Abstract
Monolayer epithelial cells interact constantly with the substrate they reside on and their surrounding neighbors. As such, the properties of epithelial cells are profoundly governed by the mechanical and molecular cues that arise from both the substrate and contiguous cell neighbors. Although both cell-substrate and cell-cell interactions have been studied individually, these results are difficult to apply to native confluent epithelia, in which both jointly regulate the cell phenotype. Specifically, it remains poorly understood about the intertwined contributions from intercellular adhesion and substrate stiffness on cell morphology and gene expression, two essential microenvironment properties. Here, by adjusting the substrate modulus and altering the intercellular adhesion within confluent kidney epithelia, we found that cell-substrate and cell-cell interactions can mask each other's influence. For example, we found that epithelial cells exhibit an elongated morphological phenotype only when the substrate modulus and intercellular adhesions are both reduced, whereas their motility can be upregulated by either reduction. These results illustrate that combinatorial changes of the physical microenvironment are required to alter cell morphology and gene expression.
Significance
Although cell-substrate and cell-cell interactions have been studied individually, these results are difficult to apply to native confluent epithelia, in which both are important. To understand how epithelial cells respond to their microenvironment, it is critical to understand the contributions from cell-substrate and cell-cell interactions to cell phenotype. Here, we found full confluency to mask differences in single or colony epithelial phenotype in response to the cell-substrate interaction. Despite being grown on substrates of variable stiffness, confluent epithelium exhibited very little change in morphology and gene expression. However, cells began to exhibit differential phenotype in response to cell-cell contact inhibition, suggesting that confluent cells are a separate model of study. This work helps understand the biomechanics of confluent epithelium.
Introduction
Epithelial cells form continuous, protective layers that line organ surfaces throughout the body (1). They perform vital physiological functions including adsorption, secretion, and sensory reception (2,3). They are also related to many fatal diseases including drug-induced injuries, polycystic diseases, and 90% of cancers (4, 5, 6). Therefore, understanding the factors influencing epithelial cell behavior is of importance in basic biology research, therapeutic developments, and tissue engineering. Mechanical rigidity of the substrate and intercellular interactions have been shown to be two important factors governing epithelial phenotype (7,8). Though the interplay between these interactions has been identified in previous experiments, these studies have primarily focused on cell migration (9, 10, 11, 12, 13, 14). However, how the cell morphology and gene expression, two essential phenotypes, are affected by these interactions remains not fully understood. Further studies of such intricate cell-environment interactions are essential for better utilization of physical cues for engineering the cell property.
Adjusting the substrate modulus has been shown to have a wide range of influence on the behavior of cells. For example, increasing the stiffness has been shown to cause cell spread, formation of stress fibers, and increased cell migration (15, 16, 17). This stiffness increase has also been associated with upregulated deposition of extracellular matrix proteins such as laminin and fibronectin (18,19). Furthermore, such a substrate mechanical property has been found to largely influence the malignant transformation of mammary epithelia (20,21).
Recent studies have also shown that intercellular junctions facilitate the mechanical strength of tissues and communication between neighboring cells, which substantially affect tissue development, wound healing, and cell differentiation (22, 23, 24). Specifically, adherent proteins have been shown to be responsible for contact inhibition and mesenchymal-to-epithelial transition in developing tissues (25,26). Collective motion of cells has also been shown to rely on the mechanical intercellular communication via cell-cell contact (27,28). Lastly, it has been demonstrated that disruption of different types of intercellular junctions has various effects on epithelial phenotype, such as inducing cell rounding and deteriorating tissue by disconnecting constituent cells from each other (29).
In most native tissues and in vitro models, epithelial cells are in contact with both the substrate and neighboring cells. Recent works have examined the interplay between cell-cell and cell-substrate interactions largely in the context of cell motility. In those studies, the joint activity of intercellular and tractional forces has been linked to collective motion (9, 10, 11, 12) and cancer cell invasion (13,14). However, many other essential cellular phenotypes remain to be explored. In this work, we studied how the interplay between cell-cell and cell-substrate interactions affects the morphological phenotype and gene expression of Madin-Darby Canine Kidney (MDCK) cells.
Here, we hypothesized that substrate stiffness and intercellular adhesions have an intertwined impact on epithelial cell behavior. To test this hypothesis, we fabricated polydimethylsiloxane (PDMS)-based substrates with various moduli on which to grow MDCK epithelia, which have been routinely used for mechanobiology studies and disease modeling (30, 31, 32). After previous work, we tuned the stiffness by using varying ratios of Sylgard 184 and Sylgard 527 (33,34), along with standard tissue culture plastic (TCP), to obtain substrates with stiffnesses over five orders of magnitude. To alter the intercellular interaction, we treated confluent monolayers with calcium-free media, which has been widely used to inhibit the functioning of intercellular adhesion proteins (35, 36, 37, 38). To comprehensively assess cell phenotype, we characterized the morphology, gene expression, adhesion protein localization, and motility of MDCK cells. Through this systematic investigation, we found that the combinatorial actions of cell-substrate and cell-cell interactions are responsible for determining cell morphology and gene expression of a confluent monolayer.
Materials and methods
Fabrication of substrate with tunable stiffness
To obtain a wide range of substrate stiffnesses, Sylgard 184 (Dow Corning, 11-3184-01 C) and Sylgard 527 (Dow Corning, 11-3136-01 D) were mixed to fabricate PDMS substrates. Following manufacturing directions, Sylgard 184 was prepared by mixing 10 parts base to one part curing agent, whereas Sylgard 527 was prepared by mixing equal parts of part A and part B. We tuned the elastic modulus of the material by combining Sylgard 184 and Sylgard 527 in varying ratios before casting. Samples were mixed and cast into cell culture wells (Thermo Fisher Scientific), and promptly degassed in a vacuum chamber for approximately 20 min to remove air bubbles within the substrate. All PDMS samples were then cured at 60°C overnight. To measure the PDMS modulus for each formulation, we cast the materials and then cut into thin strips with dimensions of 50 mm × 12 mm × 2mm. These samples were then heat cured and measured using an Instron 5943 by applying tensile strain (39,40). A stress-strain curve for the pure S527 sample and a bar chart detailing elastic moduli of all tested samples can be found in Fig. S1.
MDCK cell culture
MDCK cells were cultured using 1x Dulbecco's Modified Eagle media (Gibco, 11885084) supplemented with 5% fetal bovine serum (Gibco, 16000044) and 1% penicillin-streptomycin (P/S) (Gibco, 15140122). Media was replaced every 48 h and cultures were passed at 70%–80% using 0.05% trypsin-EDTA (Gibco, 25300054). All substrates used for experiments were treated with 25 μg/ml fibronectin (R&D Systems, 1030FN05M) to facilitate cell-substrate adhesion and minimize the effect of the substrate ligands. MDCK cells were seeded at a density 5000/cm2 and incubated in a CO2 incubator (Thermo Fisher Scientific, 51033557) at 37°C and 5.0% CO2. For the cell-cell adhesion reduction experiment, once cultures reached 100% confluence, experimental samples were treated with calcium-free DMEM (Gibco, 21068028) supplemented with 1% P/S for 48 h. Additional validation experiments were performed using control media supplemented with 5 μg/mL E-cadherin antibody (Invitrogen, 14-3249-82) or 1mM DTT (Millpore Sigma, 10197777001) for 48 h. For the antibody experiment, cells were first treated with Ca- media for 30 min to facilitate antibody penetration and engagement with junctional spaces (41,42).
Morphological phenotype characterization
Phase contrast images were taken using an Etaluma LS720 microscope with a 20x objective (Olympus, N5229300) five days postseeding. To obtain fluorescent images, Ca+/− treated cells were fixed and stained for nuclei (Invitrogen, R37606), actin (Invitrogen, R37110), paxillin (BD Bioscience, 610,620), YAP (Cell Signaling Technology, 14,074), and E-cadherin. Staining was performed with the following antibodies: anti-mouse IgG (Invitrogen A32766) and anti-rat IgG (Invitrogen A21247). These samples were then imaged using the Etaluma LS720 with a 40x objective (Olympus, 1-UB767). To analyze the cell morphology, acquired images were imported into Fiji ImageJ, where cells were manually traced. From the traced outlines, we calculated cell area and perimeter, which were then used to determine circularity (4πArea/Perimeter2). Aspect ratio was calculated by dividing the long axis by the short axis of individual cells derived from the same outline. A more detailed description of each metric with representative schematics can be found in Fig. S2.
Gene expression measurement
Gene expression was characterized by RNA expression profiling using the nCounter gene expression platform (NanoString Technologies, Seattle, WA) using a custom 50-gene panel (Table S1) designed to comprehensively examine multiple aspects of MDCK cell behavior. This panel includes markers for epithelial phenotype, epithelial-to-mesenchymal transition, cell adhesions, and solute transporters. RNA extraction was performed with an RNA MiniPrep Plus kit (Zymo, R2070), and samples were normalized to a minimum RNA concentration of 10 ng/μL before being transferred to NanoString Technologies. There, samples were mixed with a 3′ biotinylated capture probe and a 5′ reporter probe tagged with a fluorescent barcode from the custom gene expression probe set. Samples were then hybridized at 65°C for 18 h and run on the Nanostring nCounter preparation station. Excess probes were removed and hybridized samples were immobilized on a streptavidin-coated cartridge. Finally, samples were scanned and individually counted on the nCounter Digital Analyzer.
Time-lapse cell motility and particle image velocimetry analysis
Time-lapse images were obtained and used for particle image velocimetry (PIV) analysis for cell motility measurements. Upon the corresponding calcium+/− treatment, samples were placed on the Etaluma LS720 and imaged over 48 h with a 20x objective. Registered regions of interest were imaged at a regular interval of ∼4 min. PIV analysis of time-lapse images was performed using the ImageJ PIV plugin. Cells were analyzed for movement with interrogation window sizes of 64 pixels and search window sizes of 128 pixels. Before the PIV analysis, all time-lapse images were aligned to remove any potential drift using the ImageJ plugin StackReg (43).
Statistical analysis
Data are reported as mean values ± standard deviation (SD). Statistical analysis was performed using Microsoft Excel, and statistical significance was determined using 1-tailed paired t-tests. Hierarchical average clustering was performed using the ClustVis web tool (44). Different significance levels are indicated with asterisks in each figure caption. A p value of less than 0.05 was considered significant.
Results and discussion
Influence of substrate stiffness on cell morphology
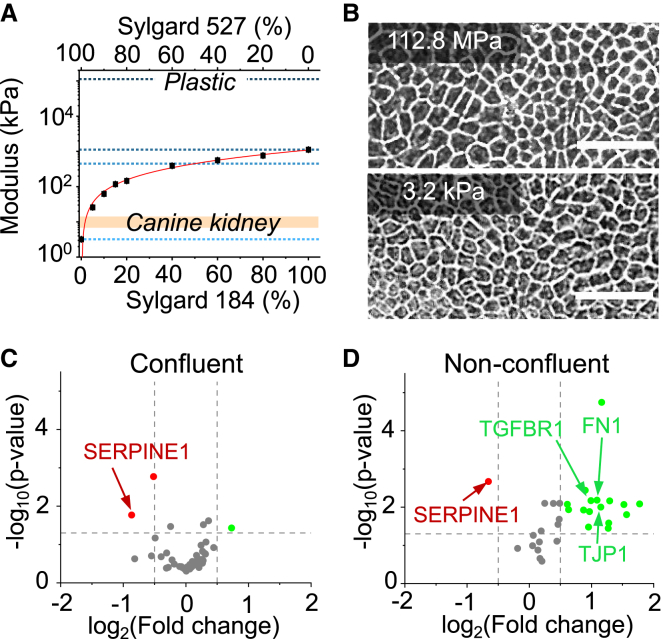
To investigate the effect of the substrate modulus on confluent epithelia, we cultured MDCK cells on a soft substrate (S527, 3.2 kPa) and compared the corresponding morphology with cells grown on a TCP (112.8 MPa). TCP was chosen for its importance in bioengineering settings such as standard cell culture, synthetic tissue scaffolds (45, 46, 47), and reconstructive and implant surgery (47,48). Understanding how cells interact with plastic-based materials is an important biological question in many technological applications. Cells were first seeded at a density of 5000 cells/cm2, typically forming a fully confluent monolayer within five days. We then characterized cell morphology by acquiring phase contrast images 24 h after samples reached full confluence to allow full establishment of intracellular adhesion (49,50). Despite nearly five orders of magnitude difference in the substrate stiffness, both confluent samples exhibit virtually indistinguishable morphology (Fig. 1 B). To validate this finding, we systematically varied the substrate stiffness by mixing different ratios of S527 and S184 to obtain three intermediate stiffnesses 3.2 kPa, 450.7 kPa, and 1123.2 kPa, with TCP as the control. We characterized the cell morphology by measuring the cell aspect ratio, perimeter, circularity, and area (Fig. S2), in which we found no significant differences in any of these morphological features between all tested stiffnesses (Fig. S3). In addition, confluent monolayers were found to establish similar levels of junctional E-cadherin enrichment despite differences in substrates stiffness, further supporting this result (Fig. S4). Specifically, we found that cells became morphologically indistinguishable upon 17 h postconfluence, in which the cell density reaches ∼570k cells/cm2 (Fig. S5). This observed unchanged phenotype is in contrast to the substantial morphological change found in previous studies that used nonconfluent cells (15,51,52). In particular, single-cell experiments have highlighted phenotypic differences associated with the substrate mechanical properties (15,53). Stiffer substrates tend to cause single cells to exhibit increased spreading and elongation, whereas cells grown on softer substrates are more cuboidal (54,55). Our experiments suggest that these differences are negligible in a highly packed monolayer.
Figure 1.
Substrates were produced by mixing various ratios of Sylgard 184 (S184) and Sylgard 527 (S527), and their stiffnesses were characterized. (A) These stiffnesses show a logarithmic trend as the percentage of S184 was increased. Pure S527 was shown to have a similar stiffness to in vivo kidney tissue (∼6 kPa). Tissue culture plastic (TCP) was also plotted for comparison. From top to bottom, the dotted lines denote substrate moduli for substrates of TCP (112.8 MPa), S184 (1123.2 kPa), 1:1 blend of S184 and S527 (450.7 kPa), and S527 (3.2 kPa). (B) Phase contrast images of confluent MDCK layers, five days postseeding, grown on TCP and S527 show no significant morphological change despite the five orders magnitude difference in the stiffness. Scale bar represents 30 μm. Volcano plots of log2 ratio versus p value compare genes expressed by cells grown on TCP and S527 for confluent (C) and nonconfluent, 24 h postseeding (D) conditions. For confluent culture, the substrate stiffness induced virtually no differences in gene expression. In contrast, nonconfluent cells grown on the soft substrate show upregulation of genes related to EMT, epithelial phenotype, cell adhesion, and solute transporters. The vast differences between confluence conditions suggest that intercellular contact may be a factor in determining cell phenotype. For (C)–(D), n = 6.
Influence of substrate stiffness on gene expression
To further study how a confluent MDCK monolayer responds to a substrate stiffness change, we quantified the expression level (mRNA) using a custom 50-gene panel to characterize the essential properties of MDCK cells (Table S1). This gene panel was designed to report the epithelial characteristics, epithelial-to-mesenchymal transition (EMT), intercellular adhesion, and expression of transporters. We compared mRNA expression between cells grown on TCP and S527. We found that altering the substrate stiffness had a minor effect on gene expression (Fig 1 C), in which ∼94% of tested genes did not show significant change. In contrast, nonconfluent cells (Fig. S6) grown on S527 showed a noticeable change of gene expression, where ∼40% of tested genes were upregulated (Fig. 1 D). These genes are related to cell adhesion (TJP1, LAMB1, LAMC1, CDH6, and CTNNB1), mesenchymal phenotype (SMAD7, TGFBR1, FN1, and SKIL), and expression of solute transporters (ABCC4, ABCA5, and SLC1A1) (Table S2). Many tissue engineering approaches have hypothesized that matching the substrate moduli to in vivo values should substantially improve cell phenotype and function (56, 57, 58). However, our data suggests that such a hypothesis might be oversimplified. For example, the tested confluent monolayers have a minimal response to substrate adjustments. Also, for nonconfluent layers, some mesenchymal-related markers can be upregulated by stiffness close to the physiological value (∼6kPa) (59), which suggests higher plasticity for proliferating MDCK cells. Collectively, our gene expression analysis suggests that intercellular adhesions may mask MDCK's response to the substrate stiffness, consistent with our morphology experiment.
Influence of intercellular adhesion on MDCK behavior
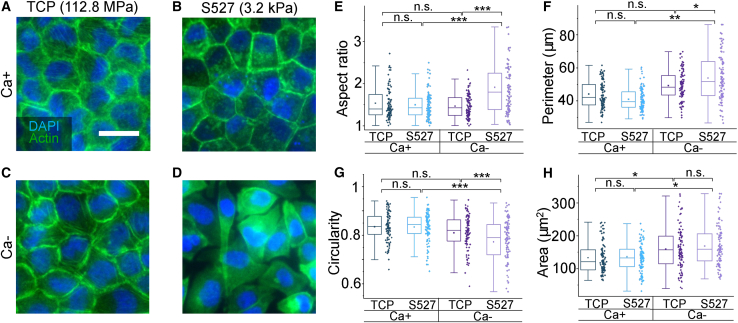
To identify the effect that intercellular adhesions have on MDCK monolayers, we treated cultures with calcium-free media, which has been widely used to reduce the cell-cell adhesions by lowering the rigidification and functional properties of junctional proteins (28,35, 36, 37, 38,60, 61, 62, 63, 64). To ensure the full effect of calcium deficiency, cells were treated with calcium-free (Ca−, experimental) or calcium-normal (Ca+, control) media for three days upon confluence. The cells were then fixed and characterized using fluorescent microscopy. As shown in Fig. 2 A–D, we found that treating the cells grown on TCP with Ca- media does not lead to a significant morphological change, suggesting that the junction perturbation alone has minimal effect on the “cobblestone” phenotype normally found in confluent monolayers. However, Ca- treated cells exhibit a drastically distinct morphology when cultured on S527 (Fig. 2 D). This morphological change was quantified by measuring the cell aspect ratio (Fig. 2 E), perimeter (Fig. 2 F), circularity (Fig. 2 G), and area (Fig. 2 H). Our data indicate that Ca- treatment of cells grown on S527 increases their aspect ratio and perimeter while decreasing their circularity, which is consistent with the cell elongation observed in Fig. 2 D. The increase in cell area due to the Ca- treatment was found to correlate with a decrease in cell density (Fig. S7).
Figure 2.
(A–D) Fluorescence images of Ca+/- treated samples grown on TCP and S527, stained for nuclei and actin, reveal significant morphological change only for MDCK cells grown on the softer substrate and subject to interference of intercellular adhesions. Scale bar represents 30 μm. Aspect ratio (E), perimeter (F), circularity (G), and cell area (H) were calculated by outlining and measuring individual cells from the obtained fluorescence images for each of the four conditions. This morphology quantification shows that Ca-/S527 cells had significant elongation, confirmed by a decrease in circularity and increases in aspect ratio and perimeter. For (E)–(H), n = 100. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001.
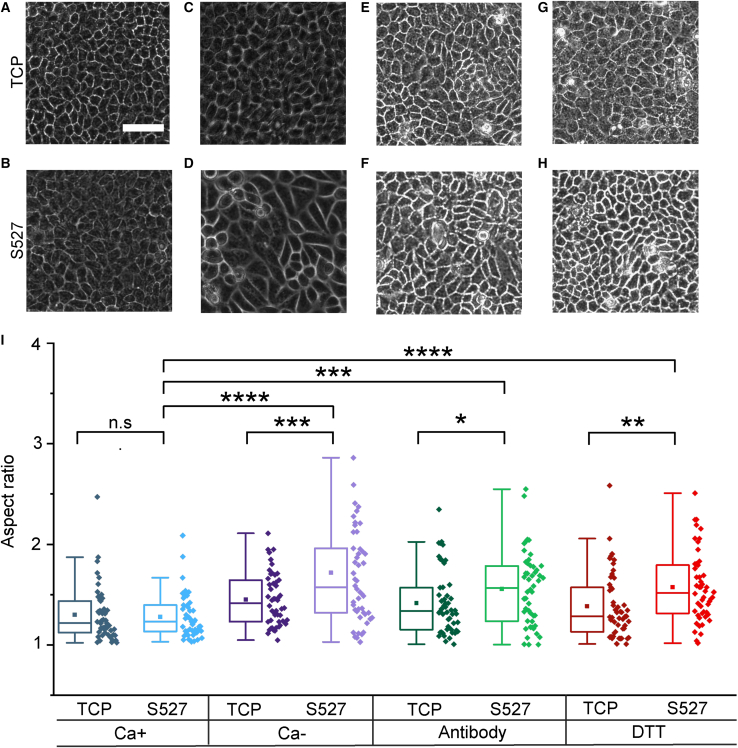
Calcium is an important mediator for virtually all cellular processes and has been shown to play significant roles in adhesion, motility, transcription, biochemical signaling, and apoptosis (65,66). Calcium depletion could have a variety of secondary effects that could impact mechanosignaling pathways, such as diminishing focal adhesion-mediated cell-substrate adhesions (67). However, we found Ca- to not significantly affect focal adhesion formation (paxillin staining) (Fig. S8) as it diminishes cell-cell interactions (E-cadherin staining) (Fig. S4). In addition, we performed experiments applying E-cadherin antibody, concentration of 5 μg/mL, to selectively block E-cadherin (Fig. 3 E and F) and 1 mM dithiothreitol (DTT) (Fig. 3 G and H) to degrade extracellular proteins via disulfide exchange (29,68,69). These additional methods have been used in previous works to repress cell-cell adhesions while retaining calcium levels (29,68,70). All three experiments exhibited similar increases in aspect ratio in cells on S527 (Fig. 3 I), validating the result of our Ca-experiment.
Figure 3.
E-cadherin antibody, which specifically blocks cellular junctions, and DTT, which inactivates extracellular adhesions through disulfide exchange, were used to perturb cell-cell adhesions while maintaining extracellular calcium to validate our Ca- results. Phase contrast images show morphology for cells treated with control media (A and B), calcium-free media (C and D), antibody (E and F), and DTT (G and H). Scale bar represents 60μm. Morphology quantification (I) reveals that morphological changes induced by both antibody and DTT are consistent with our Ca- results. Cross-category statistics for cells grown on TCP were not included as they were all found to be nonsignificant. For (G), n = 50. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Overall, our results suggest that altering the intercellular adhesion greatly impacts the cell morphology response to substrate stiffness. The important role of intercellular adhesion in regulating cell-substrate interactions is also illustrated in our confluence experiment, in which aspect ratio differences between substrate conditions persisted until the 17-h time point (Fig. S5). This finding suggests that higher cell packing, and thus a further establishment of intercellular junctions, is required to mask the influence of substrate stiffness on morphology. Our finding is consistent with a recent study where the phenotype of smooth muscle cells was found to depend on both the matrix stiffness and cell density (71).
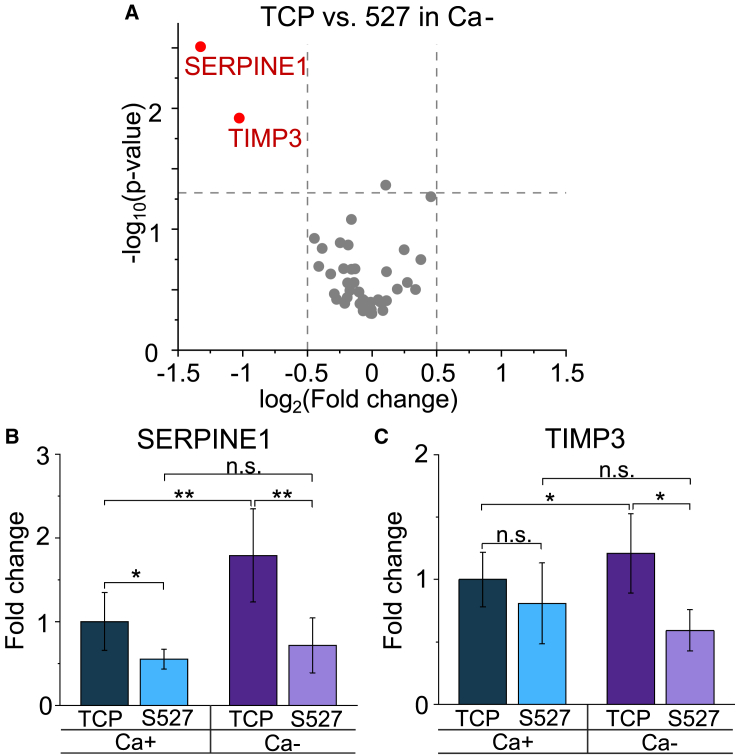
To further investigate how Ca- treatment affects the MDCK response to the substrate stiffness, we quantified gene expression using the previously described panel. Although expression of most of the genes remained the same in this measurement, we found that SERPINE1 and TIMP3 were downregulated (Fig. 4 A). The specific fold changes of these gene expressions can be more clearly observed in the bar charts, in which Ca- treatment was found to greatly exacerbate the differential expression for both genes (Fig. 4 B and C). We also found that SERPINE1 expression was downregulated by the softer substrate stiffness, regardless of intercellular adhesion condition (Figs. 1 C, D, 4 A, and B). Studies have demonstrated that a potential upregulation pathway of SERPINE1, a serin proteinase inhibitor (72,73), is through YAP signaling, which has been shown to be activated by stiff substrates (74, 75, 76, 77, 78). Therefore, we obtained immunofluorescence images of confluent Ca+/- samples stained for YAP (Fig. S9). Here, we found cells grown on S527 to exhibit decreased YAP nuclear translocation activity compared to those grown on TCP, quantified by the nucleus/cytoplasm intensity ratio (Fig. S9 E). This is consistent with the observed SERPINE1 downregulation by soft substrates (Fig. 4 B). Additionally, YAP activity was increased by application of Ca- media due to the loss of intercellular adhesions, which again supports the upregulation of SERPINE1 in cells by calcium deficiency (Fig. 4 B). Overall, our findings suggest that SERPINE1 is biomechanically regulated through YAP signaling. Unlike SERPINE1, TIMP3, a matrix metalloproteinase inhibitor (79,80), has not been well examined in a mechanobiological context. In our experiments, expression of TIMP3 was not found to strongly depend on the substrate stiffness under the normal calcium condition (Fig. 4 C, Table S2). However, upon application of Ca- media, substrate stiffness dictates a change in TIMP3 expression (Fig. 4 C). This suggests that how substrate stiffness affects TIMP3 expression may be calcium dependent.
Figure 4.
(A) Volcano plot of log2 ratio versus p value compares genes expressed by cells grown on TCP and S527 under the Ca- treatment condition. Although ∼96% of tested genes show similar expression, SERPINE1 and TIMP3 were found to be significantly downregulated by the softer substrate. These genes are both related to wound healing and cell-extracellular matrix adhesion. Bar charts for SERPINE1 (B) and TIMP3 (C), using data from Figs. 1C and 3A, provide a visual comparison of relative expression across conditions. These illustrate that Ca- treatment exacerbates the difference in gene expression derived from the substrate stiffness effect. For (A), n = 6. ∗p ≤ 0.05, ∗∗p ≤ 0.01.
To validate these findings, we performed gene expression analysis of our antibody and DTT experiments (Fig. S10). Similar to our Ca- results, both treatments show significant downregulation of SERPINE1 due to substrate stiffness. In addition, the similarity between gene expressions for the Ca- and antibody experiments likely indicates that Ca- treatment is relatively specific for inhibiting E-cadherin. Our findings collectively suggest that SERPINE1 expression in epithelial cells is primarily governed by the substrate stiffness, as the softer substrate was shown to downregulate its expression irrespective of cell-cell adhesion in all tested conditions.
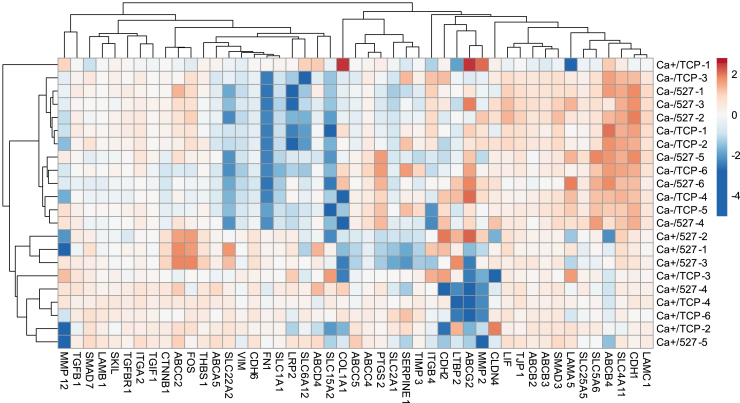
To visualize how the substrate stiffness and intercellular interaction jointly influence gene expression, we combined the data from Figs. 1 C and 3 A to perform an unsupervised hierarchical clustering analysis (Fig. 5). The Ca+ and Ca- data used in this analysis were acquired in the same experiment run to ensure the condition consistency. The clear distinction between Ca+ and Ca-clusters indicates that the influence of substrate stiffness on MDCK cell gene expression is less significant compared to that of intercellular adhesions (Fig. 5). We also found Ca- treatment to increase CDH1 transcription (Fig. 5), despite reduction of E-cadherin protein local enrichment at cellular junctions in the absence of extracellular calcium (Fig. S4 C and D). This activity may be indicative of an innate homeostatic mechanism to recover epithelial phenotype under mild perturbation.
Figure 5.
Average unsupervised hierarchical clustering analysis was performed using the same data sets used for Figs. 1C and 4A to visualize overall trends in gene expression due to substrate stiffness and cell-cell adhesion. Ca- treatment was found to downregulate FN1 and VIM and upregulate CDH1, suggesting that interference of intercellular adhesions may not always induce EMT. Extreme outliers were removed.
Cell motility response to the substrate stiffness and intercellular adhesion effect
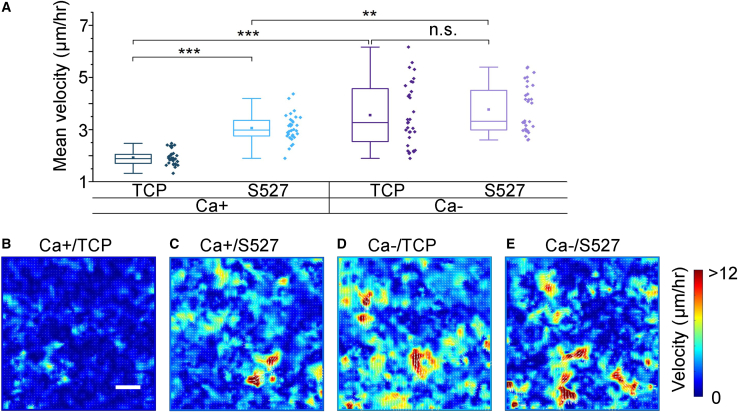
Lastly, we investigated how cell motility is influenced by the cell-substrate and cell-cell interactions. Understanding how the surrounding microenvironment governs the motility phenotype is important as such a relationship has been shown to dictate many biological processes in tissue development, injury repair, and disease progression (81, 82, 83). To characterize the cell dynamics, we acquired time-lapse videos of MDCK monolayers for 48 h post Ca+/- treatment and performed PIV to track the movements of individual cells. This method has previously been applied to epithelial monolayers to characterize the coarse-grained and instantaneous velocity field in cells within the contexts of collective migration (84), wound healing (85,86), and jamming transition (87,88). Only the last 12 h of the viewing period were used to maximize the difference in mean cell velocities, as each condition showed differential velocities over time (Fig. S11). As shown in Fig. 6 A, we found that either lowering the substrate stiffness or reducing the cell-cell adhesion approximately doubled the cell mean velocity. We also found that lowering the substrate stiffness under the Ca-condition did not further increase the cell migration speed significantly. Here, each data point in Fig. 6 A represents a mean speed averaged over a 672 μm × 672 μm field of view, corresponding to ∼2500 cells. We also found that the mobility increase is correlated with the reduced cell density (Fig. S7), consistent with previous findings in jamming transition experiments (87,89,90).
Figure 6.
(A) Cell mean velocity was measured by PIV analysis performed on time-lapse images of cell monolayers treated with Ca + or Ca- media. Regions of interest, with a field of view of 672 μm × 672 μm, were registered and imaged for 48 h post-Ca+/- treatment at an interval of ∼4 min. Both softening of the substrate modulus and disruption of cell-cell contacts were shown to increase cell motility to different degrees. (B–E) Representative heatmaps for each condition illustrate this trend. Additionally, samples with lower mean migration showed greater homogeneity in local velocity magnitudes. Scale bar represents 100 μm. For (A), n = 32. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001.
To visualize the cell motility distribution, we plotted the velocity heatmaps for all tested conditions and showed representative images in Fig. 6 B–E. By comparing these heatmaps, we readily observed the change in the mean velocity illustrated in Fig. 5 A. In addition, we observed substantial dynamic heterogeneities in the samples that exhibited an increasing motility (Fig. 6 C–E). For example, although some cells showed increased migration speeds (yellow and red regions), many cells remained relatively quiescent (blue regions). The magnitude of such a heterogeneity can be characterized by the SD of the mean velocity values, as indicated by the error bars shown in Fig. 5A. With this characterization, we found that the dynamic heterogeneity is most significant for the Ca-/TCP condition (dark purple).
Our finding of the soft substrate-induced cell motility is in contrast to previous single-cell results, where the cell speed increases with increasing substrate stiffness (91,92). In these previous experiments, the separated cells grown on a rigid substrate form rigidity-sensing stress fibers and exert stronger traction forces on the substrate, leading to a higher motility. However, these rigidity-induced cell motions are usually nondirectional in the absence of external factors (93, 94, 95) and could potentially have a small effect on the overall cell motility in a confluent culture.
Specifically, each cell in a confluent monolayer is blocked by its neighbors, requiring coordinated motion for cell migration. Although the underlying mechanism of our softness-induced motility remains unknown, it might be related to downregulated SERPINE1 expression found in Fig. 4. Further experiments such as SERPINE1 knockout tests will have to be performed to validate this hypothesis.
Conclusion
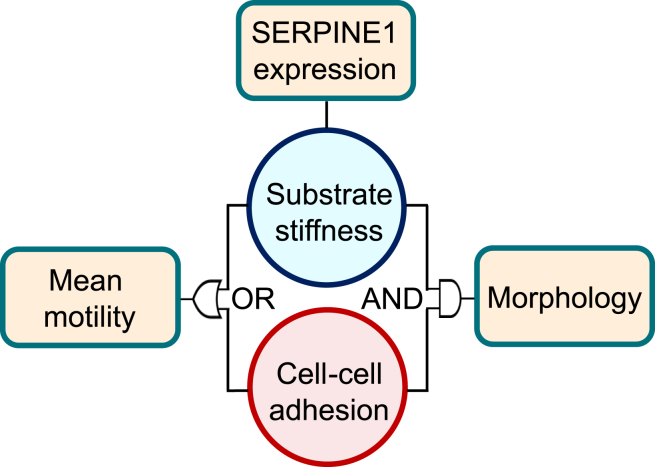
In this work, we comprehensively characterized the cell phenotype and gene expression of MDCK monolayers with systematic adjustments of the substrate stiffness and intercellular adhesion. We summarized how cell-substrate and cell-cell interactions jointly regulate cell behavior using a logic diagram (Fig. 7). First, we showed that confluent MDCK monolayers have a distinct morphological and transcriptional response to substrate stiffness compared to single cells. Specifically, in these established monolayers, intercellular interactions were shown to dominate over the diminished effect of substrate stiffness on the morphology of MDCK cells (AND gate in Fig. 7). This finding supports a central hypothesis of the vertex model, in which the cell shape is primarily determined by the cell-cell interaction. This numerical framework has been widely used to describe the morphology, migration, and mechanical phenotypes of epithelial cells within monolayers (89,96,97). We also found that either the substrate rigidity or intercellular adhesion can alter the cell motility (OR gate in Fig. 7). Furthermore, the presence of cell-cell contacts led to an unexpected increase in cell motility due to a substrate modulus reduction. Lastly, SERPINE1 expression was found to be primarily governed by substrate adjustments (upper box in Fig. 7). Our results collectively suggest that the substrate and intercellular interactions have intertwined impacts on the cells, and results identified using single-cell experiments should be individually validated before applying to confluent cultures.
Figure 7.
The present study identifies how substrate stiffness and cell-cell adhesion affect epithelial morphology, gene expression, and motility. Specifically, for morphology, we found that both factors are able to mask the influence of each other, requiring reduction of both substrate moduli and intercellular interactions for significant morphological change (AND). For motility, however, modulation of both factors was able to induce increases in cell mean velocity (OR). Lastly, SERPINE1 specifically was found to be consistently downregulated on soft substrates, suggesting that the intercellular adhesions are not as influential on SERPINE1 expression as the substrate.
Author contributions
D.C., Z.G., S.Y.H., and N.Y.C.L designed research. D.C., Z.G., S.Y.H., and A.B. performed research. D.C., Z.G., S.Y.H., and N.Y.C.L analyzed data. D.C., Z.G., S.Y.H., and N.Y.C.L wrote the manuscript.
Acknowledgments
We thank Professors Dapeng Max Bi, Amy Rowat, and Jimmy Hu for insightful discussions and guidance.
This work was funded by the Broad Stem Cell Research Center at the University of California – Los Angeles and the UCLA SPORE in Prostate Cancer Grant (P50 CA092131).
Editor: Philip LeDuc.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.11.2887.
Supporting material
References
- 1.Kagnoff M.F., Eckmann L. Epithelial cells as sensors for microbial infection. J. Clin. Invest. 1997;100:6–10. doi: 10.1172/JCI119522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okumura R., Takeda K. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp. Mol. Med. 2017;49:e338. doi: 10.1038/emm.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guguen-Guillouzo C., Clément B., et al. Guillouzo A. Maintenance and reversibility of active albumin secretion by adult rat hepatocytes co-cultured with another liver epithelial cell type. Exp. Cell Res. 1983;143:47–54. doi: 10.1016/0014-4827(83)90107-6. [DOI] [PubMed] [Google Scholar]
- 4.Proud D., Leigh R. Epithelial cells and airway diseases. Immunol. Rev. 2011;242:186–204. doi: 10.1111/j.1600-065X.2011.01033.x. [DOI] [PubMed] [Google Scholar]
- 5.Wilson P.D. Polycystic kidney disease. N. Engl. J. Med. 2004;350:151–164. doi: 10.1056/NEJMra022161. [DOI] [PubMed] [Google Scholar]
- 6.Elenbaas B., Spirio L., et al. Weinberg R.A. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez-Boulan E., Nelson W.J. Morphogenesis of the polarized epithelial cell phenotype. Science. 1989;245:718–725. doi: 10.1126/science.2672330. [DOI] [PubMed] [Google Scholar]
- 8.Tian Y.-C., Fraser D., et al. Phillips A.O. TGF-β1-mediated alterations of renal proximal tubular epithelial cell phenotype. Am. J. Physiol. Renal Physiol. 2003;285:F130–F142. doi: 10.1152/ajprenal.00408.2002. [DOI] [PubMed] [Google Scholar]
- 9.Tambe D.T., Hardin C.C., et al. Trepat X. Collective cell guidance by cooperative intercellular forces. Nat. Mater. 2011;10:469–475. doi: 10.1038/nmat3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayor R., Etienne-Manneville S. The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 2016;17:97–109. doi: 10.1038/nrm.2015.14. [DOI] [PubMed] [Google Scholar]
- 11.Sunyer R., Conte V., et al. Trepat X. Collective cell durotaxis emerges from long-range intercellular force transmission. Science. 2016;353:1157–1161. doi: 10.1126/science.aaf7119. [DOI] [PubMed] [Google Scholar]
- 12.Ladoux B., Mège R.-M. Mechanobiology of collective cell behaviours. Nat. Rev. Mol. Cell Biol. 2017;18:743–757. doi: 10.1038/nrm.2017.98. [DOI] [PubMed] [Google Scholar]
- 13.Friedl P., Locker J., et al. Segall J.E. Classifying collective cancer cell invasion. Nat. Cell Biol. 2012;14:777–783. doi: 10.1038/ncb2548. [DOI] [PubMed] [Google Scholar]
- 14.Friedl P., Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 15.Yeung T., Georges P.C., et al. Janmey P.A. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 16.Tee S.-Y., Fu J., et al. Janmey P.A. Cell shape and substrate rigidity both regulate cell stiffness. Biophys. J. 2011;100:L25–L27. doi: 10.1016/j.bpj.2010.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saez A., Ghibaudo M., et al. Ladoux B. Rigidity-driven growth and migration of epithelial cells on microstructured anisotropic substrates. Proc. Natl. Acad. Sci. U S A. 2007;104:8281–8286. doi: 10.1073/pnas.0702259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei S.C., Fattet L., et al. Yang J. Matrix stiffness drives epithelial–mesenchymal transition and tumour metastasis through a TWIST1–G3BP2 mechanotransduction pathway. Nat. Cell Biol. 2015;17:678–688. doi: 10.1038/ncb3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisenberg J.L., Safi A., et al. Jones J.C. Substrate stiffness regulates extracellular matrix deposition by alveolar epithelial cells. Res. Rep. Biol. 2011;2011:1. doi: 10.2147/RRB.S13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaudhuri O., Koshy S.T., et al. Mooney D.J. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat. Mater. 2014;13:970–978. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- 21.Ondeck M.G., Kumar A., et al. Engler A.J. Dynamically stiffened matrix promotes malignant transformation of mammary epithelial cells via collective mechanical signaling. Proc. Natl. Acad. Sci. U S A. 2019;116:3502–3507. doi: 10.1073/pnas.1814204116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams C.L., Chen Y.-T., et al. James Nelson W. Mechanisms of epithelial cell–cell adhesion and cell compaction revealed by high-resolution tracking of E-cadherin–green fluorescent protein. J. Cell Biol. 1998;142:1105–1119. doi: 10.1083/jcb.142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desai R.A., Gao L., et al. Chen C.S. Cell polarity triggered by cell-cell adhesion via E-cadherin. J. Cell Sci. 2009;122:905–911. doi: 10.1242/jcs.028183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao A.S., Shin J.-W., Mooney D.J. Effects of substrate stiffness and cell-cell contact on mesenchymal stem cell differentiation. Biomaterials. 2016;98:184–191. doi: 10.1016/j.biomaterials.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim N.-G., Koh E., et al. Gumbiner B.M. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. U S A. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Auersperg N., Pan J., et al. Roskelley C.D. E-cadherin induces mesenchymal-to-epithelial transition in human ovarian surface epithelium. Proc. Natl. Acad. Sci. U S A. 1999;96:6249–6254. doi: 10.1073/pnas.96.11.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rappel W.-J. Cell–cell communication during collective migration. Proc. Natl. Acad. Sci. U S A. 2016;113:1471–1473. doi: 10.1073/pnas.1524893113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vedula S.R.K., Leong M.C., et al. Ladoux B. Emerging modes of collective cell migration induced by geometrical constraints. Proc. Natl. Acad. Sci. U S A. 2012;109:12974–12979. doi: 10.1073/pnas.1119313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brückner B.R., Janshoff A. Importance of integrity of cell-cell junctions for the mechanics of confluent MDCK II cells. Sci. Rep. 2018;8:1–11. doi: 10.1038/s41598-018-32421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Praetorius H., Spring K. Bending the MDCK cell primary cilium increases intracellular calcium. J. Membr. Biol. 2001;184:71–79. doi: 10.1007/s00232-001-0075-4. [DOI] [PubMed] [Google Scholar]
- 31.Takada K., Kawakami C., et al. Kawaoka Y. A humanized MDCK cell line for the efficient isolation and propagation of human influenza viruses. Nat. Microbiol. 2019;4:1268–1273. doi: 10.1038/s41564-019-0433-6. [DOI] [PubMed] [Google Scholar]
- 32.Imai M., Furusawa K., et al. Haga H. Three-dimensional morphogenesis of MDCK cells induced by cellular contractile forces on a viscous substrate. Sci. Rep. 2015;5:1–10. doi: 10.1038/srep14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saxena N., Jadhav S., Sen S. Fabrication of a microfluidic device for studying the combinatorial effect of physical and chemical cues on cell migration. STAR Protoc. 2021;2:100310. doi: 10.1016/j.xpro.2021.100310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palchesko R.N., Zhang L., et al. Feinberg A.W. Development of polydimethylsiloxane substrates with tunable elastic modulus to study cell mechanobiology in muscle and nerve. PLoS one. 2012;7:e51499. doi: 10.1371/journal.pone.0051499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hinck L., Nelson W.J., Papkoff J. Wnt-1 modulates cell-cell adhesion in mammalian cells by stabilizing beta-catenin binding to the cell adhesion protein cadherin. J. Cell Biol. 1994;124:729–741. doi: 10.1083/jcb.124.5.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heimark R.L., Degner M., Schwartz S.M. Identification of a Ca2 (+)-dependent cell-cell adhesion molecule in endothelial cells. J. Cell Biol. 1990;110:1745–1756. doi: 10.1083/jcb.110.5.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirano S., Nose A., et al. Takeichi M. Calcium-dependent cell-cell adhesion molecules (cadherins): subclass specificities and possible involvement of actin bundles. J. Cell Biol. 1987;105:2501–2510. doi: 10.1083/jcb.105.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Damsky C.H., Richa J., et al. Buck C.A. Identification and purification of a cell surface glycoprotein mediating intercellular adhesion in embryonic and adult tissue. Cell. 1983;34:455–466. doi: 10.1016/0092-8674(83)90379-3. [DOI] [PubMed] [Google Scholar]
- 39.Jacot J.G., McCulloch A.D., Omens J.H. Substrate stiffness affects the functional maturation of neonatal rat ventricular myocytes. Biophys. J. 2008;95:3479–3487. doi: 10.1529/biophysj.107.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown X.Q., Ookawa K., Wong J.Y. Evaluation of polydimethylsiloxane scaffolds with physiologically-relevant elastic moduli: interplay of substrate mechanics and surface chemistry effects on vascular smooth muscle cell response. Biomaterials. 2005;26:3123–3129. doi: 10.1016/j.biomaterials.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Noren N.K., Niessen C.M., et al. Burridge K. Cadherin engagement regulates Rho family GTPases. J. Biol. Chem. 2001;276:33305–33308. doi: 10.1074/jbc.C100306200. [DOI] [PubMed] [Google Scholar]
- 42.Zantek N.D., Azimi M., et al. Kinch M.S. E-cadherin regulates the function of the EphA2 receptor tyrosine kinase. Cell Growth Differ. 1999;10:629–638. [PubMed] [Google Scholar]
- 43.Thevenaz P., Ruttimann U.E., Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 44.Metsalu T., Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhandayuthapani B., Yoshida Y., et al. Kumar D.S. Polymeric scaffolds in tissue engineering application: a review. Int. J. Polym. Sci. 2011;2011:290602. [Google Scholar]
- 46.Meng Z., Wang Y., et al. Zheng Y. Electrospinning of PLGA/gelatin randomly-oriented and aligned nanofibers as potential scaffold in tissue engineering. Mater. Sci. Eng. C. 2010;30:1204–1210. [Google Scholar]
- 47.Maitz M.F. Applications of synthetic polymers in clinical medicine. Biosurf. Biotribol. 2015;1:161–176. [Google Scholar]
- 48.Ulery B.D., Nair L.S., Laurencin C.T. Biomedical applications of biodegradable polymers. J. Polym. Sci. B Polym. Phys. 2011;49:832–864. doi: 10.1002/polb.22259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hogan C., Serpente N., et al. Fujita Y. Rap1 regulates the formation of E-cadherin-based cell-cell contacts. Mol. Cell. Biol. 2004;24:6690–6700. doi: 10.1128/MCB.24.15.6690-6700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ando-Akatsuka Y., Yonemura S., et al. Tsukita S. Differential behavior of E-cadherin and occludin in their colocalization with ZO-1 during the establishment of epithelial cell polarity. J. Cell. Physiol. 1999;179:115–125. doi: 10.1002/(SICI)1097-4652(199905)179:2<115::AID-JCP1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 51.Bhana B., Iyer R.K., et al. Radisic M. Influence of substrate stiffness on the phenotype of heart cells. Biotechnol. Bioeng. 2010;105:1148–1160. doi: 10.1002/bit.22647. [DOI] [PubMed] [Google Scholar]
- 52.Zarkoob H., Bodduluri S., et al. Sander E.A. Substrate stiffness affects human keratinocyte colony formation. Cell Mol. Bioeng. 2015;8:32–50. doi: 10.1007/s12195-015-0377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mak M., Spill F., et al. Zaman M.H. Single-cell migration in complex microenvironments: mechanics and signaling dynamics. J. Biomech. Eng. 2016;138:021004. doi: 10.1115/1.4032188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim M., Gweon B., et al. Shin J.H. Matrix stiffness induces epithelial mesenchymal transition phenotypes of human epidermal keratinocytes on collagen coated two dimensional cell culture. Biomed. Eng. Lett. 2015;5:194–202. [Google Scholar]
- 55.Kass L., Erler J.T., et al. Weaver V.M. Mammary epithelial cell: influence of extracellular matrix composition and organization during development and tumorigenesis. Int. J. Biochem. Cell Biol. 2007;39:1987–1994. doi: 10.1016/j.biocel.2007.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mennens S.F., Bolomini-Vittori M., et al. van den Dries K. Substrate stiffness influences phenotype and function of human antigen-presenting dendritic cells. Sci. Rep. 2017;7:1–14. doi: 10.1038/s41598-017-17787-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen J., Backman L.J., et al. Danielson P. Regulation of keratocyte phenotype and cell behavior by substrate stiffness. ACS Biomater. Sci. Eng. 2020;6:5162–5171. doi: 10.1021/acsbiomaterials.0c00510. [DOI] [PubMed] [Google Scholar]
- 58.Semler E.J., Ranucci C.S., Moghe P.V. Mechanochemical manipulation of hepatocyte aggregation can selectively induce or repress liver-specific function. Biotechnol. Bioeng. 2000;69:359–369. doi: 10.1002/1097-0290(20000820)69:4<359::aid-bit2>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 59.Emelianov S., Lubinski M., et al. O’Donnell M. Reconstructive ultrasound elasticity imaging for renal transplant diagnosis: kidney ex vivo results. Ultrason. Imaging. 2000;22:178–194. doi: 10.1177/016173460002200303. [DOI] [PubMed] [Google Scholar]
- 60.Sotomayor M., Schulten K. The allosteric role of the Ca2+ switch in adhesion and elasticity of C-cadherin. Biophys. J. 2008;94:4621–4633. doi: 10.1529/biophysj.107.125591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim S.A., Tai C.-Y., et al. Schuman E.M. Calcium-dependent dynamics of cadherin interactions at cell–cell junctions. Proc. Natl. Acad. Sci. U S A. 2011;108:9857–9862. doi: 10.1073/pnas.1019003108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vedula S.R., Hirata H., et al. Ladoux B. Epithelial bridges maintain tissue integrity during collective cell migration. Nat. Mater. 2014;13:87–96. doi: 10.1038/nmat3814. [DOI] [PubMed] [Google Scholar]
- 63.Jain S., Cachoux V.M.L., et al. Ladoux B. The role of single-cell mechanical behaviour and polarity in driving collective cell migration. Nat. Phys. 2020;16:802–809. doi: 10.1038/s41567-020-0875-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Volksdorf T., Heilmann J., et al. Brandner J.M. Tight junction proteins claudin-1 and occludin are important for cutaneous wound healing. Am. J. Pathol. 2017;187:1301–1312. doi: 10.1016/j.ajpath.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 65.Carafoli E., Krebs J. Why calcium? How calcium became the best communicator. J. Biol. Chem. 2016;291:20849–20857. doi: 10.1074/jbc.R116.735894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clapham D.E. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 67.Sjaastad M.D., Nelson W.J. Integrin-mediated calcium signaling and regulation of cell adhesion by intracellular calcium. Bioessays. 1997;19:47–55. doi: 10.1002/bies.950190109. [DOI] [PubMed] [Google Scholar]
- 68.Ozawa M., Hoschützky H., et al. Kemler R. A possible new adhesive site in the cell-adhesion molecule uvomorulin. Mech. Dev. 1990;33:49–56. doi: 10.1016/0925-4773(90)90134-8. [DOI] [PubMed] [Google Scholar]
- 69.Sevier C.S., Kaiser C.A. Formation and transfer of disulphide bonds in living cells. Nat. Rev. Mol. Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 70.Li L., Hartley R., et al. Zhao M. E-cadherin plays an essential role in collective directional migration of large epithelial sheets. Cell Mol. Life Sci. 2012;69:2779–2789. doi: 10.1007/s00018-012-0951-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stasiak S.E., Jamieson R.R., et al. Parameswaran H. Intercellular communication controls agonist-induced calcium oscillations independently of gap junctions in smooth muscle cells. Sci. Adv. 2020;6:eaba1149. doi: 10.1126/sciadv.aba1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou A., Huntington J.A., et al. Read R.J. How vitronectin binds PAI-1 to modulate fibrinolysis and cell migration. Nat. Struct. Mol. Biol. 2003;10:541–544. doi: 10.1038/nsb943. [DOI] [PubMed] [Google Scholar]
- 73.Mimuro J., Loskutoff D.J. Purification of a protein from bovine plasma that binds to type 1 plasminogen activator inhibitor and prevents its interaction with extracellular matrix: evidence that the protein is vitronectin. J. Biol. Chem. 1989;264:936–939. [PubMed] [Google Scholar]
- 74.Liu F., Lagares D., et al. Tschumperlin D.J. Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 2015;308:L344–L357. doi: 10.1152/ajplung.00300.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saito A., Nagase T. Hippo and TGF-β interplay in the lung field. Am. J. Physiol. Lung Cell Mol. Physiol. 2015;309:L756–L767. doi: 10.1152/ajplung.00238.2015. [DOI] [PubMed] [Google Scholar]
- 76.Kong H.-J., Kwon E.-J., et al. Cha H.-J. Crosstalk between YAP and TGFβ regulates SERPINE1 expression in mesenchymal lung cancer cells. Int. J. Oncol. 2021;58:111–121. doi: 10.3892/ijo.2020.5153. [DOI] [PubMed] [Google Scholar]
- 77.Marquard S., Thomann S., et al. Breuhahn K. Yes-associated protein (YAP) induces a secretome phenotype and transcriptionally regulates plasminogen activator Inhibitor-1 (PAI-1) expression in hepatocarcinogenesis. Cell Commun. Signal. 2020;18:1–15. doi: 10.1186/s12964-020-00634-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Girard C.A., Lecacheur M., et al. Tartare-Deckert S. A feed-forward mechanosignaling loop confers resistance to therapies targeting the MAPK pathway in BRAF-mutant melanoma. Cancer Res. 2020;80:1927–1941. doi: 10.1158/0008-5472.CAN-19-2914. [DOI] [PubMed] [Google Scholar]
- 79.Baker A.H., Zaltsman A.B., George S.J., Newby A.C. Divergent effects of tissue inhibitor of metalloproteinase-1,-2, or-3 overexpression on rat vascular smooth muscle cell invasion, proliferation, and death in vitro. TIMP-3 promotes apoptosis. J. Clin. Invest. 1998;101:1478–1487. doi: 10.1172/JCI1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lambert E., Dassé E., et al. Petitfrère E. TIMPs as multifacial proteins. Crit. Rev. Oncol. Hematol. 2004;49:187–198. doi: 10.1016/j.critrevonc.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 81.Kurten R.C., Chowdhury P., et al. Jones S.M. Coordinating epidermal growth factor-induced motility promotes efficient wound closure. Am. J. Physiol. Cell Physiol. 2005;288:C109–C121. doi: 10.1152/ajpcell.00024.2003. [DOI] [PubMed] [Google Scholar]
- 82.Thiery J.P., Acloque H., et al. Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 83.Detrich H.W., Kieran M.W., et al. Zon L.I. Intraembryonic hematopoietic cell migration during vertebrate development. Proc. Natl. Acad. Sci. U S A. 1995;92:10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Angelini T.E., Hannezo E., et al. Weitz D.A. Glass-like dynamics of collective cell migration. Proc. Natl. Acad. Sci. U S A. 2011;108:4714–4719. doi: 10.1073/pnas.1010059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Park J.A., Kim J.H., et al. Fredberg J.J. Unjamming and cell shape in the asthmatic airway epithelium. Nat. Mater. 2015;14:1040–1048. doi: 10.1038/nmat4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Poujade M., Grasland-Mongrain E., et al. Silberzan P. Collective migration of an epithelial monolayer in response to a model wound. Proc. Natl. Acad. Sci. U S A. 2007;104:15988–15993. doi: 10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Garcia S., Hannezo E., et al. Gov N.S. Physics of active jamming during collective cellular motion in a monolayer. Proc. Natl. Acad. Sci. U S A. 2015;112:15314–15319. doi: 10.1073/pnas.1510973112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Puliafito A., Hufnagel L., et al. Shraiman B.I. Collective and single cell behavior in epithelial contact inhibition. Proc. Natl. Acad. Sci. U S A. 2012;109:739–744. doi: 10.1073/pnas.1007809109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bi D., Yang X., et al. Manning M.L. Motility-driven glass and jamming transitions in biological tissues. Phys. Rev. X. 2016;6:021011. doi: 10.1103/PhysRevX.6.021011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mongera A., Rowghanian P., et al. Campàs O. A fluid-to-solid jamming transition underlies vertebrate body axis elongation. Nature. 2018;561:401–405. doi: 10.1038/s41586-018-0479-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ulrich T.A., de Juan Pardo E.M., Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69:4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li S., Guan J.-L., Chien S. Biochemistry and biomechanics of cell motility. Annu. Rev. Biomed. Eng. 2005;7:105–150. doi: 10.1146/annurev.bioeng.7.060804.100340. [DOI] [PubMed] [Google Scholar]
- 93.Huttenlocher A. Cell polarization mechanisms during directed cell migration. Nat. Cell Biol. 2005;7:336–337. doi: 10.1038/ncb0405-336. [DOI] [PubMed] [Google Scholar]
- 94.Jiang X., Bruzewicz D.A., et al. Whitesides G.M. Directing cell migration with asymmetric micropatterns. Proc. Natl. Acad. Sci. U S A. 2005;102:975–978. doi: 10.1073/pnas.0408954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roussos E.T., Condeelis J.S., Patsialou A. Chemotaxis in cancer. Nat. Rev. Cancer. 2011;11:573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nagai T., Honda H. A dynamic cell model for the formation of epithelial tissues. Philos. Mag. B. 2001;81:699–719. [Google Scholar]
- 97.Fletcher A.G., Osterfield M., et al. Shvartsman S.Y. Vertex models of epithelial morphogenesis. Biophys. J. 2014;106:2291–2304. doi: 10.1016/j.bpj.2013.11.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.