Abstract
Abnormal cell proliferation caused by abnormal transcription regulation mechanism seems to be one of the reasons for the progression of breast cancer and also the pathological basis. MicroRNA-142-5p (miR-142-5p) is a low-expressed miRNA in breast cancer. The role of MKL-1’s regulation of DNMT1 in breast cancer cell proliferation and migration is still unclear. MKL-1 (myocardin related transcription factor A) can bind to the conserved cis-regulatory element CC (A/T) 6GG (called CarG box) in the promoter to regulate the transcription of miR-142-5p. The expressions of miR-142-5p and MKL-1 are positively correlated. In addition, it has been proved that DNMT1 is the target of miR-142-5p, which inhibits the expression of DNMT1 by targeting the 3′-UTR of DNMT1, thereby forming a feedback loop and inhibiting the migration and proliferation of breast cancer. Our data provide important and novel insights into the MKL-1/miR-142-5p/DNMT1/maspin signaling pathway and may become a new idea for breast cancer diagnosis, treatment, and prognosis.
Key words: DNMT1, Maspin, miR-142-5p, MKL-1, Breast cancer
INTRODUCTION
Breast cancer is a common malignant tumor with high morbidity and mortality1, and poor prognosis is the main cause of death of breast cancer patients2. Hundreds of miRNAs have been reported to be involved in the occurrence of cancer and play specific roles in tumor growth and metastasis3. In addition, it is generally believed that miRNAs can regulate the function of cell signaling pathways by binding to the pairing sites on the 3′-untranslated region (3′-UTR) of potential target mRNAs, which belongs to posttranscriptional inhibition and regulation of mRNA expression4. With the development of molecular biology of tumor genes, it has been found that epigenetic regulatory mechanisms such as DNA methylation play an important role in the occurrence and development of many tumors, including breast cancer5. DNA methyltransferase 1 (DNMT1) maintains gene methylation during DNA replication and is the main epigenetic modification pathway in mammals6. Promoter methylation of the maspin gene can cause maspin gene silencing in breast, thyroid, skin, and colon cancers7. Interestingly, pharmacological methods of DNA demethylation usually fail to activate gene expression, which indicates that the reexpression of certain genes requires the search for new transcription factors in addition to drugs that inhibit insufficient DNA methylation8. Previous studies have shown that DNMT1 is necessary to maintain tumor stem cells in a variety of cancers, including prostate cancer, pancreatic cancer, and breast cancer9. In addition to demethylation, DNMT1 has other regulatory mechanisms. miR-142-5p directly regulates the expression of DNMT1 and regulates the proliferation and migration of gastric cancer cells through DNMT110–12. The role of miR-142-5p in the development and metastasis of breast cancer has also been partially studied13. miR-142-5p can regulate the proliferation and apoptosis of breast cancer cells14, but its mechanism is still unclear. Myocardin-related transcription factor A (MKL-1), also known as MRTF-A, is a member of the myocardin-related transcription factor family and a unique coactivator of SRF (serum response factor). The protein binds to the conserved cis-regulatory element CC (A/T) 6GG (called CarG box) to regulate the transcription of target genes. It plays an important role in the growth and development of organisms15. According to reports, MKL-1 can be used as an effective inducer of growth arrest and differentiation of certain tumors and is often inhibited during malignant transformation of tumors16. We believe that the opposite mechanism may be true, and it may just be the positive and negative regulation of cell proliferation by MKL-1 under different conditions. In addition, it may be related to different upstream signals of the MKL-1 pathway. The transcription regulation of myocardium and smooth muscle cells is the current research hotspot17, and it participates in the regulation of cell proliferation and migration. However, its role in the proliferation of breast cancer development and its role in breast cancer miRNA regulation are not clear. In our previous study, we demonstrated that miR-142-5p is a down-expressed miRNA in breast cancer and significantly related to advanced tumor grade (grade III)18. The abnormal expression of miR-142-5p plays a role of tumor suppressor gene in the occurrence of breast cancer. In this study, we report that miR-142-5p modulates tumor growth in breast cancer and identify DNMT1 as a direct target of miR-142-5p. Our data reveal a novel regulatory circuit for breast cancer cell growth and prove that miR-142-5p inhibits cell invasion and migration by targeting DNMT1 in breast cancer, enriching experimental data for finding potential targets for gastric cancer treatment.
MATERIALS AND METHODS
Cell Source
MDA-MB-231, MCF-7, and 293T cell lines were purchased from Shanghai Cell Bank in China. They were frozen at −80°C in DMEM medium containing 10% fetal bovine serum (BI, Israel), and 1% penicillin–streptomycin was added using the medium. They were then cultured in humid air at 37°C, containing 5% CO2.
Luciferase Report Experiment
293T, MDA-MB-231, and MCF-7 were seeded on 24-well plates (two replicates per group) at 2 × 104/ml cells and transfected 12 h later. Forty-eight hours after transfection, a microplate reader (Promega, Madison, WI, USA) was used to measure the luciferase reporter gene. The measured fluorescence value of each sample was normalized to luciferase activity.
Antibodies, Western Blot
Antibodies: rabbit DNMT1 (ab188453) (RabMAb; Abcam, Cambridge, UK), rabbit phospho-STAT3 (Ser727) (AF3294) (PcAb; Affbiotech, Suzhou, China), rabbit MKL-1 (ab115319) (PcAb; Abcam), and mouse GAPDH (sc-51907) (mAbs; Santa Cruz, Dallas, TX, USA). The specific steps of the Western blotting experiment are as described antecedently19. The membranes were visualized by GelDoc XR+ Imaging System (Bio-Rad, Hercules, CA, USA), and the image was examined using ImageJ software. The antibodies used are as follows: GAPDH (1:1500; Santa Cruz), DNMT1 (1:1500; ABclonal, Boston, MA, USA), MKL-1 (1:1500; Cell Signaling Technology/CST, Danvers, MA, USA), and maspin (1:1500; ABclonal).
Cell Scratch Test and Transwell Test
Cell migration ability was analyzed by cell scratch test and Transwell test. Photographs of cells were sensed at 0 and 24 h after scratch test of each experiment, and gap closure was determined on phase-contrast images by using the time lapse. Cell motility was experimented in an 8-μm-pore polycarbonate membrane (Corning, Shanghai, China) by Transwell test.
Cell Proliferation and Cycle Experiments
The EdU (5-ethynyl-2′-deoxyuridine)-labeling procedure was previously reported20,21. Water-soluble CCK-8 [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium sodium salt] was used to detect cell activity at 570 nm. The cell cycle assay was performed using the Annexin V-FITC/PI Apoptosis Detection Kit (Meilunbio, Jiangsu Province, China). Experimental results were obtained using Flowjo software for analysis and mapping.
Plasmid Construction and Transfection
The coding region sequences of MKL-1 gene was amplified by polymerase chain reaction (PCR) using the cDNA of breast cancer cells as the template [MKL-1: 5′- GACGACGATGACAAAGAATTCATGCCGCCTTTGAAAAGTCCAGCCGCAT-3′ (forward) and 5′-TTGACTAGTGGATCCGAATTCCTACAAGCAGGAATCCCAGTGCAGCTGCAA-3′ (reverse)] and inserted into mammalian expression vector pcDNA3.1, respectively. The promoter sequences of miR-142-5p (−1048∼+111) were amplified by PCR [WT-miR-142-5p-Luc: 5′-CGAGCTCTTACGCGTGCTAGCAAAGCGCCTTGAAACCCTCTC-3′ (forward) and 5′-AGATCTCGAGCCCGGGCTAGCACCCTCCAGTGCTGTTAGTAGTGC-3′ (reverse)] and inserted into the luciferase vector pGL3-Basic using mammalian genome as template. MUT-miR-142-5p-Luc (the binding site of MKL-1 was mutated): 5′-ACATGGCCAAACATTTTGTTGGGATAGCCTTGGGCTCCTGCC-3′ (forward) and 5′-GGCAGGAGCCCAAGGCTATCCCAACAAAATGTTTGGCCATGT-3′ (reverse). The DNMT1 3′-UTR sequence was amplified by PCR [WT-pmir-DNMT1: 5′-ATTGCTAGCTTCTGCCCTCCCGTCACCCCTGTTT-3′ (forward) and 5′-GCCGTCTAGATGGTTTATAGGAGAGATTTA-3′ (reverse)] and inserted into dual-luciferase miRNA target expression vector pmirGLO using the mammalian genome as the template; as for MUT-pmir-DNMT1, the binding site of miR-142-5p was mutated. Since one site on DNMT1 3′-UTR bound to miR-142-5p, we made one mutation, using the following primers: MUT-pmir-DNMT1, 5′-GAGTGGAAATTAAGGTCGCCGGTAGTTTTTATATGTTGTAATATTT-3′ (forward) and 5′-ACATATAAAAACTACCGGCGACCTTAATTTCCACTCATACAGTGGTAGAT-3′ (reverse). All plasmids and vectors in the experiment were purified using commercial plasmid purification kits. MDA-MB-231 and MCF-7 cell lines (1 × 105/μl) were inoculated into a six-well plate, and when it grew to about 60%, the plasmid or si-RNA and PEI were transfected at a ratio of 1:3 into the cells.
Extraction of RNA Reverse Transcription and qPCR
The RNA was quantified, and then genomic DNA contamination removal and first-strand cDNA synthesis were performed according to the Monad reverse transcription kit. The synthesized cDNA was subjected to qPCR. The primers involved are as follows: GAPDH, 5′-CAATGACCCCTTCATTGACC-3′ (forward) and 5′-GACAAGCTTCCCGTTCTCAG-3′ (reverse); maspin, 5′-CTTGCCTGTTCCTTTTCCAC-3′ (forward) and 5′-TGGAGAGAAGAGGACATTGC-3′ (reverse); and DNMT1, 5′-CCTAGCCCCAGGATTACAAGG-3′ (forward) and 5′-ACTCATCCGATTTGGCTCTTTC-3′ (reverse).
siRNA Gene Silencing and MicroRNA Transient Transfection
The online prediction programs of microRNAs, including TargetScan (http://www.targetscan.org), PicTar (https://pictar.mdc-berlin.de/), and miRanda (https://www.microrna.org/microrna/home.do), were used to predict microRNAs associated with CX3CL1. All microRNAs, siRNAs, and their corresponding control (NC) for RNA transfection were designed and purchased from Guangzhou RIBOBIO Co. (Guangdong, China). The transfection efficiency was analyzed and confirmed by additional biological tests. The suitable transfection reagent was Lipofectamine® 2000 Reagent (Thermo Fisher, Waltham, MA, USA) according to the appropriate experimental groups and manufacturer’s protocol. A total of 1 × 106 cells were mixed with 100 pmol of siRNA in 100 μl of 37°C DMEM, and then subsequently incubated for 6 h. The media was displaced with regular culture medium for 48 h. The effectiveness of transfection was assessed by siRNA-cy3-control expression.
Immunohistochemistry (IHC) Experiment
The specific steps of the IHC experiment are as described antecedently22. We used streptavidin alkaline phosphatase (SABC-AP) immunohistochemical staining kit (BOSTER Biological, Pleasanton, CA, USA) for IHC experiments. Hematoxylin lightly counterstained the nucleus, and it was then sealed with neutral resin and observed under a microscope. All the above steps were carried out in strict accordance with the manufacturer’s instructions.
Human Tumor Xenograft Model
The animal experiment program obtained the consent of the Experimental Animal Center of Wuhan University of Science and Technology and the Committee of Experimental Animal Ethics Review. BALB/c-nu (nude) mice were obtained from Beijing Vital River (Charles River Laboratories, Xi’an, SHAANXI, China). Four-week-old nude mice were randomly divided into four groups, and 1 × 107 MCF-7 cells stably overexpressing miR-142-5p and a control group were injected subcutaneously separately; thus, we obtained three groups: group A mice were injected with MCF-7 cells stably overexpressing miR-142-5p (miR-142-5p-pLKO.1), group B mice were injected with control groups (pLKO.1), and group C mice were injected with normal saline (blank). All nude mice were euthanized after 35 days. The tumor tissue was carefully removed from the nude carcass to count the volume and weight of the tumor.
Statistical Analysis
All the experiments were completed three times independently. The values were presented as the mean ± standard deviation. Two-sided values of p < 0.05 were considered statistically significant.
RESULTS
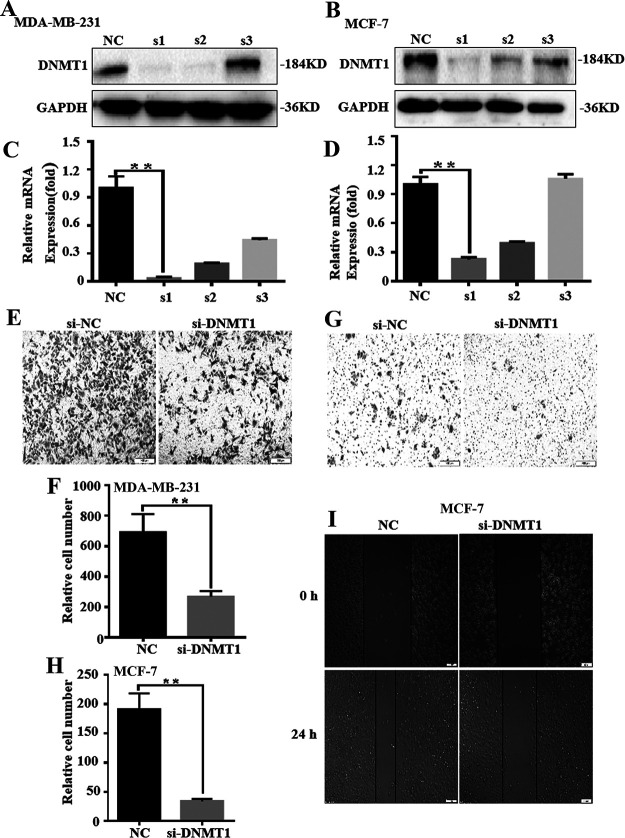
DNMT1 Inhibits the Expression of Maspin and Regulates the Proliferation and Migration of Breast Cancer Cells
The expression of DNMT1 mRNA and protein in breast cancer cells is higher than that of normal breast cells and associated with poor breast cancer survival23. The small interfering RNA (siRNA)-mediated knockdown of DNMT1 results in an increase in the expression of maspin in MDA-MB-231 cells, and confirmed that the proliferation and invasion ability of breast cancer cells were significantly inhibited by the downregulation of DNMT124. We suspect that the downregulation of DNMT1 may also affect the migration ability of breast cancer cells. To verify this conjecture, we designed three siRNAs for DNMT1 and verified the knockdown efficiency of siRNA in two breast cancer cell lines: MDA-MB-231 (Fig. 1A and C) and MCF-7 (Fig. 1B and D). The results prove that the first siRNA knockdown effect is the best in both cell lines, as shown in Figure 1A–D, so the first siRNA was selected in subsequent experiments. Transwell experiments proved that in the MCF-7 cell line, after knocking down DNMT1, the cell migration ability was significantly reduced to 20% (Fig. 1E and F). In the MDA-MB-231 cell line, after knocking down DNMT1, the cell migration ability was significantly reduced to 39% (Fig. 1G and H). The healing ability of the cells in the DNMT1 knockdown group was also significantly lower than that of the control group in the scratch test in MCF-7 (Fig. 1I). This proves that DNMT1 can indeed regulate the migration ability of breast cancer cells.
Figure 1.
DNA methyltransferase 1 (DNMT1) inhibits the expression of maspin and regulates the proliferation and migration of breast cancer cells. MDA-MB-231 and MCF-7 after serum starvation [DMEM + 0.5% fetal bovine serum (FBS)] for 48 h were transfected with si-control (si-NC) or si-DNMT1 for 48 h. (A–D) The effects of si-DNMT1 on DNMT1 expression in MDA-MB-231 and MCF-7 were detected by Western blot analysis. All experiments were repeated three times in duplicate, and the semiquantitative analysis was calculated by SPSS statistical software. **p < 0.01. (E–I) The effect of knocked down DNMT1 on MDA-MB-231 and MCF-7 migration was detected by the Transwell assay. **p < 0.01. The wound healing assay was also used to detect the migration-stimulating effects of knocked down DNMT1 on MDA-MB-231 and MCF-7. The number of cells migrated to the lower side of the Transwell chambers was counted and photographed in five fields (the upper, the lower, the left, the right, and the middle) of three independent experiments, and the migration capacity was calculated by SPSS statistical software. **p < 0.01.
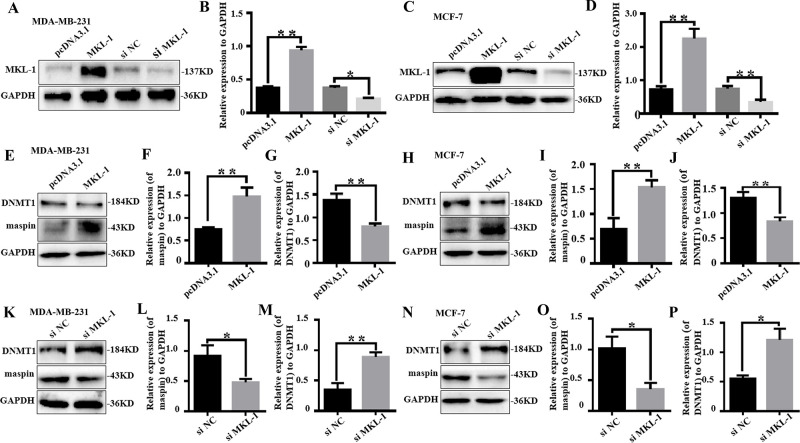
MKL-1 Affects the Expression of DNMT1 and Maspin
Studies have shown that MKL-1 inhibits cell proliferation and affects the G1–S phase of cell progression, resulting in cell growth arrest, and the expression of MKL-1 in tumor cells is significantly downregulated25. After MKL-1 was overexpressed or knocked out in two breast cancer cells, MDA-MB-231 and MCF-7, they were detected by Western blotting; the overexpression effect of MKL-1 in MDA-MB-231 and MCF-7 cells was significantly higher than that in the control group. The knockdown effect of siMKL-1 on MKL-1 is obviously in the two kinds of cells (Fig. 2A–D). Overexpression of MKL-1 in MDA-MB-231 was increased, the expression of DNMT1 protein was decreased to 54% (Fig. 2E and F), but the expression of maspin protein was increased by 2.14 times (Fig. 2E and G). Overexpression of MKL-1 in MCF-7 in the expression of maspin protein was increased by 2.06 times (Fig. 2H and I), but the expression of DNMT1 protein was decreased to 66% (Fig. 2H and J). After knocking down MKL-1 in MDA-MB-231, the protein expression of maspin decreased to 55% compared with the control group, while the protein expression of DNMT1 was upregulated 2.24 times (Fig. 2K–M). After knocking down MKL-1 in MCF-7, the protein expression of maspin was decreased to 35% compared with the control group, while the protein expression of DNMT1 was upregulated by 2.36 times. This indicates that the expression of DNMT1 and maspin in breast cancer cells may be related to MKL-1, and MKL-1 may regulate the expression of DNMT1 and maspin in some way (Fig. 2N–P).
Figure 2.
MKL-1 affects the expression of DNMT1 and maspin. MDA-MB-231 and MCF-7 after serum starvation (DMEM + 0.5% FBS) for 48 h were transfected with si-control (si-NC) or si-DNMT1 for 48 h. (A–D) The effect of MKL-1 overexpression or knock down in MDA-MB-231 and MCF-7 were detected by Western blot analysis. All experiments were repeated three times in duplicate, and the semiquantitative analysis was calculated by SPSS statistical software. *p < 0.05, **p < 0.01. (E–J) The expression of maspin and DNMT1 in MDA-MB-231 and MCF-7 transfected by MKL-1 overexpression plasmid was detected by Western blot analysis. All experiments were repeated three times in duplicate, and the semiquantitative analysis was calculated by SPSS statistical software. **p < 0.01. (K–P) The expression of maspin and DNMT1 in MDA-MB-231 and MCF-7 transfected by siMKL-1 was detected by Western blot analysis. All experiments were repeated three times in duplicate, and the semiquantitative analysis was calculated by SPSS statistical software. *p < 0.05, **p < 0.01. Data are expressed as mean ± standard deviation (SD). *p < 0.05, **p < 0.01.
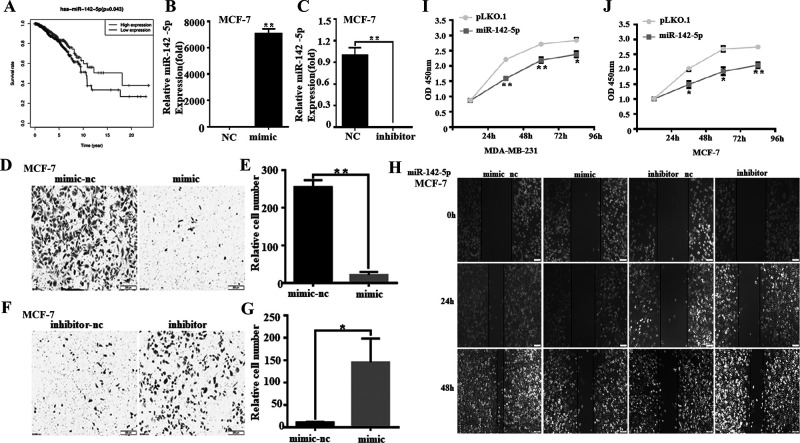
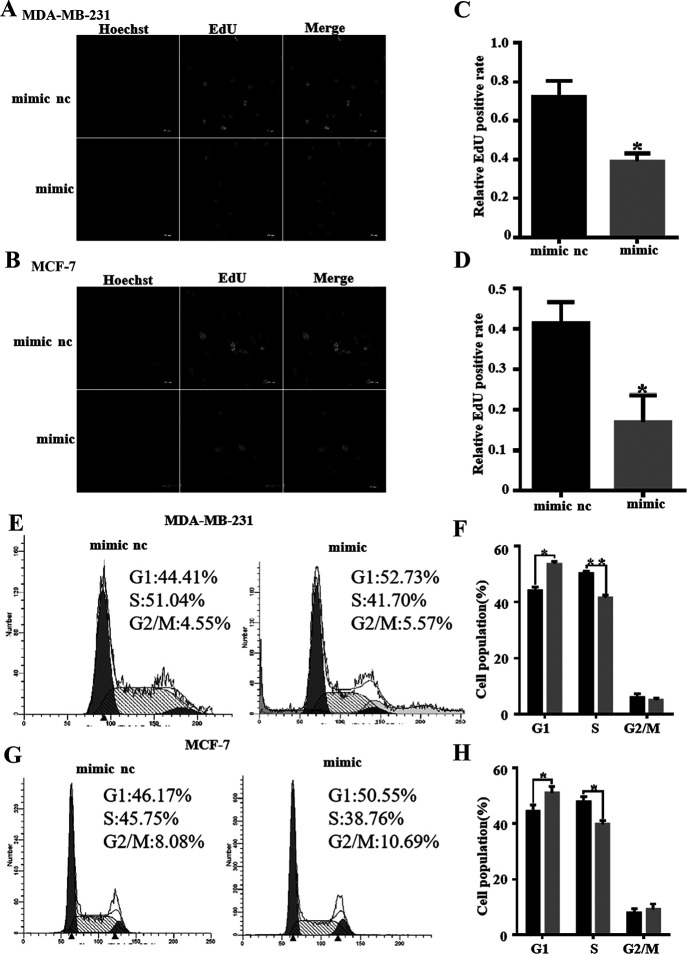
miR-142-5p Inhibits the Migration and Proliferation of Breast Cancer Cells and Regulates the Cell Cycle
Previous studies have shown that miR-142-5p acts as a tumor suppressor gene in a variety of tumors and promotes tumor metastasis of gastric cancer26. TCGA analysis found that the expression of miR-142-5p is related to the survival rate of patients, and the high expression of miR-142-5p can improve the survival rate of patients (Fig. 3A). MDA-MB-231 and MCF-7 were transfected with miR-142-5p mimic to overexpress or inhibit to knock out miR-142-5p, and the transfection efficiency was detected by miRNA reverse transcription (RT)-PCR (Fig. 3B and C). Transwell results proved that after transfection of miR-142-5p simulants in MCF-7, the mimic group’s cell migration ability was significantly reduced compared with the control group (Fig. 3D and E). The migration ability of inhibitor group cells was significantly improved (Fig. 3F and G). The number of cells migrated in the mimic group was remarkably decreased (90%), and the inhibitor group was significantly increased 6.63 times (Fig. 3H). This indicates that miR-142-5p can indeed inhibit the migration of breast cancer cells. The overexpression of miR-142-5p in these two cell lines showed that the optical density (OD) value measured after miR-142-5p overexpression was lower than that of the control group at 24, 48, and 72 h (Fig. 3I and J). The number of EdU-positive cells in the miR-142-5p-transfected mimic group was compared with that in the control group, and the decrease trend was obvious (Fig. 4A–D). Cell cycle analysis showed that overexpression of miR-142-5p resulted in a higher percentage of MDA-MB-231 and MCF-7 cells in the G0–G1 phase, and a lower percentage in the S phase, which indicates that miR-142-5p upregulation leads to G1 blockade of breast cancer cells (Fig. 4E–H).
Figure 3.
miR-142-5p inhibits the migration of breast cancer cells and regulates growth ability. (A) The high expression of miR-142-5p can improve the survival rate of patients by TCGA analysis. (B, C) After transfection with miR-142-5p analogs, the expression of miR-142-5p in the MDA-MB-231 cells was detected by miRNA reverse transcription polymerase chain reaction (RT-PCR). All experiments were repeated three times in duplicate, and the semiquantitative analysis was calculated by SPSS statistical software. **p < 0.01. (D–G) The number of cells migrated to the lower side of the Transwell chambers was counted and photographed in five fields (the upper, the lower, the left, the right, and the middle) of three independent experiments, and the migration capacity was calculated by SPSS statistical software. *p < 0.05, **p < 0.01. (H) The effect of overexpressed miR-142-5p analogs on MCF-7 migration was detected by wound healing assay. (I, J) Growth ability of MDA-MB-231 and MCF-7 cells was tested by CCK-8. At 24, 48, and 72 h, the growth ability of miR-142-5p overexpression was lower than that of the control group. Data are expressed as mean ± SD. *p < 0.05, **p < 0.01.
Figure 4.
miR-142-5p inhibits the proliferation of breast cancer cells and regulates the cell cycle. Cell proliferation of MDA-MB-231 and MCF-7 cells was measured by EdU assay. (A, B) The left panels show the DAPI staining for nuclei. The middle panels show EdU-positive cells. The right panels show double immunostaining for EdU and nuclei. Scale bar: 50 μm. (C, D) The number of EdU-positive cells of miR-142-5p overexpression was measured by ImageJ software, and the proliferation rate was calculated by SPSS statistical soft. *p < 0.05. (E–H) Cell cycle analysis showed that MDA-MB-231 and MCF-7 cells transfected by miR-142-5p simulant were detected by flow cytometry. Statistical analysis of the different phases of flow cytometry results; each group of experiments was repeated three times independently. Data are expressed as mean ± SD. *p< 0.05, **p< 0.01.
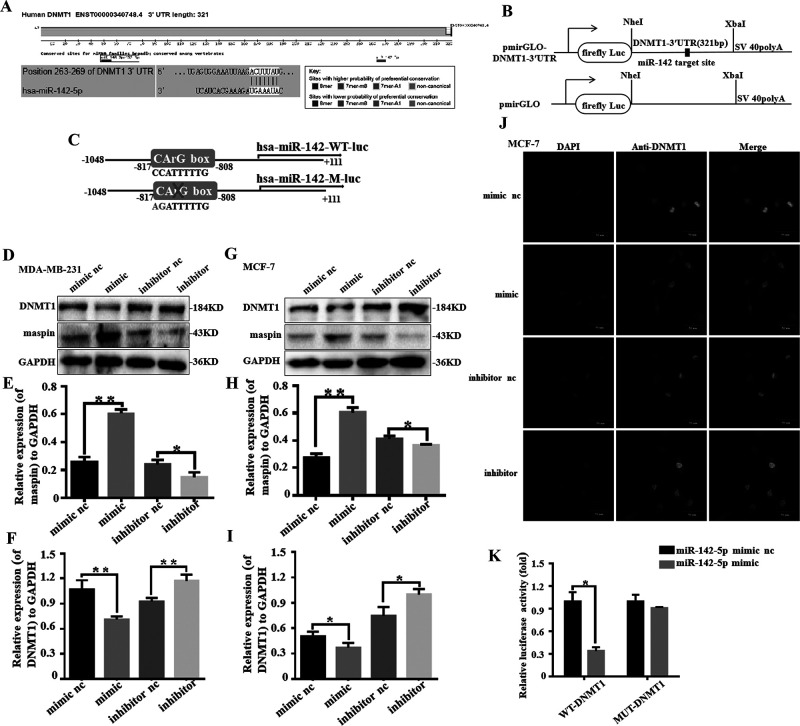
miR-142-5p Targets 3′-UTR to Inhibit DNMT1 Expression and Upregulate Maspin
We used the TargetScan database to predict the potential connection between DNMT1 and miR-142-5p (Fig. 5A and B). In order to prove that miR-142-5p can be combined with the site on the 3′-UTR of DNMT1, we constructed two plasmids containing the target fragment (DNMT1-3′-UTR-WT and DNMT1-3′-UTR-MUT) as shown in Figure 5C. By enhancing the expression of miR-142-5p in MDA-MB-231, the expression of maspin protein was upregulated about 2.22 times, while the expression of DNMT1 protein was reduced to 38% (Fig. 5D–F). By enhancing the expression of miR-142-5p in MCF-7, the expression of maspin protein was increased 2.16 times, while the DNMT1 protein expression decreased to 75% (Fig. 5G–I). We overexpressed miR-142-5p or inhibited miR-142-5p in the MCF-7, and incubated the antibody and DNMT1. The results showed that upregulation of miR-142-5p in the cells could inhibit the expression of DNMT1, and after miR-142-5p was inhibited, the expression of DNMT1 in the cells increased (Fig. 5J). The upregulation of miR-142-5p in cells can inhibit the expression of DNMT1. Luciferase reporter experiments were performed in the 293T cells. miR-142-5p overexpression can significantly reduce the luciferase activity of DNMT1-3′UTR-WT, while miR-142-5p overexpression has a negative effect on DNMT1-3′UTR-MUT (Fig. 5K).
Figure 5.
miR-142-5p targets the 3′-untranslated region (3′-UTR) to inhibit DNMT1 expression and upregulate maspin. (A) Analysis on the TargetScanHuman (http://www.targetscan.org/vert_72/) website revealed that miR-142-5p can target the 3′-UTR of DNMT1. (B, C) miR-142-5p targets the base sequence and mutant sequence of DNMT1 3′-UTR. (D–F) The effect of miR-142-5p simulant in MDA-MB-231 and MCF-7 was detected by Western blot analysis. All experiments were repeated three times in duplicate, and the semiquantitative analysis was calculated by SPSS statistical soft. *p < 0.05, **p < 0.01. (G–I) The expression of maspin and DNMT1 in MDA-MB-231 and MCF-7 transfected by miR-142-5p simulant was detected by Western blot analysis. All experiments were repeated three times in duplicate, and the semiquantitative analysis was calculated by SPSS statistical software. *p < 0.05, **p < 0.01. (J) The left panels show the DAPI staining for nuclei. The middle panels show FITC for DNMT1. The right panels show double immunostaining for DNMT1 and nuclei. Scale bar: 50 μm. (K) Luciferase reporter experiments were performed in the 293T cell, and then luciferase reporter assay was used to test the transcriptional activity of DNMT1. Values are the relative luciferase activity (100%). All experiments were repeated three times in duplicate. *p < 0.05.
MKL-1 Can Promote miR-142-5p Transcription
In order to verify whether MKL-1 affects the expression of miR-142-5p, we constructed two luciferase gene reporter plasmids, pGL3-miR-142-5p-WT-LUC and pGL3-miR-142-5p-MUT-LUC (Fig. 6A). The experimental groups were cotransformed to pcDNA3.1 + pGL3-miR-142-5p-WT-LUC (control group), pcDNA3.1-MKL-1 + pGL3-miR-142-5p-WT-LUC (experimental group), pcDNA3.1 + pGL3-miR-142-5p-MUT-LUC (mutation control group), and pcDNA3.1-MKL-1 + pGL3-miR-142-5p-MUT-LUC (mutation test group). The transfected pcDNA3.1-MKL-1 was set to a concentration gradient (0, 0.2, 0.4, 0.6, and 0.8 μg), and the results showed that the pGL-3-miR-142-5p-WT-LUC group luciferase reporter activity of MKL-1 gradually increased with the increase in the concentration of cotransfected MKL-1 (Fig. 6B). In contrast, in the mutation control group, the activity of the luciferase reporter gene did not change significantly after overexpression of MKL-1 (Fig. 6B). This indicates that overexpression of MKL-1 can promote the transcription of miR-142-5p. The expression of miR-142-5p in MDA-MB-231 cells was significantly increased by overexpressed MKL-1 (Fig. 6C). It was also found in MCF-7 cells that MKL-1 could indeed promote the transcription of miR-142-5p (Fig. 6D). In order to fully prove that MKL-1 works through miR-142-5p, we conducted a confirmation experiment in which MKL-1 was overexpressed and miR-142-5p was silenced.
Figure 6.
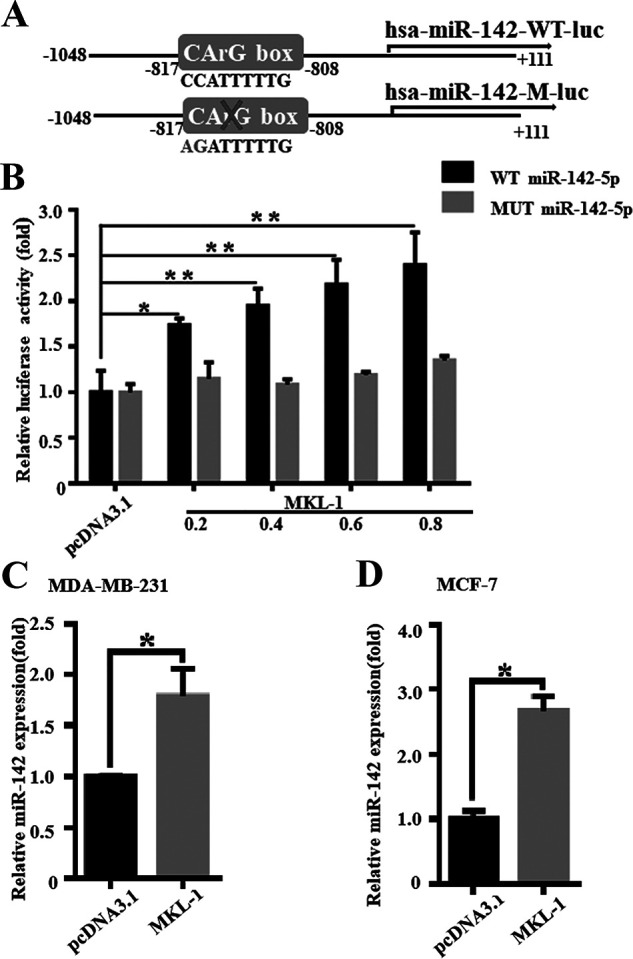
MKL-1 can promote miR-142-5p transcription. (A) In order to verify whether MKL-1 affects the expression of miR-142-5p, we constructed two luciferase gene reporter plasmids: pGL3-miR-142-5p-WT-LUC and pGL3-miR-142-5p-MUT-LUC. (B) MKL-1 or empty vector (control group) and pGL3-miR-142-5p-WT-LUC or pGL3-miR-142-5p-MUT-LUC were cotransfected into 293T cells. The transfected pcDNA3.1-MKL-1 was set to a concentration gradient (0, 0.2, 0.4, 0.6, and 0.8 μg) and transfected with MKL-1 for 48 h or a control vector (pcDNA3.1). Then the luciferase reporter assays were used to test the transactivity of miR-142-5p. Values are the relative luciferase activity (fold). All experiments were repeated three times in duplicate. *p < 0.05, **p < 0.01. (C, D) After transfection with MKL-1 overexpression plasmid, the expression of miR-142-5p in the MDA-MB-231 and MCF-7 cells was detected by miRNA RT-PCR. All experiments were repeated three times in duplicate, and the semiquantitative analysis was calculated by SPSS statistical software. *p < 0.05.
miR-142-5p Inhibits Tumor Growth of Breast Cancer Cells In Vivo
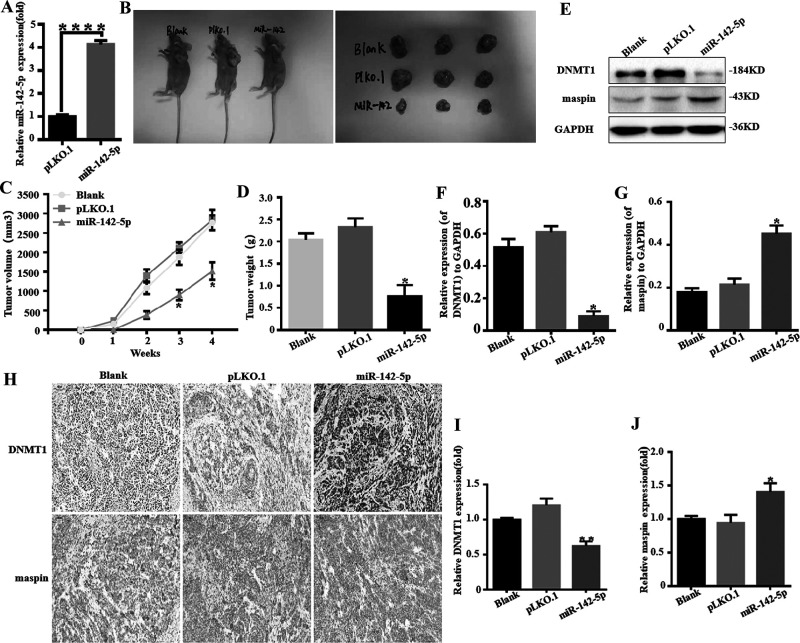
In order to determine the effect of miR-142-5p on tumor growth in vivo, we constructed MCF-7 cells stably overexpressing miR-142-5p with an empty plasmid (pLKO.1) as a control. We tested the transfection efficiency of the stably transfected cell line (Fig. 7A). The results showed that the tumors from miR-142-5p-overexpressed cells were smaller and lighter than the control, while their tumor volume growth was also slower (Fig. 7B–D). Western blotting results showed that the protein expression of DNMT1 in tumor tissues with high expression of miR-142-5p was lower than that of the control and blank groups, while the protein expression of maspin was higher than that of the control and blank groups (Fig. 7E–G). IHC analysis further confirmed that the upregulation of miR-142-5p can reduce the expression of DNMT1 in tumor tissues, and at the same time upregulate the expression of maspin in tumor tissues (Fig. 7H). We also proved by real-time fluorescence quantitative PCR that the mRNA of DNMT1 in tumor tissues overexpressed by miR-142-5p was lower than that of the control and blank groups, and the mRNA expression of maspin was higher than that of the control and blank groups (Fig. 7I and J).
Figure 7.
miR-142-5p inhibits tumor growth of breast cancer cells in vivo. (A) In order to determine the effect of miR-142-5p on tumor growth in vivo, we constructed MCF-7 cells stably overexpressing miR-142-5p with empty plasmid (pLKO.1) as control. We tested the transfection efficiency of the stably transfected cell line. ****p < 0.00001. (B–D) Nude mice injected with different groups of MCF-7 cells after 28 days. The images show the results for three mice. Image shows tumor tissue isolated from nude mice with three mice in each group. Tumor growth curves of nude mice injected with MCF-7 cells after 28 days, and the tumor volume was estimated using calipers. The image shows the statistics of the weight of tumor tissue isolated from nude mice; the weight of the tumor tissue is measured with a balance. *p < 0.05. (E–G) After being injected with different groups of MCF-7 cells after 28 days, the expressions of DNMT1 and maspin were detected by Western blot. All experiments were repeated three times in duplicate, and the semiquantitative analysis was calculated by SPSS statistical software. *p < 0.05. (H–J) Immunohistochemical analysis of tumor tissue isolated from nude mice; the image shows the results of in situ analysis of DNMT1 and maspin protein. Tumor tissues of miR-142-5p and pLKO.1 (control group) isolated from nude mice were analyzed for protein expression levels of DNMT1 and maspin by Western blot. *p < 0.05, **p < 0.01.
DISCUSSION
In recent years, studies have shown that epigenetic changes are critical to the occurrence and development of cancers including breast cancer, and they also provide new ideas for the treatment and positive prognosis of breast cancer. DNA methylation is the most common mammalian epigenetic modification27. It is believed that the occurrence and development of human cancers are driven by epigenetic and genetic changes, which activate multiple steps in the cancerous process28. DNA hypermethylation inhibits tumor suppressor genes and leads to insensitivity to growth inhibitory signals and avoids programmed cell death. This is the main epigenetic feature that distinguishes cancer cells from normal cells29. DNA hypermethylation is closely related to the occurrence of breast cancer and cell immortalization and is often caused by abnormal expression of DNMT, including DNMT1, DNMT3a, and DNMT3b30. DNMT1 and BCL11A can coregulate the function of thyroid cells by silencing BCL11A31. More importantly, a recent study showed that DNMT1 is upregulated in breast tumors. DNMT1 regulates the number of cancer stem cells (CSCs) in breast cancer cells by inhibiting or inducing Islet-1 (ISL1) hypermethylation/downregulation32. By downregulating DNMT1, the expression of tumor suppressor genes can be partially increased to reduce tumor formation. Compared with nondividing cells, DNMT1 is most abundantly expressed in dividing cells and has become the main therapeutic target for inhibiting methylation of cancer cells33. In addition, it has also been reported that inhibition of DNMT1 expression can inhibit cell proliferation and induce apoptosis34. Dhawan et al. proved that DNMT1 can be used as an emerging target for the treatment of invasive bladder cancer in a dog model35. Previous studies have shown knockdown of DNMT1 in breast cancer cells, the expression of the tumor suppressor gene maspin is upregulated36, and the proliferation and invasion ability of cancer cells are significantly inhibited by the downregulation of DNMT137. We inhibited the expression of DNMT1 in breast cancer cells. After DNMT1 was inhibited, the migration ability of breast cancer tumor cells was significantly downregulated. This shows that DNMT1 plays a vital role in breast cancer migration. Although we prove that DNMT1 can inhibit the migration of tumor cells, the role of transcription factors in tumor cells is a complicated process.
MicroRNAs are small noncoding RNAs that directly bind to the 3′-UTR of the target messenger RNA (mRNA) in a sequence-specific manner at the posttranscription level to regulate gene expression38. Many cellular processes that miRNA participates in are related to the occurrence and development of cancer39. miR200 promotes fibroblast heterogeneity and immunosuppression in ovarian cancer through targeted regulation of CXCL12β40. miRNA-143-3p inhibits the growth and invasion of melanoma cells by targeting cyclooxygenase 2 and is inversely related to the progression of malignant melanoma41. miR-31 promotes breast stem cell expansion and breast tumorigenesis by inhibiting Wnt signaling antagonists42. Genome analysis shows that in most cancer types, miRNA expression is controlled by a variety of mechanisms. Defects, amplification, and deletion of miRNA biological genetic mechanisms will cause abnormal miRNA local transcription control. There is strong evidence that miRNAs in cancer may also get out of control due to abnormal CpG methylation and/or histone modification43. On the other hand, some miRNAs are not only regulated by epigenetic mechanisms but also play an active role in epigenetic mechanisms, resulting in highly adjustable feedback pathways that can fine-tune gene expression. The target DNMT1 is abnormally upregulated in breast cancer and is negatively correlated with miRNA expression44. In our research, we can prove that DNMT1 is a functional target for miR-142-5p to regulate breast cancer cells. According to predictive target software analysis, miR-142-5p can inhibit the expression of DNMT1 by targeting the 3′-UTR region that binds to DNMT1. In this way, a signaling pathway is formed to promote the downregulation of DNMT1, and the downregulation of DNMT1 promotes the reexpression of the tumor suppressor gene maspin, inhibiting the migration and proliferation of tumor cells. Our nude mice tumorigenesis experiments also proved that miR-142-5p targeting DNMT1 can be a feasible tumor treatment strategy. MKL-1 plays a key role in cell proliferation, differentiation, migration, and apoptosis45. It is worth noting that recent breast cancer genomics studies have identified susceptibility SNPs (single nucleotide polymorphisms) located in the MKL-1 intron46, which implies the well-known muscle cell regulator MKL-1 with potential relevance in the development of breast cancer. MKL-1 is gradually becoming a prognostic indicator and treatment target for cancer patients. We analyzed the potential miRNAs that may be regulated by MKL-1 through bioinformatics correlation and checked whether there is a CarG box site in the promoter region of these miRNAs. According to these criteria, miR-142-5p was identified as a potential target. The analysis of the luciferase reporter gene showed that MKL-1 can indeed regulate the transcription of miR-142-5p through the promoter binding site of miR-142-5p, thereby inhibiting the expression of the downstream target gene DNMT1 of miR-142-5p. The downstream oncogene maspin is reexpressed, thereby regulating the migration and proliferation of breast cancer cells.
Previous studies have reported that the abnormal silencing of the antimetastatic tumor suppressor gene maspin in breast epithelial tumor cells is related to DNA hypermethylation and histone hypoacetylation of its promoter47. Studies have shown that the expression of maspin is negatively correlated with the adverse clinical progression of breast cancer48. DNMT1 can recognize the region rich in CpG islands in the maspin promoter to promote the abnormal methylation of the maspin promoter, thereby silencing its gene expression. miR-142-5p can target the 3′-UTR of DNMT1 to inhibit translation, and MKL-1 can bind to the CarG site on the miR-142-5p promoter to promote miR-142-5p transcription. This shows that MKL-1/miR-142-5p can indirectly regulate the expression level of maspin through DNMT1, restore the expression of silenced tumor suppressor genes, and promote the growth and metastasis of cancer cells.
Our data show that MKL-1 promotes its transcription by targeting the miR-142-5p promoter and attenuates the migration of MCF-7 and MDA-MB-231 cells induced by DNMT1. The expressions of miR-142-5p and MKL-1 are positively correlated. In addition, DNMT1 has been proven to be the target of miR-142-5p, which inhibits the expression of DNMT1 by targeting the 3′-UTR of DNMT1, thereby inhibiting the migration and proliferation of breast cancer. As the upstream regulator of DNMT1, miR-142-5p can promote the reexpression of maspin silenced by DNMT1. Animal tumorigenesis experiments also prove that miR-142-5p targeting DNMT1 can become a viable tumor treatment plan. Therefore, our data provide important and novel insights into the MKL-1/miR-142-5p/DNMT1/maspin signaling pathway and may become a new idea for breast cancer diagnosis, treatment, and prognosis.
ACKNOWLEDGMENTS
The authors acknowledge the technical assistance of Dr. Xing-Hua Liao and Dr. Chao-Jiang Gu. This work was financially supported by the Natural Science Foundation of Hubei Province (2018CFB369), China Postdoctoral Science Foundation Grant (2018M632927), Applied Basic Research Project of Wuhan City (No. 2017060201010193), and Hubei Provincial Plan of Natural Science Foundation of China (No. ZRMS2019000926). The authors declare no conflicts of interest.
REFERENCES
- 1. Greaney ML, Sprunck-Harrild K, Ruddy KJ, Ligibel J, Barry WT, Baker E, Meyer M, Emmons KM, Partridge AH. 2015. Study protocol for young & strong: A cluster randomized design to increase attention to unique issues faced by young women with newly diagnosed breast cancer. BMC Public Health 15:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DeSantis C, Ma J, Bryan L, Jemal A. 2014. Breast cancer statistics, 2013. CA Cancer J Clin. 64(1):52–62. [DOI] [PubMed] [Google Scholar]
- 3. Greenberg MVC, Bourc’his D. 2019. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 20(10):590–607. [DOI] [PubMed] [Google Scholar]
- 4. Wu CH, Chen CY, Yeh CT, Lin KH. 2020. Radiosensitization of hepatocellular carcinoma through targeting radio-associated microRNA. Int J Mol Sci. 9;21(5):1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Grosselin K, Durand A, Marsolier J, Poitou A, Marangoni E, Nemati F, Dahmani A, Lameiras S, Reyal F, Frenoy O, Pousse Y, Reichen M, Woolfe A, Brenan C, Griffiths AD, Vallot C, Gérard A. 2019. High-throughput single-cell chip-seq identifies heterogeneity of chromatin states in breast cancer. Nat Genet. 51(6):1060–1066. [DOI] [PubMed] [Google Scholar]
- 6. Zhang S, Wang Y, Gu Y, Zhu J, Ci C, Guo Z, Chen C, Wei Y, Lv W, Liu H, Zhang D, Zhang Y. 2018. Specific breast cancer prognosis-subtype distinctions based on DNA methylation patterns. Mol Oncol. 12(7):1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Esopi D, Graham MK, Brosnan-Cashman JA, Meyers J, Vaghasia A, Gupta A, Kumar B, Haffner MC, Heaphy CM, De Marzo AM, Meeker AK, Nelson WG, Wheelan SJ, Yegnasubramanian S. 2020. Pervasive promoter hypermethylation of silenced tert alleles in human cancers. Cell Oncol. (Dordr) 43(5):847–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gowher H, Jeltsch A. 2018. Mammalian DNA methyltransferases: New discoveries and open questions. Biochem Soc Trans. 46(5):1191–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bettstetter M, Woenckhaus M, Wild PJ, Rümmele P, Blaszyk H, Hartmann A, Hofstädter F, Dietmaier W. 2005. Elevated nuclear maspin expression is associated with microsatellite instability and high tumour grade in colorectal cancer. J Pathol. 205(5):606–614. [DOI] [PubMed] [Google Scholar]
- 10. Li YQ, Wang K, Ao L, Lu XL, Shi JP. 2019. Research on the mechanism of Mir-142-5p inhibiting the proliferation and migration of gastric cancer cells through DNA methyltransferase 1. Chin Clin Pharm Thera. 24(12):1328–1334. [Google Scholar]
- 11. Angeloni A, Bogdanovic O. 2019. Enhancer DNA methylation: Implications for gene regulation. Essays Biochem. 63(6):707–715. [DOI] [PubMed] [Google Scholar]
- 12. Sengupta D, Deb M, Rath SK, Kar S, Parbin S, Pradhan N, Patra SK. 2016. DNA methylation and not H3K4 trimethylation dictates the expression status of miR-152 gene which inhibits migration of breast cancer cells via DNMT1/CDH1 loop. Exp Cell Res. 346(2):176–187. [DOI] [PubMed] [Google Scholar]
- 13. Yu W, Li D, Zhang Y, Li C, Zhang C, Wang L. 2019. Mir-142-5p acts as a significant regulator through promoting proliferation, invasion, and migration in breast cancer modulated by targeting sorbs1. Technol Cancer Res Treat. 18:1533033819892264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu W, Wang W. 2018. MicroRNA-142-5p modulates breast cancer cell proliferation and apoptosis by targeting phosphatase and tensin homolog. Mol Med Rep. 17(6):7529–7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zagorac S, Alcala S, Fernandez Bayon G, Bou Kheir T, Schoenhals M, González-Neira A, Fernandez Fraga M, Aicher A, Heeschen C, Sainz B Jr. 2016. DNMT1 inhibition reprograms pancreatic cancer stem cells via upregulation of the miR-17-92 cluster. Cancer Res. 76(15):4546–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quinn SR, Mangan NE, Caffrey BE, Gantier MP, Williams BR, Hertzog PJ, McCoy CE, O’Neill LA. 2014. The role of Ets2 transcription factor in the induction of microRNA-155 (miR-155) by lipopolysaccharide and its targeting by interleukin-10. J Biol Chem. 289(7):4316–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yan J, Yang B, Lin S, Xing R, Lu Y. 2019. Downregulation of miR-142-5p promotes tumor metastasis through directly regulating CYR61 expression in gastric cancer. Gastric Cancer 22(2):302–313. [DOI] [PubMed] [Google Scholar]
- 18. Ozawa PMM, Vieira E, Lemos DS, Souza ILM, Zanata SM, Pankievicz VC, Tuleski TR, Souza EM, Wowk PF, Urban CA, Kuroda F, Lima RS, Almeida RC, Gradia DF, Cavalli IJ, Cavalli LR, Malheiros D, Ribeiro E. 2020. Identification of miRNAs enriched in extracellular vesicles derived from serum samples of breast cancer patients. Biomolecules 10(1):150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parmacek MS. 2010. Myocardin-related transcription factor-A: Mending a broken heart. Circ Res. 107(2):168–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chehrehasa F, Meedeniya ACB, Dwyer P, Abrahamsen G, Mackay-Sim A. 2009. Edu, a new thymidine analogue for labelling proliferating cells in the nervous system. J Neurosci Methods 177(1):122–130. [DOI] [PubMed] [Google Scholar]
- 21. Limsirichaikul S, Niimi A, Fawcett H, Lehmann A, Yamashita S, Ogi T. 2009. A rapid non-radioactive technique for measurement of repair synthesis in primary human fibroblasts by incorporation of ethynyl deoxyuridine (edu). Nucleic Acids Res. 37(4):e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang HM, Li H, Wang GX, Wang J, Xiang Y, Huang Y, Shen C, Dai ZT, Li JP, Zhang TC, Liao XH. 2020. MKL1/miR-5100/CAAP1 loop regulates autophagy and apoptosis in gastric cancer cells. Neoplasia (New York, NY). 22(5):220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wong KK. 2021. DNMT1: A key drug target in triple-negative breast cancer. Semin Cancer Biol. 72:198–213. [DOI] [PubMed] [Google Scholar]
- 24. Jin X, Li Y, Guo Y, Jia Y, Qu H, Lu Y, Song P, Zhang X, Shao Y, Qi D, Xu W, Quan C. 2019. ERα is required for suppressing OCT4-induced proliferation of breast cancer cells via DNMT1/ISL1/ERK axis. Cell Prolif. 52(4):e12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jehanno C, Percevault F, Boujrad N, Le Goff P, Fontaine C, Arnal JF, Primig M, Pakdel F, Michel D, Métivier R, Flouriot G. 2021. Nuclear translocation of MRTFA in MCF7 breast cancer cells shifts ERα nuclear/genomic to extra-nuclear/non genomic actions. Mol Cell Endocrinol. 530:111282. [DOI] [PubMed] [Google Scholar]
- 26. Li M, Cai O, Tan S. 2019. LOXL1-AS1 drives the progression of gastric cancer via regulating miR-142-5p/PIK3CA axis. Onco Targets Ther. 12:11345–11357. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27. Sadakierska-Chudy A, Filip M. 2015. A comprehensive view of the epigenetic landscape. Part II: Histone post-translational modification, nucleosome level, and chromatin regulation by ncRNAs. Neurotox Res. 27(2):172–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Easwaran H, Tsai HC, Baylin SB. 2014. Cancer epigenetics: Tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell. 54(5):716–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sher G, Salman NA, Khan AQ, Prabhu KS, Raza A, Kulinski M, Dermime S, Haris M, Junejo K, Uddin S. 2020. Epigenetic and breast cancer therapy: Promising diagnostic and therapeutic applications. Semin Cancer Biol. S1044-579X(20)30181-4. [DOI] [PubMed]
- 30. Zhang W, Chang Z, Shi KE, Song L, Cui LI, Ma Z, Li X, Ma W, Wang L. 2016. The correlation between DNMT1 and ERα expression and the methylation status of ERα, and its clinical significance in breast cancer. Oncol Lett. 11(3):1995–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu J, Bauer DE, Kerenyi MA, Vo TD, Hou S, Hsu YJ, Yao H, Trowbridge JJ, Mandel G, Orkin SH. 2013. Corepressor-dependent silencing of fetal hemoglobin expression by Bcl11a. Proc Natl Acad Sci USA 110(16):6518–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S, Song W, Li X, Li L, Du Z, Jia L, Zhou L, Li W, Hoffman AR, Hu JF, Cui J. 2018. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 19(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wong KK. 2020. DNMT1 as a therapeutic target in pancreatic cancer: Mechanisms and clinical implications. Cell Oncol. (Dordr) 43(5):779–792. [DOI] [PubMed] [Google Scholar]
- 34. Han G, Wei Z, Cui H, Zhang W, Wei X, Lu Z, Bai X. 2018. NUSAP1 gene silencing inhibits cell proliferation, migration and invasion through inhibiting DNMT1 gene expression in human colorectal cancer. Exp Cell Res. 367(2):216–221. [DOI] [PubMed] [Google Scholar]
- 35. Dhawan D, Ramos-Vara JA, Hahn NM, Waddell J, Olbricht GR, Zheng R, Stewart JC, Knapp DW. 2013. DNMT1: An emerging target in the treatment of invasive urinary bladder cancer. Urol Oncol. 31(8):1761–1769. [DOI] [PubMed] [Google Scholar]
- 36. Wozniak RJ, Klimecki WT, Lau SS, Feinstein Y, Futscher BW. 2007. 5-Aza-2’-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene. 26(1):77–90. [DOI] [PubMed] [Google Scholar]
- 37. Li W, Wang Q, Feng Q, Wang F, Yan Q, Gao SJ, Lu C. 2019. Oncogenic KSHV-encoded interferon regulatory factor upregulates HMGB2 and CMPK1 expression to promote cell invasion by disrupting a complex lncRNA-OIP5-AS1/miR-218-5p network. PLoS Pathog. 15(1):e1007578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ha M, Kim VN. 2014. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 15(8):509–524. [DOI] [PubMed] [Google Scholar]
- 39. Takahashi RU, Prieto-Vila M, Hironaka A, Ochiya T. 2017. The role of extracellular vesicle microRNAs in cancer biology. Clin Chem Lab Med. 55(5):648–656. [DOI] [PubMed] [Google Scholar]
- 40. Givel AM, Kieffer Y, Scholer-Dahirel A, Sirven P, Cardon M, Pelon F, Magagna I, Gentric G, Costa A, Bonneau C, Mieulet V, Vincent-Salomon A, Mechta-Grigoriou F. 2018. MiR200-regulated CXCL12β promotes fibroblast heterogeneity and immunosuppression in ovarian cancers. Nat Commun. 9(1):1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Panza E, Ercolano G, De Cicco P, Armogida C, Scognamiglio G, Botti G, Cirino G, Ianaro A. 2018. MicroRNA-143-3p inhibits growth and invasiveness of melanoma cells by targeting cyclooxygenase-2 and inversely correlates with malignant melanoma progression. Biochem Pharmacol. 156:52–59. [DOI] [PubMed] [Google Scholar]
- 42. Lv C, Li F, Li X, Tian Y, Zhang Y, Sheng X, Song Y, Meng Q, Yuan S, Luan L, Andl T, Feng X, Jiao B, Xu M, Plikus MV, Dai X, Lengner C, Cui W, Ren F, Shuai J, Millar SE, Yu Z. 2017. MiR-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing WNT signaling antagonists. Nat Commun. 8(1):1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Karimzadeh MR, Pourdavoud P, Ehtesham N, Qadbeigi M, Asl MM, Alani B, Mosallaei M, Pakzad B. 2020. Regulation of DNA methylation machinery by epi-miRNAs in human cancer: Emerging new targets in cancer therapy. Cancer Gene Ther. 28(3–4):157–174. [DOI] [PubMed] [Google Scholar]
- 44. Suzuki H, Maruyama R, Yamamoto E, Kai M. 2012. DNA methylation and microRNA dysregulation in cancer. Mol Oncol. 6(6):567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu Q, Jiang Y, Yin Y, Li Q, He J, Jing Y, Qi YT, Xu Q, Li W, Lu B, Peiper SS, Jiang BH, Liu LZ. 2013. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 5(1):3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiang Y, Liao XH, Yu CX, Yao A, Qin H, Li JP, Hu P, Li H, Guo W, Gu CJ, Zhang TC. 2017. MiR-93-5p inhibits the EMT of breast cancer cells via targeting MKL-1 and STAT3. Exp Cell Res. 357(1):135–144. [DOI] [PubMed] [Google Scholar]
- 47. Shankar E, Pandey M, Verma S, Abbas A, Candamo M, Kanwal R, Shukla S, MacLennan GT, Gupta S. 2020. Role of class I histone deacetylases in the regulation of maspin expression in prostate cancer. Mol Carcinog. 59(8):955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bernardo MM, Dzinic SH, Matta MJ, Dean I, Saker L, Sheng S. 2017. The opportunity of precision medicine for breast cancer with context-sensitive tumor suppressor maspin. J Cell Biochem. 118(7):1639–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]