Abstract
Malignant melanoma is a highly aggressive skin cancer characterized by an elevated grade of tumor cell plasticity. Such plasticity allows adaptation of melanoma cells to different hostile conditions and guarantees tumor survival and disease progression, including aggressive features such as drug resistance. Indeed, almost 50% of melanoma rapidly develop resistance to the BRAFV600E inhibitor vemurafenib, with fast tumor dissemination, a devastating consequence for patients’ outcomes. Vasculogenic mimicry (VM), the ability of cancer cells to organize themselves in perfused vascular-like channels, might sustain tumor spread by providing vemurafenib-resistant cancer cells with supplementary ways to enter into circulation and disseminate. Thus, this research aims to determine if vemurafenib resistance goes with the acquisition of VM ability by aggressive melanoma cells, and identify a driving molecule for both vemurafenib resistance and VM. We used two independent experimental models of drug-resistant melanoma cells, the first one represented by a chronic adaptation of melanoma cells to extracellular acidosis, known to drive a particularly aggressive and vemurafenib-resistant phenotype, the second one generated with chronic vemurafenib exposure. By performing in vitro tube formation assay and evaluating the expression levels of the VM markers EphA2 and VE-cadherin by Western blotting and flow cytometer analyses, we demonstrated that vemurafenib-resistant cells obtained by both models are characterized by an increased ability to perform VM. Moreover, by exploiting the CRISPR-Cas9 technique and using the urokinase plasminogen activator receptor (uPAR) inhibitor M25, we identified uPAR as a driver of VM expressed by vemurafenib-resistant melanoma cells. Thus, uPAR targeting may be successfully leveraged as a new complementary therapy to inhibit VM in drug-resistant melanoma patients, to counteract the rapid progression and dissemination of the disease.
Key words: Drug resistance, Vasculogenic mimicry, Extracellular acidosis, Melanoma, Urokinase plasminogen activator receptor (uPAR)
INTRODUCTION
Malignant melanoma is the most lethal skin cancer whose incidence continues to increase1. It is a very heterogeneous cancer due to the high mutation rate, especially BRAF point mutations2,3. However, it is very hard to find a driver mutation, for instance, limited to metastatic dissemination. Thus, multiple cell phenotypes that characterize every melanoma lesion must depend not only on changes in gene expression but also on new activated transcriptional programs. This means that changes in transcriptional regulators are responsible for continuous adaptive phenotype switching, critical to survival: this is the so-called phenotype plasticity. The plasticity of melanoma cells plays a critical role in therapy response. Upon the finding of BRAFV600E point mutation, a targeted compound, namely, vemurafenib (also known as PLX4032), was generated4. Despite its initial important benefits, almost 50% of patients rapidly relapse, developing drug resistance5,6. Behind tumor relapse, the acquisition of vasculogenic mimicry (VM) ability might play an important role. VM, firstly described by Maniotis and colleagues in 19997, consists in the de novo formation of a perfused vasculogenic-like network by aggressive tumor cells, characterized by features proper of both tumor and endothelial cells8–11. VM provides sufficient oxygen and nutrient supply required for tumor sustainment and growth. VM is closely associated with distant metastasis, a higher recurrence rate, and a shorter survival rate in many tumor types including melanoma11,12. In a meta-analysis of 22 clinical studies that enrolled 3,062 patients across 15 different types of cancers, the 5-year overall survival of the VM+ cancer patients was 31%, whereas the overall survival for VM− cancer patients was found to be around 56%. Moreover, the relative risk of relapse of the VM+ patients in 5 years was significantly higher than that of the VM− cancer patients13.
This study aims to determine if vemurafenib-resistant melanoma cells are also capable of VM, which can contribute to the dissemination of this subset of highly aggressive and resistant cancer cells, and thus disease progression. The identification of a molecule driving both these aggressive features of cancer cells would be crucial for developing new therapeutic strategies to counteract tumor progression and dissemination. Here, to test the hypothesis that melanoma cells undergoing drug resistance also acquire VM ability, we have used two different experimental models of drug-resistant melanoma cells. The first one comes from our previous study demonstrating the acquired vemurafenib resistance of acid-adapted melanoma cells. Acidosis of tumor microenvironment represents a new hallmark of cancer able to elicit in melanoma cells a well-established aggressive phenotype characterized by stem-like features, resistance to apoptosis, anoikis, and drug treatment—this last feature overcome only by everolimus treatment14,15. The second model instead consists of vemurafenib-resistant melanoma cells obtained in vitro by a continuous drug treatment until the recovering, in the presence of vemurafenib, of a growth rate comparable to control cells grown in the absence of the BRAF inhibitor. Using these experimental models, we clearly demonstrated that resistant melanoma cells are also endowed with VM ability and identified the urokinase plasminogen activator receptor (uPAR) as the driving molecule of both these features. Indeed, uPAR inhibition not only restored vemurafenib sensitivity in resistant melanoma cells16 but also abrogated their ability to organize themselves in vascular-like channels in vitro used as a critical marker of VM.
MATERIALS AND METHODS
Cell Culture
Melanoma cell lines A375 [American Type Culture Collection (ATCC), Rockville, MD, USA], A375-M614, and WM266-4 (ATCC) were cultured in DMEM 4.5 g/L glucose, 2 mM L-glutamine, and 10% fetal bovine serum (FBS) (Euroclone, Milan Italy). Extracellular acidosis was mimicked in vitro by culturing melanoma cells in pH 6.7 ± 0.1 medium for at least 3 months before use (chronic acidosis). The acidified medium was obtained by directly adding 1 N HCl in complete culture medium to reach pH 6.7, and pH value was monitored by using Orion pH meter 520A-1. As previously reported, acid-adapted melanoma cells show comparable proliferative levels to control15. Vemurafenib-resistant A375-M6 (VEM-R) cells were obtained by chronic administration of 2 μM vemurafenib (PLX4032; MedChemtronica AB, Stockholm, Sweden) until cells regained their proliferative activity (about 3 months from the beginning of the treatment)16. A375-M6 and WM266-4 grown in standard condition at pH 7.4 ± 0.1 have been used as control and referred to as A375-M6 wild type (WT) and WM266-4 WT. Everolimus (MedChemtronica) was used at 10 μM for 24 h. uPAR knockout (KO) A375-M6 and A375 cells were obtained via CRISPR-Cas9 technique as previously reported17, while uPAR rescue cells via stably transfecting uPAR KO cells with an Okayama–Berg vector containing uPAR cDNA18. The pharmacological uPAR blockage was obtained by treating for 48 h melanoma cells with 50 μM M25 (amino acidic sequence STYHHLSLGYMYTLN), a peptide produced in collaboration with the Peptide Facility at Biotechnology Center University of Padova (CRIBI), able to impair the integrin α chain–uPAR interaction, and thus blocking the subsequent signaling cascade16. As the control, cells were treated with a scrambled peptide (amino acidic sequence LSLYNMTSHTHGLYY).
Cell Viability
Trypan Blue Exclusion Assay
A375-M6 WT and VEM-R (1.5 × 105) were seeded in six-well plates and, the day after, treated with increasing doses of vemurafenib as indicated. Ninety-six hours later, 20 μl of cell suspension was incubated for 3 min at room temperature with an equal volume of 0.4% (w/v) trypan blue solution prepared in 0.81% NaCl and 0.06% (w/v) dibasic potassium phosphate. Viable (trypan blue-negative) and nonviable (trypan blue-positive) cells were counted at a light microscope using a dual-chamber hemocytometer.
Annexin V/Propidium Iodide (PI) Flow Cytometer Assay
A375-M6 WT and A375-M6 adapted to chronic acidosis were grown for 24 h in the presence or absence of 10 μM everolimus (MedChemtronica). Cells were harvested with Accutase (Euroclone), collected in flow cytometer tubes (1 × 105 cells/tube), washed in phosphate-buffered saline (PBS), and incubated for 15 min at 4°C in the dark with 100 μl of Annexin Binding buffer (100 mM HEPES, 140 mM NaCl, 25 mM CaCl2, pH 7.4), 1 μl of 100 μg/ml PI (Sigma-Aldrich, St. Louis, MO, USA) working solution, and 5 μl annexin V fluorescein isothiocyanate (FITC) conjugated (Immunotools, GmbH). Each sample was added with Annexin Binding Buffer to reach 500 μl volume/tube. Samples were then analyzed at BD FACSCanto II (BD Biosciences, Milan, Italy). Cellular distribution depending on annexin V and/or PI positivity allowed the measurement of the percentage of viable cells (annexin V- and PI-negative cells), early apoptosis (annexin V-positive and PI-negative cells), late apoptosis (annexin V- and PI-positive cells), and necrosis (annexin V-negative and PI-positive cells).
Cell Cycle Analysis
A375-M6 WT and A375-M6 VEM-R were enzymatically collected in a tube, centrifuged, and stained with a mixture of 50 μg/ml PI (Sigma-Aldrich), 0.1% trisodium citrate, and 0.1% NP-40 (or Triton X-100) in the dark at 4°C for 30 min. The stained cells were evaluated by flow cytometry (BD FACSCanto II) using red propidium–DNA fluorescence, and data were analyzed with ModFit analysis software.
In Vitro Tube Formation Assay
A total of 0.2 × 105 melanoma cells/well were plated in 2% FBS medium on 50 μl Matrigel (Miscio Rappresentanze S.A.S., Perugia, Italy)-precoated 96-well plate, incubated at 37°C up to 24 h, and pictures were acquired at regular intervals at EVOS optical microscope (Thermo Fisher Scientific, Monza, Italy). The Angiogenesis Analyzer tool of ImageJ software19 provided the statistical analysis for each experimental condition tested. “Nodes” are identified as pixels that had at least three neighbors, corresponding to a bifurcation. “Junctions” are elements composed of several nodes. “Segments” are elements delimited by two junctions. “Peaces” are the sum of the number of segments, isolated elements, and branches (where branches are defined as elements delimited by a junction and one extremity).
Western Blot Analysis
A375-M6 cells (WT, Chr.ac., VEM-R, uPAR KO, and uPAR-rescue) were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Merck Millipore, Milan, Italy) containing Pierce Protease Inhibitor Tablets (Thermo Fisher Scientific), protein concentration was measured with Bradford reagent (Merck Millipore), and equal amounts of protein were separated in Laemmli buffer on 8%–12% (v/v) SDS-PAGE gel (Thermo Fischer Scientific) and transferred to a polyvinylidene difluoride (PVDF) membrane using the iBlot 2 System (Thermo Fischer Scientific). Following 1 h of blocking with Odyssey blocking buffer (LI-COR® Bioscience, Milan, Italy), membranes were probed overnight at 4°C with the anti-EphA2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), the anti-GAPDH (Abcam, Cambridge, UK), and the anti-tubulin (Sigma-Aldrich, Milan, Italy). After that, membranes were incubated for 1 h at room temperature with goat anti-mouse IgG Alexa Fluor 680 antibody (Invitrogen) or goat anti-rabbit IgG Alexa Flour 750 antibody (Invitrogen, Life Technologies, Milan, Italy). Membranes were visualized by the Odyssey Infrared Imaging System (LI-COR® Bioscience).
Flow Cytometry Analysis
Cells were harvested by using Accutase (Euroclone), collected in flow cytometer tubes (2 × 105 cells/tube), and stained 1 h at 4°C with anti-uPAR R3 (Thermofisher Scientific) or VE-cadherin F-8 (sc-9989; Santa Cruz Biotechnology) (for VE-cadherin, a prior permeabilization step with 0.25% Triton X-100 PBS was required). Cells were washed in PBS and incubated for 1 h in the dark at 4°C with secondary antibodies conjugated with FITC (Merck Millipore). Samples were washed in PBS and then analyzed at BD FACSCanto II (BD Biosciences). The flow cytometer was calibrated using cells incubated with FITC-conjugated irrelevant IgG and a gate delineated to discriminate positive (on the right side of the gate) and negative (on the left side of the gate) cells. The whole VE-cadherin–FITC–positive cellular population has been considered for the quantification. For each sample, 1 × 104 events were analyzed.
Quantitative Real-Time Polymerase Chain Reaction (qPCR)
Total RNA was prepared using Tri Reagent (Sigma-Aldrich), agarose gel checked for integrity, and reverse-transcribed with iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. Selected genes were evaluated by a real-time qPCR with 7500 Fast Real-Time PCR System (Applied Biosystems, Monza, Italy). Fold change was determined by the comparative Ct method calculating the average of β-actin, β2-microglobulin, TATA-Box Binding Protein (TBP), and 18s, used as reference genes. Amplification was performed with the PCR setting: 40 cycles of 95°C for 15 s and 60°C for 60 s using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific). Primer sequences (IDT, Tema Ricerca, Bologna, Italy) are the following: uPAR, 5′-GGTCACCCGCCGCTG-3′ (forward) and 5′-CCACTGCGGGTACTGGACA-3′’ (reverse); β-actin, 5′-TCGAGCCATAAAAGGCAACT-3′ (forward) and 5′-CTTCCTCAATCTCGCTCTCG-3′ (reverse); β2-microglobulin, 5′-GCCGTGTGAACCATGTGACT-3′ (forward) and 5′-GCTTACATGTCTCGATCCCACTT-3′ (reverse); TBP, 5′-CAACAGCCTGCCACCTTAC-3′ (forward) and 5′-CTGAATAGGCTGTGGGGTC-3′ (reverse); 18s, 5′-CGCCGCTAGAGGTGAAATTCT-3′ (forward) and 5′-CGAAC-CTCCGACTTTCGTTCT-3′ (reverse).
Colony Formation Assay
A375-M6 WT and A375-M6 adapted to chronic acidosis were treated for 24 h with 2 μM vemurafenib and 50 μM M25 as a single or combined therapy. Cells (2 × 102) were seeded in a six-well plate and grown for 7 days. Developed colonies were fixed for 20 min in 4% paraformaldehyde at 4°C and stained for 30 min with crystal violet solution at room temperature.
Statistical Analysis
The experiments were performed at least four times for a reliable application of statistics. All samples used were included in the statistical analysis. Statistical analysis was performed by t-test, one-way analysis of variance (ANOVA), and two-way ANOVA with GraphPad Prism 6 software, as specified in each figure legend. Values are presented as mean ± standard deviation (SD). The p values are presented as *p < 0.05, **p < 0.01, ***p < 0.001. Values are presented as mean of independent experiments ± SD.
RESULTS
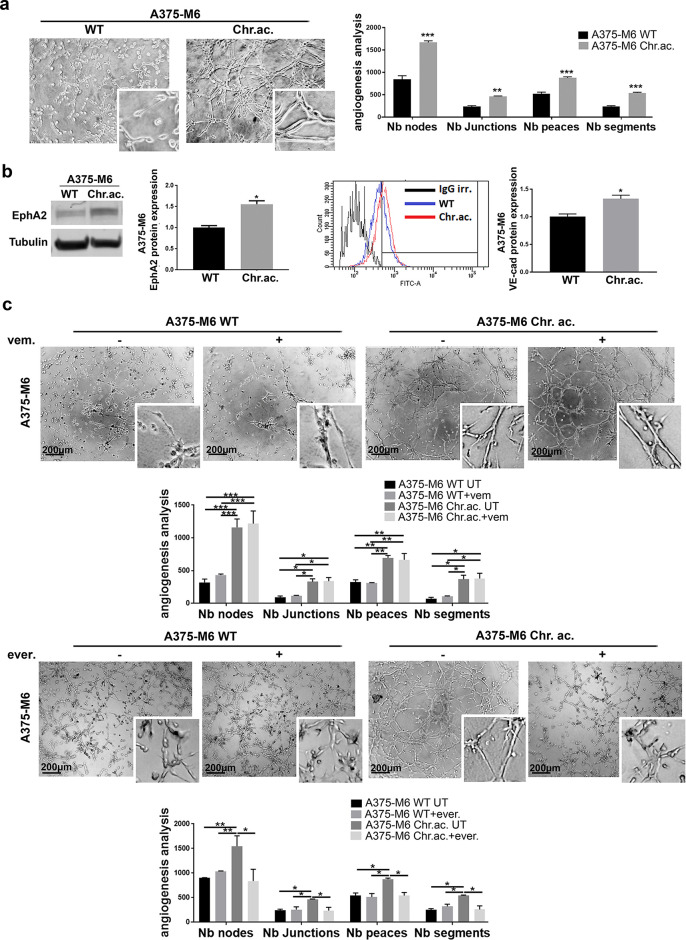
Extracellular Acidosis of Tumor Microenvironment Promotes VM in Melanoma Cells
To verify the connection between drug resistance and VM, we decided to study VM of melanoma cells exposed to an environmental condition known to induce drug resistance (i.e., extracellular acidosis). Indeed, extracellular acidosis, which characterizes the microenvironment of almost all solid cancers20, has been demonstrated to drive the acquisition of high-malignant tumor cell phenotype able to resist chemotherapy, radiotherapy, and target therapy, including vemurafenib15. By using an in vitro model of A375-M6 cells chronically adapted to extracellular acidosis, we observed that VM ability is strongly associated with acid-adapted cells, as reported by the increased number, compared to A375-M6 WT, of the nodes, junctions, peaces, and segments, which are parameters related to the vascular-like channel formation under each experimental condition (Fig. 1a). In line with this result, we observed an increased expression level of the VM markers EphA2 and VE-cadherin in acid-adapted A375-M6 cells compared to control cells (Fig. 1b). As expected, with acidic cancer cells being vemurafenib-resistant15, the treatment with vemurafenib did not affect VM in acid-adapted A375-M6, without affecting even the very low VM ability of sensitive A375-M6 WT (Fig. 1c, upper panel). Based on our experience, we decided to test the anti-VM ability of the mTOR inhibitor everolimus, since it has been already shown to target the acid-adapted vemurafenib-resistant melanoma cells15. Being an inhibitor of the PI3K/AKT/mTOR pathway, the effects exerted by everolimus on VM can also help the identification of the signaling pathway exploited by melanoma cells undergoing this alternative vessel formation program. Treating melanoma cells with a sublethal dose of the mTOR inhibitor everolimus, we obtained a significant VM impairment in acid-adapted A375-M6 (Fig. 1c, lower panel), together with a lowering of the expression of EphA2 and VE-cadherin (Supplementary Fig. S1, available at https://www.sbsc.unifi.it/vp-351-supplementary-material-rev.html). We ascertained that the VM inhibitory effect exerted by everolimus was not due to a cell viability impairment, as shown in Supplementary Fig. S2 (available at https://www.sbsc.unifi.it/vp-351-supplementary-material-rev.html) by annexin V/PI flow cytometer assay after 24 h of treatment. These findings were confirmed in the WM266-4 melanoma cell line, which when chronically exposed to extracellular acidosis acquired VM ability (completely absent in control cells) (Supplementary Fig. S3a, available at https://www.sbsc.unifi.it/vp-351-supplementary-material-rev.html). Everolimus treatment was able to reduce acidosis-induced VM even in WM266-4 cells, without altering at all control cells, unable to perform VM (Supplementary Fig. S3b, available at https://www.sbsc.unifi.it/vp-351-supplementary-material-rev.html). These data evidence that acid-adapted melanoma cells are capable of VM, suggesting a liaison between drug resistance and tumor-derived vascular channel formation. These results also let us hypothesize that VM might require the PI3K/AKT/mTOR pathway, selectively targeted by everolimus, rather than the BRAF/MEK/ERK one, impaired instead by vemurafenib.
Figure 1.
Extracellular acidosis induces VM in melanoma cells. (a) Representative pictures and relative quantification chart of capillary morphogenesis assay of A375-M6 wild type (WT) or chronically exposed to extracellular acidosis (chr.ac.). Two-way analysis of variance (ANOVA), GraphPad Prism. (b) Western blot (left) and flow cytometer analysis (right) with relative quantification charts of VM markers EphA2 and VE-cadherin of A375-M6 WT or chronically exposed to extracellular acidosis (chr.ac.). t-test, GraphPad Prism. (c) Representative pictures and relative quantification chart of capillary morphogenesis assay of A375-M6 WT or acid-adapted treated or not with 2 μM vemurafenib or 10 μM everolimus for 24 h. Two-way ANOVA, GraphPad Prism. Scale bar: 200 μm. *p < 0.05, **p < 0.01, ***p < 0.001.
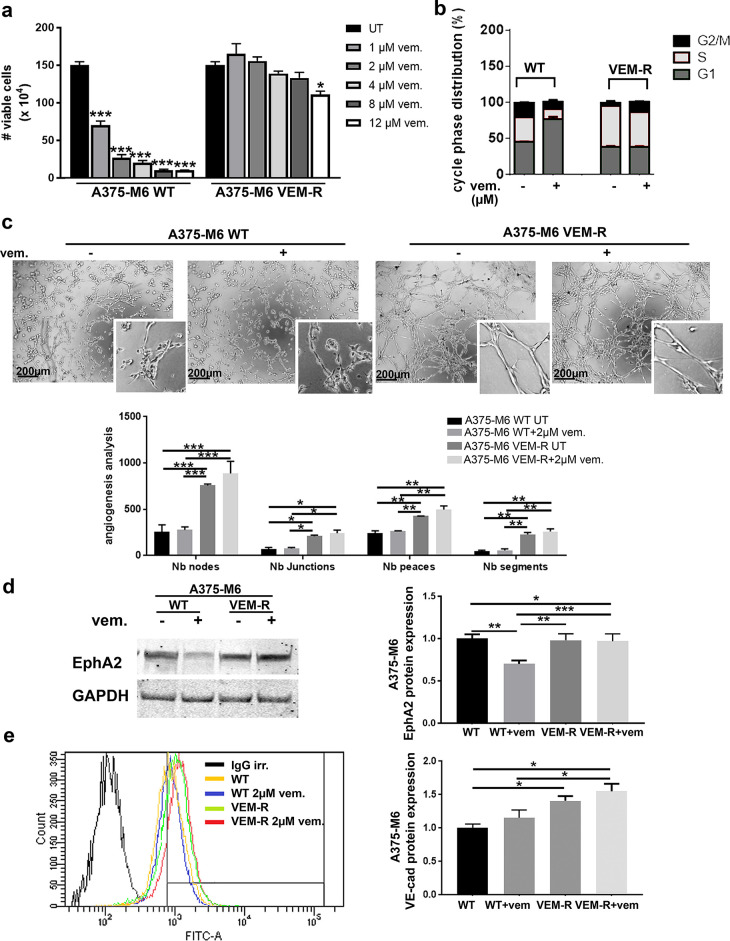
Vemurafenib-Resistant A375-M6 Cells Are Endowed With VM Ability
To verify the association of drug resistance and VM, we used a supplementary cellular model of induced vemurafenib resistance. Vemurafenib-resistant A375-M6 cells (hereafter called VEM-R), deeply characterized by Laurenzana and colleagues16, were tested for viability and cell cycle distribution upon vemurafenib treatment. VEM-R cells showed nonimpaired viability when treated with increasing doses of vemurafenib up to 8 μM, while a slight reduction was observed with 12 μM, which is far from the dose already effective on A375-M6 WT cells (Fig. 2a). Indeed, the cell cycle distribution shows that 2 μM vemurafenib treatment induced a cell cycle arrest (increased G1 with decreased S and G2/M phases) in A375-M6 WT cells, without significantly affecting VEM-R cells (Fig. 2b). These data confirm the goodness of A375-M6 VEM-R as an experimental model of vemurafenib resistance. We thus verified in vitro that VEM-R cells are endowed with a significantly higher ability to organize themselves in the capillary-like network compared to A375-M6 WT cells; see the increased number of angiogenesis parameters exploited by the ImageJ software (i.e., nodes, junctions, peaces, and segments) (Fig. 2c). The treatment with 2 μM vemurafenib did not alter the in vitro VM ability of both A375-M6 WT and A375-M6 VEM-R cells compared to the relatively untreated (UT) (Fig. 2c). A375-M6 WT and VEM-R cells showed comparable levels of EphA2 but responded differently to vemurafenib treatment since treated A375-M6 WT cells suffered a decreased EphA2 expression, while treated A375-M6 VEM-R displayed unaltered EphA2 levels compared to relative UT (Fig. 2d). VE-cadherin was instead upregulated in A375-M6 VEM-R compared to A375-M6 WT cells, and vemurafenib treatment did not affect its expression in both the cell lines used (Fig. 2e). Above all, these findings indicate that vemurafenib resistance in A375-M6 cells is accompanied by VM.
Figure 2.
Vemurafenib-resistant A375-M6 melanoma cells are capable of vasculogenic mimicry (VM). (a) Viability assay of A375-M6 WT and VEM-R cells treated with increasing doses of vemurafenib. One-way ANOVA, GraphPad Prism. (b) Cell cycle analysis of A375-M6 WT and VEM-R cells treated with 2 μM vemurafenib. One-way ANOVA, GraphPad Prism. (c) Representative pictures and relative quantification chart of capillary morphogenesis assay of A375-M6 WT and VEM-R cells in the presence or absence of 2 μM vemurafenib. Scale bar: 200 μm. Two-way ANOVA, GraphPad Prism. (d) Western blot of EphA2 and (e) flow cytometer analysis of VE-cadherin of A375-M6 WT and VEM-R cells treated or not with 2 μM vemurafenib. One-way ANOVA, GraphPad Prism. *p < 0.05, **p < 0.01, ***p < 0.001.
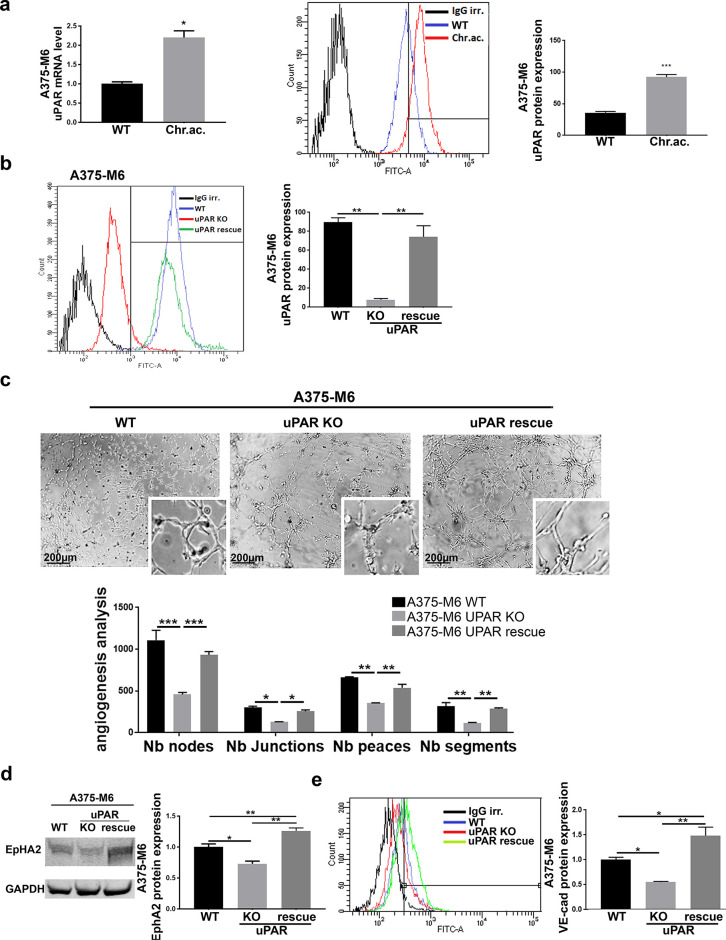
uPAR as a Central Driver of VM Acquisition by Drug-Resistant Melanoma Cells
As we recently reported, uPAR has been demonstrated to significantly contribute to the development of drug resistance in melanoma, and its loss of function is sufficient to restore vemurafenib sensitiveness in resistant cells16. Here we wondered whether uPAR is also involved in VM associated with drug resistance in our experimental conditions. First of all, we observed that uPAR is overexpressed in A375-M6 melanoma cells chronically exposed to extracellular acidosis (Fig. 3a), already shown to be vemurafenib-resistant15, reinforcing the evidence provided by Laurenzana and colleagues, linking uPAR with vemurafenib resistance. uPAR, despite not varying at the basal level, has indeed been shown to be differently expressed in WT and VEM-R cells after vemurafenib treatment: in particular, while WT cells significantly decreased uPAR protein expression upon vemurafenib treatment, VEM-R cells showed comparable uPAR level compared to the untreated counterpart16. Acidosis-induced uPAR overexpression was confirmed in WM266-4 melanoma cells (Supplementary Fig. S3c-d, available at https://www.sbsc.unifi.it/vp-351-supplementary-material-rev.html). To disclose the real involvement of uPAR in VM, we tested uPAR KO A375-M6 cells, genetically edited with the CRISPR/Cas9 technique17, and thus showing an abrogated expression of uPAR, that was then restored via a stable overexpression of the uPAR coding gene PLAUR in uPAR rescue A375-M6 cell line (Fig. 3b). This is a completely different cell model compared to the previous ones described above (i.e., acid-adapted A375-M6 and VEM-R A375-M6 cells), obtained by the uPAR gene editing of A375-M6 WT cells. By using this model, we observed an almost total abrogation of vascular-like channel formation in vitro in uPAR KO cells, which was instead restored along with uPAR expression in uPAR rescue cells (Fig. 3c). This finding was confirmed by the decreased expression of VM markers EphA2 (Fig. 3d) and VE-cadherin (Fig. 3e) in uPAR KO A375-M6 cells, which went back to the control level—and ever higher- in uPAR rescue cells. Similar effects were observed in the A375 cell line, where uPAR KO was accompanied with a significantly decreased vascular-like channel formation in vitro and then restored in the uPAR rescue A375 cell line (Supplementary Fig. S4, available at https://www.sbsc.unifi.it/vp-351-supplementary-material-rev.html). These results highlight a central role of uPAR, known especially for its proangiogenic functions, in VM, disclosing another possible way for uPAR to sustain tumor mass perfusion.
Figure 3.
Urokinase plasminogen activator receptor (uPAR) expression is required for VM in melanoma cells. (a) Real-time polymerase chain reaction (PCR) (left) and flow cytometer analysis (right) of uPAR expression in A375-M6 WT and acid-adapted cells. t-test, GraphPad Prism. (b) uPAR flow cytometer analysis (one-way ANOVA, GraphPad Prism) and (c) capillary morphogenesis assay (representative pictures on the top and quantification chart on the bottom; two-way ANOVA, GraphPad Prism) of WT, uPAR knockout (KO), and uPAR rescue A375-M6 cells maintained in standard condition. Scale bar: 200 μm. (d) Western blot of EphA2 (one-way ANOVA, GraphPad Prism) and (e) flow cytometer analysis of VE-cadherin (one-way ANOVA, GraphPad Prism) of WT, uPAR KO, and uPAR rescue A375-M6 cells. *p < 0.05, **p < 0.01, ***p < 0.001.
To verify the uPAR involvement in VM associated with drug resistance, we also tested the effects of a synthetic uPAR-blocking peptide called M25, acting as an uncoupler of integrin–uPAR interaction and thus preventing the subsequent signaling cascade. We tested M25 in both our experimental models of drug-resistant cells. We observed that M25 was able to abrogate the VM of VEM-R melanoma cells (Fig. 4a) and selectively inhibit tube formation in acidic A375-M6 cells, without affecting the VM of control cells (Fig. 4b). This was expected given the different uPAR expression in acid-adapted A375-M6 compared to standard cells. Further consideration should be done for the scenario presented by VEM-R cells, where uPAR expression level alone appears insufficient to explain differences in VM ability compared to WT: as previously reported, WT and VEM-R cells in basal condition, despite showing different VM ability, displayed similar uPAR level, but in the presence of vemurafenib treatment, WT almost halved uPAR expression while VEM-R maintained its levels high16. In this context, besides uPAR expression per se, there would be likely crucial interactions of uPAR with lateral partners, and this could be the reason why the M25 succeeded in preventing VM. Together with VM inhibition, M25 was able to restore drug sensitivity in resistant melanoma cells. This has been deeply demonstrated by Laurenzana and colleagues for the VEM-R A375-M6 melanoma cells16 and here confirmed even in the acidosis-induced resistance model. Both VEM-R (Fig. 4c) and acid-adapted (Fig. 4d) A375-M6 cells were indeed resensitized to the cytostatic effect of vemurafenib in the presence of M25, as shown by the number of viable cells and representative pictures of colony formation (colony formation assay of VEM-R cells is shown in Laurenzana et al.16). These data demonstrate that uPAR plays a central role in the VM associated with drug resistance in melanoma.
Figure 4.
uPAR inhibition by M25 blocking peptide impairs VM and drug resistance in resistant melanoma cells. (a) Representative pictures of capillary morphogenesis assay with relative quantification chart of A375-M6 VEM-R treated with vemurafenib and M25 peptide alone or in combination. Scale bar: 200 μm. Two-way ANOVA, GraphPad Prism. (b) Representative pictures of capillary morphogenesis assay with relative quantification chart of A375-M6 WT or acid-adapted treated with scramble or uPAR-blocking peptide M25. Scale bar: 200 μm. Two-way ANOVA, GraphPad Prism. (c) Cell growth of A375-M6 VEM-R cells treated 24 h with vemurafenib and M25 peptide alone or in combination. Two-way ANOVA, GraphPad Prism. (d) Cell growth (upper) and representative pictures of colony formation assay (lower) of A375-M6 WT and acid-adapted cells treated for 24 h with vemurafenib and M25 peptide as a single or combined therapy. Two-way ANOVA, GraphPad Prism. *p < 0.05, **p < 0.01, ***p < 0.001.
DISCUSSION
Prognosis of advanced malignant melanoma is always poor due to the development of chemo- and target therapy resistance and the concomitant wide tumor cell dissemination. The most predominant genetic alteration in melanoma cells is the V600E activating mutation in the BRAF gene, and the efficacy of vemurafenib (from V600E mutated BRAF inhibition) proves the critical role of this mutation in sustaining melanoma aggressiveness. Nevertheless, after a very limited period of therapy response, most patients rapidly relapse with a lethal drug-resistant disease5, so understanding such resistant phenotypes and their peculiarities is critical to offer a therapeutic option for these patients.
Very impressed by the devastating and rapid clinical relapse of resistant patients, we decided to investigate whether the vemurafenib-resistant melanoma phenotype drives VM ability, which is critical for cancer cell growth and dissemination and always correlates with poor prognosis11–13,21. Both drug resistance and VM can indeed be crucial features for the plasticity of advanced melanoma required for tumor progression toward metastatic disease. Vartanian and colleagues showed a clear association of VM with Anaroza resistance in melanoma since its inhibition was able to restore melanoma cell sensitiveness22. Similarly, Hori and colleagues recently reported an association between VM and trastuzumab (Tmz) resistance in HER2+ breast cancer, demonstrating that the loss of Tmz sensitivity leads to VM in breast cancer cells23. To test the association between vemurafenib resistance and VM, we used two independent human melanoma experimental models, the first one induced by a chronic adaptation of melanoma cells to low pH medium, and the second one by a continuous vemurafenib administration. Extracellular acidosis, a characteristic shared by almost all solid tumors20 and mainly caused by the Warburg metabolism of cancer cells24, is indeed known to favor, along with other aggressive features25–28, the development of therapy resistance in tumors; in particular, we recently reported that it induces vemurafenib resistance in BRAFV600E melanoma cells15. We now provide evidence that both the vemurafenib-resistant models used are characterized by high VM ability. To our knowledge, although much evidence is highlighting the role of tumor environmental components including hypoxia12,29,30, no prior studies have examined the contribution of the acidic tumor microenvironment to VM. We previously reported that the acidic tumor microenvironment stimulates in melanoma cells VEGF-c production, a factor that is strongly associated with lymphangiogenesis and lymph node metastasis, but also a key marker of VM31. Alongside the in vitro capillary morphogenesis assay, we evaluated the expression level of well-known VM markers, such as EphA2 and VE-cadherin32. In particular, VE-cadherin and EphA2 act in a coordinated manner as key regulatory elements in the VM formation in melanoma12,33. VE-cadherin indeed mediates the EphA2 localization at the intercellular junctions between VM tube-forming tumor cells. The kinase EphA2 activates through FAK, PI3K, and ERK1/2 pathways, which besides being associated with survival, proliferation, and migration, are also important in the VM process. Indeed, PI3K mediates MMP-14-driven activation of MMP-2, which in turn, by cutting laminin 5γ2, produces γ2 and γ2x fragments, which promote cell migration, crucial for tube formation34. Also, the PI3K/AKT signaling regulates the MMP-9 activity, contributing to the extracellular matrix (ECM) remodeling toward VM35. Hence, inhibiting the PI3K/AKT pathway may provide a new target for anti-VM therapy.
In line with these observations, our data suggest that VM might not strictly depend on the ERK pathway; VM would rather need the PI3K/AKT/mTOR pathway—still active in VEM-R16 and upregulated in acid-adapted melanoma cells15—since the treatment with the mTOR inhibitor everolimus can pull down the tube formation in the acidic tumor model. Interestingly, very recently, we also reported that the AKT pathway is significantly downregulated in uPAR KO melanoma cells17, which indeed have been shown here to be almost unable for VM, suggesting a key role of uPAR in VM. We further confirmed uPAR involvement in melanoma VM by using the blocking peptide M25, which showed an efficient VM inhibition in both the experimental models used. Accordingly, it has been reported that uPAR+ tumor cells promoted VM formation and tumor dissemination in lung cancer36, and collagen XVI NC11 domain to trigger VM in oral squamous cell carcinoma via uPAR upregulation37. This could represent a complementary role for the well-known proangiogenic activity of uPAR, since, by triggering also VM, it can better accomplish tumor mass sustainment even when/where endothelial cell-driven angiogenesis is impaired or just delayed. It is indeed known that VM can exist as it is or provide tumor perfusion just for the period needed by endothelial angiogenesis to substitute VM-derived vessels and perfuse the extremely rapidly growing tumor mass. A model has been proposed in which VM seems to play a major role in providing blood supply especially in the early stages; then, as the mass grows, tumor cells lining the VM vessel wall are little by little—but completely—replaced by endothelial cells, creating a middle transitional phase where both tumor and endothelial cells line the vessels (known as mosaic vessels)11. Besides its involvement in VM, we recently proved that uPAR overexpression in melanoma cells reduces the sensitivity to BRAF inhibition, and by targeting uPAR and EGFR interaction with the blocking peptide M25, we restored vemurafenib responsiveness in melanoma-resistant cells16. uPAR was also found to be inversely associated with disease-free survival of papillary thyroid cancer-bearing patients harboring BRAFV600E mutation, as uPAR correlates with lymph node metastasis, tumor node metastasis stage, and disease recurrences38.
We identify uPAR as the key mediator of the high aggressive melanoma phenotype endowed with vemurafenib resistance and VM ability, which we suggest as the “two sides of the same coin.” We thus purpose to consider uPAR targeting for the development of complementary therapy to restore drug sensitivity and abolish VM-driven tumor mass perfusion in resistant melanoma patients.
ACKNOWLEDGMENTS
This research was funded by Istituto Toscano Tumori. E.A. and A.B. were supported by Fondazione Associazione Italiana per la Ricerca sul Cancro AIRC fellowship for Italy.
REFERENCES
- 1. Davey MG, Miller N, McInerney NM. 2021. A review of epidemiology and cancer biology of malignant melanoma. Cureus 13(5):e15087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bisschop C, Ter Elst A, Bosman LJ, Platteel I, Jalving M, van den Berg A, Diepstra A, van Hemel B, Diercks GFH, Hospers GAP, Schuuring E. 2018. Rapid BRAF mutation tests in patients with advanced melanoma: Comparison of immunohistochemistry, droplet digital PCR, and the idylla mutation platform. Melanoma Res. 28(2):96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. 2002. Mutations of the BRAF gene in human cancer. Nature 417(6892):949–954. [DOI] [PubMed] [Google Scholar]
- 4. Sanchez JN, Wang T, Cohen MS. 2018. BRAF and MEK inhibitors: Use and resistance in BRAF-mutated cancers. Drugs 78(5):549–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arozarena I, Wellbrock C. 2017. Overcoming resistance to BRAF inhibitors. Ann Transl Med. 5(19):387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kakadia S, Yarlagadda N, Awad R, Kundranda M, Niu J, Naraev B, Mina L, Dragovich T, Gimbel M, Mahmoud F. 2018. Mechanisms of resistance to BRAF and MEK inhibitors and clinical update of US Food and Drug Administration-approved targeted therapy in advanced melanoma. Onco Targets Ther. 11:7095–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maniotis AJ, Folberg R, Hess A, Seftor EA, Gardner LM, Pe’er J, Trent JM, Meltzer PS, Hendrix MJ. 1999. Vascular channel formation by human melanoma cells in vivo and in vitro: Vasculogenic mimicry. Am J Pathol. 155(3):739–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seftor RE, Seftor EA, Koshikawa N, Meltzer PS, Gardner LM, Bilban M, Stetler-Stevenson WG, Quaranta V, Hendrix MJ. 2001. Cooperative interactions of laminin 5 gamma2 chain, matrix metalloproteinase-2, and membrane type-1-matrix/metalloproteinase are required for mimicry of embryonic vasculogenesis by aggressive melanoma. Cancer Res. 61(17):6322–6327. [PubMed] [Google Scholar]
- 9. Seftor EA, Meltzer PS, Schatteman GC, Gruman LM, Hess AR, Kirschmann DA, Seftor RE, Hendrix MJ. 2002. Expression of multiple molecular phenotypes by aggressive melanoma tumor cells: Role in vasculogenic mimicry. Crit Rev Oncol Hematol. 44(1):17–27. [DOI] [PubMed] [Google Scholar]
- 10. Maniotis AJ, Chen X, Garcia C, DeChristopher PJ, Wu D, Pe’er J, Folberg R. 2002. Control of melanoma morphogenesis, endothelial survival, and perfusion by extracellular matrix. Lab Invest. 82(8):1031–1043. [DOI] [PubMed] [Google Scholar]
- 11. Zhang X, Zhang J, Zhou H, Fan G, Li Q. 2019. Molecular mechanisms and anticancer therapeutic strategies in vasculogenic mimicry. J Cancer 10(25):6327–6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Delgado-Bellido D, Serrano-Saenz S, Fernández-Cortés M, Oliver FJ. 2017. Vasculogenic mimicry signaling revisited: Focus on non-vascular VE-cadherin. Mol Cancer 16(1):65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cao Z, Bao M, Miele L, Sarkar FH, Wang Z, Zhou Q. 2013. Tumour vasculogenic mimicry is associated with poor prognosis of human cancer patients: A systemic review and meta-analysis. Eur J Cancer 49(18):3914–3923. [DOI] [PubMed] [Google Scholar]
- 14. Andreucci E, Peppicelli S, Ruzzolini J, Bianchini F, Biagioni A, Papucci L, Magnelli L, Mazzanti B, Stecca B, Calorini L. 2020. The acidic tumor microenvironment drives a stem-like phenotype in melanoma cells. J Mol Med. (Berl) 98(10):1431–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ruzzolini J, Peppicelli S, Andreucci E, Bianchini F, Margheri F, Laurenzana A, Fibbi G, Pimpinelli N, Calorini L. 2017. Everolimus selectively targets vemurafenib resistant BRAFV600E melanoma cells adapted to low pH. Cancer Lett. 408:43–54. [DOI] [PubMed] [Google Scholar]
- 16. Laurenzana A, Margheri F, Biagioni A, Chillà A, Pimpinelli N, Ruzzolini J, Peppicelli S, Andreucci E, Calorini L, Serratì S, Del Rosso M, Fibbi G. 2019. EGFR/uPAR interaction as druggable target to overcome vemurafenib acquired resistance in melanoma cells. EBioMedicine 39:194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Biagioni A, Laurenzana A, Chillà A, Del Rosso M, Andreucci E, Poteti M, Bani D, Guasti D, Fibbi G, Margheri F. 2020. uPAR knockout results in a deep glycolytic and OXPHOS reprogramming in melanoma and colon carcinoma cell lines. Cells 9(2):308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pucci M, Fibbi G, Magnelli L, Del Rosso M. 2001. Regulation of urokinase/urokinase receptor interaction by heparin-like glycosaminoglycans. J Biol Chem. 276(7):4756–4765. [DOI] [PubMed] [Google Scholar]
- 19. Carpentier G, Berndt S, Ferratge S, Rasband W, Cuendet M, Uzan G, Albanese P. 2020. Angiogenesis analyzer for ImageJ—A comparative morphometric analysis of “endothelial tube formation assay” and “fibrin bead assay”. Sci Rep. 10(1):11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reshetnyak YK, Yao L, Zheng S, Kuznetsov S, Engelman DM, Andreev OA. 2011. Measuring tumor aggressiveness and targeting metastatic lesions with fluorescent pHLIP. Mol Imaging Biol. 13(6):1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Treps L, Faure S, Clere N. 2021. Vasculogenic mimicry, a complex and devious process favoring tumorigenesis—Interest in making it a therapeutic target. Pharmacol Ther. 223:107805. [DOI] [PubMed] [Google Scholar]
- 22. Vartanian A, Baryshnikova M, Burova O, Afanasyeva D, Misyurin V, Bely vsky A, Shprakh Z. 2017. Inhibitor of vasculogenic mimicry restores sensitivity of resistant melanoma cells to DNA-damaging agents. Melanoma Res. 27(1):8–16. [DOI] [PubMed] [Google Scholar]
- 23. Hori A, Shimoda M, Naoi Y, Kagara N, Tanei T, Miyake T, Shimazu K, Kim SJ, Noguchi S. 2019. Vasculogenic mimicry is associated with trastuzumab resistance of HER2-positive breast cancer. Breast Cancer Res. 21(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vander Heiden MG, Cantley LC, Thompson CB. 2009. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324(5930):1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Webb BA, Chimenti M, Jacobson MP, Barber DL. 2011. Dysregulated pH: A perfect storm for cancer progression. Nat Rev Cancer 11(9):671–677. [DOI] [PubMed] [Google Scholar]
- 26. Corbet C, Feron O. 2017. Tumour acidosis: From the passenger to the driver’s seat. Nat Rev Cancer 17(10):577–593. [DOI] [PubMed] [Google Scholar]
- 27. Gatenby RA, Gillies RJ. 2008. A microenvironmental model of carcinogenesis. Nat Rev Cancer 8(1):56–61. [DOI] [PubMed] [Google Scholar]
- 28. Lardner A. 2001. The effects of extracellular pH on immune function. J Leukoc Biol. 69(4):522–530. [PubMed] [Google Scholar]
- 29. Wang HF, Wang SS, Zheng M, Dai LL, Wang K, Gao XL, Cao MX, Yu XH, Pang X, Zhang M, Wu JB, Wu JS, Yang X, Tang YJ, Chen Y, Tang YL, Liang XH. 2019. Hypoxia promotes vasculogenic mimicry formation by vascular endothelial growth factor A mediating epithelial–mesenchymal transition in salivary adenoid cystic carcinoma. Cell Prolif. 52(3):e12600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang JG, Zhou HM, Zhang X, Mu W, Hu JN, Liu GL, Li Q. 2020. Hypoxic induction of vasculogenic mimicry in hepatocellular carcinoma: Role of HIF-1α, RhoA/ROCK and Rac1/PAK signaling. BMC Cancer 20(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peppicelli S, Bianchini F, Contena C, Tombaccini D, Calorini L. 2013. Acidic pH via NF-κB favours VEGF-C expression in human melanoma cells. Clin Exp Metastasis 30(8):957–967. [DOI] [PubMed] [Google Scholar]
- 32. Kirschmann DA, Seftor EA, Hardy KM, Seftor RE, Hendrix MJ. 2012. Molecular pathways: Vasculogenic mimicry in tumor cells: Diagnostic and therapeutic implications. Clin Cancer Res. 18(10):2726–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Topczewska JM, Postovit LM, Margaryan NV, Sam A, Hess AR, Wheaton WW, Nickoloff BJ, Topczewski J, Hendrix MJ. 2006. Embryonic and tumorigenic pathways converge via Nodal signaling: Role in melanoma aggressiveness. Nat Med. 12(8):925–932. [DOI] [PubMed] [Google Scholar]
- 34. Andonegui-Elguera MA, Alfaro-Mora Y, Cáceres-Gutiérrez R, Caro-Sánchez CHS, Herrera LA, Díaz-Chávez J. 2020. An overview of vasculogenic mimicry in breast cancer. Front Oncol. 10:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chiablaem K, Lirdprapamongkol K, Keeratichamroen S, Surarit R, Svasti J. 2014. Curcumin suppresses vasculogenic mimicry capacity of hepatocellular carcinoma cells through STAT3 and PI3K/AKT inhibition. Anticancer Res. 34(4):1857–1864. [PubMed] [Google Scholar]
- 36. Li Y, Sun B, Zhao X, Zhang D, Wang X, Zhu D, Yang Z, Qiu Z, Ban X. 2015. Subpopulations of uPAR+ contribute to vasculogenic mimicry and metastasis in large cell lung cancer. Exp Mol Pathol. 98(2):136–144. [DOI] [PubMed] [Google Scholar]
- 37. Bedal KB, Grässel S, Spanier G, Reichert TE, Bauer RJ. 2015. The NC11 domain of human collagen XVI induces vasculogenic mimicry in oral squamous cell carcinoma cells. Carcinogenesis 36(11):1429–1439. [DOI] [PubMed] [Google Scholar]
- 38. Ulisse S, Baldini E, Sorrenti S, Barollo S, Prinzi N, Catania A, Nesca A, Gnessi L, Pelizzo MR, Mian C, De Vito C, Calvanese A, Palermo S, Persechino S, De Antoni E, D’Armiento M. 2012. In papillary thyroid carcinoma BRAFV600E is associated with increased expression of the urokinase plasminogen activator and its cognate receptor, but not with disease-free interval. Clin Endocrinol. (Oxf) 77(5):780–786. [DOI] [PubMed] [Google Scholar]