Summary
Background
Rheumatoid arthritis (RA) is an inflammatory disease that manifests as a preclinical stage of systemic autoimmunity followed by chronic progressive synovitis. Disease-associated genetic SNP variants predominantly map to non-coding, regulatory regions of functional importance in CD4 T cells, implicating these cells as key regulators. A better understanding of the epigenome of CD4 T cells holds the promise of providing information on the interaction between genetic susceptibility and exogenous factors.
Methods
We mapped regions of chromatin accessibility using ATAC-seq in peripheral CD4 T cell subsets of patients with RA (n=18) and compared them to T cells from patients with psoriatic arthritis (n=11) and age-matched healthy controls (n=10). Transcripts of selected genes were quantified using qPCR.
Findings
RA-associated epigenetic signatures were identified that in part overlapped between central and effector memory CD4 T cells and that were to a lesser extent already present in naïve cells. Sites more accessible in RA were highly enriched for the motif of the transcription factor (TF) CTCF suggesting differences in the three-dimensional chromatin structure. Unexpectedly, sites with reduced chromatin accessibility were enriched for motifs of TFs pertinent for T cell function. Most strikingly, super-enhancers encompassing RA-associated SNPs were less accessible. Analysis of selected transcripts and published DNA methylation patterns were consistent with this finding. The preferential loss in accessibility at these super-enhancers was seen in patients with high and low disease activity and on a variety of immunosuppressive treatment modalities.
Interpretation
Disease-associated genes are epigenetically less poised to respond in CD4 T cells from patients with established RA.
Funding
This work was supported by I01 BX001669 from the Veterans Administration.
Keywords: Rheumatoid arthritis, Super-enhancer, Susceptibility loci, Epigenome, Chromatin accessibility
Abbreviations: ATAC-seq, assay for transposase-accessible chromatin using sequencing; CDAI, clinical disease activity index; CM, central memory; CRP, C-reactive protein; DA, differentially accessible; EM, effector memory; FACS, fluorescence activated cell sorting; GO, gene ontology; GREAT, genomic regions enrichment of annotations tool; HC, healthy individuals; MTX, methotrexate; PBMCs, peripheral blood mononuclear cells; PCA, principal component analysis; PsA, psoriatic arthritis; qPCR, quantitative polymerase chain reaction; RA, rheumatoid arthritis; SNP, single nucleotide polymorphisms; SRA, sequence read archive; TF, transcription factor; TFBS, transcription factor binding site; UMAP, uniform manifold approximation and projection; vst, variance stabilized transformation)
Research in context.
Evidence before this study
Disease-associated SNPs in RA are enriched in super-enhancers that are important for the CD4 T cell lineage. Differences in DNA methylation have been described in circulating mononuclear cells from RA patients, indicating a pathogenetic role of factors driving epigenetic remodelling.
Added value of this study
Chromatin accessibility maps of purified CD4 T cells are provided that allow conclusions on upstream regulators of RA-associated epigenetic differences and on the downstream genes regulated by the differentially regulated sites. Disease risk genes were found to be epigenetically less accessible in established disease, consistent with a state of immunodeficiency or exhaustion.
Implications of all the available evidence
Epigenetic modifications preferentially silence regulatory regions of disease risk genes in CD4 memory and effector T cells, providing a new view to our understanding of their pathogenetic role. It remains to be seen whether this immunodeficient state of a large fraction of the T cell population predates synovitis or is a consequence of chronic inflammation and treatment.
Alt-text: Unlabelled box
Introduction
Rheumatoid arthritis (RA) is a complex inflammatory disease that progresses from a preclinical stage with autoantibody production to eventually relentless and destructive synovitis.1 Autoantibodies are preferentially directed towards post-translational protein modifications and have the features of a T cell-driven adaptive immune response.2,3 However, the recognized autoantigen does not provide information on the nature of the tolerance defect and does not explain the eventual tissue tropism towards diarthrodial joints. At the later stage, multiple cell populations and pathways are involved to establish chronic and progressive synovitis.4
Several lines of evidence implicate T lymphocytes and in particular CD4 T cells as key players in all stages of the disease process.5 Early studies have identified allelic polymorphisms of the HLA-DRB1 alleles that are involved in peptide binding and T cell receptor recognition, as the most prominent genetic risk factor. Likewise, a sequence polymorphism of PTPN22 associated with RA is involved in regulation of T cell receptor signalling.6 Over the last three decades, more than 120 susceptibility loci have been identified, most of them in non-coding regions.7 Quantitative trait loci frequently map to regulatory regions of CD4 T cells, in particular disease-associated SNPs are enriched in super-enhancers functioning primarily in CD4 T cells compared to other cell types.8,9
In addition to the genetic predispositions implicating T cells, cell-intrinsic features have been identified that are already apparent in naïve CD4 T cells of RA patients subsequent to activation.10 RA CD4 T cells have a propensity to differentiate into short-lived effector T cells that are tissue-invasive and cause synovitis.5 Key to this differentiation deviation is a reprogramming of metabolic pathways, manifesting as increased biosynthesis programs through activation of the pentose phosphatase pathway in conjunction with mitochondrial defects resulting in ATP depletion and defects in DNA repair.11, 12, 13, 14 This bias is not limited to a small T cell subpopulation that could be antigen-specific but is seen with bulk naïve CD4 T cells and therefore indicates a global intrinsic T cell defect. The finding that this T cell defect is not related to risk genes raises the possibility that it is acquired and therefore epigenetically encoded.
Indeed, DNA methylation studies have identified RA-associated signatures in T cells. Pitaksalee et al. found differential DNA methylation in peripheral CD4 T cells of RA patients in the promoter of genes contributing to disease-relevant pathways.15 Differential methylation was found in naïve as well as memory CD4 T cells and the disease-related difference was five-fold higher in T cells than in monocytes, emphasizing the role of CD4 T cells in RA. Genome-wide methylation differences in T and B lymphocytes from RA patients were subsequently described by other groups.16, 17, 18, 19 A recent, large and comprehensive study examined differentially expressed genes and differentially methylated regions in CD4 T cells of RA patients and analyzed quantitative trait loci for expression and methylation.20 The results suggested that methylation differences shaped the expression of a fraction of differentially expressed genes, frequently related to RA risk genes. These studies established the existence of disease-associated epigenetic signatures for CD4 T cells; however, they mostly did not control for differentiation states of CD4 T cells.
In addition to DNA methylation, epigenetic signatures can be revealed from genome-wide mapping of histone modifications or of chromatin accessibility. The recent development of the assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) has enabled epigenetic studies on small sample sizes of cells. ATAC-seq employs the transposase Tn5 to fragment chromatin and integrate adapters for next generation sequencing into open chromatin regions. The assay provides information on open chromatin, nucleosome positioning, and transcription factor (TF) occupancy, thereby enabling conclusions on functional consequences. Here, we used ATAC-seq to map chromatin accessibility in peripheral, purified naïve, central memory (CM) and effector memory (EM) CD4 T cells from RA patients and age-matched healthy individuals and identified the corresponding genes predicted to be regulated by these differentially accessible regions. The number of differentially accessible sites increased from naïve to CM and EM T cells, consistent with the interpretation that RA-associated epigenetic signatures on the global T cell population are acquired during differentiation. Regions with increased accessibility were highly enriched for the binding motif of CTCF, indicating differences in the three-dimensional genomic architecture. Unexpectedly, we did not find increased accessibility to regulatory regions of genes that drive an inflammatory response. On the contrary, regulatory regions of functionally relevant genes were predominantly less open in RA CM and EM compared to control CD4 T cell subsets. In particular, regions with reduced accessibility mapped to super-enhancers that included RA-associated SNPs and that therefore regulate disease risk genes. Treatment or disease activity had an influence on epigenetic signatures only in CD4 effector T cells but did not account for the reduced accessibility at disease risk genes.
Methods
Ethics
The protocol was approved by the Stanford University Institutional Review Board (protocol #45993). All participants gave written, informed consent for this study.
Study population
Peripheral blood mononuclear cells (PBMCs) were obtained for ATAC-seq from 18 patients with RA, 11 patients diagnosed with psoriatic arthritis (PsA) and 10 healthy individuals (HC) matched for the same age range. RA patients fulfilled the American College of Rheumatology diagnostic criteria for RA, were positive for rheumatoid factor and/or anti-citrullinated protein antibodies and had longstanding disease of more than 5 years duration. Demographic and clinical data are given in Table 1. We used the clinical disease activity index (CDAI) as disease activity marker because C-reactive protein (CRP) was not available for all patients at the time of the visit. Peripheral blood from an additional 23 RA patients and 23 HCs were obtained for transcriptome studies. RA and PsA patients were predominantly male reflecting the gender distribution at the Palo Alto Veterans Administration Rheumatology Clinic. All patients were recruited from the same clinic and represented the spectrum of patients seen there. Healthy individuals did not have a personal or family history of autoimmune disease and had no history of cancer. Chronic diseases such as hypertension or diabetes mellitus were permitted as long as controlled on oral medication.
Table 1.
Demographic and Clinical Characteristics of Patient Populations.
| Rheumatoid arthritis (RA) | Psoriatic Arthritis (PsA) | |
|---|---|---|
| Demographic parameters | ||
| No. of subjects | 18 | 11 |
| Sex (F/M) | 4/14 | 1/10 |
| Age (mean ± SEM [years]) | 60.38±3.48 | 57.44±4.89 |
| Clinical parameters | ||
| Tender joint count (mean ± SEM) | 3.69±1.37 | 1.4±0.83 |
| Swollen joint count (mean ± SEM) | 3.94±1.22 | 1.3±0.5 |
| CDAI (mean ± SEM) | 13.44±3.46 | 7.18±2.17 |
| CDAI < 2.8 (% patients) | 22.22 | 45.45 |
| CDAI 2.8 – 10 (% patients) | 38.89 | 27.27 |
| CDAI 10 – 22 (% patients) | 11.11 | 18.18 |
| CDAI > 22 (% patients) | 27.78 | 9.09 |
| No DMARD (% patients) | 11.11 | 0 |
| Medications at the time of study, including those on combination therapies (% of patients) | ||
| Corticosteroids | 5.56 | 9.1 |
| Methotrexate | 55.56 | 36.36 |
| Anti-TNFα | 27.78 | 81.82 |
| Other DMARDs | 44.44 | 9.1 |
T cell subset purification
T cells were isolated from peripheral blood using Human T Cell Enrichment Cocktail (STEMCELL Technologies, Canada). CD4 naïve (CD3+CD4+CD62L+CD45RA+CD28+), CM (CD3+CD4+CD62L+CD45RA–CD28+), and EM (CD3+CD4+CD62L-CD45RA–CD28+) T cells were further isolated by fluorescence activated cell sorting (FACS) with a BD Aria 3 cell sorter. The gating strategy is shown in Figure S1. Subset purity was >95%. We used CD62L and not CCR7 as marker for naïve and CM because the antibody allowed for better separation between positive and negative cells in non-frozen PBMCs.
ATAC-seq library preparation
ATAC-seq libraries were generated on naïve, CM and EM CD4 T cells following the standard protocol described previously.21,22 Briefly, 50,000 sorted T cells were washed with cold PBS and RSB buffer (10mM Tris-HCl (pH 7.4), 10mM NaCl, 3mM MgCl2) followed by washing with RSB buffer containing 0.1% NP-40 and 0.1% Tween 20. Cell pellets were resuspended in the transposase reaction mix (25μL 2 × TD buffer, 2.5μL transposase (Illumina) and 22.5μL nuclease-free water) and incubated at 37°C for 30 min. Fragmented genomic DNA was further purified with a Qiagen MiniElute kit and library amplification was performed using Nextera PCR primers. The quality of the libraries was confirmed using a 2100 Bioanalyzer (Agilent Technologies), and libraries were sequenced on an Illumina NextSeq 500 by the Stanford Functional Genomics Facility.
Quantitative PCR
CD4 memory T cells were isolated from PBMC from age-matched healthy donors and RA patients by using the EasySep™ Human Memory CD4 T Cell Enrichment Kit (STEMCELL Technologies, Catalog #19157). Total RNA was extracted from 0.5 million cells using the RNeasy Plus Micro Kit (Qiagen, 74034). 20ng RNA were used to synthesize cDNA by using the High-Capacity RNA-to cDNA Kit (Applied Biosystems, 4387406). qPCR was performed in duplicates in 384-well plates using the ABI 7900HT System with PowerUp SYBR Green Master Mix (ThermoFisher, A25742). The reagents are listed in Table S1 and primers used can be found in Table S2.
Statistics
ATAC-seq preprocessing
Sequencing reads were processed and normalized as described before.21,22 The samples were sequenced in 6 batches. To maintain stringency, an aggregate peak set was generated by including sites present in at least 3 or more samples followed by exclusion of sites with low read counts using the filterByExpr function across sample groups.
Uniform manifold approximation and projection (UMAP) and principal component analysis (PCA)
UMAP and PCA was performed on read counts normalized using variance stabilized transformation (vst) from the DESeq2 package in R. Counts were further filtered to exclude regions contributing to sequencing batch effects using “removeBatchEffect” function from limma package in R. Top 5000 most variable regions, representing a reasonable subsample of the total number of peaks, were selected to perform clustering using UMAP and PCA. The distribution of samples in the PCA were similar when the total peak set was included.
Differential accessibility analysis
We used batch normalization to account for technical effects with the assumption that the donor effect normalization (duplicateCorrelation) from limma23 normalized most of the effects across donors (HC, PsA, RA). We further removed low QC samples defined as those which have a transcription start site enrichment score < 8 and did not show nucleosome specific periodicity in fragment size distribution. The resulting design model was as follows: Group (disease) + Lineage+ Batch along with duplicate correlation to control for donor-specific effects. The data were normalized for GC bias by calculating the offsets and estimating common dispersions as described in the cqn R package.24 Differential sites were inferred using “voom” normalization in limma. Sites/peaks were identified to be differential based on 0.05 adjusted (Benjamini and Hochberg) p-value cutoff.
Transcription factor binding site (TFBS) prediction and pathway analysis
TFBS prediction was performed using HOMER25 on sites identified to be differential across corresponding subset comparisons. Sites were linked to the presumptive gene using GREAT26 followed by determining the frequency of differential peaks per gene. Gene ontology analysis was carried out using DAVID27 and pathway analysis and chromatin state enrichment were done using ChIPseeker package in R.28 Upstream regulator analysis was performed in iPathwayGuide.29 The prediction of upstream regulators was based on two types of information: i) the enrichment of differentially accessible (DA) genes from the experiment and ii) a network of regulatory interactions from iPathwayGuide's proprietary knowledge base. The network is a directed graph, in which the nodes represent genes and the edges represent regulatory interactions between two genes. The results are represented as a two-way plot showing the upstream regulators predicted as activated or inhibited. Dots representing upstream regulators are positioned using P-zscore on the horizontal axis and P-act (activation)/ P-inh (inhibition) on the vertical axis. P-act/P-inh is the p-value based on the number of DA targets consistent with the type of the incoming signal and with expected hypothesis of activation/inhibition. Upstream regulators with a significant combined uncorrected p-value are shown in red, whereas the ones in gray are non-significant. The size of each dot represents the number of DA genes for that regulator.
k-means clustering
Read counts normalized as described above for PCA and UMAP were further median normalized for each T cell subset in HC, PsA and RA. Sites determined to be differentially accessible in RA T cell subsets compared to those of HC were subjected to k-mean clustering. Gap statistic was used to determine the optimal number of clusters.
Statistical comparisons of groups
Group comparisons (selected transcripts in RA and HC T cells, peak sets for different principal components, peak sets for RA patients with different disease activities or drug treatment) were performed using a nonparametric Mann-Whitney/Wilcoxon rank-sum test. For the ATAC-seq comparisons, sites/peaks were identified to be differential based on 0.05 Benjamini-Hochberg adjusted p-value cutoff after fitting a linear model as described above.
Sample sizes were chosen to ensure 80% power with a level of significance of 5% when the difference in their means would be 1.5 standard deviation (n≥10).
Role of funders
Funding agencies providing financial support did not participate in the design, data analyses, interpretation, or writing of this study.
Data deposition
ATAC-Sequencing reads were submitted to SRA (Accession number: PRJNA686153)
Results
Epigenetic changes in circulating CD4 T cells from RA patients
To determine whether T cells from RA patients have distinct chromatin accessibilities, setting them up for different transcriptional response patterns, we performed ATAC-seq of peripheral CD4 T cell subsets including naïve, CM and EM cells from 18 RA patients and 10 HC. Eleven patients with PsA served as disease control. All patients had long-standing disease of more than 5 years duration, and all RA patients had antibodies to citrullinated antigens and/or rheumatoid factor. As shown in Table 1, most patients were on combinations of different immunosuppressive treatments at the time of the blood collection with variable control of disease activity. Two RA patients were non-compliant and off treatment and had active disease. Four RA patents and five PSA patients fulfilled the criteria for remission on treatment with a CDAI of <2.8.
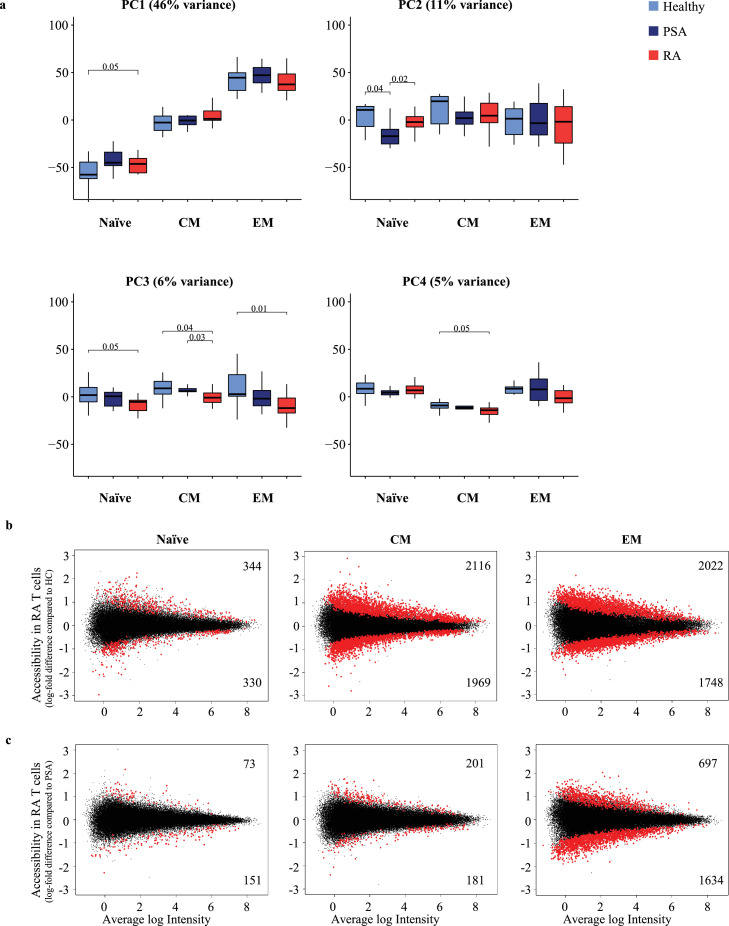
As expected, PCA based on the top 5000 sites most variable in chromatin accessibility showed clear clusters corresponding to differentiation states in PC1, re-emphasizing the need to separately analyze functional subsets in such studies (46% of variance, Figure 1a). Very subtle disease-associated changes in PC1 indicated accelerated differentiation in RA. PC2 did not show any correlation with differentiation or disease. In contrast, PC3 (6%) showed separation of samples of RA patients from HC across all three differentiation states (Naïve p=0.05, CM p=0.04, EM p=0.01) and PC4 (5%) for CM (p=0.05) and less for EM (p=0.09). For both PC3 and PC4, T cells from PsA patients mapped between RA patients and HC. Since differentiation states accounted for a high proportion of the most variable sites, we further selected the top 5000 most variable sites within each T cell subset to delineate disease-specific effects and performed clustering using uniform manifold approximation and projection (UMAP) (Figure S2a). While there was no clear disease-specific clustering for any of the T cell subsets, the majority of RA patients trended to be more distant from HC with PSA patients being scattered.
Figure 1.
Differences in chromatin accessibility in CD4 T cell subsets from patients with rheumatoid arthritis (RA) compared to healthy controls (HC) and psoriatic arthritis (PsA). (a) Principal component analysis (PC) for 5000 top variable peaks; box plots showing contribution of Naïve, CM and EM CD4 T cell subsets in HC (light blue), PsA (dark blue) and RA patients (red) to PC1-4. P-values were determined using a nonparametric Mann-Whitney/Wilcoxon rank-sum test. (b) MA plots showing the distribution of log-fold differences between RA and HC or (c) RA and PsA samples in peak sets of each T cell subset. Differential peaks (red) were determined using adjusted (Benjamini and Hochberg) p-value < 0.05. Numbers of peaks that are more (top) or less accessible (bottom) in T cells from RA patients.
Calling differential sites in RA compared to HC individuals for each cell subset separately provided evidence that disease-specific patterns in chromatin accessibility are present in circulating T cells, more so in the more differentiated CM and EM than in naïve CD4 T cells (Figure 1b). 674 significant differences were identified for naïve CD4 T cells, much less than for CM (n=4085) and EM T cells (n=3770). About equal proportions of differentially accessible sites were more or less accessible in RA T cells. Fewer significant differences in chromatin accessibility were observed when T cell subsets from PsA patients and HC were compared (Figure S2), in particular EM of PsA patients were similar to HC EM but differed from RA EM at 2331 sites (Figure 1c). Given that ATAC-seq was performed on total circulating CM and EM T cells that are highly diverse expressing more than one million unique T cell receptors,30 the epigenetic signatures in RA cannot be accounted for by infrequent antigen-specific T cells. Rather, they reflect a global differentiation bias of RA CD4 T cells.
Functional implications of disease-specific epigenetic alterations in RA
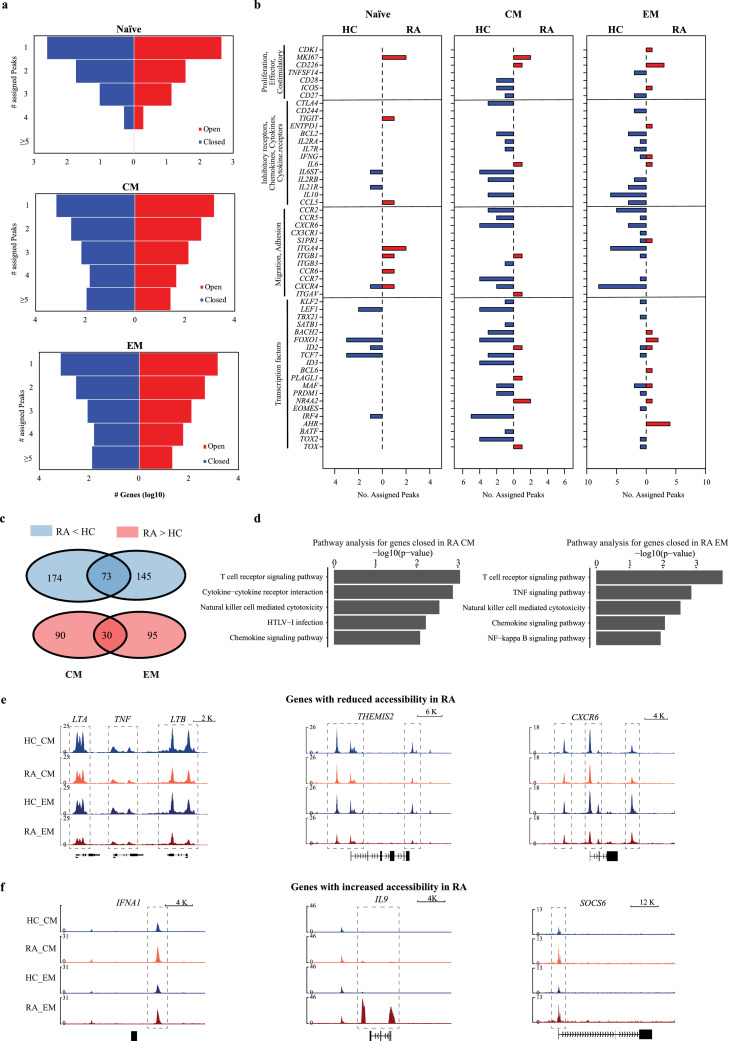
To predict functional implications of differentially accessible sites, we used Genomic Regions Enrichment of Annotations Tool (GREAT)26 to assign the sites to genes based on their location in regulatory regions. For naïve CD4 T cells, only few genes were identified that had >2 differentially accessible regulatory regions. In contrast, the memory cell subsets showed a high number of genes associated with multiple less or more accessible regions (Figure 2a). These genes included many molecules associated with immunological functions. Surprisingly, most of these genes were associated with regulatory regions that were more closed in RA compared to HC T cells (Figure 2b). Findings were very similar for CM and EM T cells, with an overlap in genes that had multiple regulatory sites (>2, Figure 2c). Moreover, changes in regulatory regions frequently exhibited the same directionality in CM and EM cells (Figure 2b). Similar results were obtained when EM T cells from RA and PsA patients were compared (Figure S3a). Genes with lesser accessibility were significantly enriched for T cell receptor, cytokine and chemokine signalling pathways for both CM and EM subsets as determined by DAVID27 pathway annotation (Figure 2d). In contrast, no significant pathway enrichment was found for genes associated with more accessible sites. Figure 2e, 2f and S3b depict accessibility in representative genomic tracks, comparing naïve, CM and EM CD4 T cells from HC (blue) and RA patients (red). Patterns of reduced accessibility were frequently found across both differentiation states. Remarkably, the entire LTA-TNF-LTB region was less accessible in RA. Many of the closed genes are involved in T cell regulation (THEMIS2, CD2, CD48, SLAMF1, SLAMF6, PRKCQ) predicting a complex T cell defect. For the chemokine receptors CCR2 and CXCR6, CM and EM from RA T cells had failed to increase accessibility with differentiation and appeared to be more like naïve T cells. An extended region including TF SMAD3 exhibited reduced accessibility. Also, PFKFB3 was less accessible, a gene which reduced expression level has been implicated in metabolic defects in activated RA T cells.13 In contrast, only few immune-related genes displayed convincingly increased accessibility, including IFNA1 and SOCS6. Of note, SOCS6 is a negative STAT signalling regulator, overexpression is therefore fitting to the theme of reduced T cell responsiveness. One notable exception of this theme was IL9, which was more accessible exclusively in EM T cells of RA patients. Accessibility was highly variable; a few patients showed very high levels of accessibility to the IL9 locus compared to HC, while in most patients the increase was detectable but less striking.
Figure 2.
Identification of genes corresponding to regulatory regions differentially accessible in CD4 T cells from RA patients. (a) Genes predicted to be regulated by differentially accessible regions were determined by GREAT57. Y-axis denotes the number of differentially accessible peaks associated to each gene. X-axis denotes log10 of the total number of genes with regulatory sites more open (red) or more closed in RA (blue). (b) Selected genes of potential immunological relevance and multiple differentially accessible regions are shown for naïve, CM and EM T cells. Bar graphs depict the number of differentially open gene regulatory regions that are more accessible in HC (left, blue) or RA (right, red). (c) Venn diagrams showing that genes associated with >2 differentially accessible regions partially overlap between CM and EM subsets. (d) Top enriched pathways determined by DAVID26 for genes associated with > 2 regulatory regions less accessible in RA. A pathway enriched in genes with more accessible regulatory sites was not identified. (e) Representative tracks for genes with regulatory regions less accessible in RA. Results are shown for CM and EM T cells from HC (blue) and RA patients (red). Additional tracks are shown in Fig. S3b. (f) Representative tracks for genes with regulatory regions more accessible in RA.
Upstream regulators of altered chromatin accessibility in RA
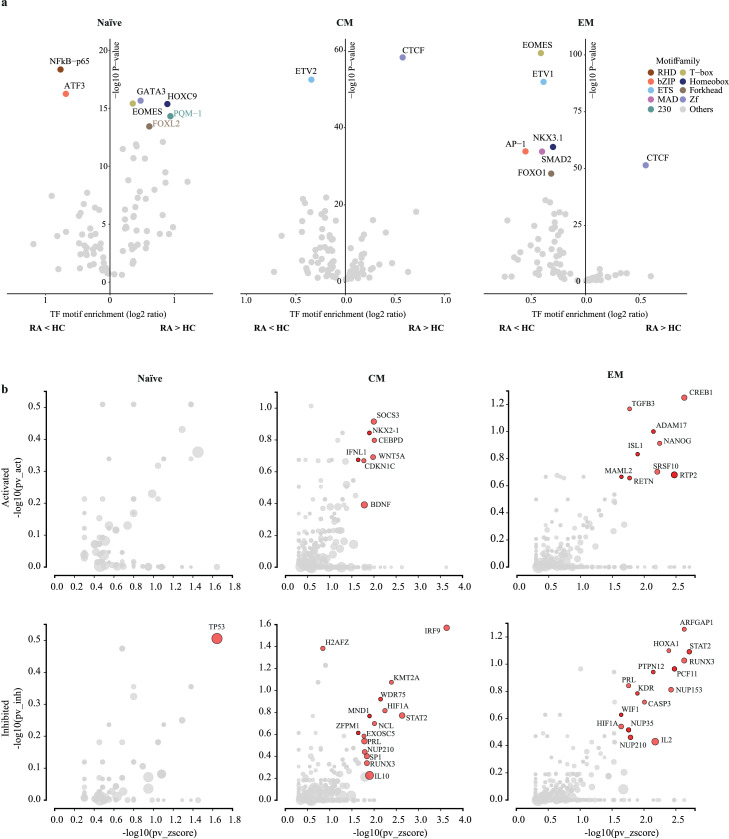
To infer TF networks differentially regulating RA and HC CD4 T cells, we predicted the TF binding motifs enriched at sites differentially accessible using HOMER (Figure 3a).25 RHD (NF-κB-P65) and bZIP (ATF3) family TFs were among the top motifs more closed in RA naïve cells whereas Zf (GATA3), T-box (EOMES) and Homeobox (HOXC9) family TFs were more open. For both, the CM and EM subsets, CTCF motifs were by far the most enriched motif at the differentially more open sites, while ETS (ETV1/2) family members were more enriched at less accessible sites. In addition, motifs for differentiation- and proliferation-related TFs like bZIP (AP-1), T-box (EOMES), Forkhead (FOXO1), MAD (SMAD2) and Homeobox (NKX3.1) were less accessible in the EM cell subset. In summary, TF networks that were predicted to be involved in determining the distinct chromatin accessibility patterns were similar in CM and EM with CTCF motifs characteristic for sites with increased accessibility in RA and several TFs involved in T cell differentiation/activation at sites more open in HC.
Figure 3.
Upstream regulators of differentially accessible regulatory sites in RA CD4 T cells. (a) Identification of transcription factor (TF) motifs enriched at sites more (right) or less open (left) in RA for each T cell subset. For each motif family, only the top ranked TF is shown. X-axis denotes enrichment of each motif over background, Y-axis the enrichment log10 p-value as reported by HOMER. (b) Upstream regulator genes predicted to be activated (top) or inhibited (bottom) in RA compared to HC for each T cell subset. Y-axis compares the number of targets consonant with the prediction to the total number of targets. X-axis denotes the overrepresentation of these targets compared to the number of target genes expected just by chance. Upstream regulators with a significant combined p-value < 0.05 are labelled (red) as reported by iPathwayGuide.
To identify upstream regulators, we analyzed genes identified for each CD4 T cell subset by GREAT as differentially regulated using iPathway Guide. Regulators that were activated in RA were few and included WNT5A, SOCS3 and BDNF in the memory subsets (Figures 3b, S4). Even of those, SOCS3 is a negative regulator as its activity results in suppression of cytokine signalling. Regulators that accounted for reduced accessibility in regulatory regions of target genes in RA included IRF9, STAT2, IL2, RUNX3 and HIF1A (Figure 3b, Figure S4). In particular, upstream regulators STAT2 and IRF9 shared loss of accessibility of many downstream targets in RA CD4 memory T cells, including closure of classic interferon-response genes. Inhibition in targets downstream of IL2, RUNX3 and HIF1α was consistent with the interpretation that T cell activation/differentiation-related genes were less accessible in RA.
Sites with differential chromatin accessibility in RA T cells change with differentiation
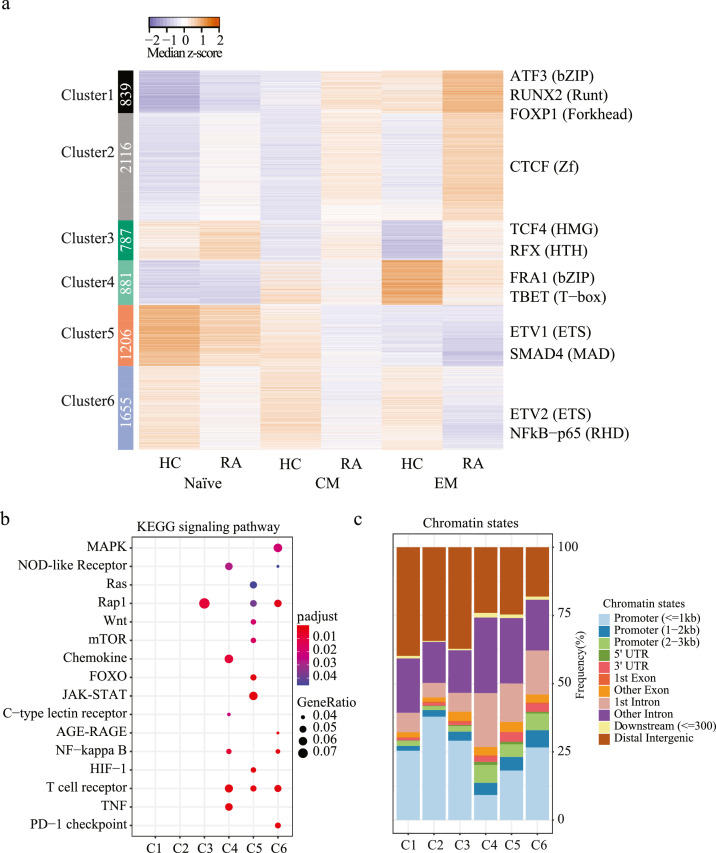
We applied k-means clustering to peaks identified to be significantly different between RA and HC for any of the T cell subset comparisons. A total of 6 clusters were identified with clusters 1-3 showing higher and clusters 4-6 showing lower accessibility in RA (Figure 4a). The pattern was consistent for all three differentiation states, i.e., although differences between HC and RA T cells were more prominent for EM, they were also present for naïve and CM CD4 T cells. With the exception of cluster 6, sites in all clusters were dependent on differentiation states. In particular sites in clusters 1 and 4 opened up with differentiation and were accordingly enriched for bZIP, T-box and Runt family motifs. Since sites in cluster 1 were more and sites in cluster 4 less accessible in RA, the data suggest a distinct differentiation process for HC and RA. Similarly, clusters 3 and 5 were different in RA and HC and closed with differentiation, however involving different TFs. A unique scenario was cluster 2 that progressively opened with differentiation only in RA T cells and was enriched for CTCF motifs.
Figure 4.
k-means clustering of sites differentially open in naïve, CM or EM T cells from RA compared to HC. (a) Heatmap shows median normalized z-scores grouped according to k-means clustering (left). TF family motifs mostly enriched at peaks included in each cluster were identified (right); highest ranked family member and family is given (right) (b) KEGG Pathway enrichment analysis58 identifying signalling pathways for genes associated to peaks in each cluster. (c) Distribution of peaks across chromatin state for each cluster as reported by ChIPseeker.
Although the peak set in the k-means analysis was chosen based on differential accessibility between RA and HC, there were some similarities to PsA. Figure S5a shows the heat plot with the data from the corresponding peak sets of PsA patients included. The PsA samples followed the same clustering, with accessibility in PsA samples frequently in between those of RA and HC T cells.
KEGG signalling pathway analysis identified enrichment of functional pathways nearly exclusively only for sites closing in RA (Figure 4b). The only exception was Rap1 signalling enriched in cluster 3 with sites opening in RA. Consistent with a lack in pathway enrichment, many of the sites in clusters 1 to 3 mapped to distal intergenic regions (Figure 4c). In contrast, clusters encompassing sites closing in RA were functionally important involving many KEGG signalling pathways. DAVID functional annotation analysis showed common enrichment of all three clusters 4, 5 and 6 for the T cell receptor signalling pathway (Figure S5b).
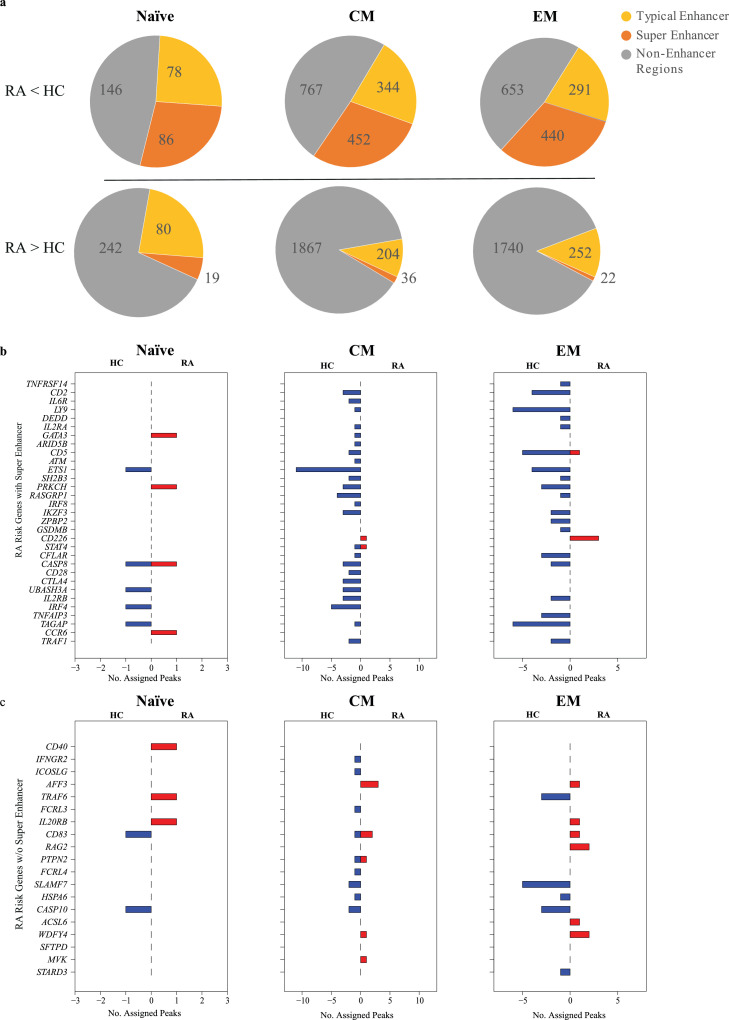
Reduced accessibility to RA risk genes with super-enhancers
Consistent with the lack in pathway enrichment, the majority of sites more open in CD4 T cell subsets from RA patients did not map to enhancer regions (Figure 5a). In contrast, about 50% of sites with reduced accessibility included enhancers. Especially striking was the frequent finding of super-enhancers. Vahedi et al. have previously reported that RA-associated SNPs are highly enriched at super-enhancers of CD4 T cells.8 To explore the relationship between chromatin accessibility and disease-associated SNPs, we examined disease risk genes for the number of differentially accessible sites in regulatory regions (Figure 5b). Most of the RA risk genes with super-enhancers were associated with sites that were closing in RA in CM and EM subsets. In contrast, accessibilities to regulatory regions of RA risk genes that did not have a super-enhancer were not much different between RA and HC (Figure 5c).
Figure 5.
Sites with decreased accessibility in RA overlap with super-enhancers encompassing disease-associated SNP. (a) Pie charts showing the number of sites that can be mapped to CD4 T cell typical enhancers (yellow) or super-enhancers (orange) as identified by Vahedi et. al8 for sites that are more closed (top) or more open (bottom) in RA T cells compared to HC. (b) Bar charts showing the number of assigned peaks to RA risk genes that have super-enhancers including RA-associated SNPs8 (blue - more open in HC, red - more open in RA T cells). (c) Bar charts showing the number of assigned peaks to RA risk genes that do not have a super-enhancer.
To confirm the functional relevance of the reduced accessibility, we determined transcript levels in total memory CD4 T cells for selected representative genes. ETS1, STAT4, SH2B3 and CD2 all have super-enhancers with disease-associated SNPs and showed lower accessibility in RA (Figure 5b). Three out of these four also had reduced transcript levels (ETS1 p=0.02, STAT4 p=0.03 and CD2 p=0.02) (Figure S6). BACH2 and BCL6 are TF genes with less accessible regulatory sites (Figure 2). BACH2 (p=0.04) was also less transcribed, while BCL6 (p=0.2) showed a trend (Figure S6).
To determine whether the association with selected super-enhancers was disease-specific, we analyzed sites differentially accessibility in PsA compared to HC T cell subsets for their relationship to super-enhancers. As already shown in Figure S2b, sites with differential accessibility in PsA were mostly found for CD4 CM T cells. Surprisingly and very much in contrast to RA, only few sites were significantly different in CD4 EM T cells when compared to HC. Accordingly, only few genes with differential accessibility in super-enhancers were identified for PsA naïve and EM T cells (Figure S7a). In case of PsA CM CD4 T cells, the number of RA risk genes with differential accessibility was low as was the number of differentially closed regulatory sites per gene irrespectively of whether they had super-enhancers or not (Figure S7a, b). In contrast, PsA EM T cells, hardly distinguishable from HC, were highly different from RA EM T cells. The patterns of chromatin accessibility for genes with super-enhancers in RA vs PSA EM T cells was reminiscent of the comparison RA vs HC; most of these genes were associated with sites closing in RA (Figure S7c).
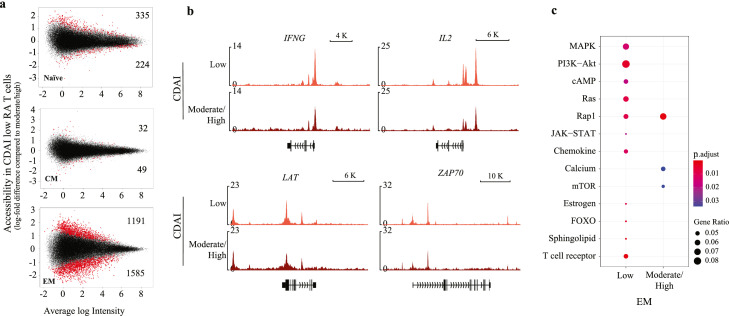
Effect of disease activity and treatment on chromatin accessibility in RA
To explore whether disease activity had an effect on chromatin accessibility, we compared RA patients with low and moderate/high CDAI. We observed minimal changes in naïve (n=559) and CM (n=81) subsets, but clear differences in the EM subset (n=2776) (Figure 6a). Several examples of chromatin accessibility influenced by disease activity are shown in Figure 6b. IFNG and IL2 were less accessible in patients with high disease activity. Genes involved in TCR signalling like LAT and ZAP70 were more closed in RA in general with more prominent reduction in patients with higher CDAI. Pathway analysis (KEGG in Figure 6c and DAVID in Figure S8a) revealed enrichment for the T cell receptor, PI3K-Akt and FOXO signalling pathways for sites more open in patients with low disease activity. Conversely sites more open in patients with moderate/high disease activity were enriched for RAP1, WNT and mTOR signalling pathways. TF-motif enrichment provided further classification of upstream regulators (Figure S8b). Motifs belonging to IRF family TFs were exclusively enriched at sites opening in patients with low disease activity, while motifs of a large variety of TFs including MAD, T-box, Homeobox, HLH and Zf family TFs were enriched at sites more accessible in patients with higher CDAIs. Taken together, the chromatin appeared to be more poised in EM T cells with lower disease activity.
Figure 6.
Influence of disease activity on chromatin accessibility. (a) MA plots comparing chromatin accessibility in RA patients with low (n=11) versus medium/high CDAI (n=7). Sites with significantly differential accessibility are indicated by red (top more accessible in RA patients with low disease activity). Differential peaks (red) were determined using adjusted (Benjamini and Hochberg) p-value < 0.05. (b) Representative tracks for genes showing differences in accessibility in EM cells of patients with different disease activity. (c) KEGG pathway enrichment for genes associated with sites differentially accessible in EM T cells of patients with different disease activity.
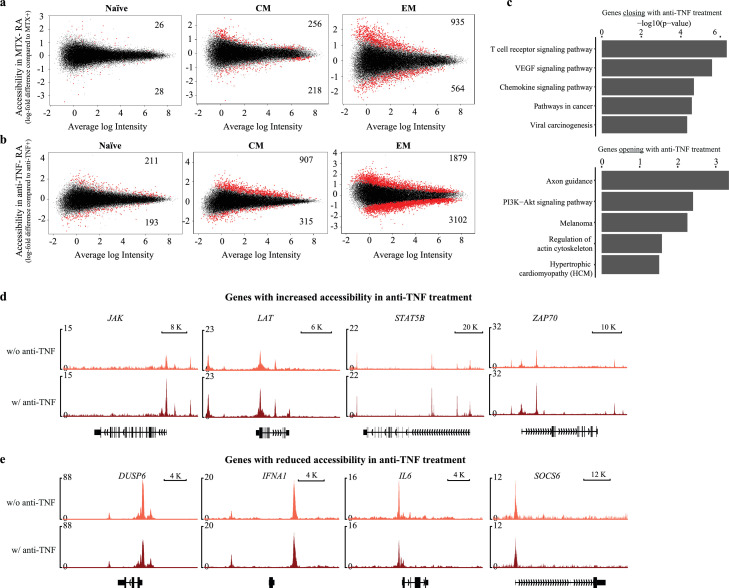
Patients were on a variety of treatment combinations, limiting the ability to assess the impact of treatment on chromatin structures. We observed minimal differences in the naïve (n=54) and CM (n=474) and a slightly higher number of differentially accessible sites in the EM subset (n=1499) in patients on treatment with methotrexate (MTX) vs those on other modalities (Figure 7a). Pathway analysis for the genes associated with differential sites associated with MTX treatment did not identify any significant enrichment. Larger differences were associated with anti-TNF treatment (Figure 7b). Similar to MTX, we observed the largest effect for the EM subset (n=4981). Sites closing with anti-TNF treatment in EM were enriched for T cell receptor and chemokine signalling pathways (Figure 7c). Genes with sites closing in patients with anti-TNF treatment include IFNA1 as well as IL6. Conversely, PI3K-Akt signalling was enriched for sites more open with anti-TNF treatment. Sites more open in patients with anti-TNF treatment include molecules involved in JAK-STAT signalling (JAK, STAT5B) and T cell receptor signalling (LAT, ZAP70) (Figure 7d). Small sample sizes precluded definite conclusions, but TNF inhibition appeared to have complex consequences on EM cells enabling as well as inhibiting inflammatory pathways.
Figure 7.
Influence of treatment on chromatin accessibility maps of RA T cells. (a) MA plots comparing chromatin accessibility in RA patients on treatment with MTX (n=9) versus those not on MTX (n=9). (b) MA plots comparing chromatin accessibility in RA patients on treatment with a TNF inhibitor (n=5) versus those not on a TNF inhibitor (n=13). Differential peaks (red) were determined using adjusted (Benjamini and Hochberg) p-value < 0.05. (c) DAVID pathway enrichment analysis for genes associated to sites with higher accessibility in EM cells of patients on TNF inhibitor treatment. (d, e) Representative tracks of genes with different chromatin accessibility in EM cells depending on TNF inhibitor treatment.
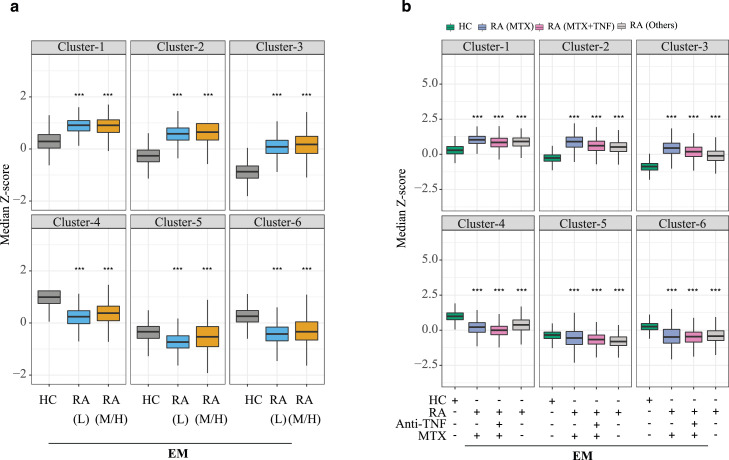
RA-associated epigenetic signatures are not accounted for by treatment- and disease activity-related effects
As shown in Figures 6 and 7, disease activity and treatment, in particular with TNF inhibitors, induced chromatin changes in EM T cells. To determine whether the disease-related epigenetic signature described in Figures 1 to 5 is due to these confounding variables or whether it is an inherent feature of the disease itself, we reanalyzed the peak set defined in Figure 1b separately for different subgroups of patients. Figure 8a shows median z-scores of EM cells as box plots for the clusters shown in Figure 4a. Median z-scores did not differ between T cells from patients with low (CDAI < 10, n=11) and moderate/high disease activity (CDAI > 10, n=7); both of them were equally different from those of HC for all 6 clusters. Similarly, we separated the RA patients into three groups based on whether they were on MTX only (n=6), MTX in combination with anti-TNF inhibitors (n=3) or neither (n=7). Again, RA-associated shifts in z-scores were similar for all three treatment groups although individual group sizes were too small to assess significance levels. (Figure 8b). Taken together, with few exceptions, there was little similarity in RA-associated differences in chromatin accessibility and disease activity-induced or treatment-related changes, suggesting that the RA-associated signature is largely independent of these confounding variables.
Figure 8.
Contribution of disease activity and treatment to chromatin differences identified in RA. (a) RA patients were segregated into patients with low (<10, n=11) and moderate/severe CDAI (>10, n=7). Boxplots show z-scores of sites differentially accessible between patients and HC as shown in Figure 4a but depicting patients with low and moderate/high CDAI as separate boxplots for each cluster. (b) Sites differentially accessible between RA and HC T cells as shown in Figure 4a were separately analysed for RA patients on methotrexate (MTX, n=6), on a TNF inhibitor plus MTX (n=3) or on neither (n=7). Results are shown as box plots of median z-scores for each k-means cluster. P-values were determined using a nonparametric Mann-Whitney/Wilcoxon rank-sum test. *** P < 0.001.
Discussion
Here, we show that peripheral CD4 T cells from RA patients epigenetically differ from those of age-matched, healthy controls. Differences in chromatin accessibility were already present in unstimulated naïve CD4 T cells but were more pronounced in CM and EM T cells, consistent with the model that naïve CD4 T cells from RA patients are poised to differentiate distinctly from those of healthy adults.5 Since these differences were identified at a population level, they represent a polyclonal feature that cannot be explained by the activation and expansion of infrequent autoantigen-specific CD4 T cells. The sequence and the genomic location of the differentially accessible region allowed conclusions on the TF networks driving the epigenetic signatures as well as predicting differences in cells’ responsiveness to stimuli. Surprisingly, sites more accessible in RA were not enriched for motifs of TF known to be involved in T cell activation or lineage determination, such as bZIP or T-box family members.31 Particularly, promoter regions of inflammatory mediators were not more accessible in T cells from RA patients, with the notable exception of IL9 in a subset of patients and IFNA. On the contrary, super-enhancers of genes implicated in RA were less accessible in CM and EM T cells.
Genome-wide association studies have identified more than 100 SNP that are associated with RA7. The majority of disease-associated variants map to non-coding, regulatory regions and in particular super-enhancers.32 About one quarter of RA-associated SNP locate to super-enhancers compared to less than 10% to typical enhancers.8 Since super-enhancers regulate genes that are important for cell identity,33 this information allows implicating the cell type likely involved in pathogenesis. While super-enhancers in muscle cells have little association with RA-risk genes, the strongest association was seen for CD4 T cells with about half of the disease-associated SNP mapping to CD4 super-enhancers. In the current study, we therefore focused on CD4 T cells of RA patients and mapped chromatin accessibility in comparison to CD4 T cells from age-matched healthy controls.
Disease-associated signatures in the epigenome can provide information on how environmental stimuli and genetic predisposition interact to contribute to the pathogenesis of an autoimmune disease such as RA. In the adaptive immune system, memory cells reflect the history of antigen encounters. Distinct T cell differentiation states grossly differ in epigenetic structures.34,35 We therefore controlled for differentiation states by separately examining naïve, CM and EM CD4 T cells for chromatin accessibility. We found a higher number of differentially accessible, disease-associated sites in both memory subsets compared to naïve T cells. This was consistent with the PC analysis, with PC3 segregating patients and controls irrespective of the differentiation state and PC4 only for memory T cells. These data support the notion of global shifts in memory cell differentiation in RA patients irrespective of the nature of the antigen and is reminiscent of in vitro studies that show a preferential differentiation of naïve CD4 T cells into short-lived effector T cells producing a variety of pro-inflammatory cytokines.11, 12, 13,36, 37, 38, 39 However, the observed chromatin accessibility pattern was distinct from accelerated effector cell differentiation. Differentially accessible sites were partially overlapping for central memory and effector memory T cells and TF motifs enriched at these sites did not include those of lineage determining TF such as those of the TH1 or TH17 lineages.40 In fact, the motif that was by far and most significantly enriched was CTCF indicating a change in the three-dimensional genome structure, thereby possibly allowing distal enhancers to abnormally influence gene expression.41,42
Surprisingly, sites that were less accessible in RA CD4 memory T cells had numerous features of RA-relevant genes. Less accessible genes included IKFZ1 and PFKB3, which were previously found to be less expressed in naïve CD4 T cells from RA patients. The reduced expression of these genes endowed T cells with enhanced ability to cause synovial inflammation.43,44 In contrast, most of the less accessible genes are predicted to be supportive of T cell function. Most compellingly, these sites are enriched for super-enhancers encompassing disease-associated SNPs. This epigenetic state of the disease risk genes indicates that they are less poised to respond. Consistent with this interpretation, constitutive transcript expression of 3 of 4 disease risk genes with less accessible super-enhancers was significantly lower in RA T cells (Figure S6). Moreover, KEGG pathway as well as Gene Ontology (GO) term analysis of genes regulated by less accessible sites yielded enrichment for pathways important for T cell activation including T cell receptor, JAK-STAT, MAPK and NF-kB signalling (Figure 2d, 4b, S5b). Also, TF motifs enriched at these sites include many that are known to be involved in T cell activation and differentiation (Figure 3a).
Epigenetic studies in RA have so far been limited to comparing DNA methylation. Early studies suggested global DNA demethylation in PBMC of RA patients.44 Subsequent studies showed that differences in DNA methylated sites were highest for CD4 T cells, including naïve CD4 T cells, when compared to myeloid cells.15 The general assumption has been that disease-relevant genes are hypomethylated. In a very comprehensive study, Ha et al. profiled genome-wide differential gene expression and DNA methylation in CD4 T cells and related them to RA-associated genetic variants in a Korean population.20 The authors concluded that methylomic differences involved genomic regions including RA-associated SNP and were related to differential gene expression in CD4 T cells from RA patients. We re-analyzed the methylation data to determine how far the differential accessibility data in our study correlated with differential DNA methylation, particularly whether loss in accessibility for disease risk genes with super-enhancers is associated with hypermethylation. Only a small fraction of the large number of differentially methylated sites was also differentially accessible. Of the 452 and 440 less accessible sites that mapped to disease risk genes with super-enhancers in CM and EM RA T cells, respectively, more than twice as many were hyper- rather than hypomethylated (Table S3). Conversely, there was no significant overlap of increased accessibility and DNA methylation pattern. Limitations in this comparison are that the DNA methylome was obtained on total CD4 T cells, irrespective of the differentiation state and that RA-associated methylation patterns in total CD4 T cells in general did not correlate well with differential chromatin accessibility. Still, the data are highly suggestive that hypermethylation dominated at super-enhancers with reduced chromatin accessibility in CM and EM CD4 T cells from RA patients.
Our epigenetic findings are reminiscent of functional studies that showed defective activation of RA T cells, also coined as T cell anergy.45 Several models can be considered to explain these findings. Loss in accessibility could be a result of treatment. Previous studies have shown that CD4 T cell super-enhancers are maintained by baseline JAK-STAT activity that occurs even in the absence of exogenous cytokines.8 However, RA patients in our study population, with one exception, were not on a JAK inhibitor. The two most common treatment regimens were methotrexate and a TNF inhibitor. These treatment modes had impact on epigenetic signatures, but only in effector memory and not in central memory T cells. Although a final determination will have to await longitudinal studies in patients before and after initiation of treatment, results shown in Figure 8 suggest that the loss in accessibility is present irrespective of the type of treatment. Also, epigenetic signatures in PsA patients, who were on similar treatment regimens, were mostly different. Similar to treatment, disease activity had an impact on the epigenome of effector T cells but did not explain the epigenetic RA signature in EM CD4 T cells (Figure 8).
The loss in chromatin accessibility could occur as an attenuation to constant exposure to chronic inflammation.46 This model is reminiscent of T cell exhaustion,47 although epigenetic signatures of RA T cells do not resemble those of antigen-specific exhausted cells.22,48, 49, 50 Oligoclonality within the peripheral T cell repertoire has been a consistent feature of RA patients, and the extensive replicative history associated with oligoclonal expansion could cause a quasi-exhausted state with loss of chromatin accessibility. In the initial studies, RA patients were found to have clonal effector T cell populations that have later been coined as TEMRA cells.51 More systematic repertoire studies have found a contraction in T cell receptor diversity in RA.52 More recently, repertoires of clonally expanded T cells were found to be shared between the EM and the TH17 peripheral blood population of RA patients.53 Similarly, general defects, also coined as immune paralysis, are found in the post-septic environment and include impaired antigen-specific expansion and effector functionality of memory CD4 and CD8 T cells.54 Upstream regulators of the less poised genes in RA CD4 T cells included molecules of the JAK-STAT, which may be a result of chronic cytokine exposure. As discussed above, the suppression of JAK-STAT signalling would result in loss of accessibility to super-enhancers.
Finally, loss of accessibility could also be a primary event and not a consequence of disease. There is evidence that immunodeficiency can predispose for a hyper-inflammatory state. Hereditary immunodeficiency syndromes are frequently associated with an autoimmune syndrome.55 Also, immune defects in animal models can lead to RA-like clinical presentation. For example, the SKG mouse, one of the better mouse models of RA, has a Zap-70 mutation that impairs T cell responses and increases the susceptibility to infections while causing arthritis and other organ inflammation.56 Clinical observations suggest a similar constellation for human disease. Cohort studies have shown that RA is associated with an increased risk of serious infection.57 This elevated susceptibility of patients with RA is not fully explained by comorbid conditions or the immunosuppressive treatment; rather, the pathobiology of RA itself appears to be an important contributor. Irrespective of whether the observed epigenetic signature is primary or secondary, our findings indicate that disease susceptibility genes are less poised in patients with established RA, which will impact their overall ability to generate a protective immune response.
One limitation of our study is that it only relied on chromatin accessibility data. In contrast to DNA methylation data, ATAC-seq allows us to propose upstream regulators based on predicted binding motifs but cannot prove their involvement. Moreover, functional consequences are difficult to predict because the differentially accessible sites frequently map to regulatory regions, whose functional role is often poorly defined. In contrast to many epigenetic studies, we controlled for differences in cell types and differentiation states by isolating and comparing highly purified T cell subsets. However, we did not control for clinical covariables that may have affected our findings. We were strict on only enrolling RA patients with positive serology, but otherwise our study population represented a sample of patients seen in a continuity clinic with a longer disease duration. In studying patients with a chronic disease, conclusions whether a finding is primary or secondary are difficult to be drawn. Future studies will have to assess patients with early disease, and longitudinal studies have to be performed to assess the influence of disease activity and treatment.
Contributors
RRJ, BH, WJG, CMW and JJG designed the research. RRJ and BH contributed equally. BH, XL and ZY performed the experimental work. RRJ performed the bioinformatics analysis on the data. RRJ, BH, WJG and JJG analyzed and interpreted data. KS and JJG recruited patients. CMW and JJG acquired funding. RRJ and JJG wrote the manuscript. RRJ, BH and JG verified the underlying data. All authors read and approved the final version of the manuscript.
Declaration of interests
W.J.G. declared royalties or licenses and stock holding with 10x genomics. The other authors did not indicate any potential conflicts of interest.
Acknowledgments
Acknowledgments
This work was supported by the Veterans Administration I01 BX001669 to JJG, with resources and the use of facilities at the Palo Alto Veterans Administration Healthcare System, and funds from the National Institutes of Health (R01 AR042527, R01 AI108906 to CMW and U19 AI057266 to JJG), and the Praespero Foundation to JJG and CMW. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Data sharing statement
The supporting data for this work is available from the corresponding author upon request.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2022.103825.
Appendix. Supplementary materials
References
- 1.Firestein GS, McInnes IB. Immunopathogenesis of rheumatoid arthritis. Immunity. 2017;46(2):183–196. doi: 10.1016/j.immuni.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Delft MAM, Huizinga TWJ. An overview of autoantibodies in rheumatoid arthritis. J Autoimmun. 2020;110 doi: 10.1016/j.jaut.2019.102392. [DOI] [PubMed] [Google Scholar]
- 3.Scherer HU, Huizinga TWJ, Kronke G, Schett G. Toes REM. The B cell response to citrullinated antigens in the development of rheumatoid arthritis. Nat Rev Rheumatol. 2018;14(3):157–169. doi: 10.1038/nrrheum.2018.10. [DOI] [PubMed] [Google Scholar]
- 4.Wei K, Korsunsky I, Marshall JL, et al. Notch signalling drives synovial fibroblast identity and arthritis pathology. Nature. 2020;582(7811):259–264. doi: 10.1038/s41586-020-2222-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weyand CM, Goronzy JJ. The immunology of rheumatoid arthritis. Nat Immunol. 2021;22(1):10–18. doi: 10.1038/s41590-020-00816-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mustelin T, Bottini N, Stanford SM. The contribution of PTPN22 to rheumatic disease. Arthritis Rheumatol. 2019;71(4):486–495. doi: 10.1002/art.40790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okada Y, Wu D, Trynka G, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vahedi G, Kanno Y, Furumoto Y, et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520(7548):558–562. doi: 10.1038/nature14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, McGovern A, Martin P, et al. Analysis of chromatin organization and gene expression in T cells identifies functional genes for rheumatoid arthritis. Nat Commun. 2020;11(1):4402. doi: 10.1038/s41467-020-18180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weyand CM, Goronzy JJ. Immunometabolism in the development of rheumatoid arthritis. Immunol Rev. 2020;294(1):177–187. doi: 10.1111/imr.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Shen Y, Hohensinner P, et al. Deficient activity of the nuclease MRE11A induces t cell aging and promotes arthritogenic effector functions in patients with rheumatoid arthritis. Immunity. 2016;45(4):903–916. doi: 10.1016/j.immuni.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Shen Y, Oishi H, et al. Restoring oxidant signaling suppresses proarthritogenic T cell effector functions in rheumatoid arthritis. Sci Transl Med. 2016;8(331):331ra38. doi: 10.1126/scitranslmed.aad7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z, Fujii H, Mohan SV, Goronzy JJ, Weyand CM. Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. J Exp Med. 2013;210(10):2119–2134. doi: 10.1084/jem.20130252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu B, Zhao TV, Jin K, et al. Mitochondrial aspartate regulates TNF biogenesis and autoimmune tissue inflammation. Nat Immunol. 2021;22(12):1551–1562. doi: 10.1038/s41590-021-01065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pitaksalee R, Burska AN, Ajaib S, et al. Differential CpG DNA methylation in peripheral naive CD4(+) T-cells in early rheumatoid arthritis patients. Clin Epigenetics. 2020;12(1):54. doi: 10.1186/s13148-020-00837-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glossop JR, Nixon NB, Emes RD, et al. Epigenome-wide profiling identifies significant differences in DNA methylation between matched-pairs of T- and B-lymphocytes from healthy individuals. Epigenetics. 2013;8(11):1188–1197. doi: 10.4161/epi.26265. [DOI] [PubMed] [Google Scholar]
- 17.Glossop JR, Emes RD, Nixon NB, et al. Genome-wide DNA methylation profiling in rheumatoid arthritis identifies disease-associated methylation changes that are distinct to individual T- and B-lymphocyte populations. Epigenetics. 2014;9(9):1228–1237. doi: 10.4161/epi.29718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhead B, Holingue C, Cole M, et al. Rheumatoid arthritis naive t cells share hypermethylation sites with synoviocytes. Arthritis Rheumatol. 2017;69(3):550–559. doi: 10.1002/art.39952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guderud K, Sunde LH, Flam ST, et al. Rheumatoid arthritis patients, both newly diagnosed and methotrexate treated, show more dna methylation differences in CD4(+) memory than in CD4(+) naive T cells. Front Immunol. 2020;11:194. doi: 10.3389/fimmu.2020.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ha E, Bang SY, Lim J, et al. Genetic variants shape rheumatoid arthritis-specific transcriptomic features in CD4(+) T cells through differential DNA methylation, explaining a substantial proportion of heritability. Ann Rheum Dis. 2021;80(7):876–883. doi: 10.1136/annrheumdis-2020-219152. [DOI] [PubMed] [Google Scholar]
- 21.Hu B, Jadhav RR, Gustafson CE, et al. Distinct age-related epigenetic signatures in CD4 and CD8 T cells. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.585168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jadhav RR, Im SJ, Hu B, et al. Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc Natl Acad Sci U S A. 2019;116(28):14113–14118. doi: 10.1073/pnas.1903520116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen KD, Irizarry RA, Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 2012;13(2):204–216. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLean CY, Bristor D, Hiller M, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang DW, Sherman BT, Tan Q, et al. DAVID bioinformatics resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31(14):2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 29.Ahsan S, Draghici S. Identifying significantly impacted pathways and putative mechanisms with ipathwayguide. Curr Protoc Bioinformatics. 2017;57 doi: 10.1002/cpbi.24. 7 15 1–7 30. [DOI] [PubMed] [Google Scholar]
- 30.Qi Q, Liu Y, Cheng Y, et al. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci USA. 2014;111(36):13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang JT, Wherry EJ, Goldrath AW. Molecular regulation of effector and memory T cell differentiation. Nat Immunol. 2014;15(12):1104–1115. doi: 10.1038/ni.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamagata K, Nakayamada S, Tanaka Y. Critical roles of super-enhancers in the pathogenesis of autoimmune diseases. Inflamm Regen. 2020;40:16. doi: 10.1186/s41232-020-00124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weng NP, Araki Y, Subedi K. The molecular basis of the memory T cell response: differential gene expression and its epigenetic regulation. Nat Rev Immunol. 2012;12(4):306–315. doi: 10.1038/nri3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moskowitz DM, Zhang DW, Hu B, et al. Epigenomics of human CD8 T cell differentiation and aging. Sci Immunol. 2017;2(8) doi: 10.1126/sciimmunol.aag0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen Y, Wen Z, Li Y, et al. Metabolic control of the scaffold protein TKS5 in tissue-invasive, proinflammatory T cells. Nat Immunol. 2017;18(9):1025–1034. doi: 10.1038/ni.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen Z, Jin K, Shen Y, et al. N-myristoyltransferase deficiency impairs activation of kinase AMPK and promotes synovial tissue inflammation. Nat Immunol. 2019;20(3):313–325. doi: 10.1038/s41590-018-0296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Shen Y, Jin K, et al. The DNA repair nuclease MRE11A functions as a mitochondrial protector and prevents T Cell pyroptosis and tissue inflammation. Cell Metab. 2019;30(3):477–492. doi: 10.1016/j.cmet.2019.06.016. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu B, Qiu J, Zhao TV, et al. Succinyl-CoA ligase deficiency in pro-inflammatory and tissue-invasive T Cells. Cell Metab. 2020;32(6):967–980. doi: 10.1016/j.cmet.2020.10.025. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Ruiten MS, Rowland BD. On the choreography of genome folding: A grand pas de deux of cohesin and CTCF. Curr Opin Cell Biol. 2021;70:84–90. doi: 10.1016/j.ceb.2020.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Xiang JF, Corces VG. Regulation of 3D chromatin organization by CTCF. Curr Opin Genet Dev. 2021;67:33–40. doi: 10.1016/j.gde.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye Z, Shen Y, Jin K, et al. Arachidonic acid-regulated calcium signaling in T cells from patients with rheumatoid arthritis promotes synovial inflammation. Nat Commun. 2021;12(1):907. doi: 10.1038/s41467-021-21242-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33(11):1665–1673. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- 45.Ali M, Ponchel F, Wilson KE, et al. Rheumatoid arthritis synovial T cells regulate transcription of several genes associated with antigen-induced anergy. J Clin Invest. 2001;107(4):519–528. doi: 10.1172/JCI8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cope AP. Studies of T-cell activation in chronic inflammation. Arthritis Res. 2002;4(Suppl 3):S197–S211. doi: 10.1186/ar557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kahan SM, Wherry EJ, Zajac AJ. T cell exhaustion during persistent viral infections. Virology. 2015;479-480:180–193. doi: 10.1016/j.virol.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sen DR, Kaminski J, Barnitz RA, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354(6316):1165–1169. doi: 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan O, Giles JR, McDonald S, et al. TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature. 2019;571(7764):211–218. doi: 10.1038/s41586-019-1325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng Z, Wei F, Ren X. Exhausted T cells and epigenetic status. Cancer Biol Med. 2020;17(4):923–936. doi: 10.20892/j.issn.2095-3941.2020.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt D, Goronzy JJ, Weyand CM. CD4+ CD7- CD28- T cells are expanded in rheumatoid arthritis and are characterized by autoreactivity. J Clin Invest. 1996;97(9):2027–2037. doi: 10.1172/JCI118638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner UG, Koetz K, Weyand CM, Goronzy JJ. Perturbation of the T cell repertoire in rheumatoid arthritis. Proc Natl Acad Sci U S A. 1998;95(24):14447–14452. doi: 10.1073/pnas.95.24.14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang X, Wang S, Zhou C, et al. Comprehensive TCR repertoire analysis of CD4(+) T-cell subsets in rheumatoid arthritis. J Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102432. [DOI] [PubMed] [Google Scholar]
- 54.Jensen IJ, Sjaastad FV, Griffith TS, Badovinac VP. Sepsis-induced T cell immunoparalysis: the ins and outs of impaired T cell immunity. J Immunol. 2018;200(5):1543–1553. doi: 10.4049/jimmunol.1701618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidt RE, Grimbacher B, Witte T. Autoimmunity and primary immunodeficiency: two sides of the same coin? Nat Rev Rheumatol. 2017;14(1):7–18. doi: 10.1038/nrrheum.2017.198. [DOI] [PubMed] [Google Scholar]
- 56.Takeuchi Y, Hirota K, Sakaguchi S. Impaired T cell receptor signaling and development of T cell-mediated autoimmune arthritis. Immunol Rev. 2020;294(1):164–176. doi: 10.1111/imr.12841. [DOI] [PubMed] [Google Scholar]
- 57.Listing J, Gerhold K, Zink A. The risk of infections associated with rheumatoid arthritis, with its comorbidity and treatment. Rheumatology (Oxford) 2013;52(1):53–61. doi: 10.1093/rheumatology/kes305. [DOI] [PubMed] [Google Scholar]
- 58.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.