Abstract
The fliD gene encoding the flagellar cap protein (FliD) of Clostridium difficile was studied in 46 isolates belonging to serogroups A, B, C, D, F, G, H, I, K, X, and S3, including 30 flagellated strains and 16 nonflagellated strains. In all but three isolates, amplification by PCR and reverse transcription-PCR demonstrated that the fliD gene is present and transcribed in both flagellated and nonflagellated strains. PCR-restriction fragment length polymorphism (RFLP) analysis of amplified fliD gene products revealed interstrain homogeneity, with one of two major patterns (a and b) found in all but one of the strains, which had pattern c. A polyclonal monospecific antiserum raised to the recombinant FliD protein reacted in immunoblots with crude flagellar preparations from 28 of 30 flagellated strains but did not recognize FliD from nonflagellated strains. The fliD genes from five strains representative of the three different RFLP groups were sequenced, and sequencing revealed 100% identity between the strains with the same pattern and 88% identity among strains with different patterns. Our results show that even though FliD is a structure exposed to the outer environment, the flagellar cap protein is very well conserved, and this high degree of conservation suggests that it has a very specific function in attachment to cell or mucus receptors.
Clostridium difficile is an opportunistic human pathogen that causes nosocomial infections such as antibiotic-associated colitis (pseudomembranous colitis) and diarrhea. Its pathogenicity is mediated by two exotoxins, toxins A (308 kDa) and B (270 kDa), both of which damage the human colonic mucosa and are potent cytotoxic enzymes (4). Before these events take place, C. difficile must be implanted in the gut and colonize suitable epithelial cells which are protected by a layer of dense mucus. Confirmed and putative accessory virulence factors that could play a role in adhesion and in intestinal colonization have been identified, including the capsule (8), proteolytic enzymes (28, 30), and adhesins involved in the association with mucus and the cell (5, 13, 17, 35). During the colonization process, the bacterium penetrates the mucus layer and attaches to enterocytes; at these different stages flagella are suspected to play a role, but this has yet to be proven. In some bacterial species flagella have been implicated in adherence to mucus and cells and in colonization and virulence; these include Pseudomonas aeruginosa (2), Vibrio cholerae (29), Vibrio anguillarum (21), Helicobacter pylori (12), Burkholderia pseudomallei (6), Campylobacter jejuni (16), Xenorhabdus nematophilus (15), Salmonella enterica serovar Typhi (20), and Proteus mirabilis (22).
A bacterial flagellum consists of a basal body in the membrane, the hook, and a helicoidal filament. The major structural component of the filament, the flagellin FliC, is assembled in subunits. Proteins called hook-associated proteins (HAP1, HAP2, and HAP3) are required to join the filament to the hook and to cap the distal tip of the filament. The fliD gene encodes HAP2, which functions as a capping structure at the distal end of the filament. It has been shown to have a function in mucin attachment by P. aeruginosa (1, 3), and H. pylori (18) and virulence in P. mirabilis (22).
We are interested in finding out whether flagella play a role in C. difficile intestinal attachment. Earlier studies from our laboratory have allowed characterization of the 39-kDa flagellin protein. The flagellin gene (fliC) was cloned and sequenced, and the recombinant protein was characterized (31). The diversity of the fliC gene among different isolates was studied, and it was found that the gene is present and expressed in both flagellated and nonflagellated strains (32).
In the study described here, in order to complete the findings concerning the proteins of the flagellar filament in C. difficile, we have characterized the fliD gene and its corresponding protein at the molecular level. We have investigated its presence and its variability among a series of C. difficile isolates from different serogroups and of various origins.
Forty-six C. difficile isolates belonging to 12 different serogroups (serogroups A1, A10, B, C, D, F, G, H, I, K, S3, and X) were selected at the Microbiology Unit of the Catholic University of Louvain, Brussels, Belgium, with care taken to choose strains isolated from several geographical locations (32). Clostridium sordellii (Institut Pasteur, Paris, France) was used as a negative control. All strains were grown under anaerobic conditions as described previously (32).
The primers used for amplification of the fliD genes from various C. difficile isolates were fliD-Nter (5′-ATGTCAAGTATAAGTCCAGTAAG-3′) and fliD-Cter (5′-TTAATTACCTTGTGCTTGTG-3′), corresponding to the 5′- and 3′-end sequences of the fliD gene of strain C. difficile 630, respectively, the genome sequence of which is now available on the Internet (www.sanger.ac.uk). Amplification was performed as described previously (32). At the end of the amplification, 5 μl of each of the samples was digested with the restriction enzymes AccI, DraI, EcoRI, HincII, HinfI, MboII, and XbaI (Amersham-Pharmacia Biotechnology).
The PCR products of strain 79685. reference strains of serogroups A, B, and C, and strain EX482 were purified with the QIAquick PCR purification kit (Qiagen). The nucleotide sequences of both strands were analyzed with an ABI PRISM 310 genetic analyzer (Perkin-Elmer), as described previously (32). Protein sequence alignments were performed with the DNA Strider software and the CLUSTAL W program (33). Homologies with sequences stored in GenBank were searched for by using Fasta3 (European Bioinformatics Institute) or Blast (National Center for Biotechnology Information) software.
RNA was extracted from 10 ml of an 8-h C. difficile anaerobic culture as described previously (32). The reverse transcription (RT)-PCR was carried out with the SuperScript one-step RT-PCR system (Life Technologies). The RNA of C. difficile 79685 was used as a positive control, and the RNA of C. sordellii was used as a negative control. The cDNA synthesis step was performed at 50°C for 30 min, and a predenaturation step was performed at 94°C for 2 min. Thirty cycles of amplification were performed in a Thermocycler 2400 instrument (Perkin-Elmer). Each cycle consisted of three steps, as described previously (32). The amplified products were subjected to electrophoresis in a standard 1% (wt/vol) agarose gel.
For the cloning of the C. difficile 79685 fliD gene into an expression vector, two oligonucleotide primers, fliD-BamHI (5′-CCCCTGGGATCCATGTCAAGTATAAGTCCAGTAAG-3′) and fliD-XhoI (5′-GGTCGACTCGAGTTAATTACCTTGTGCTTGTG-3′), which incorporated the BamHI and XhoI restriction sites, respectively, were synthesized and used to amplify by PCR the full-length coding region of the fliD gene of strain 79685 (Taq polymerase [Promega] was used at 1 U/100 μl of the reaction mixture volume). The resulting 1,524-bp DNA product was digested with BamHI and XhoI and cloned in-frame into the corresponding sites of pGEX-6P-1 (Amersham-Pharmacia Biotechnology). The nucleotide sequence of the junction between the vector and the insert was confirmed by sequencing analysis. The plasmid was transformed into Escherichia coli BL21. The expression and purification of the fusion protein were carried out as described previously (31). A polyclonal anti-FliD serum was raised against the purified recombinant FliD protein. The gel band corresponding to the purified protein was cut out, lyophilized, and injected subcutaneously into a rabbit. The polyclonal, monospecific antiserum was obtained by a previously described protocol (17) and was used at a 1:2,000 dilution in Western blots.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and immunoblotting were used to determine the presence of FliD proteins in clinical isolates. The proteins issued from the crude flagellar purification (9) were separated by SDS-PAGE (12% [wt/vol] acrylamide gel) as described by Laemmli (19). The gels were electrically transferred onto a nitrocellulose membrane for immunoblotting, and proteins were detected with the rabbit polyclonal anti-FliD serum (1:2,000 dilution) as described previously for FliC (31).
PCR amplification with the specific N-terminal and C-terminal oligonucleotide primers derived from the fliD gene of C. difficile strain 630 was carried out to study the C. difficile isolates for the presence of fliD. A single 1,524-bp amplified product was generated from 43 of the C. difficile strains studied, including strain 630, whereas no product was obtained from strains EX560, CO109, and ATCC 43604 (Table 1). These strains did not show gene amplification, despite many attempts with various parameters, such as changing the annealing temperature, MgCl2 concentration, or primers. For these strains the nonamplification of the fliD gene could result from either the absence of this gene or the presence of a genetically different gene which cannot be amplified with the primers used. The fliD gene was not amplified from the C. sordellii strain used as a negative control (data not shown). It is noteworthy that the fliD gene was present in both flagellated and nonflagellated strains.
TABLE 1.
C. difficile isolates studieda
| Strainb | Serogroup | fliD gene present (PCR) | fliD transcribed (RT-PCR) | fliD translated into protein (immunoblotting) | Flagellar structure visible by EMc | RFLP group |
|---|---|---|---|---|---|---|
| ATCC 43594d | A1 | + | NDde | + | + | b |
| 24573 | A1 | + | ND | + | + | b |
| EX482 | A1 | + | ND | + | + | c |
| SE810 | A10 | + | ND | + | + | a |
| TO005 | A10 | + | ND | + | + | b |
| 55787 | A10 | + | ND | + | + | b |
| EX560 | B | − | ND | − | − | ND |
| CO109 | B | − | ND | − | + | ND |
| ATCC 43593 | B | + | ND | + | + | b |
| ATCC 43596d | C | + | + | − | − | a |
| 54637 | C | + | + | − | − | a |
| 54828 | C | + | + | − | − | a |
| 51936 | C | + | + | − | − | a |
| 1075 | C | + | + | − | − | a |
| BR058 | D | + | ND | + | + | b |
| ATCC 43597 | D | + | + | − | − | b |
| 55944 | D | + | + | − | − | b |
| ATCC 43598d | F | + | ND | + | + | a |
| 5168 | F | + | ND | + | + | a |
| 6058 | F | + | ND | + | + | a |
| 6100 | F | + | ND | + | + | a |
| 54126 | G | + | ND | + | + | b |
| 51187 | G | + | ND | + | + | b |
| ATCC 43599d | G | + | ND | + | + | b |
| SE956 | G | + | + | − | − | b |
| ATCC 43600d | H | + | + | − | − | b |
| 50673 | H | + | ND | + | + | b |
| 53444 | H | + | ND | + | + | b |
| ATCC 43601d | I | + | + | − | − | a |
| 54823 | I | + | + | − | − | a |
| 56026 | I | + | ND | + | + | a |
| 55684 | I | + | + | − | − | a |
| 52356 | K | + | + | − | − | a |
| 51659 | K | + | ND | + | + | b |
| 48515 | K | + | ND | + | + | b |
| SE752 | K | + | ND | + | + | b |
| ATCC 43602d | K | + | ND | + | + | b |
| 79685 | S3 | + | ND | + | + | a |
| 57207 | S3 | + | + | − | − | a |
| 37561 | S3 | + | ND | + | + | b |
| EX596 | S3 | + | ND | + | + | b |
| 35962 | S3 | + | ND | + | + | b |
| 36678 | X | + | ND | + | + | a |
| 12934 | X | + | ND | + | + | a |
| 20356 | X | + | + | − | − | a |
| ATCC 43603d | X | − | ND | − | + | ND |
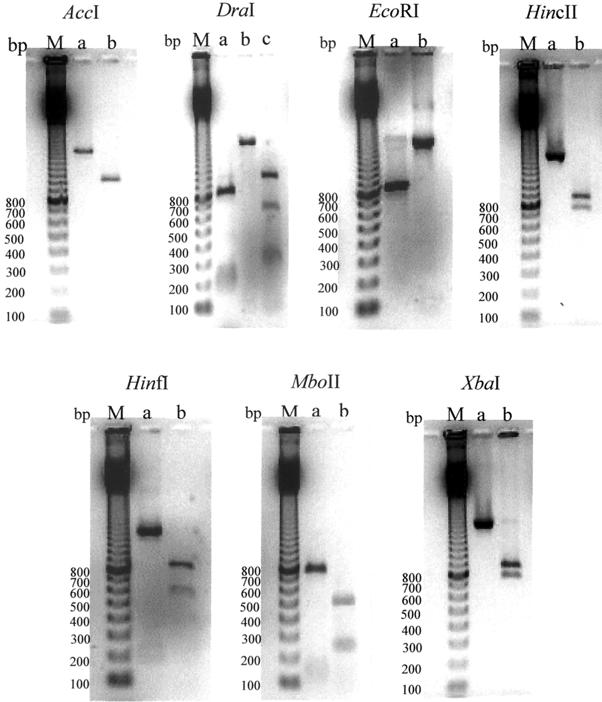
In order to study the variability of the fliD gene among C. difficile isolates, the amplified fliD gene was digested with the AccI, DraI, EcoRI, HincII, HinfI, MboII, and XbaI restriction enzymes. The different restriction patterns obtained from the C. difficile strains are shown in Fig. 1. Three different restriction profiles were obtained with the DraI enzyme (designated a, b, and c), and two different profiles (designated a and b) were obtained with the AccI, EcoRI, HincII, HinfI, MboII, and XbaI enzymes. Clinical isolates could be subdivided into two major restriction fragment length polymorphism (RFLP) groups (group a or b), each of which was represented by the profile (profiles a and b, respectively) obtained with the different restriction enzymes (Table 1; Fig. 1). The fliD RFLP analysis of strain EX482 revealed a unique RFLP group (group c), defined by profile c obtained with DraI, profile a obtained with HinfI and profile b obtained with the AccI, EcoRI, HincII, MboII, and XbaI enzymes. RFLP group a (20 strains) comprises all strains that belong to serogroups C, F, I, and X; one strain each of serogroups A10 and K; and two strains of serogroup S3. The second major RFLP group (group b; 22 strains) encompasses all strains of serogroups D, G, and H; the majority of the strains of serogroups A and K; and three strains of serogroup S3.
FIG. 1.
RFLP patterns of PCR-amplified flagellar cap genes. The amplified fliD genes of C. difficile isolates were digested with AccI, DraI, EcoRI, HincII, Hinfl, MboII, and XbaI. The different restriction profiles obtained with each enzyme were designated a, b, and c. Lanes M, 100-bp ladder (Amersham-Pharmacia Biotechnology); lanes a, profile a; lanes b, profile b; lanes c, profile c. The digested amplified products were subjected to electrophoresis in a 1.2% (wt/vol) agarose gel. The numbers next to the gels are in base pairs.
Different methods have been developed for C. difficile typing, particularly serogrouping by slide agglutination (10, 11) and comparison by PAGE of cell protein migration patterns. Newer molecular biology-based techniques have been used to study the genetic diversity of the flagellar genes, and RFLP analysis has been carried out to study the fliC genotypic variabilities in S. enterica (7), C. jejuni (24, 25, 27), P. aeruginosa (23, 36–38), H. pylori (26), and C. difficile (32). This is the first instance in which typing has been performed with the fliD gene. Since the results showed a little variability of this gene among the different isolates with two main patterns (patterns a and b), this gene is not an excellent biomarker for the study of diversity. We can note, nevertheless, that the strains belonging to the same serogroup generally exhibit the same pattern by RFLP analysis. Strains of serogroups A, G, H, and K have RFLP pattern b; interestingly, Delmée et al. (9) showed that flagellated strains of serogroup H were agglutinated by antisera raised against the flagellins of strains belonging to serogroups A, G, H, and K. The same results were observed with strains of serogroups C, F, I, and X with pattern a. It is interesting that there was a correlation between cross-agglutination of specific serogroups and RFLP profiles.
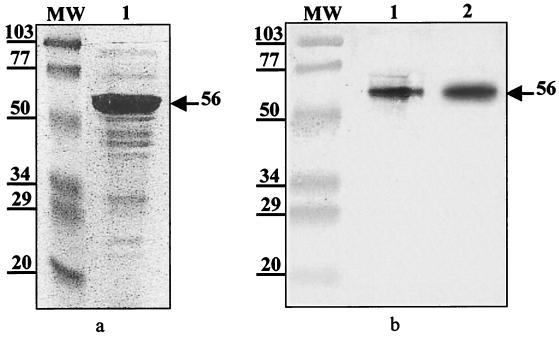
On the basis of the differences between the fliD genes obtained by RFLP analysis, we decided to sequence two strains with pattern a, two strains with pattern b, and one strain with one pattern c. Analysis of the DNA sequence of each strain revealed an open reading frame composed of 1,524 nucleotides corresponding to 507 amino acids. The C. difficile flagellar cap protein has a calculated molecular mass of 56 kDa, and thus, its mass does not differ from the estimated molecular mass of 56 kDa determined by SDS-PAGE (see Fig. 3a). The deduced amino acid sequences of the FliD proteins of strains 630 and 79685 and a reference strain of serogroup C were more than 99% identical but were different (88% identity) from those of the reference serogroup A and B strains and strain EX482, which were also more than 99% identical (data not shown). The deduced amino acid sequences of strain 79685 and the reference serogroup A strain were compared to known FliD protein sequences in GenBank. The FliD proteins of E. coli and S. enterica serovar Typhi, two genetically closed microorganisms, showed high degrees of identity (51%), whereas the degrees of identity between FliD of C. difficile and those of other bacterial genera ranged from 19 to 27%. The deduced amino acid sequences show that the structure of this protein is extremely well conserved, with no variable domains present.
FIG. 3.
(a). Purification of C. difficile 79685 FliD protein. The SDS-polyacrylamide gel shows low-molecular-mass standards of 103, 77, 50, 34, 29, and 20 kDa (Bio-Rad Laboratories) (lane mw) and FliD eluted from glutathione-Sepharose columns after digestion of GST-FliD with Prescission protease (lane 1). A major band is observed at 56 kDa (b). Immunoblotting of crude flagellar preparation of C. difficile strain 79685 reacted with a 1:2,000 dilution of polyclonal antiserum raised against purified FliD (lane 1). FliD was eluted from a glutathione-Sepharose column after digestion of GST-FliD with Prescission protease (lane 2). The arrow indicates the band corresponding to the 56-kDa flagellar cap protein.
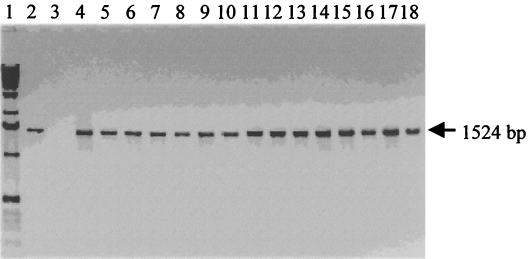
To gain insights into why certain strains are nonflagellated, we investigated the transcription of the fliD gene by detection of cap protein mRNA by RT-PCR in the nonflagellated strains. Nonflagellated strain EX560, the fliD gene of which was not amplified by PCR, was not studied. The results show that a single 1,524-bp product was obtained in all nonflagellated C. difficile strains (Fig. 2). Thus nonflagellation is not a result of the absence of transcription of the fliD gene.
FIG. 2.
RT-PCR products with specific fliD primers fliD-Nter and fliD-Cter and RNA isolated from nonflagellated C. difficile strains. Lanes: 1, 1-kb ladder (Amersham-Pharmacia Biotechnology); 2, strain 79685 (positive control); 3, RNA from C. sordellii (negative control); 4, strain ATCC 43596 (reference serogroup C. strain); 5, strain 54637; 6, strain 54828; 7, strain 51936; 8, strain 1075; 9, strain ATCC 43597 (reference serogroup D strain); 10, strain 55944; 11, strain SE956; 12, strain ATCC 43600 (reference serogroup H strain); 13, strain ATCC 43601 (reference serogroup I strain); 14, strain 54823; 15, strain 55684; 16, strain 52356; 17, strain 57207; 18, strain 20356. To confirm the purity of the RNA preparation and the specificity of the target RNA, an RNA sample treated with RNase was submitted to an RT-PCR as described in the text. Furthermore, the absence of genomic DNA contamination in the RNA samples was verified by PCR with fliD gene-specific N-terminal and C-terminal primers. No amplified products were detected in these two control experiments.
Purification of the cap protein was carried out to produce a monospecific antiserum in order to investigate the translation of the fliD gene. The fliD gene of C. difficile strain 79685 was cloned into the E. coli expression vector pGEX-6P-1, and the expression was induced with isopropyl-β-d-thiogalactopyranoside. The recombinant FliD protein was purified by affinity chromatography on glutathione-Sepharose, and the fusion protein glutathione S-transferase (GST)–FliD was cleaved with Prescission protease, as described previously (31). As shown in Fig. 3a, a major 56-kDa band free of contaminating GST was observed in the final eluate in SDS-polyacrylamide gels. Antibodies raised against the purified FliD protein recognized the purified 56-kDa protein and a protein with the same molecular mass in a crude flagellar preparation from strain 79685 (Fig. 3b). This result shows the specificity of the antiserum for FliD. In order to determine whether the fliD gene is translated in flagellated and nonflagellated strains, these antibodies were used to probe crude flagellar preparations of all C. difficile stains studied. The results showed that the antiserum recognized the 56-kDa FliD proteins of all flagellated C. difficile strains with the exception of those of strains CO109 and ATCC 43604. In contrast, no 56-kDa protein immunoreacted with the antiserum in nonflagellated strains (Table 1). This result suggests that (i) the FliD of each flagellated strain contains cross-reacting epitopes due to the presence of FliD monomers and (ii) in nonflagellated strains the absence of translation of the fliD gene could explain the lack of flagellation. We have shown previously that in strains in which no flagellar structure is visible by electron microscopy, the nonflagellated strains possess a cryptic flagellin gene (fliC) (32). The present study demonstrates that they also have a cryptic cap protein gene. Cryptic genes have been characterized in nonflagellated bacteria, and expression of surface flagella has been induced by modifying culture conditions in vitro in S. enterica serovar Pullorum (14) and in Shigella flexneri and Shigella sonnei (34). So far, little is known about the in vivo expression of flagella, and it can be hypothesized that flagellar switching on and off occurs through modification of microenvironmental factors in vivo during the host-pathogen interaction.
In conclusion, except for Arora et al. (1), who identified two distinct type of fliD genes among a group of P. aeruginosa strains, no study concerning the molecular variability of the fliD gene in other bacteria has been carried out; the protein has been studied only for its functionality. In our study, the analysis of the sequences of FliD proteins from different C. difficile strains showed scarce variability but revealed variable domains between different bacterial genera. This suggests that the FliD protein could possess specific conserved domains, which could have a function in attachment to highly specific cell or mucus receptors. The flagellar cap protein could play a role in adherence by mediating initial binding of the flagellar tip to mucin during the first stage of pathogenesis. Microenvironmental factors and host interactions could induce the production of flagella and gut colonization by C. difficile. Important questions remain to be answered concerning the exact role of the flagellar proteins in colonization and their vaccine potential.
Nucleotide sequence accession numbers.
The nucleotide sequences of the fliD loci of strains 79685, ATCC 43594, ATCC 43593, ATCC 43596, and EX482, corresponding to serogroup S3, reference strains as serogroups A, B, and C, and serogroup A1, respectively, were submitted to GenBank and were assigned accession numbers AF297024, AF297025, AF297026, AF297027, and AF297028, respectively.
Acknowledgments
This work was supported in part by the FAIR Program of the European Union (contract CT95-0433) and the ACC-SV6 program (Actions Concertées Coordonnées des Sciences du Vivant) of the Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche of France.
REFERENCES
- 1.Arora S K, Dasgupta N, Lory S, Ramphal R. Identification of two distinct types of flagellar cap proteins. FliD, in Pseudomonas aeruginosa. Infect Immun. 2000;68:1474–1479. doi: 10.1128/iai.68.3.1474-1479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora S K, Ritchings B W, Almira E C, Lory S, Ramphal R. Cloning and characterization of Pseudomonas aeruginosa fliF. necessary for flagella assembly and bacterial adherence to mucin. Infect Immun. 1996;64:2130–2136. doi: 10.1128/iai.64.6.2130-2136.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arora S K, Ritchings B W, Almira E C, Lory S, Ramphal R. The Pseudomonas aeruginosa flagellar cap protein. FliD, is responsible for mucin adhesion. Infect Immun. 1998;66:1000–1007. doi: 10.1128/iai.66.3.1000-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borriello S P, Davies H A, Kamiya S, Reed P J, Seddon S. Virulence factors of Clostridium difficile. Rev Infect Dis. 1990;12(Suppl. 2):S185–S191. doi: 10.1093/clinids/12.supplement_2.s185. [DOI] [PubMed] [Google Scholar]
- 5.Borriello S P, Welch A R, Barclay F E, Davies M A. Mucosal association by Clostridium difficile in the hamster gastrointestinal tract. J Med Microbiol. 1988;25:191–196. doi: 10.1099/00222615-25-3-191. [DOI] [PubMed] [Google Scholar]
- 6.Brett P J, Mah D C, Woods D E. Isolation and characterization of Pseudomonas pseudomallei flagellin proteins. Infect Immun. 1994;62:1914–1919. doi: 10.1128/iai.62.5.1914-1919.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dauga C, Zabrovskaia A, Grimont P A. Restriction fragment length polymorphism analysis of some flagellin genes of Salmonella enterica. J Clin Microbiol. 1998;36:2835–2843. doi: 10.1128/jcm.36.10.2835-2843.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies H A, Borriello S P. Detection of capsule in strains of Clostridium difficile of varying virulence and toxigenicity. Microb Pathog. 1990;9:141–146. doi: 10.1016/0882-4010(90)90088-8. [DOI] [PubMed] [Google Scholar]
- 9.Delmée M, Avesani V, Delferriere N, Burtonboy G. Characterization of flagella of Clostridium difficile and their role in serogrouping reactions. J Clin Microbiol. 1990;28:2210–2214. doi: 10.1128/jcm.28.10.2210-2214.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delmée M, Homel M, Wauters G. Serogrouping of Clostridium difficile strains by slide agglutination. J Clin Microbiol. 1985;21:323–327. doi: 10.1128/jcm.21.3.323-327.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delmée M, Laroche Y, Avesani V, Cornelis G. Comparison of serogrouping and polyacrylamide gel electrophoresis for typing Clostridium difficile. J Clin Microbiol. 1986;24:991–994. doi: 10.1128/jcm.24.6.991-994.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eaton K A, Suerbaum S, Josenhams C, Krakowka K S. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect Immun. 1996;64:2445–2448. doi: 10.1128/iai.64.7.2445-2448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eveillard M, Fourel V, Barc M C, Kerneis S, Coconnier M H, Karjalainen T, Bourlioux P, Servin A L. Identification and characterization of adhesive factors of Clostridium difficile involved in adhesion to human colonic enterocyte-like Caco-2 and mucus-secreting HT29 cells in culture. Mol Microbiol. 1993;7:371–381. doi: 10.1111/j.1365-2958.1993.tb01129.x. [DOI] [PubMed] [Google Scholar]
- 14.Giron J A. Expression of flagella and motility by Shigella. Mol Microbiol. 1995;18:63–75. doi: 10.1111/j.1365-2958.1995.mmi_18010063.x. [DOI] [PubMed] [Google Scholar]
- 15.Givaudan A, Lanois A. flhDC, the flagellar master operon of Xenorhabdus nematophilus: requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J Bacteriol. 2000;182:107–115. doi: 10.1128/jb.182.1.107-115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant C C, Konkel M E, Cieplak W J, Tompkins L S. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect Immun. 1993;61:1764–1771. doi: 10.1128/iai.61.5.1764-1771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karjalainen T, Barc M C, Collignon A, Trolle S, Boureau H, Cotte-Laffitte J, Bourlioux P. Cloning of a genetic determinant from Clostridium difficile involved in adherence to tissue culture cells and mucus. Infect Immun. 1994;62:4347–4355. doi: 10.1128/iai.62.10.4347-4355.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J S, Chang J H, Chung S I, Yum J S. Molecular cloning and characterization of the Helicobacter pylori fliD gene, an essential factor in flagellar structure and motility. J Bacteriol. 1999;181:6969–6976. doi: 10.1128/jb.181.22.6969-6976.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Liu S L, Ezaki T, Miura H, Matsui K, Yabuuchi E. Intact motility as a Salmonella typhi invasion-related factor. Infect Immun. 1988;56:1967–1973. doi: 10.1128/iai.56.8.1967-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGee K, Horstedt P, Milton D L. Identification and characterization of additional flagellin genes from Vibrio anguillarum. J Bacteriol. 1996;178:5188–5198. doi: 10.1128/jb.178.17.5188-5198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mobley H L, Belas R, Lockatell V, Chippendale G, Trifillis A L, Johnson D E, Warren J W. Construction of a flagellum-negative mutant of Proteus mirabilis: effect on internalization by human renal epithelial cells and virulence in a mouse model of ascending urinary tract infection. Infect Immun. 1996;64:5332–5340. doi: 10.1128/iai.64.12.5332-5340.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan J A, Bellingham N F, Winstanley C, Ousley M A, Hart C A, Saunders J R. Comparison of flagellin genes from clinical and environmental Pseudomonas aeruginosa isolates. Appl Environ Microbiol. 1999;65:1175–1179. doi: 10.1128/aem.65.3.1175-1179.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishimura M, Nukina M, Kuroki S, Obayashi H, Ohta M, Ma J J, Saida T, Uchiyama T. Characterization of Campylobacter jejuni isolates from patients with Guillain-Barré syndrome. J Neurol Sci. 1997;153:91–99. doi: 10.1016/s0022-510x(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 25.Nishimura M, Nukina M, Yuan J M, Shen B Q, Ma J J, Ohta M, Saida T, Uchiyama T. PCR-based restriction fragment length polymorphism (RFLP) analysis and serotyping of Campylobacter jejuni isolates from diarrheic patients in China and Japan. FEMS Microbiol Lett. 1996;142:133–138. doi: 10.1111/j.1574-6968.1996.tb08420.x. [DOI] [PubMed] [Google Scholar]
- 26.Ohta-Tada U, Takagi A, Koga Y, Kamiya S, Miwa T. Flagellin gene diversity among Helicobacter pylori strains and IL-8 secretion from gastric epithelial cells. Scand J Gastroenterol. 1997;32:455–459. doi: 10.3109/00365529709025080. [DOI] [PubMed] [Google Scholar]
- 27.Owen R J, Leeton S. Restriction fragment length polymorphism analysis of the flaA gene of Campylobacter jejuni for subtyping human, animal and poultry isolates. FEMS Microbiol Lett. 1999;176:345–350. doi: 10.1111/j.1574-6968.1999.tb13682.x. [DOI] [PubMed] [Google Scholar]
- 28.Poilane I, Karjalainen T, Barc M C, Bourlioux P, Collignon A. Protease activity of Clostridium difficile strains. Can J Microbiol. 1998;44:157–161. [PubMed] [Google Scholar]
- 29.Richardson K. Roles of motility and flagellar structure in pathogenicity of Vibrio cholerae: analysis of motility mutants in three animal models. Infect Immun. 1991;59:2727–2736. doi: 10.1128/iai.59.8.2727-2736.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seddon S V, Borriello S P. Proteolytic activity of Clostridium difficile. J Med Microbiol. 1992;36:307–311. doi: 10.1099/00222615-36-5-307. [DOI] [PubMed] [Google Scholar]
- 31.Tasteyre A, Barc M C, Karjalainen T, Dodson P, Hyde S, Bourlioux P, Borriello P. A Clostridium difficile gene encoding flagellin. Microbiology. 2000;146:957–966. doi: 10.1099/00221287-146-4-957. [DOI] [PubMed] [Google Scholar]
- 32.Tasteyre A, Karjalainen T, Avesani V, Delmee M, Collignon A, Bourlioux P, Barc M C. Phenotypic and genotypic diversity of the flagellin gene (fliC) among Clostridium difficile isolates from different serogroups. J Clin Microbiol. 2000;38:3179–3186. doi: 10.1128/jcm.38.9.3179-3186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tominaga A, Mahmoud M A, Mukaihara T, Enomoto M. Molecular characterization of intact, but cryptic, flagellin genes in the genus Shigella. Mol Microbiol. 1994;12:277–285. doi: 10.1111/j.1365-2958.1994.tb01016.x. [DOI] [PubMed] [Google Scholar]
- 35.Waligora A J, Barc M C, Bourlioux P, Collignon A, Karjalainen T. Clostridium difficile cell attachment is modified by environmental factors. Appl Environ Microbiol. 1999;65:4234–4238. doi: 10.1128/aem.65.9.4234-4238.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winstanley C, Coulson M A, Wepner B, Morgan J A, Hart C A. Flagellin gene and protein variation amongst clinical isolates of Pseudomonas aeruginosa. Microbiology. 1996;142:2145–2151. doi: 10.1099/13500872-142-8-2145. [DOI] [PubMed] [Google Scholar]
- 37.Winstanley C, Hales B A, Morgan J A, Gallagher M J, Puthucheary S D, Cisse M F, Hart C A. Analysis of fliC variation among clinical isolates of Burkholderia cepacia. J Med Microbiol. 1999;48:657–662. doi: 10.1099/00222615-48-7-657. [DOI] [PubMed] [Google Scholar]
- 38.Winstanley C, Morgan J A. The bacterial flagellin gene as a biomarker for detection, population genetics and epidemiological analysis. Microbiology. 1997;143:3071–3084. doi: 10.1099/00221287-143-10-3071. [DOI] [PubMed] [Google Scholar]