Abstract
Human rotavirus strains belonging to genotype G9 or P[9] were detected in a collection of stool specimens from children with diarrhea in two cities of the state of Rio de Janeiro, Brazil, between March 1997 and December 1999. G9 strains were first detected in April 1997 and remained prevalent until the end of the study, at a frequency of 15.9% (n = 157). A high percentage of VP7 nucleotide (99.0 to 99.5%) and deduced amino acid identity (98.6 to 99.1%) was found between three randomly selected Brazilian G9 strains and the American G9 strain US1205. A novel G9:P[4] genotype combination was detected in addition to G9:P[8] and G9:P[6], demonstrating that this G genotype may undergo constant genetic reassortment in nature. The P[9] rotavirus strains constituted 10.2%, the majority of which were detected between April and July 1997. The RNA electrophoretic migration pattern of the G3:P[9] strains resembled that of AU-1 virus (G3:P3[9]), suggesting a genetic similarity between the Brazilian G3:P[9] strains and the Japanese virus, which is similar to a feline rotavirus genetically.
Rotaviruses are the major etiologic agents of infantile diarrhea worldwide (15). Epidemiologic studies have demonstrated that rotavirus serotypes G1, G2, G3, and G4 are the most-common types associated with disease globally, and therefore they are the targets for vaccine development (13, 15). Recently, unusual rotavirus serotypes and genotypes have been described in association with diarrhea in various parts of the world. These include serotype G5 in Brazil (9, 17); G8 in Malawi (5), Kenya (21), South Africa (34), the United Kingdom (34), Nigeria (1), and Australia (24); and G9 in India (27), the United Sates (28), Bangladesh (36), Malawi (5), the United Kingdom, (4), Australia (25), France (2), and Ireland (22). The P specificity of a rotavirus is usually more conservative than its G specificity; P1A[8] is the most common serotype detected worldwide, followed by P1B[4] and P2A[6] (13). A rotavirus strain belonging to serotype P3[9] was first identified in Japan and was demonstrated to be closely related to feline rotavirus strains genetically (20, 35). The P[9] rotavirus strains have been detected more often in Japan (14, 37) than in other parts of the world such as Venezuela (32), Italy (32), Malaysia (29), Brazil (17), Israel (31), South Africa (33), Guinea-Bissau (7), and the United States (12). The P[9] isolates are most commonly associated with either G1 (K8-like) or G3 (AU-1-like) serotypes, except for one isolate from Guinea-Bissau which bears a G4 specificity (7).
One hundred fifty-seven (23%; n = 678) rotavirus-positive stool samples from children under 5 years of age with acute diarrhea (32 inpatients and 646 outpatients) were collected between March 1997 and December 1999, at four centers in the city of Rio de Janeiro, Brazil, and one center in the neighboring city of Niterói in the state of Rio de Janeiro. The five centers are located in areas of distinct levels of sanitation and socioeconomic backgrounds. The presence of rotavirus in those samples was determined by polyacrylamide electrophoresis analysis (PAGE) and/or latex agglutination test. Rotavirus double-stranded RNA was extracted from the stool samples and analyzed by reverse transcription-PCR for determination of G and P specificity by using primers specific for genotypes G1 to G6, G8 to G10, G12, P[4], P[6], P[8], P[9], and P[10] (6, 8, 10, 11). Twenty-five of 157 samples (15.9%) were typed as rotavirus genotype G9: 19 were single infections, while 5 were mixed infections with the G1 and G3 genotypes and 1 had a mixture of G9 and an untyped G genotype (Table 1). The G9 samples were detected in all five centers. Twenty samples were from outpatient children, and five were from inpatient children. One single strain was detected in April 1997, and the remaining 24 samples were detected between July 1998 and December 1999. Eight of the 25 G9 samples were detected in a middle-class area, 8 were detected in a wealthy area, 8 were detected in two centers that attend a poor population in the city of Rio de Janeiro, and 1 sample was detected in a middle-class child from Niterói. Nine G9 samples were detected in 1998, which constituted 25.0% of the total samples (n = 36) analyzed for that year, and fifteen G9 samples were detected in 1999, which constituted 27.7% of the total samples (n = 54) analyzed for that year, making it the second-most-prevalent G genotype in 1999 in Rio de Janeiro (data not shown).
TABLE 1.
Human rotavirus G9 genotype strains detected in the cities of Rio de Janeiro and Niterói, Brazil, from 1997 to 1999
| G:P specificitya | No. of strains detected in:
|
Total no. of strains | ||
|---|---|---|---|---|
| 1997 | 1998 | 1999 | ||
| G9:P[8] | 1 | 5 | 9 | 15 |
| G9:P[4] | 0 | 0 | 1 | 1 |
| G9:P[6] | 0 | 0 | 3 | 3 |
| G9 + G1:P[8] | 0 | 0 | 1 | 1 |
| G9 + G1:P[9] | 0 | 1 | 0 | 1 |
| G9 + G1:P[8] + P[6] | 0 | 1 | 0 | 1 |
| G9 + G3:P[9] | 0 | 1 | 0 | 1 |
| G9 + G1:P[?] | 0 | 0 | 1 | 1 |
| G9 + G?:P[9] + P? | 0 | 1 | 0 | 1 |
| Total | 1 | 9 | 15 | 25 |
?, unknown.
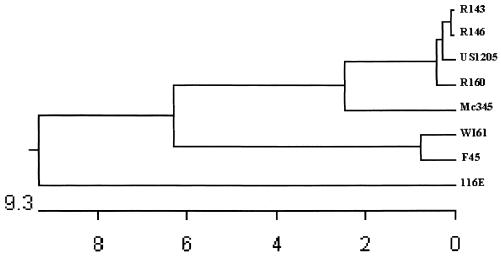
Three G9 samples (R143, R146, and R160) were randomly selected, and the VP7-encoding gene of each virus was sequenced and compared to other G9 rotavirus VP7 gene sequences available in the GenBank. The best match was with the American G9 strain US1205, which exhibited 99.0 to 99.5% nucleotide and 98.6 to 99.1% deduced amino acid identity (Table 2). The phylogenetic analysis of the nucleotide sequences revealed the existence of at least three different VP7 gene clusters among rotavirus G9 strains. One cluster was formed by the three Brazilian G9 strains, the American strain US1205, and the strain Mc345 from Thailand; the second cluster represented by the American strain WI61 and the Japanese strain F45; and the third cluster was formed by the Indian strain 116E (Fig. 1). Interestingly, the existence of minor VP7 antigenic differences has recently been reported among selected G9 viruses belonging to each of the three VP7 gene clusters reported here (3, 16).
TABLE 2.
Percentage nucleotide (top right) and deduced amino acid (bottom left) identity of VP7 sequence of human rotavirus G9 strains
| G9 strain | % identity with:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 116E | Mc345 | US1205 | F45 | WI61 | R143 | R146 | R160 | |
| 116E | 87.7 | 88.8 | 89.6 | 89.1 | 88.7 | 88.8 | 88.8 | |
| Mc345 | 88.4 | 95.1 | 88.7 | 88.5 | 95.2 | 95.3 | 95.5 | |
| US1205 | 91.7 | 94.3 | 89.0 | 89.0 | 99.4 | 99.5 | 99.0 | |
| F45 | 89.5 | 89.0 | 93.1 | 98.5 | 89.0 | 89.0 | 89.2 | |
| WI61 | 91.8 | 91.0 | 94.3 | 94.6 | 89.0 | 89.0 | 89.2 | |
| R143 | 91.8 | 94.4 | 99.1 | 92.6 | 94.4 | 99.9 | 99.3 | |
| R146 | 91.8 | 94.4 | 99.1 | 92.6 | 94.4 | 99.4 | 99.4 | |
| R160 | 91.2 | 93.8 | 98.6 | 92.1 | 93.8 | 98.9 | 98.9 | |
FIG. 1.
Phylogenetic analysis of the VP7 gene nucleotide sequences of human rotavirus G9 strains R143, R146, R160, US1205, Mc345, WI61, F45, and 116E. The dendrogram was constructed by the Clustal method using the DNASTAR program. The units at the bottom of the tree indicate distance between sequence pairs.
A large diversity was observed with regard to the VP4 specificity of the Brazilian G9 strains. Although the majority of the G9 isolates carried P[8] specificity, strains bearing P[4] or P[6] specificity were also detected (Table 1). Thus far, rotavirus serotype G9 has been associated with VP4 genotype P[6], P[8], P[11], or P[19] (2, 6, 23, 25, 28). Here we found a G9 rotavirus isolate bearing the P[4] genotype. It is believed to be the first detection of such a G:P genotype combination. Rotavirus strains bearing G9 specificity have been previously described in Brazil on only three occasions: one isolate was detected in an infant (18), and the other isolates were detected in pigs (26, 30). The detection of rotavirus genotype G9 strains with such remarkable frequency among diarrheal patients (15.9%) in the last 2 years demonstrates that such a rotavirus genotype is an emerging pathogen in this country.
Sixteen of 157 (10.2%) samples were typed as rotavirus genotype P[9]: seven single infections and nine mixed infections (Table 3), all from outpatients. Twelve strains were detected in 1997, eleven of them being detected during the period from April to July and one being detected in October. Five of 12 samples were from one center in Niterói, and seven were from a center in a wealthy area in the city of Rio de Janeiro. Three samples were detected between July and September of 1998 in one center in Rio de Janeiro located in a middle-class area; one sample was detected in January 1999 in a center in Rio de Janeiro that attends the poor population of a slum.
TABLE 3.
Rotavirus P[9] genotype strains detected in the cities of Rio de Janeiro and Niterói, Brazil, from 1997 to 1999
| P:G specificity | 1997 | 1998 | 1999 | Total |
|---|---|---|---|---|
| P[9]:G1 | 1 | 0 | 0 | 1 |
| P[9]:G3 | 5 | 0 | 1 | 6 |
| P[9]:G1 + G3 | 2 | 0 | 0 | 2 |
| P[9]:G1 + G9 | 0 | 1 | 0 | 1 |
| P[9]:G3 + G9 | 0 | 1 | 0 | 1 |
| P[9] + P[8]:G3 | 1 | 0 | 0 | 1 |
| P[9] + P[4]:G1 + G3 | 1 | 0 | 0 | 1 |
| P[9] + P[8] + P[4]:G3 | 1 | 0 | 0 | 1 |
| P[9] + P[8] + P[4]:G? | 1 | 0 | 0 | 1 |
| P[9] + P?:G9 + G? | 0 | 1 | 0 | 1 |
| Total | 12 | 3 | 1 | 16 |
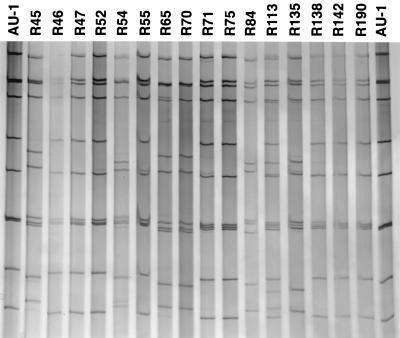
The 16 P[9] samples were adapted to growth in primary African green monkey kidney cells (BioWhittaker, Walkersville, Md.) and double-stranded RNAs of such viruses were analyzed by PAGE. Interestingly, 10 of 16 viruses demonstrated a characteristic AU-1-like electropherotype (i.e., widely spaced genes 5 and 6 and genes 10 and 11) or a mixture of an AU-1-like pattern plus an additional RNA pattern (Fig. 2). This observation suggests a genetic similarity between the Brazilian G3:P[9] strains and the Japanese AU-1 virus (G3:P3[9]), which is similar to feline rotavirus genetically (19). Since (i) 6 of 16 P[9] samples had an AU-1-like PAGE pattern with G3:P[9] specificity and (ii) 4 of 16 P[9] samples had a mixture of an AU-1-like PAGE pattern plus an additional PAGE pattern, it is possible that the remaining 6 P[9] samples which did not show the AU-1-like pattern may be naturally occurring reassortants between the AU-1-like virus and non-AU-1-like virus.
FIG. 2.
Electrophoretic migration pattern of RNAs of the 16 culture-adapted Brazilian P[9] rotavirus strains and the Japanese AU-1 strain. The viral RNAs were analyzed by electrophoresis in a 10% polyacrylamide gel and visualized by staining with silver nitrate.
Rotavirus P3[9] strains have been detected most commonly among nonhospitalized children with diarrhea (7, 31). Silberstein and colleagues (31) speculate that genotype P[9] viruses (i) infect children as a consequence of zoonotic infection by circulating feline rotaviruses or by reassortants formed between feline and human group A rotaviruses such as K8 strain and thus (ii) are attenuated to some extent in the human host and cause sporadic and mild infections in children. Unfortunately, we have no access to the clinical data of patients who shed P[9] viruses in this study, which might shed light on the severity of diarrhea caused by such a P genotype. However, it is noteworthy that none of 16 patients who shed P[9] viruses in this study required hospitalization. Before this study, the detection of the rotavirus P[9] genotype was reported in Brazil only once, where three isolates were detected in a collection of fecal samples obtained between 1982 and 1994 in the state of São Paulo (17). The detection of 11 samples in a short period of time, 4 consecutive months within the same year, suggests the possibility of the occurrence of one or maybe two (since the samples came from two different cities) small outbreaks.
Of note is the finding that no G5 rotavirus strains were detected during the 3-year study period (1997 to 1999) in two cities of the state of Rio de Janeiro. This was particularly surprising since the incidence of this genotype appeared to be increasing in Brazil in the last decade (9). Is the disappearance of the genotype G5 virus in this study area somehow related to the emergence of the genotype G9 virus? It will be important to continue rotavirus strain surveillance in this and other regions of Brazil to determine whether a similar phenomenon occurs in the country. The results described here should reinforce the importance of rotavirus G9 as an epidemiologically important serotype and intensify the need of considering it as a vaccine candidate.
Nucleotide sequence accession numbers.
The sequences of the R143, R146, and R160 isolates were deposited in GenBank under the accession numbers AF274969, AF274970, and AF274971.
Acknowledgments
We thank Maria Odete O. Carvalho and Giovani C. V. Costa for supplying the stool samples used in this work; Ronald Jones, Jerri Ross, and Mariam Wagner for their expert technical assistance; and Albert Kapikian for encouragement throughout the study.
This study was partially supported by CNPq, FINEP, FAPERJ, and FUJB, Brazil.
REFERENCES
- 1.Adah M I, Rohwedder A, Olaleyly O D, Werchau H. Nigerian rotavirus serotype G8 could not be typed by PCR due to nucleotide mutation at the 3′ end of the primer binding site. Arch Virol. 1997;142:1881–1887. doi: 10.1007/s007050050206. [DOI] [PubMed] [Google Scholar]
- 2.Bon F, Fromantin C, Aho S, Pothier P, Kohli E The Azay Group. G and P genotyping of rotavirus strains circulating in France over a three-year period: detection of G9 and P[6] strains at low frequencies. J Clin Microbiol. 2000;38:1681–1683. doi: 10.1128/jcm.38.4.1681-1683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coulson B, Gentsch J R, Das B K, Bhan M K, Glass R I. Comparison of enzyme immunoassay and reverse transcriptase PCR for identification of serotype G9 rotaviruses. J Clin Microbiol. 1999;37:3187–3193. doi: 10.1128/jcm.37.10.3187-3193.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cubitt W D, Steele A D, Iturriza M. Characterization of rotavirus from children treated at a London hospital during 1996: emergence of strains G9P2A[6] and G3P2A[6] J Med Virol. 2000;61:150–154. doi: 10.1002/(sici)1096-9071(200005)61:1<150::aid-jmv24>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 5.Cunliffe N A, Gondwe J S, Broadhead R L, Molyneux M E, Woods P A, Bresee J S, Glass R I, Gentsch J R, Hart C A. Rotavirus G and P types in children with acute diarrhea in Blantyre, Malawi, from 1997 to 1998: predominance of novel P[6]G8 strains. J Med Virol. 1999;57:308–312. [PubMed] [Google Scholar]
- 6.Das B K, Gentsch J R, Cicirelo H G, Woods P A, Gupta A, Ramachandran M, Kumar R, Bhan M K, Glass R I. Characterization of rotavirus strains from newborns in New Delhi, India. J Clin Microbiol. 1994;32:1820–1822. doi: 10.1128/jcm.32.7.1820-1822.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer T K, Steinsland H, Molbak K, Ca R, Gentsch J R, Valentiner-Branth P, Aaby P, Sommerfelt H. Genotype profiles of rotavirus strains from children in a suburban community in Guinea-Bissau, Western Africa. J Clin Microbiol. 2000;38:264–267. doi: 10.1128/jcm.38.1.264-267.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gentsch J R, Glass R I, Woods P, Gouvea V, Gorziglia M, Flores J, Das B K, Bhan M K. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gouvea V, Santos N. Rotavirus serotype G5: an emerging cause of epidemic childhood diarrhea. Vaccine. 1999;17:1291–1292. doi: 10.1016/s0264-410x(98)00378-8. [DOI] [PubMed] [Google Scholar]
- 10.Gouvea V, Glass R I, Woods P, Taniguchi K, Clark H F, Forrester B, Fang Z-Y. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouvea V, Santos N, Timenetsky M C. Identification of bovine and porcine G types by PCR. J Clin Microbiol. 1994;32:1338–1340. doi: 10.1128/jcm.32.5.1338-1340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin D D, Kirkwood C D, Parashar U D, Woods P A, Bresee J S, Glass R I, Gentsch J R The National Rotavirus Strain Survelillance System Collaborating Laboratories. Surveillance of rotavirus strains in the United States: identification of unusual strains. J Clin Microbiol. 2000;38:2784–2787. doi: 10.1128/jcm.38.7.2784-2787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoshino, Y., and A. Z. Kapikian. Rotavirus serotypes: classification and importance in rotavirus epidemiology, immunity and vaccine development. J. Health Popul. Nutr., in press. [PubMed]
- 14.Iizuka M, Chiba M, Masamune O, Kaga E, Nakagomi T, Nakagomi O. A highly conserved genomic RNA constellation of Japanese isolates of human rotavirus carrying G serotype 3 and P serotype 3. Res Virol. 1994;145:21–24. doi: 10.1016/s0923-2516(07)80003-3. [DOI] [PubMed] [Google Scholar]
- 15.Kapikian A Z. Overview of viral gastroenteritis. Arch Virol. 1996;12(Suppl.):7–19. doi: 10.1007/978-3-7091-6553-9_2. [DOI] [PubMed] [Google Scholar]
- 16.Kirkwood C D, Gentsch J R, Hoshino Y, Clark H F, Glass R I. Genetic and antigenic characterization of a serotype P[6]G9 human rotavirus strain isolated in the U. S Virology. 1999;256:45–53. doi: 10.1006/viro.1998.9591. [DOI] [PubMed] [Google Scholar]
- 17.Leite J P G, Alfieri A A, Woods P A, Glass R I, Gentsch J R. Rotavirus G and P types circulating in Brazil: characterization by RT-PCR, probe hybridization, and sequence analysis. Arch Virol. 1996;141:2365–2374. doi: 10.1007/BF01718637. [DOI] [PubMed] [Google Scholar]
- 18.Linhares A C, Gabbay Y B, Mascarenhas J D P, de Freitas R B, Oliveira C S, Bellesi N, Monteiro T A F, Lins-Laison Z, Ramos F L P, Valente S A. Immunogenicity, safety and efficacy of tetravalent rhesus-human, reassortant rotavirus vaccine in Belém, Brazil. Bull W H O. 1996;74:491–500. [PMC free article] [PubMed] [Google Scholar]
- 19.Nakagomi O, Nakagomi T, Hoshino Y, Flores J, Kapikian A Z. Genetic analysis of a human rotavirus that belongs to subgroup I but has an RNA pattern typical of subgroup II human rotaviruses. J Clin Microbiol. 1987;25:1159–1164. doi: 10.1128/jcm.25.7.1159-1164.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagomi O, Nakagomi T. RNA-RNA hybridization identifies a human rotavirus that is genetically related to feline rotavirus. J Virol. 1989;63:1431–1434. doi: 10.1128/jvi.63.3.1431-1434.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakata S, Gatheru Z, Ukae S, Adachi N, Kobayashi N, Honma S, Muli J, Ogaja P, Nyangao J, Kiplagat E, Tukei P M, Chiba S. Epidemiological study of the G serotype distribution of group A rotaviruses in Kenya from 1991 to 1994. J Med Virol. 1999;58:296–303. doi: 10.1002/(sici)1096-9071(199907)58:3<296::aid-jmv17>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 22.O'Halloran F, Lynch M, Cryan B, O'Shea H, Fanning S. Molecular characterization of rotavirus in Ireland: detection of novel strains circulating in the population. J Clin Microbiol. 2000;38:3370–3374. doi: 10.1128/jcm.38.9.3370-3374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okada J, Urasawa T, Kobayashi N, Taniguchi K, Hasegawa A, Mise K, Urasawa S. New P serotype of group A human rotavirus closely related to that of a porcine rotavirus. J Med Virol. 2000;60:63–69. [PubMed] [Google Scholar]
- 24.Palombo E, Clark R, Bishop R F. Characterization of a “European-like” serotype G8 human rotavirus isolated in Australia. J Med Virol. 2000;60:56–62. [PubMed] [Google Scholar]
- 25.Palombo E, Masendycz P J, Bugg H C, Bogdanovic-Sakran N, Barnes G L, Bishop R F. Emergence of serotype G9 human rotavirus in Australia. J Clin Microbiol. 2000;38:1305–1306. doi: 10.1128/jcm.38.3.1305-1306.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rácz M L, Kroeff S S, Munford V, Caruzo T A R, Durigon E L, Hayashi Y, Gouvea V, Palombo E. Molecular characterization of porcine rotaviruses from the Southern region of Brazil: characterization of an atypical genotype G[9] strain. J Clin Microbiol. 2000;38:2443–2446. doi: 10.1128/jcm.38.6.2443-2446.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramachandran M, Das B K, Vij A, Kumar R, Bhambal S S, Kesari N, Rawt H, Bahl L, Thakur S, Woods P A, Glass R I, Bhan M K, Gentsch J R. Unusual diversity of human rotavirus G and P genotypes in India. J Clin Microbiol. 1996;34:436–439. doi: 10.1128/jcm.34.2.436-439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramachandran M, Gentsch J R, Parashar U D, Jin S, Woods P, Holmes L, Kirkwood C D, Bishop R F, Greenberg H B, Urasawa S, Gerna G, Coulson B S, Taniguchi K, Bresse J S, Glass R I. Detection and characterization of novel rotavirus strains in the United States. J Clin Microbiol. 1998;36:3223–3229. doi: 10.1128/jcm.36.11.3223-3229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasool N B, Larralde G, Gorziglia M I. Determination of human rotavirus VP4 using serotype-specific cDNA probes. Arch Virol. 1993;133:275–282. doi: 10.1007/BF01313768. [DOI] [PubMed] [Google Scholar]
- 30.Santos N, Lima R C C, Nozawa C M, Linhares R E, Gouvea V. Detection of porcine rotavirus type G9 and a mixture of types G1 and G5 associated with Wa-like VP4 specificity: evidence for natural human-porcine genetic reassortment. J Clin Microbiol. 1999;37:2734–2736. doi: 10.1128/jcm.37.8.2734-2736.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silberstein I, Shulman L M, Mendelson E, Shif I. Distribution of both rotavirus VP4 genotypes and VP7 serotypes among hospitalized and nonhospitalized Israeli children. J Clin Microbiol. 1995;33:1421–1422. doi: 10.1128/jcm.33.5.1421-1422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steele A D, Garcia D, Sears J, Gerna G, Nakagomi O, Flores J. Distribution of VP4 gene alleles in human rotaviruses by using probes to the hyperdivergent region of the VP4 gene. J Clin Microbiol. 1993;31:1735–1740. doi: 10.1128/jcm.31.7.1735-1740.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steele A D, van Niekerk M C, Mphahlele M J. Geographic distribution of human rotavirus VP4 genotypes and VP7 serotypes in five South African regions. J Clin Microbiol. 1995;33:1516–1519. doi: 10.1128/jcm.33.6.1516-1519.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steele D A, Parker S P, Peenze I, Pager C T, Taylor M B, Cubitt W D. Comparative studies of human rotavirus serotype G8 strains recovered in South Africa and the United Kingdom. J Gen Viol. 1999;80:3029–3034. doi: 10.1099/0022-1317-80-11-3029. [DOI] [PubMed] [Google Scholar]
- 35.Taniguchi K, Nishikawa K, Urasawa T, Urasawa S, Midthun K, Kapikian A Z, Gorziglia M. Complete nucleotide sequence of the gene encoding VP4 of a human rotavirus (strain K8) which has unique VP4 neutralization epitopes. J Virol. 1989;63:4101–4106. doi: 10.1128/jvi.63.9.4101-4106.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unicomb L E, Podder G, Gentsch J R, Woods P A, Hasan K Z, Faruque A S G, Albert M J, Glass R I. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: emergence of type G9 in 1995. J Clin Microbiol. 1999;37:1885–1891. doi: 10.1128/jcm.37.6.1885-1891.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, H., K. Taniguchi, F. Wakasugi, S. Ukae, S. Chiba, M. Ohseto, A. Hasegawa, T. Urasawa, and S. Urasawa. Survey on the distribution of the gene 4 allele of human rotaviruses by polymerase chain reaction. Epidemiol. Infect. 112:615–622. [DOI] [PMC free article] [PubMed]