Highlights
-
•
Natural compound library screening identified Sanguinarine chloride as a tumor suppressor in SCLC.
-
•
Sanguinarine chloride exhibits anti-SCLC ability in vitro and in vivo.
-
•
Sanguinarine chloride upregulates CDKN1A to suppress SCLC development.
-
•
Sanguinarine chloride enhances the anti-tumor effect of panobinostat in SCLC by promoting cell cycle arrest.
Keywords: Natural compound, SCLC, Sanguinarine chloride, Panobinostat, CDKN1A
Abstract
Objectives
Small cell lung cancer (SCLC) is notorious for aggressive malignancy without effective treatment, and most patients eventually develop tumor progression with a poor prognosis. There is an urgent need for discovering novel antitumor agents or therapeutic strategies for SCLC.
Materials and methods
We performed a screening method based on CCK-8 assay to screen 640 natural compounds for SCLC. The effects of Sanguinarine chloride on SCLC cell proliferation, colony formation, cell cycle, apoptosis, migration and invasion were determined. RNA-seq and bioinformatics analysis was performed to investigate the anti-SCLC mechanism of Sanguinarine chloride. Publicly available datasets and samples were analyzed to investigate the expression level of CDKN1A and its clinical significance. Loss of functional cancer cell models were constructed by shRNA-mediated silencing. Quantitative RT-PCR and Western blot were used to measure gene and protein expression. Immunohistochemistry staining was performed to detect the expression of CDKN1A, Ki67, and Cleaved caspase 3 in xenograft tissues.
Results
We identified Sanguinarine chloride as a potential inhibitor of SCLC, which inhibited cell proliferation, colony formation, cell cycle, cell migration and invasion, and promoted apoptosis of SCLC cells. Sanguinarine chloride played an important role in anti-SCLC by upregulating the expression of CDKN1A. Furthermore, Sanguinarine chloride in combination with panobinostat, or THZ1, or gemcitabine, or (+)-JQ-1 increased the anti-SCLC effect compared with either agent alone treatment.
Conclusions
Our findings identified Sanguinarine chloride as a potential inhibitor of SCLC by upregulating the expression of CDKN1A. Sanguinarine chloride in combination with chemotherapy compounds exhibited strong synergism anti-SCLC properties, which could be further clinically explored for the treatment of SCLC.
Introduction
Small cell lung cancer (SCLC) is a highly malignant neuroendocrine tumor, which is characterized by a short cell doubling time, high growth fraction, rapid progression and early occurrence of widespread metastasis [1,2]. Although SCLC is highly sensitive to initial chemotherapy and radiotherapy, standard radiotherapy and chemotherapy have not significantly prolonged the overall survival (OS) of SCLC patients over the past decades, and many patients die of cancer recurrence and metastasis [3]. The OS of SCLC patients is not very optimistic at present [4]. Therefore, searching for new and effective therapeutic drugs to improve the clinical efficacy is of great importance.
Increasing evidence has supported that many naturally occurring products possess remarkable anti-tumor activities, or enhance the efficacy of chemotherapy [5]. Natural compounds are stable and safe as they are toxin-free, non-hazardous, residue-free, and pollution-free, provide opportunities for the management of cancer in the future [6]. In purpose of searching for effective pharmaceutical of SCLC, we performed high-throughput screening with a natural compound library, and identified Sanguinarine chloride (Sg) with excellent anti-SCLC activity. Sg is a natural compound derived from Macleaya cordata and recent studies have highlighted that Sg have different degrees of anti-tumor effects on prostate cancer [7,8], cervical cancer [9], breast cancer [10], [11], [12], colorectal cancer [13], gastric cancer [14], melanoma [15,16], pancreatic cancer [17] and non-small cell lung cancer (NSCLC) [18]. But the role of Sg in SCLC has not been elucidated.
Our studies revealed Sg with excellent anti-SCLC activities, including inhibition of tumor cell proliferation, colony formation, cell cycle, migration and invasion, and acceleration of apoptosis by upregulating the expression of cyclin dependent kinase inhibitor 1A (CDKN1A). CDKN1A negatively regulates cell cycle progression as a member of the cyclin-dependent kinase inhibitors (CDKIs) family, and targeting CDKN1A is an important strategy for cancer therapy [19]. Our results showed that the expression of CDKN1A was upregulated in SCLC cells with the treatment of Sg and functioned as a tumor suppressor in SCLC. Furthermore, Sg in combination with panobinostat (Pa), or THZ1, or gemcitabine (GEM), or JQ1 increased the anti-SCLC effect compared with either agent alone treatment, which demonstrated that combination therapy of Sg with chemotherapy agents was a potentially effective treatment for SCLC.
Materials and methods
Cell culture and reagents
Human SCLC cell line NCI-H1688 and human lung bronchial epithelial cell line BEAS-2B were obtained and authenticated from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). NCI-H82, NCI-H526 and 16HBE were purchased and authenticated from the ATCC. All cell lines were maintained in either RPMI-1640 medium or DMEM (Thermo Fisher Scientific, MA, USA) medium supplemented with 10% fetal bovine serum. Cells were cultured at 37 °C in a humidified atmosphere with 5% CO2. The natural compound library containing 640 natural compounds and panobinostat(T2383), THZ1(T3664), Gemcitabine(T0251), (+)-JQ-1(T2110) were purchased from Target Mol (Shanghai, China).
Cell viability assay
Cell proliferation was determined using Cell Counting Kit-8 (CCK-8) kit (Dojindo Molecular Technologies, Inc) according to the manufacturer's protocol.
Colony formation assay
For the formation assay, a total of 1000 cells were seeded into six-well plates and cultured in RPMI-1640 medium containing various concentrations of Sg (0, 0.5, 1, 1.5 µM). After 24 h treatment, cells were changed to fresh complete medium and cultured for 2 weeks. The colonies were then fixed with methanol and stained with a 0.1% crystal violet solution.
Cell cycle assay
Cells were collected after 24 h treatment with different concentrations of Sg. The collected cells were fixed in 70% ethanol at 4 °C overnight and stained with Propidium Iodide (P4170, Sigma). Cell cycle were then analyzed by flow cytometry (Accuri C6, BD Biosciences).
Apoptosis assay
Cells were collected after 24 h treatment with different concentrations of Sg. Apoptosis was determined using FITC Annexin-V apoptosis detection kit (556547, BD Biosciences) according to the manufacturer's protocol. Apoptotic cells were analyzed by flow cytometry (Accuri C6, BD Biosciences).
Wound healing assay
3 × 105 cells were seeded in six-well plates and grown to 90% confluency in an incubator, followed by 48 h of starvation in serum-free medium. The culture medium was removed and the monolayers were scratched using a 200 μl pipette to create a uniform cell-free wound area. Debris was removed by gently washing with PBS. Cell migration into the wounded area was monitored and photographed at each designated time point under a microscope.
Transwell cell migration and invasion assay
Cell migration assay was carried out in a 24-well plate containing a polycarbonate membrane with a pore size of 8 mm. For cell invasion assay, the top side of the membrane was coated with Matrigel and was placed in the upper chamber. Cells were seeded into the upper chambers. The lower chambers were filled with complete medium. After 24 h, the chambers were removed, and inner side was wiped with cotton swaps. The cells were fixed with 4% formaldehyde, stained with 0.1% crystal violet and counted under light microscope.
High-throughput screening
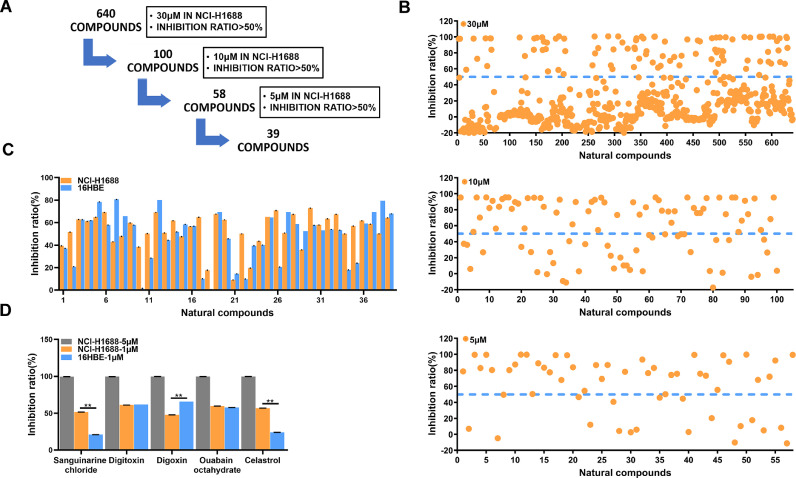
A library of 640 natural compounds (Target Mol) were prepared as stock solutions in DMSO at a concentration of 10 mM for use in this screening. For first round screeding, NCI-H1688 cells were treated with each compound at a concentration of 30 µM. After 48 h, the proliferation of NCI-H1688 cells was measured by CCK-8 assay. 100 compounds with an inhibition rate greater than 50% were selected for the second-round screening and cells were treated with each compound at 10 µM. Next, 58 compounds with an inhibition rate greater than 50% were selected for the next screening process and cells treated with each compound at 5 µM.
Plasmids and cell transfections
Short hairpin RNA (shRNA) sequences were cloned into psiF-copGFP vectors (System Biosciences, Mountain View, CA). The shRNA sequences for CDKN1A were as follows: 5′-GACACCACTGGAGGGTGACTT-3′(#1), 5′-GACAGCAGAGGAAGACCATGT-3′(#2), 5′-ACAGATTTCTACCACTCCAA-3′(#3). The sequence for negative control(shControl) was 5′-GGTGTGCAGTTGGAATGTA-3′. To establish stably transfected cell lines, lentivirus was produced by HEK-293T cells with the second-generation packaging system pMD2.G and psPAX2, and harvested at 48 h after transfection. SCLC cell lines were transduced with virus in the presence of 8 μg/mL polybrene and screened them with puromycin for 7 days.
Western blot
Cells were lysed with RIPA buffer (Cell Signaling) supplemented with protease inhibitor, and boiled at 95 °C for 5 min. Equal amount of protein were added to sodium dodecyl sulfate polyacrylamide-polyacrylamide gel electrophoresis, then transferred to polyvinylidene difluoride membrane. After membrane transferred, the membrane was blocked with 5% nonfat dry milk for 1 h at room temperature, and then incubated with the primary antibodies at 4 °C overnight. Antibodies for CDKN1A (A19094), CDKN1B (A19095), CDKN1C (A2060) and p53 (A0263) were purchased from ABclonal Technology. β-actin antibody (H11459) was purchased from Sigma as a loading control. Next day, the membranes were incubated for 1 h at room temperature with an HRP-conjugated anti-rabbit or anti-mouse secondary antibody, The immunoreactive proteins were visualized with SuperSignal West Dura Chemiluminescent (Thermo Fisher).
Quantitative real-time PCR (qPCR)
Total RNA was isolated by using Trizol reagent and reverse transcribed with a StarScript II First-strand cDNA Synthesis Kit (GenStar, A212-05) in accordance with a standard protocol. QPCR was performed using the SYBR Green Supermix (Bio-Rad, CA, USA) and CFX96 real-time PCR detection system (Bio-Rad). The mRNA expression of genes was evaluated by using the 2−△△Ct method. The primers for CDKN1A were: 5′-GGAAGGGACACACAAGAAGAAG-3′ and 5′-AGCCTCTACTGCCACCATCTTA-3′; the primers for CDKN1B were: 5′-GCTTCTTGGGCGTCTGCTC-3′ and 5′-TAACCCGGGACTTGGAGAAG-3′; the primers for CDKN1C were: 5′-GCGGCGATCAAGAAGCTGT-3′ and 5′-ATCGCCCGACGACTTCTCA-3′; the primers for β-actin were: 5′-TCGTGCGTGACATTAAGGAG-3′ and 5′-ATGCCAGGGTACATGGTGGT-3′.
Xenograft model
Animal experiments were performed according to protocols approved by the ethical committee of Guangzhou University of Chinese Medicine Institutional Animal Care and Use Committee. Five-week-old BALB/C nude mice were used for in vivo anti-SCLC effect of Sg. 2 × 106 NCI-H1688 cells were injected subcutaneously into athymic nude mice. Tumor volume was measured as [(length × width × height)/2] every 3 days. When the tumor volume reached ∼30 mm3, treatments were carried out via intraperitoneal injection. After treatment for 21 days, the mice were sacrificed by cervical dislocation method, and tumor tissues were collected for immunohistochemical staining (IHC) and Western blot.
Immunohistochemistry
Tumors were collected at the indicated times and fixed with 10% neutral buffered formalin. After fixation for 24 h, the tissues were paraffin-embedded, sectioned (4 μm). For immunohistochemistry staining, deparaffinized and rehydrated sections were boiled in Na-citrate buffer (10 mM, pH 6.0) for 30 min for antigen retrieval. The sections were incubated with primary antibodies and developed by using the Ultra Vision Detection System (Thermo Fisher Scientific). Images were acquired using an Olympus IX51 microscope and processed using cellSens Dimension software.
RNA-sequencing and bioinformatics analysis
NCI-H1688 cells were treated with Sg at 1 µM. After 24 h, total RNA was extracted for RNA-sequencing (RNA-Seq) by using the TRI Reagent and Direct-zol RNA kit (Zymo Research, Irvine, CA, USA) following the manufacturer's instructions. The sequencing was performed by a sequencing service company (Novogene, Beijing, China) using the Illumina sequencing platform. Log2|Fold change| ≥ 1 and P < 0.05 were used as a cut-off to define overexpression or downregulation. Ingenuity Pathway Analysis (IPA) was used to identify enriched molecular pathways, gene networks, canonical pathways, and cellular biological consequences. Protein-protein interaction (PPI) analysis of genes was processed by STRING (https://string-db.org/) and visualized by Cytoscape. The hub genes in the network defined as possessing a connective degree >10.
Gene expression analysis from the public database
Data from the Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo) was used to analyze CDKN1A mRNA expression in SCLC, GSE6044 [20] and GSE40275 [21], a total of 19 normal lung and 17 SCLC samples were included in the present study. Patient data from George [22] was used to conduct OS analysis, in brief, patients were grouped into the higher (CDKN1A-H) and lower (CDKN1A-L) group based on their relative expression of CDKN1A.
Statistical analysis
Data are presented as the means ± SD. All statistical analyses were carried out using SPSS software (version 22). An unpaired two-tailed Student's t-test was performed for two-group comparisons and one-way ANOVA analysis (Post hoc: LSD) was performed for multiple group comparisons. For the survival analysis, Kaplan-Meier curves were analyzed by log rank test. The statistical significance was set at a p-value <0.05.
Results
Natural compounds screening identified Sanguinarine chloride as a potential SCLC inhibitor
To identify small molecules that suppress SCLC cell growth, we performed a high-throughput screening of a compound library composed of 640 natural compounds in human SCLC cell NCI-H1688 (Fig. 1A). In brief, all compounds were initially screened at a concentration of 30 µM and the cytotoxicity against NCI-H1688 was measured by CCK-8 assay. One hundred compounds elicited greater than 50% reduction in cellular growth and were triaged for further inspection. Fifty-eight compounds had an IC50 at the concentration of 10 µM, and thirty-nine had an IC50 at 5 µM (Fig. 1B). These thirty-nine compounds were divided into several categories based on their putative targets, which including microtubules/Tubulin (9 compounds), Na+/K+ATPase (5 compounds), Topoisomerase (4 compounds) and other targets (21 compounds) (Supplementary Fig. 1 and Table S1). Next, the top five ranked natural compounds that reduced cellular growth against NCI-H1688 were used to treat both NCI-H1688 and normal bronchial epithelial cell line 16HBE, at the concentration of 1 µM or 5 µM for 48 h. Compared with the other compounds, Sg could not only inhibited SCLC cells at the concentration of 1 µM, but also had a lower lethality rate against 16HBE (Fig. 1C and D). These results indicate that Sg is an ideal candidate compound for treating SCLC.
Fig. 1.
Natural compounds screening identified Sanguinarine chloride as a potential SCLC inhibitor. (A) Natural compound screening process. (B) The inhibition ratio of NCI-H1688 treated with 640 natural compounds at a concentration of 30 µM, 10 µM, 5 µM. (C) The inhibition ratio of NCI-H1688 and 16HBE after 48 h incubation with top 39 ranked natural compounds at 5 µM or 1 µM, respectively. (D) The inhibition ratio of top 5 ranked natural compounds against NCI-H1688 and 16HBE after 48 h incubation at 1 µM and NCI-H1688 at 5 µM, respectively. All data are presented as mean ± SD of 3 replicates. *p < 0.05, **p < 0.01, vs. DMSO.
Sanguinarine chloride inhibited SCLC cells in vitro and in vivo
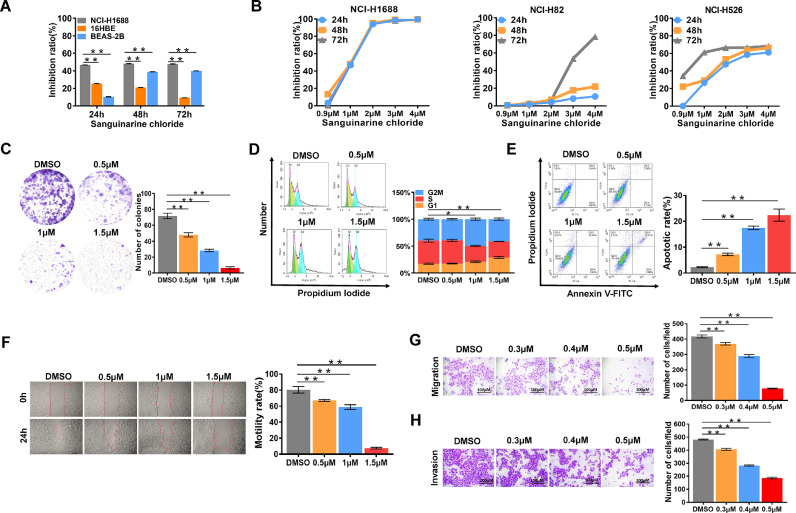
Firstly, we sought to determine the anti-SCLC activity of Sg in vitro. NCI-H1688, 16HBE and BEAS-2B were treated with 1 µM Sg for 24, 48 and 72 h. We observed that the inhibitory effect of Sg on NCI-H1688 was significantly stronger than that on 16HBE and BEAS-2B (Fig. 2A). Three SCLC cell lines, NCI-H1688, NCI-H82 and NCI-H526, were treated with Sg at different concentrations for 24 h, 48 h and 72 h respectively. As shown in Fig. 2B, dose and time-dependent growth inhibition was observed in three SCLC cell lines with the treatment of Sg (Fig. 2B). In addition, we found the viability of SCLC cells was effectively reduced with Sg in a dose-dependent manner by using the colony formation assay (Fig. 2C). To further explore the role of Sg on cell cycle and apoptosis in SCLC cells, NCI-H1688 were treated with Sg at various concentrations for 24 h and then analyzed by flow cytometry. Results from flow cytometry showed that Sg arrested cell cycle in S phase (Fig. 2D), and increased apoptotic cell death in NCI-H1688 (Fig. 2E). Furthermore, wound-healing and trans-well assay demonstrated that Sg impaired the migration and invasion capability of NCI-H1688 (Fig. 2F–H).
Fig. 2.
Sanguinarine chloride inhibited cell proliferation, colony formation, cell cycle, migration, invasion and promoted cellular apoptosis. (A) The inhibitory effects of Sg (1 µM) on the cell proliferation of NCI-H1688, 16HBE and BEAS-2B cells at 24 h, 48 h, 72 h. (B) Dose- and time-dependent inhibitory effects of Sg on the growth and proliferation of NCI-H1688, NCI-H82 and NCI-H526 cells. (C) Colony formation assay of the dose-dependent inhibitory effect of Sg on NCI-H1688 cells. Cell viability was measured using CCK-8 assay while the number of colonies was counted using a microscope. (D) Evaluating the numbers of NCI-H1688 cells during G1/S/G2-M phase after 24 h treatment with Sg at 0.5, 1, 1.5 µM or DMSO respectively. (E) NCI-H1688 were treated with Sg at 0.5, 1, 1.5 µM or DMSO for 24, followed by Annexin V-FITC and PI staining and flow cytometry analysis. Both Annexin V-FITC and PI double positive cells represent apoptotic cells. (F) The scratch area was measured at 0 and 24 h after treatment with Sg at 0.5, 1, 1.5 µM or DMSO, respectively. (G) The effect of Sg on transwell migration and invasion (H) of NCI-H1688 cells after 24 h treatment with Sg at 0.3, 0.4, 0.5 µM or DMSO, respectively. Data are presented as mean ± SD of 3 replicates, *p < 0.05, **p < 0.01, vs. DMSO.
Next, we established mouse xenograft models using NCI-H1688 cells to determine the anti-SCLC of Sg in vivo. Significantly smaller tumor volumes and lower tumor weigh were observed in the Sg treated group than vehicle group (Fig. 3A and B). Immunohistochemical results demonstrated that the expression level of cell proliferation marker Ki67 was significantly decreased, while the apoptotic marker Cleaved-caspase 3 was markedly increased in Sg-treated tumors compared to vehicle-treated tumors (Fig. 3C and D). These results indicate that Sg inhibits the development of SCLC in vitro and in vivo, which demonstrate Sg as an ideal candidate compound for further mechanistic studies in SCLC.
Fig. 3.
Sanguinarine chloride inhibited SCLC cells in vivo. (A) Xenograft growth in nude mice injected with NCI-H1688 cells and treated with or without Sg (2.5 mg/kg•bw, every three days for three weeks, n = 4 mice per group) and tumor volumes in the mice was showed. (B)Tumor weight in the mice. Immunohistochemical staining for Ki-67(C) and Cleaved caspase 3(D), Red arrows mark Cleaved caspase 3 positive, *p < 0.05, **p < 0.01, ***p < 0.001, vs. Vehicle.
Bioinformatic analysis of the molecular mechanism of Sanguinarine chloride against SCLC
To explore the mechanism underlying Sg inhibited the growth of SCLC cells, we performed RNA-sequencing (RNA-seq) on NCI-H1688 cells treated with Sg or dimethyl sulfoxide (DMSO) for 24 h. A total of 891 differently expressed genes (DEGs) were identified (Fig. 4A). Ingenuity Pathway Analysis (IPA) revealed the most enriched canonical pathways involved in the inhibition of MMPs, fatty acid α-oxidation, sirtuin signaling pathway, oxidative phosphorylation and the methylglyoxal degradation Ⅲ (Fig. 4B). And the top three enrichment of IPA molecular and cellular functions analysis were protein trafficking, cellular movement and cell cycle (Fig. 4C). Considering the effect of Sg on the cell cycle of SCLC cells in vitro, we next focused on those forty-seven DEGs enriched in cell cycle. The expression levels of the forty-seven DEGs in RNA-seq data were detected, and most of them were upregulated in NCI-H1688 cells with the treatment of Sg, including CDKN1A and CDKN1C, which were two members of the cyclin-dependent kinase inhibitors (CDKIs) family (Fig. 4D and Supplementary Table S2). Notably, IPA interaction network analysis showed CDKN1A was present in the core modules in the merged network (Fig. 4E). Furthermore, protein-protein interaction (PPI) analysis of the forty-seven DEGs also revealed that CDKN1A and CDKN1C were the two most enriched core genes with a connective degree >10 (Fig. 4F). Collectively, these data indicate that CDKN1A and CDKN1C might positively correlate with the anti-SCLC activity of Sg.
Fig. 4.
Bioinformatics analysis of the molecular mechanism of Sanguinarine chloride in SCLC. (A) Volcano plot showing the differentially expressed genes of NCI-H1688 cells treated with Sg (1 µM) or DMSO. Significantly differentially expressed genes were determined based on Log2|Fold change| ≥ 1 and P < 0.05. (B) IPA Canonical pathways analysis of the DEGs in NCI-H1688 after 24 h treatment with Sg (1 µM). (C) The top 5 ranked molecular and cellular functions in IPA analysis. (D) Heatmap shows the expression of the 47 DEGs in RNA-seq data. (E) IPA interaction network analysis of the 47 DEGs related to cell cycle. (F) PPI analysis of the most core 15 nodes (connective degree >10) in the cell cycle-related molecular network.
CDKN1A functions as a potential tumor suppressor gene against SCLC
To validate the positively correlation of CDKN1A, CDKN1B and CDKN1C with anti-SCLC activity of Sg, we first measured the expression level of them in SCLC cells treated with Sg. CDKN1A and CDKN1B, but not CDKN1C, were significantly upregulated in NCI-H1688 and NCI-H82 cells treated with Sg compared to vehicle treatment (Fig. 5A and B). We also found that the protein level of p53 was not altered in cells treated with Sg. Correspondingly, Sg treatment markedly upregulated the CDKN1A expression in tumors from SCLC mouse xenograft models (Fig. 5C). By data mining GSE6044 and GSE40275, we confirmed the lower expression of CDKN1A in tumors from SCLC patients compared to their normal tissues (Fig. 5D). And the CDKN1A expression in three SCLC tumor cell lines were all significantly lower than the normal cell line 16HBE in both mRNA and protein level as expected (Fig. 5E and F). Moreover, overall survival (OS) analysis revealed a correlation between low CDKN1A expression level and poor OS (P = 0.0176) (Fig. 5G). To further investigate the role of CDKN1A in SCLC, we used loss-of-function method to evaluate the impact of CDKN1A on NCI-H1688 cells (Fig. 5H), and found that CDKN1A silencing effectively enhanced the ability of colony formation and promoted cell proliferation of NCI-H1688 (Fig. 5I and J). Collectively, our results suggest that Sg plays an important role of anti-SCLC by stimulating the expression of CDKN1A and stagnating cell cycle.
Fig. 5.
CDKN1A is a potential tumor suppressor gene in SCLC. (A) and (B) CDKN1A, CDKN1B, CDKN1C and p53 expression level in NCI-H1688 and NCI-H82 were measured by qPCR (A) and Western blot (B) after treated with 0.5, 0.75, 1 µM Sg or DMSO for 24 h. (C) Representative samples showing high and low intensity CDKN1A staining in tumor tissues derived from mice with Sg or DMSO treatment. (D) GEO database(GSE6044 and GSE40275) was used to analysis the expression of CDKN1A in SCLC and normal tissues. (E) qPCR and western blot (F) analyzed the expression level of CDKN1A in 16HBE, NCI-H1688, NCI-H82, and NCI-H526 cells. (G) OS analysis of SCLC patients based on CDKN1A expression, Patients were grouped into the higher (CDKN1A-H) and lower (CDKN1A-L) group based on their relative expression of CDKN1A. (H) Effect of shRNA knockdown on CDKN1A translation in NCI-H1688 cells. (I) The colony forming abilities of NCI-H1688 cells after stably knockdown of CDKN1A (shCDKN1A #3). (J) Cell viability of NCI-H1688 cells after knockdown of CDKN1A (shCDKN1A #3). Data are presented as mean ± SD of 3 replicates, *p < 0.05, **p < 0.01, vs. DMSO.
Sanguinarine chloride synergistically interacts with chemotherapy agents in SCLC cells
Several studies have shown that combination therapy is a potentially effective treatment for SCLC [23] and Sg was the best synergist in comparison with other alkaloids, phenolics and terpenoids [24]. Thus, we explored the potential synergistic anti-SCLC activity of Sg with four well known chemotherapeutic compounds. Firstly, the IC30 of panobinostat (Pa), THZ1, gemcitabine (GEM) and (+)-JQ-1 (JQ1) in NCI-H1688 were determined to be 60 nM, 60 nM, 3 μM and 28 μM respectively, and this concentration was used for the following experiments (Fig. 6A). Compared with each single-agent treatment, all the combination treatment was found to significantly decrease the cell viability of NCI-H1688 (Fig. 6B–E). Sg combined with Pa, JQ1 and GEM showed the higher and durable anti-SCLC activities according to the Combination index (CI) (Fig. 6F). Furthermore, the combination treatment of Sg and Pa resulted in a significantly decreased cellular number in both G2M and S phase (Fig. 6G). Correspondingly, the expression of CDKN1A was significantly increased in three SCLC cell lines treated with Pa or Sg compared to vehicle treatment (Fig. 6H). Interestingly, our data showed that Sg and Pa induced high level of CDKN1B in NCI-H1688 and NCI-H82, but not in NCI-H69, which may suggest the complicated distinct subtypes of SCLC.
Fig. 6.
Sanguinarine chloride synergistically interacts with chemotherapy molecules in SCLC cells. (A) Cell proliferation was measured by using CCK-8 inNCI-H1688 cells after 48 h treatment with different concentrations of Pa, THZ1, GEM and JQ1, and the IC30 of the corresponding drugs were determined. (B) - (E) The dose-dependent anti-proliferative activity of Sg in combination with Pa (B), THZ1 (C), GEM (D) and JQ1 (E) in NCI-H1688 after 24, 48 and 72 h incubation. (F) Combination index (CI) was determined using CalcuSyn software. (G) The cell cycle distribution of NCI-H1688 cells treated with Sg (0.5 µM) and Pa (60 nM), compared with single agent alone. Histograms illustrating the percentage of cells in each cell cycle phase. (H) Western blotting was performed to analyze the expression of CDKN1A, CDKN1B and CDKN1C in NCI-H1688, NCI-H82 and NCI-H69 cells treated with Sg (0.5 µM) and/or -Pa (60 nM) for 24 h. Data are presented as mean ± SD of 3 replicates. *p < 0.05, **p < 0.01, vs. DMSO.
Discussion
Unlike the increasingly personalized therapy targets and compounds against lung adenocarcinoma, SCLC lacks identifying targets, tumor specific somatic mutants and effective therapies drugs. Standard chemotherapy regiment for SCLC, cisplatin combined with etoposide, has been used over the past four decades, however, only a small minority of patients benefit from surgery or concomitant chemotherapy. FDA approved a topoisomerase inhibitor, topotecan for recurrent SCLC, which has substantial toxicities in approximately 25% patients. Notably, natural-based alternatives with high biological activity and less toxicity have become more important against malignant tumors. Numerous bioactive natural compounds have been shown to be useful in the prevention and therapy of cancer by targeting various signaling molecules and pathways [25,26]. In this study, we performed high-throughput screening to identify promising natural compounds with anti-SCLC activity. We found the top 39 effective compounds mostly target Na+/K+ ATPase, topoisomerase and Microtubule/Tubulin pathways, which represent prototype drug targets for SCLC therapy. We further identified Sanguinarine chloride as an ideal candidate for the treatment of SCLC. In vitro data and results from murine tumor models indicate that SCLC is sensitive to Sg treatment.
The encouraging results prompted us to investigate the anti-SCLC mechanism of Sg. IPA and PPI analysis utilized RNA-seq data and subsequently validation preliminarily revealed that CDKN1A was a candidate target gene of Sg. CDKN1A, also known as P21/WAF1/CIP1, which encodes a potent CDKI and the encoded protein functions as a regulator of cell cycle progression at G1 [27]. Given that the most promising clinical investigation in SCLC currently focuses on agents targeting DNA damage repair and cell cycle checkpoints, we speculate that CDKN1A could be a therapeutic potential target in SCLC. Results from public database demonstrated that the CDKN1A expression in both SCLC cell lines and tumors from patients was significantly lower than vehicle control, and this lower CDKN1A level had a correlation with poor OS of SCLC patients. After treated with Sg, CDKN1A expression was increased in SCLC cell lines. Following silencing of CDKN1A effectively enhanced the ability of colony formation and promoted cell proliferation in SCLC cell lines. We demonstrate that Sg stimulating the expression of CDKN1A to induce cell cycle arrest in SCLC, which ascertains that CDKN1A is a therapeutic target against SCLC.
It has been reported that Sg sensitized doxorubicin, paclitaxel and cisplatin to multiple cancers, including colon, prostate, breast, ovarian and lung cancers [24]. Histone deacetylase inhibitors (HDACi) are anti-cancer agents and commonly used in chemotherapy for solid tumors and hematological cancers [28]. Panobinostat (Pa), a pan-HDACi, has been proved the anti-tumor effects in SCLC preclinical models, but only modest single-agent activity was found in clinical trials [29]. Thus, combination strategy for Pa with other agents is a way to enhance its anti-tumor effects in SCLC [30]. Our data showed that Sg synergistically enhanced the anti-tumor activity of four chemotherapy compounds, including panobinostat, gemcitabine, THZ1 and JQ1 in SCLC. Especially, Sg combined with Pa, or GEM or JQ1 exhibits strong synergism anti-SCLC properties in cell culture experiments. When treated with Sg or panobinostat, SCLC cells displayed higher level of CDKN1A and increased number of G1 cells, which certainly confirmed the tumor suppressor role of CDKN1A. Moreover, CDKN1A was also robustly induced by panobinostat, and Sg combined with panobinostat could not further increase the expression level of CDKN1A compared to panobinostat alone treatment, which suggested that additional signaling pathway and targets were involved in the synergist effective role of Sg and panobinostat against SCLC. In the future studies, it will be critical to determine whether additional therapeutic targets and precise mechanism could be identified to explain the strong synergistic action of Sg and panobinostat in vitro and in vivo experiments.
Availability of data and materials
The data that support the findings of this study were submitted to the Gene Expression Omnibus Database (Accession: GSE182821). And the data are available from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE182821.
CRediT authorship contribution statement
Mingtian Zhong: Writing – original draft, Formal analysis, Conceptualization. Xun Li: Writing – original draft, Investigation, Formal analysis. Fengyun Zhao: Writing – original draft, Validation, Data curation. Yanni Huang: Data curation. Yihao Long: Methodology. Kaizhao Chen: Investigation. Xuemei Tian: Validation. Ming Liu: Project administration, Resources. Xiaodong Ma: Project administration, Supervision, Funding acquisition, Conceptualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by the grants from National Natural Science Foundation of China (Nos.81772533 and 81773249), Public Welfare Scientific Research Project of Zhongshan City (No.2019B1015), and a grant from the State Key Lab of Respiratory Disease, Guangzhou Medical University (SKLRD-OP-202101) and Natural Science Foundation of Guangdong Province (No.2020A1515011384).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101345.
Contributor Information
Ming Liu, Email: mingliu128@hotmail.com.
Xiaodong Ma, Email: sciencema@hotmail.com.
Appendix. Supplementary materials
References
- 1.Govindan R., Page N., Morgensztern D., Read W., Tierney R., Vlahiotis A., Spitznagel E.L., Piccirillo J. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J. Clin. Oncol. 2006;24(28):4539–4544. doi: 10.1200/JCO.2005.04.4859. [DOI] [PubMed] [Google Scholar]
- 2.Karachaliou N., Pilotto S., Lazzari C., Bria E., de Marinis F., Rosell R. Cellular and molecular biology of small cell lung cancer: an overview. Transl. Lung Cancer Res. 2016;5(1):2–15. doi: 10.3978/j.issn.2218-6751.2016.01.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jett J.R., Schild S.E., Kesler K.A., Kalemkerian G.P. Treatment of small cell lung cancer: diagnosis and management of lung cancer, 3rd ed: american college of chest physicians evidence-based clinical practice guidelines. Chest. 2013;143(5 Suppl):e400S–e419S. doi: 10.1378/chest.12-2363. [DOI] [PubMed] [Google Scholar]
- 4.Sabari J.K., Lok B.H., Laird J.H., Poirier J.T., Rudin C.M. Unravelling the biology of SCLC: implications for therapy. Nat. Rev. Clin. Oncol. 2017;14(9):549–561. doi: 10.1038/nrclinonc.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mondal A., Gandhi A., Fimognari C., Atanasov A.G., Bishayee A. Alkaloids for cancer prevention and therapy: current progress and future perspectives. Eur. J. Pharmacol. 2019;858 doi: 10.1016/j.ejphar.2019.172472. [DOI] [PubMed] [Google Scholar]
- 6.Luo H., Vong C.T., Chen H., Gao Y., Lyu P., Qiu L., Zhao M., Liu Q., Cheng Z., Zou J., Yao P., Gao C., Wei J., Ung C.O.L., Wang S., Zhong Z., Wang Y. Naturally occurring anti-cancer compounds: shining from Chinese herbal medicine. Chin. Med. 2019;14:48. doi: 10.1186/s13020-019-0270-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adhami V.M., Aziz M.H., Reagan-Shaw S.R., Nihal M., Mukhtar H., Ahmad N. Sanguinarine causes cell cycle blockade and apoptosis of human prostate carcinoma cells via modulation of cyclin kinase inhibitor-cyclin-cyclin-dependent kinase machinery. Mol. Cancer Ther. 2004;3(8):933–940. [PubMed] [Google Scholar]
- 8.Huh J., Liepins A., Zielonka J., Andrekopoulos C., Kalyanaraman B., Sorokin A. Cyclooxygenase 2 rescues LNCaP prostate cancer cells from Sanguinarine-induced apoptosis by a mechanism involving inhibition of nitric oxide synthase activity. Cancer Res. 2006;66(7):3726–3736. doi: 10.1158/0008-5472.CAN-05-4033. [DOI] [PubMed] [Google Scholar]
- 9.Ding Z., Tang S.C., Weerasinghe P., Yang X., Pater A., Liepins A. The alkaloid Sanguinarine is effective against multidrug resistance in human cervical cells via bimodal cell death. Biochem. Pharmacol. 2002;63(8):1415–1421. doi: 10.1016/s0006-2952(02)00902-4. [DOI] [PubMed] [Google Scholar]
- 10.Guo W., Zhu T., Dong Z., Cui L., Zhang M., Kuang G. Decreased expression and aberrant methylation of Gadd45G is associated with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Clin. Exp. Metastasis. 2013;30(8):977–992. doi: 10.1007/s10585-013-9597-2. [DOI] [PubMed] [Google Scholar]
- 11.Kalogris C., Garulli C., Pietrella L., Gambini V., Pucciarelli S., Lucci C., Tilio M., Zabaleta M.E., Bartolacci C., Andreani C., Giangrossi M., Iezzi M., Belletti B., Marchini C., Amici A. Sanguinarine suppresses basal-like breast cancer growth through dihydrofolate reductase inhibition. Biochem. Pharmacol. 2014;90(3):226–234. doi: 10.1016/j.bcp.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Choi W.Y., Kim G.Y., Lee W.H., Choi Y.H. Sanguinarine, a benzophenanthridine alkaloid, induces apoptosis in MDA-MB-231 human breast carcinoma cells through a reactive oxygen species-mediated mitochondrial pathway. Chemotherapy. 2008;54(4):279–287. doi: 10.1159/000149719. [DOI] [PubMed] [Google Scholar]
- 13.Han M.H., Kim G.Y., Yoo Y.H., Choi Y.H. Sanguinarine induces apoptosis in human colorectal cancer HCT-116 cells through ROS-mediated Egr-1 activation and mitochondrial dysfunction. Toxicol. Lett. 2013;220(2):157–166. doi: 10.1016/j.toxlet.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 14.Choi W.Y., Jin C.Y., Han M.H., Kim G.Y., Kim N.D., Lee W.H., Kim S.K., Choi Y.H. Sanguinarine sensitizes human gastric adenocarcinoma AGS cells to TRAIL-mediated apoptosis via down-regulation of AKT and activation of caspase-3. Anticancer Res. 2009;29(11):4457–4465. [PubMed] [Google Scholar]
- 15.Hammerova J., Uldrijan S., Taborska E., Slaninova I. Benzo[c]phenanthridine alkaloids exhibit strong anti-proliferative activity in malignant melanoma cells regardless of their p53 status. J. Dermatol. Sci. 2011;62(1):22–35. doi: 10.1016/j.jdermsci.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Burgeiro A., Bento A.C., Gajate C., Oliveira P.J., Mollinedo F. Rapid human melanoma cell death induced by Sanguinarine through oxidative stress. Eur. J. Pharmacol. 2013;705(1–3):109–118. doi: 10.1016/j.ejphar.2013.02.035. [DOI] [PubMed] [Google Scholar]
- 17.Ahsan H., Reagan-Shaw S., Breur J., Ahmad N. Sanguinarine induces apoptosis of human pancreatic carcinoma AsPC-1 and BxPC-3 cells via modulations in Bcl-2 family proteins. Cancer Lett. 2007;249(2):198–208. doi: 10.1016/j.canlet.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 18.Jang B.C., Park J.G., Song D.K., Baek W.K., Yoo S.K., Jung K.H., Park G.Y., Lee T.Y., Suh S.I. Sanguinarine induces apoptosis in A549 human lung cancer cells primarily via cellular glutathione depletion. Toxicol. Vitro. 2009;23(2):281–287. doi: 10.1016/j.tiv.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz G.K., Shah M.A. Targeting the cell cycle: a new approach to cancer therapy. J. Clin. Oncol. 2005;23(36):9408–9421. doi: 10.1200/JCO.2005.01.5594. [DOI] [PubMed] [Google Scholar]
- 20.Rohrbeck A., Neukirchen J., Rosskopf M., Pardillos G.G., Geddert H., Schwalen A., Gabbert H.E., von Haeseler A., Pitschke G., Schott M., Kronenwett R., Haas R., Rohr U.P. Gene expression profiling for molecular distinction and characterization of laser captured primary lung cancers. J. Transl. Med. 2008;6:69. doi: 10.1186/1479-5876-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kastner S., Voss T., Keuerleber S., Glockel C., Freissmuth M., Sommergruber W. Expression of G protein-coupled receptor 19 in human lung cancer cells is triggered by entry into S-phase and supports G(2)-M cell-cycle progression. Mol. Cancer Res. 2012;10(10):1343–1358. doi: 10.1158/1541-7786.MCR-12-0139. [DOI] [PubMed] [Google Scholar]
- 22.George J., Lim J.S., Jang S.J., Cun Y., Ozretic L., Kong G., Leenders F., Lu X., Fernandez-Cuesta L., Bosco G., Muller C., Dahmen I., Jahchan N.S., Park K.S., Yang D., Karnezis A.N., Vaka D., Torres A., Wang M.S., Korbel J.O., Menon R., Chun S.M., Kim D., Wilkerson M., Hayes N., Engelmann D., Putzer B., Bos M., Michels S., Vlasic I., Seidel D., Pinther B., Schaub P., Becker C., Altmuller J., Yokota J., Kohno T., Iwakawa R., Tsuta K., Noguchi M., Muley T., Hoffmann H., Schnabel P.A., Petersen I., Chen Y., Soltermann A., Tischler V., Choi C.M., Kim Y.H., Massion P.P., Zou Y., Jovanovic D., Kontic M., Wright G.M., Russell P.A., Solomon B., Koch I., Lindner M., Muscarella L.A., la Torre A., Field J.K., Jakopovic M., Knezevic J., Castanos-Velez E., Roz L., Pastorino U., Brustugun O.T., Lund-Iversen M., Thunnissen E., Kohler J., Schuler M., Botling J., Sandelin M., Sanchez-Cespedes M., Salvesen H.B., Achter V., Lang U., Bogus M., Schneider P.M., Zander T., Ansen S., Hallek M., Wolf J., Vingron M., Yatabe Y., Travis W.D., Nurnberg P., Reinhardt C., Perner S., Heukamp L., Buttner R., Haas S.A., Brambilla E., Peifer M., Sage J., Thomas R.K. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudin C.M., Poirier J.T., Byers L.A., Dive C., Dowlati A., George J., Heymach J.V., Johnson J.E., Lehman J.M., MacPherson D., Massion P.P., Minna J.D., Oliver T.G., Quaranta V., Sage J., Thomas R.K., Vakoc C.R., Gazdar A.F. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat. Rev. Cancer. 2019;19(5):289–297. doi: 10.1038/s41568-019-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Achkar I.W., Mraiche F., Mohammad R.M., Uddin S. Anticancer potential of Sanguinarine for various human malignancies. Future Med. Chem. 2017;9(9):933–950. doi: 10.4155/fmc-2017-0041. [DOI] [PubMed] [Google Scholar]
- 25.Yuan R., Hou Y., Sun W., Yu J., Liu X., Niu Y., Lu J.J., Chen X. Natural products to prevent drug resistance in cancer chemotherapy: a review. Ann. N. Y. Acad. Sci. 2017;1401(1):19–27. doi: 10.1111/nyas.13387. [DOI] [PubMed] [Google Scholar]
- 26.Boulos J.C., Rahama M., Hegazy M.F., Efferth T. Shikonin derivatives for cancer prevention and therapy. Cancer Lett. 2019;459:248–267. doi: 10.1016/j.canlet.2019.04.033. [DOI] [PubMed] [Google Scholar]
- 27.Holy J., Lamont G., Perkins E. Disruption of nucleocytoplasmic trafficking of cyclin D1 and topoisomerase II by Sanguinarine. BMC Cell Biol. 2006;7:13. doi: 10.1186/1471-2121-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suraweera A., O'Byrne K.J., Richard D.J. Combination therapy with histone deacetylase inhibitors (HDACi) for the treatment of cancer: achieving the full therapeutic potential of HDACi. Front. Oncol. 2018;8:92. doi: 10.3389/fonc.2018.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crisanti M.C., Wallace A.F., Kapoor V., Vandermeers F., Dowling M.L., Pereira L.P., Coleman K., Campling B.G., Fridlender Z.G., Kao G.D., Albelda S.M. The HDAC inhibitor panobinostat (LBH589) inhibits mesothelioma and lung cancer cells in vitro and in vivo with particular efficacy for small cell lung cancer. Mol. Cancer Ther. 2009;8(8):2221–2231. doi: 10.1158/1535-7163.MCT-09-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Y., Sun Y., Yue S., Wang Y., Lu F. Histone deacetylase inhibitors in cancer therapy. Curr. Top. Med. Chem. 2018;18(28):2420–2428. doi: 10.2174/1568026619666181210152115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study were submitted to the Gene Expression Omnibus Database (Accession: GSE182821). And the data are available from https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE182821.