Abstract
Although life-history trade-offs are central to life-history evolution, their mechanistic basis is often unclear. Traditionally, trade-offs are understood in terms of competition for limited resources among traits within an organism, which could be mediated by signal transduction pathways at the level of cellular metabolism. Nevertheless, trade-offs are also thought to be produced as a consequence of the performance of one activity generating negative consequences for other traits, or the result of genes or pathways that simultaneously regulate two life-history traits in opposite directions (antagonistic pleiotropy), independent of resource allocation. Yet examples of genes with antagonistic effects on life-history traits are limited. This study provides direct evidence for a gene—RLS1, that is involved in increasing survival in nutrient-limiting environments at a cost to immediate reproduction in the single-celled photosynthetic alga, Chlamydomonas reinhardtii. Specifically, we show that RLS1 mutants are unable to properly suppress their reproduction in phosphate-deprived conditions. Although these mutants have an immediate reproductive advantage relative to the parental strain, their long-term survival is negatively affected. Our data suggest that RLS1 is a bona fide life-history trade-off gene that suppresses immediate reproduction and ensures survival by downregulating photosynthesis in limiting environments, as part of the general acclimation response to nutrient deprivation in photosynthetic organisms.
Keywords: Chlamydomonas reinhardtii, life-history trade-off, antagonistic pleiotropy, acclimation, phosphate-deprivation, RLS1
1. Introduction
Although life-history trade-offs are central to life-history evolution [1–3], their mechanistic basis is often unclear (e.g. [3–6]). Traditionally, trade-offs have been understood in terms of competition for limited resources among traits (e.g. reproduction, somatic growth, maintenance) within an organism (aka adaptive resource allocation [7]). Reduced nutrient availability is known to substantially magnify an apparent trade-off, while increased nutrient availability can diminish (or remove) negative interactions between traits (e.g. [8]). Such plastic responses are thought to be determined by priority rules that determine how resources are allocated in response to different nutrient inputs [7]. In some animal lineages, laboratory and field experiments showed that under nutrient-limiting or stressful conditions, energy and resources were directed to maintenance or storage functions, taking precedence over reproduction [7].
Trade-offs can occur between physiological traits that are expressed at the same or at different times of the life cycle, and can result from genetic factors (pleiotropy), environmental factors, or combinations of these two types of factors that result in negative interactions between traits [7,9]. Although artificial selection and experimental evolution have shown that trade-offs can have a genetic basis (see [10]), few studies have been able to pinpoint the underlying molecular mechanisms [6]. For instance, while trade-offs could be the result of adaptive resource allocation at the organismal level [2,11], this differential allocation can be caused by a trade-off between protein biosynthesis (growth) and energy metabolism (survival)—probably mediated by signal transduction pathways at the level of cellular metabolism [2]. Trade-offs can also be produced as a consequence of the performance of one activity generating negative consequences for other traits. For example, aerobic metabolism generates reactive oxygen species (ROS) that, if not fully neutralized, can be damaging to biological molecules and negatively affect other activities or life traits, such as lifespan [5,12,13]. Furthermore, trade-offs could be the result of signalling genes or pathways that simultaneously regulate two life-history traits in opposite directions, independent of resource allocation [6,14]. For instance, yeast mutants in the Ras2/cAMP pathway cannot arrest their reproduction in limiting environments and are more sensitive to stress due to their inability to induce the transcription of stress-related genes (e.g. [15]). Nevertheless, examples of genes with antagonistic effects on two life-history traits are limited [16].
Because the loci involved in life-history trade-offs are expected to affect traits in opposite ways (i.e. show antagonistic pleiotropy [1]), life-history trade-offs are generally thought to limit the set of possible trait combinations, and thus restrict the range of possible evolutionary outcomes [10]. However, during transitions in individuality—such as the transition to multicellularity and eusociality, trade-offs constraining the evolutionary trajectories of solitary individuals can be uncoupled through the evolution of specialized cells in multicellular individuals [17,18] and castes in eusocial insects [19]. Specifically, in unicellular individuals, the same cell contributes to both survival and reproduction, but these activities are not performed at the same time; the cell switches from investing in survival or in reproduction (i.e. a trade-off). In multicellular individuals, however, cells can specialize in either activity (the trade-off is broken), and this leads to the differentiation of survival-enhancing cells (soma) and reproductive cells (germ). At a mechanistic level, we argued that the evolution of soma and germ involved the co-option of mechanisms underlying survival-reproduction trade-offs in unicellular lineages, by changing their expression from a temporal into a spatial context [20,21].
To explore this possibility, we are using the volvocine lineage—a group of haploid green algae that comprises both unicellular species (e.g. Chlamydomonas reinhardtii) and multicellular species with fully differentiated somatic and germ lines, such as Volvox carteri. In V. carteri, a single gene—known as regA, is both necessary and sufficient to determine somatic cell fate [22]. regA is only expressed in somatic cells and codes for a putative transcription factor that is thought to repress the expression of nuclear-encoded chloroplast proteins [23,24], which in turn will negatively affect photosynthesis and thus the cell growth and division of somatic cells. We have previously suggested that regA evolved from a life-history gene that was involved in trading off reproduction for survival in its unicellular ancestors [20]. In this context, we showed that regA's closest homolog in the unicellular C. reinhardtii (known as RLS1; RegA-Like-Sequence 1) is expressed under conditions that require the suppression of immediate reproduction to ensure survival [20,21]. However, the direct role of this putative life-history trade-off gene in the survival and reproduction of C. reinhardtii and its ability to trade off these two life-history traits has not been examined. Here, we directly tested the hypothesis that RLS1 is a life-history trade-off gene by investigating the reproduction and long-term survival of a C. reinhardtii RLS1 mutant and its wild-type parental strain under limiting environmental conditions.
2. Material and methods
(a) . Strains and growth conditions
A C. reinhardtii genomic RLS1 mutant (LMJ.RY0402.057072) and its wild-type parental strain (CC-4533 cw15 mt-) were obtained from the Chlamydomonas Resource Center (https://www.chlamycollection.org/). The RLS1 mutant was generated by random insertion of a paromomycin resistance cassette [25]. Stock cultures of both strains were grown in Tris-Acetate-Phosphate (TAP) medium (https://www.chlamycollection.org/methods/) on a rotary shaker (100 r.p.m.) at 25°C, under a 12 h light (150 µmol quanta m−2 s−1): 12 h dark regime.
Experimental cultures were grown in either the normal light : dark (LD) cycle or in continuous dark (CD). The LD regime ensures the population is synchronized, as cells grow during the light phase (when they perform photosynthesis) and divide in the dark. Cultures grown in CD are strictly heterotrophic, allowing us to assess the impact of light and photosynthesis on cell growth and division in optimal and nutrient-limiting conditions. Nutrient-deprived cultures were grown in phosphate-depleted medium (Tris-Acetate; TA) prepared by substituting potassium phosphate with potassium chloride [26]. Experimental cultures were initiated at 5 × 105 cells ml−1, with three biological replicates (see electronic supplementary material for additional detail). Population growth was assessed daily (at 2.5 h into the light cycle) by counting the cells using a haemocytometer; for each biological replicate, four technical replicates (i.e. four aliquots per culture) were averaged.
(b) . Cell viability
Dead and live cells were assessed using a fluorescent exclusion dye (SYTOX Green; excitation/emission = 504/523 nm; Invitrogen). To evaluate long-term viability (i.e. cells that are both alive and able to exit quiescence and reproduce when optimal conditions return) we used a simplified serial dilution agar plating method [27] (see electronic supplementary material for additional detail). Briefly, equivalent culture aliquots containing 250 cells were plated on TAP agar plates and the visible growing colonies (corresponding to the cells that were both alive and viable) were counted for each plate. The number of non-viable cells (alive but not able to produce colonies) was calculated as the difference between the number of live cells (as assessed using the SYTOX green) and the number of colonies (viable cells) [28]. Viable, non-viable and dead cells were then expressed as percentage of the total number of cells in the culture.
(c) . Statistical analyses
Statistical analyses were performed using GraphPad Prism 9 (www.graphpad.com/). To assess the significance of the potential differences in number of cells (i) between the wild-type and RLS1 mutant strain at each time point and (ii) over time (for each strain), we used two-way ANOVA analyses (95% confidence interval; Tukey's multiple comparison tests). For the viability analyses, we performed unpaired two-sample t-tests (95% confidence interval).
(d) . Identification of the RLS1 mutation
To confirm the disruption of RLS1 and characterize the specific RLS1 mutation, clonal cultures were established and DNA was extracted using the Qiagen DNA Plant Extraction Kit. The genomic area assumed to be affected was amplified using primers corresponding to the RLS1 regions flanking the insertion cassette as well as cassette-specific primers (following the protocol provided at https://www.chlamylibrary.org) (electronic supplementary material, figures S1 and S2).
3. Results and discussion
(a) . Genetic and phenotypic characterization of the RLS1 mutant
PCR amplification and sequencing confirmed that the RLS1 coding region is disrupted in the mutant strain. Specifically, the cassette is inserted three nucleotides upstream of the 3′-end of the first coding exon, resulting in a truncated mutant protein containing part of the cassette (figure 1a). Notably, the insertion interrupts the SAND domain—a conserved DNA binding domain responsible for the function of SAND-containing transcription factors [29].
Figure 1.
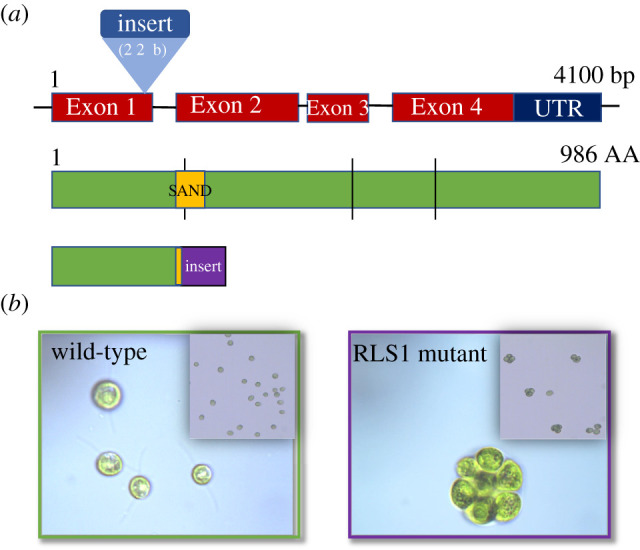
Genetic (a) and phenotypic (b) characterization of the Chlamydomonas reinhardtii RLS1 mutant. (a). RLS1 gene structure (top row) showing the location of the paromomycin resistance cassette [25]; the insertion results in a truncated protein that lacks a functional SAND domain and includes a region of the translated cassette. (b) Micrographs of the wild-type parental and the RLS1 mutant strains. (Online version in colour.)
While growing the mutant cultures, we noticed that during exponential phase, in contrast to the wild-type parental strain, the RLS1 mutant grows mainly as cell clusters (figure 1b). The phenotype appears to be due to the inability of the daughter cells to hatch out from under the mother cell wall, which results in non-flagellated clusters of cells. However, as the cultures enter the stationary phase, the populations express a mix of single cells and small clusters (see inset in figure 1b), and revert to single-celled individuals in late stationary phase.
(b) . RLS1 suppresses reproduction in phosphate-deprived environments
In C. reinhardtii—as in other photosynthetic organisms, the lack of nutrients (e.g. phosphate, sulfur, nitrogen) limits the consumption of NADPH and ATP generated via photosynthesis, due to the slow-down of anabolic processes and the decreased demand for reductant [30]. Consequently, the photosynthetic electron transport chain becomes reduced and the redox potential of the cell increases [30,31]. Excessive reduction of the electron transport chain can lead to over-excitation of chlorophyll molecules and the accumulation of high potential electrons that can interact with oxygen and create ROS [32]. Although ROS are by-products of normal metabolism and act as secondary messengers in various signal transduction pathways (e.g. [33–35]), increased intracellular levels of ROS (oxidative stress) can alter cellular functions and damage many biological structures, most importantly, DNA (e.g. [33]). Thus, it has been suggested that in order to decrease the potential damaging effect of excess light energy under nutrient limitation—and increase survival, photosynthesis needs to be downregulated [30,32,36]. This acclimation process is a well-known general stress response that coordinates nutrient availability with the metabolism of the photosynthetic cell and its growth and division potential, resulting in a temporary inhibition of cell division (cell cycle arrest), and thus a decrease in reproduction [30,31]. We predicted that if RLS1 acted as a life-history gene that increases survival at a cost to immediate reproduction (as we previously hypothesized; [20]), loss of RLS1 should reflect in differences between the RLS1 mutant and its parental wild-type strain in the ability to both suppress reproduction and survive in nutrient-deprived environments.
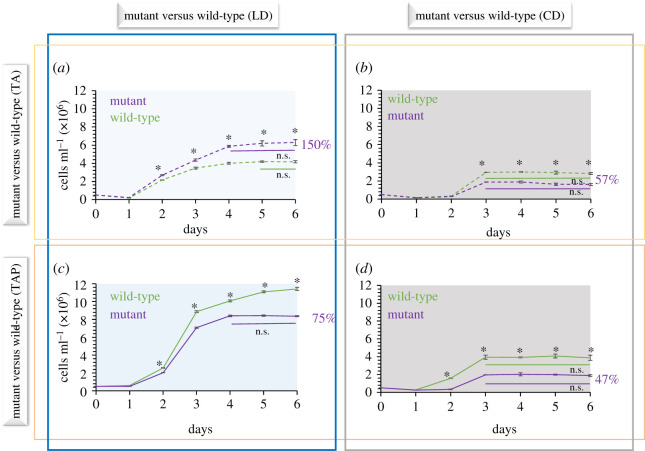
First, to address the role of RLS1 in suppressing the reproduction of C. reinhardtii in nutrient-limited conditions, we compared the population growth of the two strains grown in phosphate-depleted media (figure 2 and electronic supplementary material, figure S3). As predicted, in a phosphate-deprived medium under an LD regime, the RLS1 mutant reproduced more than the wild-type strain and the final population exceeded the size of its wild-type counterpart by 50% (i.e. 6.2 × 106 versus 4.1 × 106 cells ml−1; p < 0.0001; figure 2a). However, the reproductive advantage of RLS1 mutant was lost when cultures were maintained in constant dark (figure 2b); in fact, its population size only reached 57% of that of the wild-type strain (1.6 × 106 versus 2.8 × 106 cells ml−1; p < 0.0001). These differences in the RLS1 mutant's response to phosphate deprivation between LD and CD regimes are consistent with RLS1's involvement in the response to nutrient deprivation being linked to photosynthetic activities. Other mutants in genes involved in acclimation to nutrient stress in C. reinhardtii are also known to respond differently in light and dark. For instance, mutants that are unable to downregulate the photosynthetic electron transport during nutrient deprivation die sooner than the wild-type when grown in the light due to accumulation of photo-oxidative damage; but when maintained in the dark, they can survive nutrient deprivation as well as the wild-type strains do [32,36,37].
Figure 2.
Comparisons between the population growth of wild-type and RLS1 mutant strains under either LD (a,c) or CD (b,d) regimes, in phosphate-depleted (TA; a,b) or phosphate-replete (TAP; c,d) media. Solid and dashed lines indicate phosphate-replete and phosphate-depleted media, respectively. Error bars represent 2xSE (three biological replicates); asterisks denote significant differences (p < 0.0001) between the two strains at each corresponding time point. Straight lines below growth curves indicate time periods characterized by non-significant (n.s.) growth (i.e. stationary phase) for each strain. Percentages indicate the final mutant population size relative to that of the parental strain. (Online version in colour.)
Interestingly, the reproductive advantage of RLS1 mutants was also lost in phosphate-replete medium; in fact, the loss of RLS1 had a negative impact on population growth in cultures grown under either an LD regime or CD (figure 2c,d). Specifically, under an LD regime, the mutant population achieved only around 75% of the final wild-type population size (8.3 × 106 cells ml−1 versus 1.1 × 107; p < 0.0001; figure 2c). Also, the mutant population entered the stationary phase on day 4 (i.e. no difference in population size between days 4 and 5, and days 5 and 6; p = 0.7414 and p = 0.4813, respectively) while the wild-type population continued to grow from day 4 to 5 and 6 (p < 0.0001 and p = 0.0015, respectively). In CD, the difference in population growth between the two strains was even more pronounced, as the mutant only achieved 47% of the wild-type population size (1.8 × 106 versus 3.8 × 106 cells ml−1; p < 0.0001; figure 2d). The more significant negative effect associated with the loss of RLS1 in cultures grown in CD suggests that RLS1 is also involved—directly or indirectly, in regulating reproduction in the dark. Such a possibility is supported by the fact that RLS1 was found to be induced in cultures maintained in the dark [20,21]. Since the expression of RLS1 in the dark coincided with the downregulation of a chloroplast light-harvesting protein, it has been suggested that RLS1 might be involved in the suppression of chloroplast biosynthesis in the dark to avoid investment of resources and energy in a structure whose function is limited in the absence of light [20]. Consequently, those resources and energy can be re-allocated to growth and reproduction. In this scenario, the loss of RLS1 will prevent the reallocation of resources from chloroplast biosynthesis to cell growth and reproduction, which can explain the lower population levels of the RLS1 mutant grown in either CD or a 12 h light : 12 h dark regime (figure 2c,d).
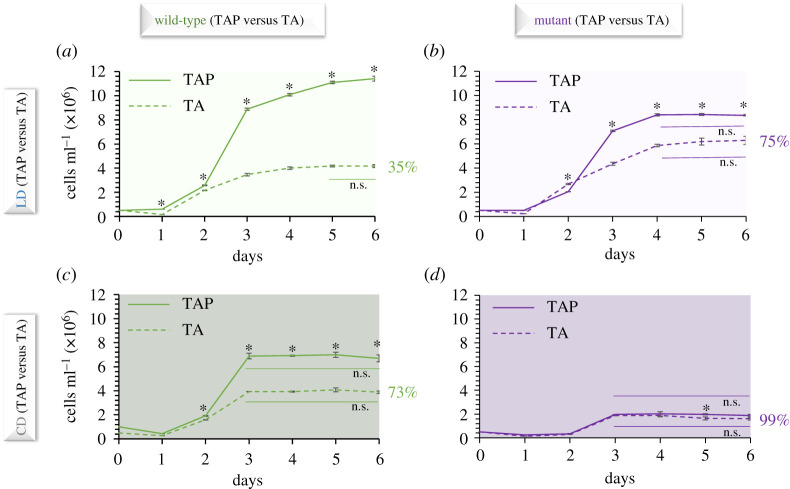
To further address the exact role of RLS1 in suppressing the reproduction of C. reinhardtii in nutrient-limiting environments, we compared the growth curves of the wild-type and RLS1 mutant strains in phosphate-depleted media in either LD or CD regimes (figure 3; electronic supplementary material, figure S4). Phosphate is an important macronutrient for all organisms, and C. reinhardtii grown in phosphate-depleted media was shown to have a significantly lower population growth [38,39]. As expected, under an LD regime, the wild-type responded to phosphate deprivation by suppressing its reproduction such that the maximal population size after 6 days was only ca. 35% of the level achieved in phosphate-replete medium (1.14 × 107 versus 4.16 × 106 cells ml−1; p < 0.0001; figure 3a). However, under the same conditions, the RLS1 mutant population continued to grow and reached up to 75% of its growth capacity in optimal conditions (8.36 × 106 versus 6.28 × 106 cells ml−1; p < 0.0001; figure 3b), confirming that the mutant is less able to suppress its reproduction under phosphate deprivation. Also, as expected if the suppression of reproduction under phosphate deprivation was related to photosynthetic activities, the response of the wild-type to the lack of phosphate was less pronounced in CD (figure 3c). Specifically, in phosphate-depleted media, compared to only 35% in the LD regime (figure 3a), the wild-type strain maintained approximately 73% of its growth capacity in the dark (3.87 × 106 versus 2.82 × 106 cells ml−1; p < 0.0001; figure 3c; electronic supplementary material, figure S5c). On the other hand, compared to the LD regime (figure 3b), the mutant showed almost no response to phosphate deprivation when grown in CD (95%; p = 0.0002; figure 3d; electronic supplementary material, figure S5d). Overall, the differences in the way the wild-type and RLS1 mutant strains responded to phosphate deprivation in both LD and D regimes indicate that the involvement of RLS1 in suppressing reproduction in a diurnal cycle is linked to changes in photosynthetic activities that are known to be associated with the acclimation to nutrient deprivation [30]. This is consistent with the fact that RLS1 can also be induced by a photosynthetic electron transport inhibitor that elicits acclimation-like responses [21]. Furthermore, the differences between the RLS1 mutant and the wild-type populations when grown in the dark (in either phosphate-replete or depleted media) are consistent with an additional role of RLS1 in optimizing reproduction in the dark. In fact, when grown in the dark, the RLS1 mutant is not able to maintain the same relative population size as the wild-type strain (see electronic supplementary material, figure S5).
Figure 3.
Comparisons between the responses to phosphate-deprivation in a LD regime (a,b) and in CD (c,d) of the C. reinhardtii wild-type (a,c) and RLS1 mutant (b,d). Solid and dashed lines indicate phosphate-replete (TAP) and phosphate-depleted (TA) media, respectively. Error bars represent 2xSE (three biological replicates); asterisks denote significant differences p < 0.0001 between the two strains at each corresponding time point. Straight lines below growth curves indicate time periods characterized by non-significant (ns) growth (i.e. stationary phase) for each strain. Percentages indicate the final population size grown in phosphate-deplete relative to phosphate-replete media. (Online version in colour.)
(c) . RLS1 ensures long-term survival
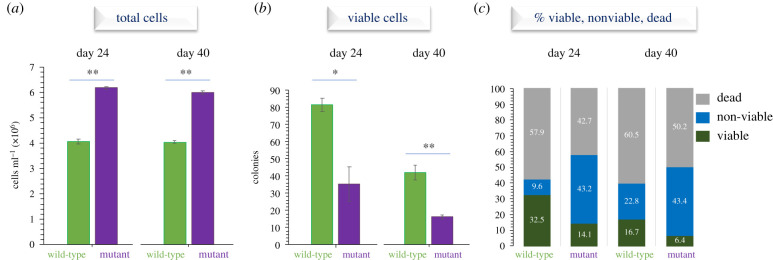
Under nutrient deprivation, C. reinhardtii individuals enter a resting/quiescent state that is excited when optimal conditions are restored. This non-reproductive and lowered metabolic state is thought to prevent oxidative damage and ensure long-term survival [40,41]. To address if RLS1 is a bona fide life-history gene that trades off reproduction for survival in limiting conditions, we compared the long-term viability (i.e. the ability of cells to successfully exit dormancy and reproduce) of the wild-type and RLS1 mutant strains in stationary cultures grown in phosphate-depleted media for up to 40 days (figure 4; electronic supplementary material, figure S6). First, we found that although the RLS1 mutant continued to maintain a higher population size relative to the wild-type over a long period of time (up to 40 days; figure 4a; p < 0.0001), a lower number of cells formed colonies (e.g. 36 versus 82 at Day 24, and 16 versus 42 at Day 40; p < 0.01 and p < 0.0001, respectively) when the same number of cells (i.e. 250) were plated on TAP agar plates (figure 4b; electronic supplementary material, figure S7a). The number of growing colonies reflects the proportion of viable cells in each population.
Figure 4.
Comparisons between the C. reinhardtii wild-type and RLS1 mutant populations grown for 24 and 40 days in phosphate-depleted media, in terms of (a) population size (i.e. total number of cells), (b) long-term viability (i.e. viable cells estimated as the number of growing colonies from a total of 250 plated cells), and (c) the relative proportion of viable (alive and reproducing), non-viable (alive but non-reproducing/senescent) and dead cells. Asterisks (*) and (**) denote p < 0.01 and p < 0.0001, respectively. (Online version in colour.)
To address if the cells that did not resume reproduction were dead or permanently arrested (non-viable; senescent), we calculated the difference between the number of live cells (assessed using a viability dye) and the number of viable cells (inferred from the number of colonies) [28]. Interestingly, we found that the lower proportion of viable cells in the RLS1 mutant populations is paralleled by a higher proportion of non-viable/senescent cells (figure 4c and electronic supplementary material, figure S7b). During quiescence, photosynthetic cells need to downregulate photosynthetic activities to avoid potential photo-oxidative damage [28,41]. The possibility that the decreased ability to resume reproduction (and thus cell viability) in the RLS1 mutant population RLS1 is due to the accumulation of photo-oxidative damage is consistent with a role for RLS1 in increasing long-term survival by downregulating photosynthesis.
(d) . RLS1 is a life-history gene with antagonistic effects on reproduction and survival in limiting environments
Overall, our data provide strong evidence that RLS1 is directly involved in the ability to trade off reproduction for survival in nutrient-limiting conditions. Specifically, we found that, compared to the parental strain, RLS1 mutants are less able to suppress their reproduction in phosphate-depleted media (figure 3). In fact, in a normal LD regime, the RLS1 mutants maintain higher population densities relative to the wild-type strain (figure 2a), which could provide them with a reproductive competitive advantage in phosphate-deprived environments. However, this reproductive advantage is not only lost in optimal conditions (figure 2c), but is also counteracted by a reduced long-term viability (figure 4). The opposite effects that the loss of RLS1 has on the reproduction and survival of C. reinhardtii grown in phosphate-depleted media provide direct evidence that RLS1 is a life-history trade-off gene with antagonistic effects on the two fitness components—reproduction and survival, under nutrient-limiting conditions.
We have previously shown that RLS1 was induced in both phosphate- and sulfur-deprived media [21]. Here we focused on phosphate deprivation as, compared to sulfur, it has a less immediate effect on reproduction [21]. However, many aspects of the general responses to nutrient deprivation—including the downregulation of photosynthesis and temporary cessation of reproduction, are similar during both phosphate- and sulfur-deprivation [30,31]. Nevertheless, the specific role of RLS1 in other nutrient-deprived environments (including nitrogen—which can also induce sexual differentiation in C. reinhardtii) needs to be fully investigated.
Interestingly, RLS1's antagonistic pleiotropic effects have been maintained in its closest homologue in V. carteri (i.e. regA). A functional RegA protein not only suppresses the reproduction of somatic cells but also protects them against stress, as RegA mutant cells have been found to be more sensitive to stress relative to their wild-type counterparts [42]. Furthermore, RLS1 mutants are not only more affected by long-term nutrient deprivation, but they are more sensitive to heat stress as well (electronic supplementary material, figure S8). This difference in sensitivity to heat stress between the wild-type and RLS1 mutant strains is also analogous to that observed between somatic cells (expressing regA) and gonidia (not expressing regA) in V. carteri [43]. But how can RLS1 regulate these two life-history traits? Below we suggest that RLS1's role as a life-history trade-off gene involves the downregulation of photosynthesis (and thus growth and reproduction) as part of the general acclimation response to increase survival in limiting conditions.
(e) . RLS1's life-history trade-off activity is linked to photosynthesis
In unicellular organisms, reproduction is linked to cell growth, which is dependent on nutrient availability. Under nutrient deprivation, cell growth and reproduction are repressed, culminating with the induction of quiescence/dormancy that will ensure long-term survival [40,44–46]. In unicellular photosynthetic organisms, cell growth is dependent not only on nutrients (inorganic substrates) but also on light. In C. reinhardtii, when nutrients (e.g. phosphorus, sulfur, nitrogen) are limited (including during stationary phase and conditional senescence), imbalances between excitation energy and cell's reducing power result in the downregulation of photosynthesis, as an adaptive response to avoid potential light-induced, oxidative damage [31,36,47,48]. For instance, a 75% decrease in maximal in vivo photosynthetic O2 evolution was observed within 4 days of phosphate deprivation or 1 day of sulfur deprivation in C. reinhardtii [30]. In addition to downregulating photosynthesis, the general acclimation response in C. reinhardtii also involves the temporary cessation of reproduction [31]. However, the signalling pathways coordinating these two processes that underlie a basic life-history trade-off are not well understood.
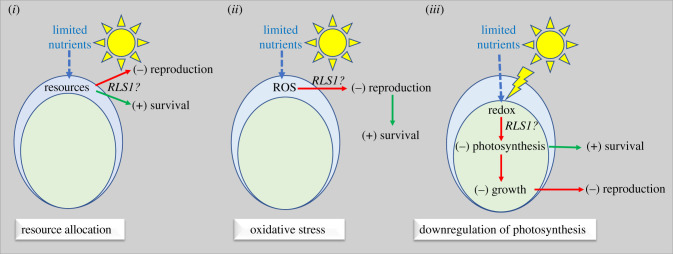
Theoretically, the suppression of reproduction in nutrient-limiting conditions can involve three hypothetical distinct scenarios. First (figure 5, scenario (i)), the suppression of reproduction and increased survival can be a direct response to the re-allocation of nutrients, possibly involving a trade-off between protein biosynthesis (growth) and energy metabolism (survival). Alternatively (figure 5, scenario (ii)), reproduction could be suppressed in response to nutrient stress-production of ROS; such oxidative stress could induce a temporary cell cycle arrest (suppression of reproduction) to repair the ROS-induced DNA damage, which will result in an increase in survival. Lastly (figure 5, scenario (iii)), redox signals associated with imbalances between excitation energy and reducing power under nutrient deprivation will induce the downregulation of photosynthesis—a response that can avoid potential photo-oxidative damage (and increase survival), but will also limit cell growth and thus reproduction.
Figure 5.
Three hypothetical mechanistic scenarios that could account for the involvement of RLS1 in the reproduction-survival trade-off in C. reinhardtii in response to nutrient-deprivation. Scenario (i) is independent of light and involves allocation of resources to survival (at a cost to reproduction). Scenario (ii) assumes ROS produced in response to nutrient stress having a negative effect on reproduction by inducing cell cycle arrest, which will have a positive effect on survival by repairing the DNA damage. Scenario (iii) envisions downregulation of photosynthesis having antagonistic effects on survival and reproduction. See text for discussion. (Online version in colour.)
These three hypothetical scenarios reflect the different proposed mechanisms underlying life-history trade-offs, and make distinct predictions as far as the role of RLS1 in the survival–reproduction trade-off under nutrient limitation. The first scenario is consistent with resource allocation and is independent of light; consequently, if RLS1 is involved in this resource-based trade-off, the loss of RLS1 should have the same effect on reproduction in both LD and CD regimes. The second scenario envisions that RLS1 is activated by ROS and acts as a general cell cycle regulator (similar to the animal p53, for instance) that induces a temporary cell cycle arrest. Loss of RLS1 would result in increased reproduction (as in cancer cells) and potentially lower long-term survival due to accumulation of unrepaired damage/mutations. However, its effect on reproduction should be similar in both LD and CD regimes. In the third scenario, redox signals associated with imbalances between excitation energy and availability of reducing power under nutrient limitation [21] would trigger the expression of RLS1. As a transcription factor, RLS1 could act to induce the downregulation of photosynthetic activities, which would have an antagonistic pleiotropic effect on the two life-history traits; that is, it will promote survival at a cost to immediate reproduction. As the postulated redox signal requires light, the loss of RLS1 should not increase reproduction in a CD regime. Our data are consistent with this scenario.
Overall, our findings and the models discussed above suggest that RLS1 acts as a master acclimation regulator that downregulates photosynthesis in response to nutrient-limiting conditions to ensure survival, though at a cost to immediate reproduction. Consistent with this possibility, RLS1 was shown to be expressed not only in phosphate-depleted media but also under sulfur-deprivation as well as in cultures in the stationary phase, at the time when reproduction declined [21]. Furthermore, expression of RLS1 can be induced by inhibitors of the photosynthetic electron transport chain that trigger acclimation-like responses, and coincides with the downregulation of a light-harvesting chloroplast protein [21]. Notably, the homologue of RLS1 in V. carteri (regA) can also be induced by environmental stress associated with light exposure after prolonged dark periods and its loss results in decreased viability [42]. By contrast, the loss of genes shown to be involved in specific responses to nutrient deprivation (i.e. nutrient acquisition/scavenging) in C. reinhardtii are known to negatively affect population growth in limiting environments [32,39,49].
Nevertheless, in addition to contributing to survival in nutrient-deprived environments by avoiding photo-oxidative damage in the light (as part of the general photosynthetic acclimation response), RLS1 might also optimize resource allocation in the dark (if alternative sources of carbon are available) by reducing the unnecessary investment in the photosynthetic apparatus, which will optimize reproduction in the dark. Interestingly, in V. carteri, regA is also thought to downregulate the expression of chloroplast proteins in somatic cells [23,24]. However, since V. carteri is unable to use organic substrates, the inability to express chloroplast proteins results in a permanent repression of cell growth and reproduction—that is, somatic cell differentiation [50]. Altogether, our data suggest that RLS1 plays an important role in balancing reproduction and survival to increase fitness in limiting conditions, which argues for this gene acting as a bona fide life-history trade-off gene.
4. Conclusion
This study suggests that single-celled species are good model-systems to investigate the fundamental genetic and molecular mechanisms underlying life-history trade-offs. In addition to their suitability for experimental work, in these lineages the organismal and cellular level coincide, and thus cellular-level responses (e.g. cell cycle arrest) are directly reflected in organismal-level processes (e.g. cessation of reproduction). Our data show that in the unicellular species, C. reinhardtii, a single gene—RLS1, can adaptively adjust both survival and reproduction as part of the general acclimation response that ensures survival at a cost to immediate reproduction by regulating photosynthetic activities in response to nutrient and light availability.
Genes with potential antagonistic pleiotropic effects (independent of resource allocation) might also be responsible for some of the observed life-history trade-offs in multicellular organisms. For instance, in the nematode Caenorhabditis elegans, a natural variant (associated with a small deletion in the cis-region of eak 3—a gene involved in the synthesis and activity of a hormone) that has increased sensitivity to dauer induction (a dormant stage induced in unfavourable conditions) is also negatively affected in reproduction [16]. Furthermore, similar to our RLS1 mutant, eak-3 variants have been shown to have a fitness advantage in stressful environments (through increased dauer production), but be outcompeted in favourable environments (because of their decreased reproduction). Deciphering the specific pathways regulated by genes with antagonistic pleiotropic effects on fitness will provide a deeper understanding of the general mechanisms underlying life-history trade-offs.
Supplementary Material
Data accessibility
Supporting data are provided as electronic supplementary material.
Authors' contributions
R.M.S.S.: conceptualization, data curation, formal analysis, investigation, methodology, visualization, writing—review and editing; C.W.J.L. and I.C.W.C.: formal analysis, investigation, methodology, visualization, writing—review and editing; D.G.D.: conceptualization, data curation, formal analysis, funding acquisition, project administration, resources, supervision, validation, writing—review and editing; A.M.N.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, visualization, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by grants from the Natural Sciences and Engineering Research Council (NSERC) of Canada to A.M.N. and D.G.D. C.W.J.L. was supported by an NSERC Undergraduate Summer Research Award (USRA).
References
- 1.Stearns SC. 1989. Trade-offs in life-history evolution. Funct. Ecol. 3, 259-268. ( 10.2307/2389364) [DOI] [Google Scholar]
- 2.Bochdanovits Z, De Jong G.. 2004. Antagonistic pleiotropy for life-history traits at the gene expression level. Proc. R. Soc. Lond. B 271, S75-S78. ( 10.1098/rsbl.2003.0091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flatt T. 2020. Life-history evolution and the genetics of fitness components in Drosophila melanogaster. Genetics 214, 3-48. ( 10.1534/genetics.119.300160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roff DA, Fairbairn DJ. 2007. The evolution of trade-offs: where are we? J. Evol. Biol. 20, 433-447. ( 10.1111/j.1420-9101.2006.01255.x) [DOI] [PubMed] [Google Scholar]
- 5.Monaghan P, Metcalfe NB, Torres R. 2009. Oxidative stress as a mediator of life history trade-offs: mechanisms, measurements and interpretation. Ecol. Lett. 12, 75-92. ( 10.1111/j.1461-0248.2008.01258.x) [DOI] [PubMed] [Google Scholar]
- 6.Flatt T, Heyland A. 2011. Mechanisms of life history evolution: the genetics and physiology of life history traits and trade-offs. Oxford, UK: Oxford University Press. [Google Scholar]
- 7.Zera AJ, Harshman LG. 2001. The physiology of life history trade-offs in animals. Annu. Rev. Ecol. Syst. 32, 95-126. ( 10.1146/annurev.ecolsys.32.081501.114006) [DOI] [Google Scholar]
- 8.Villellas J, García MB. 2018. Life-history trade-offs vary with resource availability across the geographic range of a widespread plant. Plant Biol. 20, 483-489. ( 10.1111/plb.12682) [DOI] [PubMed] [Google Scholar]
- 9.Hughes KA, Leips J. 2017. Pleiotropy, constraint, and modularity in the evolution of life histories: insights from genomic analyses. Ann. NY Acad. Sci. 1389, 76-91. ( 10.1111/nyas.13256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roff DA. 2007. Contributions of genomics to life-history theory. Nat. Rev. Genet. 8, 116-125. ( 10.1038/nrg2040) [DOI] [PubMed] [Google Scholar]
- 11.St-Cyr J, Derome N, Bernatchez L. 2008. The transcriptomics of life-history trade-offs in whitefish species pairs (Coregonus sp.). Mol. Ecol. 17, 1850-1870. ( 10.1111/j.1365-294X.2008.03696.x) [DOI] [PubMed] [Google Scholar]
- 12.Koch RE, et al. 2021. Integrating mitochondrial aerobic metabolism into ecology and evolution. Trends Ecol. Evol. 36, 321-332. ( 10.1016/j.tree.2020.12.006) [DOI] [PubMed] [Google Scholar]
- 13.Hood WR, Zhang Y, Mowry AV, Hyatt HW, Kavazis AN. 2018. Life history trade-offs within the context of mitochondrial hormesis. Integr. Comp. Biol. 58, 567-577. ( 10.1093/icb/icy073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leroi AM. 2001. Molecular signals versus the Loi de Balancement. Trends Ecol. Evol. 16, 24-29. ( 10.1016/S0169-5347(00)02032-2) [DOI] [PubMed] [Google Scholar]
- 15.Stanhill A, Schick N, Engelberg D. 1999. The yeast Ras/Cyclic AMP pathway induces invasive growth by suppressing the cellular stress response. Mol. Cell. Biol. 19, 7529-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Billard B, Vigne P, Braendle C. 2020. A natural mutational event uncovers a life history trade-off via hormonal pleiotropy. Curr. Biol. 30, 4142-4154.e9. ( 10.1016/j.cub.2020.08.004) [DOI] [PubMed] [Google Scholar]
- 17.Michod RE, Viossat Y, Solari CA, Hurand M, Nedelcu AM. 2006. Life-history evolution and the origin of multicellularity. J. Theor. Biol. 239, 257-272. ( 10.1016/j.jtbi.2005.08.043) [DOI] [PubMed] [Google Scholar]
- 18.Michod RE. 2006. The group covariance effect and fitness trade-offs during evolutionary transitions in individuality. Proc. Natl Acad. Sci. USA 103, 9113-9117. ( 10.1073/pnas.0601080103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roux EA, Roux M, Korb J. 2009. Selection on defensive traits in a sterile caste - caste evolution: a mechanism to overcome life-history trade-offs? Evol. Dev. 11, 80-87. ( 10.1111/j.1525-142X.2008.00304.x) [DOI] [PubMed] [Google Scholar]
- 20.Nedelcu AM, Michod RE. 2006. The evolutionary origin of an altruistic gene. Mol. Biol. Evol. 23, 1460-1464. ( 10.1093/molbev/msl016) [DOI] [PubMed] [Google Scholar]
- 21.Nedelcu AM. 2009. Environmentally induced responses co-opted for reproductive altruism. Biol. Lett. 5, 805-808. ( 10.1098/rsbl.2009.0334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirk MM, Stark K, Miller SM, Müller W, Taillon BE, Gruber H, Schmitt R, Kirk DL. 1999. regA, a Volvox gene that plays a central role in germ-soma differentiation, encodes a novel regulatory protein. Development 126, 639-647. [DOI] [PubMed] [Google Scholar]
- 23.Meissner M, Stark K, Cresnar B, Kirk DL, Schmitt R. 1999. Volvox germline-specific genes that are putative targets of RegA repression encode chloroplast proteins. Curr. Genet. 36, 363-370. ( 10.1007/s002940050511) [DOI] [PubMed] [Google Scholar]
- 24.Tam LW, Kirk DL. 1991. Identification of cell-type-specific genes of Volvox carteri and characterization of their expression during the asexual life cycle. Dev. Biol. 145, 51-66. ( 10.1016/0012-1606(91)90212-L) [DOI] [PubMed] [Google Scholar]
- 25.Li X, et al. 2019. A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis. Nat. Genet. 51, 627-635. ( 10.1038/s41588-019-0370-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quisel JD, Wykoff DD, Grossman AR. 1996. Biochemical characterization of the extracellular phosphatases produced by phosphorus-deprived Chlamydomonas reinhardtii. Plant Physiol. 111, 839-848. ( 10.1104/pp.111.3.839) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomasiewicz DM, Hotchkiss DK, Reinbold GW, Read RB, Hartman PA. 1980. The most suitable number of colonies on plates for counting. J. Food Prot. 43, 282-286. ( 10.4315/0362-028x-43.4.282) [DOI] [PubMed] [Google Scholar]
- 28.Takeuchi T, Sears BB, Lindeboom C, Lin YT, Fekaris N, Zienkiewicz K, Zienkiewicz A, Poliner E, Benninga C. 2020. Chlamydomonas CHT7 is required for an effective quiescent state by regulating nutrient-responsive cell cycle gene expression. Plant Cell 32, 1240-1269. ( 10.1105/TPC.19.00628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bottomley MJJ, Collard MWW, Huggenvik JII, Liu Z, Gibson TJJ, Sattler M. 2001. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat. Struct. Biol. 8, 626-633. ( 10.1038/89675) [DOI] [PubMed] [Google Scholar]
- 30.Wykoff DD, Davies JP, Melis A, Grossman AR. 1998. The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 117, 129-139. ( 10.1104/pp.117.1.129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grossman A. 2000. Acclimation of Chlamydomonas reinhardtii to its nutrient environment. Protist. 151, 201-224. ( 10.1078/1434-4610-00020) [DOI] [PubMed] [Google Scholar]
- 32.Davies JP, Yildiz FH, Grossman A. 1996. Sac1, a putative regulator that is critical for survival of Chlamydomonas reinhardtii during sulfur deprivation. EMBO J. 15, 2150-2159. ( 10.1002/j.1460-2075.1996.tb00568.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anthony JR, Warczak KL, Donohue TJ. 2005. A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc. Natl Acad. Sci. USA 102, 6502-6507. ( 10.1073/pnas.0502225102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foyer CH, Noctor G. 2005. Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell. 17, 1866-1875. ( 10.1105/tpc.105.033589) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eberhard S, Finazzi G, Wollman F-A. 2008. The dynamics of photosynthesis. Annu. Rev. Genet. 42, 463-515. ( 10.1146/annurev.genet.42.110807.091452) [DOI] [PubMed] [Google Scholar]
- 36.González-Ballester D, Casero D, Cokus S, Pellegrini M, Merchant SS, Grossman AR. 2010. RNA-Seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22, 2058-2084. ( 10.1105/TPC.109.071167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moseley JL, Chang CW, Grossman AR. 2006. Genome-based approaches to understanding phosphorus deprivation responses and PSR1 control in Chlamydomonas reinhardtii. Eukaryot. Cell 5, 26-44. ( 10.1128/EC.5.1.26-44.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K. 1999. Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc. Natl Acad. Sci. USA 96, 15 336-15 341. ( 10.1073/pnas.96.26.15336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bajhaiya AK, Dean AP, Zeef LAH, Webster RE, Pittman JK. 2016. PSR1 is a global transcriptional regulator of phosphorus deficiency responses and carbon storage metabolism in Chlamydomonas reinhardtii. Plant Physiol. 170, 1216-1234. ( 10.1104/pp.15.01907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeuchi T, Benning C. 2019. Nitrogen-dependent coordination of cell cycle, quiescence and TAG accumulation in Chlamydomonas. Biotechnol. Biofuels. 12, 292. ( 10.1186/s13068-019-1635-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai CH, Uygun S, Roston R, Shiu SH, Benning C. 2018. Recovery from N deprivation is a transcriptionally and functionally distinct state in Chlamydomonas. Plant Physiol. 176, 2007-2023. ( 10.1104/PP.17.01546) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.König SG, Nedelcu AM. 2020. The genetic basis for the evolution of soma: mechanistic evidence for the co-option of a stress-induced gene into a developmental master regulator. Proc. R. Soc. B 287, 20201414. ( 10.1098/rspb.2020.1414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nedelcu AM. 2006. Evidence for p53-like-mediated stress responses in green algae. FEBS Lett. 580, 3013-3017. ( 10.1016/j.febslet.2006.04.044) [DOI] [PubMed] [Google Scholar]
- 44.Rittershaus ESC, Baek SH, Sassetti CM. 2013. The normalcy of dormancy: common themes in microbial quiescence. Cell Host Microbe. 13, 643-651. ( 10.1016/j.chom.2013.05.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. 2004. ‘Sleeping beauty’: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68, 187-206. ( 10.1128/mmbr.68.2.187-206.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herman PK. 2002. Stationary phase in yeast. Curr. Opin. Microbiol. 5, 602-607. ( 10.1016/S1369-5274(02)00377-6) [DOI] [PubMed] [Google Scholar]
- 47.Meagher E, Rangsrikitphoti P, Faridi B, Zamzam G, Durnford DG. 2021. Photoacclimation to high-light stress in Chlamydomonas reinhardtii during conditional senescence relies on generating pH-dependent, high-quenching centres. Plant Physiol. Biochem. 158, 136-145. ( 10.1016/j.plaphy.2020.12.002) [DOI] [PubMed] [Google Scholar]
- 48.Damoo DY, Durnford DG. 2021. Long-term survival of Chlamydomonas reinhardtii during conditional senescence. Arch. Microbiol. 203, 5333-5344. ( 10.1007/S00203-021-02508-Y) [DOI] [PubMed] [Google Scholar]
- 49.Chang CW, Moseley JL, Wykoff D, Grossman AR. 2005. The LPB1 gene is important for acclimation of Chlamydomonas reinhardtii to phosphorus and sulfur deprivation. Plant Physiol. 138, 319-329. ( 10.1104/pp.105.059550) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kirk DL. 2001. Germ-soma differentiation in Volvox. Dev. Biol. 238, 213-223. ( 10.1006/dbio.2001.0402) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supporting data are provided as electronic supplementary material.