Abstract
Pancreatic adenocarcinoma is one of the most lethal cancers with rising incidence. Despite progress in its treatment, with the introduction of more effective chemotherapy regimens in the last decade, prognosis of metastatic disease remains inferior to other cancers with long term survival being the exception. Molecular characterization of pancreatic cancer has elucidated the landscape of the disease and has revealed common lesions that contribute to pancreatic carcinogenesis. Regulation of proteostasis is critical in cancers due to increased protein turnover required to support the intense metabolism of cancer cells. The proteasome is an integral part of this regulation and is regulated, in its turn, by key transcription factors, which induce transcription of proteasome structural units. These include FOXO family transcription factors, NFE2L2, hHSF1 and hHSF2, and NF-Y. Networks that encompass proteasome regulators and transduction pathways dysregulated in pancreatic cancer such as the KRAS/ BRAF/MAPK and the Transforming growth factor beta/SMAD pathway contribute to pancreatic cancer progression. This review discusses the proteasome and its transcription factors within the pancreatic cancer cellular micro-environment. We also consider the role of stemness in carcinogenesis and the use of proteasome inhibitors as therapeutic agents.
Keywords: Pancreatic adenocarcinoma, Proteasome, Transcription factors, Regulation, Unfolded protein response, Cancer stem cells
Core Tip: Pancreatic adenocarcinoma is a gastrointestinal cancer with high incidence and bleak outcomes. The molecular pathways involved in the pathogenesis of the disease have been increasingly clarified in recent years. This article reviews the role of proteostasis regulation through the proteasome in pancreatic cancer. Major molecular pathways affected in pancreatic cancer closely interconnect with regulators of the proteasome.
INTRODUCTION
Pancreatic cancer (PC) is a prevalent digestive system malignancy with a 5-year survival rate of less than 5%, a median survival period of 6 mo and, in 2019, accounted for 45750 deaths in the United States alone[1]. Despite representing the thirteenth most prevalent cancer, accounting for 458000 cases worldwide, it is the seventh most deadly cancer[2], and affects men slightly more than women. Unfortunately, PC tends to exhibit a distinct resistance to radiotherapy and chemotherapy, contributing to increased mortality. Understanding how particular lesions can induce and maintain pancreatic carcinogenesis, as well as how the molecular mechanisms both upstream and downstream of the initial pathology contribute to morbidity, treatment resistance, and mortality will ultimately inform clinical strategies to combat this deadly disease.
Pancreatic adenocarcinoma is a cancer with low overall tumor mutation burden (TMB). The majority of pancreatic cancer cases in The Cancer Genome Atlas series display less than 50 mutations, and only few cancers have 50 to 80 mutations and even fewer cases have a TMB above 80 mutations[3]. Few cancer-associated genes display recurrent mutations in pancreatic adenocarcinoma and include activating mutations in the oncogene KRAS which are observed in most cases of these cancers, and mutations in tumor suppressor TP53 encoding for the p53 protein which are observed in about two thirds of cases. Two additional tumor suppressors, cell cycle inhibitor p16 and the Transforming growth factor beta (TGFβ) pathway signal transducer SMAD4, are mutated in about 20% of cases in pancreatic cancers[3,4]. p16 is encoded by gene CDKN2A which is located at chromosome 9p. The same locus also encodes for the p53 positive regulator p14ARF. The locus is deleted in 10% to 25% of pancreatic cancer cases. Besides these four recurrent gene abnormalities no other oncogenes or tumor suppressors are commonly altered in pancreatic cancers. Studies of genomic profiles of pancreatic cancers have shown that the disease is heterogeneous and different sub-types exist, similar with other cancers. A most recent genomic classification, for example, assigns pancreatic cancer in four types: squamous, pancreatic progenitor, immunogenic and aberrant differentiation endocrine-exocrine[5].
Cancer cells rely on proteasome activity for regulation of their increased metabolism[6,7]. In addition, specific cancer-associated pathways are dependent on proteasome degradation of tumor suppressors or for neutralization of inhibitors of oncogenes. As examples, activation of the NF-κB pathway requires proteasome degradation of inhibiting I-κB protein and tumor suppressor p53 is tightly controlled through ubiquitination by ubiquitin ligase MDM2 and other ligases for proteasome degradation[8]. Another example is provided by kinase GSK3β, which phosphorylates substrates such as oncogenic β-catenin for subsequent ubiquitination and proteasome degradation. This process is frequently debilitated in cancers[9]. Pharmacologic proteasome inhibition leads to pancreatic cancer cells growth arrest and decreased viability in vitro and in vivo, especially in combination therapies[10]. In contrast, pancreatic cancer cells that acquire the ability to shut down protein translation in conditions of proteasome inhibition are resistant to proteasome inhibitors[11].
Given the importance of proteostasis regulation in cancer and the increased metabolism of cancer cells, proteasome sub-unit production and availability as building blocks of the organelle are regulated at the transcriptional level by several factors[12]. The current paper will discuss the regulation of proteasome by these factors in cancer with a focus on pancreatic cancer.
PROTEASOME
Proteasomes are multi-subunit proteolytic organelles that recognize, unfold, and digest unneeded or damaged proteins within the cell. In mammals, the most common variant is the cytosolic 26S proteosome (2-MDa) that consists of a core protease (750 kDa) with a Svedberg sedimentation coefficient (rate under acceleration) or S-value of 20 (or 20 × 108 cm2/s). The core is flanked by two, ATP-dependent 19 S regulatory subunits (700 kDa, called PA700) that unfold substrates and direct them toward the core for degradation[13]. Together, they form a tube-like structure through which linearized proteins pass and get cleaved within the center-most lumen of the complex. Unlike lysosomes, which are primarily activated under stress conditions, proteasomes (which can also become activated under stress conditions) regulate normative protein turnover associated with basal metabolic conditions. The process is highly selective, owing to the identification of targets by a three-step ubiquitination process. Briefly, E1 ubiquitin-activating enzymes capture ubiquitin which are then transferred to E2 ubiquitin-conjugating enzymes which then, in coordination with E3 ubiquitin ligases, catalyze bonds between the C-terminal glycine of the ubiquitin molecule and a lysine molecule within the substrate protein. The net result of the E1-E2-E3 cascade is the addition of one or more ubiquitin molecules to the substrate protein which are then recognized by ubiquitin-binding sites located at the 19S caps of the proteosome. Proteasome recognition requires a chain of at least four ubiquitin molecules tagged to a target protein, usually through lysines at position 48 of each ubiquitin molecule[14]. Once ubiquitinated proteins are unfolded and directed toward the central protease by the 19S subunits, proteolysis proceeds whereby the inner β subunit rings of the core trigger a threonine-dependent nucleophilic attack[15]. Specifically, β1, β2, and β5 subunits within the inner-most domains of the core generate caspase-like, trypsin-like, and chymotrypsin-like activities, respectively. As proteins pass through the core, they are cleaved into short polypeptides typically consisting of 3 to 15 amino-acid residues each which are then recycled by hydrolysis.
The ubiquitin-proteasome proteolytic pathway has, unsurprisingly, been identified as a conserved cell function that can be exploited to combat cancer. The discovery that caspases–a family of proteases–were the chief executors of apoptosis spurred initial interest in the proteasome as a potential mediator of similar functions relevant to the control of programmed cell death. Further, its role as a regulator of the tumor suppressor protein p53, Bcl-2 family apoptosis inhibitors, and cyclin-dependent kinase (CDK) inhibitors reflected its potential as a target for cancer therapy[16,17]. Indeed, proteasome inhibitors, either alone or in combination with other anti-cancer therapies, have confirmed anti-tumor properties in hematologic cancers[18,19]. Antineoplastic activity derives from diverse actions such as blocking antiapoptotic genes, down-regulating survival signals, preventing efflux of cytotoxic agents, promoting DNA stabilization for cleaving, and by other mechanisms. The potential of proteasome inhibition has been translated to successful therapies in hematologic malignancies including multiple myeloma and mantle cell lymphoma[20,21]. However, in solid tumors, proteasome inhibition has not met with similar success and no benefit has been shown, despite extensive clinical investigations[22,23]. Development, on most occasions, has followed a one-size-fit-all model with no attempt to tailor treatment to sub-sets with possible sensitivity, despite pre-clinical evidence for the existence of such molecular sub-sets in various cancers[24]. Thus, a targeted development of proteasome inhibitors based on biomarkers deserves further investigation in solid tumors, including pancreatic cancer.
REGULATORS OF PROTEASOME WITH FOCUS ON PANCREATIC CANCER
Regulation of proteostasis is an important part of metabolism in active cells including cancer cells which possess an increased proteasome activity[25]. Proteasome regulation at the transcriptional level is effectuated in various cellular contexts by coordinated transcription of the proteasome multiple sub-units by a panel of transcription factors that include FOXO family members, NFE2L2 (also called NRF2), heat shock factors 1 and 2 (hHSF1 and hHSF2) and NF-Y (Figure 1). Activity of these factors vary in cancer cells but results in the overall increased proteasome availability and function that is necessary for their increased protein turn-over associated with cellular processes. Increased proteasome activity in bulk cancer cells contrast with reduced activity in cancer stem cells which are metabolically less active or quiescent[12,26]. Despite lower proteasome activity, cancer stem cells still depend on proteostatic controls for their basic functions and are sensitive to proteasome inhibition. Proteasome inhibitor, carfilzomib sensitivity has been described in the squamous/ cornified subtype of pancreatic cancer[27]. This subtype presents with down-regulated epithelial mesenchymal transition (EMT) genes which are associated with cancer stem characteristics, suggesting a lower abundance of stem cell sub-population. Although no correlation with proteasome activity was found in squamous/cornified pancreatic cancers, a correlation with unfolded protein response genes ATF4 and CHOP expression and de novo RNA and protein synthesis was discerned, confirming the role of proteostasis[27]. The following sub-sections will discuss the regulation of the proteasome by the major transcription factors that govern their sub-units’ transcription.
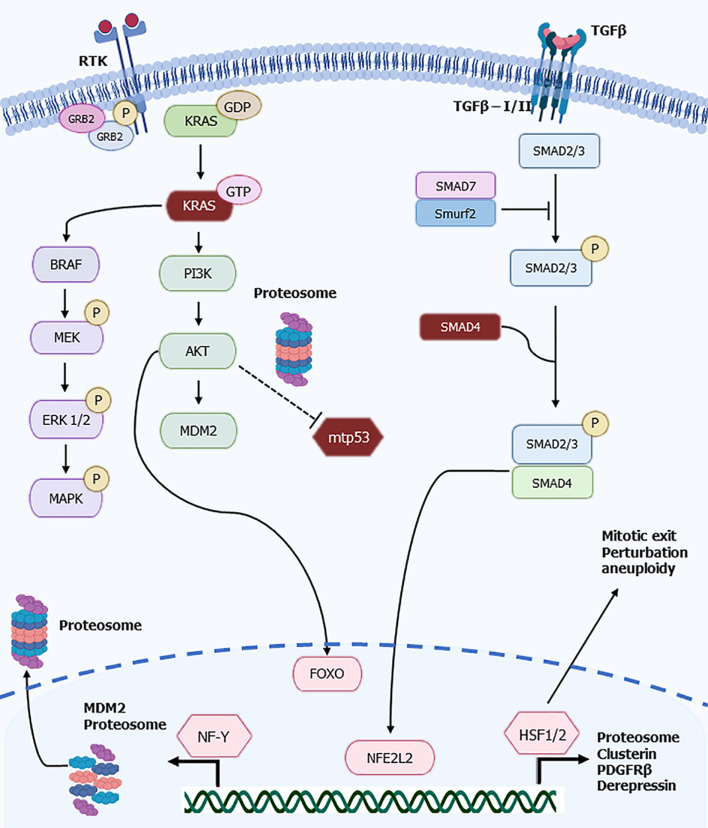
Figure 1.
Schematic of receptor tyrosine kinases/KRAS and transforming growth factor beta/SMAD signaling pathways and downstream key transcription factors regulating the proteosome including FOXO, NF-Y, NFE2L2, and heat shock factors 1/2. Stabilized mutant p53 is involved in proteostasis transcription factors deregulation. For details see text. RTK: Receptor tyrosine kinases; TGFβ: Transforming growth factor beta; hHSF1/2: Heat shock factors 1/2; mtp53: Mutant p53.
FOXO
FOXO family transcription factors regulate key carcinogenesis processes such as apoptosis and proliferation, acting as tumor suppressors. Thus, suppression of FOXO factors is important during carcinogenesis and is accomplished through activation of kinase Akt, the major negative regulator of FOXOs, which is commonly activated down-stream of receptor tyrosine kinases, by KRAS and PI3K kinase cascades in various cancers[28] (Figure 1). FOXO transcription factors have a positive effect in proteasome regulation through activation of transcription of proteasome sub-units[12]. A pathophysiologic role of FOXO transcription factors in muscle atrophy through proteasome genes induction, and reversal of atrophy by protein kinase A inhibition of FOXO members FOXO1 and FOXO3 has been described[29]. Transcription co-activator Peroxisome Proliferator Activated Receptor gamma Co-activator 1alpha (PGC-1α) counteracts the atrophy promoting effects of FOXO3 and reduces denervation-induced muscle atrophy and cancer cachexia in mice[30]. In pancreatic cancer, PGC-1α, acting in concert with the nuclear receptor family transcription factors PPARγ and RXRα, has a tumor suppressing effect through induction of phosphatase PTEN and inhibition of Akt[31]. Given that p53 is a negative regulator of PCG-1α, PCG-1α may be up-regulated in pancreatic cancers with TP53 mutations[32]. Whether PCG-1α negatively regulates FOXO activity in pancreatic cancer, similarly to the effects in muscle remains unstudied. Such a negative regulation would synergize with the activity of the mutant KRAS/ PI3K/ Akt cascade.
The family member FOXO3 has an indirect negative effect on proteasome regulation through induction of protein Keap1, a negative regulator of NFE2L2[33]. Moreover, FOXO3 activated by cGMP has a role in maintenance of pancreatic cancer stem cells with CD44+ phenotype[34]. Keap1 induction due to FOXO3 activation would suppress proteasome transcription in these cells through NFE2L2 down-regulation. Cancer stem cells are, in general, less active metabolically and present a lower proteasome activity[12]. Also relevant for cancer stem cells, FOXO activity in proteasome regulation is counter-acted by transcription regulator ZEB1[35]. ZEB1 is a zinc finger protein that belongs to the core regulators of EMT, a process involved in metastasis and associated with stemness[36,37]. The dual role, direct and indirect, of FOXO family members in the transcription of proteasome sub-units may decrease the importance of FOXO transcriptional activity for maintenance of proteasome function in cancer cells. Suppression of FOXO family activity through upstream overactivated KRAS and PI3K/ Akt signaling, in addition to other pro-carcinogenic effects, down-regulates Keap1 leading to an eventual stabilization and increased activity of NFE2L2, which is a key factor for proteasome regulation and reactive oxygen species detoxification[38].
A reciprocal regulation of FOXO transcription factors by the proteasome is mediated by ubiquitination and proteasome degradation through the action of ubiquitin ligase MDM2, which also ubiquitinates p53 and the other proteasome regulating transcription factor NFE2L2[39].
NFE2L2 (NRF2)
NFE2L2 is a transcription factor that serves as a bulwark against oxidative damage following injury or inflammation by regulating the expression of antioxidant proteins[40]. It has also been implicated as a major participant in regulatory networks that control cell metabolism, autophagy, mitochondrial function, and proteolysis, including upregulated expression of the catalytic subunits of the 26S proteasome[41]. As the accumulation of oxidated and polyubiquitinated protein aggregates are major correlates of aging and senescence[42,43], regulation of NFE2L2 demonstrates therapeutic potential for neurodegenerative and cardiac disease as well as cancer[41].
NFE2L2 is overexpressed and predicts anti-cancer drug resistance[44,45]. The neoplastic progression to pancreatic ductal adenocarcinoma is induced by a signaling pathway that is potentiated by stress involving NFE2L2, E3 ubiquitin-protein ligase MDM2 (a negative regulator of p53), and ubiquitin-binding protein p62[46]. The mechanism is thought to involve the accumulation of p62 which triggers NFE2L2-MDM2 to modulate p53 and the morphogen Notch which induce conversions of acinar cells to progenitor-like cells, which then accelerate lesions toward malignant phenotypes. Pancreatic carcinogenesis can be experimentally suppressed by selective deletion of NFE2L2 in a mutant K-ras and p53 model[47], highlighting the intrinsic links between oxidative stress, NFE2L2, proteostasis, and cancer. Mice with mutations in K-ras and p53 display decreased pancreatic tumor formation and progression when NFE2L2 is lost compared with animals with intact NFE2L2. Loss of NFE2L2 results in down-regulation of several oxidative detoxification enzymes, such as glutathione S-transferases and UDP glucuronosyltransferases. In addition, transporters of the ABC family are down-regulated leading to increased gemcitabine sensitivity[48]. However, the pro-carcinogenic effect of NFE2L2 is tumor environment-specific, given that in the absence of p53 mutations animals with K-ras mutations and NFE2L2 activation develop pancreatic parenchyma atrophy[47].
hHSF1 and hHSF2
hHSF1 belongs, together with hHSF2, hHSF3 and hHSF4, to a family of transcription factors involved in proteostasis and is upregulated after proteasome inhibition and under other stress conditions[34,49]. hHSF1 is the founding member of the family and is homologous to the fly heat shock factor. In this organism only one HSF protein exists[50]. Following hyperthermia, oxidative stress or proteasome inhibition hHSF1 dissociates from chaperone HSP90 and trimerizes. The trimeric form is capable of DNA binding to Heat Shock Elements (HSE), the target sequence of Heat Shock Factors and initiation of target gene transcription, that include chaperone proteins, ubiquitin and proteasome sub-nits[50,51]. hHSF2 binds DNA as a homotrimer or heterotrimer with hHSF1, with the activation capacity depending on the consistency of the trimer[52]. Cells that lack HSF2 display decreased expression of proteasome sub-units and display p53 stabilization due to decreased proteasome activity[53]. In addition, hHSF1 and hHSF2 participate in proteotoxic stress response by up-regulating transcription of chaperone protein clusterin (also called apolipoprotein J), which possesses a noncanonical HSE in its promoter[54,55]. Thus, HSFs constitute part of a feedback proteostasis response whence proteostasis perturbations lead to up-regulation of these factors that help re-establish the balance of cellular proteins metabolism[56].
Besides the proteostasis response, hHSF1 and hHSF2 have a broader role in cancer that derives from additional functions of these transcription factors in cancer-associated processes, such as proliferation, inhibition of apoptosis and metastasis[51]. hHSF1 promotes the cell cycle through a direct interaction with ubiquitin ligase APC/C co-factor cdc20, which leads to inhibition of cyclin B and securin degradation[57]. Phosphorylation of hHSF1 by mitotic kinase PLK1 is required for mitotic exit inhibition and is observed in cancer cells with mutated p53, which have defective PLK1 function regulation[58]. As a result of mitotic perturbation, aneuploidy ensues with micronuclei formation, a hallmark of chromosomal instability. The role of hHSF1 in metastasis is exemplified by induction of EMT core transcription regulator Slug in breast cancer cells that depends on hHSF1 activation by kinase Akt[59]. hHSF2 deficiency affects cell-cell adhesion cadherins, whose downregulation leads to loss of cellular adhesions and cell demise, possibly due to anoikis[60]. Absence of cell-cell adhesion is associated with intolerance of prolonged proteotoxic stress[60]. This could explain the association of cancer stemness and stem cells, which have decreased proteasome activity with the ability to undergo EMT, which requires dissolving cell adhesions[12]. Indeed, hHSF1 activity endows cancer cells with stem cell properties, at least in the case of breast cancer[61]. hHSF1 induction in breast cancer cells increases cells with the stem cell phenotype and chemotherapy resistance while knockdown of hHSF1 reduces stem cells.
Interestingly, hHSF1 shares with FOXO transcription factors a role in longevity[50]. This could be the result of their also shared role in proteostasis, as improved protein handling provides a benefit in cellular function which would be expected to provide a cell survival advantage. The benefit is usurped by cancer cells, the ultimate immortal cells. A similar dual effect in pancreatic cancer is at play for another proteostatic mechanism related to longevity, autophagy, which is beneficial in established pancreatic cancers[62]. Increased autophagic flux is beneficial and promotes longevity in cells where the mitochondrial membrane potential is preserved through a closed mitochondrial permeability transition pore (mPTP), while apoptosis ensues in cells with high autophagic flux but an open mPTP[63]. KRAS mutations as observed in pancreatic cancer sensitize cells to mitochondrial membrane potential destabilization[64].
NF-Y
The CCAAT-binding factor, also known as NF-Y, is a transcription factor with well-established gene regulatory properties and serves as a safeguard against abnormal translation by maintaining nucleosome-depleted regions at gene promoters[65]. NF-Y is a trimeric complex consisting of NF-YA, NF-YB and NF-YC[66]. The NF-YA sub-unit is the DNA binding partner and mutations in the DNA binding domain in its carboxyterminus lead to inability of the trimer to bind promoters of target genes. NF-Y has been shown to regulate transcription of cell cycle-related genes including CDKs, embryonic differentiation and morphogenesis by activation of SOX genes, and cancer-related genes including the tumor suppressor TβRII among others[67]. Indeed, the integrity of CCAAT boxes and functional NF-Y complexes are vital to cell cycle progression as a response to DNA damage[68,69]. Several proteasome genes carry CCAAT boxes in their promoters and are regulated by NF-Y[70]. Interestingly, CCAAT boxes are disproportionately represented in gene promoters that are overexpressed in cancers and NF-Y activity is critical to both cell transformation and proliferation by way of interactions with p53[66]. In addition, there is a strong metabolic component to the NF-Y regulome, with de novo biosynthesis of lipids, purines, and polyamines, as well as glycolysis and activation of glutamine pathway[71]. NF-Y therefore appears to represent a watershed factor at the intersection of cell transformation, proliferation, and metabolism–a deadly combination when co-opted by cancer. In pancreatic cancer, disruption by mutant p53 of the p73/NF-Y complex initiates the transcription of platelet-derived growth factor receptor β (PDGFRβ) and potentiates invasion and metastasis[72]. Thus, NF-Y has been identified as a promising target for anti-cancer therapies. For example, indirubin derivatives, which are known to exert anti-tumor effects by inhibiting CDKs, also act to inhibit binding of NF-Y to DNA[73]. Computational drug repositioning techniques indicate that other candidate anti-cancer drugs are likely to impact NF-Y transcription[74].
REGULATION OF PROTEASOME REGULATORS BY MOLECULAR LESIONS OF PANCREATIC CANCER
As mentioned above, only a few recurrent molecular abnormalities in oncogenes and tumor suppressors are present in pancreatic cancer[3]. Up to 80%-90% of human pancreatic adenocarcinomas bear classic activating mutations at codons 12, 13 or 61 in oncogene KRAS and about two thirds of pancreatic cancers display mutations in tumor suppressor p53. Two other tumor suppressors, SMAD4 and p16 are mutated in a sizeable minority of pancreatic cancers. Lesions in these proteins that affect important molecular pathways of pancreatic carcinogenesis also have repercussions for proteasome master regulators and their function and they will be discussed in this section. Moreover, transcription factors-regulators of the proteasome expression are involved in pathways regulated by common pancreatic cancer molecular lesions and reciprocal relationships exist, constituting an elaborate network.
KRAS
RAS proteins are single-subunit small GTPases that are fundamental to cell signaling pathways, regulating proliferation, cell adhesion, apoptosis, migration, and differentiation. Perhaps the most well-studied and indispensable pathway is the mitogen-activated protein kinase (MAPK) cascade, which potentiates downstream gene transcription involved in proliferation and growth. RAS signals also through the PI3K Akt pathway that regulates metabolism, cell growth and survival[75]. The human genome contains 3 Ras gene isoforms-HRAS, KRAS, and NRAS-which are notably the most predominant oncogenes involved in cancer. KRAS mutations are found in almost all (95%) cases of pancreatic ductal adenocarcinoma, dwarfing the mutation frequencies of HRAS (< 1%) and NRAS (< 1%)[76]. Incidentally, the same pattern of KRAS mutation frequency dominance over other RAS isoforms is observed in colorectal cancer, renal cell carcinoma, and stomach cancer, though to lesser degrees. In lung cancer, polymorphisms associated with the NF-Y binding site within the KRAS gene may increase the likelihood of developing the malignancy[77]. The KRAS oncogene mutation was found to abnormally activate several pathways including the PI3K-AKT-mTOR and Ras-MAPK pathways in both human pancreatic cancers and mouse models of pancreatic adenocarcinoma[78]. Using a mouse model of pancreatic adenocarcinoma, LSL-KrasG12D/+, that recapitulates the development of pancreatic cancer, it was recently demonstrated that the heat shock factor 1 (HSF1) and epidermal growth factor receptor (EGFR) pathway is a major determinant of progression[79]. HSF1 is an important regulator of proteostasis in cancers, exerting control over metabolism and cancer-promoting signals[80,81]. Thus, increased activity emanating from a mutated KRAS has the potential to increase proteasome transcription through regulation of several core regulators including NFE2L2, FOXO family members and hHSF1.
Mutant p53
Proteasome plays a key role in regulation of the physiologic function of wild type p53, as the tumor suppressor turnover needs to be tightly controlled to avoid untimely cell cycle arrest or cell demise from its accumulation in normal cells[9]. Pharmacologic inhibition of the proteasome with bortezomib results in cell death associated with p53 accumulation[82]. The oncogene c-Myc also accumulates in this model which may also result in indirect induction of p53 through p14ARF activation. c-Myc activation in normal cells in stress conditions leads to cell cycle arrest and apoptosis through activation of the p14ARF/ p53 axis and thus, in cancer, neutralization of this axis is a prerequisite for tolerance of c-Myc activation, which is present in a sub-set of pancreatic adenocarcinomas[83].Pancreatic cancers with mutations in TP53 rely less on proteasome activity to neutralize p53 activity, while the sub-set with intact TP53 may particularly benefit from the increased proteasome activity that results from up-regulation of proteasome sub-units. In addition, mutant TP53 pancreatic cancers carry more often than TP53 wild-type cancers lesions of the CDKN2A locus, encoding for p14ARF and p16, which may impair further residual proteasomal degradation of p53 due to impaired ubiquitination by MDM2[3].
Mutated p53 may play a role in stabilization of NFE2L2 through an interplay between HSP90 that provides a feedback antioxidant response, involving the interaction with p62/sequestrome and stabilization of NFE2L2[84]. This response protected pancreatic cancer cells from excess reactive oxygen species through up-regulation of antioxidant enzymes, in vitro. However, whether proteasome was up-regulated in these cells was not examined in this study[84]. Mutated p53 co-operates with NFE2L2 in transcription of targets genes with ARE sequences in their promoters such as the thioredoxin gene[85]. In addition, the mutant p53 transcriptional program included proteasome genes and induced proteasome inhibitor resistance in another study[86].
Another gain of function of mutated p53 involves promotion of the transcriptional program of transcription factor hHSF1 by interacting directly with hHSF1 phosphorylated at S326[87]. This interaction favors binding of hHSF1 to HSE DNA target sequences and induces cancer cell resistance to proteotoxic stress, by upregulation of genes such as the chaperone HSP90. Increased chaperone activity favors stability of mutant oncoproteins, including mutated p53 itself, in a positive feed-forward loop. Besides chaperone stabilizing activity for mutated p53, HSP90 interacts and interferes with the function of ligase MDM2 and another p53 ligase, CHIP (Carboxyterminus of HSP70 Interacting Protein), preventing mutated p53 ubiquitination and degradation by the proteasome[88]. Thus, chaperoning of mutated p53 by HSP90 instead of HSP70, that interacts with CHIP promotes stability of the mutant protein altering its fate of degradation[89]. In addition, hHSF1 facilitates nuclear localization of p53, where the two factors can continue their co-operation on the transcription from HSE containing promoters[90]. Phosphorylation of hHSF1 at the serine of position 326 is executed by activation of MAPK and PI3K cascades and thus may be a direct effect downstream of activated KRAS[90].
Similar to the interaction with hHSF1, mutant p53 binds to the proteasome regulator NF-Y on CCAAT target sequences and alters the transactivation capability of this regulator[91]. Following exposure to DNA damaging agents, NF-Y bound to mutant p53 interacts with acetyltransferase p300 and activates cell cycle genes, instead of its interaction with HDACs when bound to wild-type p53, which leads to gene suppression and cell cycle arrest[91,92]. In pancreatic cancer, mutant p53 replaces the family member p73 from complexes with NF-Y factors resulting in derepression of the promoter of receptor tyrosine kinase PDGFRβ gene[72]. PDGFRβ signaling contributes to increased metastatic potential in this model. Mutant p53 could contribute also to up-regulation of proteasome genes which are targets of NF-Y.
TGFβ / SMAD
TGFβ is one of three cytokines belonging to the transforming growth factor family. TGFβ is most notably involved in immunosuppression, angiogenesis, metabolic activity, and cell-cycle control. Upon interaction with the TGFβ receptor (type II), this cytokine triggers a signaling cascade which phosphorylates receptor-activated Smad proteins which in turn form complexes, translocate to the nucleus, and induce gene transcription. Inactivation of SMAD4, which mediates pancreatic cell apoptosis and proliferation, is observed in half of advanced pancreatic cancers[93]. While SMAD4-regulated genes associated with the TGFβ pathway are normally tumor-suppressive in pancreatic epithelial cells[94], pancreatic tumors often display increased expression of TGFβ which promotes a tissue microenvironment of paracrine-like signaling that becomes tumorigenic when SMAD4 incurs mutations or complete deletion[95,96]. Further, TGFβ signaling can become deregulated by the genetic state of KRAS, becoming pro-carcinogenic[97]. In addition, specific SMAD4 mutants encountered in pancreatic and colon cancers show an increased phosphorylation by GSK3β kinase and MAPK and subsequent ubiquitination and proteasome degradation[98]. Both GSK3β and MAPK activities are modulated by activated KRAS cascades. Interestingly, pancreatic cancer development is tied to the negative regulation of E-cadherin expression by ZEB2 (also known as SIP1- Smad-interacting protein 1) and is associated with migration and invasiveness[99]. TGFβ’s dual, context-dependent relationship as a tumor suppressor or promoter is further complicated when considering that during pancreatic carcinogenesis there is crosstalk with NFE2L2, contributing to malignant transformation[100]. In pancreatic cancer cells, TGFβ signaling up-regulates NFE2L2 and suppresses E-cadherin expression, contributing to invasion (Figure 1). In contrast, in premalignant human pancreatic duct cells, TGFβ is not able to affect NFE2L2. Knockdown of NFE2L2 decreases the potential of TGFβ to induce invasion of pancreatic cancer cells[100]. TGFβ signaling regulates also nuclear localization of the NF-YA sub-unit of NF-Y factor[101]. In cells exposed to TGFβ, NF-YA localizes to the nucleus and binds target gene promoters in a manner that depends on the activity of MAPK kinases but is independent of the SMAD factors. The baseline activity of MAPK kinases in various cell types affects the kinetics of NF-YA nuclear localization and activity[101]. The intermediate signal from TGFβ to MAPK for the activation of NF-Y may be carried by MAPKKK TGFβ Activated Kinase 1 (TAK1)[102]. These data suggest that the micro-environment of cancer cells rewires TGFβ transduction that may underline proteasome levels and activity[103].
p16 mutations and deep deletions of the p16/ p14 locus
p16 (INK4a) is a cyclin dependent kinase inhibitor that inhibits the Cyclin Dependent Kinase 4 and 6 (CDK4/6)/Cyclin D complex activity resulting in cell cycle arrest[104]. Cyclin D is a proteasome substrate and is degraded when p16 prevents interaction of the CDK4/Cyclin D complex with the Retinoblastoma (Rb) protein. Cells with activated KRAS/PI3K/AKT pathway and with increased KRAS/BRAF/MEK activity undergo oncogene induced senescence through up-regulation of p16 and the related cyclin dependent kinase inhibitor p15 and thus, derive benefit from absence of p16[105,106]. The benefit of inactivating p16 in KRAS mutated cancers is also observed in colorectal cancers where the mechanism is promoter methylation at the CDKN2A gene locus that encodes for p16[107]. Inactivation of the fail-safe mechanism of senescence induction in KRAS mutant cells may be accomplished in different cancers by different mechanisms such as mutations, deletion of the locus and promoter methylation[108]. A subset of pancreatic cancers bears mutations in the CDKN2A gene. Other pancreatic cancer cases display deletions of the CDKN2A locus, leading, besides the absence of p16, to absence of expression of p14ARF protein, a positive regulator of p53. p14ARF protein is transcribed from the same locus with an overlapping sequence but an alternative reading frame. Absence of p14ARF allows MDM2 to ubiquitinate wild type p53 for proteasomal degradation. In pancreatic cancers with p16 mutations or deletions, the absence of p16 function allows proliferation despite high proteasome activity[109]. In addition, p14ARF has regulatory activities beyond induction of p53, among which is a negative regulation of NFE2L2[110]. p14ARF prevents NFE2L2 from activating targets genes such as SLC7A11. Thus, deletion of p14ARF in pancreatic cancer may contribute to NFE2L2 activation and increased target gene expression, including proteasome component genes. On the other hand, cell cycle inhibitor p16 is a target gene of NFE2L2 and as a result deletion of the locus prevents up-regulation that would be an effect of NFE2L2 activation[6].
p16 also regulates the transcriptional activity of transcription factor NF-Y through inhibition of cyclin dependent kinases[111]. The transcription of human thymidine kinase gene which possesses a CAATT boxes in its promoter and is activated by NF-Y is reduced in cervical cancer cells transfected with p16. Thus, p16 has a broad effect in proteasome factors regulation and its absence or deregulation in pancreatic cancer may contribute to increased proteasomal activity.
The tumor suppressor role of p16 relates to cell cycle inhibition and permanent exit from cycling, associated with induction of senescence[112]. Hence the frequent occurrence of neutralization of p16 in cancers offers proliferation advantage. In normal cells, protracted activation of p16 also induces senescence and contributes to aging. A related effect of p16 is in prevention of reprogramming and pluripotency. p16 is a major roadblock in the induction of stem cells from adult differentiated cells. In contrast, p16 inhibition favors reprogramming[113]. In pancreatic cancer, expression of p16 by immunohistochemistry, suggesting an intact protein, was associated with an improved prognosis in patients who had undergone surgical resection[114]. Moreover, patients with absence of lesions in any of the other three common molecular alterations of pancreatic cancer had better survival outcomes. This associations together with the roles of common pancreatic cancer-related pathways in proteasome expression and function further argues for the important role of the proteasome in the disease.
THE PROTEASOME AND ITS REGULATION IN EMT AND STEMNESS AND THERAPEUTIC IMPLICATIONS IN PANCREATIC CANCER
Proteasome as a major regulator of proteostasis is important for the function of any cell, including cancer cells. Regulation of proteasomes is critical for the normal function of cells and needs to be tightly modulated to reflect the metabolic needs of host cells. During embryonic development, the proteasome is fundamental for embryonic cell proliferation and regulated apoptosis, both co-operating in organismal morphogenesis. Up-regulation of proteasome production induced by transcription factor NFE2L2 and high levels of proteasome activity are present in human embryonic stem cells, which confers structural and functional plasticity[115]. Similarly, cancer cells are characterized by significant intratumoral plasticity that is integral to their neoplastic state and derives from the inherent instability of their genomes[116]. Indeed, pancreatic adenocarcinomas and several other cancers contain, for example, inactivating mutations of the guardian of the genome p53. Further, cancers contain cells with variable proliferation status ranging from a usually smaller subset with lower proliferation and stem cell characteristics and a bulk cancer cell component that are more differentiated and possess higher proliferative activity. Between these extremes, a cell compartment with variable differentiation and proliferation states fills the spectrum. These compartments are not static and cells transition between different proliferative and differentiation states, which is facilitated by their genomic instability. Thus, the stemness of cancer, although it shares characteristics with development, is more fluid than the embryonic stem cell state where the directionality is solely towards differentiation, and typically an ontogenetic goal state set by the body plan. In addition, the cancer stem cell state is less proliferative than embryonic stem cells, where the basic functional output is to maintain cancer cell supply and protect the tumor from external toxins, such as chemotherapy. In contrast, the primary functional output of embryonic stem cells is exactly timed and drives the development of a whole organism from one to several cells. Thus, in contrast to embryonic stem cells, cancer stem cells are for protracted periods quiescent and have low metabolic activity. As a result, in contrast to embryonic stem cells and to bulk cancer cells, their proteasome activity is low[26].
The plasticity of cancer stemness endows cancer stem cells with another property derived from embryonic development, EMT and the reverse process, MET. Cancer stem cells have the ability to access both an epithelial and a mesenchymal state using the two processes appropriated from development. When directed towards differentiation stem cells lose this ability and are locked, albeit not irreversibly, into the epithelial state of the bulk stem cells. Notably, mesenchymal stem cell function declines with senescence, which is linked to proteasome dysfunction, where stemness can be enhanced by core subunit β5-overexpression-induced proteasomal re-activation[117]. Fluctuations of proteasome levels and function follow and are regulated by the state of the cancer cell with low levels being adequate in cancer stem cells, but high levels required in proliferating bulk cancer cells with high metabolic and protein turn over. Thus, tight regulation of proteasome levels is expected to be critical for cancer cells and fluctuations of proteasome availability are integrated in the cancer signaling programs with key players being parts of major cancer pathways as detailed in the previous paragraphs. Unsurprisingly, proteasomal regulators such as the deubiquitinating enzyme USP21, which is frequently amplified in 22% of pancreatic adenocarcinomas, when overexpressed, promotes stemness in cancer cells, enhances tumor growth, and drives progression from the precursor pancreatic intraepithelial neoplasia (PanIN) to pancreatic adenocarcinoma[118].
Despite the critical role of proteasome in cancer, therapeutic exploitation of proteasome inhibition has not been successful in solid tumors, in stark contrast to specific hematologic malignancies, such as multiple myeloma where proteasome inhibitors are successfully integrated in the therapeutic armamentarium[119]. In pancreatic cancer, proteasome inhibitors have produced disappointing results in clinical trials and development is not actively in pursuit[120]. However, it is clear from pre-clinical investigations that subsets of pancreatic cancer cells are sensitive to proteasome inhibition. Pancreatic cancers with oncogene c-Myc constitute a subset with such vulnerability[121]. Although c-Myc remains not directly targetable, high activity confers sensitivity to proteotoxic stress and pharmacologic proteasome inhibition. Interestingly, c-Myc amplifications in pancreatic cancer are observed exclusively in cases with p53 mutations, consistent with the induction of oncogenic stress-induced apoptosis if p53 is intact.
Differential regulation of protein translation may also confer proteasome inhibition sensitivity in subsets of pancreatic adenocarcinomas. Pancreatic cancer cells exposed to bortezomib up-regulated the kinase EIF2AK1 (eukaryotic translation Initiation Factor 2 alpha Kinase 1, also known as HRI- Heme Regulated Inhibitor) and are resistant to the drug[11]. Knockdown of EIF2AK1 led to increased protein translation and accumulation resulting to cell death after bortezomib exposure. Another study in pancreatic cancer cell lines that were resistant to 5-FU chemotherapy showed that these lines displayed stem cell and EMT markers expression and high activity of the proteasome master regulator NFE2L2[122]. Knockdown of NFE2L2 in this model sensitized cells to 5-FU.
mTOR inhibitors are drugs that directly affect proteostasis by interfering with protein translation through inhibition of the translation initiation complex[123]. Everolimus, an mTOR inhibitor, provides an example of the challenges and disappointments the pancreatic cancer therapeutics field has faced. The drug is successfully used in combination with hormonal therapy for the treatment of metastatic Estrogen Receptor positive breast cancer[124]. Everolimus has been investigated in pancreatic adenocarcinoma both alone and in combination with various other drugs with the aim to inhibit protein production in proliferating cancer cells. Clinical translation of initial positive pre-clinical data has provided disappointing results[125,126]. As monotherapy, everolimus produces no responses and adding it to chemotherapy or targeted therapies has not provided any benefit. However, the clinical development of the drug has followed the usual chemotherapy development paradigm without any attempt to identify and include sub-sets of pancreatic cancer patients with probable sensitivity to everolimus based on underlying molecular defects. In addition, inhibition of protein translation may not be a good target in pancreatic cancer altogether, based on the above discussion, given that protein production shut-down may alleviate proteostatic stress and could counter-intuitively promote the ability of cancer cells to cope with this stress. In addition, decreased protein turn over could allow for a decreased proteasome activity compatible with acquiring a cancer stem cell phenotype and drug resistance. Other studies have shown that resistance to everolimus results from a feed-back activation of up-stream receptor tyrosine kinases. Nevertheless, attempts to address this resistance mechanism with combinations of everolimus and EGFR kinase inhibitor erlotinib were not met with success[127].
CONCLUSION
It is evident that challenges in pancreatic cancer therapeutics remain. However, targeted exploitation of pancreatic cancer vulnerabilities stemming from proteostasis dysregulation is possible and could promote therapeutics in well-defined molecular sub-sets of patients. Further development of proteasome inhibitors, possibly in combination with other molecularly defined targeted therapies, relies on the discovery of synthetic vulnerabilities. A strategy for drug development of proteasome inhibitors in pancreatic cancer should identify vulnerable cell lines in vitro, examine their molecular make-up and subsequently examine whether patient-derived xenografts with similar molecular lesions are indeed sensitive to these drugs or their combinations with other candidate drugs, before testing the drug(s) in pancreatic cancer patients bearing tumors with the same underlying driver molecular defects.
Footnotes
Conflict-of-interest statement: The authors declare no conflict of interest regarding this manuscript.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Peer-review started: February 23, 2021
First decision: March 29, 2021
Article in press: December 2, 2021
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Canada
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Jiang W S-Editor: Wu YXJ L-Editor: A P-Editor: Wu YXJ
Contributor Information
Nirosha J Murugan, Department of Biology, Algoma University, Sault Sainte Marie P6A3T6, ON, Canada.
Ioannis A Voutsadakis, Department of Medical Oncology, Sault Area Hospital, Sault Sainte Marie P6A3T6, ON, Canada. ivoutsadakis@yahoo.com.
References
- 1.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32:185–203.e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, Hellmann MD, Barron DA, Schram AM, Hameed M, Dogan S, Ross DS, Hechtman JF, DeLair DF, Yao J, Mandelker DL, Cheng DT, Chandramohan R, Mohanty AS, Ptashkin RN, Jayakumaran G, Prasad M, Syed MH, Rema AB, Liu ZY, Nafa K, Borsu L, Sadowska J, Casanova J, Bacares R, Kiecka IJ, Razumova A, Son JB, Stewart L, Baldi T, Mullaney KA, Al-Ahmadie H, Vakiani E, Abeshouse AA, Penson AV, Jonsson P, Camacho N, Chang MT, Won HH, Gross BE, Kundra R, Heins ZJ, Chen HW, Phillips S, Zhang H, Wang J, Ochoa A, Wills J, Eubank M, Thomas SB, Gardos SM, Reales DN, Galle J, Durany R, Cambria R, Abida W, Cercek A, Feldman DR, Gounder MM, Hakimi AA, Harding JJ, Iyer G, Janjigian YY, Jordan EJ, Kelly CM, Lowery MA, Morris LGT, Omuro AM, Raj N, Razavi P, Shoushtari AN, Shukla N, Soumerai TE, Varghese AM, Yaeger R, Coleman J, Bochner B, Riely GJ, Saltz LB, Scher HI, Sabbatini PJ, Robson ME, Klimstra DS, Taylor BS, Baselga J, Schultz N, Hyman DM, Arcila ME, Solit DB, Ladanyi M, Berger MF. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, Nourse C, Murtaugh LC, Harliwong I, Idrisoglu S, Manning S, Nourbakhsh E, Wani S, Fink L, Holmes O, Chin V, Anderson MJ, Kazakoff S, Leonard C, Newell F, Waddell N, Wood S, Xu Q, Wilson PJ, Cloonan N, Kassahn KS, Taylor D, Quek K, Robertson A, Pantano L, Mincarelli L, Sanchez LN, Evers L, Wu J, Pinese M, Cowley MJ, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chantrill LA, Mawson A, Humphris J, Chou A, Pajic M, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Lovell JA, Merrett ND, Toon CW, Epari K, Nguyen NQ, Barbour A, Zeps N, Moran-Jones K, Jamieson NB, Graham JS, Duthie F, Oien K, Hair J, Grützmann R, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Rusev B, Capelli P, Salvia R, Tortora G, Mukhopadhyay D, Petersen GM Australian Pancreatic Cancer Genome Initiative, Munzy DM, Fisher WE, Karim SA, Eshleman JR, Hruban RH, Pilarsky C, Morton JP, Sansom OJ, Scarpa A, Musgrove EA, Bailey UM, Hofmann O, Sutherland RL, Wheeler DA, Gill AJ, Gibbs RA, Pearson JV, Waddell N, Biankin AV, Grimmond SM. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 6.Hashimoto-Hachiya A, Tsuji G, Furue M. Antioxidants cinnamaldehyde and Galactomyces fermentation filtrate downregulate senescence marker CDKN2A/p16INK4A via NRF2 activation in keratinocytes. J Dermatol Sci. 2019;96:53–56. doi: 10.1016/j.jdermsci.2019.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Shirazi F, Jones RJ, Singh RK, Zou J, Kuiatse I, Berkova Z, Wang H, Lee HC, Hong S, Dick L, Chattopadhyay N, Orlowski RZ. Activating KRAS, NRAS, and BRAF mutants enhance proteasome capacity and reduce endoplasmic reticulum stress in multiple myeloma. Proc Natl Acad Sci U S A. 2020;117:20004–20014. doi: 10.1073/pnas.2005052117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voutsadakis IA. Pathogenesis of colorectal carcinoma and therapeutic implications: the roles of the ubiquitin-proteasome system and Cox-2. J Cell Mol Med. 2007;11:252–285. doi: 10.1111/j.1582-4934.2007.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voutsadakis IA. The ubiquitin-proteasome system in colorectal cancer. Biochim Biophys Acta. 2008;1782:800–808. doi: 10.1016/j.bbadis.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Belalcazar A, Shaib WL, Farren MR, Zhang C, Chen Z, Yang L, Lesinski GB, El-Rayes BF, Nagaraju GP. Inhibiting heat shock protein 90 and the ubiquitin-proteasome pathway impairs metabolic homeostasis and leads to cell death in human pancreatic cancer cells. Cancer. 2017;123:4924–4933. doi: 10.1002/cncr.30944. [DOI] [PubMed] [Google Scholar]
- 11.White MC, Schroeder RD, Zhu K, Xiong K, McConkey DJ. HRI-mediated translational repression reduces proteotoxicity and sensitivity to bortezomib in human pancreatic cancer cells. Oncogene. 2018;37:4413–4427. doi: 10.1038/s41388-018-0227-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voutsadakis IA. Proteasome expression and activity in cancer and cancer stem cells. Tumour Biol. 2017;39:1010428317692248. doi: 10.1177/1010428317692248. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka K, Yoshimura T, Kumatori A, Ichihara A, Ikai A, Nishigai M, Kameyama K, Takagi T. Proteasomes (multi-protease complexes) as 20 S ring-shaped particles in a variety of eukaryotic cells. J Biol Chem. 1988;263:16209–16217. [PubMed] [Google Scholar]
- 14.Grice GL, Nathan JA. The recognition of ubiquitinated proteins by the proteasome. Cell Mol Life Sci. 2016;73:3497–3506. doi: 10.1007/s00018-016-2255-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka K. The proteasome: overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Almond JB, Cohen GM. The proteasome: a novel target for cancer chemotherapy. Leukemia. 2002;16:433–443. doi: 10.1038/sj.leu.2402417. [DOI] [PubMed] [Google Scholar]
- 17.Ciznadija D, Zhu XH, Koff A. Hdm2- and proteasome-dependent turnover limits p21 accumulation during S phase. Cell Cycle. 2011;10:2714–2723. doi: 10.4161/cc.10.16.16725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujita E, Mukasa T, Tsukahara T, Arahata K, Omura S, Momoi T. Enhancement of CPP32-like activity in the TNF-treated U937 cells by the proteasome inhibitors. Biochem Biophys Res Commun. 1996;224:74–79. doi: 10.1006/bbrc.1996.0986. [DOI] [PubMed] [Google Scholar]
- 19.Imajoh-Ohmi S, Kawaguchi T, Sugiyama S, Tanaka K, Omura S, Kikuchi H. Lactacystin, a specific inhibitor of the proteasome, induces apoptosis in human monoblast U937 cells. Biochem Biophys Res Commun. 1995;217:1070–1077. doi: 10.1006/bbrc.1995.2878. [DOI] [PubMed] [Google Scholar]
- 20.Lee HC, Cerchione C. How I treat relapsed and/or refractory multiple myeloma. Hematol Rep. 2020;12:8955. doi: 10.4081/hr.2020.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott K, Hayden PJ, Will A, Wheatley K, Coyne I. Bortezomib for the treatment of multiple myeloma. Cochrane Database of Systematic Reviews, 2013, John Wiley & Sons, Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Infante JR, Mendelson DS, Burris HA 3rd, Bendell JC, Tolcher AW, Gordon MS, Gillenwater HH, Arastu-Kapur S, Wong HL, Papadopoulos KP. A first-in-human dose-escalation study of the oral proteasome inhibitor oprozomib in patients with advanced solid tumors. Invest New Drugs. 2016;34:216–224. doi: 10.1007/s10637-016-0327-x. [DOI] [PubMed] [Google Scholar]
- 23.Roth P, Mason WP, Richardson PG, Weller M. Proteasome inhibition for the treatment of glioblastoma. Expert Opin Investig Drugs. 2020;29:1133–1141. doi: 10.1080/13543784.2020.1803827. [DOI] [PubMed] [Google Scholar]
- 24.Chattopadhyay N, Berger AJ, Koenig E, Bannerman B, Garnsey J, Bernard H, Hales P, Maldonado Lopez A, Yang Y, Donelan J, Jordan K, Tirrell S, Stringer B, Xia C, Hather G, Galvin K, Manfredi M, Rhodes N, Amidon B. KRAS Genotype Correlates with Proteasome Inhibitor Ixazomib Activity in Preclinical In Vivo Models of Colon and Non-Small Cell Lung Cancer: Potential Role of Tumor Metabolism. PLoS One. 2015;10:e0144825. doi: 10.1371/journal.pone.0144825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vlashi E, Lagadec C, Chan M, Frohnen P, McDonald AJ, Pajonk F. Targeted elimination of breast cancer cells with low proteasome activity is sufficient for tumor regression. Breast Cancer Res Treat. 2013;141:197–203. doi: 10.1007/s10549-013-2688-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vlashi E, Kim K, Lagadec C, Donna LD, McDonald JT, Eghbali M, Sayre JW, Stefani E, McBride W, Pajonk F. In vivo imaging, tracking, and targeting of cancer stem cells. J Natl Cancer Inst. 2009;101:350–359. doi: 10.1093/jnci/djn509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraunhoffer NA, Abuelafia AM, Bigonnet M, Gayet O, Roques J, Telle E, Santofimia-Castaño P, Borrello MT, Chuluyan E, Dusetti N, Iovanna J. Evidencing a Pancreatic Ductal Adenocarcinoma Subpopulation Sensitive to the Proteasome Inhibitor Carfilzomib. Clin Cancer Res. 2020;26:5506–5519. doi: 10.1158/1078-0432.CCR-20-1232. [DOI] [PubMed] [Google Scholar]
- 28.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 29.Silveira WA, Gonçalves DA, Machado J, Lautherbach N, Lustrino D, Paula-Gomes S, Pereira MG, Miyabara EH, Sandri M, Kettelhut IC, Navegantes LC. cAMP-dependent protein kinase inhibits FoxO activity and regulates skeletal muscle plasticity in mice. FASEB J. 2020;34:12946–12962. doi: 10.1096/fj.201902102RR. [DOI] [PubMed] [Google Scholar]
- 30.Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci U S A. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farrow B, Evers BM. Activation of PPARgamma increases PTEN expression in pancreatic cancer cells. Biochem Biophys Res Commun. 2003;301:50–53. doi: 10.1016/s0006-291x(02)02983-2. [DOI] [PubMed] [Google Scholar]
- 32.Deng X, Li Y, Gu S, Chen Y, Yu B, Su J, Sun L, Liu Y. p53 Affects PGC1α Stability Through AKT/GSK-3β to Enhance Cisplatin Sensitivity in Non-Small Cell Lung Cancer. Front Oncol. 2020;10:1252. doi: 10.3389/fonc.2020.01252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan L, Zhang L, Gong Z, Hou X, Xu Y, Feng X, Wang H, You H. FoxO3 inactivation promotes human cholangiocarcinoma tumorigenesis and chemoresistance through Keap1-Nrf2 signaling. Hepatology. 2016;63:1914–1927. doi: 10.1002/hep.28496. [DOI] [PubMed] [Google Scholar]
- 34.Kawazoe Y, Nakai A, Tanabe M, Nagata K. Proteasome inhibition leads to the activation of all members of the heat-shock-factor family. Eur J Biochem. 1998;255:356–362. doi: 10.1046/j.1432-1327.1998.2550356.x. [DOI] [PubMed] [Google Scholar]
- 35.Ninfali C, Siles L, Darling DS, Postigo A. Regulation of muscle atrophy-related genes by the opposing transcriptional activities of ZEB1/CtBP and FOXO3. Nucleic Acids Res. 2018;46:10697–10708. doi: 10.1093/nar/gky835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Battula VL, Evans KW, Hollier BG, Shi Y, Marini FC, Ayyanan A, Wang RY, Brisken C, Guerra R, Andreeff M, Mani SA. Epithelial-mesenchymal transition-derived cells exhibit multilineage differentiation potential similar to mesenchymal stem cells. Stem Cells. 2010;28:1435–1445. doi: 10.1002/stem.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morel AP, Lièvre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy SK, Srivastava RK, Shankar S. Inhibition of PI3K/AKT and MAPK/ERK pathways causes activation of FOXO transcription factor, leading to cell cycle arrest and apoptosis in pancreatic cancer. J Mol Signal. 2010;5:10. doi: 10.1186/1750-2187-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu W, Ma Q, Chen L, Li P, Zhang M, Ramamoorthy S, Nawaz Z, Shimojima T, Wang H, Yang Y, Shen Z, Zhang Y, Zhang X, Nicosia SV, Pledger JW, Chen J, Bai W. MDM2 acts downstream of p53 as an E3 ligase to promote FOXO ubiquitination and degradation. J Biol Chem. 2009;284:13987–14000. doi: 10.1074/jbc.M901758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He F, Ru X, Wen T. NRF2, a Transcription Factor for Stress Response and Beyond. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21134777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dodson M, de la Vega MR, Cholanians AB, Schmidlin CJ, Chapman E, Zhang DD. Modulating NRF2 in Disease: Timing Is Everything. Annu Rev Pharmacol Toxicol. 2019;59:555–575. doi: 10.1146/annurev-pharmtox-010818-021856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan JX, Short SR, Goff SA, Dice JF. Ubiquitin pools, ubiquitin mRNA levels, and ubiquitin-mediated proteolysis in aging human fibroblasts. Exp Gerontol. 1993;28:39–49. doi: 10.1016/0531-5565(93)90018-9. [DOI] [PubMed] [Google Scholar]
- 43.Pickering AM, Davies KJ. Degradation of damaged proteins: the main function of the 20S proteasome. Prog Mol Biol Transl Sci. 2012;109:227–248. doi: 10.1016/B978-0-12-397863-9.00006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong YB, Kang HJ, Kwon SY, Kim HJ, Kwon KY, Cho CH, Lee JM, Kallakury BV, Bae I. Nuclear factor (erythroid-derived 2)-like 2 regulates drug resistance in pancreatic cancer cells. Pancreas. 2010;39:463–472. doi: 10.1097/MPA.0b013e3181c31314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lister A, Nedjadi T, Kitteringham NR, Campbell F, Costello E, Lloyd B, Copple IM, Williams S, Owen A, Neoptolemos JP, Goldring CE, Park BK. Nrf2 is overexpressed in pancreatic cancer: implications for cell proliferation and therapy. Mol Cancer. 2011;10:37. doi: 10.1186/1476-4598-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.You A, Nam CW, Wakabayashi N, Yamamoto M, Kensler TW, Kwak MK. Transcription factor Nrf2 maintains the basal expression of Mdm2: An implication of the regulation of p53 signaling by Nrf2. Arch Biochem Biophys. 2011;507:356–364. doi: 10.1016/j.abb.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 47.Hamada S, Shimosegawa T, Taguchi K, Nabeshima T, Yamamoto M, Masamune A. Simultaneous K-ras activation and Keap1 deletion cause atrophy of pancreatic parenchyma. Am J Physiol Gastrointest Liver Physiol. 2018;314:G65–G74. doi: 10.1152/ajpgi.00228.2017. [DOI] [PubMed] [Google Scholar]
- 48.Hamada S, Taguchi K, Masamune A, Yamamoto M, Shimosegawa T. Nrf2 promotes mutant K-ras/p53-driven pancreatic carcinogenesis. Carcinogenesis. 2017;38:661–670. doi: 10.1093/carcin/bgx043. [DOI] [PubMed] [Google Scholar]
- 49.Mathew A, Mathur SK, Morimoto RI. Heat shock response and protein degradation: regulation of HSF2 by the ubiquitin-proteasome pathway. Mol Cell Biol. 1998;18:5091–5098. doi: 10.1128/mcb.18.9.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem. 2011;80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- 51.Dong B, Jaeger AM, Thiele DJ. Inhibiting Heat Shock Factor 1 in Cancer: A Unique Therapeutic Opportunity. Trends Pharmacol Sci. 2019;40:986–1005. doi: 10.1016/j.tips.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 52.Puustinen MC, Sistonen L. Molecular Mechanisms of Heat Shock Factors in Cancer. Cells. 2020;9 doi: 10.3390/cells9051202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pincus D. Regulation of Hsf1 and the Heat Shock Response. Adv Exp Med Biol. 2020;1243:41–50. doi: 10.1007/978-3-030-40204-4_3. [DOI] [PubMed] [Google Scholar]
- 54.Björk JK, Sistonen L. Clustering of heat-shock factors. Biochem J. 2006;395:e5–e6. doi: 10.1042/BJ20060071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loison F, Debure L, Nizard P, le Goff P, Michel D, le Dréan Y. Up-regulation of the clusterin gene after proteotoxic stress: implication of HSF1-HSF2 heterocomplexes. Biochem J. 2006;395:223–231. doi: 10.1042/BJ20051190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santopolo S, Riccio A, Rossi A, Santoro MG. The proteostasis guardian HSF1 directs the transcription of its paralog and interactor HSF2 during proteasome dysfunction. Cell Mol Life Sci. 2021;78:1113–1129. doi: 10.1007/s00018-020-03568-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee YJ, Kim EH, Lee JS, Jeoung D, Bae S, Kwon SH, Lee YS. HSF1 as a mitotic regulator: phosphorylation of HSF1 by Plk1 is essential for mitotic progression. Cancer Res. 2008;68:7550–7560. doi: 10.1158/0008-5472.CAN-08-0129. [DOI] [PubMed] [Google Scholar]
- 58.Kim EH, Lee YJ, Bae S, Lee JS, Kim J, Lee YS. Heat shock factor 1-mediated aneuploidy requires a defective function of p53. Cancer Res. 2009;69:9404–9412. doi: 10.1158/0008-5472.CAN-09-1411. [DOI] [PubMed] [Google Scholar]
- 59.Frezzato F, Raggi F, Martini V, Severin F, Trimarco V, Visentin A, Scomazzon E, Accordi B, Bresolin S, Piazza F, Facco M, Basso G, Semenzato G, Trentin L. HSP70/HSF1 axis, regulated via a PI3K/AKT pathway, is a druggable target in chronic lymphocytic leukemia. Int J Cancer. 2019;145:3089–3100. doi: 10.1002/ijc.32383. [DOI] [PubMed] [Google Scholar]
- 60.Joutsen J, Da Silva AJ, Luoto JC, Budzynski MA, Nylund AS, de Thonel A, Concordet JP, Mezger V, Sabéran-Djoneidi D, Henriksson E, Sistonen L. Heat Shock Factor 2 Protects against Proteotoxicity by Maintaining Cell-Cell Adhesion. Cell Rep. 2020;30:583–597.e6. doi: 10.1016/j.celrep.2019.12.037. [DOI] [PubMed] [Google Scholar]
- 61.Wang B, Lee CW, Witt A, Thakkar A, Ince TA. Heat shock factor 1 induces cancer stem cell phenotype in breast cancer cell lines. Breast Cancer Res Treat. 2015;153:57–66. doi: 10.1007/s10549-015-3521-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma S, Kong D, Fu X, Liu L, Liu Y, Xue C, Tian Z, Li L, Liu X. p53-Induced Autophagy Regulates Chemotherapy and Radiotherapy Resistance in Multidrug Resistance Cancer Cells. Dose Response. 2021;19:15593258211048046. doi: 10.1177/15593258211048046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaushik S, Tasset I, Arias E, Pampliega O, Wong E, Martinez-Vicente M, Cuervo AM. Autophagy and the hallmarks of aging. Ageing Res Rev. 2021;72:101468. doi: 10.1016/j.arr.2021.101468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu PK, Hong SK, Starenki D, Oshima K, Shao H, Gestwicki JE, Tsai S, Park JI. Mortalin/HSPA9 targeting selectively induces KRAS tumor cell death by perturbing mitochondrial membrane permeability. Oncogene. 2020;39:4257–4270. doi: 10.1038/s41388-020-1285-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oldfield AJ, Henriques T, Kumar D, Burkholder AB, Cinghu S, Paulet D, Bennett BD, Yang P, Scruggs BS, Lavender CA, Rivals E, Adelman K, Jothi R. NF-Y controls fidelity of transcription initiation at gene promoters through maintenance of the nucleosome-depleted region. Nat Commun. 2019;10:3072. doi: 10.1038/s41467-019-10905-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gurtner A, Manni I, Piaggio G. NF-Y in cancer: Impact on cell transformation of a gene essential for proliferation. Biochim Biophys Acta Gene Regul Mech. 2017;1860:604–616. doi: 10.1016/j.bbagrm.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 67.Ly LL, Yoshida H, Yamaguchi M. Nuclear transcription factor Y and its roles in cellular processes related to human disease. Am J Cancer Res. 2013;3:339–346. [PMC free article] [PubMed] [Google Scholar]
- 68.Chae HD, Kim J, Shin DY. NF-Y binds to both G1- and G2-specific cyclin promoters; a possible role in linking CDK2/Cyclin A to CDK1/Cyclin B. BMB Rep. 2011;44:553–557. doi: 10.5483/bmbrep.2011.44.8.553. [DOI] [PubMed] [Google Scholar]
- 69.Yun J, Chae HD, Choy HE, Chung J, Yoo HS, Han MH, Shin DY. p53 negatively regulates cdc2 transcription via the CCAAT-binding NF-Y transcription factor. J Biol Chem. 1999;274:29677–29682. doi: 10.1074/jbc.274.42.29677. [DOI] [PubMed] [Google Scholar]
- 70.Xu H, Fu J, Ha SW, Ju D, Zheng J, Li L, Xie Y. The CCAAT box-binding transcription factor NF-Y regulates basal expression of human proteasome genes. Biochim Biophys Acta. 2012;1823:818–825. doi: 10.1016/j.bbamcr.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 71.Benatti P, Chiaramonte ML, Lorenzo M, Hartley JA, Hochhauser D, Gnesutta N, Mantovani R, Imbriano C, Dolfini D. NF-Y activates genes of metabolic pathways altered in cancer cells. Oncotarget. 2016;7:1633–1650. doi: 10.18632/oncotarget.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weissmueller S, Manchado E, Saborowski M, Morris JP 4th, Wagenblast E, Davis CA, Moon SH, Pfister NT, Tschaharganeh DF, Kitzing T, Aust D, Markert EK, Wu J, Grimmond SM, Pilarsky C, Prives C, Biankin AV, Lowe SW. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor β signaling. Cell. 2014;157:382–394. doi: 10.1016/j.cell.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanaka T, Ohashi S, Saito H, Higuchi T, Tabata K, Kosuge Y, Suzuki T, Miyairi S, Kobayashi S. Indirubin derivatives alter DNA binding activity of the transcription factor NF-Y and inhibit MDR1 gene promoter. Eur J Pharmacol. 2014;741:83–89. doi: 10.1016/j.ejphar.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 74.Gayvert KM, Dardenne E, Cheung C, Boland MR, Lorberbaum T, Wanjala J, Chen Y, Rubin MA, Tatonetti NP, Rickman DS, Elemento O. A Computational Drug Repositioning Approach for Targeting Oncogenic Transcription Factors. Cell Rep. 2016;15:2348–2356. doi: 10.1016/j.celrep.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3K Pathway in Human Disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lanfredini S, Thapa A, O'Neill E. RAS in pancreatic cancer. Biochem Soc Trans. 2019;47:961–972. doi: 10.1042/BST20170521. [DOI] [PubMed] [Google Scholar]
- 77.Antontseva EV, Matveeva MY, Bondar NP, Kashina EV, Leberfarb EY, Bryzgalov LO, Gervas PA, Ponomareva AA, Cherdyntseva NV, Orlov YL, Merkulova TI. Regulatory single nucleotide polymorphisms at the beginning of intron 2 of the human KRAS gene. J Biosci. 2015;40:873–883. doi: 10.1007/s12038-015-9567-8. [DOI] [PubMed] [Google Scholar]
- 78.Mann KM, Ying H, Juan J, Jenkins NA, Copeland NG. KRAS-related proteins in pancreatic cancer. Pharmacol Ther. 2016;168:29–42. doi: 10.1016/j.pharmthera.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 79.Qian W, Chen K, Qin T, Xiao Y, Li J, Yue Y, Zhou C, Ma J, Duan W, Lei J, Han L, Li L, Shen X, Wu Z, Ma Q, Wang Z. The EGFR-HSF1 axis accelerates the tumorigenesis of pancreatic cancer. Journal of experimental & clinical cancer research: CR. 2021;40:25. doi: 10.1186/s13046-020-01823-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ciocca DR, Arrigo AP, Calderwood SK. Heat shock proteins and heat shock factor 1 in carcinogenesis and tumor development: an update. Arch Toxicol. 2013;87:19–48. doi: 10.1007/s00204-012-0918-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang Z, Dai S, He Y, Doty RA, Shultz LD, Sampson SB, Dai C. MEK guards proteome stability and inhibits tumor-suppressive amyloidogenesis via HSF1. Cell. 2015;160:729–744. doi: 10.1016/j.cell.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen S, Blank JL, Peters T, Liu XJ, Rappoli DM, Pickard MD, Menon S, Yu J, Driscoll DL, Lingaraj T, Burkhardt AL, Chen W, Garcia K, Sappal DS, Gray J, Hales P, Leroy PJ, Ringeling J, Rabino C, Spelman JJ, Morgenstern JP, Lightcap ES. Genome-wide siRNA screen for modulators of cell death induced by proteasome inhibitor bortezomib. Cancer Res. 2010;70:4318–4326. doi: 10.1158/0008-5472.CAN-09-4428. [DOI] [PubMed] [Google Scholar]
- 83.Campaner S, Doni M, Hydbring P, Verrecchia A, Bianchi L, Sardella D, Schleker T, Perna D, Tronnersjö S, Murga M, Fernandez-Capetillo O, Barbacid M, Larsson LG, Amati B. Cdk2 suppresses cellular senescence induced by the c-myc oncogene. Nat Cell Biol. 2010;12:54–9; sup pp 1-14. doi: 10.1038/ncb2004. [DOI] [PubMed] [Google Scholar]
- 84.Gilardini Montani MS, Cecere N, Granato M, Romeo MA, Falcinelli L, Ciciarelli U, D'Orazi G, Faggioni A, Cirone M. Mutant p53, Stabilized by Its Interplay with HSP90, Activates a Positive Feed-Back Loop Between NRF2 and p62 that Induces Chemo-Resistance to Apigenin in Pancreatic Cancer Cells. Cancers (Basel) 2019;11 doi: 10.3390/cancers11050703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lisek K, Campaner E, Ciani Y, Walerych D, Del Sal G. Mutant p53 tunes the NRF2-dependent antioxidant response to support survival of cancer cells. Oncotarget. 2018;9:20508–20523. doi: 10.18632/oncotarget.24974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Walerych D, Lisek K, Sommaggio R, Piazza S, Ciani Y, Dalla E, Rajkowska K, Gaweda-Walerych K, Ingallina E, Tonelli C, Morelli MJ, Amato A, Eterno V, Zambelli A, Rosato A, Amati B, Wiśniewski JR, Del Sal G. Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat Cell Biol. 2016;18:897–909. doi: 10.1038/ncb3380. [DOI] [PubMed] [Google Scholar]
- 87.Li D, Yallowitz A, Ozog L, Marchenko N. A gain-of-function mutant p53-HSF1 feed forward circuit governs adaptation of cancer cells to proteotoxic stress. Cell Death Dis. 2014;5:e1194. doi: 10.1038/cddis.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li D, Marchenko ND, Schulz R, Fischer V, Velasco-Hernandez T, Talos F, Moll UM. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res. 2011;9:577–588. doi: 10.1158/1541-7786.MCR-10-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li Q, Feldman RA, Radhakrishnan VM, Carey S, Martinez JD. Hsf1 is required for the nuclear translocation of p53 tumor suppressor. Neoplasia. 2008;10:1138–1145. doi: 10.1593/neo.08430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Toma-Jonik A, Vydra N, Janus P, Widłak W. Interplay between HSF1 and p53 signaling pathways in cancer initiation and progression: non-oncogene and oncogene addiction. Cell Oncol (Dordr) 2019;42:579–589. doi: 10.1007/s13402-019-00452-0. [DOI] [PubMed] [Google Scholar]
- 91.Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, Blandino G, Piaggio G. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 92.Peart MJ, Prives C. Mutant p53 gain of function: the NF-Y connection. Cancer Cell. 2006;10:173–174. doi: 10.1016/j.ccr.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 93.Xia X, Wu W, Huang C, Cen G, Jiang T, Cao J, Huang K, Qiu Z. SMAD4 and its role in pancreatic cancer. Tumour Biol. 2015;36:111–119. doi: 10.1007/s13277-014-2883-z. [DOI] [PubMed] [Google Scholar]
- 94.Saiki Y, Horii A. Molecular pathology of pancreatic cancer. Pathol Int. 2014;64:10–19. doi: 10.1111/pin.12114. [DOI] [PubMed] [Google Scholar]
- 95.Biswas S, Criswell TL, Wang SE, Arteaga CL. Inhibition of transforming growth factor-beta signaling in human cancer: targeting a tumor suppressor network as a therapeutic strategy. Clin Cancer Res. 2006;12:4142–4146. doi: 10.1158/1078-0432.CCR-06-0952. [DOI] [PubMed] [Google Scholar]
- 96.Jakowlew SB. Transforming growth factor-beta in cancer and metastasis. Cancer Metastasis Rev. 2006;25:435–457. doi: 10.1007/s10555-006-9006-2. [DOI] [PubMed] [Google Scholar]
- 97.Vivekanandhan S, Mukhopadhyay D. Genetic status of KRAS influences Transforming Growth Factor-beta (TGF-β) signaling: An insight into Neuropilin-1 (NRP1) mediated tumorigenesis. Semin Cancer Biol. 2019;54:72–79. doi: 10.1016/j.semcancer.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Demagny H, De Robertis EM. Point mutations in the tumor suppressor Smad4/DPC4 enhance its phosphorylation by GSK3 and reversibly inactivate TGF-β signaling. Mol Cell Oncol. 2016;3:e1025181. doi: 10.1080/23723556.2015.1025181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Imamichi Y, Menke A. Signaling pathways involved in collagen-induced disruption of the E-cadherin complex during epithelial-mesenchymal transition. Cells Tissues Organs. 2007;185:180–190. doi: 10.1159/000101319. [DOI] [PubMed] [Google Scholar]
- 100.Arfmann-Knübel S, Struck B, Genrich G, Helm O, Sipos B, Sebens S, Schäfer H. The Crosstalk between Nrf2 and TGF-β1 in the Epithelial-Mesenchymal Transition of Pancreatic Duct Epithelial Cells. PLoS One. 2015;10:e0132978. doi: 10.1371/journal.pone.0132978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roy SK, Wang J, Yang P. Dexamethasone inhibits transforming growth factor-beta receptor (Tbeta R) messenger RNA expression in hamster preantral follicles: possible association with NF-YA. Biol Reprod. 2003;68:2180–2188. doi: 10.1095/biolreprod.102.013011. [DOI] [PubMed] [Google Scholar]
- 102.Eggen BJ, Benus GF, Folkertsma S, Jonk LJ, Kruijer W. TAK1 activation of the mouse JunB promoter is mediated through a CCAAT box and NF-Y. FEBS Lett. 2001;506:267–271. doi: 10.1016/s0014-5793(01)02928-3. [DOI] [PubMed] [Google Scholar]
- 103.Dardare J, Witz A, Merlin JL, Gilson P, Harlé A. SMAD4 and the TGFβ Pathway in Patients with Pancreatic Ductal Adenocarcinoma. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21103534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Voutsadakis IA. Atlas of Genetics and Cytogenetics in Oncology and Haematology. 2011. Ubiquitin, ubiquitination and the ubiquitin-proteasome system in cancer. [Google Scholar]
- 105.Cisowski J, Sayin VI, Liu M, Karlsson C, Bergo MO. Oncogene-induced senescence underlies the mutual exclusive nature of oncogenic KRAS and BRAF. Oncogene. 2016;35:1328–1333. doi: 10.1038/onc.2015.186. [DOI] [PubMed] [Google Scholar]
- 106.El Maï M, Marzullo M, de Castro IP, Ferreira MG. Opposing p53 and mTOR/AKT promote an in vivo switch from apoptosis to senescence upon telomere shortening in zebrafish. Elife. 2020;9 doi: 10.7554/eLife.54935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kohonen-Corish MR, Tseung J, Chan C, Currey N, Dent OF, Clarke S, Bokey L, Chapuis PH. KRAS mutations and CDKN2A promoter methylation show an interactive adverse effect on survival and predict recurrence of rectal cancer. Int J Cancer. 2014;134:2820–2828. doi: 10.1002/ijc.28619. [DOI] [PubMed] [Google Scholar]
- 108.Singh SK, Ellenrieder V. Senescence in pancreatic carcinogenesis: from signalling to chromatin remodelling and epigenetics. Gut. 2013;62:1364–1372. doi: 10.1136/gutjnl-2012-302793. [DOI] [PubMed] [Google Scholar]
- 109.Qie S, Diehl JA. Cyclin D degradation by E3 ligases in cancer progression and treatment. Semin Cancer Biol. 2020;67:159–170. doi: 10.1016/j.semcancer.2020.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen D, Tavana O, Chu B, Erber L, Chen Y, Baer R, Gu W. NRF2 Is a Major Target of ARF in p53-Independent Tumor Suppression. Mol Cell. 2017;68:224–232.e4. doi: 10.1016/j.molcel.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chang ZF, Huang DY, Hu SF. NF-Y-mediated trans-activation of the human thymidine kinase promoter is closely linked to activation of cyclin-dependent kinase. J Cell Biochem. 1999;75:300–309. [PubMed] [Google Scholar]
- 112.LaPak KM, Burd CE. The molecular balancing act of p16(INK4a) in cancer and aging. Mol Cancer Res. 2014;12:167–183. doi: 10.1158/1541-7786.MCR-13-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hayashi Y, Hsiao EC, Sami S, Lancero M, Schlieve CR, Nguyen T, Yano K, Nagahashi A, Ikeya M, Matsumoto Y, Nishimura K, Fukuda A, Hisatake K, Tomoda K, Asaka I, Toguchida J, Conklin BR, Yamanaka S. BMP-SMAD-ID promotes reprogramming to pluripotency by inhibiting p16/INK4A-dependent senescence. Proc Natl Acad Sci U S A. 2016;113:13057–13062. doi: 10.1073/pnas.1603668113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schlitter AM, Segler A, Steiger K, Michalski CW, Jäger C, Konukiewitz B, Pfarr N, Endris V, Bettstetter M, Kong B, Regel I, Kleeff J, Klöppel G, Esposito I. Molecular, morphological and survival analysis of 177 resected pancreatic ductal adenocarcinomas (PDACs): Identification of prognostic subtypes. Sci Rep. 2017;7:41064. doi: 10.1038/srep41064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kneissig M, Bernhard S, Storchova Z. Modelling chromosome structural and copy number changes to understand cancer genomes. Curr Opin Genet Dev. 2019;54:25–32. doi: 10.1016/j.gde.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 116.Jang J, Wang Y, Kim HS, Lalli MA, Kosik KS. Nrf2, a regulator of the proteasome, controls self-renewal and pluripotency in human embryonic stem cells. Stem Cells. 2014;32:2616–2625. doi: 10.1002/stem.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McKenna DJ, Nazarali AJ, Himeno A, Saavedra JM. Chronic treatment with (+/-)DOI, a psychotomimetic 5-HT2 agonist, downregulates 5-HT2 receptors in rat brain. Neuropsychopharmacology. 1989;2:81–87. doi: 10.1016/0893-133x(89)90010-9. [DOI] [PubMed] [Google Scholar]
- 118.Hou P, Ma X, Zhang Q, Wu CJ, Liao W, Li J, Wang H, Zhao J, Zhou X, Guan C, Ackroyd J, Jiang S, Zhang J, Spring DJ, Wang YA, DePinho RA. USP21 deubiquitinase promotes pancreas cancer cell stemness via Wnt pathway activation. Genes Dev. 2019;33:1361–1366. doi: 10.1101/gad.326314.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kumar SK, Callander NS, Hillengass J, Liedtke M, Baljevic M, Campagnaro E, Castillo JJ, Chandler JC, Cornell RF, Costello C, Efebera Y, Faiman M, Garfall A, Godby K, Holmberg L, Htut M, Huff CA, Kang Y, Landgren O, Malek E, Martin T, Omel J, Raje N, Sborov D, Singhal S, Stockerl-Goldstein K, Tan C, Weber D, Johnson-Chilla A, Keller J, Kumar R. NCCN Guidelines Insights: Multiple Myeloma, Version 1.2020. J Natl Compr Canc Netw. 2019;17:1154–1165. doi: 10.6004/jnccn.2019.0049. [DOI] [PubMed] [Google Scholar]
- 120.Alberts SR, Foster NR, Morton RF, Kugler J, Schaefer P, Wiesenfeld M, Fitch TR, Steen P, Kim GP, Gill S. PS-341 and gemcitabine in patients with metastatic pancreatic adenocarcinoma: a North Central Cancer Treatment Group (NCCTG) randomized phase II study. Ann Oncol. 2005;16:1654–1661. doi: 10.1093/annonc/mdi324. [DOI] [PubMed] [Google Scholar]
- 121.Lankes K, Hassan Z, Doffo MJ, Schneeweis C, Lier S, Öllinger R, Rad R, Krämer OH, Keller U, Saur D, Reichert M, Schneider G, Wirth M. Targeting the ubiquitin-proteasome system in a pancreatic cancer subtype with hyperactive MYC. Mol Oncol. 2020;14:3048–3064. doi: 10.1002/1878-0261.12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim EJ, Kim YJ, Lee HI, Jeong SH, Nam HJ, Cho JH. NRF2 Knockdown Resensitizes 5-Fluorouracil-Resistant Pancreatic Cancer Cells by Suppressing HO-1 and ABCG2 Expression. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21134646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Babiker HM, Karass M, Recio-Boiles A, Chandana SR, McBride A, Mahadevan D. Everolimus for the treatment of advanced pancreatic ductal adenocarcinoma (PDAC) Expert Opin Investig Drugs. 2019;28:583–592. doi: 10.1080/13543784.2019.1632289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Royce M, Bachelot T, Villanueva C, Özgüroglu M, Azevedo SJ, Cruz FM, Debled M, Hegg R, Toyama T, Falkson C, Jeong J, Srimuninnimit V, Gradishar WJ, Arce C, Ridolfi A, Lin C, Cardoso F. Everolimus Plus Endocrine Therapy for Postmenopausal Women With Estrogen Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Advanced Breast Cancer: A Clinical Trial. JAMA Oncol. 2018;4:977–984. doi: 10.1001/jamaoncol.2018.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wolpin BM, Hezel AF, Abrams T, Blaszkowsky LS, Meyerhardt JA, Chan JA, Enzinger PC, Allen B, Clark JW, Ryan DP, Fuchs CS. Oral mTOR inhibitor everolimus in patients with gemcitabine-refractory metastatic pancreatic cancer. J Clin Oncol. 2009;27:193–198. doi: 10.1200/JCO.2008.18.9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kordes S, Richel DJ, Klümpen HJ, Weterman MJ, Stevens AJ, Wilmink JW. A phase I/II, non-randomized, feasibility/safety and efficacy study of the combination of everolimus, cetuximab and capecitabine in patients with advanced pancreatic cancer. Invest New Drugs. 2013;31:85–91. doi: 10.1007/s10637-012-9802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Javle MM, Shroff RT, Xiong H, Varadhachary GA, Fogelman D, Reddy SA, Davis D, Zhang Y, Wolff RA, Abbruzzese JL. Inhibition of the mammalian target of rapamycin (mTOR) in advanced pancreatic cancer: results of two phase II studies. BMC Cancer. 2010;10:368. doi: 10.1186/1471-2407-10-368. [DOI] [PMC free article] [PubMed] [Google Scholar]