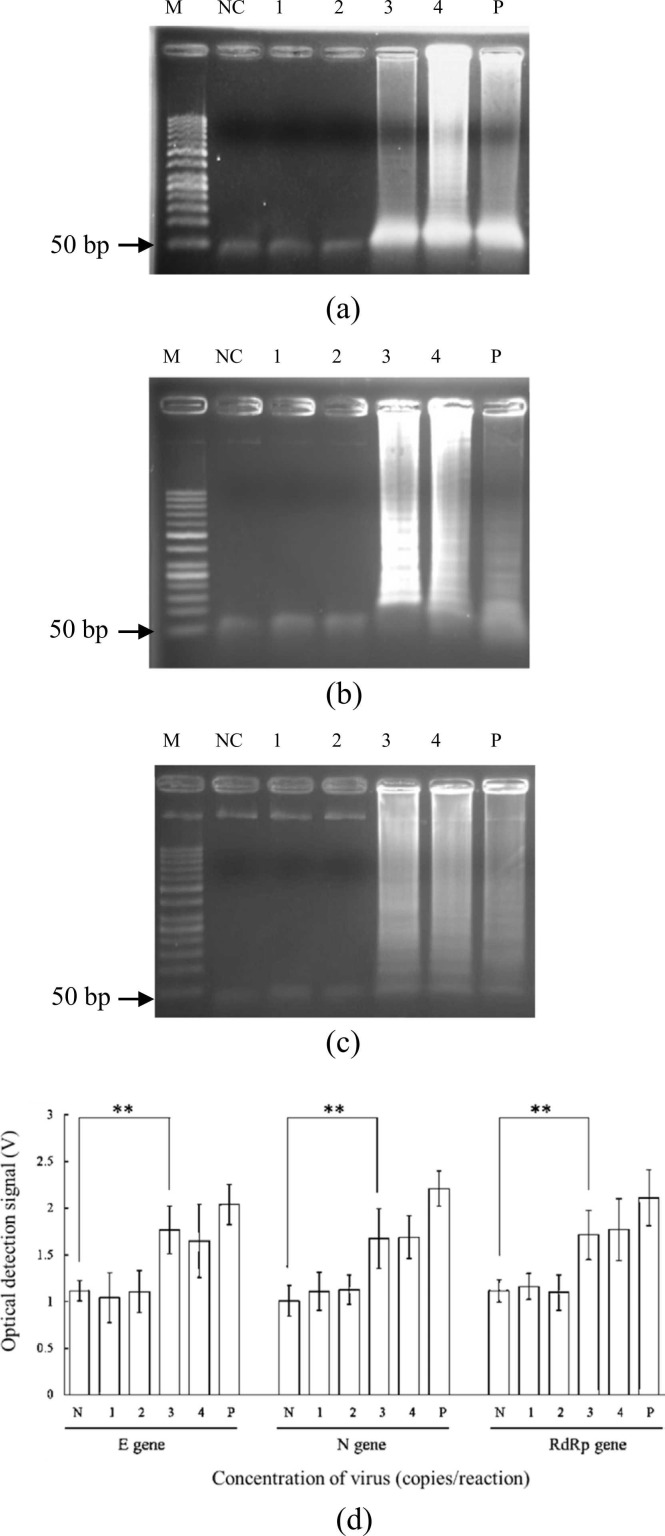
The assay was then used with serially diluted inactive viruses (5 × 104 to 5 × 101 copies/reaction), and each gene was successfully amplified after on-chip viral lysis+RNAextraction+RT-LAMP ( Fig. 7); LODs were 5 × 103 copies/reaction (i.e., 2 × 102 copies/μL) for all genes (Fig. 7a-c), lower than the 5 × 104 copies/μL value of a previous work [20] with the S and orf 8 genes. Note that the LOD of the N gene was only 50 copies/μL in that work, though the associated device could not detect all three genes simultaneously. The corresponding fluorescence detection results (Fig. 7d) also highlight the high performance of our novel device. Similarly, the two-tailed student t test revealed a significant difference (p < 0.05) for LOD determination for each gene with a concentration higher than 5 × 103 copies/reaction. Hence, the optical detection could be conducted accurately.
Fig. 7.
Gel electrophoresis results of viral lysis, RNA extraction, and RT-LAMP with inactive virus samples on-chip for the (a) E, (b) N, and (c) RdRp genes. (d) Relationship between the concentration and the voltage of the optical detection module. M: 50-bp DNA ladder; NC: Negative control (ddH2O mixed with RT-LAMP reagents); 1: 5 × 101 copies/reaction; 2: 5 × 102 copies/reaction; 3: 5 × 103 copies/reaction; 4: 5 × 104 copies/reaction; P: Positive control (5 × 105 copies/reaction). (d) Plot of RT-LAMP optical detection signal versus starting concentrations for the three target genes, negative control, and positive control. Error bars represent standard error of the mean (n = 3). * * was shown as p < 0.05 by the two-tailed student t test assay. The p values for E, N, and RdRp genes were calculated to be 0.0148, 0.033, and 0.0230, respectively.
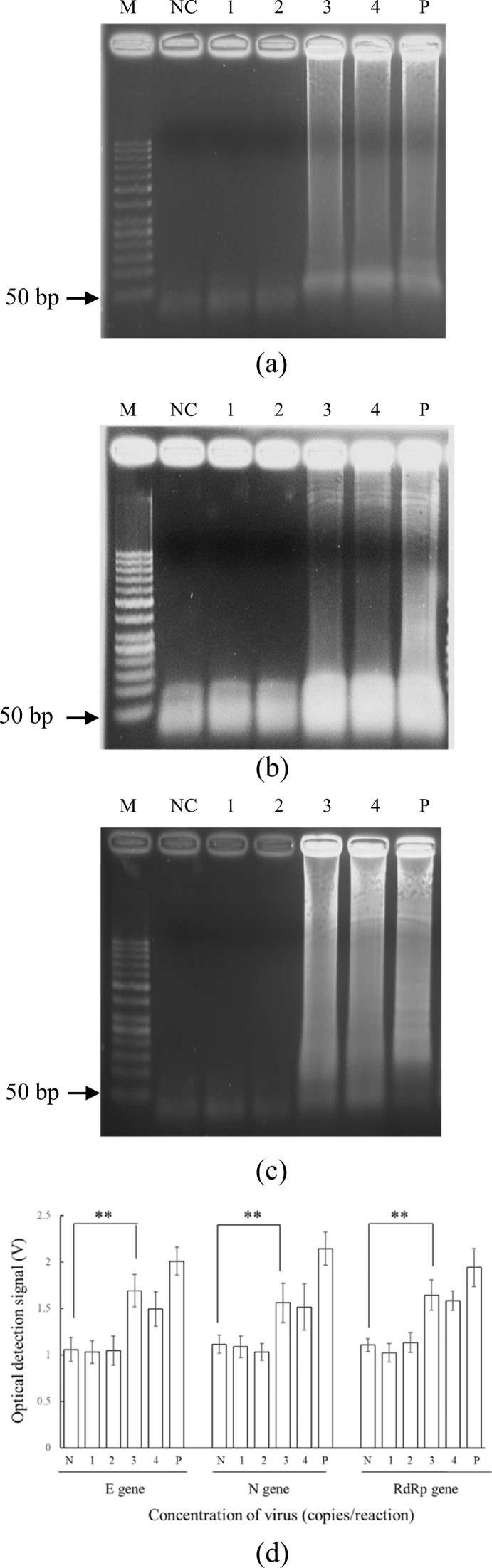
For clinical samples of lineage B.1.1.7. (i.e., the original COVID-19 virus) provided by NCKUH, samples were lysed, and viral RNAs were 1) extracted using protocols previously described [19] and 2) diluted from 5 × 104 to 5 × 101 copies/reaction. All three genes were successfully amplified ( Fig. 8), and on-chip LOD was 5 × 103 copies/reaction (i.e., 2 × 105 copies/mL) for each gene (the same as for synthesized RNA & inactive viruses). The LOD of the developed device was measured to be 5 × 103 copies/reaction (i.e., 2 ×105 copies/mL or Ct of 32.6 [28]) for each gene. According to the previous work [28], threshold (Ct) values > 30 or copy numbers < 106 copies/mL are defined as “non-infectious,” meaning that our device can detect clinically meaningful levels of viruses; this is further evidenced in Fig. 8d, in which fluorescence differences can be observed across dilutions. Moreover, as for each gene, there was a significant difference (p < 0.05) for LOD determination in the two-tailed student t test. Therefore, the device could detect each gene.
Fig. 8.
Gel electrophoresis results of clinical samples from National Cheng Kung University Hospital (NCKUH) on-chip for the (a) E, (b) N, and (c) RdRp genes. (d) The relationship between the concentration and the voltage of the optical detection signal on the developed device. M: 50-bp DNA ladder; NC: Negative control (ddH2O mixed with RT-LAMP reagents); 1: 5 × 101 copies/reaction; 2: 5 × 102 copies/reaction; 3: 5 × 103 copies/reaction; 4: 5 × 104 copies/reaction; P: Positive control (5 × 105 copies/reaction). (d) Plot of RT-LAMP optical detection signal versus starting concentrations for the three target genes, negative control, and positive control. Error bars represent standard error of the mean (n = 3). * * indicated p < 0.05 by the two-tailed student t test assay. The p values for E, N, and RdRp genes were determined to be 0.0293, 0.0063, and 0.0073, respectively.
Furthermore, in Fig. 7(d) and 8(d), the optical detection signals of lanes 3, 4, and P were for samples with three different concentrations. Samples with higher concentrations should yield higher optical detection signals. Theoretically, the signal at lane 4 should be higher than the one at lane 3. However, the optical detection signals in Fig. 7(d) and 8(d) referred to “end-point” detection after 60 min of the LAMP process. It indicated that the signals after saturation may be deteriorated, thus resulting a lower signal. Therefore, the end-point detection approach could be only used for qualitative analysis, not quantitative analysis.
3.3. Real-time quantification
For real-time detection, synthesized RNAs, inactive viruses, and RNA extracts from clinical samples were prepared in concentrations ranging from 5 × 107 to 5 × 103 copies/reaction, with optical detection monitored over time ( Fig. 9a) to generate standard curves of threshold time versus copy number for the three target genes (Fig. 9b); all trends were relatively linear with similar slopes. Because the LAMP chamber for the RdRp gene was farthest from the air injection hole and had a larger area for optical detection (Fig. 2a), only this gene was tested as a demonstration for real-time detection by creating standard curves of threshold time value from theoretical copy numbers of synthesized RNAs, inactive viruses, and clinical samples (Fig. 9c). Note that this real-time approach could quantify the initial concentration of the virus samples when compared with the end-point detection approach, as shown in Fig. 7(d) and 8(d). Experimental results showed that our optical detection module was capable of real-time, automatic detection of virus molecules, and the new laser blocker provided a shorter response time (2 s) than a previous designed featuring pneumatic air cylinders [25]. Therefore, the optical detection module could provide more reliable optical detection than in previous works [19], [25], and three COVID-19 genes could be detected in only 90 min. This platform could consequently revolutionize COVID-19 diagnostics.
Fig. 9.
(a) Relationship between the time and the optical detection signal of the optical detection module on the microfluidic device for the RdRp gene with synthesized RNA. (b) Standard curves between the copy number and the threshold time for three genes with clinical samples. (c) Standard curves between the copy number and the threshold time for the RdRp gene with synthesized RNA, inactive virus, and clinical samples. Different optical detection signals were approximated by the output voltage of the optical signals on the developed device. Error bars represent standard error of the mean (n = 3).
4. Conclusions
In this study, an automatic real-time RT-LAMP microfluidic system has been demonstrated. The system was consisted of 3 modules, including fluidic control, temperature control, and optical detection modules, which have been integrated in a compact packaging case. The entire process for viral lysis, RNA extraction, RT-LAMP, and optical detection/quantification could be automated. It could simultaneously amplify E, N, and RdRp genes in SARS-CoV-2 and the LOD was found to be 5 × 103 copies/reaction (i.e. 2 × 105 copies/mL) for all three genes. With this approach, the rapid detection of three genes could be completed in 90 min. To the best of our knowledge, it is the first time that three genes of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) could be detected and quantified on an integrated microfluidic platform by automating the entire process (including rival lysis, RNA extraction, RT-LAMP and optical quantification). The device could also conduct real-time detection. The initial copy number of viruses could be determined accordingly. Therefore, the developed platform should meet the need of rapid detection of COVID-19.
CRediT authorship contribution statement
You-Ru Jhou: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – original draft. Chih-Hung Wang: Conceptualization, Methodology, Writing – review & editing. Huey-Pin Tsai: Resources, Writing – review & editing. Yan-Shen Shan Conceptualization, Resources, Writing – review & editing. Gwo-Bin Lee: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Writing – proofreading, Writing – review & editing.
Declaration of Competing Interest
There is no conflict of interests for this article.
Acknowledgments
The authors thank the Ministry of Science and Technology (MOST) of Taiwan funded this work (MOST 109B0247J3, 109-2224-E-007-002, 109-3114-Y-001-001, 109Q22803E1, & 110Q2804E1). Partial financial support from the National Health Research Institutes of Taiwan (NHRI-EX110-11020EI) is also greatly appreciated.
Biographies

You-Ru Jhou received his B.S. degrees from the Department of Mechanical Engineering at National Taipei University of Technology, Taiwan and M.S. degree from the Department of Power Mechanical Engineering at National Tsing Hua University, Taiwan. His research specialties are automatic control, programming, and microfluidic device development.

Chih-Hung Wang received his M.S. degrees from the Institute of Molecular Biology at National Chung Hsing University, Taiwan. He completed his Ph.D. study at the Institute of Basic Medical Science of National Cheng Kung University, Taiwan. Currently, he is a postdoctoral researcher in the Department of Power Mechanical Engineering at National Tsing Hua University, Taiwan. His research specialties are molecular diagnosis, microbiology and nanobiotechnology.
Huey-Pin Tsai received her BS and MS degree from the Department of Medical Biotechnology at National Cheng Kung University in 1993 and 2004 respectively. Now, she is a Clinical Assistant Professor in Department of Medical Biotechnology at National Cheng Kung University and Leader of the clinical virology lab in Department of Pathology at National Cheng Kung University Hospital. She makes great work in clinical virology and published several reputed papers in detection virus.

Yan-Shen Shan has completed his MD in 1993. After the residency training in the Department of Surgery, National Cheng Kung University Hospital, Tainan, Taiwan, he also got the PhD in 2004 from Institute of Clinical Medicine, National Cheng Kung University. He was awarded as the distinguished professor since 2016 Aug and elected to be chief of the Institute of Clinical Medicine, NCKU from Aug 2017 to Jul 2019. From Aug 2019, He was elected to be the Dean of College of Medicine, NCKU. He has published more than 190 papers in reputed journals. He devotes in management of biliopancreatic cancer and gastric cancer, the translational study of biliary tract cancer, pancreatic cancer, gastric cancer and GIST. He also performs several IIT and attends international trials in stomach, biliary, and pancreatic cancer.

Gwo-Bin Lee received his BS and MS degrees from the Department of Mechanical Engineering at National Taiwan University in 1989 and 1991, respectively. He received his PhD from the Department of Mechanical & Aerospace Engineering at the University of California, Los Angeles, USA in 1998. He is a Tsing Hua Chair Professor at the Department of Power Mechanical Engineering, National Tsing Hua University, Taiwan. His research interests are on microfluidics, bio-sensing, nanobiotechnology and its biomedical applications.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.snb.2022.131447.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
References
- 1.Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Tan K.S., Wang D.Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil. Med. Res. 2020;7:11–21. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan X., Chen D., Xia Y., Wu X., Li T., Ou X., Zhou L., Liu J. Asymptomatic cases in a family cluster with SARS-CoV-2 infection. Lancet. 2020;20:410–411. doi: 10.1016/S1473-3099(20)30114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H., Li X., Li T., Zhang S., Wang L., Wu X., Liu J. The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur. J. Clin. Microbiol Infect. Dis. 2020;39:1629–1635. doi: 10.1007/s10096-020-03899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fehr A.R., Perlman S. Coronaviruses: an oerview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eur. Surveill. 2020;25:3–11. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H.L., Li G., Zhao J., Li Y.J., Ai Y.S. An overview of nucleic acid testing for the novel coronavirus SARS-CoV-2. Front. Med. 2021;7:1–7. doi: 10.3389/fmed.2020.571709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A., Tan J., Bhavsar D., Capuano C., Kirkpatrick E., Meade P., Brito R.N., Teo C., McMahon M., Simon V., Krammer F. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr. Protoc. Microbiol. 2020;57:100–114. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long Q.X., Liu B.Z., Deng H.J., Wu G.C., Deng K., Chen Y.K., Liao P., Qiu J.F., Lin Y., Cai X.F., Wang D.Q., Hu Y., Ren J.H., Tang N., Xu Y.Y., Yu L.H., Mo Z., Gong F., Zhang X.L., Tian W.G., Hu L., Zhang X.X., Xiang J.L., Du H.X., Liu H.W., Lang C.H., Luo X.H., Wu S.B., Cui X.P., Zhou Z., Zhu M.M., Wang J., Xue C.J., Li X.F., Wang L., Li Z.J., Wang K., Niu C.C., Yang Q.J., Tang X.J., Zhang Y., Liu X.M., Li J.J., Zhang D.C., Zhang F., Liu P., Yuan J., Li Q., Hu J.L., Chen J., Huang A.L. Antibody Responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 10.Jääskeläinen A.J., Korhonen E.M., Huhtamo E., Lappalainen M., Vapalahti O., Kallio-Kokko H. Validation of serological and molecular methods for diagnosis of Zika virus infections. J. Virol. Methods. 2019;263:68–74. doi: 10.1016/j.jviromet.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Lu R., Wu X., Wan Z., Li Y., Jin X., Zhang C. A novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Int J. Mol. Sci. 2020;21:2826–2835. doi: 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori Y., Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and xost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 2009;vol 15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang W.H., Yang S.Y., Wang C.H., Tsai M.A., Wang P.C., Chen T.Y., Chen S.C., Lee G.B. Rapid isolation and detection of aquaculture pathogens in an integrated microfluidic system using loop-mediated isothermal amplification. Sens. Actuators B Chem. 2003;180:96–106. [Google Scholar]
- 14.Zhang H., Xu Y., Fohlerova Z., Chang H., Iliescu C., Neuzil P. LAMP-on-a-chip: revising microfluidic platforms for loop-mediated DNA amplification. Trends Anal. Chem. 2019;113:44–53. doi: 10.1016/j.trac.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asiello P.J., Baeumner A.J. Miniaturized isothermal nucleic acid amplification: a review. Lab a Chip. 2011;11:1420–1430. doi: 10.1039/c0lc00666a. [DOI] [PubMed] [Google Scholar]
- 16.Fang X., Liu Y., Kong J., Jiang X. Loop-mediated isothermal amplification integrated on microfluidic chips for point-of-care quantitative detection of pathogens. Anal. Chem. 2010;82:3002–3006. doi: 10.1021/ac1000652. [DOI] [PubMed] [Google Scholar]
- 17.Kaarj K., Akarapipad P., Yoon J.Y. Simpler, faster, and sensitive Zika virus assay using smartphone detection of loop-mediated isothermal amplification on paper microfluidic chips. Sci. Rep. 2018;8:12438–12448. doi: 10.1038/s41598-018-30797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucchi N.W., Demas A., Narayanan J., Sumari D., Kabanywanyi A., Kachur S.P., Barnwell J.W., Udhayakumar V. Real-time fluorescence loop mediated isothermal amplification for the diagnosis of malaria. PLoS One. 2010;5:13733–13739. doi: 10.1371/journal.pone.0013733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y.D., Li K.H., Chen Y.H., Lee Y.M., Chou S.T., Lai Y.Y., Huang P.C., Ma H.P., Lee G.B. A Sample-to-answer, portable platform for rapid detection of pathogens with a smartphone interface. Lab Chip. 2019;19:3804–3814. doi: 10.1039/c9lc00797k. [DOI] [PubMed] [Google Scholar]
- 20.Ganguli A., Mostafa A., Berger J., Aydin M.Y., Sun F., Ramirez S.A.S., Valera E., Cunningham B.T., King W.P., Bashir R. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc. Natl. Acad. Sci. 2020;117:22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Oliveira K.G., Estrela P.F.N., Mendes G.M., Dos Santos C.A., Silveira-Lacerda E.P., Duarte G.R.M. Rapid molecular diagnostics of COVID-19 by RT-LAMP in a centrifugal polystyrene-toner based microdevice with end-point visual detection. Analyst. 2021;146:1178–1187. doi: 10.1039/d0an02066d. [DOI] [PubMed] [Google Scholar]
- 22.Soares R.R.G., Akhtar A.S., Pinto I.F., Lapins N., Barrett D., Sandh G., Yin X., Pelechano V., Russom A. Sample-to-answer COVID-19 nucleic acid testing using a low-cost centrifugal microfluidic platform with bead-based signal enhancement and smartphone read-out. Lab Chip. 2021;21:2932–2944. doi: 10.1039/d1lc00266j. [DOI] [PubMed] [Google Scholar]
- 23.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: The disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 24.Hsu K.W., Lee W.B., You H.L., Lee M.S., Lee G.B. An automated and portable antimicrobial susceptibility testing system for urinary tract infections. Lab Chip. 2021;21:755–763. doi: 10.1039/d0lc01315c. [DOI] [PubMed] [Google Scholar]
- 25.Chang W.H., Yu J.C., Yang S.Y., Lin Y.C., Wang C.H., You H.L., Wu J.J., Lee M.S., Lee G.B. Vancomycin-resistant gene identification from live bacteria on an integrated microfluidic system by using low temperature lysis and loop-mediated isothermal amplification. Biomicrofluidics. 2017;11:24101–24112. doi: 10.1063/1.4977439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao Y., Wang L., Duan L., Li J., Ma J., Xie S., Shi L., Li H. Development of a real-time fluorescence loop-mediated isothermal amplification assay for rapid and quantitative detection of Ustilago maydis. Sci. Rep. 2017;7:13394–13405. doi: 10.1038/s41598-017-13881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z., Zu X., Du Z., Hu Z. Research on magnetic bead motion characteristics based on magnetic beads preset technology. Sci. Rep. 2021;11:19995–20007. doi: 10.1038/s41598-021-99331-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Platten M., Hoffmann D., Grosser R., Wisplinghoff F., Wisplinghoff H., Wiesmüller G., Schildgen O., Schildgen V. SARS-CoV-2, CT-values, and infectivity-conclusions to be drawn from side observations. Viruses. 2021;13:1459–1464. doi: 10.3390/v13081459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material