Abstract
Nucleic acid testing (NAT) is important for the identification and quantification of specific nucleic acid targets, both DNA and RNA, in life sciences and clinical diagnostics. Nucleic acid amplification can be a time-consuming step in NAT using the polymerase chain reaction (PCR) assay. Therefore, this study aimed to develop a simple method to reduce the amplification time while maintaining the PCR system. The three-step process of a general qPCR was reduced to a two-step process. The annealing/extension temperatures were increased to minimize the differences between the denaturation temperature and the annealing/extension temperatures. Subsequently, the time for each of these steps was reduced and, finally, the denaturation temperature was lowered. Taq polymerase was replaced with SD polymerase because it has strand displacement activity and is efficient in amplifying partial dsDNA at lower denaturation temperatures. In the two-step qPCR of genomic DNA using SD polymerase, the final conditions included an initial denaturation at 92 °C for 2 min, and 1 s at each cycling step with a denaturation temperature of 87 °C and an annealing/extension temperature of 72 °C. Amplification of the nucleocapsid (N) gene of SARS-CoV-2 RNA virus was evaluated at a template concentration as low as 10 copies. This method, named SF-qPCR (strand displacement-based fast quantitative polymerase chain reaction), can stably detect less than 10 copies of DNA and RNA within 25–40 min. This new protocol allows for sensitive and rapid detection of important DNA and RNA targets in clinical diagnosis.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13206-021-00044-x.
Keywords: Quantitative PCR, SF-qPCR, SD polymerase, Strand displacement activity, BRCA, SARS-CoV-2
Introduction
Nucleic acid testing (NAT) is an important tool for the identification and quantification of specific nucleic acid targets in the life sciences and clinical diagnostics [1, 2]. Nucleic acid targets are broadly divided into DNA and RNA. DNA targets are mainly used for cancer diagnosis, such as the BRCA1 and BRCA2 genes, which are associated with breast, ovarian, pancreatic, and prostate cancers [3, 4]. Accurate BRCA testing is essential for early detection, identifying gene mutations, and selection of treatments [5, 6]. RNA viruses include Zika [7], Ebola [8], and influenza [9, 10], as well as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [11], first identified in December 2019. The outbreak of each of these viruses caused an international public health emergency due to its large spread. Rapid and accurate diagnosis of RNA viruses allows the identification of patients infected with the virus at an early stage, thereby preventing the spread of infection.
One of the important elements of NAT is amplification; polymerase chain reaction (PCR) and quantitative polymerase chain reaction (qPCR) are the gold standards for exponentially amplifying trace amounts of nucleic acid targets. However, conventional PCR/qPCR has limitations. The first limitation is sensitivity. The commonly used Taq polymerase has limitations in amplifying GC-rich sequences or complex structures such as stem-loop (hairpin) structures [12–14]. These structures may prematurely terminate polymerase extension and may lead to inaccurate amplification owing to mispriming and misannealing between the template and the complementary strand [15, 16]. This may reduce the sensitivity of the diagnosis. Taq polymerase can be replaced with SD polymerase, which successfully and efficiently amplifies templates with a wide range of secondary structures. SD polymerase is a novel, mutant Taq polymerase with high thermal stability and strand displacement activity. The strong strand displacement activity of SD polymerase results in a higher product yield than that using Taq polymerase in the amplification of templates that are GC-rich or have complex secondary structures. The strong polymerase activity enables high-efficiency PCR and long-range PCR (LR PCR), and significantly improves the sensitivity and efficiency in polymerase chain displacement reaction (PCDR) and real-time quantitative polymerase chain displacement reaction (qPCDR) [17, 18].
The second limitation of conventional PCR/qPCR is the time taken. PCR/qPCR takes 1.5–2 h, which is relatively long; therefore, it is not suitable for situations that require rapid diagnosis of a large number of samples, such as SARS-CoV-2 detection at an airport or point-of-care (POC) testing. SARS-CoV-2 detection using RT-qPCR takes 3–4 h from sampling to diagnosis [19]. This could lead to high chances of quarantine breach. Therefore, minimizing the diagnosis time is important for establishing a quarantine system. The time required for PCR/qPCR could be reduced by improving heat transfer and thermal equilibrium by lowering the PCR reaction volume to a nanoliter level or supplementing the thermal cycler to speed up temperature changes [20–24]. However, these are difficult to establish in routine laboratories and are proposals that deviate from the existing established PCR system.
This study aimed to develop a simple method that can reduce the time for NAT while maintaining the PCR system as much as possible. First, the three-step process of a general qPCR was reduced to a two-step process. The annealing/extension temperatures were increased to minimize differences between the denaturation temperature and the annealing/extension temperatures. Subsequently, the time for the denaturation step and the annealing/extension steps was reduced and, finally, the denaturation temperature was lowered. It is difficult to implement all of these strategies successfully using conventional Taq polymerase. Nevertheless, it is possible to reduce the three-step process to a two-step process using Taq polymerase. If the time within the denaturation and annealing/extension step is reduced, it is not possible to know exactly when hybridization and extension of the primers occur. For successful amplification, the time lost in each step must be made up during extension, even when the temperature is not ideal. Amplification should be possible even with partially formed dsDNA, but Taq polymerase without strand displacement activity may not be suitable (Fig. 1). Thus, a polymerase with strand displacement activity is required because partial dsDNA can be formed at lowered denaturation temperatures. Therefore, SD polymerase was introduced to implement the proposed strategy successfully. This strategy maintained sensitivity even at a low starting copy number of fewer than 10 copies and shortened the total detection time from 1.5–2 h to 25–40 min, when applied to the detection of BRCA and the SARS-CoV 2 RNA virus. Therefore, this method allows for the rapid detection of genomic DNA and RNA.
Fig. 1.
Mechanism of Taq polymerase and SD polymerase
Materials and Methods
Enzymes and Reagents
SD HotStart DNA polymerase (10 U/mL), 10 × SD polymerase reaction buffer, and MgCl2 were purchased from Bioron GmbH (Römerberg, Germany). dNTPs (10 mM each) were purchased from New England BioLabs (NEB; Ipswitch, MA, USA). Syto82 was purchased from Thermo Fisher Scientific (Waltham, MA, USA). The LunaScript® RT SuperMix Kit was purchased from New England BioLabs. Genomic DNA was purchased from Novagen. Quantitative synthetic SARS-CoV-2 RNA ORF, E, and N were purchased from ATCC. Primers were designed using the NCBI primer-BLAST program and synthesized by Integrated DNA Technology (IDT; Coralville, IA, USA). Real-time PCR reactions were carried out using the CFX96-IVD real-time PCR detection system (Bio-Rad Laboratories, Inc.).
Three-Step qPCR Assay
The genomic DNA template was amplified using SD HotStart DNA polymerase (2 U) in a reaction mixture consisting of 1 × SD polymerase reaction buffer, 3 mM MgCl2, 0.2 mM of each dNTP, 0.1 ng template DNA, 0.2 mM each of the primers F1 and R1, and 1 μM syto82 dye, and the volume was made up to 25 μl with distilled water. The reaction mixture without template was used as the negative control. The thermocycling conditions were as follows: preheating at 92 °C for 2 min, followed by 50 cycles at 92 °C for 30 s, 60 °C for 30 s, and 68 °C for 30 s.
Two-Step qPCR Assay
The genomic DNA template was amplified using SD HotStart DNA polymerase (2 U) in a reaction mixture consisting of 1 × SD polymerase reaction buffer, 3 mM MgCl2, 0.2 mM of each dNTP, 0.1 ng template DNA, 0.2 mM each of the primers F1 and R1, and 1 μM syto82 dye, and the volume was made up to 25 μl with distilled water. The reaction mixture without template was used as the negative control. The thermocycling conditions were as follows: preheating at 92 °C for 2 min, followed by 50 cycles at 92 °C for 30 s and 60 °C for 30 s.
SARS-CoV-2 RNA Virus RT-qPCR Assay
The LunaScript RT SuperMix was used for the RT reaction. The RT reaction mixture (10 µl) contained 2 µl of 1 × LunaScript RT SuperMix, 1 µl of RNA template, and 7 µl of nuclease-free water. The thermocycling conditions were as follows: primer annealing at 25 °C for 2 min, cDNA synthesis at 55 °C for 10 min, and heat inactivation at 95 °C for 1 min.
The qPCR mixture consisted of SD HotStart DNA polymerase (2 U), 1 × SD polymerase reaction buffer, 3 mM MgCl2, 0.2 mM of each dNTP, cDNA (5 µl), 0.2 mM each of the primers F1 and R1, and 1 μM syto82 dye, and the volume was made up to 25 μl with distilled water. The reaction mixture without template was used as the negative control. The thermocycling conditions were as follows: preheating at 92 °C for 2 min, followed by 50 cycles at 89 °C for 1 s and 72 °C for 1 s.
Results and Discussion
Reducing a Three-Step qPCR to a Two-Step qPCR
General PCR and qPCR using Taq polymerase consist of three steps: denaturation, annealing, and extension, which together take approximately 1.5–2 h. Two-step qPCR using Taq polymerase is a well-known approach [25, 26]; therefore, for better time management, the three-step process was modified into a two-step process (Fig. 2A).
Fig. 2.
Comparison of three- and two-step qPCR. A Scheme for reducing the three-step qPCR to two-step qPCR. B Detection time of the three- and two-step qPCR processes
To show that two-step PCR is possible with SD polymerase, we used human genomic DNA as the template and a primer set that amplified a BRCA-related gene (Online Resource Table 1) [5]. To compare the sensitivity of the three-step qPCR and two-step qPCR processes, serial dilutions of the DNA template with 300, 30, 3, and 0 copies were tested in duplicate assays. The uniformly increasing Ct value with a decrease in the target amount enabled quantification; the results were obtained two to three cycles earlier in the two-step PCR, compared with that in the three-step PCR, at the same concentration (Fig. 2B; Online Resources Fig. S1 and S2). The sensitivity was maintained even at a low starting copy number of three, despite the reduction to a two-step process. The total detection time was reduced by 23 min when the three-step PCR was reduced to a two-step qPCR.
Optimization of Annealing/Extension Temperature
Minimizing the difference in temperature between the denaturation step and the annealing/extension steps decreases the time consumed for temperature changes, thereby reducing the total detection time. To reduce the difference between denaturation and annealing/extension temperature, higher annealing/extension temperatures were optimized (Fig. 3A). Two-step qPCR was performed using two types of primers with different melting temperatures (Tm). In the case of the primer with a Tm of 65 °C, amplification was possible up to annealing/extension temperatures of 66 °C. To assess amplification at a higher temperature, a primer with a Tm of 70 °C was designed (Online Resource Table 1); the annealing/extension temperatures were increased to 68, 70, 72, and 74 °C. Amplification was observed at all temperatures (Fig. 3B; Online Resource Figs. S3 and S4). We chose 72 °C, within a Ct value of 40, as the annealing/extension temperature, and showed that the detection time was shortened by approximately 5 min.
Fig. 3.
Optimization of annealing/extension temperatures for the two-step qPCR. A Optimization scheme. B Detection time of the two-step qPCR for a primer with a Tm of 65 °C and a primer with a Tm of 70 °C
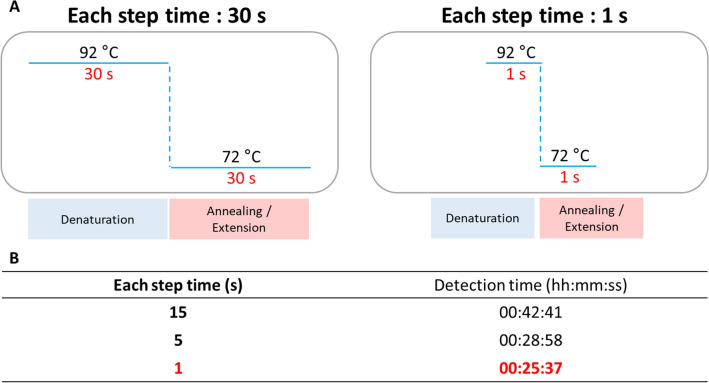
Optimization of Time for Each Step
We assessed the possibility of reducing the reaction time in the denaturation step and the annealing/extension steps (Fig. 4A). Sensitivity was examined by reducing the time to 15, 5, and 1 s for the conditions of denaturation at 92 °C and annealing/extension at 72 °C, respectively. To optimize the conditions for maintaining sensitivity, qPCR was performed using 3 and 0 copies of template DNA. Sufficient amplification was observed at 15, 5, and 1 s; the shortest time among them, 1 s, was selected because the Ct value fell within 40 (Fig. 4B and Online Resource Fig. S5). Through these optimization steps, the time taken for the PCR was shortened by approximately 35 min when compared to that using the conventional standard condition of 30 s.
Fig. 4.
Optimization of time for each step of the two-step qPCR. A Optimization scheme. B Detection time at each step time (15, 5, and 1 s)
Optimization of the Denaturation Conditions
The time taken per cycle was reduced by minimizing the difference in temperature between steps by lowering the temperature of the denaturation step (Fig. 5A). The previously established conditions (two-step, annealing/extension temperature: 72 °C, duration of each step: 1 s) were modified to lower the denaturation temperature from 91 to 85 °C. Amplification was performed within 40 cycles at a denaturation temperature of 86 °C. However, the denaturation temperature was optimized at 87 °C, because it was more stable than 86 °C (Online Resource Fig. S6); the time was reduced by approximately 3 min (Fig. 5B).
Fig. 5.
Optimization of denaturation conditions for the two-step qPCR. A Optimization scheme. B Detection time of the denaturation temperature range of 91–85 °C
Comparison of SD Polymerase and Taq Polymerase
In the two-step qPCR of genomic DNA using SD polymerase, the final conditions were an initial denaturation at 92 °C for 2 min, and 1 s at each cycling step with a denaturation temperature of 87 °C and an annealing/extension temperature of 72 °C. This method was named strand displacement-based fast quantitative polymerase chain reaction (SF-qPCR), and the total detection time using these conditions was approximately 23 min, which was significantly shorter than that of the conventional protocol (88 min).
We evaluated the use of Taq polymerase in the SF-qPCR process. The template and primers used were the same as that used with SD polymerase. The three-step qPCR conditions with Taq polymerase were as follows: preheating at 95 °C for 2 min, followed by 50 cycles at 95 °C for 30 s, 60 °C for 30 s, and 68 °C for 1 min, final extension at 68 °C for 5 min. The two-step qPCR conditions were as follows: preheating at 95 °C for 2 min, followed by 50 cycles at 95 °C for 30 s and 60 °C for 30 s. To compare the sensitivity of the three-step qPCR and two-step qPCR processes, serial dilutions of the DNA template with 30, 3, and 0 copies were tested in duplicate assays. When reducing from the three-step to two-step process using a primer with a Tm of 65 °C, dimer formation was observed in the two-step process; therefore, accurate comparison was difficult (Online Resource Fig. 7, 8). The two-step process was further optimized to increase the annealing/extension temperature, using a primer with a Tm of 70 °C; stable amplification was possible up to an annealing/extension temperature of 74 °C (Online Resource Fig. 9). However, in the experiments for reducing the reaction time within the denaturation step and the annealing/extension step to 15 s, 5 s, and 1 s; the process using 5 s lacked sensitivity because of a large delay in Ct value; there was no amplification at 1 s (Online Resource Fig. 10). Therefore, it is possible to reduce the three-step process to a two-step process using Taq polymerase; however, reducing the time taken for each step compromises the sensitivity, making it difficult to realize this strategy with Taq polymerase.
Diagnosis of SARS-CoV-2 RNA Virus via SF-qPCR
To verify that the amplified protocol optimized using SD polymerase applies equally to DNA as well as RNA templates, N gene of SARS-CoV-2 RNA virus was selected as the target. The entire reaction was performed in the step of generating cDNA through reverse transcription and the step of SF-qPCR (Fig. 6A). The cDNA synthesis was carried out as per the kit guidelines. The SF-qPCR was optimized for the SARS-CoV-2N gene cDNA clone; the two-step qPCR was performed using denaturation at 92 °C and an annealing/extension temperature of 68, 70, 72, or 74 °C, and the template concentration was 6 × 109 copies; the optimal annealing/extension temperature was determined to be 72 °C (Online Resource Fig. S11). The two-step qPCR was performed at template concentrations of 6000, 600, 60, or 6 copies with denaturation at 92 °C and annealing/extension at 72 °C; amplification was positive at a template concentration as low as 6 copies (Online Resource Fig. S12). The two-step qPCR was evaluated by lowering the denaturation temperature using temperatures from 91 to 86 °C; the denaturation temperature was optimized to 89 °C (Online Resource Fig. S13). Using the protocol determined through the optimization process, the RNA template was serially diluted to 1000, 100, 10, and 0 copies; amplification was observed even when using as low as 10 copies (Online Resource Fig. S14). The total detection time was approximately 40 min, including the reverse transcription process (Fig. 6B); this is shorter, considering the time taken for other processes. However, combining the RT and SF-qPCR steps would enable further reduction in the process time.
Fig. 6.
Diagnosis of SARS-CoV-2 RNA virus using SF-qPCR. A Diagnostic scheme using two-step qPCR. B Detection time of amplification of the SARS-CoV-2 RNA viral template. C Real-time fluorescence curve of the amplification of SARS-CoV-2 RNA virus at template concentrations of 1000 (violet), 100 (red), 10 (yellow), and 0 (black) copies
Conclusion
To reduce the time required for qPCR, three strategies were established using SD polymerase, which has thermal stability and strand displacement activity. The total detection time was reduced, while maintaining high sensitivity; the three-step process of general qPCR was reduced to a two-step process, the difference in temperature between the steps was minimized, and the reaction time of each step was minimized. This method, named SF-qPCR, can stably detect less than 10 copies of DNA and RNA within 25–40 min. This new protocol allows for sensitive and rapid detection of important DNA and RNA targets in clinical diagnosis.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the Technology Innovation Program [20009356] funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea), the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science & ICT (2019M3A9H1103633), a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) [2021R1C1C1004147], and the Cooperative Research Program for Agriculture Science and Technology Development (PJ014912032021).
Declarations
Conflict of Interest
The authors declare no conflict of interest.
Approval by Institutional Research Ethics Committee
No ethical approval was required.
Informed Consent
No informed consent was required.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Craw P, Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12:2469–2486. doi: 10.1039/C2LC40100B. [DOI] [PubMed] [Google Scholar]
- 2.Tang Y-W, Procop GW, Persing DH. Molecular diagnostics of infectious diseases. Clin. Chem. 1997;43:2021–2038. doi: 10.1093/clinchem/43.11.2021. [DOI] [PubMed] [Google Scholar]
- 3.Pilarski R. The role of BRCA testing in hereditary pancreatic and prostate cancer families. Am. Soc. Clin. Oncol. Educ. Book. 2019;39:79–86. doi: 10.1200/EDBK_238977. [DOI] [PubMed] [Google Scholar]
- 4.Nelson HD, et al. Risk assessment, genetic counseling, and genetic testing for BRCA-related cancer in women: a systematic review to update the US Preventive Services Task Force recommendation. Ann. Internal Med. 2014;160:255–266. doi: 10.7326/M13-1684. [DOI] [PubMed] [Google Scholar]
- 5.Jung C, Yim S-C, Cho D-Y, Chang HN, Park HG. Microarray-based detection of Korean-specific BRCA1 mutations. Anal. Bioanal. Chem. 2008;391:405–413. doi: 10.1007/s00216-008-1988-x. [DOI] [PubMed] [Google Scholar]
- 6.Wallace AJ. New challenges for BRCA testing: a view from the diagnostic laboratory. Eur. J. Hum. Genet. 2016;24:S10–S18. doi: 10.1038/ejhg.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petersen LR, Jamieson DJ, Powers AM, Honein MA. Zika virus. N. Engl. J. Med. 2016;374:1552–1563. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- 8.Baize S, et al. Emergence of Zaire Ebola virus disease in Guinea. N. Engl. J. Med. 2014;371:1418–1425. doi: 10.1056/NEJMoa1404505. [DOI] [PubMed] [Google Scholar]
- 9.Claas EC, et al. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 10.Ellis JS, Zambon MC. Molecular diagnosis of influenza. Rev. Med. Virol. 2002;12:375–389. doi: 10.1002/rmv.370. [DOI] [PubMed] [Google Scholar]
- 11.Zhang S, et al. Nucleic acid testing for coronavirus disease 2019: demand, research progression, and perspective. Crit. Rev. Anal. Chem. 2020 doi: 10.1080/10408347.2020.1805294. [DOI] [PubMed] [Google Scholar]
- 12.Henke W, Herdel K, Jung K, Schnorr D, Loening SA. Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res. 1997;25:3957–3958. doi: 10.1093/nar/25.19.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, et al. Enhanced amplification of GC-rich DNA with two organic reagents. Biotechniques. 2009;47:775–779. doi: 10.2144/000113203. [DOI] [PubMed] [Google Scholar]
- 14.Strien J, Sanft J, Mall G. Enhancement of PCR amplification of moderate GC-containing and highly GC-rich DNA sequences. Mol. Biotechnol. 2013;54:1048–1054. doi: 10.1007/s12033-013-9660-x. [DOI] [PubMed] [Google Scholar]
- 15.Singh VK, Govindarajan R, Naik S, Kumar A. The effect of hairpin structure on PCR amplification efficiency. Mol. Biol. Today. 2000;1:67–69. [Google Scholar]
- 16.Loewen PC, Switala J. Template secondary structure can increase the error frequency of the DNA polymerase from Thermus aquaticus. Gene. 1995;164:59–63. doi: 10.1016/0378-1119(95)00383-H. [DOI] [PubMed] [Google Scholar]
- 17.Ignatov KB, et al. A strong strand displacement activity of thermostable DNA polymerase markedly improves the results of DNA amplification. Biotechniques. 2014;57:81–87. doi: 10.2144/000114198. [DOI] [PubMed] [Google Scholar]
- 18.Harris CL, Sanchez-Vargas IJ, Olson KE, Alphey L, Fu G. Polymerase chain displacement reaction. Biotechniques. 2013;54:93–97. doi: 10.2144/000113951. [DOI] [PubMed] [Google Scholar]
- 19.Lai C-C, Wang C-Y, Ko W-C, Hsueh P-R. In vitro diagnostics of coronavirus disease 2019: technologies and application. J. Microbiol. Immunol. Infect. 2021;54:164–174. doi: 10.1016/j.jmii.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Son JH, et al. Ultrafast photonic PCR. Light Sci. Appl. 2015;4:e280–e280. doi: 10.1038/lsa.2015.53. [DOI] [Google Scholar]
- 21.Neuzil P, Zhang C, Pipper J, Oh S, Zhuo L. Ultra fast miniaturized real-time PCR: 40 cycles in less than six minutes. Nucleic Acids Res. 2006;34:e77–e77. doi: 10.1093/nar/gkl416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheeler E, et al. Under-three minute PCR: probing the limits of fast amplification. Analyst. 2011;136:3707–3712. doi: 10.1039/C1AN15365J. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Enzelberger M, Quake S. A nanoliter rotary device for polymerase chain reaction. Electrophoresis. 2002;23:1531–1536. doi: 10.1002/1522-2683(200205)23:10<1531::AID-ELPS1531>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 24.Roper MG, Easley CJ, Landers JP. Advances in polymerase chain reaction on microfluidic chips. Anal. Chem. 2005;77:3887–3894. doi: 10.1021/ac050756m. [DOI] [PubMed] [Google Scholar]
- 25.Jin HX, et al. Differences of PCR efficiency between two-step PCR and standard three-step PCR protocols in short tandem repeat amplification. Aust. J. Forensic Sci. 2014;46:80–90. doi: 10.1080/00450618.2013.788681. [DOI] [Google Scholar]
- 26.Jing C-Q, et al. Amplification of deoxyribonucleic acid (DNA) fragment using two-step polymerase chain reaction (PCR) Afr. J. Biotechnol. 2011;10:2838–2843. doi: 10.5897/AJB10.18. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.