Summary
Background
Mitochondrial DNA (mtDNA) encodes 37 genes necessary for synthesizing 13 essential subunits of the oxidative phosphorylation system. mtDNA alterations are known to cause mitochondrial disease (MitD), a clinically heterogeneous group of disorders that often present with neuropsychiatric symptoms. Understanding the nature and frequency of mtDNA alterations in health and disease could be a cornerstone in disentangling the relationship between biochemical findings and clinical symptoms of brain disorders. This systematic review aimed to summarize the mtDNA alterations in human brain tissue reported to date that have implications for further research on the pathophysiological significance of mtDNA alterations in brain functioning.
Methods
We searched the PubMed and Embase databases using distinct terms related to postmortem human brain and mtDNA up to June 10, 2021. Reports were eligible if they were empirical studies analysing mtDNA in postmortem human brains.
Findings
A total of 158 of 637 studies fulfilled the inclusion criteria and were clustered into the following groups: MitD (48 entries), neurological diseases (NeuD, 55 entries), psychiatric diseases (PsyD, 15 entries), a miscellaneous group with controls and other clinical diseases (5 entries), ageing (20 entries), and technical issues (5 entries). Ten entries were ascribed to more than one group. Pathogenic single nucleotide variants (pSNVs), both homo- or heteroplasmic variants, have been widely reported in MitD, with heteroplasmy levels varying among brain regions; however, pSNVs are rarer in NeuD, PsyD and ageing. A lower mtDNA copy number (CN) in disease was described in most, but not all, of the identified studies. mtDNA deletions were identified in individuals in the four clinical categories and ageing. Notably, brain samples showed significantly more mtDNA deletions and at higher heteroplasmy percentages than blood samples, and several of the deletions present in the brain were not detected in the blood. Finally, mtDNA heteroplasmy, mtDNA CN and the deletion levels varied depending on the brain region studied.
Interpretation
mtDNA alterations are well known to affect human tissues, including the brain. In general, we found that studies of MitD, NeuD, PsyD, and ageing were highly variable in terms of the type of disease or ageing process investigated, number of screened individuals, studied brain regions and technology used. In NeuD and PsyD, no particular type of mtDNA alteration could be unequivocally assigned to any specific disease or diagnostic group. However, the presence of mtDNA deletions and mtDNA CN variation imply a role for mtDNA in NeuD and PsyD. Heteroplasmy levels and threshold effects, affected brain regions, and mitotic segregation patterns of mtDNA alterations may be involved in the complex inheritance of NeuD and PsyD and in the ageing process. Therefore, more information is needed regarding the type of mtDNA alteration, the affected brain regions, the heteroplasmy levels, and their relationship with clinical phenotypes and the ageing process.
Funding
Hospital Universitari Institut Pere Mata; Institut d'Investigació Sanitària Pere Virgili; Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación (PI18/00514).
Keywords: Mitochondrial DNA, Mitochondrial diseases, Neurological diseases, Psychiatric diseases, Ageing, Postmortem
Abbreviations: mtDNA, Mitochondrial DNA; MitD, Mitochondrial disease/s; NeuD, Neurological disease/s; PsyD, Psychiatric disease/s; pSNV, Pathogenic single nucleotide variant; CN, copy number; DEL, deletion
Research in context.
Evidence before this study
The human mitochondrial genome consists of a 16,569 bp molecule present in almost all cell types with some exceptions, the most significant being erythrocytes. On average, there are approximately 1,000 mtDNA molecules in a human cell, but the specific mtDNA CN depends on the energy requirements of each cell. Brain tissue has a high energy requirement, which leads to a large number of mitochondria in the brain.1 It is known that alterations in mtDNA cause MitD, a clinically heterogeneous group of disorders that arise as a consequence of mitochondrial respiratory chain dysfunction. The clinical characteristics of MitD show enormous variability, and although they can affect a single organ, most of them involve multiple organ systems often presenting with neurological disturbances. Moreover, there is increasing evidence of mitochondrial dysfunction in neurodegeneration,2 in the development of psychiatric symptoms,3 and in the brain ageing process.4,5 The aims of this study were a) to determine the specific diagnoses or health conditions in which the presence of mtDNA alterations has been assessed in the human postmortem brain and b) to identify and summarize the specific mtDNA defects reported. For these purposes, we conducted a PubMed and Embase search for articles published before June 10, 2021, using several search strings and keywords, including postmortem, brain, neuron, glia, mtDNA, variant, mutation, and deletion.
Added value of this study
Our systematic review identified that mtDNA alterations have been investigated in human postmortem brain samples associated with ageing and disease, mostly in individuals with MitD, neurological, or psychiatric diagnoses. We report a comprehensive summary of the identified mtDNA alterations organized into three clinical categories (MitD, NeuD and PsyD) plus a miscellaneous clinical group and two further categories, including ageing and technical issues. pSNVs, alterations in mtDNA CN and mtDNA deletions have been reported in MitD. In NeuD, most of the studies investigated the presence of mtDNA deletions or differences in mtDNA CN between affected and nonaffected individuals, with conflicting results, while few studies evaluated mtDNA pSNVs. Similarly, pSNVs and altered mtDNA CNs were not consistently evaluated in PsyD; in contrast, several studies identified mtDNA deletions in some patients. Finally, mtDNA deletions have been recurrently associated with ageing. We also identified some studies that have explored mtDNA gene expression, oxidation and methylation.
Implications of all the available evidence
Among the three types of well-known mtDNA alterations (pSNVs, mtDNA CN and mtDNA rearrangements) that are associated with mitochondrial dysfunction, only one or two of them were investigated in most studies. Most studies reported mtDNA alterations, demonstrating their presence in the postmortem brain of patients with MitD, NeuD and PsyD and the ageing process. With the currently available molecular techniques and bioinformatic tools, it is crucial to further investigate the presence of all types of mtDNA alterations in postmortem brain samples of patients with MitD, NeuD and PsyD in all age groups and in healthy individuals to shed light on the role of mtDNA in brain function, disease development and the ageing process. These studies should be conducted with current validated techniques to obtain unambiguous data regarding mtDNA alterations and associated heteroplasmy levels and are particularly relevant when measuring mtDNA CN. Because mitochondria can be acknowledged as a therapeutic target for ameliorating brain function, it is crucial to decipher the role of all types of mtDNA variations in health and disease.
Alt-text: Unlabelled box
Introduction
Mitochondrial DNA (mtDNA)
Mitochondria are membrane-bound organelles that generate most of the chemical energy needed to power the cell's biochemical reactions; this energy is stored as adenosine triphosphate (ATP) molecules. Two distinct bilayer membranes separate the matrix of the mitochondria from the cytosol—the smooth outer membrane and the highly folded inner membrane—forming invaginated structures called cristae. In these cristae, ATP synthesis takes place by the oxidative phosphorylation system (OXPHOS) through oxidoreductase complexes I-IV of the electron transport chain (ETC) and the ATP synthase enzyme of complex V. Mitochondria act as a signalling hub regulating cellular processes relevant to cell differentiation, cell proliferation, apoptosis and the immune response.6, 7, 8, 9, 10 The mitochondrial proteome is estimated to contain 1,500 proteins, most of which are under nuclear genome control.
Human mtDNA consists of a 16,569 bp circular DNA molecule that is maternally inherited and whose sequence and gene organization were published in 1981.11,12 It encodes 37 genes, including 13 protein subunits of the respiratory chain, 2 ribosomal units (rRNAs 12S and 16S) and 22 transfer (tRNA) genes. mtDNA also contains a noncoding control region of approximately 1,200 bp in length and is known as the displacement loop (D-loop) region, which regulates mtDNA transcription and replication. The 13 resulting proteins are crucial for the proper function of the respiratory chain even though they represent only a small fraction of the ∼100 subunits that constitute complexes I-V.13 Another peculiar feature of mtDNA is polyploidy. A uniform collection of mtDNA copies—either completely normal mtDNA or completely mutant mtDNA—is known as homoplasmy, while heteroplasmy refers to different proportions of normal and mutant mtDNA in a mitochondrion, cell, organ or tissue. The amount of mtDNA in a cell, known as the mtDNA content or mtDNA CN, usually varies from hundreds to thousands14 and depends on the cell function and the cell response to endogenous and exogenous agents.15 Tissues with high energy requirements contain large amounts of mitochondria in their cells and, accordingly, a high mtDNA CN. The central nervous system, cardiac and skeletal muscles, endocrine system, and liver and renal systems are among those with the highest energy requirements.10
mtDNA is composed of double-stranded DNA, comprising heavy strands (H) and light strands (L). The H-strand, which is guanine-rich, encodes 28 genes, while the L-strand, which is cytosine-rich, encodes the remaining 9 genes. Several characteristics differ between the nuclear and mitochondrial genomes: mtDNA is circular, small, has no introns, is not enveloped with proteins and is maternally inherited; in contrast, nuclear DNA (nDNA) is linear, has a large number of nucleotides (∼3.3 billion bp), has introns, is packaged into chromatin, undergoes recombination and is biparentally inherited. Both mtDNA and nDNA use the same deoxynucleotide triphosphates (dNTPs) for DNA replication, and although mtDNA follows the universal codon usage rules when coding sequences are translated into proteins, there are some specific deviations: UGA codes for tryptophan instead of a stop codon, AGA and AGG are also stop codons, and AUA codes for methionine. Additionally, some nucleotide bases exhibit functional overlap between two genes, as they are the last base of one gene and the first base of the next gene.16 In the mitochondrial matrix, mtDNA forms nucleoids with mitochondrial transcription factor A, which acts to provide structure to the mtDNA genome.
Finally, the mtDNA mutation rate (the speed at which mutations are introduced) is much higher than that of nDNA. In animals, it is estimated that the mutation rate in mtDNA is ∼25-fold higher than that in nDNA.17 In humans, based on the appearance of de novo mtDNA variants in human pedigree studies, an ∼10-fold higher rate in mtDNA than in nDNA has been suggested.18 The molecular damage to mtDNA is thought to be due to the high levels of reactive oxygen species present in the mitochondrion, the high mtDNA replication levels, and the high coding rate of mtDNA, which is ∼93%.19 Additionally, this higher mutation rate suggests that many mtDNA variants may be subjected to poor selection, which can occur at the germline level or at the somatic level throughout life, implying that even though a specific variant is not detected in blood, a tissue commonly used in genetic testing, it cannot be ruled out that the variant is not present in other tissues or organs. Correspondingly, mtDNA alterations can lead to cellular energy impairment that might cause a disease or be implicated in the physiopathology of age-associated diseases or the ageing process itself.20
mtDNA and human ageing
The decline of mitochondrial functioning has been largely implicated in the ageing process and is characterized by a reduced density of mitochondria and reduced mitogenesis.21 In fact, the ageing process is strongly linked to noninherited mtDNA changes, mainly point variants and large deletions, that increase in frequency with age.22, 23, 24 Such changes, which originate as replication errors, accumulate in postmitotic tissues during ageing, leading to increased proportions of impaired mitochondria that may differ between cells and tissues.25 In the ageing brain, dysfunctional synaptic mitochondria leading to impaired neurotransmission and cognitive failure26 have been amply demonstrated,27,28 and mtDNA deletions correlate with mitochondrial respiratory chain malfunction.27,28 Thus, elucidating the temporal and spatial distribution of mutated mtDNA in the brain might resolve important questions regarding the importance of mtDNA changes in the ageing process.
mtDNA and disease
Given the nature of the genetic material and the dual genomic control of mitochondria, alterations occurring either in the nDNA or mtDNA sequence can potentially cause mitochondrial functional defects. Indeed, some are known to cause very heterogeneous diseases that together are called primary mitochondrial disease (MitD). More than 300 nuclear genes are known to cause MitD, although most adult patients exhibit variants in mtDNA. The mechanisms involved in these nuclear genes involve assembly factors, mitochondrial structure, coenzyme Q biosynthesis, protein synthesis, and mtDNA maintenance.29,30 However, this review focuses on MitDs associated with mtDNA pathogenic variants that include pathogenic single nucleotide variants (pSNVs), mtDNA rearrangements (mostly deletions), and altered mtDNA CNs. They can be either maternally inherited or occur de novo, and their pathogenic role can be established by taking advantage of the association between each variant and a specific phenotype.31 This was investigated by a recent analysis of 265 mtDNA SNVs in 483,626 individuals from the United Kingdom (UK) biobank, which has allowed the identification of 260 new mtDNA-phenotype associations, including type 2 diabetes, multiple sclerosis, adult height, and liver and renal biomarkers. Notably, this study identified a key role for mtDNA common and rare SNV variation (only homoplasmic) in many quantitative human traits and disease risks, with a particular emphasis on cardiometabolic and neurodegenerative diseases.32 mtDNA pSNVs can be observed in homoplasmy or heteroplasmy, while mtDNA deletions are always heteroplasmic, since within the deleted region, mtDNA consistently contain one or more tRNAs indispensable for the translation of the protein-associated mtDNA genes, which are essential for life. Known mtDNA pSNVs lead to syndromes such as mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes (MELAS), myoclonic epilepsy with ragged red fibres (MERRF), neuropathy, ataxia and retinitis pigmentosa (NARP), and Leigh syndrome (LS).33, 34, 35, 36 mtDNA rearrangements are responsible for Kearns–Sayre syndrome (KSS), progressive external ophthalmoplegia (PEO), and Pearson's syndrome.37 These are considered the most typical MitDs associated with mtDNA alterations; however, there are many other diseases and human characteristics related to mtDNA variation, such as diabetes and hypertension, as well as ageing.31 In fact, it has been estimated that 1 out of 3,500–6,000 individuals are affected by or are at risk of developing a MitD.38,39 Human phenotypes associated with mtDNA alterations include extremely severe diseases that can be present from infancy to adulthood and can affect a single organ or multiple tissues, and most of them are included as rare diseases (ORPHANET, https://www.orpha.net/). In June 2021, the Online Mendelian Inheritance in Man (https://omim.org) catalogue40 included 33 phenotypic descriptions (Supplementary Table 1) in which the molecular basis is known to be associated with mtDNA alterations. The catalogue also included the 37 mtDNA genes (Supplementary Table 2) containing several allelic variants associated with human conditions. In addition, the human mitochondrial genome database (https://www.mitomap.org41) includes a large and growing number of variants, some of which are related to a wider constellation of phenotypes. Among the reported mtDNA base substitution disease variants, 431 entries are located in rRNA or tRNA genes, and 481 are located in coding regions or the noncoding (D-loop) control region. These include 52 rRNA/tRNA and 43 coding/D-loop variants that have been confirmed as pathogenic (on March 2021).41, 42, 43, 44 However, it is worth mentioning that most of the variants seem to have no effect and have been widely used as haplotype markers in evolutionary anthropology and population history, genetic genealogy, and forensic science in addition to medical genetics.45 Moreover, a group of phenotypes known as mtDNA maintenance defects or mitochondrial depletion syndromes must be noted; these are characterized by mtDNA depletion and/or the presence of multiple mtDNA deletions, resulting in inadequate energy production.46 These mtDNA defects are caused by pathogenic variants located in one of the 20 nuclear-encoded genes that are involved in mtDNA maintenance.47, 48, 49 The involvement of nuclear gene defects causing mitochondrial depletion syndromes is beyond the scope of this review and is discussed elsewhere.50,51 Another aspect that must be considered is that an altered mtDNA CN is associated with mitochondrial function and dysfunction, and consequently, several conditions have been associated with either increases or decreases in the mtDNA content.52
MitD refers to a heterogeneous group of phenotypes; the common clinical features include ptosis, external ophthalmoplegia, proximal myopathy and exercise intolerance, cardiomyopathy, sensorineural deafness, optic atrophy, pigmentary retinopathy, and diabetes mellitus. Regarding the central nervous system, phenotypes include fluctuating encephalopathy, seizures, dementia, migraine, stroke-like episodes, ataxia, and spasticity. Some MitD types affect only a single organ, while many others involve multiple organ systems,36,37 leading to clinical heterogeneity, a hallmark of MitD. The organs/tissues most often affected in MitD are the brain and skeletal muscle, but the heart, liver, peripheral nerves, gastrointestinal tract and endocrine system can also be involved. Therefore, it is relevant to identify the mtDNA defects present in the brain, their nature and prevalence, and the correlation between the genetic defects and the postmortem neuropathologic features to advance our understanding of the underlying mechanisms of mitochondrial function in disease. Within this systematic review, we aim to summarize the main mtDNA alterations reported in postmortem human brain samples and the related phenotypes.
Methods
Search strategy and selection criteria
We examined PubMed and Embase for English language articles published from inception to June 10, 2021 using two search strings combining the following keywords: postmortem/post-mortem, brain, mitochondrial DNA/mtDNA, mutation, variant, deletion, neuron and glia, according to the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines.53 The specific search strings in both databases are shown in Figure 1. We examined the retrieved titles and abstracts and selected empirical studies based on the eligibility criteria. The literature search strategy, data collection, data extraction and appraisal were conducted independently by three authors (AV-P, JT and BKB). When there was no agreement, a fourth author (LM) contributed to gain consensus. The abstraction and summary of the main results of the studies were first performed independently by one of the three main authors and revised by the remaining authors.
Figure 1.
PRISMA flow diagram for selecting published articles for review. The two search strategies used, the number of articles with PMID numbers obtained for each of them, and the result of combining them are shown. Those that met the inclusion criteria and those that were excluded and the groups to which they were assigned are indicated. The final number of articles included in each group after manual inclusion of references is indicated in brackets.
Inclusion and exclusion criteria. Studies were included only if they reported results of mtDNA analyses in postmortem human brain tissue and were written in English. The exclusion criteria were as follows: i) studies that did not report the results of mtDNA analyses in postmortem human brain tissue, ii) cell models, iii) animal models; iv) review studies; v) studies focused on brain tumours or cancer, vi) studies that reported duplicated data; or vii) forensic studies. No other restrictions were applied.
Review process
The combined search yielded 637 potentially eligible studies. Abstracts or full articles (if the eligibility criteria were not clearly stated in the abstract) were screened to decipher eligibility. Figure 1 shows the PRISMA flow chart depicting the specific information at the different stages of the systematic review, and Supplementary Table 3 provides the PRISMA checklist of items to include when reporting a systematic review.53 We discarded 479 reports, and a detailed examination was performed on the 158 remaining records and on others obtained from hand-searching references.
Data extraction
From eligible articles, we recorded the name of the first author, PMID number, publication year, number of patients and controls analysed, age, sex, disease or condition, brain region, technique used, main results regarding mtDNA variants, mtDNA CN and/or mtDNA rearrangements reported, and additional information.
Pathogenicity assignment of mtDNA variants
Pathogenicity status was collected based on the information present in the Mitomap and ClinVar databases when available.
Role of the funding source
The funders had no role in the study design, in the collection and interpretation of data, in the report writing, or in the decision to submit the manuscript for publication.
Results
Characteristics of the examined studies
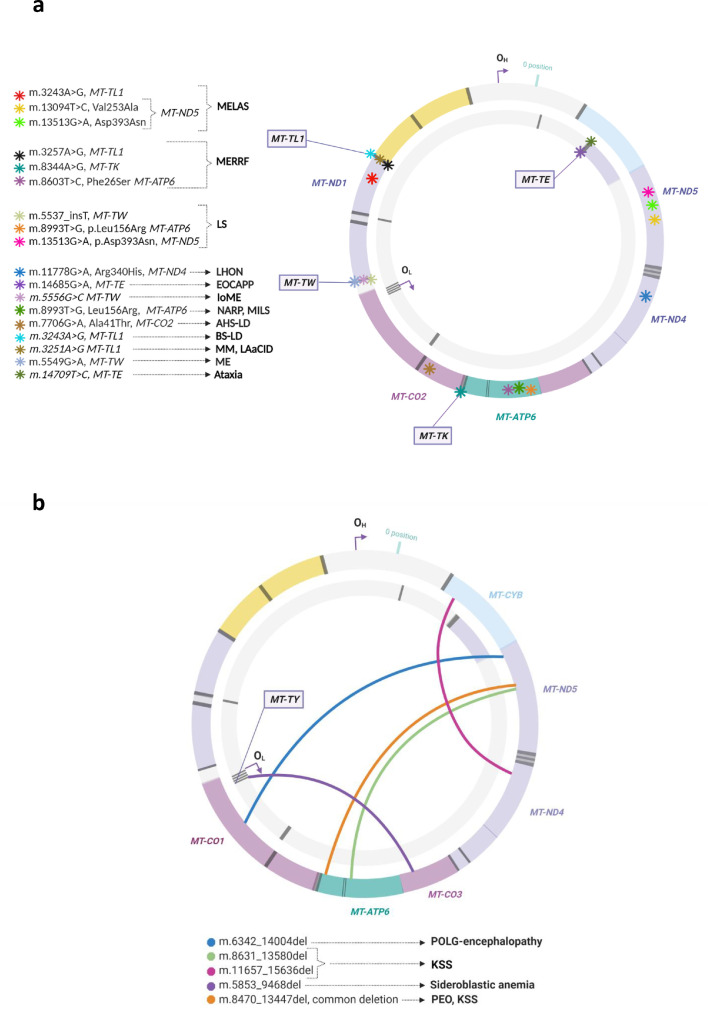
The manuscripts fulfilling the inclusion criteria could be ascribed to the following 6 categories: 1) mitochondrial diseases (MitD), 2) neurological diseases (NeuD), 3) psychiatric diseases (PsyD), 4) other clinical conditions included in a miscellaneous group, 5) ageing, and 6) technical issues. Some reports could be ascribed to two groups, as they presented results from more than one category, and others were manually included after reading specific references of the included studies. A summary of the relevant data, number of evaluated subjects, age, sex, brain region/s studied, techniques used, and mtDNA alterations reported are presented in Table 1, Table 2, Table 3, Table 4, Table 5, while the technical issues are summarized at the end of this section. We screened the variants for putative pathogenic characteristics, and a selection of presumably pathogenic mtDNA variants is shown in Table 6, with pathogenicity information obtained from public databases.
Table 1.
Studies reporting the results of mtDNA analyses in postmortem brain samples of patients with mitochondrial diseases (MitD).
| Study reference (PMID) |
Patient/control characteristics | Disease (nuclear gene) |
Brain region | Technique | mtDNA alteration in brain | Additional information | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| N P/C |
Age (y) in P/C |
Sex in P/C |
Variant | mtDNA CN | Rearrangements | |||||
| Laine-Menéndez et al. 2021 (34070501) |
1/1 | 43/37 | F/F | MM (TK2, c.323C>T, p.Thr108Met) |
FCtx, TCtx, OCtx, Hi, Amg, Th, Hy, Cb, pons, spinal |
qPCR | NR | No differences in mtDNA CN between P and C in brain tissue | NR | Low mtDNA content was observed in skeletal muscle, liver, kidney, small intestine, and particularly in the diaphragm. Heart and brain tissue did not show differences. mtDNA deletions were observed in skeletal muscle and diaphragm |
| Scholle et al. 2020 (32085658) |
1 | 46 | F | Optic neuropathy, sensorineural hearing loss and diabetes mellitus type I. Not MELAS | Pu, Cd, Mb, Th, Hy, Gic, GP, visual Ctx, pons, medulla oblongata, Ver, spinal, WM | RFLP, qPCR |
m.3243A>G, MT-TL1 Mutation load: >75% |
Higher heteroplasmy levels significantly correlated with lower mtDNA CN | NR | 60% level of heteroplasmy in the biceps brachii muscle |
| Geffroy et al. 2018 (29454073) |
1 | 32 | F | MELAS | NA | Fluorescence RFLP | m.3243A>G, MT-TL1 Mutation load: 92.1% |
NR | NR | - |
| Gramegna et al. 2018 (29348134) |
1/3 | 36/ Age-matched |
1/NA | MNGIE (TYMP, c.457G>A, p.G153S) |
FCtx, SN | NA | NR | mtDNA depletion in frontal GM and WM, and SN | NR | Severe mtDNA depletion in the P, also in smooth muscle and endothelial cells |
| Lax et al. 2016 (25786813) |
5 | 45 | 4M, 6F | MELAS MERRF |
FCtx, TCtx, OCtx | Pyrosequencing | MELAS, m.3243A>G, MT-TL1 MERRF, m.8344A>G, MT-TK Mutation load range (%): MELAS 79–94; MERRF 87–93 |
NR | NR | Mutation load did not vary between brain regions |
| Tzoulis et al. 2014 (24841123) |
8/15 | Infantile and adult/ age-matched C |
NA | ME (POLG) |
SN, SNC | qPCR, Long-range PCR, NGS |
The overall burden of mtDNA point mutations present at a frequency >0.2% was higher in P than in C in MT-HV2 and MT-CO3 and associated with disease duration | In homogenate tissue, apparent 20–30% mtDNA depletion was present in infantile P. Microdissected SN neurons showed a 40% lower mtDNA CN than the neurons of age-matched C | Neurons of both P and C harbored mtDNA DELs in the homogenate. In microdissected neurons mtDNA DELs were more prominent in P with longer disease duration | mtDNA analyses conducted in laser microdissected neurons and homogenate tissue |
| Giordano et al. 2014 (24369379) |
4/8 | 71/68 | NA | LHON | Optic nerve | NA | m.11778G>A, p.Arg340His, MT-ND1, present in affected P and unaffected carriers | Unaffected female carrier had a higher mtDNA CN than two C | NR | Unaffected mutation carriers showed higher mitochondrial DNA CN than their affected relatives and C |
| Lax et al. 2013 (23334599) |
1 | 45 | F | Early-onset cataracts, ataxia and progressive paraparesis | Optic nerve, basal ganglia, Cb, medulla oblongata, pons |
Sequencing, pyrosequencing |
m.14685G>A, MT-TE. Mutation load range (%): medulla oblongata 82, basal ganglia 68, Cb 58, pons 51, optic nerve 44 |
NR | NR | - |
| Tzoulis et al. 2013 (23625061) |
2/4 | 34/58 | NA | ME (POLG) |
Mesencephalon pons, Th, Str | Long-range PCR, nested PCR, qPCR |
NR | P neurons contained 50–60% lower mtDNA CNs than age-matched C neurons | DELs were detectable in P and C, and appeared to be more prominent in P | - |
| Lax et al. 2012 (22491194) |
3/1 | 53/- | M/- | KSS and ME (POLG, p.A467T, p.X1240Q; POLG, p.G848S, p.W748S) |
Dt WM and GM | Long-range PCR, sequencing, qPCR | NR | NR | The P with KSS showed the single m.11657_15636del (3978 bp) and heteroplasmy levels in WM were higher than the 60% threshold, thus considered pathogenic. In this P, heteroplasmy levels in WM where higher than in GM. The P with POLG mutations showed multiple mtDNA DELs, and heteroplasmy levels were lower than in the P with KSS |
mtDNA analyses conducted in laser microdissected regions. The P with KSS showed decreased immunoreactivity of complex I and COX compared with complex II |
| Lax et al. 2012 (22249460) |
14/- | 42/- | 5M/9F | MELAS (7), MERRF (1), MELAS/LS (1), KSS (1), Ataxia-retinopathy, arPEO (3) | Dt, CbCtx, IO | qPCR, pyrosequencing | m.3243A>G; m.8344A>G; m.14709T>C; m.13094T>C. Mutation load range %: IO 44–93, CbCtx 47–95, Dt 51–95 | NR | Single and multiple large-scale mtDNA DELs. Mutation load %: IO 68, CbCtx 20–68, Dt 25–44 | Neuronal cell loss occurred independently of the level of mutated mtDNA present within surviving neurons |
| Brinckmann et al. 2010 (20976001) |
1/4 | 16/16-32 | F/NA | MERRF | Mcer, Ox, Mb, Th, Cd, GP, Pu, Ic, Hy, Hi, sensomotor Ctx, visual Ctx, WM, Chpx, Ver, folia of Cb, medulla of Cb, pons, SN | qPCR, pyrosequencing | m.8344A>G, MT-TK. Mutation load (%) Mcer 98, Ox 97, Mb 94, Th 89, Cd 95, GP 94, Pu 95, Ic 95, Hy 94, Hi 94, sensomotor Ctx 96, visual Ctx 96, white matter 95, Chpx 100, Ver 97, folia of Cb 97, medulla of Cb 93, pons 95, SN 91 |
mtDNA CN increased between 3–7-fold in affected brain tissues compared to nonaffected tissues | NR | Mutation load (%): Skeletal muscles 97–98, diaphragm 97, external ocular muscles 95, heart 93, renal cortex 99, renal medulla 95, adrenal cortex 100, lung 98, liver 67, pancreas 73, spleen 98, stomach 98, ileum 97, bladder 100, uterus 100, ovary 100, adipose tissue 99, skin 100 |
| Sanaker et al. 2010 (19744136) |
1 | 64 | M | loME | OCtx, TCtx, CbCtx | Sequencing, PCR-RFLP | m.5556G>C, MT-TW. Mutation load (%): Octx 34, TCtx 32, CbCtx 12 |
NR | NR | Mutation load (%): skeletal muscle 70, heart muscle 27, kidney 26, liver 27, adrenal cortex 5, pancreas 8, lung 2, spleen 4 |
| Zsurka et al. 2008 (18716558) |
1/30 | 17/28 | M/NA | ME (POLG) |
NA | Long-range PCR, qPCR |
NR | Progressive decrease of mtDNA CN in the disease course in P but not significantly different from that in C | Presence of the m.6342_14004del (7662 bp) | - |
| Götz et al. 2008 (18819985) |
2/2 | NA | NA | MDS (Patient 1: TK2, c.739C>T, p.R172W Patient 2: TK2, c.898C>T, p.R225W) |
Ctx, Cb, Basal ganglia | qPCR | NR | Severe brain mtDNA depletion in patients with R172W but not with R225W mutation. Higher mtDNA content in the Cb of the patient with R2 25W mutation |
NR | mtDNA depletion present in muscle and liver in all patients |
| Rojo et al. 2006 (16525806) |
1 | 64/- | M | NARP-MILS | Pu, brain stem, Th, Ctx | Southern blot, PCR-RFLP |
m.8993T>G, p.Leu156Arg, MT-ATP6 Mutation load (%): brain stem and Ctx 89, Pu and Cb 90, Th 91 |
NR | NR | Mutation load (%): blood 75, muscle 87 |
| Betts et al. 2006 (16866982) |
2 | 47 | F | MELAS | Chpx, Hi, CbCtx, Dt, GPI, OCtx, FCtx, GPI |
PCR-RFLP using radiolabeled nucleotides |
MELAS, m.3243A>G, MT-TL1 Mutation load in patient 1 and 2, respectively (%): chpx 81 and NA, Hi 79 and 92, CbCtx 54 and 96, Dt 75 and NA, GPI 70 and 93, Octx 69 and 93, FCtx NA and 92 |
NR | NR | Mutation load was higher in COX-deficient than in COX-positive microdissected cells from Hi, chpx and skeletal muscle. Mutation load varied considerably between different cells of the same region |
| Matthes et al. 2006 (16856911) |
1 | 19 | M | Sideroblastic anemia | NA | Long-range PCR, sequencing, qPCR | NR | NR | Presence of the m.5853_9468del (3614 bp). Mutation load (%): 70 |
Mutation load (%): Liver 90, skeletal muscle 75, pancreas 85, peripheral blood 80 |
| Ferrari et al. 2005 (15689359) |
1/2 | 19/age-matched | M/- | AHS (POLG1, c.1399G>A, p.A467T) |
NA | NA | NR | 30% mtDNA content reduction | NR | - |
| Pistilli et al. 2003 (14608542) |
1 | 36/- | F | KSS | FCtx, PCtx, TCtx, OCtx, Cb, basal ganglia, brain stem | Sequencing, Southern blot, competitive PCR using radiolabeled nucleotides | NR | NR | Presence of the m.8631_13580del (4949 bp). DEL load (%): FCtx 54; Octx 54; TCtx 58; PCtx 59; basal ganglia 75; Cb 88 |
- |
| Uusimaa et al. 2003 (12612282) |
1 | 7/- | F | AHS-like disease | NA | CSGE + sequencing, PCR-RFLP using radiolabeled nucleotides |
m.7706G>A, p.Ala41Thr, MT-CO2. Mutation load: 91% |
NR | NR | Mutation load (%): blood 87, heart 87 |
| Kirby et al. 2003 (14520659) |
3 | 31 | 2M, 1F | LS | Medulla oblongata, pons, Cb, basal ganglia, Th, midbrain, Hi, FCtx, Chpx | PCR-RFLP with radiolabeled nucleotides | m.13513G>A, p.Asp393Asn, MT-ND5 Mutation load (%): 73 |
NR | NR | - |
| Jiang et al. 2002 (12174968) |
1 | - | - | LS | NA | NA | m.8993T>G, Leu156Arg, MT-ATP6 Mutation load (%): 89 |
NR | NR | Mutation load (%): muscles 85, lymphocytes 72 |
| De Kremer et al. 2001 (11241464) |
1/20 | 4/NA | 1/NA | Barth syndrome-like disorder | NA | PCR-RFLP sequencing |
m.3243A>G, MT-TL1. The mutation was heteroplasmic in brain but not quantified |
NR | NR | The mutation was also heteroplasmic in all the tissues analyzed: blood, skeletal muscle, cardiac muscle, and liver |
| Moslemi et al. 1999 (10408540) |
1 | 61 | F | PEO | WM, FCtx, Th, Pu Cd, SN, CbCtx | Long-range PCR, Southern blot | NR | NR | Multiple DELs (25), most of them were less than 8 kb. The CbCtx showed the lowest mutation load %. The common DEL was found in all the specimens | DELs present in all brain regions, skeletal muscle samples and myocardium |
| Nagashima et al. 1999 (10208283) |
1 | 43 | F | LS | FCtx | PCR-RFLP | m.8993T>G, p.Leu156Arg, MT-ATP6. Mutation load (%): 95 |
NR | NR | Mutation load (%): liver 98, muscle 90, heart 91, kidney 99, pancreas 93 |
| Santorelli et al. 1998 (9851442) |
1 | 12 | F | MERRF | NA | Sequencing with radiolabeled nucleotides | m.8344A>G, MT-TK, mutation load (%): 92%. m.8603T>C, Phe26Ser, MT-ATP6, m.3257A>G MT-TL1 |
NR | NR | - |
| Di Trapani et al. 1997 (9266144) |
1 | 27/- | M | MELAS | NA | PCR-RFLP | m.3243A>G, MT-TL1. Mutation load: 84% |
NR | NR | Mutation load (%): liver 79, kidney 86, skeletal muscle 83, cardiac muscle 83 |
| Zhou et al. 1997 (9315896) |
1 | 14 | F | MERRF | CbCtx, Dt, IO, FLV | PCR-RFLP | m.8344A>G, MT-TK. Mutation load (%): between 81 and 98. Similar % in soma, neuropil, glia and homogenate tissue. |
NR | NR | Analyses of homogenate issue and individual neurons |
| Suomalainen et al. 1997 (9153451) |
2 | 67 | F | adPEO | FCtx, TCtx, Cd, Cb | Southern blot | NR | NR | Multiple DELs ranging from 0.5 kb to 10.0 kb DEL load (%): FCtx 50, TCtx 40, Cd 60, Cb 10 |
Mutation load (%): Skeletal muscle 50, kidney 10, liver 10 |
| Santorelli et al. 1997a (9299505) |
1 | 45 | M | MELAS | NA | PCR-RFLP | m.13513G>A, p.Asp393Asn, MT-ND5. Mutation load (%): 73 |
NR | NR | Mutation load (%): muscle 68 |
| Santorelli et al. 1997b (9266739) |
1 | 1.4 | M | MM, LS | Basal ganglia, SN, brain stem | Southern blot PCR-RFLP sequencing |
m.5537_insT MT-TW. Mutation load (%): ≥98 |
NR | NR | Mutation load (%) in other tissues: >95 |
| Kaido et al. 1996 (8870835) |
1 | 53/- | F | MELAS | FCtx, Pu, GP | Southern blot | m.3243A>G, MT-TL1. Mutation load (%): FCtx 89, Pu 86, GP 80 |
NR | NR | - |
| Sanger et al. 1996 (8652018) |
1 | 14/- | F | MERRF | CbCtx, FCtx, Cd, Pu | PCR-RFLP using radiolabeled-nucleotides |
m.8344A>G, MT-TK. Mutation load (%): CbCtx 97, FCtx 88, Cd 88, Pu 88 |
NR | NR | Mutation load (%): blood 75, muscle 86–93, spinal cord 83, medulla 87 |
| Melberg et al. 1996 (8937533) |
1 | 20 | M | MELAS-like syndrome | FCtx | Southern blot for variants m.3243A>G and m.8344A>G | No presence of these two variants | NR | NR | - |
| Houshmand et al. 1996 (8786060) |
1 | 14 | F | MM, lactic acidosis and complex I deficiency | NA | PCR-RFLP | m.3251A>G MT-TL. Mutation load (%): 90 |
NR | NR | Mutation load (%): muscle 94, fibroblast 93, heart 79, liver 80 |
| Oldfors et al. 1995 (8525809) |
1 | 18/- | M | MERRF | FCtx, PCtx, TCtx, OCtx, frontal WM, Pu, pallidum, Th, pons, IO, CbCtx, Dt | PCR-RFLP using radiolabeled-primer | m.8344A>G, MT-TK. Mutation load (%): FCtx 95, PCtx 95, TCtx 95, Octx 95, Pu 96, pallidum 96, Th 94, pons 94, IO 94, low. Brain stem 94, CbCtx 95, Dt 93 |
NR | NR | Mutation load (%): skeletal muscle 97, myocardium 97, aorta 97, subcutaneous adipose tissue 95, liver 95, pancreas 91, spleen 95, lymph node 93, bone marrow 94, testis 99, adrenal gland 97, thyroid gland 98 |
| Nelson et al. 1995 (7695240) |
1 | 53/- | M | ME | Ctx, Cb | Southern blot | m.5549G>A, MT-TW Mutation load (%): Ctx 87, Cb 88 |
NR | NR | Mutation load (%): blood 40, muscle 83–86, myocardium 93, kidney 93, lung 79, liver 77, optic nerve 51 |
| Brockington et al. 1995 (7561952) |
1 | 41 | M | KSS | TCtx, OCtx, Cb | Southern blot | NR | NR | Presence of the common DEL. DEL load (%): TCtx 62, OCtx 70, Cb 17 |
Mutation load (%): quadriceps 91, psoas 72, diaphragm 75, cardiac muscle 25, kidney 21, liver 84, lung 18, spleen 0, testis 0, blood 0 |
| Sweeney et al. 1994 (8133313) |
1 | 18 | M | LS | Cb, CbCtx | PCR | m.8993T>G, p.Leu156Arg, MT-ATP6 Mutation load (%): Cb 97, CbCtx 97 |
NR | NR | Mutation load (%): blood 81, quadriceps muscle 99, extraocular muscle 97, cardiac muscle 97, liver 99, kidney 98, blood 72 |
| MacMillan et al. 1993 (8351017) |
2 | 24 | F | MELAS | PCtx GM, Octx GM, PCtx WM, Cd, Cb, pons, | PCR-RFLP | m.3243A>G, MT-TL1. Mutation load (%): PCtx GM 79, Octx GM 80, PCtx WM 62, Cd 73, Cb 72, pons 70 |
NR | NR | Mutation load (%): psoas muscle 82, oculomotor muscle 59, cardiac atrium 86, myocardium 67, oesophagus 84 |
| Love et al. 1993 (8326463) |
2 | 23, 16 | F | MELAS | FCtx, TCtx, pons, PCtx, OCtx | PCR-RFLP, sequencing with radiolabeled primers | Case 1: m.3243A>G, MT-TL1. Mutation load (%): FCtx 46, TCtx 40, PCtx 33, pons 30. Not detected in OCtx. Case 2: neither m.3243A>G nor m.3271T>C, MT-TL1, was present. |
NR | NR | Mutation load (%) in case 1: liver 78, thenar muscle 77, myocardium 73. Poor correlation between mutation load and distribution of histological lesions |
| Tanno et al. 1993 (8170566) |
2 | 29, 30 | M, F | MERRF | FCtx, TCtx, CbCtx | PCR-RFLP | 8344A>G, MT-TK. Mutation load (%): FCtx 97, TCtx 98, CbCtx 97 |
NR | NR | Mutation load (%): heart 96, kidney 96, adrenal gland 93, liver 94, muscle 96–99, leukocytes 93 |
| Shiraiwa et al. 1993 (8138807) |
1 | 27 | F | MELAS | FCtx, TCtx, PCtx, OCtx, Cd, Pu, pallidum, Th, frontal WM, Pit, CbCtx, Dt | PCR | m.3243A>G, MT-TL1. Mutation load (%): Pit 95; FCtx, TCtx, PCtx and Octx 85–88; Cd, Pu, pallidum and WM 73–79 |
NR | NR | - |
| Tatuch et al. 1992 (1550128) |
1 | 0.6 | F | LS | NA | Southern blot PCR-RFLP |
m.8993T>G, p.Leu156Arg, MT-ATP6. Mutation load (%): >95 |
NR | NR | Mutation load (%): >95 in fibroblasts, kidney and liver |
| Suomalainen et al. 1992 (1634620) |
1 | 60 | F | PEO and MDD | FCtx, basal ganglia | Southern blot PCR |
NR | NR | Presence of several DELs. Most of the DEL breakpoints were between ∼m.11,900 and m.12,600. DEL sizes between ∼2.0 to 10 kb | Mutation load (%): kidney 10, liver 20, extraocular muscle 40, heart 40, vastus lateralis muscle 60. No DELs in blood |
| Lombes et al. 1991 (1849240) |
3 | 5, 2, 0.5 | M | LS and COX deficiency | NA | Southern blot Northern blot |
NR | mtDNA/nDNA content was higher in P than in C (in P3, 4.6 times higher) | No mtDNA DEL detected | mRNA levels of the mtDNA encoded COX subunits was decreased compared to the nDNA encoded subunits |
| Ciafoloni et al. 1991 (1922812) |
1 | 26 | M | MELAS | NA | Southern blot PCR-RFLP |
m.3243A>G, MT-TL1 Mutation load (%): 84 |
NR | NR | Mutation load (%): muscle 83, liver 79, heart 83, kidney 86 |
| Enter et al. 1991 (1684568) |
1 | 12 | F | MELAS | NA | Southern blot PCR-RFLP |
m.3243A>G, MT-TL1 Mutation load (%): 80 |
NR | NR | Mutation load (%): cardiac muscle 70, skeletal muscle 30, liver 70, diaphragm 60 |
| Bordarier et al. 1990 (2359483) |
1 | 17 | F | KSS | NA | Southern blot | NR | NR | DEL located between positions ∼ m.8,200 and m.13,000 (±400 bp) | The DEL was also found in muscle and spinal cord |
C: control; DEL: deletion; F: female; GM: gray matter; M: male; mtDNA CN: mitochondrial DNA copy number; N: number of subjects; NA: information not available; NR: not reported; P: patient; y: years; WM: white matter.
ad/arPEO: autosomal dominant/autosomal recessive progressive external ophthalmoplegia; AHS: Alpers-Huttenlocher syndrome; KSS: Kearns-Sayre syndrome; LHON: Leber hereditary optic neuropathy; LoME: late onset mitochondrial encephalomyopathy; LS: Leigh's syndrome; MDD: major depressive disorder; MDS: mitochondrial depletion syndrome; ME: mitochondrial encephalomyopathy; MELAS: mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes; MERRF: myoclonic epilepsy with ragged-red fibers; MILS: maternally inherited LS; MM: mitochondrial myopathy; MNGIE: mitochondrial neurogastrointestinal encephalopathy; NARP: neuropathy, ataxia, retinitis pigmentosa; POLG: DNA polymerase subunit gamma, TK2: thymidine kinase 2.
Amg: amigdala; Cb: cerebellum; CbCtx: cerebellar cortex; Cd: caudate nucleus; Chpx: choroid plexus; Ctx: cortex; Dt: dentate nucleus; FCtx: frontal cortex; FLV: frontal horn of the lateral ventricle; Gic: internal capsula, genu; GP: globus pallidus; GPI: internal globus pallidus; Hi: hippocampus; Hy: hypothalamus; Ic: internal capsule; IO: inferior olive; Mb: mammillary bodies; Mcer: middle cerebral artery; OCtx: occipital cortex; Ox: optic chiasm; PCtx: parietal cortex; Pit: pituitary gland; Pu: putamen; SN: substantia nigra; SNC: substantia nigra, pars compacta; Spinal: spinal cord; Str: striatum; TCtx: temporal cortex; Th: thalamus; Ver: vermis of cerebellum.
CSGE: conformation-sensitive gel electrophoresis; NGS: next-generation sequencing; qPCR: quantitative real-time polymerase chain reaction; RFLP: restriction fragment length polymorphism.
ATP6: ATP synthase subunit 6; CO2: cytochrome c oxidase II; CO3: cytochrome c oxidase III; HV2: hypervariable segment 2; MT-: mitochondrially encoded gene; ND1: NADH-ubiquinone oxidoreductase subunit 1; TE: tRNA-Glu; TK: tRNA-Lys; TL1: tRNA-Leu 1; TW: tRNA-Trp.
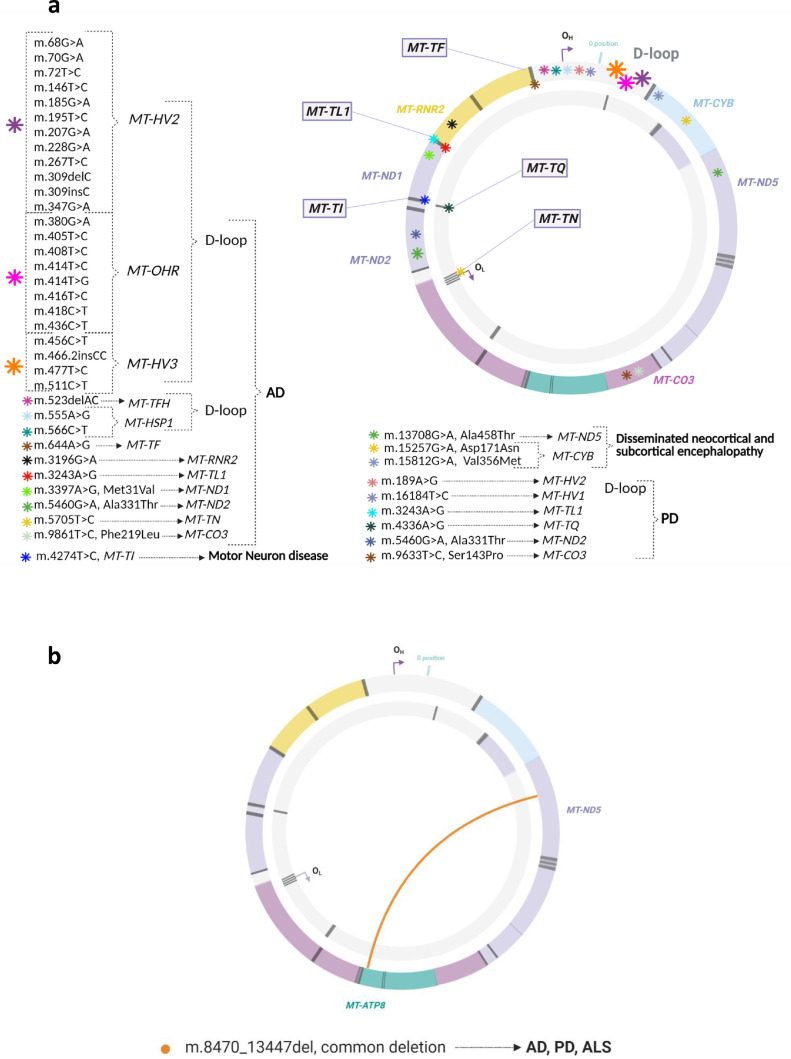
Table 2.
Studies reporting results of the mtDNA analyses in postmortem brain samples of patients with neurological diseases (NeuD).
| Study reference (PMID) |
Patient/control characteristics |
Disease (N) |
Brain region | Technique | mtDNA alteration | Additional findings | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| N P/C |
Age (y) in P/C | Sex (M/F) |
Variant | mtDNA CN | Rearrangements | |||||
| Castora et al. 2020 (31561357) |
40/40 | NA | NA | AD (40) | PCtx, TCtx, Cd, Hi | PCR-RFLP | 6 AD P showed the m.9861T>C variant compared to none in C. Mutation load %: between 11 and 95 |
NR | NR | m.9861T>C was found in multiple regions, with the highest mutation load % occurring in PCtx and TCtx |
| Chen et al. 2020 (31689514) |
20/15 | 49-81/75 | 21/14 | MD (12) PD (8) C (15) |
SN | qPCR | NR | High mtDNA CN in healthy aged TH-positive neurons of C, despite showing similar DEL levels as P with PD. mtDNA CN was significantly lower in neurons of P with MD showing mtDNA point mutations or multiple deletions and in P with PD. Data showed evidence of mtDNA depletion in PD neurons, with large individual diversity in cases. |
DELs in MT-ND4 were detected in healthy TH-positive neurons of aged C. Comparable DEL levels in MT-ND4 were found in the cases with multiple mtDNA DELs and in P with PD. Prominent MT-ND1 DELs were detected in neurons from cases with multiple DELs. They were also present in older C and P with PD; however, with lower levels. |
- |
| Kim et al. 2020 (32005289) |
34/25 | NA | NA | ALS (34) AD (10) C (15) |
Ctx | Aldehyde reactive probe-based assay, ELISA |
NR | NR | NR | Apurinic/apyrimidinic sites (abasic sites) did not differ between ALS and C. Levels of oxidized mtDNA did not differ between ALS and C. mtDNA analyses were conducted in laser-microdissected neurons |
| Thubron et al. 2019 (31388037) |
44/30 | 79/77 | 36/38 | AD (34) MCI (10) NCI (30) |
PCtx, FCtx, Cb | qPCR | NR | 48% mtDNA CN reduction in PCtx of nondiabetic AD vs. nondiabetic NCI. No reduction observed in diabetic AD compared to diabetic NCI |
NR | The lowest mtDNA CN level was observed in Cb, the highest in PCtx, both in NCI. Higher mtDNA CN was observed in all brain regions of diabetic vs. nondiabetic cases, irrespective of cognitive status |
| Alvarez-Mora et al. 2019 (30887649) |
2/3 | 93,97/73-83 | NA | FXTAS (1) no-FXTAS (1) C (3) | Ver, Dt, PCtx, TCtx, Th, Cd, Hi | ddPCR | NR | mtDNA CN in Ver, Dt, PCtx and TCtx was descreased in FMR1 premutation carriers with FXTAS than in C but no differences were detected in Th, Cd and Hi. P with FXTAS showed lower mtDNA CN than Cs | NR | - |
| Soltys et al. 2019 (30359878) |
10/20 | 82/80 | 11/19 | AD (10) HpC (10) C (10) |
Cb, TCtx | RMC assay, ddPCR |
mtDNA mutation frequencies were similar in all three groups in both brain regions | mtDNA CN depletion observed in TCtx of AD P vs. HpC or C. No mtDNA CN variation in Cb between all three groups |
NR | - |
| Strobel et al. 2019 (30475765) |
22/10 | 75/70 | 16/16 | AD (22) C (10) |
Hi, Cb | qPCR | NR | NR | In C, higher rates of mtDNA DELs were observed in astrocytes and microglia of the Hi compared to brain stem and Cb | - |
| Bury et al. 2017 (29149768) |
6/6 | 76/84 | Ratio 5:1 | PD (6) C (6) |
PPN | qPCR | NR | Significant mtDNA CN increase in PPN cholinergic neurons from PD P (10.7%) vs. those from C (7.0%) | mtDNA DEL levels were significantly increased in PPN cholinergic neurons from PD P (21.6%) vs. those from C (17.2%) | mtDNA DELs correlated with Braak staging in PD |
| Wei et al. 2017 (28153046) |
902/ 461 |
69/59 | 765/ 598 |
AD (282) CJD (181) DLB-PD (89) FTD-ALS (236) Other (114) C (461) |
Cb, Ctx, other | Exome sequencing | No evidence of disease association with homoplasmic or heteroplasmic rare variants | mtDNA CN was significantly lower in AD and CJD P. Positive correlation between age and mtDNA CN in CJD P |
NR | No correlation between mean levels of heteroplasmy, total number of heteroplasmic variants or variant pathogenicity score with age or disease group |
| Nido et al. 2018 (29257976) |
2/NA | NA | NA | PD (2) | SN | NGS | Sequence heteroplasmy was significantly different between deleted and nondeleted mtDNA populations for 4/55 of the DELs. 11.3% of the analysed positions had a heteroplasmic frequency significantly different between deleted and nondeleted mtDNA populations |
NR | The 20 microdissected neurons showed 373 unique DELs (only 31 previously annotated). Each neuron contained on average 38.2 ± 29.8 distinct DELs. Mean size deletion was 5080 ± 2367 bp. The common DEL was detected in 15/17 neurons and was the most prevalent DEL in 6 neurons | 20 single laser-microdissected neurons were evaluated |
| Flones et al. 2017 (29270838) |
18/11 | 79/74 | 17/12 | PD (18) C (11) |
FCtx, SN, Cb, Hi, Pu | qPCR | NR | There was no significant difference between C and PD for mtDNA CN in Pu or Hi | Only dopaminergic neurons from the SN harboured significantly higher mtDNA DEL levels | 1189 single laser-microdissected neurons were evaluated. Neuronal complex I deficiency did not correlate with mtDNA damage |
| Dölle et al. 2016 (27874000) |
10/22 | 82/56 | 21/11 | PD (10) C (22) |
SN, FCtx, CbCtx | qPCR, NGS-(4,767 bp) | The mean load of heteroplasmic SNVs was 33.14%/1,000bp per neuron, and most of these clustered in the low-frequency spectrum in both PD and C. The overall burden of heteroplasmic SNVs was similar in PD and C. The proportion of G:C to T:A transversions was also similar in the two groups |
mtDNA CN was similar in PD and C in SN; however, the subset of neurons with high mtDNA depletion (<10,000 copies/cell) was 14% in PD and 2.7% in C (p=0.01). No differences were observed in FCtx or CbCtx |
SN neurons from PD contained significantly higher mtDNA DEL levels than C. The proportion of neurons with DEL levels exceeding 60%, was 21.4% in PD and 10.8% in C. mtDNA DEL levels were generally low in frontal neurons and Cb of both PD and C |
871 single laser-microdissected neurons were evaluated. In SN but not in FCtx or CbCtx, DELs and mtDNA CN showed a positive correlation with age; DEL was a predictor of mtDNA CN |
| Chen et al. 2016 (27299301) |
13/12 | 81/82 | NA | AD (13) C (12) |
FCtx | NGS, qPCR |
Similar heteroplasmy levels were observed in some mtDNA positions in AD P and C | NR | DELs were increased in AD P (9%) vs. C (2%). Rearrangement rate was higher in AD P (18%) than in C (7%). The common DEL was detected in most samples but at low % (1.5%) |
Different numbers and types of mtDNA rearrangement fragments were detected depending on the sequencing coverage depth |
| Gatt et al. 2016 (26853899) |
81/33 | 79/80 | 67/47 | PD (41) PDD (40) C (33) |
PFCtx | qPCR | NR | mtDNA CN was significantly reduced (18%) in PDD vs. C. Nonsignificant decrease was observed in PD |
NR | Although mtDNA CN was reduced in PDD, mitochondrial biogenesis was unaffected, as the expression of mitochondrial proteins in PDD and C was similar |
| Blanch et al. 2016 (26776077) |
26/18 | NA | 23/21 | AD (16) PD (10) C (18) |
Ent, SN | PQ, qPCR |
NR | NR | NR | Increased 5-methylcytosine levels observed in the D-loop in Ent of AD P vs. C. Lower 5-methylcytosine levels observed in the D-loop in SN of PD P vs. C |
| Coxhead et al. 2016 (26639157) |
180/40 | 78/77 | 128/92 | IPD (180) C (40) |
SNC, FCtx | NGS | The mean heteroplasmic variant burden differed between PD P and C in both SNC and FCtx | NR | NR | Nonsignificant correlation of heteroplasmy with age. Increased heteroplasmic variation was observed in COX genes |
| Grunewald et al. 2016 (26605748) |
10/10 | 76/75 | 5/5 | IPD (10) C (10) |
SN | qPCR | NR | mtDNA CN was reduced (73.1%) in IPD P vs. C. 7S D-loop mtDNA CN was reduced (91%) in IPD P vs. C |
mtDNA DEL prevalence did not differ between IPD P and C | - |
| Rice et al. 2014 (24448779) |
10/9 | 79/59 | 12/22 | AD (10) | Hi | qPCR | NR | mtDNA CN was significantly reduced in PyNs from AD. CN in other neuronal cells was not significantly different between P/C | Deletion levels were not significant | - |
| Azakli et al. 2014 (23872536) |
1/0 | 36 | 0/1F | MTLE-HS (1) | Hi (6 regions) | Pyrosequencing | The number of heteroplasmic variants was higher in the CA2 region and accumulated in MT-ND2, MT-ND3 and MT-ND5 genes. m.3563insC, p.Trp86>fs was suggested to be studied in other MTLE-HS patients |
NR | NR | Hi contained more heteroplasmic variants than blood |
| Müller et al. 2013 (23566333) |
14/14 | 78/73 | NA | PD (7) AD (7) C (14) |
SN, Hi | qPCR | NR | In PD, mtDNA CN did not differ significantly between LB+ and LB- neurons. In AD, mtDNA CN did not differ significantly between tau protein+ and tau protein- neurons |
In SN, DEL levels differed between P with LB+ neurons (40.5%), LB- neurons (31.8%) and C (25.6%), p<0.005. In Hi, DEL levels did not differ between groups, independent of disease status and cell type (tau protein + or -) |
2–6 single laser-microdissected neurons were evaluated per patient/control |
| Krishnan et al. 2012 (21925769) |
10/6 | 76/76 | NA | AD (10) C (6) |
Hi | qPCR, Long-range PCR + sequencing |
NR | NR | mtDNA DEL levels were higher in COX-deficient neurons (57%) than in COX-normal neurons (9%) in AD P and in C (48% and 24%, respectively). No differences were observed in COX-deficient neurons from AD P and C. mtDNA DELs ranged in size from 3670 to 6088 bp. mtDNA DELs accumulated with age |
- |
| Keeney et al. 2010 (20504367) |
10/7 | NA | NA | ALS (10) AD (1) C (6) |
Cb, Ctx | qPCR | NR | There were no differences in ND2 CN between P and C | There was an approximate doubling of DELs for ND4 and a consistent increase for CO3. Two P showed greater abundance in ND4 DELs with higher CN levels. Purkinje neurons showed low levels of DELs | Single laser-microdissected neurons from cervical spinal cord, anterior gray matter and Cb were evaluated |
| Naydenov et al. 2010 (20740286) |
32/31 | 75/71 | NA | PD (32) | Pu, Cb | qPCR | NR | Dopamine led to a 25% downregulation of mtDNA levels and L-Dopa caused an increase in mtDNA levels. In dyskinetic-PD, lower mtDNA levels in early disease processes were found. Pu showed a 50% reduction and a significant negative correlation with L-DOPA/year |
Pu from dyskinetic-PD showed a higher % of deletions and a correlation with mtDNA levels (↓mtDNA levels↑mtDNA deletions in PD and dyskinetic-PD) | - |
| Coskun et al. 2010 (20463402) |
38/25 | NA | NA | AD (13) DS (11) DSAD (14) C (25) |
FCtx | PNA campling PCR, qPCR, sequencing-(1,115 bp) | The frequency of mtDNA control region mutations was significantly higher in AD than in C. m.414T>G/ was found in 65% of AD but not in C, and in 57% of DSAD but not in DS. Other heteroplasmic mutations common in AD reported: m.68G>A, m.70G>A, m.72T>C, m.185G>A, m.207G>A, m.228G>A, m.309delC, m.309insC, m.408T>C, m.414T>C, m.418C>T, m.466.2insCC |
mtDNA CN of AD brains was significantly lower than age-matched C. Additionally, mtDNA CN of DSAD was significantly lower than age-matched C. mtDNA CN declined with age in C |
NR | AD-like neuropathology was present in AD and DSAD but not in DS. The frequency of somatic mutations in the regulatory control region increased with age in the normal brain (p=0.0029). The MT-ND6 to MT-ND2 transcript ratio did not change with age but was significantly lower in AD, DSAD and DS compared to C. Thus, reduced mtDNA L-strand transcription level was associated with intellectual disability and dementia. β-Secretase activity was associated with some mtDNA alterations |
| Campbell et al. 2010 (21446022) |
13/10 | 55/74 | NA | MS (13) C (10) |
Ctx | Long-range PCR, qPCR, sequencing | NR | NR | DELs were evident in 66% of normal appearing gray matter regions and in 53% of lesioned regions of MS P and in 16% of C. Multiple DELs were observed t in respiratory-deficient laser microdissected pooled neurons when compared to neurons with intact complex IV activity. Heteroplasmy DEL levels were also increased in respiratory-deficient neurons compared to neurons with intact complex IV activity | Single laser-microdissected neurons from WM and GM were evaluated. No differences in DEL heteroplasmy levels between WM and GM. DELs were supposed to be pathogenic because they contained MT-CO1 and MT-CO2 catalytic subunits |
| Arthur et al. 2009 (19775436) |
8/10 | 78/67 | 12/6 | sPD (8) C (10) |
FCtx and SN | Surveyor nuclease assay, qPCR | Nonsignificant variation in heteroplasmy levels between sPD P and C | No differences in mtDNA CN between sPD P and C | NR | - |
| Aliev et al. 2008 (18827923) |
NA | NA | NA | AD | Ctx, Hi, Endothelial cells |
Cytological in situ hybridization | NR | NR | 5 kb mtDNA DEL was localised in lysosomes of P but not in neuronal cell bodies. The main location of these DELs was in lysosomes, but not in other neuronal cell compartments |
DEL detection was achieved by electron microscopy ultrastructural visualisation of immune-positive gold particle clusters |
| Bender et al. 2008 (18604467) |
9/8 | 76/71 | 10/7 | AD (9) C (10) |
Pu, FCtx, SN | qPCR | NR | NR | DEL levels were higher in AD SN (32%) than in FCtx (13%) and Pu (14%) but did not differ from that of C (SN 35%, FCtx 14% and Pu 14%) | 1530 single laser-microdissected neurons were evaluated. There was no difference in mtDNA DEL levels per brain region between groups |
| Hakonen et al. 2008 (18775955) |
4/9 | 23/44 | 9/6 | IOSCA (4) C (9) (CFTR, c.1523A>G, p.Y508C) |
Ctx, Cb | Long-range PCR, qPCR, Southern blot, sequencing | IOSCA P did not show increased mtDNA point mutation load in affected tissues | mtDNA depletion was present in Ctx and Cb of IOSCA P | No mtDNA DEL was detected in IOSCA P | mtDNA CN decreased by 5–20% in brain and 10–70% in liver of IOSCA P compared to C but similar amounts were observed in skeletal muscle |
| Reeve et al. 2008 (18179904) |
6/5 | 77/78 | NA | PD (5) PEO (1) C (5) |
SN | Long-range PCR | NR | NR | Various DELs were found in P and C, there was no difference in the distribution nor in the types of DEL breakpoints detected between groups | Single laser-microdissected neurons from SN were evaluated |
| Blokhin et al. 2008a (18566918) |
5/9 | 38-53/ 34-80 |
8/6 | MS (5) C (9) |
FCtx, PCtx, OCtx | qPCR | NR | No differences in mtDNA CN between COX+ neurons, COX- neurons and neurons of chronic active plaques in MS. mtDNA CN in WM did not differ between MS P and C. In MS, mtDNA CN in neurons of GM was higher than in cells of other regions |
NR | mtDNA CN decreased with age in both MS P and C |
| Blokhin et al. 2008b (18286391) |
5/12 | 38-53/ 34-80 |
8/6 | MS (5) C (12) |
FCtx, PCtx, OCtx | qPCR | NR | NR | No pathology-related accumulation of mtDNA DELs was observed when comparing distinct brain specimens from MS or when comparing MS and C. The rate of mtDNA DELs was higher in COX- than in COX+ cells |
Proportion of mtDNA DELs correlated with age |
| Borthwick et al. 2006 (16358336) |
1/0 | 73 | M | MND (1) | FCtx, Pons, spinal cord | Sequencing, PCR-RFLP with radiolabeled nucleotides | m.4274T>C in MT-TI. Mutation load %: FCtx 45%, Pons 37%. | NR | NR | Single laser-microdissected motor neurons were evaluated. Mutation levels were significantly higher in all COX-deficient than in COX-positive motor neurons |
| Bender et al. 2006 (16604074) |
15/8+8 | 76/77 | NA | PD (15) C (8+8) |
Hi, SN | Long-range qPCR, sequencing | Nonpathogenic m.189A>G, m.16184T>C and m.9633T>C variants were detected | NR | In SN, mtDNA DEL levels did not differ between PD (52.3%) and aged C (43.3%); however, they differed in the Hi (17.8% in PD and 14.3% in C), p=0.0002. DEL levels correlated with age | 50 single laser-microdissected neurons were evaluated. The level of mtDNA DELs was significantly greater in COX-deficient neurons |
| Wang et al. 2006 (16405502) |
8/6 | 90/81 | NA | MCI (8) C (6) |
FCtx, TCtx, PCtx, Cb | GC/MS-SIMA | NR | NR | NR | Levels of multiple oxidised bases were significantly higher in FCtx, TCtx and PCtx of MCI P than in C |
| Wang et al. 2005 (15857398) |
8/8 | 85/84 | 8/8 | AD (8) C (8) |
FCtx, TCtx, PCtx, Cb | GC/MS-SIMA | NR | NR | NR | Levels of multiple oxidised bases were significantly higher in FCtx, TCtx and PCtx of AD P than in C. mtDNA had ∼10-fold higher levels of oxidised bases than nDNA |
| Aliyev et al. 2005 (15760652) |
NA | NA | NA | AD | Hi | Cytological in situ hybridization | NR | NR | 5 kb mtDNA DELs were mostly localised in lysosomal structures but not in neuronal cell bodies. There was a 3-fold increase of mtDNA DEL in AD compared to C |
DEL detection was achieved in neurons by electron microscopy ultrastructural visualization of immune-positive gold particle clusters |
| Coskun et al. 2004 (15247418) |
23/40 | NA | NA | AD (23) C (40) |
FCtx | PNA-clamped PCR, RT-qPCR, sequencing-(669 bp) | Frequency of heteroplasmic variants in the mtDNA control region showed a 63% increase in P with AD compared to C (p<0.01). In P 80 years and older this increase was 130%. m.414T>G proved to be specific for AD brains. m.146T>C, m.195T>C and m.477T>C showed heteroplasmic levels up to 70–80% in P with AD aged between 74 and 83 years |
There was a significant 50% mtDNA CN reduction in AD compared to C | NR | Paper focused on studying the mtDNA control region. Variants identified in brains of P with AD were preferentially located in known functional transcription and replication elements and were also frequently present at exceptionally high proportions. P with AD showed reduced MT-ND6 mRNA expression |
| Mawrin et al. 2004 (15036587) |
4/4 | 61/57 | 3/5 | AD (1) PD (1) FTD-MND (1) DLB (1) C (4) |
FCtx, OCtx, TCtx, Hi, SN, Cb, basal ganglia, brain stem | qPCR | NR | NR | In FTD-MND, high levels of the common DEL were found in the brain stem, FCtx and TCtx. In DLB, they were observed in OCtx. In PD and AD, no increase of the common DEL was observed. In PD, high levels of common DEL were observed in SN after normalisation for the mtDNA deletion in Cb. The lowest levels of the common DEL were found in the Cb |
Levels of the common DEL were markedly raised with increasing age |
| Mawrin et al. 2003 (12924443) |
7/3 | NA | NA | ALS (7) C (3) |
Ctx, brain stem | PCR | NR | NR | Common DEL levels did not differ between ALS and C. Common DEL levels increased with age |
Single laser-microdissected neurons were evaluated |
| Aliev et al. 2003 (14503022) |
NA | 57-93/54-85 | NA | AD | Hi, TCtx, Cb, FCtx | In situ hybridization | NR | NR | mtDNA DEL showed a 3-fold increase in AD compared to C. The majority of these DELs were found in mitochondria-derived lysosomes |
DEL detection was achieved by electron microscopy ultrastructural visualisation of immune-positive gold particle clusters |
| Gu et al. 2002 (12125742) |
24/4 | 77/81 | 17/11 | PD (8) AD (6) DLB (6) MSA (4) C (4) |
SN, Ctx, Hi, Cb (in 5 PD) Hi (in 5 AD) |
Long-range PCR + FIGE Southern blot |
NR | NR | The number of mtDNA DEL/rearrangements in SN of P with PD was significantly higher than that in other groups. In PD, the number of mtDNA DEL/rearrangements did not differ between the other brain regions studied. No significant increase was observed in the total number of DEL/rearrangements in the Hi of AD P |
The average number of rearranged forms in patients with PD, AD, MSA, DLB and C was 10.8, 5.7, 4.8, 4.8 and 6.0, respectively |
| Zhang et al. 2002 (12039426) |
26/21 | NA | NA | PD (7) MSP-A (4) PSP (4) DLB (4) AD (7) C (21) | SN, other midbrain regions | ISH | NR | NR | Common DEL accumulated in neurons but not glia in both the SN and other midbrain regions with no differences between P and age-matched controls | - |
| Simon et al. 2001 (11352572) |
38/44 | NA | NA | AD (8) PD (27) MSA (4) C (44) |
Ctx, FCtx, OCtx, TCtx, PCtx | PCR-RFLP, sequencing | No presence of the m.414T>G variant. mtDNA variants reported in AD: m.267T>C, m.347G>A, m.380G>A, m.405T>C, m.416T>C, m.436C>T, m.456C>T, m.511C>T, m.523delAC, m.555A>G, m.566C>T and m.644A>G |
NR | NR | No further investigation in the pathogenicity of these mutations was made |
| de la Monte et al. 2000 (10950123) |
37/25 | 76/78 | NA | AD (37) C (25) |
TCtx | PCR with radiolabeled nucleotides | NR | mtDNA CN levels were significantly lower in AD P than in C although several AD cases showed high levels of mtDNA CN | NR | - |
| Chang et al. 2000 (10873587) |
20/20 | 82/71 | NA | AD (20) C (20) |
PCtx, Hi, Cb | PCR with radiolabeled nucleotides, PCR-RFLP | Point mutation frequencies were 2 to 3-fold higher in PCtx, Hi and Cb of AD P than in C. Point mutation frequencies did not differ between brain regions |
NR | Common DEL levels of AD did not differ from the C. Common DEL frequency was 15 to 25-fold lower in Cb than Ctx in both AD and C |
In blood, point mutation frequencies were not elevated in AD. No correlation was found between age and common DEL frequency in AD and C |
| Dhaliwal et al. 2000 (10943712) |
6/4 | 72/83 | NA | ALS (6) C (4) |
FCtx, TCx | PCR | NR | NR | Common DEL levels were higher in FCtx than in TCtx in both ALS and C. The relative difference in the two brain regions was >11-fold higher in ALS than in C | In ALS, there was no correlation between duration of disease and common DEL levels in the two brain regions |
| Ito et al. 1999 (10051601) |
1/NA | 81/NA | 1M/NA | AD (1) | SN, GP | PCR | NR | NR | The common DEL was detected in SN and GP | Percentage load of the m.3243A>G, MT-TL1 was 0.04% and 0.05% in cybrids obtained from SN and GP, respectively. m.3243A>G, MT-TL1 and the common DEL was not detected in blood |
| Hatanpaa et al. 1998 (9729244) |
5/4 | 83/72 | 2M/2M | AD (5) C (4) |
Motor Ctx, midtemporal Ctx | Northern blot | NR | NR | NR | COXIII mRNA expression was lower in midtemporal Ctx of AD P than in C |
| Haferkamp et al. 1998 (9561330) |
2/0 | 54/0 | 0/2 | Disseminated neocortical and subcortical encephalopathy | Brain stem | PCR | Both patients revealed the same homoplasmic variants at positions m.13708G>A and m.15257G>A. Patient 2 also carried an homoplasmic variant at position m.15812G>A | NR | NR | - |
| Ozawa et al. 1997 (9196054) |
1/1 | 65/65 | 1F/1F | PD (1) ALS (1) |
Str | PCR | NR | NR | Presence of 134 DEL in the P with PD and 98 DEL in the P with ALS | The ALS patient was considered a C individual in this study |
| Hamblet & Castora. 1997 (9357554) |
9/9 | 68/66 | NA | AD (9) C (9) |
TCtx | PCR Southern blot |
NR | NR | Mean % of the Common DEL in AD P and C was 0.059 and 0.009, respectively, a 6.5-fold significant change | - |
| Kösel et al. 1997 (9380043) |
4/4 | 74/73 | 7/1 | PD (4) C (4) |
SN, Ctx, CbCtx, Pu, Cb | PCR, PCR-RFLP | m.4336A>G homoplasmic in 1PD. m.5460G>A heteroplasmic (95% mutation load) in 1C |
NR | DEL levels in CbCtx were 50 to 200-fold lower compared to SN, and 12 to 23-fold greater in Pu than in Cb | - |
| Hutchin et al. 1997 (9425253) |
65/76 | 76/71 | NA | AD (65) C (76) |
NA | PCR-RFLP | Frequencies of the analysed variants: m.3196G>A, m.3397A>G and m.8021A>G were not detected in P or in C, m.4336A>G was only present in 1.7% of the C, m.5460G>A was present in 3.1% of P and 2.6% of C, m.5705T>C was present in 1.5% of AD P but not in C |
NR | NR | - |
| Janetzky et al. 1996 (8738945) |
48/19 | 75/71 | NA | AD (48) C (19) |
FCtx, Ent, Hi | Nested, allele-specific PCR, sequencing |
m.5460G>A, p.Ala331Thr, MT-ND2 was present in 6 AD cases (4 homo- and 2 heteroplasmic, with a mutation load of 5%). Not present in C |
NR | NR | - |
| Schnopp et al. 1996 (8937782) |
2/2 | NA | NA | PD (2) C (2) |
FCtx, PCtx, TCtx, OcCtx, Hi, Pu, Th, Cd, CbCtx, CC, SN, among others | PCR-RFLP | Ratios of the m.5460G>A, varied between 44% and 98% in the brain regions studied. No differences were observed when comparing WM and GM, or between PD P and C | NR | NR | - |
| Kösel et al. 1996 (8723226) |
21/77 | 74/72 | 45/53 | PD (21) C (77) |
SN, Ctx, Cb, Pu | PCR-RFLP | m.5460G>A heteroplasmic variant was found in 4/21 PD P and in 5/77 C. m.4336A>G homoplasmic variant was present in 1 PD P |
NR | NR | - |
| Chen et al. 1995 (7599213) |
3/3 | 27-42/ 27-42 |
3/3 | HD (3) C (3) |
Pu, OCtx, Cd | Competitive PCR | NR | NR | Similar levels of the common DEL in the three regions when comparing HD P and C. Lower levels of the common DEL in Pu and OCtx |
- |
| Cavelier et al. 1995 (8530074) |
33/9 | 80/72 | 20/22 | AD (33) C (9) |
FG, Cd, FCtx, OCtx, PCtx | Competitive PCR | NR | NR | Similar % of the common DEL in AD P (0.01–2.9) and C (0.003–2.0). Levels of the common DEL were higher in Cd than in FG |
No correlation between COX activity and the common DEL levels |
| Mecocci et al. 1994 (7979220) |
13/12 | 71/75 | 15/10 | AD (13) C (12) |
FCtx, TCtx, PCtx, Cb | PCR | NR | NR | NR | The amount of oxidised mtDNA showed a threefold increase in PCtx of AD P vs. C |
| Reichmann et al. 1993 (8393094) |
7/7 | 77/77 | 8/6 | AD (7) C (7) |
PCtx, Ent | PCR | NR | NR | DELs larger than 500 bp were discarded | - |
| DiDonato et al. 1993 (8232940) |
1/1 | 72/62 | 1M/1M | PD (1) C (1) |
SN, Hi, OCtx, Th, FCtx, Pu, GP, CbCtx, R, LC | qPCR | NR | NR | The SN showed the highest proportion of the common DEL (3.16%), while CbCtx showed the lowest (0.02%) | - |
| Blanchard et al. 1993 (8347829) |
6/6 | 80/64 | 7/5 | AD (6) C (6) |
FCtx | PCR | NR | NR | Similar mtDNA DEL levels were observed in AD (0.14%) and C (0.12%) | - |
| Lestienne et al. 1991 (2013767) |
1/1 | NA | NA | PD (1) C (1) |
Pu, SN, Ctx | PCR | NR | NR | The common DEL was present in PD P and in C | - |
| Lestienne et al. 1990 (2120389) |
15/5 | 60–85/NA | NA | PD (15) C (5) |
Pu. SN, FCtx | Southern blot | NR | NR | No DELs were identified | - |
| Schapira et al. 1990 (1979656) |
6/6 | NA | NA | PD (6) C (6) |
SN | RFLP Hybridization using a radiolabeled probe | NR | NR | No DELs were identified | - |
| Ikebe et al. 1990 (2390073) |
5/6 | 68/55 | 6/5 | PD (5) C (6) |
FCtx, Str | PCR | NR | NR | Proportion of deleted mtDNA to normal mtDNA was lower in FCtx than in the Str of both PD and C | - |
C: control; DEL: deletion; F: female; GM: gray matter; M: male; mtDNA CN: mitochondrial DNA copy number; N: number of subjects; NA: information not available; NR: not reported; P: patient; TH: tyrosine hydroxylase; y: years; WM: white matter.
AD: Alzheimer's disease; ALS: amyotrophic lateral sclerosis; CJD: Creutzfeldt-Jakob disease; DLB: dementia with Lewy bodies; DS: Down's syndrome; DSAD: Down's syndrome and dementia; FTD: frontotemporal dementia; HD: Huntington disease; HpC: high-pathology control subjects, individuals who meet criteria for high AD neuropathologic changes but remain cognitively normal; IOSCA: infantile onset spinocerebelar ataxia; IPD: idiopathic Parkinson's disease; LB: Lewy bodies; MCI: mild-cognitively impaired; MD: mitochondrial disease; MND: motor neuron disease; MS: multiple sclerosis; MSA: multiple system atrophy; MSA-P: MSA-parkinsonian type; MTLE-HS: mesial temporal lobe epilepsy-hippocampal sclerosis; NCI: noncognitively impaired controls; NFT: neurofibrillary tangles; PD: Parkinson's disease; PDD: PD with dementia; sPD: sporadic PD; PEO: progressive external ophthalmoplegia; PSP: progressive supranuclear palsy.
Cb: cerebellum; CbCtx: cerebellar cortex; CC: corpus callosum; Cd: caudate nucleus; Ctx: cortex; Ent: entorhinal cortex; FCtx: frontal cortex; FG: frontal gyrus; GP: globus pallidus; Hi: hippocampus; IO: inferior olive; LC: locus coeruleus; PCtx: parietal cortex; PFCtx: prefrontal cortex; PPN: pedunculopontine nucleus; SN: substantia nigra; SNC: substantia nigra pars compacta; OCtx: occipital cortex; Pu: putamen; R: red nucleus; Str: striatum; TCtx: temporal cortex; Th: thalamus; Ver: vermis of cerebellum.
ddPCR: digital-droppled PCR; ELISA: enzyme-linked immunosorbent assay; FIGE: field inversion gel electrophoresis; GC/MS-SIMA: gas chromatography/mass spectrometry with selective ion monitoring analysis; NGS: next-generation sequencing; PQ: pyrosequencing; qPCR: quantitative real-time polymerase chain reaction; RMC: random mutation capture; RFLP: restriction fragment length polymorphism; RT-qPCR: reverse transcription qPCR.
COX: cytochrome c oxidase; D-loop: displacement loop; MT-: mitochondrially encoded gene; ND2: NADH-ubiquinone oxidoreductase subunit 2; TL1: tRNA-Leu 1.
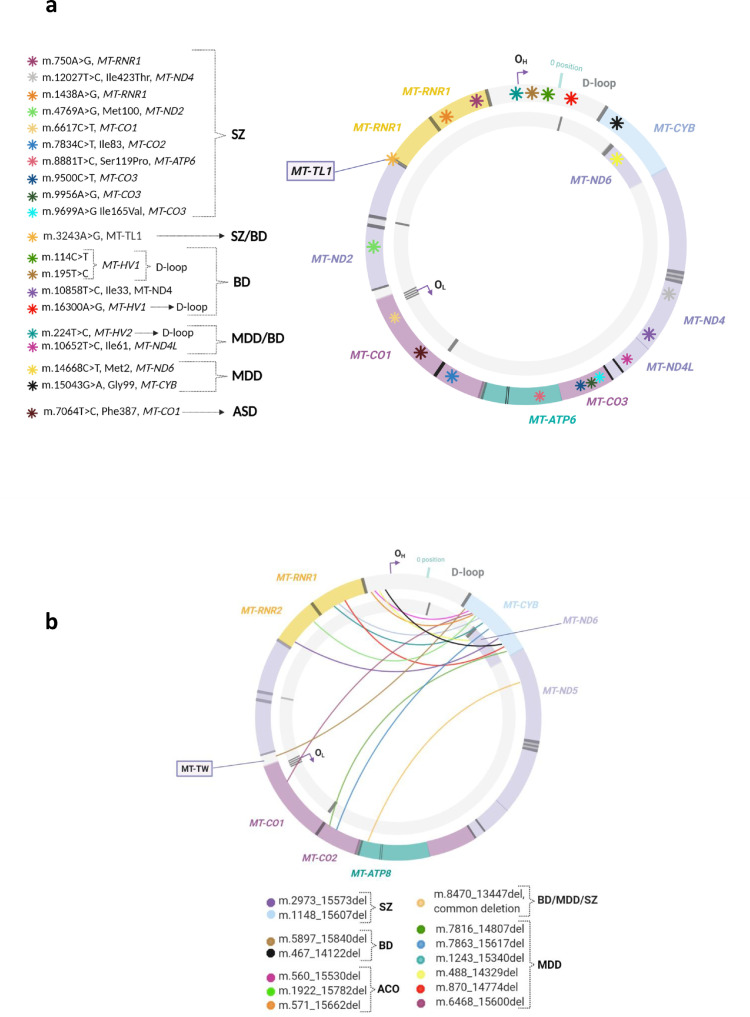
Table 3.
Studies reporting the results of mtDNA analyses in postmortem brain samples of patients with psychiatric diseases (PsyD).
| Study reference (PMID) |
Patient/Control characteristics | Disease (N) |
Brain region | Technique | mtDNA alteration in brain | Additional information | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| N P/C |
Age (y) in P/C |
Sex M/F |
Variant | mtDNA CN | Rearrangements | |||||
| Fries et al. 2019 (31746071) |
32/32 | 45/47 | P: 15/17 C: 19/13 |
BD (32) | Hi | qPCR | NR | Less mtDNA CN in BD P vs. C. Significant correlations between telomere length and mtDNA CN |
NR | mtDNA CN was not correlated with chronological age |
| Hjelm et al. 2019 (30869147) |
39/2 | 46 | 33/8 | SZ (12) BD (10) MDD (9) ADO (8) C (2) |
ACCtx, DLPFCtx, Hi, Pu, Cd |
Long-range PCR and NGS, exome sequencing, splice-break pipeline, qPCR, Sanger sequencing |
NR | NR | The study identified 4489 DELs: 513 with size range 7–8 kb and 127 with size of approximately 50 bp; 346 unique; 12 out of the 30 most frequent DELs were previously described. 13 high-impact DELs (read rate ≥ 5%) were present in 14 brain tissue samples of 9 subjects with psychiatric diagnoses. The group of subjects with SZ or BD had a higher DLPFCtx/ACCtx DEL ratio than the group of C, MDD or ADO subjects. The highest DEL burdens occurred in two subjects with MDD. The common DEL was neither the most frequent nor the most abundant |
Brain samples contained significantly more DELs and higher cumulative read percentages than blood samples. The 13 high-impact DELs detected in the brain were not detected in the blood. Many individual DELs had significant positive correlations with age The highest DEL burdens were observed in MDD P, at higher levels than KKS muscle |
| Bodenstein et al. 2019 (31797868) |
66/37 | Hi C: 60 BD: 59 SZ: 62 BA24 and Cb C: 71 BD: 72 SZ:69 PFCtx C: 72 BD: 65 SZ: 71 |
NA | SZ (35) BD (31) C (37) |
Hi, BA24, Cb, PFCtx | qPCR | NR | BD group had significantly higher mtDNA content for MT- ND4 and MT-ND5 in Hi tissue compared to C. Cb of patients with BD or SZ also had higher mtDNA CN than BA24 region of the same patients | No significant alteration in the accumulation of the common mtDNA DEL across the brain regions and groups | BA24 and Cb tissues came from the same patients |
| Otsuka et al. 2017 (28600518) |
20/25 | SV: 52 C: 58 |
SV:11/9 C: 19/6 |
SV (20) C (25) |
DLPFCtx | qPCR | NR | Significantly lower mtDNA CN in the DLPFC of SV compared to C (p = 0.0044) | NR | In the DLPFCtx of SV, significantly shorter telomere length was reported (p = 0.0014) |
| Rollins et al. 2018 (29594135) |
53/41 | BD: 46 SZ: 41 C: 58 |
BD: 13/13 SZ: 24/3 C: 35/6 |
BD (26) SZ (27) C (41) |
PFCtx | qPCR with SYBR green and TaqMan probes | NR | BD and SZ groups displayed a significant increase in mtDNA CN (p= 0.028 and p= 0.025, respectively) | No significant decrease in mtDNA common DEL in PFCtx of patients with SZ. Female subjects displayed 60% increase in the accumulation of the common DEL |
Complex I ELISA data indicated that the brains of SZ and BD had fewer functional mitochondria with larger amounts of mtDNA compared to controls |
| Mamdani et al. 2014 (25270547) |
30/10 | MDD: 48 BD: 52 SZ: 47 C: 46 |
MDD: 3/7 BD: 5/5 SZ: 5/5 C: 7/3 |
MDD (10) BD (10) SZ (10) C (10) |
ACCtx, Amg, Cd, DLPFCtx, Hi, Ac, OFCtx, Pu, SN, and Th | qPCR, Sanger sequencing | NR | NR | Common DELs in SZ were significantly decreased, mostly in dopaminergic regions, compared to MDD, BD and C. | Possible impacts of antipsychotic and antidepressant medications were not quantified. |
| Torrell et al. 2013 (23355257) |
45/15 | SZ: 45 BD: 43 MDD: 47 C: 48 |
SZ: 9/6 BD: 9/6 MDD: 9/6 C: 9/6 |
SZ (15) BD (15) MDD (15) C (15) |
OCtx | qPCR | NR | No differences in mtDNA CN were observed between study groups | NR | MT-ND1 gene expression was increased in BD P vs. C |
| Gu et al. 2013 (24002085) |
14/12 | ASD: 11 C: 11 |
ASD: 11/3 C: 9/3 |
ASD (14) C(12) |
FCtx | RT-PCR | NR | A significant increase of mtDNA CN in ASD subjects compared with C. | MT-ND4 deletion was found in 44% of the ASD group, and 33% of them also had Cyt B deletion. | |
| Tang et al. 2013 (23333625) |
20/25 | ASD: 2–67 C: 2–60 |
ASD: 18/3 C: 22/2 |
ASD (20) C (25) |
TL | MitoChip assay, qPCR | m.7064T>C, Phe387, MT-CO1 | No changes in mtDNA CN (studied in 8 ASD P and 8 C) | No presence of large-scale DELs or duplications. | |
| Sequeira et al. 2012 (22723804) |
3 cohorts Co 1: 4/7 Co 2: 29/6 Co 3: 6/17 |
Co 1: SZ: 53 C: 57 Co 2: SZ: 44 BD: 50 MDD: 51 C: 53 Co 3: SZ: 41 BD: 57 MDD: 41 C: 52 |
Co 1: SZ: 2/3 C: 2/4 Co 2: SZ: 11/3 BD: 9/3 MDD: 11/4 C: 60/16 Co 3: SZ: 2/2 BD: 3/1 MDD: 4/1 C: 16/7 |
Co 1: SZ (5) C (6) Co 2: SZ (14) BD (11) MDD (15) C (76) Co 3: SZ (4) BD (4) MDD (5) C (10) |
Co 1: ACCtx, Amg, Cd, DLPFCtx, Ac, OFCtx, Pu, SN, Th Co 2: DLPFCtx Co 3: DLPFCtx |
NGS, Affymetrix 6.0 SNP chip, qPCR |
149 homoplasmic novel or rare variants: 7 not previously reported (5 synonymous variants, 1 in the D-loop, 1 in a tRNA). 88% transitions and 11% transversions. Higher number of transitions and transversions in MDD vs. C and vs. SZ or BD. The m.195T>C, MT-HV2 and m.16519T>C, D-Loop were underrepresented in pooled SZ and BD vs. C. |
NR | In the DLPFCtx, the common DEL levels were significantly increased in P with BD, marginally increased in P with MDD and not increased in P with SZ vs. C. Higher levels of the common DEL in SN and Cd. The common DEL levels were correlated with age |
Certain brain regions accumulated somatic mutations at higher levels than the blood. |
| Ichikawa et al. 2012 (22030097) |
28 | 63.1 | 20/8 | SZ | FCtx | Sanger sequencing, allele-specific PCR | Homoplasmic substitutions m.8881T>C and m.9699 A>G were detected exclusively in SZ patients. Unregistered m.6617 C>T and m.9500 C>T substitutions with 50% heteroplasmy were found in a brain sample from a single patient. m.9956 A>G was also found as a homoplasmic variant in a brain sample of an SZ patient, while it was heteroplasmic in controls. | NR | NR | m.7196C>A was detected in SZ patients' brain tissue in both a homoplasmic and heteroplasmic state; it was also detected in blood samples of SZ patients and in blood samples of controls as a homoplasmic variant. |
| Rollins et al. 2009 (19290059) |
41/36 | BD: 50 SZ:45 MDD: 51 C: 53 |
SZ: 11/3 BD: 9/3 MDD: 11/4 C: 31/5 |
SZ (14) BD (12) MDD (15) C (36) |
DLPFCtx | Affymetrix mtDNA resequencing array, SNaPshot and allele-specific RT- PCR |
The rate of synonymous base pair substitutions in the coding regions of the mtDNA was 22% higher in P with SZ vs. C. One MDD P carried a homoplasmic mutation in DLPFC at m.10652T>C, Ile61, MT-ND4L, and two P with SZ showed less than 1% heteroplasmy. Low levels of heteroplasmic nonsynonymous mutations were found in the brain. Several mtDNA variants were significantly associated with specific psychiatric disorders: m.114T>C, m.195C>T, m.10652T>C and m.16300G>A with BD; m.750A>G, 1438A>G and 4769A>G with SZ; and 10652C>T, 14668T>C and 15043A>G with MDD. |
NR | NR | Brain pH was significantly associated with super haplogroup U, K and UK |
| Fuke et al. 2008 (18514404) |
99/48 | NA | NA | SZ (50) BD (49) C (48) |
FCtx | RT-qPCR with SYBR Green | NR | NR | Age- and sex-dependent accumulation of the common DEL, independent of the diagnosis. One P with SZ showed high levels of the common DEL |
|
| Sabunciyan et al. 2007 (17195919) |
100/44 | ≤ 68 | NA | SZ (45) BD (40) MDD (15) C (44) |
PFCtx | qPCR, RT-PCR with TagMan and SYBR Green | NR | No significant difference in mtDNA CN level between C and P | No significant difference in the amount of the common DEL between C and P with SZ or BD | Female BD patients had significantly less common DEL (p=0.03) compared with male patients. |
| Vawter et al. 2006 (16636682) |
20/20 | BD: 54 MDD: 51 C: 53 |
P: 14/6 C: 14/6 |
BD (9) MDD (11) C (20) |
ACCtx, DLPFCtx, Cb |
qPCR | NR | NR | NR | mtDNA gene expression increased with agonal duration |
| Munakata et al. 2005 (15737668) |
43/14 | SZ: 44 BD: 42 MDD: 47 C:49 |
SZ: 7/6 BD: 9/6 MDD: 9/6 C: 9/5 |
SZ (13) BD (15) MDD (15) C (14) |
PFCtx | PNA-clamped PCR-RFLP | m.3243A>G, MT-TL1, detected in two BD (mutation load range 0.90–1.00%) and one SZ (0.60%). | NR | NR | LARS2 was upregulated in cybrids carrying the m.3243A>G, MT-TL1 |
| Marchbanks et al. 2003 (14623372) |
15/9 | 70/69 | SZ: 10/5 C: 5/4 |
SZ (15) | NA | PCR-RFLP | m.12027T>C, Ile423Thr, MT-ND4 mutation load of 54% in SZ P vs. 58% in C. | NR | NR | NR |
| Kato et al. 1997 (9359971) |
16/9 | BD: 45 SV: 39 C: 40 |
BD: 3/4 SV: 4/5 C: 4/5 |
BD (7) SV (9) C (9) |
CbCtx | qPCR | NR | NR | The ratio of the common DEL was significantly higher in BD (0.23) compared with that in age-matched C (0.06; p < 0.05). | No significant difference in the common DEL ratio between SVs and controls. Antidepressant was found in the blood of 5 SVs. |
| Cavelier et al. 1995 (8530074) |
13/9 | 80/73 | SZ: 7/6 C: 5/4 |
SZ (13) | FG, Cd | Competitive PCR | NR | NR | No accumulation of the common DEL with age in Cd and a decrease in FG. Lack of age-related accumulation of the DEL. Higher DEL levels in Cd compared to FG. |
No correlation between COX activity and levels of common DEL. |
C: control; Co: cohort; DEL: deletion; F: female; M: male; mtDNA CN: mitochondrial DNA copy number; N: number of subjects; NA: information not available; NR: not reported; P: patient; y: years.
ADO: alcohol/drug abuse/other psychiatric symptoms; ASD: autism spectrum disorder; BD: bipolar disorder; MDD: major depressive disorder; SV: suicide victims; SZ: schizophrenia; KSS: Kerns-Sayre syndrome.
Ac: nucleus accumbens; ACCtx: anterior cingulate cortex; Amg: amigdala; BA24: Brodmann area 24; Cb: cerebellum; CbCtx: cerebellar cortex; Cd: caudate nucleus; DLPFCtx: dorsolateral prefrontal cortex; FCtx: frontal cortex; FG: frontal gyrus; Hi: hippocampus; OCtx: occipital cortex; OFCtx: orbitofrontal cortex; PFCtx: prefrontal cortex; Pu: putamen; SN: substantia nigra; Th: thalamus; TL: temporal lobe.
NGS: next-generation sequencing; PNA: peptide nucleic acid; qPCR: quantitative real-time polymerase chain reaction; RFLP: restriction fragment length polymorphism; RT-qPCR: reverse transcription qPCR.
CO1: cytochrome c oxidase I; COX: cytochrome c oxidase; D-loop: displacement loop; HV2: hypervariable segment 2; LARS2: Leucyl-tRNA synthetase 2; MT-: mitochondrially encoded gene; ND1: NADH-ubiquinone oxidoreductase subunit 1; ND4: NADH-ubiquinone oxidoreductase subunit 4; ND4L: NADH-ubiquinone oxidoreductase subunit 4L; TL1: tRNA-Leu 1.
Table 4.
Studies reporting the results of the mtDNA analyses in postmortem brain samples of individuals with a diagnosis not included in Table 1, Table 2, Table 3.
| Study reference (PMID) |
Patient/control characteristics |
Disease or condition (N) |
Brain region | Technique | mtDNA alteration | Additional findings | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| N P/C |
Age (y) in P/C | Sex (M/F) |
Variant | mtDNA CN | Rearrangements | |||||
| Wnek et al. 2016 (27457581) |
3/5 | 64/76 | 5/3 | HSE (3) C (5) |
FCtx, Amg, Hi, CiG and ICtx | Microarray, qPCR | NR | MT-CO1 exhibited lower abundance in CiG, Amg and FCtx in P than C | NR | Greater decline in P than C in mtDNA-encoded compared to nDNA-encoded transcripts. |
| Var et al. 2016 (26807965) |
27/30 | 48/51 | 57/0 | HIV+METH+ (16) HIV+METH- (11) C (30) |
Ctx: Brodmann areas 7, 8, 9, 46 | ddPCR | NR | HIV+METH+ group had higher mtDNA CN compared with HIV+METH- and HIV-METH- (WM from area 8) | Higher abundance of the common DEL was associated with increasing age. | A higher proportion of the common DEL was associated with lower neurocognitive function in HIV+METH but higher in HIV+METH- |
| Naue et al. 2014 (25526677) |
0/98 | 52 | 67/31 | C (100) | NA | qPCR, sequencing, minisequencing, NGS | Heteroplasmies were observed in 37% of the individuals (47 observations). 13 of the 98 samples showed 1 bp deletion between positions 66 and 71 |
NR | NR | The highest relative number of heteroplasmies was detected in muscle and liver (79%, 69%), followed by brain, hair, and heart (36.7%–30.2%). Bone (19.8%), blood (18%), lung (17%), and buccal cells (16.2%) showed a comparatively low number of heteroplasmies |
| Lynn et al. 2003 (12627331) |
1/0 | 46 |
1/0 |
Diabetes and recurrent stroke-like episodes, seizures and cognitive decline | Cb, OCtx | Hot last cycle PCR, radioactive PCR | m.3243A>G mutation load %: OCtx 78, Cb 66 |
NR | NR | Mutation load %: skeletal muscle 60, liver 60, pancreas 31, kidney 75, myocardium 58, blood 8 |
| Nádasi et al. 2003 (14711030) |
15/8 | <4mth/66 | 13/10 | Deceased neonates, newborns and infants (15), adults (8) |
FCtx, TCtx, Cb,Cd, Th, Hi | PCR | NR | NR | The common DEL was present in all brain samples from all individuals. The ratio of the common DEL/wild-type mtDNA was lower in the infant group than in adults |
The ratio of the common DEL/wild-type mtDNA was lower in blood than in brain |
C: control; DEL: deletion; F: female; M: male; mtDNA CN: mitochondrial DNA copy number; mth: month; N: number of subjects; NR: not reported; P: patients; y: year; WM: white matter.
HSE: Herpes simplex virus type-1 encephalitis; HIV: human immunodeficiency virus infection; METH: methamphetamine use.
Amg: amigdala; Cb: cerebellum; Cd: caudate nucleus; CiG: cingulate gyrus; Ctx: cortex; FCtx: frontal cortex; Hi: hippocampus; ICtx: insular cortex; OCtx: occipital cortex; TCtx: temporal cortex; Th: thalamus.
ddPCR: digital-droplet PCR; NGS: next-generation sequencing; qPCR: quantitative real-time polymerase chain reaction.
MT-CO1: mitochondrially encoded cytochrome c oxidase I gene.
Table 5.
Studies reporting the results of mtDNA analyses in postmortem brain samples in ageing.
| Study reference (PMID) |
Patient/Control characteristics | Condition (N) |
Brain region | Technique | mtDNA alteration in brain | Additional information | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| N P/C |
Age (y) In P/C |
Sex M/F |
Variant | mtDNA CN | Rearrangements | |||||
| Roca-Bayerri et al. 2020 (32722761) |
52/40 | NA | NA | PLWH (52) HIV-negative C (40) |
FCtx, FL GM | qPCR, long-range PCR, NGS | Mutations accumulated in the mtDNA noncoding D-loop were significantly associated with age | The mtDNA CN in FCtx decreased with age | An increase of the mutation load of the common DEL was associated with increasing age | The observed effects of HIV were calculated as equal to approximately 32 years for mtDNA CN, and approximately 12 years for the common DEL in advancing age |
| Dölle et al. 2016 (27874000) |
21 | 11–87 | 13/8 | C with no neurological disease | SN, FCtx, Cb | dPCR, qPCR, NGS | NR | Total mtDNA CN increased with age (p=0.0004) | Major arc DEL showed a significant positive correlation with age in SN neurons | In FCtx neurons and Purkinje cells, the level of the common DEL was generally low and did not increase with age. Ageing upregulated mtDNA CN in SN neurons, and this correlated with the levels of somatic mtDNA DELs in cells |
| Taylor et al. 2014 (23911137) |
21 | 15–80 | NA | NA | NA | 3D, ddPCR, NGS | NR | NR | The deletion load increased with age, while the number and diversity of unique deletions remained constant | The analysis was based on over 8 billion mitochondrial genomes |
| Kennedy et al. 2013 (24086148) |
10 | YI:<1 AI: 79–90 |
NA | YI (5) and AI (5) individuals without known brain pathology |
PFCtx | qPCR, duplex sequencing | A significant (5-fold) increase in mutation rate was reported in AI. 78.3% of mutations were nonsynonymous and predicted to be more deleterious. Most of them accumulated in the D-loop during ageing. | NR | NR | No significant increase in G>T mutations, considered the hallmark of oxidative damage to DNA with age |
| Coskun et al. 2010 (20463402) |
38/25 | DSAD: 40–62 DS: Newborn-45 AD: 55–90 C: Newborn-95 |
M:F ratio equal in both groups | DSAD (14) DS (11) AD (13) C (25) |
FCtx | PNA-clamp PCR, sequencing, qPCR, RT-qPCR | Mutations in the regulatory coding region of mtDNA increased with age in C and were significantly elevated in AD and DSAD relative to age-matched C and DS | mtDNA CN declined in C brains after age 65 in parallel with the increased mtDNA mutation rate | NR | ND6 (L-strand)/ND2 (H-strand) mRNA ratio did not change significantly with age in C, while it was significantly lower in both AD and DSAD |
| Meissner et al. 2008 (18439778) |
92 | 0.2–102 | NA | Acute or peracute cause of death with no known brain pathology | SN, Cd | PCR-CE | NR | NR | A positive correlation between the common DEL amount and ageing, and a strong interindividual variability were detected. Abundance of the common DEL varied with tissue type: SN>Cd>Pu>FL>Cb | The common DEL was detectable in individuals as young as 10 and 12 years |
| Bender et al. 2006 (16604074) |
15/16 | PD: 76 C: 77 AI:19–51 |
NA | PD (15) C (8) AI (8) |
SN, Hi | Long-range PCR, qPCR | NR | NR | High levels of mtDNA DELs were observed (52.3% ± 9.3% in SN of individuals with PD, and 43.3% ± 9.3% in AI; p = 0.06). The level of mtDNA DELs increased linearly with age. These DELs were characterised as the common DEL and the m.7409_13687 DEL | The level of mtDNA DELs accumulated in Hi neurons was significantly lower than in SN neurons in both groups |
| Kraytsberg et al. 2005 (16604072) |
1/8 | AD: 80 C: 31–102 |
NA | AD (1) C (8) |
SN | Single molecule PCR | NR | NR | The number of mtDNA DELs was significantly different between old and young tissues. The was a very high absolute prevalence of mtDNA DELs in aged SN | mtDNA DELs were directly involved in the development of COX defects in aged SN |
| Frahm et al. 2005 (16099018) |
50 | 0.2–93 | NA | Acute or peracute cause of death with no neurological disease | Cd, FCtx, CbCtx, | qPCR | NR | mtDNA CN in three age groups (0–30, 31–59 and >60 years) revealed no significant age-dependent increase | NR | In skeletal muscle and heart muscle tissue samples, mtDNA CN did not show any significant change |
| Cantuti-Castelvetri et al.2005 (16243605) |
6 | 40–69 | 4/2 | C without neurological disease | SN | Single-cell, allele-specific PCR, cloning-sequencing | Somatic mtDNA point mutations were distributed throughout tRNAs for Thr and Pro and in portions of the MT-CYB and D-loop in both glia and neurons. Higher mutation levels were detected in aged neurons | NR | NR | Mean number of somatic point mutations per mitochondrial genome was 3.3 for single neurons and 2.2 for single glia |
| Mawrin et al. 2004 (15036587) |
4/4 | FTD-MND: 33 AD: 84 DLB: 74 PD: 54 C: 35–75 |
FTD-MND: M AD: F DLB: F PD: F C: 2/2 |
FTD-MND (1) AD (1) DLB (1) PD (1) |
FL, TL, OL, Hi, SN, Cb | qPCR | NR | NR | The common DEL ratio increased with age. In the basal ganglia, it reached the highest level |
The lowest common DEL levels in individual cases were reported in Cb, with no age-related increase |
| Simon et al. 2004 (14675733) |
16/32 | C Group 1: 1–4 C Group 2: 12–24 C Group 3: 65–91 PD: 71–86 |
NA | C Group 1 (5) C Group 2 (10) C Group 3 (17) PD (16) |
FCtx, SN | PCR, sequencing | Accumulation of G>C to T>A and T>A to G>C transversions and all point mutations increased with age in FCtx. No significant differences in somatic mutation level between PD patients and age-matched controls |
NR | NR | SN of young individuals had similar accumulation of point mutations as in the SN and FCtx of older individuals |
| Storm et al. 2002 (12559408) |
5 YI 5 AI |
YI: 16–25 AI: 80–91 |
YI: 4/1 AI: 1/4 |
Diverse causes of death with no neuropathology | FL | Single-cell dPCR | NR | NR | Significant differences in the relative distribution of the common DEL levels were not identified, neither between astrocytes and neurons, nor between healthy YI and AI | The frequency of the common DEL in YI was 15.4% in astrocytes and 21.6% in neurons. In AI, 21.6% of the astrocytes and 17.8% of the neurons carried the common DEL |
| Murdock et al. 2000 (11058135) |
16 | 23–93 | 7/9 | NA | Cb,Cd,EC, FCtx,TCtx, SN, Pu | Competitive PCR, PNA-directed PCR clamping | m.3243A>G, m.8344A>G and m.414T>G mutations did not accumulate with age to levels >1/1000 in brain | NR | NR | The accumulation of m.414T>G mutation was identified in muscle samples of aged individuals |
| Chang et al. 2000 (10873587) |
20/20 | 82–71 | NA | AD (20) | Cb, PG, Hi | PCR, RFLP | No significant correlation with age for the frequency of m.16390 G>A in both AD and C groups | NR | No significant increase in the common DEL levels was detected with age | - |
| Lezza et al. 1999 (10336891) |
7/6 | AD: 51–79 C: 63–86 |
AD: 4/3 C: 3/3 |
AD (7) | FCtx, PCtx | Kinetics PCR | NR | NR | The common DEL levels increased with age in C. The common DEL % in AD was 3-fold lower than in C. DEL levels were much lower in younger AD than in older AD |
HPLC-EC revealed a positive correlation between the common DEL and OH8dG levels in aging human brain. Lower levels of common DEL in the presence of a higher content of OH8dG in AD compared with that of C |
| McDonald et al. 1999 (10501524) |
67/43 | STS-CI: 50 LTS-HI:56 C:50 |
NA |
STS-CI (53) LTS-HI (14) |
TL, Hi | PCR, RFLP | NR | NR | The common DEL was found in 54% of C, 57% of SE, and 21% of LS. Additionally, the common DEL was more prevalent in older individuals among C. A trend toward increased prevalence of the m.8649_16084del (7436 bp DEL) in older patients in all groups was observed |
A strong correlation was found in the presence of the common DEL and 7436 bp DEL within individual cases |
| Melov et al. 1999 (10638530) |
5 Y 6 E |
Y: 23–44 E: 51–79 |
NA | No histopathological abnormalities. No history of neurodegenerative disease |
Cb, FCtx, Pu, ECtx, SN, Cd, TCtx | Long-range PCR, sequencing | NR | NR | An increase in the number and the variety of mtDNA rearrangements in aged brains was detected. In the FCtx of 79-year-old subjects, a unique m.1989_14366del (12 kb) was reported |
- |
| Kösel et al. 1997 (9380043) |
4/4 | 57–87/55–84 | 3/1 4/0 |
PD (4) | SN, CbCtx | Competitive PCR, PNA-directed PCR clamping | NR | NR | The common DEL levels increased with age in C | - |
| Kapsa et al. 1996 (8994125) |
2/2 | C: 83–75 PD: 74–75 |
C: 1/1 PD: 2/0 |
PD (2) | SN | PCR, sequencing |
23 missense, 2 tRNA and one nonsense polymorphism were detected. Eight missense polymorphisms caused nonconservative amino acid replacements at evolutionary constrained sites |
NR | Several multiple DELs (4.5–7.1 kb) were detected in the SN of both aged P and C groups | - |
| Merril et al. 1996 (8579367) |
41/22 | 34–73/ (Age-comparable individuals) |
NR | SZ (13) Neuroleptic Suicides (14) ADO (5) C (9) |
FG, Pu | PCR | NR | NR | mtDNA DELs were associated with chronic hypoxia conditions rather than ageing | Neuroleptic drugs had little or no effect on the observed levels of deleted mtDNA |
| Jazin et al. 1996 (8901590) |
3/4 | SZ: 99 EAD: 72 LAD: 95 C: 60 |
NA | SZ (1) EAD (2) LAD (1) C (4) |
FG, Cd | PCR, sequencing | The overall heteroplasmy level in D-loop was 2.2-fold higher in two aged individuals (96 and 99) compared with a 28-year-old individual | NR | A 7.7-fold increase of small insertions and deletions was detected in aged individuals | Substitutions did not show any significant increase with age |
| Corral-Debrinski et al. 1992 (1303288) |
7 | 24–94 | NA | No history of neurodegenerative disease | FCtx, TCtx, OL, Pu, Cb | Dilution-PCR, PCR-RFLP | NR | NR | A significant increase in the common DEL ratio was detected in Ctx and Pu but not in Cb. In Ctx it ranged from 0.00023 to 0.012 in 67–77-year-olds and up to 0.034 in those over 80. In Pu: 0.0016 to 0.010 in 66–77-year-olds and up to 0.12 in those over 80. Age-related accumulation of the 7436 bp DEL was also detected with similar changes |
Some adult subjects presented neurofibrillary tangles and amyloid plaques consistent with their age |
| Mann et al. 1992 (1544498) |
6/6 | NA | NA | PD (6) | SN | Southern blot | NR | NR | No significant difference in % of the common DEL among patients and age-matched controls (0.01–0.02%) | No correlation between complex I activity and the common DEL levels |
| Soong et al. 1992 (1303287) |
7 | 0.3–82 | 4/3 | No neuropathology. | Cd, Pu, SN, GP, Th, Ctx GM and WM, Cb GM | PCR, qPCR with radiolabelled primers |
NR | NR | In neonatal brain regions, the common DEL level was 0.0004%. An age-related significant increase in DEL levels among adults was reported with 0.94 correlation efficiency in SN |
Adult Cd, Pu and SN had an average of 355, 204 and 121 times higher DEL level, respectively; GP, Th, neocortical GM and WM showed ratios 10–83 times higher than in Cb GM |
| Zhang et al. 1992 (1551433) |
1 | 69 | F | Patients with primary carcinoma of splenic flexure of bowel | NA | PCR, sequencing |
NR | NR | The common DEL and the 7436 bp DEL were detected in brain tissue | Heart and skeletal muscle samples were examined, and multiple DELs were also detected in these tissues, suggesting that accumulation of multiple DELs is a general phenomenon during normal ageing |
| Lestienne et al. 1991 (2013767) |
1/1 | 60–85 | NA | PD (1) C (1) |
SN | PCR shift assay | NR | NR | Low levels of the common DEL were detected in SN of the aged control without neurological or psychiatric antecedents and the PD P | |
| Cortopassi et al. 1990 (2263455) |
22 | Adults: 27–104, Stillborn: 32, 40 weeks Spontaneous abortions: 22, 29 weeks Newborn: 4d |
10/3 NA (9) |
No neuropathology | Cb | PCR, RFLP, nested PCR, dilution PCR | NR | NR | The common DEL was detected in the brain of older individuals while it was not observed in foetal brains. DELs in foetal tissues were estimated to be 1/100 to 1/100,000 times less than in adults |
The heart tissue of 7 adults and 5 foetuses were also examined, and the common DEL was only found in aged adults |
| Ikebe et al. 1990 (2390073) |
5/6 | 51–77/38–73 | 2/3 4/2 |
PD (5) | Str, FCtx |
PCR, sequencing |
NR | NR | Accumulation of the common DEL was reported in both PD and aged C. The DEL load % was higher in Str than in Fctx in both PD and aged C | The common DEL seemed to selectively accumulate in the nigrostriatal pathway |
AI: aged individuals; C: control; DEL: deletion; E: elderly; F: female; M: male; mtDNA CN: mitochondrial DNA copy number; N: number of subjects; NA: information not available; NR: not reported; P: patient; y: years; YI: young individuals.
ADO: alcohol/drug abuse/other psychiatric symptoms; AD: Alzheimer's disease; DS: Down's syndrome; DSAD: Down's syndrome and dementia; DLB: dementia with Lewy bodies; EAD: early-onset Alzheimer's disease; FTD-MND: frontotemporal dementia with motor neuron disease-like inclusions; LoAD: late-onset Alzheimer's disease; LTS-HI: long-term survivor of head injury; PD: Parkinson's disease; PLWH: people living with HIV; STS-CI: short-term survivor of cerebral ischaemia; SZ: schizophrenia.
Cb: cerebellum; CbCtx: cerebellar cortex; Cd: caudate nucleus; Ctx: cortex; ECtx: entorhinal cortex; FCtx: frontal cortex; FG: frontal gyrus; FL: frontal lobe; GM: gray matter; GP: globus pallidus; Hi: hippocampus; OCtx: occipital cortex; OL: occipital lobe; PFCtx: prefrontal cortex; PG: parietal gyrus; Pu: putamen; SN: substantia nigra; Str: striatum; TCtx: temporal cortex; Th: thalamus; TL: temporal lobe; WM: white matter
3D; digital deletion detection; dPCR: duplex PCR; ddPCR: digital-droplet PCR; HPLC-EC: high-performance liquid chromatography with electrochemical detection; NGS: next-generation sequencing; PNA: peptide nucleic acid; qPCR: quantitative real-time polymerase chain reaction; RFLP: restriction fragment length polymorphism; RT-qPCR: reverse transcription qPCR.
COX: cytochrome c oxidase; CYB: cytochrome B; D-loop: displacement loop; MT-: mitochondrially encoded gene; ND6: NADH-ubiquinone oxidoreductase core subunit 6; ND2: ADH-ubiquinone oxidoreductase core subunit 2; OH8dG: 8’-hydroxy-2′-deoxyguanosine.
Table 6.
mtDNA disease-related variants with pathogenicity information retrieved from public databases.
| Locus | Nucleo-tide position | Nucleo-tide change | Variant type | Pathogenicity status/TOOLS |
GB Freq (%) |
Reported phenotype (Homo-/heteroplasmy) |
Cell or tissue type of reported mtDNA somatic variant (Homo-/heteroplasmy) |
Database | Related clinical features in this systematic review |
|---|---|---|---|---|---|---|---|---|---|
|
MT-HV2, MT-ATT, MT-CR, MT-7S |
68 | G>A | Noncoding | NR | 0.021 | NR | NR (NA) | MITOMAP | AD |
| 70 | G>A | Noncoding | NR | 0.073 | NR | NR (NA) | MITOMAP | AD | |
| 72 | T>C | Noncoding | NR/NA | 1.792 | NR | Aging brains, POLG/PEO & control muscle, normal tissues (-/+) | MITOMAP | AD | |
|
MT-HV2, MT-OHR, MT-ATT, MT-CR, MT-7S |
114 | C>T | Noncoding | R/NA | 0.442 | BD-associated (+/-) | POLG/PEO muscle, bladder tumour back-mutation (-/+) | MITOMAP | BD |
| 146 | T>C | Noncoding | R/NA | 19.510 | Absence of endometriosis (+/-) | Elderly fibroblasts, elderly/AD brains, POLG/PEO & control muscle, various tumours (+/+) | MITOMAP | AD | |
| 185 | G>A | Noncoding | R/NA | 3.999 | Low VO2 max response (+/-) | POLG/PEO muscle, thyroid tumour, glioblastoma (+/+) | MITOMAP | AD | |
| 189 | A>G | Noncoding | NR | 5.436 | NR | Elderly muscle & brains, myocyte, POLG/PEO muscle & fibroblasts, various tumours (-/+) | MITOMAP | PD | |
|
MT-HV2, MT-OHR, MT-ATT, MT-CR |
195 | T>C | Noncoding | R/NA | 19.228 | BD-associated/melanoma (+/+) | Elderly fibroblasts, elderly/AD brains, tumours: lung, thyroid, ovarian, prostate, glioblastoma (+/+) | MITOMAP | AD/BD |
| 207 | G>A | Noncoding | NR | 4.645 | NR | Oral, prostate & thyroid tumours/OPA1 defect (+/+) | MITOMAP | AD | |
| MT-HV2, MT-OHR, MT-CSB1, MT-ATT, MT-CR | 224 | T>C | Noncoding | NR | 0.012 | NR | NR | MITOMAP | BD/MDD |
| 228 | G>A | Noncoding | R/NA | 2.579 | Low VO2 max response (+/-) | NR | MITOMAP | AD | |
|
MT-HV2, MT-OHR, MT-ATT, MT-CR |
267 | T>C | Noncoding | NR | 0.027 | NR | NR | MITOMAP | AD |
| MT-HV2, MT-OHR, MT-CSB2, MT-ATT, MT-CR | 309 | delC | Noncoding | NR | 0.000 | NR | buccal cell, colonic crypt (-/+) | MITOMAP | AD |
| insC | Noncoding | R/NA | 1.142 | AD-weakly associated (NR) | NR | MITOMAP | AD | ||
| MT-HV2, MT-OHR, MT-CSB3, MT-ATT, MT-CR | 347 | G>A | Noncoding | NR | 0.000 | NR | NR | MITOMAP | AD |
|
MT-OHR, MT-ATT, MT-CR |
380 | G>A | Noncoding | NR | 0.004 | NR | NR | MITOMAP | AD |
|
MT-OHR, MT-LSP, MT-ATT, MT-CR |
405 | T>C | Noncoding | NR | 0.000 | NR | NR | MITOMAP | AD |
| 408 | T>C | Noncoding | NA | 0.004 | NR | NR | MITOMAP | AD | |
| 414 | T>C | Noncoding | NR/NA | 0.002 | NR | AD brains/POLG OPA1 and control samples (-/+) | MITOMAP | AD | |
| 414 | T>G | Noncoding | NR/NA | 0.029 | NR | Elderly fibroblasts, elderly muscle, POLG/PEO, DS, AD brains, oocytes, normal tissues (-/+) | MITOMAP | AD | |
| 416 | T>C | Noncoding | NR | 0.000 | NR | NR | MITOMAP | AD | |
| MT-OHR, MT-LSP, MT-TFL, MT-ATT, MT-CR | 418 | C>T | Noncoding | NR | 0.114 | NR | NR | MITOMAP | AD |
| MT-OHR, MT-TFL, MT-ATT, MT-CR | 436 | C>T | Noncoding | NR | 0.000 | NR | NR | MITOMAP | AD |
|
MT-HV3, MT-ATT, MT-CR |
456 | C>T | Noncoding | NR/NA | 2.450 | NR | Thyroid tumour (+/-) | MITOMAP | AD |
| 466 | 2insCC | Noncoding | NR | NA | NR | NR | MITOMAP | AD | |
| 477 | T>C | Noncoding | NR/NA | 0.938 | NR | AD brains, ovarian tumour (-/+) | MITOMAP | AD | |
| MT-HV3, MT-CR | 511 | C>T | Noncoding | NR | 0.143 | NR | NR | MITOMAP | AD |
|
MT-HV3, MT-TFH, MT-CR |
523 | delAC | Noncoding | NR | 0.019 | NR | NR | MITOMAP | AD |
|
MT-HV3, MT-HSP1, MT-CR |
555 | A>G | Noncoding | NR | 0.000 | NR | NR | MITOMAP | AD |
| 566 | C>T | Noncoding | NR | 0.000 | NR | NR | MITOMAP | AD | |
| MT-TF | 644 | A>G | (tRNA) | NR/Likely benign | 0.050 | NR | NR | MITOMAP | AD |
| Benign | NA | Juvenile MELAS | NR | ClinVar | |||||
| MT-RNR1 | 750 | A>G | (rRNA) | R/NA | 98.277 | NR | NR | MITOMAP | SZ |
| R/NA | NR | Not provided | NR | ClinVar | |||||
| 1438 | A>G | (rRNA) | NR/NA | 94.853 | NR | NR | MITOMAP | SZ | |
| Benign | NR | Not provided, not specified | NR | ClinVar | |||||
| MT-RNR2 | 3196 | G>A | (rRNA) | R/NA | 0.025 | ADPD (+/+) | NR | MITOMAP | AD |
| MT-TER, MT-TL1 | 3243 | A>G | (tRNA) | Confirmed pathogenic | 0.019 | MELAS/Leigh syndrome/DMDF/MIDD/SNHL/CPEO/MM/FSGS/ASD/cardiac multi-organ dysfunction (-/+) | NR | MITOMAP | MELAS/BS-LD/AD/PD/BD/SZ |
| Pathogenic | NR | BD- and SZ-associated | NR | ClinVar | |||||
| 3251 | A>G | (tRNA) | R/Possibly benign | 0.000 | MM/MELAS with chorea-ballism (-/+) | NR | MITOMAP | MM/LAaCID | |
| Pathogenic | NR | Juvenile MELAS/PEO, proximal myopathy, and sudden death | NR | ClinVar | |||||
| 3257 | A>G | (tRNA) | NR | NA | NR | NR | MITOMAP | MERRF | |
| MT-ND1 | 3397 | A>G | Met31Val | R/Likely pathogenic | 0.305 | ADPD/possibly LVNC cardiomyopathy-associated/resistance to high altitude pulmonary oedema (+/-) | NR | MITOMAP | AD |
| Benign | NR | PD, LoLS, AD | NR | ClinVar | |||||
| MT-TI | 4274 | T>C | (tRNA) | R/Likely pathogenic | 0.000 | CPEO/motor neuron disease (-/+) | NR | MITOMAP | Motor neuron disease |
| MT-TQ | 4336 | A>G | (tRNA) | Unclear/Possibly benign | 0.839 | ADPD/hearing loss & migraine/ASD/ID (+/+) | NR | MITOMAP | PD |
| Conflicting reports | NA | Sensorineural deafness and migraine/juvenile MELAS | NR | ClinVar | |||||
| MT-ND2 | 4769 | A>G | Met100 | R/NA | 97.604 | NR | NR | MITOMAP | SZ |
| R/NA | NR | Not provided | NR | ClinVar | |||||
| 5460 | G>A | Ala331Thr | Conflicting reports/Possibly benign | 6.904 | AD/PD/LHON (+/+) | NR | MITOMAP | AD/PD | |
| Benign | NR | LS | NR | ClinVar | |||||
| MT-TW | 5537 | insT | (tRNA) | R/NA | 0.000 | LS (-/+) | NR | MITOMAP | LS |
| Pathogenic | NR | LS/ME | NR | ClinVar | |||||
| 5549 | G>A | (tRNA) | R/Likely pathogenic | 0.000 | Dementia and chorea (-/+) | NR | MITOMAP | ME | |
| Pathogenic | NR | ME | NR | ClinVar | |||||
| 5556 | G>C | (tRNA) | R/Possibly benign | 0.000 | Encephalomyopathy (-/+) | NR | MITOMAP | LoME | |
| MT-TN | 5705 | T>C | (tRNA) | NR | 0.017 | NR | NR | MITOMAP | AD |
| MT-CO1 | 6617 | C>T | Phe238 | NR/NA | 0.015 | NR | NR | MITOMAP | SZ |
| 7064 | T>C | Phe387 | NR/NA | 0.062 | NR | NR | MITOMAP | ASD | |
| MT-CO2 | 7706 | G>A | Ala41Thr | R/Possibly benign | 0.015 | AHS-like (+/+) | NR | MITOMAP | AHS-like disease |
| 7834 | C>T | Ile83 | NR/NA | 0.004 | NR | NR | MITOMAP | SZ | |
| MT-TK | 8344 | A>G | (tRNA) | Confirmed pathogenic | 0.008 | MERRF; Other-LD/DMD/leukoencephalopathy/HiCM (-/+) | Bone marrow, elderly muscle (-/+) |
MITOMAP | MERRF |
| Pathogenic | NR | LS/MERRF/PD/juvenile MELAS | NR | ClinVar | |||||
| MT-ATP6 | 8603 | T>C | Phe26Ser | NR/Possibly benign | 0.336 | NR | NR | MITOMAP | MERRF |
| Benign | NR | LS | NR | ClinVar | |||||
| 8881 | T>C | Ser119Pro | R/Possibly benign | 0.002 | Patient with suspected mitochondrial disease (NR/NR) | NR | MITOMAP | SZ | |
| 8993 | T>G | Leu156Arg | Confirmed/Likely pathogenic | 0.012 | NARP/LS/MILS/other (+/+) | NR | MITOMAP | LS/NARP/MILS | |
| Pathogenic | NR | Mitochondrial complex V (ATP synthase) deficiency, mitochondrial type 1/LS/NARP/other | NR | ClinVar | |||||
| MT-CO3 | 9500 | C>T | Phe98 | NR/NA | 0.008 | NR | NR | MITOMAP | SZ |
| 9633 | T>C | Ser143Pro | R/Possibly benign | 0.000 | NR | NR | MITOMAP | PD | |
| Likely benign | NR | NR | NR | ClinVar | |||||
| 9699 | A>G | Ile165Val | NR/Possibly benign | 0.008 | NR | NR | MITOMAP | SZ | |
| Uncertain significance | NR | LS | NR | ClinVar | |||||
| 9861 | T>C | Phe219Leu | R/Possibly benign | 0.220 | AD (+/-) | NR | MITOMAP | AD | |
| Benign/Likely benign | NR | LS | NR | ClinVar | |||||
| 9956 | A>G | Leu250 | NR/NA | 0.006 | NR | NR | MITOMAP | SZ | |
| MT-ND4L | 10652 | T>C | Ile61 | R/NA | 0.104 | BD/MDD-associated (-/+) | NR | MITOMAP | BD/MDD |
| MT-ND4 | 10858 | T>C | Ile33 | NR/NA | 0.033 | NR | NR | MITOMAP | BD |
| 11778 | G>A | Arg340His | Confirmed/Possibly pathogenic | 0.357 | LHON/progressive dystonia (+/+) | NR | MITOMAP | LHON | |
| Pathogenic | NR | LHON | NR | ClinVar | |||||
| 12027 | T>C | Ile423Thr | R/Possibly benign | 0.004 | SZ-associated (NR/NR) | NR | MITOMAP | SZ | |
| MT-ND5 | 13094 | T>C | Val253Ala | Confirmed/Likely pathogenic | 0.002 | Ataxia + PEO/MELAS, LD, LHON, myoclonus, fatigue (+/+) | NR | MITOMAP | MELAS |
| Pathogenic | NR | Juvenile MELAS | NR | ClinVar | |||||
| 13513 | G>A | Asp393Asn | Confirmed/Likely pathogenic | 0.002 | LS/MELAS/LHON-MELAS overlap syndrome/negative association with carotid atherosclerosis (-/+) | NR | MITOMAP | MELAS/LS | |
| Pathogenic | NR | Mitochondrial diseases/LS/LS due to CID/Juvenile MELAS | NR | ClinVar | |||||
| MT-ND6 | 14668 | C>T | Met2 | R/NA | 3.951 | Depressive disorder-associated (+/-) | NR | MITOMAP | MDD |
| MT-TE | 14685 | G>A | (tRNA) | R/Likely pathogenic | 0.000 | Cataracts with spastic paraparesis & ataxia (-/+) | NR | MITOMAP | ECOAPP |
| 14709 | T>C | (tRNA) | Confirmed pathogenic | 0.000 | MM+DMDF/encephalomyopathy/dementia + diabetes + ophthalmoplegia (+/+) | NR | MITOMAP | Ataxia | |
| Pathogenic/Likely pathogenic | NR | Mitochondrial diseases/MI DMDF/Juvenile MELAS/MM | NR | ClinVar | |||||
| MT-CYB | 15043 | G>A | Gly99 | R/NA | 23.640 | MDD-associated/possible role in high-altitude sickness (+/-) | NR | MITOMAP | MDD |
| Likely pathogenic | NR | Familial breast cancer | NR | ClinVar | |||||
|
MT-HV1, MT-ATT, MT-CR, MT-7S |
16184 | C>T | Noncoding | NR/NA | 0.735 | NR | Colonic mucosa (-/+) | MITOMAP | PD |
| 16300 | A>G | Noncoding | R/NA | 0.536 | BD-associated (+/-) | Head/neck tumour (+/-) | MITOMAP | BD |
NA: not available; NR: not reported; R: reported.
AD: Alzheimer's disease; ADPD: Alzheimer's disease and Parkinson's; AHS: Alpers-Huttenlocher syndrome; ASD: autism spectrum disorder; BD: bipolar disorder; BS-LD: Barth syndrome-like disorder; CPEO: chronic progressive external ophthalmoplegia; CID: combined immunodeficiency; DMD: depressive mood disorder; DMDF: diabetes mellitus + deafness; ECOAPP: early-onset cataracts, ataxia and progressive paraparesis; DS: Down's syndrome; FSGS: focal segmental glomerulosclerosis; HiCM: histiocytoid cardiomyopathy; LAaCID: lactic acidosis and complex I deficiency; LD: learning disabilities; LHON: Leber's hereditary optic neuropathy; LoLS: late-onset Leigh syndrome; LoME: late-onset mitochondrial encephalomyopathy; LVNC: left ventricular non-compaction; ME: mitochondrial encephalopathy; MELAS: mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes; MERRF: myoclonic epilepsy with ragged red fibres; MDD: major depressive disorder; MIDD: maternally inherited diabetes and deafness; MILS: maternally inherited Leigh syndrome; MM: mitochondrial myopathy; NARP: neuropathy, ataxia, and retinitis pigmentosa; PEO: progressive external ophthalmoplegia; PD: Parkinson's disease; SNHL: sensorineural hearing loss; SZ: schizophrenia; VO2 max: maximum rate of oxygen consumption.
TOOLS: If available, predictive data of pathogenicity are obtained from the tools MitoTIP, HmtVar and/or APOGEE (https://www.mitomap.org/foswiki/bin/view////Main/SearchAllele); del: deletion; GB Freq: The frequency data derived from 51836 GenBank sequences with sizes greater than 15.4 kbp.
The frequency data and disease-associated phenotypes were retrieved from the MITOMAP and ClinVar databases in June 2021.
mtDNA analysis in MitD
We included 50 reports referring to MitD (Table 1). Figure 2 shows the variants reported in a varied number of phenotypes, with the most reported being MELAS (13 reports); MERRF (8 reports); LS (7 reports); KSS (5 reports); mitochondrial encephalomyopathy (ME) (6 reports) and PEO (4 reports); in addition to optic neuropathy, sensorineural hearing loss and diabetes mellitus type I; Leber hereditary optic neuropathy (LHON); early-onset cataracts, ataxia and progressive paraparesis; mitochondrial depletion syndrome; neuropathy, ataxia, retinitis pigmentosa and maternally inherited LS; sideroblastic anaemia; and Alpers-Huttenlocher syndrome (AHS). Among the 50 reports, eight reported variants in the nDNA that ultimately produced mtDNA alterations. Most of the studies were case reports; only 11 were carried out after 2010, and only one study analysed the three different types of mtDNA alterations: pSNV, mtDNA CN variations, and deletions. Seven studies analysed two types of alterations, and 41 investigated just one, mostly pSNV.
Figure 2.
Map of human mtDNA with variants (a) and deletions (b) identified in postmortem brain samples of patients with MitD. mtDNA replication initiates within the D-loop region and proceeds from the origin of heavy-strand replication (OH) until the origin of light-strand replication (OL). The positions of variants are represented by asterisks, while deletions are represented by circles. MitD diagnoses are indicated in boldface. MELAS: mitochondrial encephalopathy, lactic acidosis and stroke-like episodes; ME: mitochondrial encephalomyopathy; AHS: Alpers-Huttenlocher syndrome; MERRF: myoclonic epilepsy with ragged-red fibres; NARP: neuropathy with ataxia and retinitis pigmentosa; MILS: maternally inherited Leigh's syndrome; LHON: Leber hereditary optic neuropathy; EOCAPP: early-onset cataracts, ataxia and progressive paraparesis; POLG: DNA polymerase gamma gene; KSS: Kearns-Sayre syndrome. MT-: mitochondrially encoded gene; TL1: tRNA-Leu 1; TW: tRNA-Trp; CO2: cytochrome c oxidase II; TK: tRNA-Lys; ATP6: ATP synthase subunit 6; ND4: NADH-ubiquinone oxidoreductase subunit 4; TE: tRNA-Glu; CO1: cytochrome c oxidase I.
mtDNA analyses in MELAS
The m.3243A>G variant in the MT-TL1 gene coding for tRNA-Leu is the most reported variant in postmortem brain samples of patients with MELAS,54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64 although one study also identified the presence of m.13513G>A, p. Asp393Asn, located in the MT-ND5 gene.65 All the studies except one60 reported mutation loads greater than 70% in the brain and similar mutation loads in other evaluated tissues.54,55,57, 58, 59,61, 62, 63, 64, 65, 66 However, some discrepancies were also observed—while some studies reported that mutation loads did not vary between brain regions,55 others reported high mutation load variability between different cells of the same region.57 None of the studies on MELAS analysed mtDNA CN or the presence of mtDNA deletions in the brain. The most frequent m.3243A>G variant associated with MELAS was also present in the brain of a 4.5-year-old child with a lethal MitD, with a Barth syndrome-like presentation. This child showed the m.3243A>G variant in all of the analysed tissues, including blood, skeletal muscle, cardiac muscle, and liver. Additionally, in the peripheral blood mtDNA of the mother, as well as in four of the 5 siblings, heteroplasmy percentages were not reported.67 m.3243A>G was also detected in a patient with optic neuropathy, sensorineural hearing loss and diabetes mellitus type 1, but not MELAS, with a mutation load greater than 75% in white and grey matter, putamen, caudate, pons, visual cortex, among other brain areas and 60% in the biceps muscle.68
mtDNA analyses in MERRF
The m.8344A>G variant in the MT-TK gene coding for tRNA-Lys was reported in all eight studies evaluating postmortem brain samples of patients with MERRF.55,56,69, 70, 71, 72, 73, 74 Moreover, one of these studies also reported m.8603T>C, p.Phe26Ser, in the MT-ATP6 gene and m.3257A>G in MT-TL1.72 Interestingly, two distinct reports from 1995 and 2010 using distinct molecular techniques that respectively evaluated an 18- and a 16-year-old patient with MERRF syndrome found similar percentages of the m.8344A>G variant that ranged between 93% and 97% across different brain regions. Both patients also showed similar heteroplasmy percentages in other tissues.69,71 Notably, one of the studies reporting the m.8344A>G variant also described a 3.7-fold increased mtDNA CN in brain-affected tissues compared to nonaffected tissues.69
mtDNA analyses in LS
Nine studies reported postmortem brain mtDNA data in LS.56,75, 76, 77, 78, 79, 80, 81, 82 The m.8993T>G, p.Leu156Arg in the MT-ATP6 gene was reported in four LS studies with mutation load percentages in the brain between 89% and 97%, and similar percentages in other tissues were lower than those in blood (72% and 81%).77,78,80,81 In addition, m.13513G>A, p.Asp393Asn, MT-ND5 was also reported in LS.76 Only one study analysed the mtDNA CN, identifying that it was 4.6 times higher in patients with LS than in controls.82
mtDNA analysis in NeuD
Table 2 includes 67 reports referring to NeuD, and Figure 2 shows the variants reported in neurodegenerative conditions, with the most reported variants being found in Alzheimer's disease (AD, 33 reports) and Parkinson's disease (PD, 27 reports). Other reported phenotypes were amyotrophic lateral sclerosis, mild cognitive impairment, Creutzfeldt-Jakob disease, dementia with Lewy bodies, frontotemporal dementia, mesial temporal lobe epilepsy-hippocampal sclerosis, Down syndrome, Down syndrome and dementia; multiple sclerosis, infantile onset spinocerebellar ataxia, motor neuron disease, multiple system atrophy, disseminated neocortical and subcortical encephalopathy, and Huntington disease. Taking all NeuD studies into account, the alterations most frequently investigated were mtDNA deletions (40 reports), followed by the presence of mtDNA variants (22 reports) and mtDNA CN (19 reports) (Figure 3).
Figure 3.
Map of human mtDNA with variants (a) and deletions (b) identified in postmortem brain samples of patients with NeuD. mtDNA replication initiates within the D-loop region and proceeds from the origin of heavy-strand replication (OH) until the origin of light-strand replication (OL). The positions of variants are represented by asterisks, while deletions are represented by circles. NeuD diagnoses are indicated in boldface. AD: Alzheimer's disease; PD: Parkinson's disease. MT-: mitochondrially encoded gene; TL1: tRNA-Leu 1; ND2: NADH-ubiquinone oxidoreductase subunit 2; ND5: NADH-ubiquinone oxidoreductase subunit 5; ATP8: ATP synthase subunit 8.
mtDNA analyses in AD
Among the AD studies reviewed, deletions were the most frequent mtDNA alterations analysed (17/33), followed by mtDNA variants (10/33) and mtDNA CN (9/33). The most frequently assessed brain tissues were the frontal cortex, hippocampus, cerebellum, temporal cortex and parietal cortex.
The results in relation to the variants are diverse; some studies agreed that the frequency of variants is similar between AD patients and controls regarding their levels of heteroplasmy,83, 84, 85 while others reported higher levels of heteroplasmy and a higher frequency of variants in the parietal cortex, hippocampus and cerebellum in AD patients.86,87 Two studies reported the m.5460G>A variant, which produces the p.Ala331Thr amino acid change in MT-ND2; this is described in MitoMap with conflicting reports regarding its pathogenicity for AD, PD and LHON. This amino acid change was reported in AD and control individuals.88 Additionally, in six patients with AD, four showed homoplasmy and two showed heteroplasmy, with a mutation load percentage of 5%. In this last study, the variant was not present in the control group.89 Similarly, the m.3243A>G variant involved in MELAS was identified in a patient with AD, although with a very low percentage (<0.05%).90 More recent studies, some of them conducted with a large number of individuals and using novel techniques, did not identify that mtDNA variation had a role in AD.83, 84, 85
Overall, most of the studies agree that the mtDNA CN levels are lower in AD patients than in controls,83,84,86,91, 92, 93, 94 although some specific hallmarks should be mentioned: 1) no difference was identified in the mtDNA CN levels between tau-positive and tau-negative neurons;95 2) focusing on brain regions, the hippocampus and the temporal cortex showed a significant mtDNA CN reduction in pyramidal neurons compared to other neuronal cells93 but not in the cerebellum,83 although a study analysing a large number of samples (282 patients and 461 control subjects) mostly obtained from the cerebellum (87.3%) was able to identify a significant reduction in the mtDNA CN levels in AD;84 3) when considering the clinical characteristics of the patients, one study observed that the mtDNA CN was reduced by 48% in nondiabetic patients with AD compared to that in nondiabetic noncognitive-impaired individuals, and this effect occurred in the parietal cortex but not in the frontal cortex or cerebellum; however, compared with nondiabetic patients, diabetic patients showed higher mtDNA CNs in the frontal cortex, parietal cortex and cerebellum;91 and 4) although a reduced mtDNA CN was reported by most of the studies, some authors highlighted that some patients with AD exhibited a high mtDNA CN.92
Regarding mtDNA rearrangements, the first studies carried out in the nineties focused on the analysis of the 4977 bp common deletion (m.8470_13477del). Similar percentages of the deletion have been reported in individuals with AD and in control individuals.87,96,97 However, the deletion was found to be more abundant in the temporal cortex of individuals with AD than in controls, although in both cases the percentage was low (<0.059%).98 Regarding the mutation load of the common deletion in the distinct brain regions, there are some aspects to note. First, higher percentages were present in the nucleus caudate than in the gyrus frontalis.96 Second, in a more recent study, a 1.5% mutation load was reported in the frontal cortex samples of patients with AD and in control individuals and, although the percentage was still low, it was higher than previously reported.85 Third, the common deletion was also present in the substantia nigra and the globus pallidus of a single individual with AD but was not detected in the blood.90 Finally, the mutation load was found to be 15 to 25-fold lower in the cerebellum than in cortices of AD and control individuals. More recent studies have investigated the presence of other rearrangements in addition to the common deletion, with controversial results. Some agree that the number of deletions is higher in AD than in controls,85,99 but they differ depending on the region studied. For instance, one of the studies did not find any significant increase in the total number of deletions in the hippocampus of patients with AD,100 while others observed higher deletion rates in both astrocytes and microglia of the hippocampus compared with the brainstem and cerebellum in control individuals.101 Additionally, a high number of deletions in cytochrome oxidase (COX)-deficient neurons of the hippocampus102 and in frontal cortices of individuals with AD85 have been reported, but there were no differences between tau-positive and tau-negative neurons.95 Reportedly, the average mtDNA deletion size is 5080±2367 bp,103 the deletion size ranges from 3670 to 6088 bp, accumulation occurs with age,102 and the number and characteristics of the rearrangements depend on the sequence coverage depth.85
mtDNA analyses in PD
mtDNA deletions were the most investigated mtDNA alterations (19/27), followed by mtDNA variants (10/27) and the CN (10/27). The brain tissue most frequently assessed was the substantia nigra, followed by the frontal cortex, cerebellum, putamen and hippocampus. mtDNA variants and heteroplasmy levels have been widely discussed regarding PD. The first study investigating mtDNA variants in PD reported no differences between variant heteroplasmy levels in PD and control individuals;104 this was also found by two additional recent studies.84,105 Another recent study investigating a large number of samples and taking advantage of next-generation sequencing found that the mtDNA mutation load in MT-CO1, MT-CO2 and MT-CYB in the substantia nigra pars compacta and in MT-CYB in the frontal cortices of patients with PD was increased compared to those in control individuals.106 A significant increase in the frequency of heteroplasmy levels in neurons with a higher number of deletions has also been reported.103 Conversely, no association was found between mtDNA variants, either homoplasmic or heteroplasmic, and PD.84
Conflicting results have also been reported regarding mtDNA CN in PD, such as 1) decreased levels in the prefrontal cortices of patients with PD and dementia and in neurons of the substantia nigra in patients with idiopathic PD;107, 108, 109 2) increased levels in cholinergic neurons of the pedunculopontine nucleus in PD patients;110 and 3) no significant differences between patients and controls.84,95,104,105,108,111
In general, patients with PD showed higher mtDNA deletion levels in the substantia nigra than in other brain regions,100,105,111,112 although some studies found a significant increase in other regions, such as the putamen or hippocampus, but not in the substantia nigra.113,114 Additionally, although most reports showed a trend or reported increased mtDNA deletion levels in PD, others did not.109,115 Notably, mtDNA deletion levels have been reported to be significantly lower in the cerebellum than in other brain regions.112,116 One of the most recent studies found that patients with PD tended to exhibit deletion levels greater than 60% in the substantia nigra, while markedly lower levels were found in the frontal cortex, cerebellum and putamen. The authors also reported that in the substantia nigra, deletion levels were a good predictor of mtDNA CN variation.105 This is in line with a reported correlation between the percentage of deletions and mtDNA CN in the putamen; the lower the CN, the higher the deletion levels.113 Some specificities regarding mtDNA deletions in PD should also be mentioned. First, increased mtDNA deletion levels were found to correlate with Braak staging,108 although these changes were not explored in other studies.109,117 Second, a few studies using laser microdissection techniques to focus on specific cell types identified 1) a significant increase in deletion levels in cholinergic neurons of the pedunculopontine nucleus;110 2) increased deletions in the dopaminergic neurons of the substantia nigra;111 3) increased deletions in the Lewy-positive neurons of the substantia nigra compared with those in Lewy-negative neurons or neurons from control individuals;95 and 4); the common deletion was also identified as the most frequent deletion in the substantia nigra of microdissected neurons.103
mtDNA analysis in PsyD
Table 3 includes 19 reports referring to PsyD, and Figure 4 shows the variants reported in various phenotypes, with the most reported being schizophrenia (SZ, N=13) and bipolar disorder (BD, N=13), followed by major depressive disorder (MDD, N=8). Other phenotypes assessed included subjects with alcohol/drug abuse and other psychiatric symptoms (ADO), suicide victims, and autism spectrum disorder (ASD). The most recurrent brain region explored was the dorsolateral prefrontal cortex (DLPFC), but many others were included in some studies. Eleven studies investigated the presence of mtDNA rearrangements, eight studies examined mtDNA CN and seven studies focused on mtDNA variants.
Figure 4.
Map of human mtDNA with variants (a) and deletions (b) identified in postmortem brain samples of patients with PsyD. mtDNA replication initiates within the D-loop region and proceeds from the origin of heavy-strand replication (OH) until the origin of light-strand replication (OL). The positions of variants are represented by asterisks, while deletions are represented by circles. PsyD diagnoses are indicated in boldface. SZ: schizophrenia; ASD: autism spectrum disorder; MDD: major depressive disorder; BD: bipolar disorder; ADO: alcohol/drug abuse and other psychiatric symptoms. MT-: mitochondrially encoded gene; RNR1: 12S rRNA; RNR2: 16S rRNA; ND4L: NADH-ubiquinone oxidoreductase subunit 4L; D-loop: displacement loop; ND4: NADH-ubiquinone oxidoreductase subunit 4; ND6: NADH-ubiquinone oxidoreductase subunit 6; CYB: cytochrome b; CO1: cytochrome c oxidase I; TL1: tRNA-Leu 1; ND2: NADH-ubiquinone oxidoreductase subunit 2; CO2: cytochrome c oxidase II; HSP1: major H-strand promoter 1; ATP8: ATP synthase subunit 8.
mtDNA analyses in SZ
A study that focused on mtDNA variants identified a total of 142 rare variants, with a minor allele frequency less than 1% based on mtDB118 and PhyloTree,45 but a relevant role in SZ was not identified.119 Similarly, in the DLPFC, a 22% higher rate of synonymous variants and an increased number of mtDNA substitutions were found in SZ patients compared with control individuals, but again, there was no major involvement in the diagnosis.120 Other rare variants (frequency <0.02%), such as m.6617C>T p.Phe238 (MT-CO1), m.8881T>C p.Ser119Pro (MT-ATP6), and m.9500C>T p.Phe98, m.9699A>G p.Ile165Val and m.9956A>G p.Leu250 (MT-CO3) were identified only in patients with SZ.121 Finally, two studies reported results regarding specific variants. The m.3243A>G variant was detected in one patient with SZ with a 60% mutation load,122 and the m.12027T>C, p.Ile423Thr (MT-ND4) was found to have no prominent effect on SZ.123
mtDNA deletions were detected in two patients with SZ, m.2973_15573del (MT-RNR2–MT-CYB) and m.1148_15607del (MT-RNR1–MT-CYB), with deletion read percentages of 31.8% and 16.8%, respectively.124 Regarding the common deletion, most studies did not observe differences between SZ patients and control individuals,96,119,125 but it has also been reported that the deletion levels highly differed among the 11 brain regions analysed, increased with age, and showed little change in blood samples.119 Additionally, one study showed a significant decrease in the accumulation of the common deletion in patients with SZ compared to MDD, BD and control subjects, mostly in dopaminergic regions.126
mtDNA analyses in BD
The synonymous variant m.10858T>C in the MT-ND4 gene was identified in a patient with BD,119 and the m.3243A>G variant was present in two patients with a low variant frequency of 0.90%-1%.122
The mtDNA CN was lower in 32 patients with BD than in 32 control individuals, and no differences were observed between subjects who died from suicide and those who did not,127 which was not observed in a previous study.128 On the other hand, high mtDNA CNs in postmortem hippocampal tissue from 47 BD patients have been described.125
Two of 10 patients showed m.5897_15840del (MT-TY/MT-CO1–MT-CYB) and m.467_14122del (D-LOOP–MT-ND5) with deletion percentages of 23.0% and 5.0%, respectively. The authors reported that patients with SZ and BD had a higher cumulative deletion ratio in the DLPFC/anterior cingulate cortex (ACCtx) than those in the MDD and ADO diagnostic groups.124 The ratio of the common deletion was significantly higher in patients with BD than in control individuals in the DLPFC119 and in the cerebellum,129 with a significant association between increased levels and advanced age.119 This age association was previously observed but was unrelated to the BD diagnosis.130 On the other hand, no significant accumulation of the common mtDNA deletion across distinct brain regions of patients with BD compared to control subjects has been reported.125
mtDNA analyses in MDD
The m.224T>C variant in the H-strand replication origin (OH) located in the D-loop region was present in one of five evaluated patients with MDD, and the m.10652T>C homoplasmic variant (which does not alter the amino acid Ile61) in the MT-ND4L gene was previously reported in another patient.120 Finally, the m.3243A>G MELAS variant identified in patients with BD was not present in 15 patients with MDD.122
Interestingly, the m.1243_15340del (MT-RNR1–MT-CYB) deletion with a high mutation load of 90.1% in the DLPFC and 85% in the ACCtx was suggested to considerably impact a 75-year-old male who had MDD and diabetes mellitus.124 Furthermore, another 46-year-old patient with MDD who committed suicide also exhibited four high-impact deletions: m.7863_15617del (MT-CO2–MT-CYB), m.7816_14807del (MT-CO2–MT-CYB), m.870_14774del (MT-RNR1-MT-CYB), and m.6468_15600del (MT-CO1–MT-CYB), with read percentages of 52.4%, 26.5%, 10.2% and 8.5%, respectively. Even though deletion mutation loads were lower than those of the former patient, the cumulative mutation load was very high. In the latter patient, five brain regions and blood samples were explored, and deletions were detected only in the caudate nucleus. Interestingly, some deletions were clonally expanded to some brain regions, while they were absent in others.124 Additionally, this study 1) did not find significant differences in the cumulative mtDNA deletion mutation load in the DLPFC or ACCtx across disorders; 2) stated that only 12 of the 30 most frequent deletions identified were previously described, 14 of which were detected only in the brain and not in other tissues from the same subjects; and 3) found that the brain contained significantly more deletions than the blood.124
mtDNA in other clinical conditions
Table 4 shows the mtDNA alterations explored in a few studies regarding other phenotypes: 1) herpes simplex virus type-1 encephalitis, 2) human immunodeficiency virus infection with or without methamphetamine use, 3) diabetes and recurrent stroke-like episodes, seizures and cognitive decline and 4) deceased neonates, newborns and infants and 5) control individuals. The most notable finding is that the common deletion was present in brain samples from stillborn individuals.131
mtDNA analyses in ageing
Table 5 presents 29 studies that analysed mtDNA in postmortem brain samples; these studies support mtDNA involvement in ageing.
mtDNA variations
Variants in the D-loop have been significantly associated with age, although when they were weighted by their heteroplasmic levels, the association was lost.132 Additionally, the overall heteroplasmy level in the D-loop was found to be higher in older individuals than in younger individuals.133 It has been reported that 78% of the accumulated variants were nonsynonymous and more deleterious in older individuals.24 In addition, a high aggregation of somatic point variants in the tRNAs for Thr and Pro, portions of the MT-CYB gene, and the D-loop region were detected in neurons of the elderly.134 The ageing process is inextricably associated with neurodegeneration. In this sense, variants in the regulatory control region were found to be increased with age in AD and Down syndrome and dementia.86 Similarly, 23 missense variants (8 of them causing nonconservative amino acid replacements at evolutionarily constrained sites), 2 tRNAs and one nonsense polymorphism were detected in the substantia nigra of elderly nonparkinsonian and idiopathic PD patients.135 Moreover, the accumulation of G>C to T>A and T>A to G>C transversions was found to increase with age in the frontal cortex of patients with PD.136
mtDNA CN
Most of the studies found that the mtDNA content in the frontal cortex decreased with age.86,132 However, in the substantia nigra, it was recently reported to be increased with age, allegedly to maintain the pool of wild-type mtDNA despite accumulating deletions.105 Additionally, no significant age-dependent increase in mtDNA CN among three age groups (0–30, 31–59 and >60 years) was identified.137
mtDNA rearrangements
Most of the studies on ageing focused on the common deletion in the brain, and they reported that the accumulation of the common deletion was associated with increasing age.108,116,132,138, 139, 140, 141, 142 According to Cortopassi et al., deletions in foetal tissues were estimated to be 1/100 to 1/100,000 times less than those in adults,143 while Soong et al. reported that the common deletion level was detected in neonatal brain regions as 4/10,000.144 The distribution of the common deletion varies among different parts of the brain, with the highest and the lowest levels reported in the substantia nigra and cerebellum, respectively.138 Similarly, the common deletion ratio has been reported to reach the highest levels in the basal ganglia with age and the lowest levels in the cerebellum without any age-related association.139 Additionally, a significant increase in the common deletion ratio was detected in the cortex and putamen with increasing age.22 Regarding clinical conditions associated with age, some PD studies also showed that the accumulation of the common deletion was more likely to be an age-related phenomenon rather than the pathogenic condition,116,117,135,139,145,146 and in AD, the common deletion levels were much lower in younger patients than in older patients.147 Similarly, small insertions and deletions were found to be significantly increased in aged individuals among controls, early- and late-onset AD patients, and SZ patients.133 On the other hand, no significant increase in common deletion with age in AD has been reported.87 Major arc deletions showed a significant positive correlation with age in nigral neurons, while the level of mtDNA deletions was commonly detected at low levels and did not increase with age in frontal neurons and Purkinje cells.105
Other mtDNA deletions have been investigated in relation to age. The accumulation of the 7436 bp deletion22,140,142 and a unique 12 kb deletion (m.1989_14366del) were also age-related in brain samples.148 In addition, several multiple deletions (4.5–7.1 kb),135 including m.7409_13687del,145 have also been reported in the substantia nigra of both aged patients with PD and controls. Apart from these findings, one study showed that mtDNA deletions were associated with chronic hypoxia conditions rather than ageing in the samples of patients with PsyD.149
mtDNA technical issues
Historically, studies exploring mtDNA variants often used radiolabelled nucleotides, primers or probes for PCR, sequencing or Southern blot techniques, while most recent studies have used exome or genome sequencing. The mtDNA CN can be assessed by quantitative real-time PCR (qPCR). Some studies explored just one region, while others investigated mtDNA alterations in distinct brain regions, in homogenate tissues, or in laser-captured single cells. Although most of the studies used molecular techniques focused on mtDNA sequences, others were based on obtaining mtDNA. In this case, the first and crucial step of mtDNA analysis is effective extraction. Phenol-chloroform DNA extraction, which isolates both nDNA and mtDNA, is a widely used method. Devall et al. performed the first systematic comparison of the effectiveness of five different mtDNA isolation protocols from frozen postmortem brain tissue.150 They reported that linear DNA digestion that leaves circular DNA (mtDNA) intact gave the lowest purity (mtDNA/nDNA), while the magnetic isolation of mitochondria using anti-human TOM22-labeled microbeads to isolate mitochondria gave the highest mtDNA enrichment.150
Although PCR-based technologies have accelerated the analysis of mtDNA deletions, their effectiveness can vary.151 Distinct results were obtained when comparing the serial dilution PCR method (in which total DNA is diluted and amplified by primers spanning the common deletion) and the kinetic PCR method (based on removing reaction tubes from 10 to 20 cycles for undeleted mtDNA and stopping reactions from 22 to 32 cycles for deleted mtDNA to obtain the ratio of deleted mtDNA to normal mtDNA). According to their serial dilution PCR results, the caudate had 10 times more deleted mtDNA than the parietal cortex, while kinetic PCR resulted in a lower difference.151 Taylor et al. used a digital deletion detection (3D) assay for absolute quantification and characterization of rare mtDNA deletions in aged human brain samples.23 This technique involves an enrichment step for deleted molecules by wild-type targeted endonucleolytic digestion, the amplification of intact mutant molecules by target-specific TaqMan probes in water-in-oil droplets, and, finally, a quantification step for chambers carrying many droplets representing thousands of single-molecule reactions.23
As an alternative to standard PCR techniques, Marquis et al. developed a novel sensitive mtDNA assay that used rolling circle amplification and sequencing (MitoRS) to detect mtDNA variants and their heteroplasmy level with high accuracy.152 In the first step, they used Phi29 polymerase (with a low error rate and strong strand displacement activity) to generate several individual mtDNA copies (mtDNA enrichment) that were not species-specific and were insensitive to nuclear mtDNA sequences and to mtDNA polymorphism priming events. Combined with high-throughput tagmentation-based library generation for next-generation sequencing (NGS), they could quantify mtDNA SNVs at the minimum 1% frequency level.152
Some additional difficulties have been reported in mtDNA analyses. The chromogen 3,3′-diaminobenzidine, a standard stain for COX activity, has a strong inhibitory effect on qPCR, thus causing significant bias in the estimation of mtDNA CN and deletion levels between COX-positive and COX-negative neurons.153 Regarding methylation, Devall et al. developed an assay to identify differentially methylated regions in mtDNA among different regions of the cortex and cerebellum by using pre-existing methylated DNA immunoprecipitation sequencing data. Interestingly, they identified 74 nominally differentially methylated regions in the mtDNA and 8 differentially methylated regions between the total cortex and cerebellum.154
Discussion
Biological processes are defined not only by cell structures but also by energy status. The brain represents between 2 and 3% of the weight of our body while consuming 20% of the total energy, which is mainly generated in the mitochondria. Unlike muscle, the brain is always highly metabolically active and thus is highly sensitive to mitochondrial functioning.36,155 For this reason, the mitochondria operate under several control mechanisms, such as mitochondrial fusion and fission, the removal of damaged proteins, and mitophagy.5 Recently, it has been suggested that if the available energy is limited, remaining below the bioenergetic threshold, neurological symptoms may appear; however, if the bioenergetic defect is subtler, the lack of energy can lead to the appearance of psychiatric symptoms.16
We collected mtDNA variants, CN and/or deletions reported in postmortem human brain samples. mtDNA variants and deletions can be inherited or occur at the germline or somatic level and have been associated with clinical and nonclinical conditions, while mtDNA CN is a proxy measure for mitochondrial function that has been associated with ageing-related diseases.156 Multiple mtDNA deletions and duplications can arise due to the accumulation of multiple errors in postmitotic tissues, often with clonal expansion of one particular mtDNA form, or be attributed to variants in nuclear genes involved in mtDNA maintenance and repair.36 Some MitD syndromes arise as a result of a sporadic large-scale single deletion that is the only deletion present. This single deletion can be of any size but there is a common deletion of 4.9kb. Notably, none of the studies in this review reported mtDNA duplications.
This review identified that many pSNVs are present in high heteroplasmy levels (generally >80%) in the postmortem brains of patients with MitD, although with variable heteroplasmy levels across brain regions and nonbrain tissues. However, the clinical condition characterized by early-onset cataracts, ataxia and progressive paraparesis showed mutation loads less than 60% in the affected tissues.66 Some pSNVs may impact the mtDNA CN in the brain, either decreasing68 or increasing69 its levels, but few studies have analysed both mtDNA alterations. A low mtDNA CN was reported in most of the studies,68,157, 158, 159 but not all,160 and mtDNA deletions have only been investigated in KSS and DNA polymerase gamma gene (POLG) encephalopathy.157,158,160,161 Only one study investigated the three types of mtDNA alterations in patients affected by POLG encephalomyopathy, and interestingly, the three types were present.157 All-encompassing mtDNA analyses were not performed in most of the reviewed studies and should be encouraged in prospective studies.
The current data from postmortem human brain samples indicate that pSNVs do not have a prominent role in NeuD or PsyD, and a reduced mtDNA CN has been extensively observed in NeuD but not PsyD. Conversely, mtDNA deletions may have a more prominent role in PsyD. This would be in accordance with the hypothesis that MitD may be underdiagnosed in some patients with PsyD162 and that bioenergetic defects can lead to the appearance of psychiatric symptoms.3,16 Furthermore, NeuD and PsyD can be accompanied by metabolic disorders that are often not considered because they are not the target of the studies. Regarding this issue, the parietal cortices of diabetic individuals showed a 48% reduction in the mtDNA CN compared with those of nondiabetic individuals.91 In nondiabetic AD subjects, the loss of mtDNA could lead to the loss of mitochondrial mass and bioenergetic capacity, whereas in diabetic AD subjects, an increased nutrient supply due to insulin resistance and hyperglycaemia could result in reduced oxidative phosphorylation and increased glycolysis, which would also lead to an energy deficit. In line with this idea, in one of the evaluated studies, a high-impact deletion was present in a patient with MDD who also had diabetes.124 Future studies should include all the phenotypic characteristics of the assessed individuals to shed light on the role of mtDNA alterations in other comorbid traits. Notably, in NeuD and PsyD, many of the early studies on human postmortem brain samples were based on the comparison of frequencies and heteroplasmy levels of specific SNV between patients and controls, and even though some reported significant differences, these results have not been confirmed in a more recent and larger dataset, which advocates that mtDNA CN rather than mtDNA SNV would have more clinical relevance in NeuD.84 However, the association of homoplasmic common and rare SNV on longevity and NeuD such as multiple sclerosis has been recently confirmed by analysing a large number of blood samples.32 An interesting study that also investigated blood samples demonstrates the involvement of the nuclear female genome in the evolution of human mtDNA variation and also suggests the different values of heteroplasmy that an individual cell, tissue or organism may exhibit during embryonic development of human germ cells.163 The different expansion of heteroplasmic variants during the ageing process could have consequences for age-associated diseases such as NeuD but also PsyD, which present in many cases within certain age ranges. However, the biological processes associated with ageing have mostly been studied in relation to NeuD.5
Different methodological issues may have interfered with the results reported in this systematic review. These include, among others, the origin of the samples and the way the DNA was obtained and stored, as some reagents and degraded samples considerably limit the PCR/qPCR technique.164 In addition, PCR-based techniques may favour the amplification of short (deleted) versus long (nondeleted) fragments resulting in inadequate detection of mtDNA fragments. NGS methodology based on previous long-range amplification is the most powerful tool to detect mtDNA alterations. It provides high and uniform coverage of each of the 16,569 bases, allowing the detection of nucleotide changes, as well as heteroplasmy levels. Moreover, it also allows the detection of small insertions/deletions (indels) and large deletions and the mapping of exact deletion breakpoints.165 Several bioinformatic tools allow a reliable analysis of large mtDNA sequences with high coverage levels for detecting mtDNA alterations.122, 166, 167, 168, 169 Additionally, current genome and exome sequencing techniques can detect heteroplasmy levels at less than 5%, and it has been reported that heteroplasmy levels depend on the coverage and the number of sequence reads. Moreover, these techniques have also been reported to be more accurate for mtDNA CN quantification than the gold standard qPCR technique.169 In any case, even with the NGS technique, it is necessary to take into account the initial enrichment strategy used and the reference used in the downstream bioinformatic analysis, as both may influence the accurate detection and quantification of mtDNA heteroplasmy levels.170
A broad understanding of brain regional variation regarding heteroplasmy levels of pSNVs, mtDNA CN and mtDNA deletions is necessary for understanding the metabolic requirements of different regions of the brain, which can improve our understanding of region-specific cell type changes and/or vulnerability to metabolic insults and related neuropathological processes. Highly automated and sensitive tools to evaluate mtDNA alterations from data obtained through exome or genome sequencing are currently available,124,166,167,171,172 and larger datasets should be assessed for mtDNA analyses, along with a full phenotypic characterization to better associate molecular mtDNA defects or variations with biochemical, metabolic and clinical or health aspects. This is supported by the recent success in identifying associations between a large number of phenotypes and homoplasmic mtDNA variants by analysing a large number of blood samples from the UK Biobank.32 In summary, most studies that explored mtDNA alterations in neurological diseases, mental illnesses and the natural ageing process have reported conflicting results. Some reasons for this are that different studies have investigated different mtDNA alterations, different diagnoses and/or different brain regions. This has made it difficult to draw conclusions. In addition, methodological issues mainly related to the techniques used but also to sample collection have led to difficulties in data interpretation. More studies are needed to identify the specific mtDNA alterations associated with health and disease. Nevertheless, the findings discussed in this systematic review argue for the involvement of mtDNA in brain disorders.
Limitations
Many of the sample sizes used to study mtDNA CN were underpowered. As an example, differences in mtDNA CN between patients with AD and control individuals were not often identified in the cerebellum58,59 until a larger number of patients and controls were screened.62 The techniques used in early mitochondrial genetics studies are not comparable to more recent technologies. It would therefore be interesting if some of the phenotypes that were analysed at the time of identifying the first mtDNA alterations could be analysed with the new technologies, especially to confirm the levels of heteroplasmy that were indicated at the time.
Conclusions
This study provides a comprehensive summary of the mtDNA alterations reported in human brain samples. The results identified in relation to MitD provide a clear idea of which genetic alteration is involved in each disorder, despite the great heterogeneity of the alterations described. Unfortunately, this is not the picture for NeuD and PsyD, where the findings are often contradictory. While mtDNA alterations have pathological implications in MitD, for most of the alterations identified in NeuD and PsyD an association has been suggested, but the pathological consequences are not yet proven. Low mtDNA CN is the most reported mtDNA alteration in NeuD, and specific mtDNA deletions may have prominent consequences for PsyD. There is also strong and abundant evidence that the ageing process is related to neurodegeneration and the loss of mtDNA integrity. Future research directions should include mtDNA analyses in larger samples and concurrent analyses of all types of mtDNA alterations and in several brain regions. Additionally, genotype-phenotype correlation studies will help to advance our understanding of the underlying molecular mechanisms and the implications of mtDNA variability in health and disease.
Declaration of interests
The authors declare that they have no conflicts of interest.
Acknowledgments
Contributors
Conceptualization and design, L.M.; Methodology, writing and editing, A.V.-P., J.T., B.K.B., and L.M.; Writing and editing, E.V., G.G., and G.M.; Acquisition and verification of reported data, A.V.-P., J.T., B.K.B. and L.M. All authors contributed to the interpretation of the findings, read and approved the final version of the manuscript, had full access to all the data and final responsibility for the decision to submit for publication.
Acknowledgments
This work was supported by the Instituto de Salud Carlos III fellowship PI18/00514 to Lourdes Martorell and Gerard Muntané and by the IISPV-Cerca Programme of the Generalitat de Catalunya, cofounded by FEDER. Alba Valiente-Pallejà was the recipient of a Talent-IISPV fellowship from the Diputació de Tarragona. Juan Tortajada and Bengisu Bulduk were the recipients of an industrial doctorate (DI-21) and a grant for the recruitment of new research staff (FI-DGR-2020) respectively, from the Generalitat de Catalunya. Figures showing mtDNA variants and deletions in the human mtDNA map were created with BioRender.com.
Data sharing statement
The data collected for this study can be provided upon reasonable request.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103815.
Appendix. Supplementary materials
References
- 1.Attwell D, Laughlin SB. An energy budget for signaling in the grey matter of the brain. J Cereb Blood Flow Metab. 2001;21:1133–1145. doi: 10.1097/00004647-200110000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Lax NZ, Gorman GS, Turnbull DM. Review: Central nervous system involvement in mitochondrial disease. Neuropathol Appl Neurobiol. 2017;43:102–118. doi: 10.1111/nan.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pei L, Wallace DC. Mitochondrial etiology of neuropsychiatric disorders. Biol Psychiatry. 2018;83:722–730. doi: 10.1016/j.biopsych.2017.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lujan SA, Longley MJ, Humble MH, et al. Ultrasensitive deletion detection links mitochondrial DNA replication, disease, and aging. Genome Biol. 2020;21:248. doi: 10.1186/s13059-020-02138-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou Y, Dan X, Babbar M, et al. Ageing as a risk factor for neurodegenerative disease. Nat Rev Neurol. 2019;15:565–581. doi: 10.1038/s41582-019-0244-7. [DOI] [PubMed] [Google Scholar]
- 6.Spinelli JB, Haigis MC. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol. 2018;20:745–754. doi: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Formosa LE, Ryan MT. Mitochondrial OXPHOS complex assembly lines. Nat Cell Biol. 2018;20:511–513. doi: 10.1038/s41556-018-0098-z. [DOI] [PubMed] [Google Scholar]
- 8.Meyer A, Laverny G, Bernardi L, et al. Mitochondria: An organelle of bacterial origin controlling inflammation. Front Immunol. 2018;9:536. doi: 10.3389/fimmu.2018.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Hattab AW, Scaglia F. Mitochondrial cytopathies. Cell Calcium. 2016;60:199–206. doi: 10.1016/j.ceca.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 10.El-Hattab AW, Craigen WJ, Scaglia F. Mitochondrial DNA maintenance defects. Biochim Biophys Acta Mol Basis Dis. 2017;1863:1539–1555. doi: 10.1016/j.bbadis.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 11.Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci U S A. 1980;77:6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 13.Schapira AH. Mitochondrial disease. Lancet. 2006;368:70–82. doi: 10.1016/S0140-6736(06)68970-8. [DOI] [PubMed] [Google Scholar]
- 14.Keogh MJ, Chinnery PF. Mitochondrial DNA mutations in neurodegeneration. Biochim Biophys Acta. 2015;1847:1401–1411. doi: 10.1016/j.bbabio.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 15.Wang L. Mitochondrial purine and pyrimidine metabolism and beyond. Vol. 35. Nucleosides Nucleotides Nucleic Acids. 2016;35:578–594. doi: 10.1080/15257770.2015.1125001. [DOI] [PubMed] [Google Scholar]
- 16.Wallace DC. A mitochondrial etiology of neuropsychiatric disorders. JAMA Psychiatry. 2017;74:863–864. doi: 10.1001/jamapsychiatry.2017.0397. [DOI] [PubMed] [Google Scholar]
- 17.Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science. 2006;311:1727–1730. doi: 10.1126/science.1118884. [DOI] [PubMed] [Google Scholar]
- 18.Howell N, Smejkal CB, Mackey DA, Chinnery PF, Turnbull DM, Herrnstadt C. The pedigree rate of sequence divergence in the human mitochondrial genome: There is a difference between phylogenetic and pedigree rates. Am J Hum Genet. 2003;72:659–670. doi: 10.1086/368264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawless C, Greaves L, Reeve AK, Turnbull DM, Vincent AE. The rise and rise of mitochondrial DNA mutations. Open Biol. 2020;10 doi: 10.1098/rsob.200061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kauppila TES, Kauppila JHK, Larsson NG. Mammalian mitochondria and aging: an update. Cell Metab. 2017;25:57–71. doi: 10.1016/j.cmet.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Sun N, Youle RJ, Finkel T. The mitochondrial basis of aging. Mol Cell. 2016;61:654–666. doi: 10.1016/j.molcel.2016.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: Regional variability and increase with advanced age. Nat Genet. 1992;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 23.Taylor SD, Ericson NG, Burton JN, et al. Targeted enrichment and high-resolution digital profiling of mitochondrial DNA deletions in human brain. Aging Cell. 2014;13:29–38. doi: 10.1111/acel.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy SR, Salk JJ, Schmitt MW, Loeb LA. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diaz F, Bayona-Bafaluy MP, Rana M, Mora M, Hao H, Moraes CT. Human mitochondrial DNA with large deletions repopulates organelles faster than full-length genomes under relaxed copy number control. Nucleic Acids Res. 2002;30:4626–4633. doi: 10.1093/nar/gkf602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo L, Tian J, Du H. Mitochondrial dysfunction and synaptic transmission failure in Alzheimer's disease. J Alzheimers Dis. 2017;57:1071–1086. doi: 10.3233/JAD-160702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ballif BC, Theisen A, Coppinger J, et al. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol Cytogenet. 2008;1:8. doi: 10.1186/1755-8166-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell GR, Ziabreva I, Reeve AK, Krishnan KJ, Reynolds R, Howell O, et al. Mitochondrial DNA deletions and neurodegeneration in multiple sclerosis. Ann Neurol. 2011;69:481–492. doi: 10.1002/ana.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schon KR, Ratnaike T, van den Ameele J, Horvath R, Chinnery PF. Mitochondrial diseases: a diagnostic revolution. Trends Genet. 2020;36:702–717. doi: 10.1016/j.tig.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 30.Schlieben LD, Prokisch H. The dimensions of primary mitochondrial disorders. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.600079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yonova-Doing E, Calabrese C, Gomez-Duran A, et al. An atlas of mitochondrial DNA genotype–phenotype associations in the UK Biobank. Nat Genet. 2021;53:982–993. doi: 10.1038/s41588-021-00868-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viscomi C, Zeviani M. Strategies for fighting mitochondrial diseases. J Intern Med. 2020;287:665–684. doi: 10.1111/joim.13046. [DOI] [PubMed] [Google Scholar]
- 34.Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: Implications for human health and disease. Nat Rev Genet. 2015;16:530–542. doi: 10.1038/nrg3966. [DOI] [PubMed] [Google Scholar]
- 35.Lott MT, Leipzig JN, Derbeneva O, et al. MtDNA variation and analysis using Mitomap and Mitomaster. Curr Protoc Bioinform. 2013;44 doi: 10.1002/0471250953.bi0123s44. 1.23.1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chinnery PF. In: GeneReviews® [Internet] Margaret P Adam, Holly H Ardinger, Roberta A Pagon, Stephanie E Wallace, Lora JH Bean, Karen Stephens AA., editors. University of Washington, Seattle; Seattle (WA): 2020. Mitochondrial disorders overview. 1993–. [Google Scholar]
- 37.Basel D. Mitochondrial DNA Depletion Syndromes. Clin Perinatol. 2020;47:123–141. doi: 10.1016/j.clp.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 38.Schaefer AM, Taylor RW, Turnbull DM, Chinnery PF. The epidemiology of mitochondrial disorders - Past, present and future. Biochim Biophys Acta. 2004;1659:115–120. doi: 10.1016/j.bbabio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 39.Majamaa K, Moilanen JS, Uimonen S, et al. Epidemiology of A3243G, the mutation for mitochondrial encephalomyopathy, lactic acidosis, and stroke like episodes: Prevalence of the mutation in an adult population. Am J Hum Genet. 1998;63:447–454. doi: 10.1086/301959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johns Hopkins University (Baltimore M. Online Mendelian Inheritance in Man; 2022. McKusick-Nathans Institute of Genetic Medicine. OMIM®. [Google Scholar]
- 41.Mitchell AL, Elson JL, Howell N, Taylor RW, Turnbull DM. Sequence variation in mitochondrial complex I genes: mutation or polymorphism? J Med Genet. 2006;43:175–179. doi: 10.1136/jmg.2005.032474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yarham JW, Al-Dosary M, Blakely EL, et al. A comparative analysis approach to determining the pathogenicity of mitochondrial tRNA mutations. Hum Mutat. 2011;32:1319–1325. doi: 10.1002/humu.21575. [DOI] [PubMed] [Google Scholar]
- 43.Wong LJ. Diagnostic challenges of mitochondrial DNA disorders. Mitochondrion. 2007;7:45–52. doi: 10.1016/j.mito.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 44.Gonzéalez-Vioque E, Bornstein B, Gallardo ME, Fernéandez-Moreno MA, Garesse R. The pathogenicity scoring system for mitochondrial trna mutations revisited. Mol Genet Genomic Med. 2014;2:107–114. doi: 10.1002/mgg3.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 46.El-Hattab AW, Craigen WJ, Wong L-JC, Scaglia F. In: Margaret P Adam, Holly H Ardinger, Roberta A Pagon, Stephanie E Wallace, Lora JH Bean, Karen Stephens AA, editor. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993– 2021.
- 47.Zeviani M, di Donato S. Mitochondrial disorders. Brain. 2004;127:2153–2172. doi: 10.1093/brain/awh259. [DOI] [PubMed] [Google Scholar]
- 48.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 49.Shoubridge EA. Nuclear genetic defects of oxidative phosphorylation. Hum Mol Genet. 2001;10:2277–2284. doi: 10.1093/hmg/10.20.2277. [DOI] [PubMed] [Google Scholar]
- 50.Rusecka J, Kaliszewska M, Bartnik E, Tońska K. Nuclear genes involved in mitochondrial diseases caused by instability of mitochondrial DNA. J Appl Genet. 2018;59:43–57. doi: 10.1007/s13353-017-0424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Almannai M, El-Hattab AW, Scaglia F. Mitochondrial DNA replication: clinical syndromes. Essays Biochem. 2018;62:297–308. doi: 10.1042/EBC20170101. [DOI] [PubMed] [Google Scholar]
- 52.Malik AN, Czajka A. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion. 2013;13:481–492. doi: 10.1016/j.mito.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 53.Shamseer L, Moher D, Clarke M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: elaboration and explanation. BMJ. 2015;350:g7647. doi: 10.1136/bmj.g7647. [DOI] [PubMed] [Google Scholar]
- 54.Geffroy G, Benyahia R, Frey S, et al. The accumulation of assembly intermediates of the mitochondrial complex I matrix arm is reduced by limiting glucose uptake in a neuronal-like model of MELAS syndrome. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1596–1608. doi: 10.1016/j.bbadis.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Lax NZ, Grady J, Laude A, et al. Extensive respiratory chain defects in inhibitory interneurones in patients with mitochondrial disease. Neuropathol Appl Neurobiol. 2016;42:180–193. doi: 10.1111/nan.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lax NZ, Hepplewhite PD, Reeve AK, et al. Cerebellar ataxia in patients with mitochondrial DNA disease: a molecular clinicopathological study. J Neuropathol Exp Neurol. 2012;71:148–161. doi: 10.1097/NEN.0b013e318244477d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Betts J, Jaros E, Perry RH, et al. Molecular neuropathology of MELAS: Level of heteroplasmy in individual neurones and evidence of extensive vascular involvement. Neuropathol Appl Neurobiol. 2006;32:359–373. doi: 10.1111/j.1365-2990.2006.00731.x. [DOI] [PubMed] [Google Scholar]
- 58.Kaido M, Fujimura H, Soga F, et al. Alzheimer-type pathology in a patient with mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS) Acta Neuropathol. 1996;92:312–318. doi: 10.1007/s004010050524. [DOI] [PubMed] [Google Scholar]
- 59.MacMillan C, Lach B, Shoubridge EA. Variable distribution of mutant mitochondrial DNAs (tRNAleu[3243]) in tissues of symptomatic relatives with MELAS: The role of mitotic segregation. Neurology. 1993;43:1586–1590. doi: 10.1212/wnl.43.8.1586. [DOI] [PubMed] [Google Scholar]
- 60.Love S, Nicoll JAR, Kinrade E. Sequencing and quantitative assessment of mutant and wild-type mitochondrial DNA in paraffin sections from cases of MELAS. J Pathol. 1993;170:9–14. doi: 10.1002/path.1711700103. [DOI] [PubMed] [Google Scholar]
- 61.Shiraiwa N, Ishii A, Iwamoto H, Mizusawa H, Kagawa Y, Ohta S. Content of mutant mitochondrial DNA and organ dysfunction in a patient with a MELAS subgroup of mitochondrial encephalomyopathies. J Neurol Sci. 1993;120:174–179. doi: 10.1016/0022-510x(93)90270-9. [DOI] [PubMed] [Google Scholar]
- 62.Ciafaloni E, Ricci E, Servidei S, et al. Widespread tissue distribution of a tRNALeu(UUR) mutation in the mitochondrial DNA of a patient with MELAS syndrome. Neurology. 1991;41:1663–1665. doi: 10.1212/wnl.41.10.1663. [DOI] [PubMed] [Google Scholar]
- 63.Enter C, Müller-Höcker J, Zierz S, et al. A specific point mutation in the mitochondrial genome of Caucasians with MELAS. Hum Genet. 1991;88:233–236. doi: 10.1007/BF00206080. [DOI] [PubMed] [Google Scholar]
- 64.Di Trapani G, Gregori B, Servidei S, Ricci E, Sabatelli M, Tonali P. Mitochondrial encephalopathy, lactic acidosis, and stroke-like episodes (MELAS) Clin Neuropathol. 1997;16:195–200. [PubMed] [Google Scholar]
- 65.Santorelli FM, Tanji K, Kulikova R, Shanske S, Vilarinho L, Hays AP, et al. Identification of a novel mutation in the mtDNA ND5 gene associated with MELAS. Biochem Biophys Res Commun. 1997;238:326–328. doi: 10.1006/bbrc.1997.7167. [DOI] [PubMed] [Google Scholar]
- 66.Lax NZ, Gnanapavan S, Dowson SJ, et al. Early-onset cataracts, spastic paraparesis, and ataxia caused by a novel mitochondrial tRNAGlu (MT-TE) gene mutation causing severe complex I deficiency: a clinical, molecular, and neuropathologic study. J Neuropathol Exp Neurol. 2013;72:164–175. doi: 10.1097/NEN.0b013e31828129c5. [DOI] [PubMed] [Google Scholar]
- 67.de Kremer RD, Paschini-Capra A, Bacman S, et al. Barth's syndrome-like disorder: a new phenotype with a maternally inherited A3243G substitution of mitochondrial DNA (MELAS mutation) Am J Med Genet. 2001;99:83–93. doi: 10.1002/1096-8628(2001)9999:9999<::aid-ajmg1136>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 68.Scholle LM, Zierz S, Mawrin C, Wickenhauser C, Urban DL. Heteroplasmy and copy number in the common m.3243a>G mutation—A post-mortem genotype– phenotype analysis. Genes. 2020;11:212. doi: 10.3390/genes11020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brinckmann A, Weiss C, Wilbert F, et al. Regionalized pathology correlates with augmentation of mtDNA copy numbers in a patient with myoclonic epilepsy with ragged-red fibers (MERRF-syndrome) PLoS ONE. 2010;5:e13513. doi: 10.1371/journal.pone.0013513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanger TD, Jain KD. MERRF syndrome with overwhelming lactic acidosis. Pediatr Neurol. 1996;14:57–61. doi: 10.1016/0887-8994(95)00226-x. [DOI] [PubMed] [Google Scholar]
- 71.Oldfors A, Holme E, Tulinius M, Larsson NG. Tissue distribution and disease manifestations of the tRNALys A»G(8344) mitochondrial DNA mutation in a case of myoclonus epilepsy and ragged red fibres. Acta Neuropathol. 1995;90:328–333. doi: 10.1007/BF00296519. [DOI] [PubMed] [Google Scholar]
- 72.Santorelli FM, Tanji K, Shanske S, et al. The mitochondrial DNA A8344G mutation in Leigh syndrome revealed by analysis in paraffin-embedded sections: revisiting the past. Annals Neurol. 1998;44:962–964. doi: 10.1002/ana.410440616. [DOI] [PubMed] [Google Scholar]
- 73.Tanno Y, Yoneda M, Tanaka K, et al. Uniform tissue distribution of tRNALys mutation in mitochondrial DNA in MERRF patients. Neurology. 1993;43:1198–1200. doi: 10.1212/wnl.43.6.1198. [DOI] [PubMed] [Google Scholar]
- 74.Zhou L, Chomyn A, Attardi G, Miller CA. Myoclonic epilepsy and ragged red fibers (MERRF) syndrome: Selective vulnerability of CNS neurons does not correlate with the level of mitochondrial tRNA(lys) mutation in individual neuronal isolates. J Neurosci. 1997;17:7746–7753. doi: 10.1523/JNEUROSCI.17-20-07746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rojo A, Campos Y, Sánchez JM, et al. NARP-MILS syndrome caused by 8993 T >G mitochondrial DNA mutation: a clinical, genetic and neuropathological study. Acta Neuropathol. 2006;111:610–616. doi: 10.1007/s00401-006-0040-5. [DOI] [PubMed] [Google Scholar]
- 76.Kirby DM, Boneh A, Chow CW, et al. Low mutant load of mitochondrial DNA G13513A mutation can cause Leigh's disease. Ann Neurol. 2003;54:473–478. doi: 10.1002/ana.10687. [DOI] [PubMed] [Google Scholar]
- 77.Nagashima T, Mori M, Katayama K, et al. Adult Leigh syndrome with mitochondrial DNA mutation at 8993. Acta Neuropathol. 1999;97:416–422. doi: 10.1007/s004010051007. [DOI] [PubMed] [Google Scholar]
- 78.Jiang YW, Qin J, Yuan Y, Qi Y, Wu XR. Neuropathologic and clinical features in eight Chinese patiens with Leigh disease. J Child Neurol. 2002;17:450–452. doi: 10.1177/088307380201700611. [DOI] [PubMed] [Google Scholar]
- 79.Santorelli FM, Tanji K, Sano M, et al. Maternally inherited encephalopathy associated with a single-base insertion in the mitochondrial tRNA(Trp) gene. Ann Neurol. 1997;42:256–260. doi: 10.1002/ana.410420220. [DOI] [PubMed] [Google Scholar]
- 80.Sweeney MG, Hammans SR, Duchen LW, et al. Mitochondrial DNA mutation underlying Leigh's syndrome: clinical, pathological, biochemical, and genetic studies of a patient presenting with progressive myoclonic epilepsy. J Neurol Sci. 1994;121:57–65. doi: 10.1016/0022-510x(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 81.Tatuch Y, Christodoulou J, Feigenbaum A, et al. Heteroplasmic mtDNA mutation (T—-G) at 8993 can cause Leigh disease when the percentage of abnormal mtDNA is high. Am J Hum Genet. 1992;50:852–858. [PMC free article] [PubMed] [Google Scholar]
- 82.Lombes A, Nakase H, Tritschler HJ, et al. Biochemical and molecular analysis of cytochrome c oxidase deficiency in Leigh's syndrome. Neurology. 1991;41:491–498. doi: 10.1212/wnl.41.4.491. [DOI] [PubMed] [Google Scholar]
- 83.Soltys DT, Pereira CPM, Rowies FT, et al. Lower mitochondrial DNA content but not increased mutagenesis associates with decreased base excision repair activity in brains of AD subjects. Neurobiol Aging. 2019;73:161–170. doi: 10.1016/j.neurobiolaging.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 84.Wei W, Keogh MJ, Wilson I, et al. Mitochondrial DNA point mutations and relative copy number in 1363 disease and control human brains. Acta Neuropathol Commun. 2017;5:13. doi: 10.1186/s40478-016-0404-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Y, Liu C, Parker WD, et al. Mitochondrial DNA rearrangement spectrum in brain tissue of Alzheimer's disease: analysis of 13 cases. PLoS ONE. 2016;11 doi: 10.1371/journal.pone.0154582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Coskun PE, Wyrembak J, Derbereva O, et al. Systemic mitochondrial dysfunction and the etiology of Alzheimer's disease and down syndrome dementia. J Alzheimers Dis. 2010;20:293–310. doi: 10.3233/JAD-2010-100351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chang SW, Zhang D, Chung HD, Zassenhaus HP. The frequency of point mutations in mitochondrial DNA is elevated in the Alzheimer's brain. Biochem Biophys Res Commun. 2000;273:203–208. doi: 10.1006/bbrc.2000.2885. [DOI] [PubMed] [Google Scholar]
- 88.Hutchin TP, Heath PR, Pearson RCA, Sinclair AJ. Mitochondrial DNA mutations in Alzheimer's disease. Biochem Biophys Res Commun. 1997;241:221–225. doi: 10.1006/bbrc.1997.7793. [DOI] [PubMed] [Google Scholar]
- 89.Janetzky B, Schmid C, Felix F, et al. Investigations on the point mutations at nt 5460 of the mtDNA in different neurodegenerative and neuromuscular diseases. Eur Neurol. 1996;36:149–153. doi: 10.1159/000117233. [DOI] [PubMed] [Google Scholar]
- 90.Ito S, Ohta S, Nishimaki K, et al. Functional integrity of mitochondrial genomes in human platelets and autopsied brain tissues from elderly patients with Alzheimer's disease. Proc Nat Acad Sci U S A. 1999;96:2099–2103. doi: 10.1073/pnas.96.5.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thubron EB, Rosa HS, Hodges A, et al. Regional mitochondrial DNA and cell-type changes in post-mortem brains of non-diabetic Alzheimer's disease are not present in diabetic Alzheimer's disease. Sci Rep. 2019;9:11386. doi: 10.1038/s41598-019-47783-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.de la Monte SM, Luong T, Neely TR, Robinson D, Wands JR. Mitochondrial DNA damage as a mechanism of cell loss in Alzheimer's disease. Lab Invest. 2000;80:1323–1335. doi: 10.1038/labinvest.3780140. [DOI] [PubMed] [Google Scholar]
- 93.Rice AC, Keeney PM, Algarzae NK, Ladd AC, Thomas RR, Bennett JP. Mitochondrial DNA copy numbers in pyramidal neurons are decreased and mitochondrial biogenesis transcriptome signaling is disrupted in Alzheimer's disease hippocampi. J Alzheimers Dis. 2014;40:319–330. doi: 10.3233/JAD-131715. [DOI] [PubMed] [Google Scholar]
- 94.Coskun PE, Beal MF, Wallace DC. Alzheimer's brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Nat Acad Sci U S A. 2004;101:10726–10731. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Müller SK, Bender A, Laub C, et al. Lewy body pathology is associated with mitochondrial DNA damage in Parkinson's disease. Neurobiol Aging. 2013;34:2231–2233. doi: 10.1016/j.neurobiolaging.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 96.Cavelier L, Jazin EE, Eriksson I, et al. Decreased cytochrome-c oxidase activity and lack of age-related accumulation of mitochondrial DNA deletions in the brains of schizophrenics. Genomics. 1995;29:217–224. doi: 10.1006/geno.1995.1234. [DOI] [PubMed] [Google Scholar]
- 97.Blanchard BJ, Park T, Fripp WJ, Lermanv LS, Ingram VM. A mitochondrial DNA deletion in normally aging and in Alzheimer brain tissue. Neuroreport. 1993;4:799–802. doi: 10.1097/00001756-199306000-00051. [DOI] [PubMed] [Google Scholar]
- 98.Hamblet NS, Castora FJ. Elevated levels of the Kearns-Sayre syndrome mitochondrial DNA deletion in temporal cortex of Alzheimer's patients. Mut Res. 1997;379:253–262. doi: 10.1016/s0027-5107(97)00158-9. [DOI] [PubMed] [Google Scholar]
- 99.Aliyev A, Chen SG, Seyidova D, et al. Mitochondria DNA deletions in atherosclerotic hypoperfused brain microvessels as a primary target for the development of Alzheimer's disease. J Neurol Sci. 2005;229-230:285–292. doi: 10.1016/j.jns.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 100.Gu G, Reyes PF, Golden GT, et al. Mitochondrial DNA deletions/rearrangements in Parkinson disease and related neurodegenerative disorders. J Neuropathol Exp Neurol. 2002;61:634–639. doi: 10.1093/jnen/61.7.634. [DOI] [PubMed] [Google Scholar]
- 101.Strobel S, Grünblatt E, Heinsen H, et al. Astrocyte- and microglia-specific mitochondrial DNA deletions levels in sporadic Alzheimer's disease. J Alzheimers Dis. 2019;67:149–157. doi: 10.3233/JAD-180661. [DOI] [PubMed] [Google Scholar]
- 102.Krishnan KJ, Ratnaike TE, de Gruyter HLM, Jaros E, Turnbull DM. Mitochondrial DNA deletions cause the biochemical defect observed in Alzheimer's disease. Neurobiol Aging. 2012;33:2210–2214. doi: 10.1016/j.neurobiolaging.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 103.Nido GS, Dölle C, Flønes I, et al. Ultradeep mapping of neuronal mitochondrial deletions in Parkinson's disease. Neurobiol Aging. 2018;63:120–127. doi: 10.1016/j.neurobiolaging.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 104.Arthur CR, Morton SL, Dunham LD, Keeney PM, Bennett JP. Parkinson's disease brain mitochondria have impaired respirasome assembly, age-related increases in distribution of oxidative damage to mtDNA and no differences in heteroplasmic mtDNA mutation abundance. Mol Neurodegener. 2009;4:37. doi: 10.1186/1750-1326-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dölle C, Flønes I, Nido GS, et al. Defective mitochondrial DNA homeostasis in the substantia nigra in Parkinson disease. Nat Commun. 2016;7:13548. doi: 10.1038/ncomms13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Coxhead J, Kurzawa-Akanbi M, Hussain R, Pyle A, Chinnery P, Hudson G. Somatic mtDNA variation is an important component of Parkinson's disease. Neurobiol Aging. 2015;38 doi: 10.1016/j.neurobiolaging.2015.10.036. 217.e1-217.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen C, Vincent AE, Blain AP, Smith AL, Turnbull DM, Reeve AK. Investigation of mitochondrial biogenesis defects in single substantia nigra neurons using post-mortem human tissues. Neurobiol Dis. 2020;134 doi: 10.1016/j.nbd.2019.104631. [DOI] [PubMed] [Google Scholar]
- 108.Gatt AP, Duncan OF, Attems J, Francis PT, Ballard CG, Bateman JM. Dementia in Parkinson's disease is associated with enhanced mitochondrial complex I deficiency. Mov Disord. 2016;31:352–359. doi: 10.1002/mds.26513. [DOI] [PubMed] [Google Scholar]
- 109.Grünewald A, Rygiel KA, Hepplewhite PD, Morris CM, Picard M, Turnbull DM. Mitochondrial DNA depletion in respiratory chain-deficient parkinson disease neurons. Ann Neurol. 2016;79:366–378. doi: 10.1002/ana.24571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bury AG, Pyle A, Elson JL, et al. Mitochondrial DNA changes in pedunculopontine cholinergic neurons in Parkinson disease. Ann Neurol. 2017;82:1016–1021. doi: 10.1002/ana.25099. [DOI] [PubMed] [Google Scholar]
- 111.Flønes IH, Fernandez-Vizarra E, Lykouri M, et al. Neuronal complex I deficiency occurs throughout the Parkinson's disease brain, but is not associated with neurodegeneration or mitochondrial DNA damage. Acta Neuropathol. 2018;135:409–425. doi: 10.1007/s00401-017-1794-7. [DOI] [PubMed] [Google Scholar]
- 112.Didonato S, Zeviani M, Giovannini P, et al. Respiratory chain and mitochondrial DNA in muscle and brain in Parkinson's disease patients. Neurology. 1993;43:2262–2268. doi: 10.1212/wnl.43.11.2262. [DOI] [PubMed] [Google Scholar]
- 113.Naydenov A v., Vassoler F, Luksik AS, Kaczmarska J, Konradi C. Mitochondrial abnormalities in the putamen in Parkinson's disease dyskinesia. Acta Neuropathol. 2010;120:623–631. doi: 10.1007/s00401-010-0740-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bender A, Schwarzkopf RM, McMillan A, et al. Dopaminergic midbrain neurons are the prime target for mitochondrial DNA deletions. J Neurol. 2008;255:1231–1235. doi: 10.1007/s00415-008-0892-9. [DOI] [PubMed] [Google Scholar]
- 115.Reeve AK, Krishnan KJ, Elson JL, et al. Nature of Mitochondrial DNA Deletions in Substantia Nigra Neurons. Am J Hum Genet. 2008;82:228–235. doi: 10.1016/j.ajhg.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kösel S, Egensperger R, Schnopp NM, Graeber MB. The “common deletion” is not increased in parkinsonian substantia nigra as shown by competitive polymerase chain reaction. Mov Disord. 1997;12:639–645. doi: 10.1002/mds.870120504. [DOI] [PubMed] [Google Scholar]
- 117.Lestienne P, Nelson I, Riederer P, Reichmann H, Jellinger K. Mitochondrial DNA in postmortem brain from patients with Parkinson's disease. J Neurochem. 1991;56 doi: 10.1111/j.1471-4159.1991.tb02087.x. 1819–1819. [DOI] [PubMed] [Google Scholar]
- 118.Rubino F, Piredda R, Calabrese FM, et al. HmtDB, a genomic resource for mitochondrion-based human variability studies. Nucleic Acids Res. 2012;40:D1150–D1159. doi: 10.1093/nar/gkr1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sequeira A, Martin MV, Rollins B, et al. Mitochondrial mutations and polymorphisms in psychiatric disorders. Front Genet. 2012;3:103. doi: 10.3389/fgene.2012.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rollins B, Martin MV, Sequeira PA, et al. Mitochondrial variants in schizophrenia, bipolar disorder, and major depressive disorder. PloS One. 2009;4:e4913. doi: 10.1371/journal.pone.0004913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ichikawa T, Arai M, Miyashita M, et al. Schizophrenia: maternal inheritance and heteroplasmy of mtDNA mutations. Mol Genet Metab. 2012;105:103–109. doi: 10.1016/j.ymgme.2011.09.034. [DOI] [PubMed] [Google Scholar]
- 122.Munakata K, Iwamoto K, Bundo M, Kato T. Mitochondrial DNA 3243A>G mutation and increased expression of LARS2 gene in the brains of patients with bipolar disorder and schizophrenia. Biol Psychiatry. 2005;57:525–532. doi: 10.1016/j.biopsych.2004.11.041. [DOI] [PubMed] [Google Scholar]
- 123.Marchbanks RM, Ryan M, Day IN, Owen M, McGuffin P, Whatley SA. A mitochondrial DNA sequence variant associated with schizophrenia and oxidative stress. Schizophr Res. 2003;65:33–38. doi: 10.1016/s0920-9964(03)00011-2. [DOI] [PubMed] [Google Scholar]
- 124.Hjelm BE, Rollins B, Morgan L, et al. Splice-Break: Exploiting an RNA-seq splice junction algorithm to discover mitochondrial DNA deletion breakpoints and analyses of psychiatric disorders. Nucleic Acids Res. 2019;47:e59. doi: 10.1093/nar/gkz164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bodenstein DF, Kim HK, Brown NC, Navaid B, Young LT, Andreazza AC. Mitochondrial DNA content and oxidation in bipolar disorder and its role across brain regions. NPJ Schizophr. 2019;5:1–8. doi: 10.1038/s41537-019-0089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mamdani F, Rollins B, Morgan L, Sequeira PA, Vawter MP. The somatic common deletion in mitochondrial DNA is decreased in schizophrenia. Schizophr Res. 2014;159:370–375. doi: 10.1016/j.schres.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fries GR, Bauer IE, Scaini G, et al. Accelerated hippocampal biological aging in bipolar disorder. Bipolar Disord. 2020;22:498–507. doi: 10.1111/bdi.12876. [DOI] [PubMed] [Google Scholar]
- 128.Torrell H, Montaña E, Abasolo N, et al. Mitochondrial DNA (mtDNA) in brain samples from patients with major psychiatric disorders: gene expression profiles, mtDNA content and presence of the mtDNA common deletion. Am J Med Genet B. 2013;162B:213–223. doi: 10.1002/ajmg.b.32134. [DOI] [PubMed] [Google Scholar]
- 129.Kato T, Stine OC, McMahon FJ, Crowe RR. Increased levels of a mitochondrial DNA deletion in the brain of patients with bipolar disorder. Biol Psychiatry. 1997;42:871–875. doi: 10.1016/S0006-3223(97)00012-7. [DOI] [PubMed] [Google Scholar]
- 130.Fuke S, Kametani M, Kato T. Quantitative analysis of the 4977-bp common deletion of mitochondrial DNA in postmortem frontal cortex from patients with bipolar disorder and schizophrenia. Neurosci Lett. 2008;439:173–177. doi: 10.1016/j.neulet.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 131.Nádasi EA, Melegh B, Seress L, Kosztolányi G. Mitochondrial DNA4977 deletion in brain of newborns died after intensive care. Acta Biol Hung. 2003;54:253–262. doi: 10.1556/ABiol.54.2003.3-4.4. [DOI] [PubMed] [Google Scholar]
- 132.Roca-Bayerri C, Robertson F, Pyle A, Hudson G, Payne BAI. Mitochondrial DNA damage and brain aging in human immunodeficiency virus. Clin Infect Dis. 2021;73:e466–e473. doi: 10.1093/cid/ciaa984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jazin EE, Cavelier L, Eriksson I, Orelandt L, Gyllensten U. Human brain contains high levels of heteroplasmy in the noncoding regions of mitochondrial DNA (displacement loop/intraindividual variation/polymorphism/oxidative phosphorylation/aging) Proc Natl Acad Sci U S A. 1996;93:12382–12387. doi: 10.1073/pnas.93.22.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cantuti-Castelvetri I, Lin MT, Zheng K, et al. Somatic mitochondrial DNA mutations in single neurons and glia. Neurobiol Aging. 2005;26:1343–1355. doi: 10.1016/j.neurobiolaging.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 135.Kapsa RM, Jean-Francois MJ, Lertrit P, et al. Mitochondrial DNA polymorphism in substantia nigra. J Neurol Sci. 1996;144:204–211. doi: 10.1016/s0022-510x(96)00247-x. [DOI] [PubMed] [Google Scholar]
- 136.Simon DK, Lin MT, Zheng L, et al. Somatic mitochondrial DNA mutations in cortex and substantia nigra in aging and Parkinson's disease. Neurobiol Aging. 2004;25:71–81. doi: 10.1016/s0197-4580(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 137.Frahm T, Mohamed SA, Bruse P, Gemünd C, Oehmichen M, Meissner C. Lack of age-related increase of mitochondrial DNA amount in brain, skeletal muscle and human heart. Mech Ageing Dev. 2005;126:1192–1200. doi: 10.1016/j.mad.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 138.Meissner C, Bruse P, Mohamed SA, et al. The 4977 bp deletion of mitochondrial DNA in human skeletal muscle, heart and different areas of the brain: a useful biomarker or more? Exp Gerontol. 2008;43:645–652. doi: 10.1016/j.exger.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 139.Mawrin C, Kirches E, Krause G, et al. Region-specific analysis of mitochondrial DNA deletions in neurodegenerative disorders in humans. Neurosci Lett. 2004;357:111–114. doi: 10.1016/j.neulet.2003.11.073. [DOI] [PubMed] [Google Scholar]
- 140.McDonald RPA, Horsburgh KJ, Graham DI, Nicoll JAR. Mitochondrial DNA deletions in acute brain injury. NeuroReport. 1999;10:1875–1878. doi: 10.1097/00001756-199906230-00014. [DOI] [PubMed] [Google Scholar]
- 141.Kraytsberg Y, Kudryavtseva E, Mckee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 142.Zhang C, Baumer A, Maxwell RJ, Linnane AW, Nagley P. Multiple mitochondrial DNA deletions in an elderly human individual. FEBS Lett. 1992;297:34–38. doi: 10.1016/0014-5793(92)80321-7. [DOI] [PubMed] [Google Scholar]
- 143.Cortopassi GA, Arnheim N. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 1990;18:6927–6933. doi: 10.1093/nar/18.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Soong NW, Hinton DR, Cortopassi G, Arnheim N. Mosaicism for a specific somatic mitochondrial DNA mutation in adult human brain. Nat Genet. 1992;2:318–323. doi: 10.1038/ng1292-318. [DOI] [PubMed] [Google Scholar]
- 145.Bender A, Krishnan KJ, Morris CM, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 146.Mann VM, Cooper JM, Schapira AHV. Quantitation of a mitochondrial DNA deletion in Parkinson's disease. FEBS Lett. 1992;299:218–222. doi: 10.1016/0014-5793(92)80118-z. [DOI] [PubMed] [Google Scholar]
- 147.Lezza AM, Mecocci P, Cormio A, et al. Mitochondrial DNA 4977 bp deletion and OH 8 dG levels correlate in the brain of aged subjects but not Alzheimer's disease patients. FASEB J. 1999;13:1083–1088. doi: 10.1096/fasebj.13.9.1083. [DOI] [PubMed] [Google Scholar]
- 148.Melov S, Schneider JA, Coskun PE, Bennett DA, Wallace DC. Mitochondrial DNA rearrangements in aging human brain and in situ PCR of mtDNA. Neurobiol Aging. 1999;20:565–571. doi: 10.1016/s0197-4580(99)00092-5. [DOI] [PubMed] [Google Scholar]
- 149.Merril CR, Zullo S, Ghanbari H, et al. Possible relationship between conditions associated with chronic hypoxia and brain mitochondrial DNA deletions. Arch Biochem Biophys. 1996;326:172–177. doi: 10.1006/abbi.1996.0062. [DOI] [PubMed] [Google Scholar]
- 150.Devall M, Burrage J, Caswell R, et al. A comparison of mitochondrial DNA isolation methods in frozen post-mortem human brain tissue—applications for studies of mitochondrial genetics in brain disorders. BioTechniques. 2015;59:241–247. doi: 10.2144/000114343. [DOI] [PubMed] [Google Scholar]
- 151.Hamblet NS, Castora FJ. Mitochondrial DNA deletion analysis: a comparison of PCR quantitative methods. Biochem Biophys Res Commun. 1995;207:839–847. doi: 10.1006/bbrc.1995.1262. [DOI] [PubMed] [Google Scholar]
- 152.Marquis J, Lefebvre G, Kourmpetis YAI, et al. MitoRS, a method for high throughput, sensitive, and accurate detection of mitochondrial DNA heteroplasmy. BMC Genomics. 2017;18:326. doi: 10.1186/s12864-017-3695-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dölle C, Bindoff LA. Tzoulis C. 3,3′-Diaminobenzidine staining interferes with PCR-based DNA analysis. Sci Rep. 2018;8:1272. doi: 10.1038/s41598-018-19745-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Devall M, Smith RG, Jeffries A, et al. Regional differences in mitochondrial DNA methylation in human post-mortem brain tissue. Clin Epigenetics. 2017;9:47. doi: 10.1186/s13148-017-0337-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sharma P, Sampath H. Mitochondrial DNA integrity: role in health and disease. Cells. 2019;8:100. doi: 10.3390/cells8020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang SY, Castellani CA, Longchamps RJ, et al. Blood-derived mitochondrial DNA copy number is associated with gene expression across multiple tissues and is predictive for incident neurodegenerative disease. Genome Res. 2021;31:349–358. doi: 10.1101/gr.269381.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tzoulis C, Tran GT, Coxhead J, et al. Molecular pathogenesis of polymerase gamma-related neurodegeneration. Ann Neurol. 2014;76:66–81. doi: 10.1002/ana.24185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tzoulis C, Tran GT, Schwarzlmüller T, et al. Severe nigrostriatal degeneration without clinical parkinsonism in patients with polymerase gamma mutations. Brain. 2013;136:2393–2404. doi: 10.1093/brain/awt103. [DOI] [PubMed] [Google Scholar]
- 159.Gramegna LL, Pisano A, Testa C, et al. Cerebral mitochondrial microangiopathy leads to leukoencephalopathy in mitochondrial neurogastrointestinal encephalopathy. Am J Neuroradiol. 2018;39:427–434. doi: 10.3174/ajnr.A5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zsurka G, Baron M, Stewart JD, et al. Clonally expanded mitochondrial DNA mutations in epileptic individuals with mutated DNA polymerase γ. J Neuropathol Exp Neurol. 2008;67:857–866. doi: 10.1097/NEN.0b013e3181839b2d. [DOI] [PubMed] [Google Scholar]
- 161.Pistilli D, di Gioia CRT, D'Amati G, et al. Detection of deleted mitochondrial DNA in Kearns-Sayre syndrome using laser capture microdissection. Hum Pathol. 2003;34:1058–1061. doi: 10.1053/s0046-8177(03)00344-7. [DOI] [PubMed] [Google Scholar]
- 162.Rosebush PI, Anglin RE, Rasmussen S, Mazurek MF. Mental illness in patients with inherited mitochondrial disorders. Schizophr Res. 2017;187:33–37. doi: 10.1016/j.schres.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 163.Wei W, Tuna S, Keogh MJ, et al. Germline selection shapes human mitochondrial DNA diversity. Science. 2019;364:eaau6520. doi: 10.1126/science.aau6520. [DOI] [PubMed] [Google Scholar]
- 164.Wai KT, Gunn P, Barash M. Development of the MitoQ assay as a real-time quantification of mitochondrial DNA in degraded samples. Int J Legal Med. 2019;133:411–417. doi: 10.1007/s00414-018-1956-8. [DOI] [PubMed] [Google Scholar]
- 165.Palculict ME, Zhang VW, Wong LJ, Wang J. Comprehensive mitochondrial genome analysis by massively parallel sequencing. Methods Mol Biol. 2016;1351:3–17. doi: 10.1007/978-1-4939-3040-1_1. [DOI] [PubMed] [Google Scholar]
- 166.Santorsola M, Calabrese C, Girolimetti G, Diroma MA, Gasparre G, Attimonelli M. A multi-parametric workflow for the prioritization of mitochondrial DNA variants of clinical interest. Hum Genet. 2016;135:121–136. doi: 10.1007/s00439-015-1615-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Calabrese C, Simone D, Diroma MA, et al. MToolBox: a highly automated pipeline for heteroplasmy annotation and prioritization analysis of human mitochondrial variants in high-throughput sequencing. Bioinformatics. 2014;30:3115–3117. doi: 10.1093/bioinformatics/btu483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Damas J, Carneiro J, Amorim A, Pereira F. MitoBreak: The mitochondrial DNA breakpoints database. Nucleic Acids Res. 2014;42:D1261–D1268. doi: 10.1093/nar/gkt982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Longchamps RJ, Castellani CA, Yang SY, et al. Evaluation of mitochondrial DNA copy number estimation techniques. PLoS ONE. 2020;15 doi: 10.1371/journal.pone.0228166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Santibanez-Koref M, Griffin H, Turnbull DM, Chinnery PF, Herbert M, Hudson G. Assessing mitochondrial heteroplasmy using next generation sequencing: A note of caution. Mitochondrion. 2019;46:302–306. doi: 10.1016/j.mito.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Goudenège D, Bris C, Hoffmann V, et al. eKLIPse: a sensitive tool for the detection and quantification of mitochondrial DNA deletions from next-generation sequencing data. Genet Med. 2019;21:1407–1416. doi: 10.1038/s41436-018-0350-8. [DOI] [PubMed] [Google Scholar]
- 172.Basu S, Xie X, Uhler JP, et al. Accurate mapping of mitochondrial DNA deletions and duplications using deep sequencing. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1009242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.