Abstract
Homeobox (HOX) genes encode highly conserved homeotic transcription factors that play a crucial role in organogenesis and tissue homeostasis. Their deregulation impacts the function of several regulatory molecules contributing to tumor initiation and progression. A functional bridge exists between altered gene expression of individual HOX genes and tumorigenesis. This review focuses on how deregulation in the HOX-associated signaling pathways contributes to the metastatic progression in cancer. We discuss their functional significance, clinical implications and ascertain their role as a diagnostic and prognostic biomarker in the various cancer types. Besides, the mechanism of understanding the theoretical underpinning that affects HOX-mediated therapy resistance in cancers has been outlined. The knowledge gained shall pave the way for newer insights into the treatment of cancer.
Graphical abstract
Keywords: HOX genes, Diagnostic and prognostic significance, Signaling pathways, Metastasis, Cancer
Introduction
Distinct classes of regulatory molecules govern the functional activities in a normal cell through the interaction with different signaling pathways. Any disruption of intercellular and intracellular signaling networks has wider implications on the cellular function resulting in many diseases including cancer (Derakhshani et al. 2020; Sever and Brugge 2015). Accumulating evidence demonstrates that homeobox (HOX) genes that function as master regulators during embryogenesis have roles in both organogenesis and oncogenesis (Smith et al. 2019). The vertebrate HOX clusters contain the protein-coding HOX genes as well as non-coding RNAs (ncRNAs) (Botti et al. 2018). Studies have demonstrated that the expression of the HOX genes is deregulated in cancer either due to temporospatial heterogeneity, gene dominance, genetic, or epigenetic mechanisms (Haria and Naora 2013; Li et al. 2019a). The aberrant expression of the HOX gene loci disrupts the dynamic process of normal intracellular signaling, thereby triggering cellular dysfunction and impaired tumor immune surveillance. The prolonged activation of crucial signaling pathways controlling cell growth, cell cycle, and cell proliferation creates a favorable microenvironment for tumor growth (Giancotti 2014). This review attempts to summarize the existing knowledge of deregulated HOX genes and their role in modulating the key signaling pathways of cancer with a special focus on metastatic progression. The molecular basis by which the tumor cells acquire different cancer hallmarks and the role of HOX genes as a prognostic marker and therapeutic target is widely explored.
HOX cluster: origin, evolution, and organization
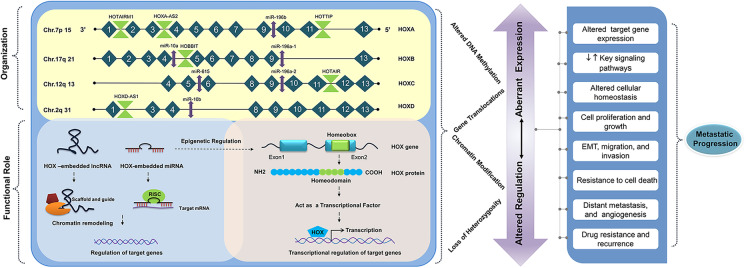
The HOX genes were first discovered in Drosophila melanogaster, due to two mutations mapped to the Antennapedia and Bithorax complex on separate chromosomes. These mutations are often referred to as homeotic transformations, specifying the change of segment identity along the anteroposterior axis. Initially, it was believed that a single proto-HOX gene of primitive organisms had undergone duplication and divergence to form a gene cluster in multicellular organisms (Ferrier and Holland 2001; Holland 2013). However, based on the two rounds (2R) of the whole-genome duplication (WGD) hypothesis, one-to-four transition in the human HOX cluster occurred by three-step sequential events of segmental duplications, independent gene duplications, and translocations (Abbasi 2015). Humans have 39 HOX genes, which are arranged in four clusters, namely, HOXA, HOXB, HOXC, and HOXD (Mallo 2018), mapped onto the specific chromosome loci (7p15, 17q21, 12q13, and 2q31). Each cluster has 13 paralog groups, each having an equivalent position in the cluster, numbered from 1 to 13 in the 3′ to 5′ direction (Apiou et al. 1996).
HOX genes are small, having two exons and a single intron. The second exon contains a homeobox region that has a high degree of homology within the paralog groups. It gets translated into a highly conserved helix-turn-helix DNA binding protein termed as “homeodomain” of 60 amino acid length (Lappin et al. 2006). Incidentally, the conservation of HOX clusters among different species is unique. Phylogenetic analysis reveals that the putative regulatory elements in the HOX cluster are evolutionarily conserved among those different species, which are separated by approximately 500 million years of evolution (Santini et al. 2003). Further, it was found that homology in non-coding sequences in different vertebrate species paved the way to explore the regulatory regions other than the protein-coding genes in the HOX cluster (Matsunami et al. 2010). Embedded within the HOX cluster are several ncRNAs. Rinn et al. described 231 ncRNAs inside the HOX cluster network (Rinn et al. 2007), of which 15% have been functionally characterized.
HOX genes and human health
Since their discovery, the regulation of the HOX genes and their function has been extensively studied. The HOX genes are first expressed in the developing embryo at the gastrulation stage (Boncinelli and Mallamaci 1995). HOX genes that are located near the 3′ of the cluster are expressed earlier than those that are located near the 5′, suggesting that the activation of HOX genes is collinear with its position, a property referred to as the principle of collinearity (Gaunt 2018). A group of cells called the functional domain requires positional information for their differentiation, which means that the HOX genes that are expressed in functional domains are committed to form specific body parts (Kmita and Duboule 2003). Besides, HOX genes have a crucial role in the formation of skeletal morphology (Song et al. 2020), limb (Pineault and Wellik 2014), and organ development (Hombría and Lovegrove 2003).
HOX genes act as transcription factors that usually complex with co-factors to activate genes involved in cell proliferation, adhesion, differentiation, cell migration, and cell death under normal physiological conditions through their homeobox domain (Hombría and Lovegrove 2003; Moens and Selleri 2006). Thus, the impact of HOX gene deregulation at various stages of development leads to abnormalities in skeletal morphology such as hand-foot-genital syndrome, syndactyly, Guttmacher syndrome, congenital heart diseases, retinal degenerative diseases, and cancer (Lescroart and Zaffran 2018; Quinonez and Innis 2014; Shah and Sukumar 2010; Zagozewski et al. 2014). This review focuses on the role of HOX gene deregulation in cancer.
Regulation of HOX genes in cancer
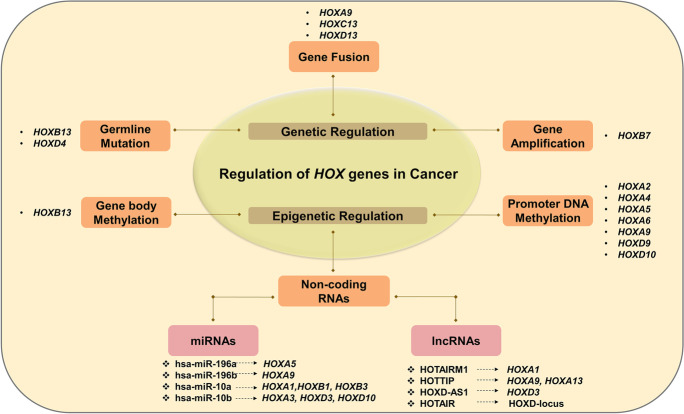
Several researchers have reviewed the role of HOX genes in the normal and tumor microenvironment (Bhatlekar et al. 2018). Meanwhile, HOX gene regulation is tissue-specific, and their deregulation varies from one cancer to another. They either act as oncogenes or tumor suppressors (Table 1) in a context-dependent manner. Functional studies have revealed that both genetic and epigenetic factors contribute to the abnormal expression of HOX genes in different cancers (Li et al. 2019a). The different modes of regulation of HOX genes in cancer have been outlined in Fig. 1.
Table 1.
The role of HOX genes in the development of human cancer
| Cluster | HOX genes | Chromosomal location | Cancer | Aberrant expression | References |
|---|---|---|---|---|---|
| Oncogenic HOX genes | |||||
| HOXA cluster | HOXA1 | chr7:27,092,993–27,096,000 | Gastric cancer, human mammary carcinoma, prostate cancer | ↑ | Hou et al. 2012; Mohankumar et al. 2007; Wang et al. 2015; Yuan et al. 2016 |
| HOXA10 | chr7:27,170,605–27,174,320 | Bladder cancer, colorectal cancer, acute myeloid leukemia, laryngeal squamous cell cancer, pancreatic cancer | ↑ | Cui et al. 2014; Guo et al. 2020; Li et al. 2020a; Liu et al. 2019a; Yuan et al. 2018 | |
| HOXA13 | chr7:27,194,364–27,200,091 | Gastric cancer, lung squamous cancer, bladder cancer, kidney renal clear-cell carcinoma, glioma | ↑ | Cui et al. 2020; Duan et al. 2015; Han et al. 2018; Hu et al. 2017; Zhang et al. 2017 | |
| HOXB cluster | HOXB2 | chr17:48,542,655–48,544,989 | Glioblastoma | ↑ | Li et al. 2020b |
| HOXB3 | chr17:48,550,006–48,590,272 | High-grade serous ovarian carcinoma | ↑ | Miller et al. 2018 | |
| HOXB4 | chr17:48,575,507–48,578,350 | Chronic myelogenous leukemia | ↑ | Wang et al. 2016 | |
| HOXB5 | chr17:48,591,257–48,593,961 | Breast cancer, gastric cancer, non-small cell lung cancer, head and neck squamous cell carcinoma, pancreatic cancer, retinoblastoma, hepatocellular carcinoma | ↑ | Gao et al. 2020; Hong et al. 2015; Lee et al. 2015; Lee et al. 2020; Su et al. 2020; Xu et al. 2018; Zhang et al. 2018 | |
| HOXB7 | chr17:48,607,232–48,611,017 | Esophageal squamous cell carcinoma, gastric cancer, glioma, cutaneous squamous cell carcinoma, breast cancer, pancreatic cancer, hepatocellular carcinoma, acute lymphoblastic leukemia, multiple myeloma, lung adenocarcinoma, colorectal cancer | ↑ | Gao and Chen 2018; He et al. 2017; Huo et al. 2019; Li et al. 2015; Liao et al. 2011; Liu et al. 2015a; Storti et al. 2011; Tsuboi et al. 2017; Wang et al. 2017; Zhong et al. 2019; Zhuang et al. 2015 | |
| HOXB8 | chr17:48,612,346–48,615,292 | Colorectal cancer, osteosarcoma | ↑ | Guo et al. 2019; Wang et al. 2019a | |
| HOXC cluster | HOXC6 | chr12:54,028,440–54,030,823 | Gastric cancer, nasopharyngeal carcinoma, cervical cancer, glioblastoma, head and neck squamous cell carcinoma | ↑ | Chang et al. 2017; Jung et al. 2020; Moon et al. 2012; Wang et al. 2019b; Yang et al. 2019 |
| HOXC8 | chr12:54,008,985–54,012,769 | Laryngeal squamous cell carcinoma, glioma, cervical cancer, non-small cell lung cancer | ↑ | de Barros E Lima Bueno et al. 2016; Huang et al. 2018; Liang et al. 2018; Liu et al. 2018 | |
| HOXC9 | chr12:53,995,509–54,003,298 | Colorectal cancer, gastric cancer, neuroblastoma, glioblastoma | ↑ | Hu et al. 2019; Kocak et al. 2013; Xuan et al. 2016; Zhao et al. 2020 | |
| HOXC10 | chr12:53,985,146–53,990,279 | Thyroid cancer, gastric cancer, glioma, breast cancer | ↑ | Feng et al. 2015; Kim et al. 2019; Li et al. 2018; Miwa et al. 2019; Sadik et al. 2016 | |
| HOXC11 | chr12:53,973,126–53,977,643 | Renal cell carcinoma | ↑ | Liu et al. 2015b | |
| HOXD cluster | HOXD3 | chr2:176,160,891–176,173,085 | Non-small cell lung cancer | ↑ | Miyazaki et al. 2002 |
| HOXD4 | chr2:176,151,550–176,153,226 | Gastric adenocarcinoma | ↑ | Liu et al. 2019b | |
| HOXD9 | chr2:176,122,719–176,124,937 | Gastric cancer | ↑ | Xiong et al. 2020 | |
| HOXD11 | chr2:176,104,216–176,109,754 | Laryngeal squamous cell carcinoma | ↑ | de Barros E Lima Bueno et al. 2016 | |
| Tumor suppressor HOX genes | |||||
| HOXA cluster | HOXA3 | chr7:27,107,012–27,140,219 | Lung adenocarcinoma | ↓ | Gan et al. 2018 |
| HOXA5 | chr7:27,141,052–27,143,681 | Cervical cancer, osteosarcoma, breast cancer, gastric cancer, non-small cell lung cancer, colorectal cancer | ↓ | Chen et al. 2004; Chen et al. 2019; Ma et al. 2020; Ordóñez-Morán et al. 2015; Peng et al. 2018; Raman et al. 2000; Wang et al. 2019c; Zhang et al. 2015 | |
| HOXD cluster | HOXD1 | chr2:176,188,668–176,190,907 | Kidney renal clear-cell carcinoma | ↓ | Cui et al. 2021 |
| HOXD8 | chr2:176,130,357–176,132,000 | Colorectal cancer | ↓ | Mansour and Senga 2017 | |
| HOXD13 | chr2:176,092,721–176,095,944 | Prostate cancer | ↓ | Xu et al. 2021 | |
| HOX genes with both oncogenic and tumor suppressor activity | |||||
| HOXA cluster | HOXA4 | chr7:27,128,507–27,130,780 | High-grade serous ovarian carcinoma | ↑ | Miller et al. 2018 |
| Lung cancer | ↓ | Cheng et al. 2018 | |||
| HOXA9 | chr7:27,162,438–27,165,537 | Acute myeloid leukemia, acute lymphoblastic leukemia, ovarian cancer | ↑ | de Bock et al. 2018; Brumatti et al. 2013; Ko et al. 2012; Zhao et al. 2015 | |
| Cervical cancer, cutaneous squamous cell carcinoma | ↓ | Alvarado-Ruiz et al. 2016; Han et al. 2019 | |||
| HOXA11 | chr7:27,181,157–27,185,232 | Laryngeal squamous cell cancer | ↑ | Li et al. 2020a | |
| Glioblastoma, breast cancer | ↓ | Se et al. 2017; Xia et al. 2017 | |||
| HOXB cluster | HOXB9 | chr17:48,621,156–48,626,358 | Breast cancer, hepatocellular carcinoma, oral squamous cell carcinoma, laryngeal squamous cell carcinoma | ↑ | Hayashida et al. 2010; Sha et al. 2015; Sun et al. 2017; Xue et al. 2017 |
| Gastric cancer | ↓ | Zhang et al. 2019 | |||
| HOXB13 | chr17:48,724,763–48,728,750 | Urinary bladder cancer, breast cancer, prostate cancer | ↑ | Kim et al. 2014; Marra et al. 2013; Shah et al. 2013 | |
| Colorectal cancer | ↓ | Jung et al. 2005 | |||
| HOXD cluster | HOXD10 | chr2:176,116,778–176,119,937 | Laryngeal squamous cell carcinoma | ↑ | de Barros E Lima Bueno et al. 2016 |
| Colon cancer, hepatocellular carcinoma, cholangiocellular carcinoma | ↓ | Guo et al. 2017; Yang et al. 2015; Yuan et al. 2019 | |||
NOTE: ↑, upregulated; ↓, downregulated
Fig. 1.
HOX gene regulation in cancer modulated by genetic and epigenetic mechanisms
Genetic regulation of HOX genes in cancer
The genetic alterations at the HOX gene loci promote cancer progression. In this section, we outline the genetic alterations associated with HOX genes upon an extensive literature search.
Missense mutations
Among the germline mutations analyzed in the HOX paralogous groups from 4 to 13, the HOXD4 missense mutation was seen in the patients of acute lymphoblastic leukemia (ALL). The transcriptional activity of p.Glu81Val mutant HOXD4 was observed to be 40% lower compared to the wild-type protein, leading to its partial loss of function causing ALL susceptibility. Due to the mutation, HOXD4 lost its ability to regulate target genes involved in hematopoietic cell proliferation and differentiation (van Scherpenzeel Thim et al. 2005).
HOXB13 is one of the widely studied genes in prostate cancer (PCa) progression (Kim et al. 2014; Yao et al. 2019). Inherited mutations in the HOXB13 gene contribute significantly to the increased risk of PCa. Screening of more than 200 germline genes at 17q21-22 loci, in 94 PCa patients, recognizes this region as cancer-susceptible loci. The occurrence of rare and recurrent mutation G84E in HOXB13 (rs138213197), along with its variants (Y88D, L144P, G216C, and R229G), has been implicated in hereditary PCa. The mutation rate was significant in men with familial PCa (3.1%) than non-familial PCa (0.6%). Overall, men with PCa carried the HOXB13-G84E allele (frequency 1.4%) when compared to normal patients (frequency 0.1%) (Ewing et al. 2012). The variant HOXB13-G84E has also shown an increased risk for leukemia and bladder cancer (Beebe-Dimmer et al. 2015). Understanding the effect of mutations in rare cases would pave the way to understand the pathways related to cancer progression. Another study examined the functional implication of HOXB13 rs138213197 (G84E) and CIP2A rs2278911 (R229Q) germline variants in PCa. The interplay between HOXB13 and CIP2A variants was most frequent in familial PCa (p < 0.001). HOXB13 protein promotes the transcription of CIP2A, by binding to its promoter region. The co-occurrence of this event is associated with enhanced cancer cell growth, proliferation, and migration. Thus, these allelic variants contribute to high cancer risk and aggressiveness (Sipeky et al. 2018).
While appraising the association between HOXB13-pGly84Glu mutation and colorectal cancer (CRC) patients in two different population registries, 0.48% of the cases had HOXB13 mutation when compared to the control group (0.17%) (Akbari et al. 2013).
Gene fusion
HOX genes act as a fusion partner of nucleoporin 98 kDa (NUP98) to drive cancer progression. The fusion protein bears the N-terminal of NUP98 with unique potential for transcriptional activation (Gough et al. 2011). The chromosomal translocation of t (7; 11) (p15; p15) results in the chimeric protein NUP98-HOXA9 (NHA9). The impact of gene fusion in driving the leukemic transformation of primary human CD34 + hematopoietic cells in acute myeloid leukemia (AML) has been studied (Takeda et al. 2006). NHA9 interacts with mixed-lineage leukemia 1 (MLL1) to promote proliferation by activating a large number of oncogenes (Takeda et al. 2006; Xu et al. 2016). Further, gene translocation at t (11;12) (p15;q13) generated a NUP98/HOXC13 fusion protein. The study showed that the fusion that occurred between the 16th exon of the NUP98 gene and the 2nd exon of HOXC13 might have pathogenic importance in AML (Panagopoulos et al. 2003). Recently, it was reported that fusion protein NUP98–HOXD13 (NHD13) promoted T cell acute lymphoblastic leukemia (T-ALL) by inducing the self-renewal of T cell progenitors in the thymus at the DN2 developmental stage of T cells (Shields et al. 2019). NHD13 was found to be regulated by T cell oncogenes LMO2 and LYL1 for self-renewal of thymocyte cells (Shields et al. 2019).
Amplification
Meanwhile, reports on copy number variations including amplifications and deletions located in HOX locus are just emerging. In breast cancer (BC), comparative genomic hybridization (CGH) was performed to identify copy number alterations. Interestingly, a novel amplicon at HOXB7 (17q21.3) was observed in 10.2% of patients whose expression was significantly correlated with poor prognosis (Hyman et al. 2002).
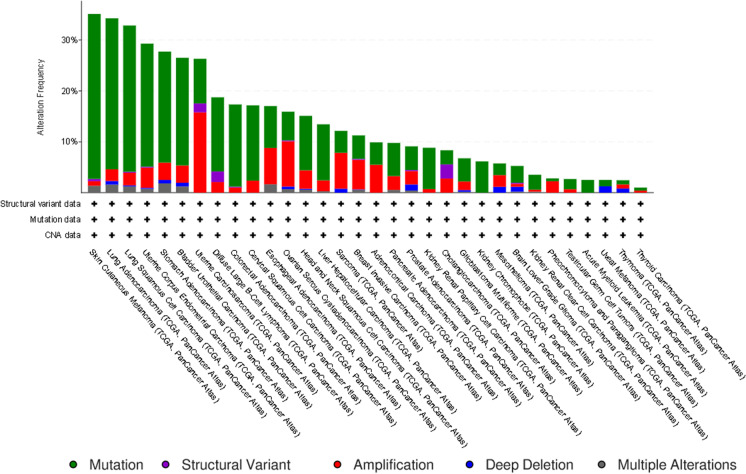
Further, the in silico pan-cancer analysis of all the 39 HOX genes, performed using cBioportal (https://www.cbioportal.org/) (Cerami et al. 2012; Gao et al. 2013), showed different kinds of genetic changes such as mutations, structural variants, amplification, and deep deletions across 32 TCGA cancer types analyzed in 10,967 tissue samples (Fig. 2).
Fig. 2.
Genetic alterations of HOX genes: the in silico pan-cancer analysis of all the 39 HOX genes, across 32 TCGA cancer types performed using cBioportal. The green bars indicate mutation, violet bars indicate structural variant, red bars indicate amplification, blue bars indicate deep deletion, and gray bars indicate multiple alterations
Epigenetic regulation of HOX genes in cancer
Epigenetic factors associated with cancer progression include aberrant methylation at CpG sites, altered chromatin modifications, and aberrant expression of ncRNAs at HOX cluster network (Li et al. 2019a). The characterization of the epigenome thus helps to predict the biological behavior of the tumor, predict disease outcome, and plan suitable treatment regimens.
Promoter DNA methylation
Aberrant gene methylation is the most common epigenetic alteration that not only promotes cancer progression but also confers resistance to anticancer therapies (Romero-Garcia et al. 2020). The promoter DNA methylation could prevent the binding of RNA polymerases and other transcription factors to suppress the transcriptional activation (Curradi et al. 2002). However, this gene silencing mechanism is context-dependent (Smith et al. 2020). Paço et al. (2020) have extensively reviewed the impact of promoter DNA methylation on the expression of HOX genes in different cancers (Paço et al. 2020b). Most recent studies that emphasized the impact of HOX gene promoter hypermethylation in different cancers have been briefly summarized in Table 2. In contrast, a study on BC has delineated the fact that HOXA9-HOXA10 methylation and their promoter–promoter interaction are responsible for cancer progression. Interestingly, the CpG islands of the HOXA10 gene functioned as an enhancer for the promoter region of the HOXA9 gene to promote its gene expression (Park et al. 2017). In addition to the promoter DNA methylation, the altered gene expression is also attributable to the gene body methylation. Su et al. in 2018 elucidated a novel mechanism of gene body hypermethylation and homeobox oncogene activation through performing the pan-cancer analysis of 4174 genome-wide datasets and whole-genome bisulfite sequencing in 30 normal tissues and 35 tumor tissues (Su et al. 2018).
Table 2.
Epigenetic regulation of HOX genes in cancer
| Promoter DNA hyper methylation | |||||
| Cancer | HOX genes | Frequency | Specificity | Role | References |
| Breast cancer | HOXA4 | 46.6% | - | Biomarker for early cancer detection | Li et al. 2019b |
| Head and neck squamous cell carcinoma | HOXA9 | - | 93.8% | Promotes cancer progression and significantly correlated with lymph node metastasis | Zhou et al. 2019 |
| Epithelial ovarian cancer | HOXA9 | 82.3% | 88.6% | Potential diagnostic serum marker for early detection | Singh et al. 2020 |
| Lung cancer | HOXA9 | - | 84.2% | Biomarker for lung cancer subtyping in liquid biopsies | Nunes et al. 2019 |
| Colorectal cancer | HOXD10 | - | - | Promotes cell proliferation, migration, and invasion | Yuan et al. 2019 |
| HOXA2, HOXA5, HOXA6 | - | - | Significant association with age, stage, lymphovascular and perineural invasion | Li et al. 2019a | |
| Cholangiocarcinoma | HOXD9 | - | 90% | A potential diagnostic marker: Promotes dedifferentiation of cholangiocytes | Wasenang et al. 2019 |
| Non-coding RNAs and chromatin modification | |||||
| LncRNA-mediated HOX gene regulation | |||||
| Cancer | HOX-embedded lncRNA | Interaction with chromatin modifiers | Target, HOX genes | Role | References |
| Breast cancer, glioblastoma multiforme | HOTAIRM1↑ | EZH2 | HOXA1↑ | Promotes resistance to tamoxifen; promotes cell proliferation, migration, and invasion and suppresses apoptosis | Kim et al. 2020; Li et al. 2018 |
| Prostate cancer, pancreatic cancer, and hepatocellular carcinoma | HOTTIP↑ | WDR5 | HOXA9, HOXA13↑ | Promotes tumorigenesis | Fu et al. 2017; Malek et al. 2017; Quagliata et al. 2014 |
| Most of the cancer types | HOTAIR↑ | PRC2, LSD1 | HOXD cluster genes↓ | Promotes cell cycle progression, migration, EMT, and metastasis | Bhan and Mandal 2015 |
| Colorectal cancer | HOXD-AS1↓ | PRC2 | HOXD3↓ | Promotes proliferation, migration, and metastasis | Yang et al. 2019 |
| miRNA-mediated HOX gene regulation | |||||
| Cancer | HOX-embedded miRNA | Target, HOX genes | Downstream targets | Role | References |
| Ovarian cancer | miR-10b↑ | HOXD10↓ | MMP14, RHOC | Induces migration and invasion; contributes to the metastatic phenotype | Nakayama et al. 2013 |
| miR-196b↑ | HOXA9↓ | - | Induces cancer cell invasion and recurrence | Chong et al. 2017 | |
| Non-small cell lung cancer | miR-196a↑ | HOXA5↓ | - | Promotes cell proliferation, migration, and invasion | Liu et al. 2012 |
| Pancreatic cancer | miR-10a↑ | HOXA1↓ | - | Promotes invasive potential | Ohuchida et al. 2012 |
| miR-10a↑ | HOXB1 and HOXB3↓ | - | Promotes metastatic behavior | Weiss et al. 2009 | |
| Glioma | miR-10b↑ | HOXB3↓ | HMGB1, RHOC, and MMP2 | Promotes cell proliferation, migration, and invasion | Li et al. 2019c |
| Endometrial cancer | miR-10b↑ | HOXB3↓ | - | Promotes cell proliferation, migration, and invasion | Chen et al. 2016 |
| Clear-cell renal cell carcinoma | miR-10b↓ | HOXA3↑ | FAK, YAP | Promotes cell invasion and metastasis | He et al. 2019 |
NOTE:YAP, Yes-associated protein; FAK, focal adhesion kinase; HOTAIR-HOX, transcript antisense RNA; HOTAIRM1-HOX, antisense intergenic RNA myeloid 1; HOXD-AS1-HOXD, cluster antisense RNA 1; HOTTIP-HOXA, distal transcript antisense RNA; EZH2, enhancer of zeste 2; PRC2, polycomb repressive complex 2; WDR5-WD, repeat domain 5; LSD1, lysine-specific histone demethylase 1A; HMGB1, high mobility group box protein 1; ↑, upregulated; ↓, downregulated
Further, in cervical cancer (CC), the methylation of the first exon of the HOXA9 gene resulted in gene repression. Ectopic expression of the HOXA9 gene significantly induced an epithelial phenotype and suppressed cell proliferation and migration (Alvarado-Ruiz et al. 2016). Hence, understanding the dynamics of methylation and the discovery of novel epigenetic drugs might be a promising strategy to treat cancer.
ncRNAs and chromatin modification
One of the integral components of epigenetic machinery regulating gene expression at the transcriptional and post-transcriptional level is the ncRNA. However, chromatin modification, DNA methylation, and ncRNAs operate in a common mechanism of gene regulation. Botti et al. (2018) have extensively reviewed the dynamic role of HOX-embedded ncRNAs such as long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) in regulating the HOX gene expression in different cancers (Botti et al. 2018). There are several highly conserved lncRNAs transcribed from specific HOX gene clusters. They control HOX gene expression in cis or trans by binding to chromatin modifiers or by sponging miRNAs (Rinn et al. 2007). The lncRNAs interact with chromatin remodeling complexes having specific enzymatic activity such as methyltransferases, demethylases, acetyltransferases, and deacetylases recruiting them to specific gene loci. Based on the complexes that are recruited to the gene loci, the target genes are either expressed or repressed. The repressing methylation marks (activating acetylation marks) at H3K27 and H3K9 positions and the activating methylation mark at H3K4 position on histone tails determine the gene expression pattern. The roles of HOX-embedded lncRNAs in regulating HOX cluster genes and their implications in the biological responses have been studied in different cancer types (Table 2).
There are six widely studied miRNAs embedded in the four HOX clusters that belong to 3 miRNA families, namely, miR-196, miR-10, and miR-615 (Tanzer et al. 2005). The impact of deregulated HOX-embedded miRNAs on HOX gene expression in different kinds of cancers has been summarized in Table 2. They bind to the 3′UTR of mRNA and facilitate transcriptional inactivation of tumor suppressor genes. Considering several genetic and epigenetic factors contribute to the aberrant expression of HOX genes in cancer, their gene expression profiling is crucial for clinical applications.
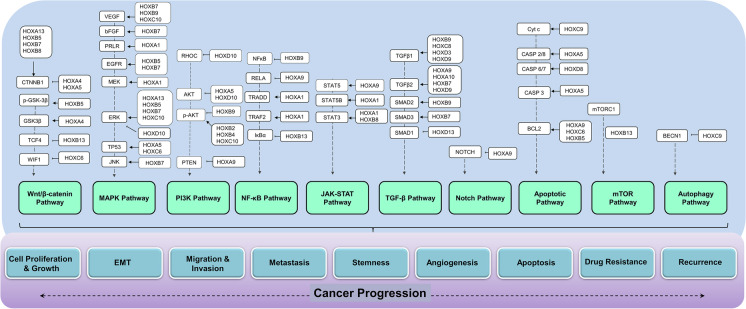
Role of HOX-targeted signaling in metastatic progression
“Metastasis” reflects the critical step of tumor progression in which the cancer cells disseminate from the circulation into the surrounding tissues to form a secondary tumor (Hanahan and Weinberg 2011; Sever and Brugge 2015). The progression to metastasis requires the cancer cells to transform into mesenchymal phenotype, leave the primary site, acquire migratory capacity, circulate and survive in the bloodstream, evade apoptosis, disseminate to the distant site, acclimate to the new site, and generate new blood vessels for their further growth. The occurrence of these cellular processes is due to the imbalance in the important signal transduction pathways (Sever and Brugge 2015). Studies have found that HOX transcription factors regulate a large number of genes that are dynamically involved in the regulation of downstream signaling molecules (Rezsohazy et al. 2015). They preserve the integrity of the cells by regulating the diverse biological processes and functional pathways (Hombría and Lovegrove 2003; Moens and Selleri 2006). Hence, the deregulation of HOX proteins is directly linked to the imbalance in the key signaling pathways, which ultimately trigger the metastatic progression (Fig. 3). In the following section, we have discussed the molecular mechanism of HOX protein-mediated signaling pathways associated with the metastatic progression in cancer.
Fig. 3.
Aberrant regulation of HOX genes causing an imbalance in the key signaling pathways leading to metastatic progression in cancer
Cell proliferation
The proliferative signal due to the prolonged activation of multiple signaling pathways is the first important property acquired by the cancer cells. Aberrantly expressed HOX proteins activate cancer cell proliferation by targeting multiple signaling pathways. The downregulated HOXA5 protein induces cell proliferation and neoplastic progression in CC via modulating the Wnt/β-catenin signaling and AKT pathway (Ma et al. 2020; Wang et al. 2019c). Gene set enrichment analysis and the TOP/FOP reporter assay confirmed that ectopic expression of HOXA5 suppresses CCND1 (cyclin D1) gene expression by inhibiting the nuclear translocation of β-catenin. It inhibits cell proliferation by upregulating the cell cycle inhibitor, p21 (CDKN1A), via transactivating TP53. The transactivation refers to the binding of HOXA5 to the 5′-TAAT-3′ motif of the TP53 promoter through its HD domain (Ma et al. 2020). Further, HOXA5 inhibits cell proliferation also by upregulating the p27 gene (CDKN1B) via controlling the phosphorylation of AKT (Wang et al. 2019c). Thus, HOXA5 has the potential to inhibit cancer stem cell proliferation and metastasis.
Ordóñez-Morán et al. (2015) found that HOXA5 is downregulated in CRC and its re-expression induces loss of cancer cell phenotype. They identified the presence of mutual antagonistic relation between HOXA5 and Wnt signaling. Wnt signaling represses HOXA5 to maintain stemness, whereas HOXA5 gets activated outside the intestinal crypt to suppress Wnt signaling and induce differentiation (Ordóñez-Morán et al. 2015).
In gastric cancer (GC), sustained proliferation is maintained by the activation of the HOXA13-induced Wnt/β-catenin signaling pathway. HOXA13 protein binds to the promoter region of the CDH17 gene (cadherin-17), a downstream effector of HOXA13. Further, knockdown studies confirmed that HOXA13 and CDH17 could also promote cell invasion and inhibit apoptosis in GC cell lines (Qu et al. 2017). The aberrant proliferation of cancer cells is due to the disruption of T cell factor-4 (TCF-4)-stimulated Wnt/β-catenin signaling pathway (Angus-Hill et al. 2011; Arce et al. 2006). There is evidence that targeting TCF-4 could restrain the progression of cancer (Shin et al. 2017). Interestingly, overexpressing HOXB13 in CRC caused growth suppression by inhibiting TCF-4 and its target c-myc at their translational level (Jung et al. 2005). Similarly, gain-of-function experiments demonstrated that HOXB13 functions as a suppressor of TCF-4 and its responsive genes (c-myc and CCND1) in PCa. This study demonstrated that HOXB13 could suppress the promoter activity of the c-myc gene and negatively regulate cell proliferation (Jung et al. 2004).
HOXB13 overexpression has been implicated in drug resistance in BC cells. Elevated HOXB13 confers resistance to tamoxifen by downregulating the protein expression of estrogen receptor-α (ER-α) in ER + BC cells. HOXB13 is involved in both estrogen-independent and mammalian target of rapamycin (mTOR)-dependent proliferation of ER + BC cells. HOXB13 driven interleukin-6 (IL-6) expression, and subsequently, IL-6-mediated activation of mTOR/AKT pathway is another mechanism of BC cell proliferation and fibroblast recruitment (Shah et al. 2013).
In addition, elevated expression of HOXB7 protein confers resistance to tamoxifen in BC cells via the activation of the epidermal growth factor receptor (EGFR) pathway (Jin et al. 2012). EGFR, a receptor tyrosine kinase (RTK), is considered as a biomarker for tumor chemoresistance (Montagut et al. 2012; Wei et al. 2013). Inappropriate activation of EGFR either by transcriptional upregulation or by ligand overactivation is the most common event in oncogenesis (Sigismund et al. 2018). In BC, HOXB7 functions as an ERα-responsive gene, which binds directly to the promoter region of EGFR to facilitate transcriptional activation. Hence, the therapeutic resistance of BC cells is linked to the HOXB7-induced crosstalk between RTKs and ERα-signaling (Jin et al. 2012).
Prolonged activation of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling has been shown to induce tumor proliferation and cell survival in solid tumors (Sau et al. 2016; Xia et al. 2018). The non-canonical NF-κB pathway is activated by tumor necrosis factor-α (TNF-α) that binds to the tumor necrosis factor receptor (TNFR) family members (Cildir et al. 2016). Upon ligand-dependent activation, they multimerize and interact with tumor necrosis factor receptor type 1-associated death domain protein (TRADD) proteins to facilitate tumor necrosis factor receptor-associated factor (TRAF) protein-mediated activation of NF-κB signaling (Cildir et al. 2016). The activation of NF-κB signaling is critical for the oncogenicity of HOXA1 in BC, which is functionally linked to the TNF/NF-κB signaling pathway. This study depicted a unique mechanism in which HOXA1 could activate TNF/NF-κB signaling pathway either by TRAF2 activation or by TRADD complex stabilization (Taminiau et al. 2016). This complex molecular interaction resulted in enhanced cell proliferation and loss of cell adhesion of BC cells.
Elevated HOXB7 has been demonstrated in the peripheral lymphocytes in ALL patients whose progression is attributable to the HOXB7-mediated activation of basic fibroblast growth factor (bFGF) and extracellular signal-regulated kinase (p-ERK1/2) and inactivation of p27 (Zhong et al. 2019). Liao et al. (2011) demonstrated that the overexpression of HOXB7 enhances growth and proliferation of CRC cells through the overactivation of the phosphatidylinositol 3-kinase (PI3K/AKT) and mitogen-activated protein kinase (MAPK) pathways. HOXB7 protein could also induce the expression of CCND1 and repress the expression of CDKN1B, leading to the acceleration of G0-G1 to S-phase transition (Liao et al. 2011). Likewise, the HOXB8-mediated Wnt/β-catenin signaling pathway has been implicated in CRC proliferation and migration. The overexpression of Wnt signaling–related genes and their downstream targets such as matrix metalloproteinase-2 (MMP2), c-Myc, CCND1, and vimentin (VIM) indicates the involvement of Wnt/β-catenin signaling in HOXB8-overexpressed CRC cells (Li et al. 2019e).
Interestingly, in vivo studies demonstrated that upregulated HOXC6 could inhibit the Wnt inhibitory factor-1 (WIF-1) expression and activate Wnt signaling pathway in glioma cells. Through xenograft assay, immunohistochemistry (IHC), and knockdown studies, it was shown that knockdown of HOXC6 could significantly block the cell cycle progression of U87 cells at G0/G1 phase and inhibit the colony formation. Hence, HOXC6 could be a suitable target for clinical therapy (Yan et al. 2018).
MAPK pathway is the most common oncogenic pathway that involves sequential coordination among RTKs, growth factor receptor binding protein-2 (GRB2), and Ras/Raf/MEK/ERK signaling molecules to drive cancer progression (Guo et al. 2020b). The key molecules of the MAPK pathway such as GRB2 and MEK1 are reported as the downstream targets of HOXA1. HOXA1 induces cell proliferation, survival, and oncogenic transformation of human mammary epithelial cells by increasing the expression of GRB2/MEK1 and facilitating the p44/42 MAPK phosphorylation (Mohankumar et al. 2007). HOXA1-mediated upregulation of PRLR (prolactin receptors) and their subsequent signal transduction is another important mechanism demonstrated in the oncogenic transformation of mammary carcinoma cells (Hou et al. 2012).
HOXA1 also regulates the signal transducer and activator of transcription (STAT) signaling pathway to facilitate the oncogenic transformation of immortalized human mammary epithelial cells (Mohankumar et al. 2008). Aberrant activation of the Janus tyrosine kinase (JAK)/STAT pathway is chiefly linked to tumor progression. Higher levels of STAT3 and STAT5 protein expression and their phosphorylation have shown to be associated with poor survival and recurrence in solid tumors (Thomas et al. 2015). In human mammary carcinoma, HOXA1 could enhance the expression and phosphorylation of STAT3 and STAT5B to promote cell survival and proliferation (Mohankumar et al. 2008).
de Bock et al. in 2018 demonstrated that HOXA9 functions as an oncogene in T-ALL and provides a platform for the JAK/STAT pathway activation and drive leukemia development. Transcriptional regulation of STAT5 by HOXA9 was further confirmed by RNA-seq, chromatin immunoprecipitation sequencing (ChIP-seq), and assay for transposase-accessible chromatin sequencing (ATAC-seq). Further, the study found PIM1 kinase (proto-oncogene serine/threonine-protein kinase) as the target gene of both HOXA9 and STAT5 (de Bock et al. 2018). In esophageal squamous cell carcinoma (ESCC), the interplay of HOX proteins and the NOTCH signaling pathway is established. HOXA9/myeloid ecotropic viral insertion site 1 (MEIS1) complex suppresses NOTCH transcription factor, mastermind-like proteins (MAML1), and hence, the complex is considered as a negative regulator of the NOTCH pathway. The downregulation of MEIS1 in ESCC enhances the NOTCH pathway to drive invasion and cancer progression (Abbaszadegan and Moghbeli 2018).
In AML, overexpressed HOXA10 functions as an autocrine stimulator to increase the cell population of myeloid progenitor cells via the transforming growth factor-beta (TGF-β) signaling pathway (Shah et al. 2011). The binding of TGF-β ligands and phosphorylation of receptors, followed by the activation of small mothers against decapentaplegic SMAD) proteins, trigger this pathway (Syed 2016). HOXA10 protein upregulates the transcription of TGFβ2 by interacting with tandem cis-elements in its promoter region and facilitate the expansion of hematopoietic stem cells and progenitor cells in AML (Shah et al. 2011).
Epithelial–mesenchymal transition (EMT), migration, and invasion
Epithelial–mesenchymal transition involves loss of epithelial cell polarity, disruption of the cell to cell adhesion, remodeling of cytoskeleton, anchorage-independent growth, acquisition of mesenchymal phenotype, and gain of migratory and invasive properties (Kalluri and Weinberg 2009). EMT induces intrinsic signals in the cancer cells to confer tumor initiation potential and therapy resistance (Dongre and Weinberg 2019).
Upregulation of HOXB7 protein enhances EMT by increasing the expression of functional EMT proteins such as N-cadherin (CDH2) and vimentin in GC cells. The regulation of two critical members of MAPK pathways (ERK, p38α) by HOXB7 at their phosphorylation level contributes to the enhanced migratory and invasive properties of cancer cells. HOXB7 regulates the invasion of GC cells via AKT pathway members. The mechanism by which HOXB7 regulates phosphatase and tensin homolog (PTEN), PI3K, MKK3/6, and RTKs needs further exploration (He et al. 2017). Additionally, HOXB5 functions as an important regulator of Wnt/β-catenin signaling in GC. The upregulation of HOXB5 in GC tissues significantly correlates with β-catenin (CTNNB1) gene expression. HOXB5 binds to the promoter region of CTNNB1 and thus functions as a transcriptional activator. Using the overexpression and knockdown models, the study confirmed that HOXB5 is involved in subsequent transcriptional activation of CTNNB1 and downstream target genes such as CCND1 and c-Myc, leading to the increased invasion and migration of GC cells (Hong et al. 2015).
The TGF-β pathway has been recognized as one of the most potent inducers of EMT (Xu et al. 2009). In GC, high expression of HOXD9 induces proliferation, migration, and invasion by controlling TGFβ1 and TGFβ2 expression (Xiong et al. 2020). Also, the overexpression of HOXA13 has been associated with proliferation, migration, and invasion in GC. Besides, HOXA13 upregulates the expression of CDH2 and VIM and downregulates the expression of CDH1 and hence plays a critical role in EMT. RNA-Seq analysis revealed that HOXA13 promotes GC progression by activation of ERK1/2. Hence, HOXA13 could thus be a novel target for anticancer therapy in GC (Qin et al. 2019).
Some HOX factors are found to be involved in inhibiting EMT and promoting mesenchymal–epithelial transition (MET) in GC. Interestingly, HOXB9 in GC cells upregulates the CDH1 expression by downregulating the N-cadherin and Snail (SNAI1) protein expression. Ectopic HOXB9 could induce apoptosis by inhibiting AKT signaling pathway and blocking the expression of p-AKT, p-glycogen synthase kinase-beta (p-GSK3β), and NF-κB (Zhang et al. 2019). It was proposed that the interaction of HOXB9-PBX1 (Pre-B cell leukemia transcription factor 1) was responsible for GC growth. When HOXB9 protein gets uncoupled from oncoprotein PBX1, it could suppress p-AKT and NF-κB-dependent SNAI1 expression to induce cell death and MET (Zhang et al. 2019). However, HOXB9 induces SNAI1 and SNAI2 (characteristic markers of EMT) and promotes invasion in oral squamous cell carcinoma (OSCC) cell lines by upregulating TGFβ1/SMAD genes (Xue et al. 2017). HOXB9 functions as a transcriptional activator and induces TGFβ1 in hepatocellular carcinoma (HCC) cells, enhancing their migratory and invasive potential (Sha et al. 2015).
In head and neck squamous cell carcinoma (HNSCC), HOXB5 functions as an oncogene, which binds directly to the promoter region of EGFR and regulates the AKT/Wnt/β-catenin synergistic signaling axis to enhance proliferation, cell migration, invasion, and EMT (Lee et al. 2020). In BC, HOXB5 is involved in the transcriptional activation of EGFR and its downstream targets to facilitate BC invasion (Lee et al. 2018). In BC and non-small cell lung cancer (NSCLC), knockdown of HOXB5 significantly suppressed the β-catenin protein and its downstream targets c-Myc and CCND1, thereby inhibiting cell proliferation, migration, invasion, and EMT of the cancer cells (Zhang et al. 2018a; Zhang et al. 2020). The anchorage-independent growth, proliferation, and migration property of NSCLC are also induced by HOXC8 through the transcriptional activation of TGFβ1. Further, the overexpression of HOXC8 results in chemoresistance to cisplatin. Targeting HOXC8 is thus a suitable therapeutic approach for the chemo-sensitization of NSCLC cells (Liu et al. 2018).
Overexpressed HOXB5 induces EMT in pancreatic cancer (PC) via the Wnt/β-catenin signaling pathway. HOXB5 inherently activates the phosphorylation of GSK-3β at Ser-9 position and facilitates the nuclear accumulation of β-catenin. HOXB5 protein upregulates the expression of mesenchymal markers such as VIM, SNAI1, and SNAI2 and downregulates the expression of CDH1, to enhance the mesenchymal transition, motility, and migratory ability of cancer cells (Gao et al. 2020). Similarly, HOXB7 activates ERK1/2 in PC to activate the downstream genes and induce the motility and invasion of cancer cells (Tsuboi et al. 2017). IHC analysis showed the localization of HOXB7 in the cell protrusions of migrating PC cells. HOXB7 could induce the phosphorylation of ERK1/2 and thereby promotes the cell protrusions to enhance the invasive potential. The invasiveness is also due to the HOXB7/ERK1/2-mediated stimulation of c-Jun N-terminal kinases (JNK) and phosphorylation of small heat shock protein (sHSP27) (Tsuboi et al. 2017). In addition to this, overexpressed HOXA10 also promotes invasiveness of PC cells via the TGFβ2-p38 MAPK pathway. It upregulates the level of TGFβ2 and MMP3 and activates p38 in PC cells to promote migration and invasion (Cui et al. 2014).
The cleavage of extracellular matrix (ECM), detachment of cancer cells from the basement membrane, and anchorage-independent growth followed by their migration and invasion are mainly due to the activity of several important proteases. In retinoblastoma (RBM), HOXB5 significantly contributes to the migration and invasion of cancer cells by stimulating the activation of the ERK1/2 pathway and subsequent production of the proteases MMP-3 and MMP-13 (Xu et al. 2018). Studies have demonstrated that aberrantly expressed HOX genes confer a growth advantage in glioblastoma (GBM). For example, HOXC10 promotes EMT by activating the PI3K pathway and inducing the expression of p-PI3K and p-AKT (Guan et al. 2019). Further, the HOXB2-induced PI3K pathway has been considered a potential therapeutic target in GBM. The knockdown of HOXB2 in GBM cell lines resulted in a reduction of proliferation and invasion via the PI3K/AKT pathway. The involvement of the PI3K pathway was confirmed by the increased levels of p-PI3K and p-AKT upon HOXB2 overexpression (Li et al. 2020c). It has also been shown that the overexpression of HOXC9 contributes to tumorigenicity in GBM. It suppresses the autophagy pathway by transcriptionally inhibiting its downstream target gene, death-associated protein kinase 1 (DAPK1), resulting in the inhibition of the DAPK1/Beclin pathway (Xuan et al. 2016). Higher levels of HOXA13 expression promote migration, invasion, and EMT of GBM cells by the activation of Wnt/β-catenin and TGF-β pathways. The small interfering RNA (siRNA)-mediated HOXA13 silencing prevents the nuclear localization of β-catenin, correlating significantly with decreased migration and invasion. Elevated levels of HOXA13 induce EMT with the stabilization of p-R-SMAD in the nucleus and activation of the TGF-β pathway (Duan et al. 2015). In another study, it was shown that the elevated HOXC6 in GBM significantly induces the expression of MAPK pathway genes including c-Myc, c-Jun, TP53, JNK, ERK, and P38 genes, promoting cell proliferation and migration (Yang et al. 2019b).
HOXD10 is downregulated in both cholangiocellular carcinoma (CCC) and CRC. Decreased HOXD10 levels mediate the activation of Ras homolog gene family, member C (RHOC), and MMPs and thus confer prolonged invasiveness and decreased apoptosis. The overexpression of HOXD10 deactivates RHOC/AKT/MAPK pathway to constrain cell migration and invasion and promote apoptosis (Yang et al. 2015; Yuan et al. 2019). The downregulation of HOXD10 is also shown to be associated with cell proliferation, cell cycle arrest, colony formation, migration, invasion, and apoptosis of HCC cells through the activation of the ERK signaling pathway (Guo et al. 2017b). Similarly, HOXA5 is underexpressed in both the CC tissues and cell lines, and its overexpression inhibits tumor progression by regulating AKT/p27 activation (Wang et al. 2019c).
Evasion of apoptosis
Following cancer cell invasion events such as intravasation, survival of tumor cells in the circulation, and resistance to cell death by immune surveillance evasion precede distant metastasis (Chiang et al. 2016). Aberrant regulation of apoptosis noted in cancer cells is due to altered homeostasis to thrive in the cellular environment (Jan and Chaudhry 2019).
Over the past decades, researchers have been extensively studying the role of HOX proteins in the regulation of key molecules of the apoptosis pathway in different cancers. They either enhance the activation of the caspase family of proteins or induce the transcription of pro-apoptotic/anti-apoptotic proteins, thereby activating two distinct and converging pathways (intrinsic and extrinsic) of apoptosis.
Aberrantly expressed HOX proteins have an inherent ability to activate the transcription of the anti-apoptotic gene B cell lymphoma 2 (BCL2) in different cancer types. In HCC, overexpressed HOXB5 inhibits apoptosis by increasing the protein levels of BCL-2 and decreasing the pro-apoptotic proteins Cyt c, BAX, and caspase-3. Further, it was demonstrated that HOXB5 could induce the expression of murine double minute 2 oncogene (MDM2) in hepatoma cells through ERK/MDM2 signaling (Su et al. 2020). In CC, elevated HOXC6 functions as a positive regulator of anti-apoptosis. However, HOXC6 knockdown results in the downregulation of BCL2 to activate the intrinsic apoptosis pathway (Wang et al. 2019b). The regulation of the apoptosis pathway by HOXC6 has also been reported in HNSCC. HOXC6 could bind to the TAAT motif of the BCL2 promoter region located − 420 bp upstream to TSS to transcriptionally upregulate its expression. The inherent anti-apoptotic property of HOXC6 is the reason for resistance to paclitaxel-induced apoptosis in HNSCC (Moon et al. 2012). A molecular link between HOXA9 and BCL2 has been established in AML. HOXA9 regulates BCL2 expression to facilitate the survival of myeloid progenitors and the immortalization of hematopoietic cells (Brumatti et al. 2013). The role of HOXA9 in influencing the tumor microenvironment has been demonstrated in ovarian cancer (OVC). The overexpressed HOXA9 activates TGFβ2 and promotes the transformation of normal peritoneal fibroblasts and normal adipose and bone marrow–derived mesenchymal stem cells (MSCs) into cancer-associated fibroblasts (CAFs) (Ko et al. 2012). In contrast to this observation, HOXA9 was significantly downregulated in cutaneous squamous cell carcinoma (CSCC). The downregulation of HOXA9 facilitates the activation of the p65-subunit of NF-κB (RELA) to activate the NF-κB pathway. The synergistic action of NF-κB, apoptosis, and autophagy pathways promotes CSCC progression. HOXA9-mediated activation of NF-κB signaling has promoted anti-apoptosis through BCL-extra large (BCL-XL) and repressed autophagy by regulating autophagy-related genes (ATG) such as ATG1, ATG3, and ATG12 (Han et al. 2019).
In GBM, the overactivation of HOXA9 is significantly correlated with the inactivation of the negative regulator of the PI3K pathway (PTEN), enhancing cell proliferation and suppressing apoptosis. However, HOXA9 inhibits apoptosis by diminishing tumor necrosis factor-related apoptosis-including ligand (TRAIL) in the GBM cell lines. Hence, reversing HOXA9 activation by PI3K inhibition may be of therapeutic significance in human GBM (Costa et al. 2010).
In BC, HOXA5 functions as a key molecule in the regulation of both p53-independent and p53-dependent apoptotic pathways (Raman et al. 2000). The study demonstrated that the p53 promoter region is enriched in HOXA5 binding sites, and hence, it regulates the endogenous synthesis of p53. Compromised HOXA5 function due to promoter methylation could lead to the loss of p53 expression in human BC (Raman et al. 2000). Further, HOXA5 could regulate the p53-independent extrinsic apoptosis pathway in BC and sensitize the cancer cells to TNF-α-induced apoptosis by regulating caspases 2 and 8 (CASP2/CASP8) (Chen et al. 2004). Likewise, HOXA5 enhances apoptosis by inducing caspase-3 (CASP3) activity in CC. The gain-of-function of HOXA5 in CC cell lines confirmed that HOXA5 could act as a tumor suppressor by inducing the expression of cell cycle inhibitor p27 and suppressing the phosphorylation of AKT (Wang et al. 2019c). Re-expression of HOXA5 could also induce apoptosis of osteosarcoma cells by inducing CASP3 activity and promoting the p53-dependent p38α MAPK pathway (Chen et al. 2019). Similarly, HOXD8 exerts a tumor suppressor role in CRC. The stable HOXD8 expression in CRC cell lines upregulated the expression of executioner caspase-6 and caspase-7 (CASP6/CASP7) and cleaved poly(ADP-ribose) polymerase (PARP) to induce apoptosis in CRC cells (Mansour and Senga 2017). In infant neuroblastoma, HOXC9 significantly triggers the intrinsic apoptosis pathway via releasing cytochrome c (Cyt c) into the cytosol. The activation of apoptosis pathway following increased expression levels of HOXC9 was significantly correlated with spontaneous regression in infant neuroblastoma (Kocak et al. 2013).
Metastasis
The dissemination of cancer cells to the surrounding tissues is facilitated by the activation of several key molecules (Fares et al. 2020). Deregulated HOX genes significantly induce distant metastasis by targeting key signaling pathways. A study on HCC demonstrated that HOXB7 is highly expressed in cells with high metastatic potential. Further, microarray data of 394 HCC tissues confirmed that HOXB7 modulates the biological functions of cancer cells via the activation of the MAPK/ERK pathway. Overexpressed HOXB7 binds to the promoter region of bFGF and induces bFGF secretion. Thus, HOXB7 functions as a molecular signature for predicting prognosis, survival, and recurrence (Wang et al. 2017). The elevated HOXB7 in lung adenocarcinoma (LAC) is linked to enhanced lymph node metastasis through the TGF-β/SMAD3 signaling pathway (Zhuang et al. 2015). The clinical relevance of the HOXB7-mediated TGF-β signaling pathway in BC demonstrated that HOXB7 activates TGFβ2 transcription by binding directly to its promoter region. Loss of HOXB7-induced TGFβ2 in BC cell lines has significantly reduced lung metastasis. The overexpression of HOXB7 promotes metastasis by inducing tumor-associated macrophage (TAM) recruitment (Liu et al. 2015a). While investigating the molecular mechanism of BC recurrence and distant metastasis after chemotherapy, researchers found that HOXC10 was responsible for suppressing apoptosis and inducing the NF-κB pathway in the BC cells treated with doxorubicin, paclitaxel, or carboplatin. Further, a unique and complex mechanism of HOXC10-mediated S-phase specific DNA damage repair was noted in the BC cells. Besides facilitating the recruitment of homologous recombination proteins (HR) to the site of DNA damage, HOXC10 reinitiates DNA replication by resolving stalled replication forks. Moreover, it could also resume RNA polymerase-II-dependent transcription by binding to the cyclin-dependent kinase 7 (CDK7). Hence, blocking the expression of HOXC10 in BC could be a useful therapeutic strategy (Sadik et al. 2016).
Researchers have determined the role of HOXB8 in CRC in promoting EMT via STAT-3 activation. The induction of EMT was characterized by the presence of mesenchymal markers such as VIM, CDH2, TWIST, ZEB1, and ZEB2. Interestingly, HOXB8 not only promotes lung metastasis but also enhances lymphatic metastasis in CRC (Wang et al. 2019a). Also, HOXB8 contributes to osteosarcoma progression and metastasis by activating the Wnt/β-catenin signaling pathway (Guo et al. 2019).
Decreased intracellular zinc concentration is the common characteristic of prostate malignancy (Huang et al. 2006). HOXB13 promotes metastasis of PCa by decreasing intracellular zinc through the NF-κB pathway. The activation of the NF-κB pathway may be due to the HOXB13-regulated nuclear localization of p65 transcription factors by reducing the levels of the NF-κB inhibitor, IκBα (Kim et al. 2014). HOXA13 and HOXC10 elevation significantly activate the MAPK pathway to induce proliferation and metastasis in GC (Guo et al. 2017a; Qin et al. 2019). However, the migration and invasion were due to the HOXA13-mediated activation of ERK1/2 and HOXC10-mediated upregulation of MAPK signaling–related genes (Guo et al. 2017a; Qin et al. 2019).
Meanwhile, HOXD3 expression is associated with an increase in the anchorage-independent growth, proliferation, and migration of NSCLC. Elevated HOXD3 reduces the expression of TGF-β-independent tumor-suppressing genes (desmoglein, desmoplakin, and plakoglobin) and upregulates the expression of TGF-β induced tumor-related genes such as MMP2, syndecan-1 (SDC1), and CD44. Thus, HOXD3 promotes the invasive and metastatic potential of cancer cells through the TGF-β-dependent and TGF-β-independent pathways (Miyazaki et al. 2002). Further, detailed information regarding the metastatic progression regulated by HOX proteins has been summarized in Table 3.
Table 3.
Contribution of HOX genes in cancer invasion and metastasis
| Cancer | Deregulated HOX genes | Signaling pathway | Targets | Role | References |
|---|---|---|---|---|---|
| Gastric cancer | ↑ HOXC10 | ↑ NF-κB | ATM | Promotes cell migration and invasion; significantly associated with TNM stage, lymph node metastasis, and distant metastasis | Yao et al. 2018 |
| Cutaneous squamous cell carcinoma | ↑ HOXB7 | ↑ Wnt/β-catenin | CTNNB1 | Enhances cell viability, migration, invasion, and metastasis and inhibits apoptosis | Gao and Chen 2018 |
| Osteosarcoma | ↑ HOXB8 | ↑ Wnt/β-catenin | CTNNB1, c-Myc, CCND1 | Promotes proliferation, migration, invasion, and metastasis | Guo et al. 2019 |
| Breast cancer | ↑ HOXB7 | ↑ TGF-β | TGFβ2 | Induces metastasis via TAM recruitment; promotes tumor progression in a cell-autonomous and non-cell-autonomous manner; and enhances migration and invasion | Liu et al. 2015 |
| ↑ HOXC10 | ↑NF-κB | - | Significantly correlated with distant metastasis after chemotherapy | Sadik et al. 2016 | |
| Hepatocellular carcinoma | ↑ HOXB5 | ↑ TGF-β-induced PI3K/AKT | - | Promotes cell migration and metastasis | (Sun et al. 2018) |
| Lung adenocarcinoma | ↑ HOXB7 | ↑TGF-β/SMAD3 | VEGFA, MMP2, SMAD3 | Significantly associated with lymph node metastasis | Zhuang et al. 2015 |
| Prostate cancer | ↑ HOXA1 | ↑ERK1/2 and AKT | ERK1/2, AKT | Promotes cell proliferation, invasion, and metastasis | Wang et al. 2015 |
| ↓ HOXD13 | ↑ TGF-β/BMP4/SMAD1 | SMAD1 | Promotes EMT and metastasis | Xu et al. 2021 | |
| Lung cancer | ↓ HOXA4 | ↑ Wnt/β-catenin | GSK3β; β-catenin, cyclin D1, c-Myc, and survivin | Promotes proliferation, migration, invasion, and lymph node metastasis and inhibits cell cycle arrest | Cheng et al. 2018 |
NOTE: TAM, tumor-associated macrophage; ATM, ataxia telangiectasia mutated; ↑, upregulated; ↓, downregulated
Angiogenesis
The metastasis of cancer cells depends mainly on the adequate supply of oxygen which can be achieved by the formation of new blood vessels (Nishida et al. 2006). HOX proteins act as an activator of angiogenic factors to bring out the aggressive phenotype of cancer cells. In BC, Carè et al. (2001) demonstrated the mechanism of stimulation of tumor invasion and neo-angiogenesis by the elevated HOXB7 levels. HOXB7 stimulates angiogenic response by inducing the important pro-angiogenic factors such as bFGF, vascular endothelial growth factor (VEGF), melanoma growth-stimulatory activity/growth-related oncogene alpha (MGSA/GRO-α), interleukin-8 (IL-8), and angiopoietin-2 (Ang-2) (Carè et al. 2001). Overexpressed HOXB9 has been involved in the stimulation of angiogenesis in high-grade BC. The pro-angiogenic response was confirmed by the induction of the expression of angiogenic molecules such as VEGF, bFGF, IL-8, and angiopoietin-like protein 2 (ANGPTL-2) upon HOXB9 overexpression. There are multiple HOX-binding sites located in these factors, indicating their activation by HOXB9. Thus, HOXB9 enriches the tumor microenvironment with these angiogenic molecules, to facilitate vascularization of tumors and distant metastasis. In addition to this, HOXB9 overexpression stimulated EMT via the generation of spindle-shaped mesenchymal cells, detachment of cells from the matrix, and formation of actin filaments. The induction of EMT was correlated with the loss of CDH1 and gain of CDH2, VIM, SNAI1, and TWIST markers (Hayashida et al. 2010).
In multiple myeloma, elevated HOXB7 expression contributed to angiogenesis by stimulating the expression of angiogenic factors. The upregulation of those angiogenic stimulators such as VEGFA, FGF2, MMP2, WNT5a, and platelet-derived growth factor subunit A (PDGFA) implicated in the blood vessel formation (Storti et al. 2011). In glioma, the vascular tube formation was enhanced by HOXC10 elevation. HOXC10 enhances the transcription of VEGFA by binding to its promoter region. Increased transcription and expression were followed by protein arginine methyltransferase 5 (PRMT5) and WD repeat domain 5 (WDR5)-mediated post-translational modification (Tan et al. 2018). Moreover, studies are required to better understand the role of HOX proteins in the regulation of angiogenesis in different types of cancer.
Clinical significance of HOX genes in cancer
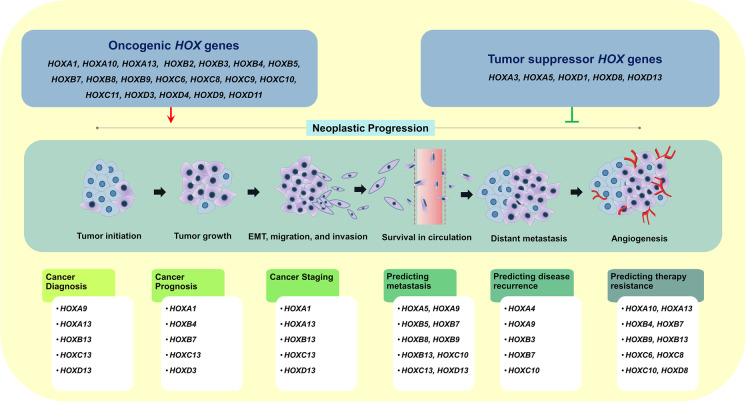
Cancer is the major cause of death globally with the majority of patients present with advanced stages of malignancy due to the absence of reliable prognostic markers. Identifying the specific oncogenic molecule that induces metastatic progression could help clinicians to diagnose cancer, stratify the stages, and suggest a suitable treatment regimen for better clinical outcomes. Understanding the role of HOX genes as a potential biomarker in cancer and exploring the utility of HOX-associated targets in various therapies may alleviate the cancer burden and reduce mortality rates (Fig. 4). The clinical significance of HOX genes in different cancer types has been summarized in Table 4.
Fig. 4.
Oncogenic and tumor suppressor HOX genes in the neoplastic progression: their biological significance in cancer diagnosis, prognosis, and therapeutic response
Table 4.
Biological role and clinical implications of HOX genes in cancer
| Deregulated HOX genes | Target | Cancer | Clinical Utility | References | |
|---|---|---|---|---|---|
| HOX gene as a prognostic marker | |||||
| HOXA5↓ | CDKN1A | Non-small cell lung cancer | Positively correlated with TNM staging | Zhang et al. 2015 | |
| HOXA5, HOXA9↑ | - | Acute myeloid leukemia | Decreases the remission rate | Zhao et al. 2015 | |
| HOXA10↑ | MMP3 | Bladder cancer | Positively associated with pathological grade and clinical stage | Liu et al. 2019 | |
| HOXA13↑ | - | Lung squamous cancer | Positively correlated with TNM stage | Zhang et al. 2017 | |
| HOXA13↑ | - | Bladder cancer | Positively correlated with lymphatic metastasis, pathological grade, and TNM stage | Hu et al. 2017 | |
| HOXB9↑ | - | Laryngeal squamous cell carcinoma | Positively correlated with histological grade | Sun et al. 2017 | |
| HOXC6↑ | Ki67 | Nasopharyngeal carcinoma | Significantly associated with tumor stage and advanced tumor status | Chang et al. 2017 | |
| HOXC9↑ | - | Colorectal cancer | Positively correlated with tumor stage, distant metastasis, and venous invasion | Hu et al. 2019 | |
| HOXC9↑ | - | Gastric cancer | Positively correlated with tumor size, lymphatic invasion, and lymph node metastasis | Zhao et al. 2020 | |
| HOXC10↑ | - | Thyroid cancer | Positively correlated with advanced and poor pathological stage | Feng et al. 2015 | |
| HOXC10↑ | - | Glioma | Positively associated high grading; induces immunosuppressive genes | Li et al. 2018 | |
| Role of HOX genes in therapy resistance | |||||
| HOXA5↑ | - | Acute myeloid leukemia | Provides resistance to cytarabine | Li et al. 2015 | |
| HOXA9↑ | BCL2 | Glioblastoma | Confers resistance to temozolomide | Pojo et al. 2015 | |
| HOXA10↑ | PTEN | Colorectal cancer | Provides resistance to 5-Fluorouracil | Yuan et al. 2018 | |
| HOXA11↓ | - | Glioblastoma | Confers resistance to radiation and temozolomidetherapy | Se et al. 2017 | |
| HOXA13↑ | - | Hepatocellular carcinoma | Provides resistance to sorafenib | Quagliata et al. 2018 | |
| HOXA13↑ | MRP1, DHRS2 | Gastric cancer | Provides resistance to 5-Fluorouracil | Han et al. 2018 | |
| HOXB cluster genes↑ | - | Breast cancer | Provides tamoxifen resistancce | Yang et al. 2018 | |
| HOXB13↑ | ERα, IL-6 | Breast cancer | Promotes tamoxifen resistancce | Shah et al. 2013 | |
| HOXC6↑ | BCL2 | Head and neck squamous cell carcinoma | Confers paclitaxel therapy resistance | Moon et al. 2012 | |
| HOXC8↑ | TGF-β1 | Non-small cell lung cancer | Provides cisplatin resistance | Liu et al. 2018 | |
| HOXC10↓ | - | Breast cancer | Confers resistance to aromatase inhibitors | Pathiraja et al. 2014 | |
| HOXD8↑ | - | Epithelial ovarian cancer | Promotes cisplatin therapy resistance | Sun et al. 2018 | |
| HOX gene as a biomarker for predicting disease recurrence | |||||
| HOXC10↑ | - | Gastric cancer | Positively associated with hepatic and peritoneal recurrence | Miwa et al. 2019 | |
| HOXA9↓ | - | Non-small cell lung cancer | Provides disease recurrence | Hwang et al. 2015 | |
| HOXA4/HOXB3↑ | - | High-grade serous ovarian carcinoma | Biomarker for recurrence after primary cytoreductive surgery and first-line adjuvant chemotherapy | Miller et al. 2018 | |
| HOXB7↑ | bFGF | Hepatocellular carcinoma | Provides disease recurrence | Wang et al. 2017 | |
NOTE: DHRS2, dehydrogenase reductase 2; CST1, cystatin SN; ↑, upregulated; ↓, downregulated
HOX gene as a diagnostic marker
To reduce the mortality of cancer and identify suitable therapy, accurate diagnosis is crucial. The existing literature supports the utility of HOX gene as a diagnostic marker. Sarno et al. (2020) established that the paralogous HOX13 group of genes (HOXA13, HOXB13, HOXC13, and HOXD13) contributes to the progression of BC and aid in histological subtyping (Sarno et al. 2020). In childhood ALL, the overexpression of HOXA9 and MEIS1 showed a significant correlation with the ALL subtypes (Adamaki et al. 2015). The overexpression of HOXA13 was correlated with the grade, size, microvascular invasion, capsular spread in the nodes, alpha-fetoprotein (AFP) level, and tumor-node-metastasis (TNM) staging in HCC. HOXA13-expressing tumors showed shorter overall survival (OS) and disease-free survival (DFS) than those with HOXA13-negative tumors, indicating its role as a prognostic marker in HCC (Pan et al. 2014). In another study, the role of HOXA13 playing a crucial role in four glioma cell lines was established. Increased cell proliferation and invasion were associated with the activation of Wnt and TGF-β-induced EMT pathways indicating its role as a diagnostic marker (Duan et al. 2015).
HOX gene as a prognostic marker
The transcriptome profiling and pathway analysis of HOXA1 expressing melanoma cells showed increased TGF-β signaling. The potential role of HOXA1 in repressing genes required for melanocyte differentiation and activating pro-invasive genes via the TGF-β signaling pathway was distinctly demonstrated. The expression of HOXA1 could stratify melanoma patients based on metastatic potential and hence function as a potential prognostic marker (Wardwell-Ozgo et al. 2014). Morgan et al. (2016) demonstrated the oncogenic nature of HOXB4 in mouse models of mesothelioma. The overexpression of HOXB4 was positively correlated with shorter OS (Morgan et al. 2016). Higher expression of HOXB7 was detected in HCC tissues, which was found to be associated with a poor prognosis. The tumor-promoting and metastatic ability of HOXB7 were validated in a xenograft tumor model and bioluminescence imaging, respectively. Interestingly, the stem cell markers such as epithelial cell adhesion molecule (EPCAM) and NANOG1 (Nanog) were found to be upregulated upon HOXB7 overactivation (Huan et al. 2017).
An extensive bioinformatic analysis in BC revealed a positive correlation between HOXC13 expression and lymph node metastasis, indicating a poor prognosis (Li et al. 2020a). Shaoqiang et al. (2013) revealed that BC patients with increased expression of HOXD3 were significantly correlated with higher histopathological grade and shorter survival time (Shaoqiang et al. 2013). Taken together, these data reflect the possibility of using the HOX cluster as a potential prognostic marker for cancer progression.
HOX gene as a biomarker for predicting metastasis
Cancer progression is mainly due to the ability of the neoplastic cells to metastasize to the distant sites of the body often via the bloodstream or lymphatics. The therapeutic goal should be the prevention of metastasis in advanced stages of malignancy. HOX-mediated invasion and metastasis in different cancer types have been reviewed recently (Paço et al. 2020a). Through the genome-wide transcriptomic and pathway analyses, Wang et al. (2015) showed that HOXA5 could suppress metastasis in NSCLC by interfering with the cytoskeleton remodeling mechanism (Wang et al. 2015a). Increased HOXA9 expression in CRC tissues was shown to be associated with lymph node metastasis (Watanabe et al. 2018).
In another study, HOXB5 was shown to transactivate metastatic-related genes such as C-X-C motif chemokine receptor 4 (CXCR4) and integrin subunit beta 3 (ITGB3) promoter and propagate distant metastasis in CRC (Feng et al. 2021). The role of the HOXB7 in progression and metastasis in LAC confirms its prognostic potential and a suitable target for therapy (Yuan et al. 2014). Also, the metastatic ability of HOXB7 in HCC cells in vivo was confirmed with bioluminescence imaging. The study demonstrated how HOXB7 could induce stem-like properties and facilitate EMT in HCC (Huan et al. 2017). In CRC, HOXB8 functioned as an oncogene whose expression was regulated by the MYC-super enhancer complex. HOXB8 has the potential ability to interact with metastatic regulator BACH1 (BTB domain and CNC homolog 1) and thereby potentiates the metastatic ability and invasive nature of cancer cells (Ying et al. 2020). For the first time, researchers demonstrated the ability of HOXB9 in promoting the metastatic nature of HCC cells. They found that the TGF-β pathway is the crucial signaling cascade in promoting EMT in HCC cells (Sha et al. 2015).
A study on OVC showed the critical role of HOXC10 in metastasis. HOXC10 could regulate the transcription of the EMT-related gene SNAI2 and could induce metastatic ability (Peng et al. 2020). By inducing the metastasis-related genes such as 3-phosphoinositide-dependent protein kinase 1 (PDPK1) and vasodilator-stimulated phosphoprotein (VASP), HOXC10 promotes metastasis in HCC (Dang et al. 2020). The overexpression of HOXC13 was observed in metastatic melanoma tissues when compared to primary melanoma tissues, implying its role as a metastatic inducer (Cantile et al. 2012).
Studies have hypothesized that targeting metastatic cancer and its related pathways might be a necessary approach for cancer therapy. As per the recent findings in PCa, HOXD13 is involved in inhibiting bone morphogenetic protein 4 (BMP4)/SMAD1-induced EMT and thus functions as a negative regulator of metastasis (Xu et al. 2021). Another study on PCa implicated HOXB13 as a metastasis promoter gene that could induce metastasis via regulating mitotic protein kinases (Yao et al. 2019). Following radical prostatectomy surgery, tumors with high expression of HOXB13 and a low expression of its binding partner MEIS1/2 were significantly associated with an increased propensity for metastasis (Weiner et al. 2020). These findings suggested the use of HOX genes as a predictive marker for cancer metastasis.
HOX gene as a biomarker for predicting disease recurrence
Despite significant advancements in oncopathology practice, the recurrence rate of many cancer types is high. Studies establishing the role of HOX genes as predictive biomarkers for cancer relapse are emerging. Among the notable ones, hypermethylated HOXA9 and its downregulation is an independent prognostic marker as it was found to be significantly associated with the disease recurrence in NSCLC (Hwang et al. 2015). Contrary to this, HOXA9 was shown to be overexpressed in patients of childhood leukemia, which showed a significant correlation with relapse (Adamaki et al. 2015).
High-grade serous ovarian cancer (HGSOC) patients who had a relapse following primary cytoreductive surgery and first-line adjuvant chemotherapy had HOXA4/HOXB3 overexpression (Miller et al. 2018). Higher expression of HOXB7 mRNA had been noted in patients diagnosed with HCC. HOXB7 promotes neoplastic progression via the bFGF-induced activation of the MAPK/ERK pathway implying a poor prognosis. The expression of HOXB7 may be a promising candidate for recurrence prediction in HCC (Wang et al. 2017). In GC, HOXC10 influences the malignant phenotype of GC cell lines, and its overexpression is correlated with hepatic and peritoneal recurrence (Miwa et al. 2019). The HOX gene expression profiling may prove to be a predictive biomarker of recurrence in various cancer subtypes.
HOX genes in therapeutic resistance
Differential expression of HOX genes in various cell types and their ability to regulate a multitude of cellular functions in cancer has been extensively studied. The relapse and recurrence secondary to therapeutic resistance may be attributed to both intrinsic (transcriptomic, proteomic, epigenetic, and genetic) and extrinsic factors (pH, hypoxia, paracrine signaling, and tumor–host interactions) (Mansoori et al. 2017). Several HOX genes have been shown to render therapeutic resistance in various cancer subtypes. The downregulation of HOXA13 enhances the effectiveness of cisplatin chemotherapy in ESCC by inhibiting the expression of EMT markers such as SNAI1 (Shi et al. 2018). However, HOXB7 has been shown to mediate cisplatin resistance in ESCC. Disrupting the HOXB7-PBX (pre-B cell leukemia transcription factor) complex by an inhibitory synthetic peptide HXR9 enhanced chemosensitivity to cisplatin proving HOXB7 to be an effective target in ESCC (Zhou et al. 2020). Likewise, HOXB4 has been shown to render multidrug resistance of human myelogenous leukemia (HML) K562/ADM cells through the prolonged activation of the PI3K/AKT pathway. Knockdown of HOXB4 could sensitize the HML cell lines by downregulating the expression of P-gp (P-glycoprotein), MRP (multidrug resistance-associated protein), and BCRP (breast cancer resistance protein) (Wang et al. 2016). Furthermore, HOXB7 and HOXB13 render BC cells resistant to tamoxifen through the stimulation of the EGFR and mTOR pathways, respectively (Jin et al. 2012; Shah et al. 2013). Another study on BC delineated the fact that overexpressed HOXD3 plays a crucial role in drug resistance and stemness via integrin β3-mediated Wnt/β-catenin signaling (Zhang et al. 2018b). Contarelli et al. (2020) reviewed that, among the genes in the HOXB cluster, HOXB9 had the capability in providing resistance to anti-angiogenic therapies in different cancer phenotypes (Contarelli et al. 2020). Moon et al. (2012) highlighted the role of HOXC6 in inducing the resistance to paclitaxel in HNSCC. The study demonstrated that HOXC6 modulates the expression of the anti-apoptotic gene BCL2 to inhibit the normal apoptotic pathway (Moon et al. 2012). In NSCLC, HOXC8 has been considered as the promising therapeutic target to sensitize the cancer cells to cisplatin. HOXC8 functions as a transcriptional activator of the TGFβ1 gene, and overexpression exaggerates normal TGF-β signaling, which could eventually promote the proliferation, migration, and growth of neoplastic cells (Liu et al. 2018). Sun et.al (2018) compared the expression levels of transcription factors that were associated with metastasis and cisplatin resistance in epithelial ovarian cancer. HOXD8 was found to be upregulated in SKOV3-DDP cisplatin-resistant cells when compared with SKOV3 cisplatin-sensitive cells. They validated this observation in clinical samples which showed the overexpression of HOXD8 in cisplatin resistance tissues when compared to primary malignant tumors, implying its potential role in cisplatin resistance (Sun et al. 2018b). Targeting the HOX genes or manipulating the expression of deregulated HOX genes is an imperative strategy for chemo-sensitization. However, HOX gene-mediated therapeutic resistance in different cancer phenotypes is poorly understood, warranting further studies.
Conclusion and future perspectives
It has been well documented that HOX transcription factors play a crucial role in organogenesis and tissue homeostasis. They regulate a large number of targets that are particularly involved in key signaling pathways. In this review, we have highlighted the impact of deregulated HOX genes in cancer development and metastatic progression. Numerous molecular and clinical studies have demonstrated that any deregulation in the genes of HOX cluster would bring an imbalance in cellular pathways, which would eventually favor the cancer cells towards their metastatic progression. The integration of multiple signaling pathways regulated by HOX genes in cancer reflects their versatility in regulating diverse targets. Current literature supports that the genetic profiling of HOX gene clusters could be utilized as a clinical biomarker in early cancer detection. Hence, molecular techniques to determine the cancer stages are advantageous over the conventional method of cancer detection. Both in vitro and in vivo studies have proved that understanding the molecular mechanisms regulated by HOX genes and modulating aberrantly expressed HOX genes that provide resistance to therapies could be the most relevant strategy to sensitize the cancer cells for a better clinical outcome. Therefore, it would be an excellent opportunity for the clinical field to establish the targeted gene therapies against oncogenic molecules in cancer.
Even though the carcinogenic mechanism regulated by HOX genes is being extensively studied, studies are required to systematically understand the multifunctional role of HOX genes in each of the cancer types. Hence, we emphasize that the following research gaps could be filled to translate the findings into the clinical field.
The genetic and epigenetic alterations in the HOX cluster genes and their mode of regulation are not studied in a variety of cancer types. Since those key alterations are responsible for driving metastasis, research should be focused on the molecular characterization of HOX genes at their transcriptional level.
In addition to the solid tumors, the determination of HOX gene expression in body fluids such as lymph, serum, and saliva would allow clinicians for early cancer detection.
To target a particular oncogenic HOX gene in cancer, more comprehensive studies are needed to understand the crosstalk between multiple signaling pathways regulated by HOX genes in cancer at both transcriptomic and proteome levels before their clinical translation.
More than 75% of the HOX cluster-embedded ncRNAs are not yet functionally characterized. Since the functional interaction between ncRNAs and HOX genes is reported in some of the cancer types, research should focus on the characterization of the entire HOX cluster network.
Acknowledgements
We thank Wellcome Trust DBT India Alliance, Government of India (Grant No. IA/CPHI/18/1/503927); Joint CSIR-UGC NET Junior Research Fellowship, Government of India (File No. 09/1165(0011)/2020-EMR-I); ICMR SRF, Government of India (2019/4115/CMB/BMS); and Manipal School of Life Sciences, MAHE, Manipal, for infrastructure support.
Abbreviations
- ALL
Acute lymphoblastic leukemia
- AML
Acute myeloid leukemia
- BC
Breast cancer
- CAFs
Cancer-associated fibroblasts
- CC
Cervical cancer
- CCC
Cholangiocellular carcinoma
- CGH
Comparative genomic hybridization
- CRC
Colorectal cancer
- CSCC
Cutaneous squamous cell carcinoma
- DFS
Disease-free survival
- ECM
Extracellular matrix
- EMT
Epithelial–mesenchymal transition
- ESCC
Esophageal squamous cell carcinoma
- GBM
Glioblastoma
- GC
Gastric cancer
- HCC
Hepatocellular carcinoma
- HGSOC
High-grade serous ovarian cancer
- HNSCC
Head and neck squamous cell carcinoma
- HOX
Homeobox
- IHC
Immunohistochemistry
- LAC
Lung adenocarcinoma
- lncRNA
Long non-coding RNA
- MET
Mesenchymal–epithelial transition
- miRNA
MicroRNA
- MLL1
Mixed-lineage leukemia 1
- ncRNAs
Non-coding RNAs
- NSCLC
Non-small cell lung cancer
- OS
Overall survival
- OSCC
Oral squamous cell carcinoma
- OVC
Ovarian cancer
- PC
Pancreatic cancer
- PCa
Prostate cancer
- RBM
Retinoblastoma
- T-ALL
T cell acute lymphoblastic leukemia
- TNM
Tumor-node-metastasis
- WGD
Whole-genome duplication
Author contribution
U. S. S. performed the literature search and wrote the manuscript; D. A. and U. S. S. designed the figures and tables. K. H., S. P. K., and R. R. edited the manuscript and did the critical revision. All the authors have read and approved the final manuscript.
Funding
Open access funding provided by Manipal Academy of Higher Education, Manipal. Wellcome Trust DBT India Alliance, Government of India (Grant No. IA/CPHI/18/1/503927).
Data availability
Not applicable.
Code availability
Not applicable.
Declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All the authors have read and approved the final manuscript.
Conflict of interest
The authors declare no competing interests.
Footnotes
Highlights
• HOX gene expression is regulated by genetic and epigenetic factors.
• HOX family of transcription factors regulate key cell signaling pathways in cancer.
• Deregulation of HOX genes is implicated in the neoplastic progression and metastasis.
• HOX genes are potential clinical biomarkers for diagnosis, prognosis, and therapy.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
3/2/2022
The original version of this paper was updated to add the missing compact agreement Open Access funding note.
References
- Abbasi AA. Diversification of four human HOX gene clusters by step-wise evolution rather than ancient whole-genome duplications. Dev Genes Evol Germany. 2015;225(6):353–357. doi: 10.1007/s00427-015-0518-z. [DOI] [PubMed] [Google Scholar]
- Abbaszadegan MR, Moghbeli M. Role of MAML1 and MEIS1 in esophageal squamous cell carcinoma depth of invasion. Pathol Oncol Res Switzerland. 2018;24(2):245–250. doi: 10.1007/s12253-017-0243-1. [DOI] [PubMed] [Google Scholar]
- Adamaki M, Lambrou GI, Athanasiadou A, Vlahopoulos S, Papavassiliou AG, Moschovi M. HOXA9 and MEIS1 gene overexpression in the diagnosis of childhood acute leukemias: significant correlation with relapse and overall survival. Leuk. Res. England. 2015;39(8):874–882. doi: 10.1016/j.leukres.2015.04.012. [DOI] [PubMed] [Google Scholar]
- Akbari MR, Anderson LN, Buchanan DD, Clendenning M, Jenkins MA, Win AK, et al. Germline HOXB13 p.Gly84Glu mutation and risk of colorectal cancer. Cancer Epidemiol. 2013;37(4):424–427. doi: 10.1016/j.canep.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado-Ruiz L, Martinez-Silva MG, Torres-Reyes LA, Pina-Sanchez P, Ortiz-Lazareno P, Bravo-Cuellar A, et al. HOXA9 is underexpressed in cervical cancer cells and its restoration decreases proliferation, migration and expression of epithelial-to-mesenchymal transition genes. Asian Pac. J. Cancer Prev. Thailand. 2016;17(3):1037–1047. doi: 10.7314/apjcp.2016.17.3.1037. [DOI] [PubMed] [Google Scholar]
- Angus-Hill ML, Elbert KM, Hidalgo J, Capecchi MR. T-cell factor 4 functions as a tumor suppressor whose disruption modulates colon cell proliferation and tumorigenesis. Proc Natl Acad Sci U S A. 2011;108(12):4914–4919. doi: 10.1073/pnas.1102300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apiou F, Flagiello D, Cillo C, Malfoy B, Poupon M-F, Dutrillaux B. Fine mapping of human HOX gene clusters. Sytogenetics Cell Genet. 1996;73(1-2):114–115. doi: 10.1159/000134320. [DOI] [PubMed] [Google Scholar]
- Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene England. 2006;25(57):7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- Beebe-Dimmer JL, Hathcock M, Yee C, Okoth LA, Ewing CM, Isaacs WB, et al. The HOXB13 G84E mutation is associated with an increased risk for prostate cancer and other malignancies. Cancer Epidemiol Biomarkers Prev. 2015;24(9):1366–1372. doi: 10.1158/1055-9965.EPI-15-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhan A, Mandal SS. LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856(1):151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatlekar S, Fields JZ, Boman BM. Role of HOX genes in stem cell differentiation and cancer. Stem Cells Int. 2018;2018:3569493. doi: 10.1155/2018/3569493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncinelli E, Mallamaci A. Homeobox genes in vertebrate gastrulation. Curr Opin. Genet. Dev. 1995;5(5):619–627. doi: 10.1016/0959-437x(95)80031-x. [DOI] [PubMed] [Google Scholar]
- Botti G, De CA, Di BM, Malzone MG, Collina F, Cantile M. Noncoding RNAs within the HOX gene network in tumor pathogenesis and progression. J Cell Physiol. 2018;234(1):395–413. doi: 10.1002/jcp.27036. [DOI] [PubMed] [Google Scholar]
- Brumatti G, Salmanidis M, Kok CH, Bilardi RA, Sandow JJ, Silke N, et al. HoxA9 regulated Bcl-2 expression mediates survival of myeloid progenitors and the severity of HoxA9-dependent leukemia. Oncotarget. 2013;4(11):1933–1947. doi: 10.18632/oncotarget.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantile M, Scognamiglio G, Anniciello A, Farina M, Gentilcore G, Santonastaso C, et al. Increased HOX C13 expression in metastatic melanoma progression. J Transl Med. 2012;10:91. doi: 10.1186/1479-5876-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carè A, Felicetti F, Meccia E, Bottero L, Parenza M, Stoppacciaro A, et al. HOXB7: a key factor for tumor-associated angiogenic switch. Cancer Res. 2001;61(17):6532–6539. [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S-L, Chan T-C, Chen T-J, Lee S-W, Lin L-C, Win KT. HOXC6 overexpression is associated with Ki-67 expression and poor survival in NPC patients. J Cancer. 2017;8(9):1647–1654. doi: 10.7150/jca.18893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chung S, Sukumar S. HOXA5-induced apoptosis in breast cancer cells is mediated by caspases 2 and 8. Mol Cell Biol. 2004;24(2):924–935. doi: 10.1128/MCB.24.2.924-935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Fan Y, Xu W, Chen J, Xu C, Wei X, et al. miR-10b inhibits apoptosis and promotes proliferation and invasion of endometrial cancer cells via targeting HOXB3. Cancer Biother Radiopharm. 2016;31(6):225–231. doi: 10.1089/cbr.2016.1998. [DOI] [PubMed] [Google Scholar]
- Chen Y-Q, Yang T-Q, Zhou B, Yang M-X, Feng H-J, Wang Y-L. HOXA5 overexpression promotes osteosarcoma cell apoptosis through the p53 and p38α MAPK pathway. Gene Netherlands. 2019;689:18–23. doi: 10.1016/j.gene.2018.11.081. [DOI] [PubMed] [Google Scholar]
- Cheng S, Qian F, Huang Q, Wei L, Fu Y, Du Y. HOXA4, down-regulated in lung cancer, inhibits the growth, motility and invasion of lung cancer cells. Cell Death Dis. 2018;9(5):465. doi: 10.1038/s41419-018-0497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang SPH, Cabrera RM, Segall JE. Tumor cell intravasation. Am J Physiol Cell Physiol. 2016;311(1):C1–14. doi: 10.1152/ajpcell.00238.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong GO, Jeon H-S, Han HS, Son JW, Lee YH, Hong DG, et al. Overexpression of microRNA-196b accelerates invasiveness of cancer cells in recurrent epithelial ovarian cancer through regulation of homeobox A9. Cancer Genomics Proteomics. 2017;14(2):137–141. doi: 10.21873/cgp.20026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cildir G, Low KC, Tergaonkar V. Noncanonical NF-κB signaling in health and disease. Trends Mol Med. 2016;22(5):414–429. doi: 10.1016/j.molmed.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Contarelli S, Fedele V, Melisi D. HOX genes family and cancer: a novel role for homeobox B9 in the resistance to anti-angiogenic therapies. Cancers (basel). 2020;12(11):3299. doi: 10.3390/cancers12113299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa BM, Smith JS, Chen Y, Chen J, Phillips HS, Aldape KD, et al. Reversing HOXA9 oncogene activation by PI3K inhibition: epigenetic mechanism and prognostic significance in human glioblastoma. Cancer Res. 2010;70(2):453–462. doi: 10.1158/0008-5472.CAN-09-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X-P, Qin C-K, Zhang Z-H, Su Z-X, Liu X, Wang S-K, et al. HOXA10 promotes cell invasion and MMP-3 expression via TGFβ2-mediated activation of the p38 MAPK pathway in pancreatic cancer cells. Dig Dis Sci. 2014;59(7):1442–1451. doi: 10.1007/s10620-014-3033-6. [DOI] [PubMed] [Google Scholar]
- Cui Y, Yan M, Zhang C, Xue J, Zhang Q, Ma S, et al. Comprehensive analysis of the HOXA gene family identifies HOXA13 as a novel oncogenic gene in kidney renal clear cell carcinoma. J Cancer Res Clin Oncol. 2020;146(8):1993–2006. doi: 10.1007/s00432-020-03259-x. [DOI] [PubMed] [Google Scholar]
- Cui Y, Zhang C, Li Y, Ma S, Cao W, Guan F. HOXD1 functions as a novel tumor suppressor in kidney renal clear cell carcinoma. Cell Biol Int. 2021;45(6):1246–1259. doi: 10.1002/cbin.11568. [DOI] [PubMed] [Google Scholar]
- Curradi M, Izzo A, Badaracco G, Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol Cell Biol. 2002;22(9):3157–3173. doi: 10.1128/MCB.22.9.3157-3173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Chen J, Feng W, Qiao C, Han W, Nie Y, et al. Interleukin 1β-mediated HOXC10 overexpression promotes hepatocellular carcinoma metastasis by upregulating PDPK1 and VASP. Theranostics. 2020;10(8):3833–3848. doi: 10.7150/thno.41712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Barros E, Lima Bueno R, Ramão A, Pinheiro DG, Alves CP, Kannen V, Jungbluth AA, et al. HOX genes: potential candidates for the progression of laryngeal squamous cell carcinoma. Tumour Biol J Int Soc Oncodev Biol Med Netherlands. 2016;37(11):15087–15096. doi: 10.1007/s13277-016-5356-8. [DOI] [PubMed] [Google Scholar]
- de Bock CE, Demeyer S, Degryse S, Verbeke D, Sweron B, Gielen O, et al. HOXA9 cooperates with activated JAK/STAT signaling to drive leukemia development. Cancer Discov. 2018;8(5):616–631. doi: 10.1158/2159-8290.CD-17-0583. [DOI] [PubMed] [Google Scholar]
- Derakhshani A, Rostami Z, Taefehshokr S, Safarpour H, Vaezi R. An overview of the oncogenic signaling pathways in different types of cancers. Cancer. 2020;26(8):1–24. [Google Scholar]
- Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20(2):69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- Duan R, Han L, Wang Q, Wei J, Chen L, Zhang J, et al. HOXA13 is a potential GBM diagnostic marker and promotes glioma invasion by activating the Wnt and TGF-β pathways. Oncotarget. 2015;6(29):27778–27793. doi: 10.18632/oncotarget.4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, et al. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366(2):141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct Target Ther. 2020;5(1):28. doi: 10.1038/s41392-020-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Li T, Liu Z, Shi Y, Peng Y. HOXC10 up-regulation contributes to human thyroid cancer and indicates poor survival outcome. Mol Biosyst. 2015;11(11):2946–2954. doi: 10.1039/c5mb00253b. [DOI] [PubMed] [Google Scholar]
- Feng W, Huang W, Chen J, Qiao C, Liu D, Ji X, et al. CXCL12-mediated HOXB5 overexpression facilitates colorectal cancer metastasis through transactivating CXCR4 and ITGB3. Theranostics. 2021;11(6):2612–2633. doi: 10.7150/thno.52199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrier DE, Holland PW. Ancient origin of the Hox gene cluster. Nat Rev Genet. 2001;2(1):33–38. doi: 10.1038/35047605. [DOI] [PubMed] [Google Scholar]
- Fu Z, Chen C, Zhou Q, Wang Y, Zhao Y, Zhao X, et al. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett Ireland. 2017;410:68–81. doi: 10.1016/j.canlet.2017.09.019. [DOI] [PubMed] [Google Scholar]
- Gan B-L, He R-Q, Zhang Y, Wei D-M, Hu X-H, Chen G. Downregulation of HOXA3 in lung adenocarcinoma and its relevant molecular mechanism analysed by RT-qPCR, TCGA and in silico analysis. Int J Oncol. 2018;53(4):1557–1579. doi: 10.3892/ijo.2018.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Chen H-Q. Specific knockdown of HOXB7 inhibits cutaneous squamous cell carcinoma cell migration and invasion while inducing apoptosis via the Wnt/β-catenin signaling pathway. Am J Physiol Cell Physiol. 2018;315(5):C675–C686. doi: 10.1152/ajpcell.00291.2017. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Fei X, Kong L, Tan X. HOXB5 promotes proliferation, migration, and invasion of pancreatic cancer cell through the activation of the GSK3β/β-catenin pathway. Anticancer Drugs. 2020;31(8):828–835. doi: 10.1097/CAD.0000000000000948. [DOI] [PubMed] [Google Scholar]
- Gaunt SJ. Hox cluster genes and collinearities throughout the tree of animal life. Int J Dev Biol. 2018;62(11–12):673–683. doi: 10.1387/ijdb.180162sg. [DOI] [PubMed] [Google Scholar]
- Giancotti FG. Deregulation of cell signaling in cancer. FEBS Lett. 2014;588(16):2558–2570. doi: 10.1016/j.febslet.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood. 2011;118(24):6247–6257. doi: 10.1182/blood-2011-07-328880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, He Y, Lv S, Hou X, Li L, Song J. Overexpression of HOXC10 promotes glioblastoma cell progression to a poor prognosis via the PI3K/AKT signalling pathway. J Drug Target. 2019;27(1):60–66. doi: 10.1080/1061186X.2018.1473408. [DOI] [PubMed] [Google Scholar]
- Guo C, Hou J, Ao S, Deng X, Lyu G. HOXC10 up-regulation promotes gastric cancer cell proliferation and metastasis through MAPK pathway. Chin J Cancer Res. 2017;29(6):572–580. doi: 10.21147/j.issn.1000-9604.2017.06.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Peng Y, Gao D, Zhang M, Yang W, Linghu E, et al. Silencing HOXD10 by promoter region hypermethylation activates ERK signaling in hepatocellular carcinoma. Clin Epigenetics. 2017;9:116. doi: 10.1186/s13148-017-0412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Zhang T, Dou D. Knockdown of HOXB8 inhibits tumor growth and metastasis by the inactivation of Wnt/β-catenin signaling pathway in osteosarcoma. Eur J Pharmacol Netherlands. 2019;854:22–27. doi: 10.1016/j.ejphar.2019.04.004. [DOI] [PubMed] [Google Scholar]
- Guo C, Ju Q-Q, Zhang C-X, Gong M, Li Z-L, Gao Y-Y. Overexpression of HOXA10 is associated with unfavorable prognosis of acute myeloid leukemia. BMC Cancer. 2020;20(1):586. doi: 10.1186/s12885-020-07088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y-J, Pan W-W, Liu S-B, Shen Z-F, Xu Y, Hu L-L. ERK/MAPK signalling pathway and tumorigenesis. Exp Ther Med. 2020;19(3):1997–2007. doi: 10.3892/etm.2020.8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Song C, Wang J, Tang H, Peng Z, Lu S. HOXA13 contributes to gastric carcinogenesis through DHRS2 interacting with MDM2 and confers 5-FU resistance by a p53-dependent pathway. Mol Carcinog. 2018;57(6):722–734. doi: 10.1002/mc.22793. [DOI] [PubMed] [Google Scholar]
- Han S, Li X, Liang X, Zhou L. HOXA9 transcriptionally promotes apoptosis and represses autophagy by targeting NF-κB in cutaneous squamous cell carcinoma. Cells. 2019;8(11):1360. doi: 10.3390/cells8111360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Haria D, Naora H. Homeobox gene deregulation: impact on the hallmarks of cancer. Cancer Hallm. 2013;1(2–3):67–76. doi: 10.1166/ch.2013.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashida T, Takahashi F, Chiba N, Brachtel E, Takahashi M, Godin-Heymann N, et al. HOXB9, a gene overexpressed in breast cancer, promotes tumorigenicity and lung metastasis. Proc Natl Acad Sci U S A. 2010;107(3):1100–1105. doi: 10.1073/pnas.0912710107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Liu Z, Xia Y, Xu J, Lv G, Wang L, et al. HOXB7 overexpression promotes cell proliferation and correlates with poor prognosis in gastric cancer patients by inducing expression of both AKT and MARKs. Oncotarget. 2017;8(1):1247–1261. doi: 10.18632/oncotarget.13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Chen Z-Y, Li Y, Yang Z-Q, Zeng F, Cui Y, et al. miR-10b suppresses cell invasion and metastasis through targeting HOXA3 regulated by FAK/YAP signaling pathway in clear-cell renal cell carcinoma. BMC Nephrol. 2019;20(1):127. doi: 10.1186/s12882-019-1322-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PWH. Evolution of homeobox genes. Wiley Interdiscip Rev Dev Biol. 2013;2(1):31–45. doi: 10.1002/wdev.78. [DOI] [PubMed] [Google Scholar]
- Hombría JC-G, Lovegrove B. Beyond homeosis-HOX function in morphogenesis and organogenesis. Differentiation. 2003;71(8):461–476. doi: 10.1046/j.1432-0436.2003.7108004.x. [DOI] [PubMed] [Google Scholar]
- Hong C-S, Jeong O, Piao Z, Guo C, Jung M-R, Choi C, et al. HOXB5 induces invasion and migration through direct transcriptional up-regulation of β-catenin in human gastric carcinoma. Biochem J. 2015;472(3):393–403. doi: 10.1042/BJ20150213. [DOI] [PubMed] [Google Scholar]
- Hou L, Xu B, Mohankumar KM, Goffin V, Perry JK, Lobie PE, et al. The prolactin receptor mediates HOXA1-stimulated oncogenicity in mammary carcinoma cells. Int J Oncol. 2012;41(6):2285–2295. doi: 10.3892/ijo.2012.1660. [DOI] [PubMed] [Google Scholar]
- Hu H, Chen Y, Cheng S, Li G, Zhang Z. Dysregulated expression of homebox gene HOXA13 is correlated with the poor prognosis in bladder cancer. Wien Klin. Wochenschr. 2017;129(11–12):391–397. doi: 10.1007/s00508-016-1108-4. [DOI] [PubMed] [Google Scholar]
- Hu M, Ou-Yang W, Jing D, Chen R. Clinical prognostic significance of HOXC9 expression in patients with colorectal cancer. Clin Lab. 2019;65(8):190. doi: 10.7754/Clin.Lab.2019.190122. [DOI] [PubMed] [Google Scholar]
- Huan H-B, Yang D-P, Wen X-D, Chen X-J, Zhang L, Wu L-L, et al. HOXB7 accelerates the malignant progression of hepatocellular carcinoma by promoting stemness and epithelial-mesenchymal transition. J Exp Clin Cancer Res. 2017;36(1):86. doi: 10.1186/s13046-017-0559-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Kirschke CP, Zhang Y. Decreased intracellular zinc in human tumorigenic prostate epithelial cells: a possible role in prostate cancer progression. Cancer Cell Int. 2006;6:10. doi: 10.1186/1475-2867-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Chen L, Guo A. Upregulated expression of HOXC8 is associated with poor prognosis of cervical cancer. Oncol Lett. 2018;15(5):7291–7296. doi: 10.3892/ol.2018.8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo X-Y, Zhang X-Y, Yuan F, Zhao X-Y, You B-A. HOXB7 promotes proliferation and metastasis of glioma by regulating the Wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci. 2019;23(6):2476–2485. doi: 10.26355/eurrev_201903_17395. [DOI] [PubMed] [Google Scholar]
- Hwang J-A, Bin LB, Kim Y, Hong S-H, Kim Y-H, Han J, et al. HOXA9 inhibits migration of lung cancer cells and its hypermethylation is associated with recurrence in non-small cell lung cancer. Mol Carcinog. 2015;54(Suppl 1):E72–80. doi: 10.1002/mc.22180. [DOI] [PubMed] [Google Scholar]
- Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62(21):6240–6245. [PubMed] [Google Scholar]
- Jan R, Chaudhry G-E-S. Understanding apoptosis and apoptotic pathways targeted cancer therapeutics. Adv. Pharm. Bull. 2019;9(2):205–218. doi: 10.15171/apb.2019.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin K, Kong X, Shah T, Penet M-F, Wildes F, Sgroi DC, et al. The HOXB7 protein renders breast cancer cells resistant to tamoxifen through activation of the EGFR pathway. Proc Natl Acad Sci U S A. 2012;109(8):2736–2741. doi: 10.1073/pnas.1018859108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C, Kim R-S, Lee S-J, Wang C, Jeng M-H. HOXB13 homeodomain protein suppresses the growth of prostate cancer cells by the negative regulation of T-cell factor 4. Cancer Res. 2004;64(9):3046–3051. doi: 10.1158/0008-5472.can-03-2614. [DOI] [PubMed] [Google Scholar]
- Jung C, Kim R-S, Zhang H, Lee S-J, Sheng H, Loehrer PJ, et al. HOXB13 is downregulated in colorectal cancer to confer TCF4-mediated transactivation. Br J Cancer. 2005;92(12):2233–2239. doi: 10.1038/sj.bjc.6602631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Jeong S, Jeong H, Oh HE, Choi J-W, Lee ES, et al. Increased HOXC6 mRNA expression is a novel biomarker of gastric cancer. PLoS ONE. 2020;15(8):e0236811. doi: 10.1371/journal.pone.0236811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-R, Kim I-J, Kang TW, Choi C, Kim KK, Kim MS, et al. HOXB13 downregulates intracellular zinc and increases NF-κB signaling to promote prostate cancer metastasis. Oncogene. 2014;33(37):4558–4567. doi: 10.1038/onc.2013.404. [DOI] [PubMed] [Google Scholar]
- Kim J, Bae D-H, Kim JH, Song K-S, Kim YS, Kim S-Y. HOXC10 overexpression promotes cell proliferation and migration in gastric cancer. Oncol Rep. 2019;42(1):202–212. doi: 10.3892/or.2019.7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CY, Oh JH, Lee J-Y, Kim MH. The LncRNA HOTAIRM1 promotes tamoxifen resistance by mediating HOXA1 expression in ER+ breast cancer cells. J Cancer. 2020;11(12):3416–3423. doi: 10.7150/jca.38728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science. 2003;301(5631):331–333. doi: 10.1126/science.1085753. [DOI] [PubMed] [Google Scholar]
- Ko SY, Barengo N, Ladanyi A, Lee J-S, Marini F, Lengyel E, et al. HOXA9 promotes ovarian cancer growth by stimulating cancer-associated fibroblasts. J Clin Invest. 2012;122(10):3603–3617. doi: 10.1172/JCI62229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocak H, Ackermann S, Hero B, Kahlert Y, Oberthuer A, Juraeva D, et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis. 2013;4(4):e586. doi: 10.1038/cddis.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappin TRJ, Grier DG, Thompson A, Halliday HL. HOX genes: seductive science, mysterious mechanisms. Ulster Med J. 2006;75(1):23–31. [PMC free article] [PubMed] [Google Scholar]
- Lee J-Y, Hur H, Yun HJ, Kim Y, Yang S, Kim SIL, et al. HOXB5 promotes the proliferation and invasion of breast cancer cells. Int J Biol Sci. 2015;11(6):701–711. doi: 10.7150/ijbs.11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-Y, Kim JM, Jeong DS, Kim MH. Transcriptional activation of EGFR by HOXB5 and its role in breast cancer cell invasion. Biochem Biophys. Res. Commun. 2018;503(4):2924–2930. doi: 10.1016/j.bbrc.2018.08.071. [DOI] [PubMed] [Google Scholar]
- Lee K, Chang JW, Oh C, Liu L, Jung S-N, Won H-R, et al. HOXB5 acts as an oncogenic driver in head and neck squamous cell carcinoma via EGFR/Akt/Wnt/β-catenin signaling axis. Eur. J. Surg. Oncol. J. Eur. Soc. 2020;46(6):1066–1073. doi: 10.1016/j.ejso.2019.12.009. [DOI] [PubMed] [Google Scholar]
- Lescroart F, Zaffran S. Hox and Tale transcription factors in heart development and disease. Int J Dev Biol. 2018;62(11–12):837–846. doi: 10.1387/ijdb.180192sz. [DOI] [PubMed] [Google Scholar]
- Li H, Shen L-Y, Yan W-P, Dong B, Kang X-Z, Dai L, et al. Deregulated HOXB7 expression predicts poor prognosis of patients with esophageal squamous cell carcinoma and regulates cancer cell proliferation in vitro and in vivo. PLoS ONE. 2015;10(6):e0130551. doi: 10.1371/journal.pone.0130551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Jia X, Wang J, Li Y, Xie S. Knockdown of homeobox A5 by small hairpin RNA inhibits proliferation and enhances cytarabine chemosensitivity of acute myeloid leukemia cells. Mol Med Rep. 2015;12(5):6861–6866. doi: 10.3892/mmr.2015.4331. [DOI] [PubMed] [Google Scholar]
- Li Q, Dong C, Cui J, Wang Y, Hong X. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer Res. 2018;37(1):265. doi: 10.1186/s13046-018-0941-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang W, Wu C, Gao H, Yu J, Wang X, et al. HOXC10 promotes proliferation and invasion and induces immunosuppressive gene expression in glioma. FEBS J. 2018;285(12):2278–2291. doi: 10.1111/febs.14476. [DOI] [PubMed] [Google Scholar]
- Li W, Li C, Xiong Q, Tian X, Ru Q. MicroRNA-10b-5p downregulation inhibits the invasion of glioma cells via modulating homeobox B3 expression. Exp Ther Med. 2019;17(6):4577–4585. doi: 10.3892/etm.2019.7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lin H, Jiang F, Lou Y, Ji L, Li S. Knock-down of HOXB8 prohibits proliferation and migration of colorectal cancer cells via Wnt/β-catenin signaling pathway. Med Sci Monit Int Med J Exp Clin Res. 2019;25:711–720. doi: 10.12659/MSM.912218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Huang Q, Wei G-H. The role of HOX transcription factors in cancer predisposition and progression. Cancers (basel). 2019;11(4):528. doi: 10.3390/cancers11040528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Bai Y, Feng Z, Li W, Yang C, Guo Y, et al. Study of promoter methylation patterns of HOXA2, HOXA5, and HOXA6 and its clinicopathological characteristics in colorectal cancer. Front Oncol. 2019;9:394. doi: 10.3389/fonc.2019.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S-Y, Wu H-C, Mai H-F, Zhen J-X, Li G-S, Chen S-J. Microarray-based analysis of whole-genome DNA methylation profiling in early detection of breast cancer. J Cell Biochem. 2019;120(1):658–670. doi: 10.1002/jcb.27423. [DOI] [PubMed] [Google Scholar]
- Li C, Cui J, Zou L, Zhu L, Wei W. Bioinformatics analysis of the expression of HOXC13 and its role in the prognosis of breast cancer. Oncol Lett. 2020;19(1):899–907. doi: 10.3892/ol.2019.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ye M, Zhou C. Expression profile and prognostic values of HOXA family members in laryngeal squamous cell cancer. Front Oncol. 2020;10:368. doi: 10.3389/fonc.2020.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wang J-F, Liu B, Wang X-M. Homeobox B2 is a potential prognostic biomarker of glioblastoma. Rev Assoc Med Bras. 2020;66(6):794–799. doi: 10.1590/1806-9282.66.6.794. [DOI] [PubMed] [Google Scholar]
- Liang T, Wang X, Li P, Cao Y, Feng E, You G. HOXC8: a predictive glioma biomarker that induces epithelia-mesenchymal transition. Chinese Neurosurg J. 2018;4:24. doi: 10.1186/s41016-018-0132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W-T, Jiang D, Yuan J, Cui Y-M, Shi X-W, Chen C-M, et al. HOXB7 as a prognostic factor and mediator of colorectal cancer progression. Clin Cancer Res. 2011;17(11):3569–3578. doi: 10.1158/1078-0432.CCR-10-2533. [DOI] [PubMed] [Google Scholar]
- Liu X, Lu K, Wang K, Sun M, Zhang E, Yang J, et al. MicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5. BMC Cancer. 2012;12:348. doi: 10.1186/1471-2407-12-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Jin K, Hui Y, Fu J, Jie C, Feng S, et al. HOXB7 promotes malignant progression by activating the TGFβ signaling pathway. Cancer Res. 2015;75(4):709–719. doi: 10.1158/0008-5472.CAN-14-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-J, Zhu Y, Yuan H-X, Zhang J-P, Guo J-M, Lin Z-M. Overexpression of HOXC11 homeobox gene in clear cell renal cell carcinoma induces cellular proliferation and is associated with poor prognosis. Tumour Biol J Int Soc Oncodevelopmental Biol Med. 2015;36(4):2821–2829. doi: 10.1007/s13277-014-2909-6. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang M, Xu S, Zhang J, Zou J, Yang C, et al. HOXC8 promotes proliferation and migration through transcriptional up-regulation of TGFβ1 in non-small cell lung cancer. Oncogenesis. 2018;7(2):1. doi: 10.1038/s41389-017-0016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Ge M, Ma J, Zhang Y, Zhao Y, Cui T. Homeobox A10 promotes the proliferation and invasion of bladder cancer cells via regulation of matrix metalloproteinase-3. Oncol Lett. 2019;18(1):49–56. doi: 10.3892/ol.2019.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Tian H, Zhao J, Jia Y. High HOXD4 protein expression in gastric adenocarcinoma tissues indicates unfavorable clinical outcomes. Saudi J. Gastroenterol. off. 2019;25(1):46–54. doi: 10.4103/sjg.SJG_105_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H-M, Cui N, Zheng P-S. HOXA5 inhibits the proliferation and neoplasia of cervical cancer cells via downregulating the activity of the Wnt/β-catenin pathway and transactivating TP53. Cell Death Dis. 2020;11(6):420. doi: 10.1038/s41419-020-2629-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek R, Gajula RP, Williams RD, Nghiem B, Simons BW, Nugent K, et al. TWIST1-WDR5-Hottip regulates Hoxa9 chromatin to facilitate prostate cancer metastasis. Cancer Res. 2017;77(12):3181–3193. doi: 10.1158/0008-5472.CAN-16-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo M. Reassessing the role of Hox genes during vertebrate development and evolution. Trends Genet. 2018;34(3):209–217. doi: 10.1016/j.tig.2017.11.007. [DOI] [PubMed] [Google Scholar]
- Mansoori B, Mohammadi A, Davudian S, Shirjang S, Baradaran B. The different mechanisms of cancer drug resistance: a brief review. Adv Pharm Bull. 2017;7(3):339–348. doi: 10.15171/apb.2017.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour MA, Senga T. HOXD8 exerts a tumor-suppressing role in colorectal cancer as an apoptotic inducer. Int J Biochem Cell Biol Netherlands. 2017;88:1–13. doi: 10.1016/j.biocel.2017.04.011. [DOI] [PubMed] [Google Scholar]
- Marra L, Cantile M, Scognamiglio G, Perdonà S, La Mantia E, Cerrone M, et al. Deregulation of HOX B13 expression in urinary bladder cancer progression. Curr Med Chem. 2013;20(6):833–839. [PubMed] [Google Scholar]
- Matsunami M, Sumiyama K, Saitou N. Evolution of conserved non-coding sequences within the vertebrate Hox clusters through the two-round whole genome duplications revealed by phylogenetic footprinting analysis. J Mol Evol. 2010;71(5–6):427–436. doi: 10.1007/s00239-010-9396-1. [DOI] [PubMed] [Google Scholar]
- Miller KR, Patel JN, Zhang Q, Norris EJ, Symanowski J, Michener C, et al. HOXA4/HOXB3 gene expression signature as a biomarker of recurrence in patients with high-grade serous ovarian cancer following primary cytoreductive surgery and first-line adjuvant chemotherapy. Gynecol. 2018;149(1):155–162. doi: 10.1016/j.ygyno.2018.01.022. [DOI] [PubMed] [Google Scholar]
- Miwa T, Kanda M, Umeda S, Tanaka H, Tanaka C, Kobayashi D, et al. Homeobox C10 influences on the malignant phenotype of gastric cancer cell lines and its elevated expression positively correlates with recurrence and poor survival. Ann Surg Oncol. 2019;26(5):1535–1543. doi: 10.1245/s10434-019-07166-5. [DOI] [PubMed] [Google Scholar]
- Miyazaki YJ, Hamada J, Tada M, Furuuchi K, Takahashi Y, Kondo S, et al. HOXD3 enhances motility and invasiveness through the TGF-beta-dependent and -independent pathways in A549 cells. Oncogene. 2002;21(5):798–808. doi: 10.1038/sj.onc.1205126. [DOI] [PubMed] [Google Scholar]
- Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291(2):193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Mohankumar KM, Xu XQ, Zhu T, Kannan N, Miller LD, Liu ET, et al. HOXA1-stimulated oncogenicity is mediated by selective upregulation of components of the p44/42 MAP kinase pathway in human mammary carcinoma cells. Oncogene. 2007;26(27):3998–4008. doi: 10.1038/sj.onc.1210180. [DOI] [PubMed] [Google Scholar]
- Mohankumar KM, Perry JK, Kannan N, Kohno K, Gluckman PD, Emerald BS, et al. Transcriptional activation of signal transducer and activator of transcription (STAT) 3 and STAT5B partially mediate homeobox A1-stimulated oncogenic transformation of the immortalized human mammary epithelial cell. Endocrinology. 2008;149(5):2219–2229. doi: 10.1210/en.2007-1320. [DOI] [PubMed] [Google Scholar]
- Montagut C, Dalmases A, Bellosillo B, Crespo M, Pairet S, Iglesias M, et al. Identification of a mutation in the extracellular domain of the epidermal growth factor receptor conferring cetuximab resistance in colorectal cancer. Nat Med. 2012;18(2):221–223. doi: 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- Moon S-M, Kim S-A, Yoon J-H, Ahn S-G. HOXC6 is deregulated in human head and neck squamous cell carcinoma and modulates Bcl-2 expression. J Biol Chem. 2012;287(42):35678–35688. doi: 10.1074/jbc.M112.361675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R, Simpson G, Gray S, Gillett C, Tabi Z, Spicer J, et al. HOX transcription factors are potential targets and markers in malignant mesothelioma. BMC Cancer. 2016;16:85. doi: 10.1186/s12885-016-2106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama I, Shibazaki M, Yashima-Abo A, Miura F, Sugiyama T, Masuda T, et al. Loss of HOXD10 expression induced by upregulation of miR-10b accelerates the migration and invasion activities of ovarian cancer cells. Int J Oncol. 2013;43(1):63–71. doi: 10.3892/ijo.2013.1935. [DOI] [PubMed] [Google Scholar]
- Nishida N, Yano H, Nishida T, Kamura T, Kojiro M. Angiogenesis in cancer. Vasc Health Risk Manag. 2006;2(3):213–219. doi: 10.2147/vhrm.2006.2.3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes SP, Diniz F, Moreira-Barbosa C, Constâncio V, Silva AV, Oliveira J, et al. Subtyping lung cancer using DNA methylation in liquid biopsies. J Clin Med. 2019;8(9):1500. doi: 10.3390/jcm8091500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchida K, Mizumoto K, Lin C, Yamaguchi H, Ohtsuka T, Sato N, et al. MicroRNA-10a is overexpressed in human pancreatic cancer and involved in its invasiveness partially via suppression of the HOXA1 gene. Ann Surg Oncol. 2012;19(7):2394–2402. doi: 10.1245/s10434-012-2252-3. [DOI] [PubMed] [Google Scholar]
- Ordóñez-Morán P, Dafflon C, Imajo M, Nishida E, Huelsken J. HOXA5 counteracts stem cell traits by inhibiting Wnt signaling in colorectal cancer. Cancer Cell. 2015;28(6):815–829. doi: 10.1016/j.ccell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Paço A, de Bessa A, Garcia S, Leitão Castro J, Costa-Pinto AR, Freitas R. Roles of the HOX proteins in cancer invasion and metastasis. Cancers. 2020;13(1):10. doi: 10.3390/cancers13010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paço A, de Bessa Garcia SA, Freitas R. Methylation in HOX clusters and its applications in cancer therapy. Cells. 2020;9(7):1613. doi: 10.3390/cells9071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T-T, Jia W-D, Yao Q-Y, Sun Q-K, Ren W-H, Huang M, et al. Overexpression of HOXA13 as a potential marker for diagnosis and poor prognosis of hepatocellular carcinoma. Tohoku J Exp Med. 2014;234(3):209–219. doi: 10.1620/tjem.234.209. [DOI] [PubMed] [Google Scholar]
- Panagopoulos I, Isaksson M, Billström R, Strömbeck B, Mitelman F, Johansson B. Fusion of the NUP98 gene and the homeobox gene HOXC13 in acute myeloid leukemia with t(11;12)(p15;q13) Genes Chromosomes Cancer. 2003;36(1):107–112. doi: 10.1002/gcc.10139. [DOI] [PubMed] [Google Scholar]
- Park S-M, Choi E-Y, Bae M, Choi JK, Kim Y-J. A long-range interactive DNA methylation marker panel for the promoters of HOXA9 and HOXA10 predicts survival in breast cancer patients. Clin Epigenetics. 2017;9:73. doi: 10.1186/s13148-017-0373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathiraja TN, Nayak SR, Xi Y, Jiang S, Garee JP, Edwards DP, et al. Epigenetic reprogramming of HOXC10 in endocrine-resistant breast cancer. Sci. Transl. Med. 2014;6(229):229ra41. doi: 10.1126/scitranslmed.3008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Zha L, Chen A, Wang Z. HOXA5 is a tumor suppressor gene that is decreased in gastric cancer. Oncol Rep. 2018;40(3):1317–1329. doi: 10.3892/or.2018.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Li Y, Li Y, Wu A, Fan L, Huang W, et al. HOXC10 promotes tumour metastasis by regulating the EMT-related gene Slug in ovarian cancer. Aging. 2020;12(19):19375–19398. doi: 10.18632/aging.103824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineault KM, Wellik DM. Hox genes and limb musculoskeletal development. Curr Osteoporos Rep. 2014;12(4):420–427. doi: 10.1007/s11914-014-0241-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pojo M, Gonçalves CS, Xavier-Magalhães A, Oliveira AI, Gonçalves T, Correia S, et al. A transcriptomic signature mediated by HOXA9 promotes human glioblastoma initiation, aggressiveness and resistance to temozolomide. Oncotarget. 2015;6(10):7657–7674. doi: 10.18632/oncotarget.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z, Chen Z, Weng J, Li S, Rong Z, Zhou C. Elevated HOXA13 expression promotes the proliferation and metastasis of gastric cancer partly via activating Erk1/2. Onco Targets Ther. 2019;12:1803–1813. doi: 10.2147/OTT.S196986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L-P, Zhong Y-M, Zheng Z, Zhao R-X. CDH17 is a downstream effector of HOXA13 in modulating the Wnt/β-catenin signaling pathway in gastric cancer. Eur Rev Med Pharmacol Sci. 2017;21(6):1234–1241. [PubMed] [Google Scholar]
- Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59(3):911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quagliata L, Quintavalle C, Lanzafame M, Matter MS, Novello C, di Tommaso L, et al. High expression of HOXA13 correlates with poorly differentiated hepatocellular carcinomas and modulates sorafenib response in in vitro models. Lab Invest. 2018;98(1):95–105. doi: 10.1038/labinvest.2017.107. [DOI] [PubMed] [Google Scholar]
- Quinonez SC, Innis JW. Human HOX gene disorders. Mol Genet Metab. 2014;111(1):4–15. doi: 10.1016/j.ymgme.2013.10.012. [DOI] [PubMed] [Google Scholar]
- Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, et al. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405(6789):974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- Rezsohazy R, Saurin AJ, Maurel-Zaffran C, Graba Y. Cellular and molecular insights into Hox protein action. Development. 2015;142(7):1212–1227. doi: 10.1242/dev.109785. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Garcia S, Prado-Garcia H, Carlos-Reyes A. Role of DNA methylation in the resistance to therapy in solid tumors. Front Oncol. 2020;10:1152. doi: 10.3389/fonc.2020.01152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadik H, Korangath P, Nguyen NK, Gyorffy B, Kumar R, Hedayati M, et al. HOXC10 expression supports the development of chemotherapy resistance by fine tuning DNA repair in breast cancer cells. Cancer Res. 2016;76(15):4443–4456. doi: 10.1158/0008-5472.CAN-16-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini S, Boore JL, Meyer A. Evolutionary conservation of regulatory elements in vertebrate Hox gene clusters. Genome Res. 2003;13(6A):1111–1122. doi: 10.1101/gr.700503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarno S, Cerrone M, Gigantino V, Scognamiglio G, Cantile M, Botti G, et al. Immunohistochemical detection of paralogous 13 HOX genes in phyllodes tumor of the breast as a useful diagnostic tool. Int J Clin Exp Pathol. 2020;13(9):2348–2351. [PMC free article] [PubMed] [Google Scholar]
- Sau A, Lau R, Cabrita MA, Nolan E, Crooks PA, Visvader JE, et al. Persistent activation of NF-κB in BRCA1-deficient mammary progenitors drives aberrant proliferation and accumulation of DNA damage. Cell Stem Cell. 2016;19(1):52–65. doi: 10.1016/j.stem.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Se Y-B, Kim SH, Kim JY, Kim JE, Dho Y-S, Kim JW, et al. Underexpression of HOXA11 is associated with treatment resistance and poor prognosis in glioblastoma. Cancer Res Treat. 2017;49(2):387–398. doi: 10.4143/crt.2016.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever R, Brugge JS. Signal transduction in cancer. Cold Spring Harb Perspect. Med. 2015;5(4):a006098. doi: 10.1101/cshperspect.a006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha L, Dong L, Lv L, Bai L, Ji X. HOXB9 promotes epithelial-to-mesenchymal transition via transforming growth factor-β1 pathway in hepatocellular carcinoma cells. Clin Exp Med. 2015;15(1):55–64. doi: 10.1007/s10238-014-0276-7. [DOI] [PubMed] [Google Scholar]
- Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- Shah CA, Wang H, Bei L, Platanias LC, Eklund EA. HoxA10 regulates transcription of the gene encoding transforming growth factor beta2 (TGFbeta2) in myeloid cells. J Biol Chem. 2011;286(4):3161–3176. doi: 10.1074/jbc.M110.183251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah N, Jin K, Cruz L-A, Park S, Sadik H, Cho S, et al. HOXB13 mediates tamoxifen resistance and invasiveness in human breast cancer by suppressing ERα and inducing IL-6 expression. Cancer Res. 2013;73(17):5449–5458. doi: 10.1158/0008-5472.CAN-13-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaoqiang C, Yue Z, Yang L, Hong Z, Lina Z, Da P, et al. Expression of HOXD3 correlates with shorter survival in patients with invasive breast cancer. Clin Exp Metastasis. 2013;30(2):155–163. doi: 10.1007/s10585-012-9524-y. [DOI] [PubMed] [Google Scholar]
- Shi Q, Shen L, Dong B, Fu H, Kang X, Dai L, et al. Downregulation of HOXA13 sensitizes human esophageal squamous cell carcinoma to chemotherapy. Thorac Cancer. 2018;9(7):836–846. doi: 10.1111/1759-7714.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields BJ, Slape CI, Vo N, Jackson JT, Pliego-Zamora A, Ranasinghe H, et al. The NUP98-HOXD13 fusion oncogene induces thymocyte self-renewal via Lmo2/Lyl1. Leukemia. 2019;33(8):1868–1880. doi: 10.1038/s41375-018-0361-0. [DOI] [PubMed] [Google Scholar]
- Shin SH, Lim DY, Reddy K, Malakhova M, Liu F, Wang T, et al. A small molecule inhibitor of the β-catenin-TCF4 interaction suppresses colorectal cancer growth in vitro and in vivo. EBioMedicine. 2017;25:22–31. doi: 10.1016/j.ebiom.2017.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund S, Avanzato D, Lanzetti L. Emerging functions of the EGFR in cancer. Mol Oncol. 2018;12(1):3–20. doi: 10.1002/1878-0261.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Gupta S, Badarukhiya JA, Sachan M. Detection of aberrant methylation of HOXA9 and HIC1 through multiplex MethyLight assay in serum DNA for the early detection of epithelial ovarian cancer. Int J Cancer. 2020;147(6):1740–1752. doi: 10.1002/ijc.32984. [DOI] [PubMed] [Google Scholar]
- Sipeky C, Gao P, Zhang Q, Wang L, Ettala O, Talala KM, et al. Synergistic interaction of HOXB13 and CIP2A predisposes to aggressive prostate cancer. Clin Cancer Res. 2018;24(24):6265–6276. doi: 10.1158/1078-0432.CCR-18-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Zyoud A, Allegrucci C. A case of identity: HOX genes in normal and cancer stem cells. Cancers (basel). 2019;11(4):512. doi: 10.3390/cancers11040512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Sen S, Weeks RJ, Eccles MR, Chatterjee A. Promoter DNA hypermethylation and paradoxical gene activation. Trends in Cancer. 2020;6(5):392–406. doi: 10.1016/j.trecan.2020.02.007. [DOI] [PubMed] [Google Scholar]
- Song JY, Pineault KM, Dones JM, Raines RT, Wellik DM. Hox genes maintain critical roles in the adult skeleton. Proc Natl Acad Sci U S A. 2020;117(13):7296–7304. doi: 10.1073/pnas.1920860117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storti P, Donofrio G, Colla S, Airoldi I, Bolzoni M, Agnelli L, et al. HOXB7 expression by myeloma cells regulates their pro-angiogenic properties in multiple myeloma patients. Leukemia. 2011;25(3):527–537. doi: 10.1038/leu.2010.270. [DOI] [PubMed] [Google Scholar]
- Su J, Huang Y-H, Cui X, Wang X, Zhang X, Lei Y, et al. Homeobox oncogene activation by pan-cancer DNA hypermethylation. Genome Biol. 2018;19(1):108. doi: 10.1186/s13059-018-1492-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B, Zhang X, Luo G. Homeobox B5 suppression attenuates proliferation and elevates apoptosis in hepatoma cell lines through ERK/MDM2 signalling. Clin Exp Pharmacol Physiol. 2020;47(6):1058–1066. doi: 10.1111/1440-1681.13278. [DOI] [PubMed] [Google Scholar]
- Sun C, Han C, Wang P, Jin Y, Sun Y, Qu L. HOXB9 expression correlates with histological grade and prognosis in LSCC. Biomed Res Int. 2017;2017:3680305. doi: 10.1155/2017/3680305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J-P, Ge Q-X, Ren Z, Sun X-F, Xie S-P. Down-regulation of HOXB5 inhibits TGF-β-induced migration and invasion in hepatocellular carcinoma cells via inactivation of the PI3K/Akt pathway. RSC Adv. 2018;8:41415–41421. doi: 10.1039/c8ra06860g. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sun P, Song Y, Liu D, Liu G, Mao X, Dong B, et al. Potential role of the HOXD8 transcription factor in cisplatin resistance and tumour metastasis in advanced epithelial ovarian cancer. Sci Rep. 2018;8(1):13483. doi: 10.1038/s41598-018-31030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed V. TGF-β Signaling in cancer. J Cell Biochem. 2016;117(6):1279–1287. doi: 10.1002/jcb.25496. [DOI] [PubMed] [Google Scholar]
- Takeda A, Goolsby C, Yaseen NR. NUP98-HOXA9 induces long-term proliferation and blocks differentiation of primary human CD34+ hematopoietic cells. Cancer Res. 2006;66(13):6628–6637. doi: 10.1158/0008-5472.CAN-06-0458. [DOI] [PubMed] [Google Scholar]
- Taminiau A, Draime A, Tys J, Lambert B, Vandeputte J, Nguyen N, et al. HOXA1 binds RBCK1/HOIL-1 and TRAF2 and modulates the TNF/NF-κB pathway in a transcription-independent manner. Nucleic Acids Res. 2016;44(15):7331–7349. doi: 10.1093/nar/gkw606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Chen K, Wu W, Zhou Y, Zhu J, Wu G, et al. Overexpression of HOXC10 promotes angiogenesis in human glioma via interaction with PRMT5 and upregulation of VEGFA expression. Theranostics. 2018;8(18):5143–5158. doi: 10.7150/thno.27310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer A, Amemiya CT, Kim C-B, Stadler PF. Evolution of microRNAs located within Hox gene clusters. J Exp Zool B Mol Dev Evol. 2005;304(1):75–85. doi: 10.1002/jez.b.21021. [DOI] [PubMed] [Google Scholar]
- Thomas SJ, Snowden JA, Zeidler MP, Danson SJ. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br J Cancer. 2015;113(3):365–371. doi: 10.1038/bjc.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi M, Taniuchi K, Shimizu T, Saito M, Saibara T. The transcription factor HOXB7 regulates ERK kinase activity and thereby stimulates the motility and invasiveness of pancreatic cancer cells. J Biol Chem. 2017;292(43):17681–17702. doi: 10.1074/jbc.M116.772780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Scherpenzeel TV, Remacle S, Picard J, Cornu G, Gofflot F, Rezsohazy R, et al. Mutation analysis of the HOX paralogous 4–13 genes in children with acute lymphoid malignancies: identification of a novel germline mutation of HOXD4 leading to a partial loss-of-function. Hum Mutat. 2005;25(4):384–395. doi: 10.1002/humu.20155. [DOI] [PubMed] [Google Scholar]
- Wang C-C, Su K-Y, Chen H-Y, Chang S-Y, Shen C-F, Hsieh C-H, et al. HOXA5 inhibits metastasis via regulating cytoskeletal remodelling and associates with prolonged survival in non-small-cell lung carcinoma. PLoS One. 2015;10(4):e0124191. doi: 10.1371/journal.pone.0124191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu G, Shen D, Ye H, Huang J, Jiao L, et al. HOXA1 enhances the cell proliferation, invasion and metastasis of prostate cancer cells. Oncol. Rep. 2015;34(3):1203–10. doi: 10.3892/or.2015.4085. [DOI] [PubMed] [Google Scholar]
- Wang H, Jia X-H, Chen J-R, Yi Y-J, Wang J-Y, Li Y-J, et al. HOXB4 knockdown reverses multidrug resistance of human myelogenous leukemia K562/ADM cells by downregulating P-gp, MRP1 and BCRP expression via PI3K/Akt signaling pathway. Int. J. Oncol. 2016;49(6):2529–37. doi: 10.3892/ijo.2016.3738. [DOI] [PubMed] [Google Scholar]
- Wang W-M, Xu Y, Wang Y-H, Sun H-X, Sun Y-F, He Y-F, et al. HOXB7 promotes tumor progression via bFGF-induced activation of MAPK/ERK pathway and indicated poor prognosis in hepatocellular carcinoma. Oncotarget. 2017;8(29):47121–47135. doi: 10.18632/oncotarget.17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Lin F, Sun X, Jiang L, Mao R, Zhou S, et al. HOXB8 enhances the proliferation and metastasis of colorectal cancer cells by promoting EMT via STAT3 activation. Cancer Cell Int. 2019;19:3. doi: 10.1186/s12935-018-0717-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang C, Liu N, Hou J, Xiao W, Wang H. HOXC6 promotes cervical cancer progression via regulation of Bcl-2. FASEB J Off Publ Fed Am Soc Exp Biol. 2019;33(3):3901–11. doi: 10.1096/fj.201801099RR. [DOI] [PubMed] [Google Scholar]
- Wang Z, Yu C, Wang H. HOXA5 inhibits the proliferation and induces the apoptosis of cervical cancer cells via regulation of protein kinase B and p27. Oncol Rep. 2019;41(2):1122–30. doi: 10.3892/or.2018.6874. [DOI] [PubMed] [Google Scholar]
- Wardwell-Ozgo J, Dogruluk T, Gifford A, Zhang Y, Heffernan TP, van Doorn R, et al. HOXA1 drives melanoma tumor growth and metastasis and elicits an invasion gene expression signature that prognosticates clinical outcome. Oncogene. 2014;33(8):1017–1026. doi: 10.1038/onc.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasenang W, Chaiyarit P, Proungvitaya S, Limpaiboon T. Serum cell-free DNA methylation of OPCML and HOXD9 as a biomarker that may aid in differential diagnosis between cholangiocarcinoma and other biliary diseases. Clin Epigenetics. 2019;11(1):39. doi: 10.1186/s13148-019-0634-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Saito M, Saito K, Matsumoto Y, Kanke Y, Onozawa H, et al. Upregulated HOXA9 expression is associated with lymph node metastasis in colorectal cancer. Oncol Lett. 2018;15(3):2756–2762. doi: 10.3892/ol.2017.7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154(6):1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AB, Faisal FA, Davicioni E, Karnes RJ, Vander Griend DJ, Lotan TL, et al. Somatic HOXB13 expression correlates with metastatic progression in men with localized prostate cancer following radical prostatectomy. Eur Urol Oncol. 2020;12:S2588. doi: 10.1016/j.euo.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss FU, Marques IJ, Woltering JM, Vlecken DH, Aghdassi A, Partecke LI, et al. Retinoic acid receptor antagonists inhibit miR-10a expression and block metastatic behavior of pancreatic cancer. Gastroenterology. 2009;137(6):2136–7. doi: 10.1053/j.gastro.2009.08.065. [DOI] [PubMed] [Google Scholar]
- Xia B, Shan M, Wang J, Zhong Z, Geng J, He X, et al. Homeobox A11 hypermethylation indicates unfavorable prognosis in breast cancer. Oncotarget. 2017;8(6):9794–9805. doi: 10.18632/oncotarget.14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Tan S, Zhou Y, Lin J, Wang H, Oyang L, et al. Role of the NFκB-signaling pathway in cancer. Onco Targets Ther. 2018;11:2063–2073. doi: 10.2147/OTT.S161109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong R, Yin T, Gao J-L, Yuan Y-F. HOXD9 activates the TGF-β/Smad signaling pathway to promote gastric cancer. Onco Targets Ther. 2020;13:2163–2172. doi: 10.2147/OTT.S234829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19(2):156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Valerio DG, Eisold ME, Sinha A, Koche RP, Hu W, et al. NUP98 fusion proteins interact with the NSL and MLL1 complexes to drive leukemogenesis. Cancer Cell. 2016;30(6):863–878. doi: 10.1016/j.ccell.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Zhao H, Yu J. HOXB5 promotes retinoblastoma cell migration and invasion via ERK1/2 pathway-mediated MMPs production. Am J Transl Res. 2018;10(6):1703–1712. [PMC free article] [PubMed] [Google Scholar]
- Xu F, Shangguan X, Pan J, Yue Z, Shen K, Ji Y, et al. HOXD13 suppresses prostate cancer metastasis and BMP4-induced epithelial-mesenchymal transition by inhibiting SMAD1. Int J cancer. 2021;148(15):3060–3070. doi: 10.1002/ijc.33494. [DOI] [PubMed] [Google Scholar]
- Xuan F, Huang M, Liu W, Ding H, Yang L, Cui H. Homeobox C9 suppresses Beclin1-mediated autophagy in glioblastoma by directly inhibiting the transcription of death-associated protein kinase 1. Neuro Oncol. 2016;18(6):819–829. doi: 10.1093/neuonc/nov281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, Zhu F-Y, Chen L, Wang K. HoxB9 promotes the migration and invasion via TGF-β1/Smad2/Slug signaling pathway in oral squamous cell carcinoma. Am J Transl Res. 2017;9(3):1151–1161. [PMC free article] [PubMed] [Google Scholar]
- Yan T-F, Wu M-J, Xiao B, Hu Q, Fan Y-H, Zhu X-G. Knockdown of HOXC6 inhibits glioma cell proliferation and induces cell cycle arrest by targeting WIF-1 in vitro and vivo. Pathol Res Pract. 2018;214(11):1818–24. doi: 10.1016/j.prp.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Yang H, Zhou J, Mi J, Ma K, Fan Y, Ning J, et al. HOXD10 acts as a tumor-suppressive factor via inhibition of the RHOC/AKT/MAPK pathway in human cholangiocellular carcinoma. Oncol Rep. 2015;34(4):1681–1691. doi: 10.3892/or.2015.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Lee J-Y, Hur H, Oh JH, Kim MH. Up-regulation of HOXB cluster genes are epigenetically regulated in tamoxifen-resistant MCF7 breast cancer cells. BMB Rep. 2018;51(9):450–5. doi: 10.5483/BMBRep.2018.51.9.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M-H, Zhao L, Wang L, Ou-Yang W, Hu S-S, Li W-L, et al. Nuclear lncRNA HOXD-AS1 suppresses colorectal carcinoma growth and metastasis via inhibiting HOXD3-induced integrin β3 transcriptional activating and MAPK/AKT signalling. Mol Cancer. 2019;18(1):31. doi: 10.1186/s12943-019-0955-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Kang W, Pan Y, Zhao X, Duan L. Overexpression of HOXC6 promotes cell proliferation and migration via MAPK signaling and predicts a poor prognosis in glioblastoma. Cancer Manag Res. 2019;11:8167–8179. doi: 10.2147/CMAR.S209904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, He L, Zhang Y, Ye L, Lai Y, Huang L, et al. HOXC10 promotes gastric cancer cell invasion and migration via regulation of the NF-κB pathway. Biochem Biophys Res Commun. 2018;501(3):628–35. doi: 10.1016/j.bbrc.2018.05.019. [DOI] [PubMed] [Google Scholar]
- Yao J, Chen Y, Nguyen DT, Thompson ZJ, Eroshkin AM, Nerlakanti N, et al. The homeobox gene, HOXB13, regulates a mitotic protein-kinase interaction network in metastatic prostate cancers. Sci Rep. 2019;9(1):9715. doi: 10.1038/s41598-019-46064-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y, Wang Y, Huang X, Sun Y, Zhang J, Li M, et al. Oncogenic HOXB8 is driven by MYC-regulated super-enhancer and potentiates colorectal cancer invasiveness via BACH1. Oncogene. 2020;39(5):1004–17. doi: 10.1038/s41388-019-1013-1. [DOI] [PubMed] [Google Scholar]
- Yuan W, Zhang X, Xu Y, Li S, Hu Y, Wu S. Role of HOXB7 in regulation of progression and metastasis of human lung adenocarcinoma. Mol Carcinog. 2014;53(1):49–57. doi: 10.1002/mc.21947. [DOI] [PubMed] [Google Scholar]
- Yuan C, Zhu X, Han Y, Song C, Liu C, Lu S, et al. Elevated HOXA1 expression correlates with accelerated tumor cell proliferation and poor prognosis in gastric cancer partly via cyclin D1. J Exp Clin Cancer Res. 2016;35:15. doi: 10.1186/s13046-016-0294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Sun S, Jiao N, Shu Y, Zhang Y. Upregulation of HOXA10 protein expression predicts poor prognosis for colorectal cancer. Genet Test Mol Biomarkers. 2018;22(6):390–7. doi: 10.1089/gtmb.2017.0240. [DOI] [PubMed] [Google Scholar]
- Yuan Y-H, Wang H-Y, Lai Y, Zhong W, Liang W-L, Yan F, et al. Epigenetic inactivation of HOXD10 is associated with human colon cancer via inhibiting the RHOC/AKT/MAPK signaling pathway. Cell Commun Signal. 2019;17(1):9. doi: 10.1186/s12964-018-0316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagozewski JL, Zhang Q, Pinto VI, Wigle JT, Eisenstat DD. The role of homeobox genes in retinal development and disease. Dev Biol. 2014;393(2):195–208. doi: 10.1016/j.ydbio.2014.07.004. [DOI] [PubMed] [Google Scholar]
- Zhang M, Nie F, Sun M, Xia R, Xie M, Lu K, et al. HOXA5 indicates poor prognosis and suppresses cell proliferation by regulating p21 expression in non small cell lung cancer. Tumour Biol. 2015;36(5):3521–31. doi: 10.1007/s13277-014-2988-4. [DOI] [PubMed] [Google Scholar]
- Zhang R, Deng Y, Zhang Y, Zhai G-Q, He R-Q, Hu X-H, et al. Upregulation of HOXA13 as a potential tumorigenesis and progression promoter of LUSC based on qRT-PCR and bioinformatics. Int J Clin Exp Pathol. 2017;10(10):10650–10665. [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Li N, Zhang H. Knockdown of homeobox B5 (HOXB5) inhibits cell proliferation, migration, and invasion in non-small cell lung cancer cells through inactivation of the Wnt/β-catenin pathway. Oncol Res. 2018;26(1):37–44. doi: 10.3727/096504017X14900530835262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang Q, Cao Z, Huang Y, Cheng S, Pang D. HOXD3 plays a critical role in breast cancer stemness and drug resistance. Cell. Physiol. Biochem. 2018;46(4):1737–47. doi: 10.1159/000489249. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wu Q, He C, Liang D, Yi Q, Shi J, et al. HOXB9 inhibits proliferation in gastric carcinoma cells via suppression of phosphorylated-Akt and NF-κB-dependent Snail expression. Dig. liver Dis. 2019;51(1):157–65. doi: 10.1016/j.dld.2018.08.018. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang S, Li X, Zhang F, Zhao L. HOXB5 promotes the progression of breast cancer through wnt/beta-catenin pathway. Pathol Res Pract. 2020;224:153117. doi: 10.1016/j.prp.2020.153117. [DOI] [PubMed] [Google Scholar]
- Zhao P, Tan L, Ruan J, Wei X-P, Zheng Y, Zheng L-X, et al. Aberrant expression of HOXA5 and HOXA9 in AML. Asian Pac J Cancer Prev. 2015;16(9):3941–4. doi: 10.7314/apjcp.2015.16.9.3941. [DOI] [PubMed] [Google Scholar]
- Zhao X-F, Yang Y-S, Park YK. HOXC9 overexpression is associated with gastric cancer progression and a prognostic marker for poor survival in gastric cancer patients. Int J Clin Oncol. 2020;25(12):2044–54. doi: 10.1007/s10147-020-01772-0. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Zhang Y, Ma D, Ren X, Xu C, Wan D. Inhibition of HOXB7 suppresses p27-mediated acute lymphoblastic leukemia by regulating basic fibroblast growth factor and ERK1/2. Life Sci Netherlands. 2019;218:1–7. doi: 10.1016/j.lfs.2018.12.011. [DOI] [PubMed] [Google Scholar]
- Zhou C, Li J, Li Q, Liu H, Ye D, Wu Z, et al. The clinical significance of HOXA9 promoter hypermethylation in head and neck squamous cell carcinoma. J Clin Lab Anal. 2019;33(5):e22873. doi: 10.1002/jcla.22873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Fu H, Dong B, Dai L, Yang Y, Yan W, et al. HOXB7 mediates cisplatin resistance in esophageal squamous cell carcinoma through involvement of DNA damage repair. Thorac Cancer. 2020;11(11):3071–3085. doi: 10.1111/1759-7714.13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang L, Li W-H, Li K, Mao Y, Gao C-L, Zhang C. Hoxb7 promotes growth and metastasis of lung adenocarcinoma cells through regulation of the tgf-β/smad3 signaling. J Biol Regul Homeost Agents. 2015;29(3):601–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.