Abstract
A new specific and sensitive 16S ribosomal DNA-based PCR assay was developed. The assay targets a 78-bp DNA fragment unique to Helicobacter bizzozeronii, Helicobacter felis, and Helicobacter salomonis and can be used with freshly frozen and formalin-fixed paraffin-embedded gastric biopsy specimens.
Virtually all cats and dogs are naturally colonized with different gastric Helicobacter species, including Helicobacter bizzozeronii, Helicobacter salomonis, and Helicobacter felis (7). Recently, an H. bizzozeronii strain, characterized on the basis of phenotypic analysis and 16S rRNA, DNA-DNA hybridization, and whole-cell protein profiling data, was isolated in vitro from the stomach of a human patient infected with “Helicobacter heilmannii”-like organisms (HHLO) (1; K. Jalava, S. L. W. On, C. S. Harrington, L. P. Andersen, M.-L. Hänninen, and P. Vandamme, Abstr. 10th Int. Workshop Campylobacter, Helicobacter Related Organisms, abstr. HD5, 1999). It has been suggested that cats and dogs could act as animal reservoirs in the transmission of HHLO to humans (9, 11). However, the difficulty in isolating HHLO from humans has hindered our understanding of the ecology and prevalence of these bacteria, thus demonstrating the need for simple and accurate diagnostic methods. It was the purpose of the study described here to develop a 16S ribosomal DNA (rDNA)-based PCR assay for the simultaneous detection of H. bizzozeronii, H. salomonis, and/or H. felis (referred to below as pet carnivore helicobacters) in both fresh and paraffin-embedded gastric biopsy specimens.
The stomachs of 21 clinically healthy, adult dogs from a local animal shelter were collected and sampled within 3 h after euthanasia. A tissue sample from the oxyntic region was removed and placed into 4% buffered formalin for 24 h. Immunohistochemical staining was performed to assess the presence of HHLO as described previously (2). For PCR analysis, a tissue sample was taken from the same region and frozen in sterile phosphate-buffered saline.
DNA was recovered from two different sources from each gastric biopsy specimen (scrapings of superficial cell layers and mucus) and from formalin-fixed paraffin-embedded biopsy specimens. DNA for use as a template was extracted from the scrapings by lysis with guanidinium isothiocyanate and was bound to silica particles by the method of Boom et al. (1). The paraffin-embedded specimens were subjected to deparaffinization and proteinase K-based lysis and were loaded onto a DNeasy tissue kit column according to the manufacturer's instructions (Qiagen, Hilden, Germany).
Primers CAR577f and CAR636r were selected from variable regions of the 16S rRNA-coding gene, which targets a 78-bp DNA fragment commonly present in H. bizzozeronii, H. salomonis, and H. felis (Table 1). Amplification was performed in a 50-μl reaction volume containing 1.8 pg of DNA, each primer (Eurogentec, Seraing, Belgium) at a concentration of 0.5 μM, 1× PCR Buffer II (Perkin-Elmer, Norwalk, Conn.), 1.5 mM MgCl2, each deoxynucleotide (Amersham Pharmacia Biotech, Uppsala, Sweden) at a concentration of 200 μM, and 1.5 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer). The PCR was performed with a model 9600 thermocycler (Perkin-Elmer). The samples were subjected to an initial 9-min preincubation step at 94°C for the activation of AmpliTaq Gold, followed by 35 amplification cycles of 30 s at 94°C, 30 s at 61°C, and 45 s at 72°C. A final primer extension at 72°C for 5 min was included. Fifteen microliters of each amplification product was analyzed by gel electrophoresis (50 min, 5 V/cm) in 3% agarose gels (Agarose MP; Boehringer Mannheim, Mannheim, Germany) and stained with ethidium bromide in Tris-borate-EDTA buffer (89 mM Tris-HCl [pH 8.0], 89 mM boric acid, 2.5 mM EDTA). DNA was visualized with a UV transilluminator. The PCR products were verified by Southern blot analysis. PCR products were transferred to a Hybond N+ nylon membrane (Amersham). The blots were prehybridized for 1 h at 42°C in a prehybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 2% blocking reagents, 0.1% N-lauroylsarcosine, 0.02% sodium dodecyl sulfate [SDS], 50% [vol/vol] formamide). A commercially synthesized 78-bp oligonucleotide, corresponding to the 16S rDNA target region (Life Technologies, Rockville, Md.), was used as a probe and was [γ-32P]ATP labeled with a Ready To Go kit (Pharmacia). The labeled oligonucleotide was then added to the prehybridization solution for 2 h. Washes were performed at 61°C in 1× SSC–0.1% SDS for 15 min. Bands were visualized overnight by autoradiography (X-Omat AR; Eastman Kodak, Rochester, N.Y.).
TABLE 1.
Oligonucleotide primers used for PCR amplification
| Primers | Sequences (5′-3′) | Polarity (direction) | Escherichia coli 16S rRNA positions |
|---|---|---|---|
| CAR577f | TGC GTA GGC GGG GTT GTA AG | Positive (forward) | 577–596 |
| CAR636r | CAG AGT TGT AG T TTC AAA TGC | Negative (reverse) | 636–656 |
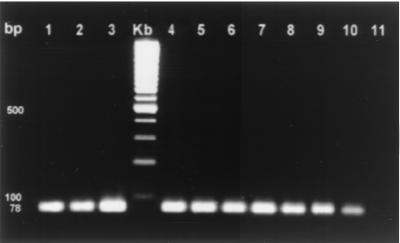
To determine the specificity of the PCR assay, the DNA of 23 different strains, representing 19 Helicobacter species and 2 non-Helicobacter species, was subjected to the PCR assay (Table 2). No DNA amplification was observed with DNA derived from Helicobacter species other than H. bizzozeronii, H. felis, and H. salomonis or with DNA from Campylobacter and Arcobacter species. The sensitivity of the assay was evaluated by using 10-fold serial dilutions of DNA extracts from the H. bizzozeronii, H. salomonis, and H. felis reference strains (0.9 μg/μl to 0.009 fg/μl) and from the fresh tissue samples of three infected dogs (0.1 μg/μl to 1 fg/μl) as templates in the PCRs. The assay was sensitive, detecting as little as 2 fg of genomic bacterial DNA. PCR products could be obtained from as little as 2 pg of DNA extracted from whole tissue (Fig. 1). The expected 78-bp PCR product was generated from 19 of 21 (90%) stomachs when the assay was applied to fresh and paraffin-embedded canine gastric biopsy samples. The amplicon was confirmed by Southern blot analysis.
TABLE 2.
Bacterial strains used for evaluation of the BSF-specific PCR
| Taxon | Source | Collection, strain, or clone no.a |
|---|---|---|
| “Candidatus Helicobacter bovis” | Cow abomasal mucosa | Clone R3XA |
| “Candidatus Helicobacter suis” | Pig gastric mucosa | Clone V2BXA |
| Helicobacter acinonychis | Cheetah gastric mucosa | LMG 12684T |
| Helicobacter bilis | Mouse bile | CCUG 38995T |
| Helicobacter bizzozeronii | Canine gastric mucosa | CCUG 35045T |
| Helicobacter canis | Canine feces | LMG 18086T |
| Helicobacter cinaedi | Human feces | LMG 7543T |
| Helicobacter felis | Feline gastric mucosa | CCUG 28539T |
| Helicobacter felis | Canine gastric mucosa | Strain 1136–7 |
| Helicobacter fennelliae | Human feces | LMG 11759 |
| Helicobacter hepaticus | Murine liver | LMG 16316T |
| Helicobacter muridarum | Murine intestinal mucosa | LMG 14378T |
| Helicobacter mustelae | Ferret gastric mucosa | LMG 18044T |
| Helicobacter pylori | Human gastric mucosa | LMG 7539T |
| Helicobacter pylori | Human gastric mucosa | Strain 23 |
| Helicobacter pullorum | Broiler chicken mucosa | LMG 16318 |
| Helicobacter salomonis | Canine gastric mucosa | CCUG 37845T |
| Helicobacter sp. strain Bird B | Bird feces | LMG 12679 |
| Helicobacter sp. strain Bird C | Bird feces | LMG 13642 |
| Helicobacter sp. strain CLO-3 | Human rectal swab | LMG 7792 |
| “Flexispira rappinii” | Human feces | LMG 13641 |
| Campylobacter jejuni | Bovine feces | LMG 8841T |
| Arcobacter butzleri | Human feces | LMG 10828 |
CCUG, Culture Collection, University of Göteborg, Göteborg, Sweden; LMG, BCCM/LMG Bacteria Collection, Laboratorium voor Microbiologie, Universiteit Gent, Ghent, Belgium.
FIG. 1.
Electrophoresis of PCR products on a 3% agarose gel. Lanes 1 to 3, positive controls (H. bizzozeronii, H. salomonis, and H. felis, respectively); lane Kb, 100-bp size markers; lanes 4 to 10, PCR products of 10-fold serial dilutions (undiluted to 10−6) of DNA extracted from the stomachs of dogs infected with gastric helicobacters; lane 11, negative control (DNA extracted from a stomach sample of a gnotobiotic piglet).
None of the available 16S rDNA-based identification methods has been evaluated for pet carnivore helicobacters (3, 8, 10). The very high degree of 16S rDNA sequence similarity between H. bizzozeronii, H. salomonis, and H. felis (>98.2%) further hinders the development of species-specific PCR assays (6). In the present study, we designed a 16S rDNA-based PCR assay that simultaneously detects H. bizzozeronii, H. salomonis, and H. felis in both freshly frozen and paraffin-embedded stomach biopsy specimens. The assay was found to be both specific and sensitive, and when it was applied to gastric biopsy specimens, the results coincided with those of immunohistochemistry analysis.
Recently, a molecular identification scheme based on 23S rRNA gene polymorphism developed by Hurtado and Owen (4) was evaluated for the identification of pet carnivore helicobacters (5). Similar to our PCR assay, it could not discriminate between H. bizzozeronii, H. salomonis, and H. felis at the species level but proved to be a useful method for the detection of these taxa as a group. Moreover, the assay of Hurtado and Owen (4) was not evaluated for use with paraffin-embedded material. Fixation of tissues prior to paraffin embedment causes fragmentation and partial destruction of the DNA, reducing DNA yields and affecting PCR efficiency (6). PCR amplification of a large, 2.6-kb DNA fragment (as in the assay of Hurtado and Owen [4]) reduces analytical sensitivity. The reduced sensitivity of the 23S rDNA assay would be compounded by the analysis of paraffin-embedded material. The amplification of a small targeted DNA fragment (78 bp) in our assay and the use of an adapted DNA extraction protocol countered these problems, enabling efficiency and applicability for the analysis of paraffin-embedded specimens.
Acknowledgments
We thank Mario Van Poucke for kind help with the Southern blot hybridization. We are also grateful to Hans Kusters and Kurt Houf for kindly providing Helicobacter, Campylobacter, and Arcobacter strains.
REFERENCES
- 1.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim van Dillen P M E, van der Noorda J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Groote D, van Doorn L J, Ducatelle R, Verschuuren A, Haesebrouck F, Quint W G V, Jalava K, Vandamme P. ‘Candidatus Helicobacter suis,’ a gastric Helicobacter from pigs, and its phylogenetic relatedness to other gastrospirilla. Int J Syst Bacteriol. 1999;49:1769–1777. doi: 10.1099/00207713-49-4-1769. [DOI] [PubMed] [Google Scholar]
- 3.Germani Y, Dauga C, Duval P, Huerre M, Levy M, Pialoux G, Sansonetti P, Grimont P A D. Strategy for the detection of Helicobacter species by amplification of 16S rRNA genes and identification of H. felis in a human gastric biopsy. Res Microbiol. 1997;148:315–326. doi: 10.1016/S0923-2508(97)81587-2. [DOI] [PubMed] [Google Scholar]
- 4.Hurtado A, Owen R J. A molecular scheme based on 23S rRNA gene polymorphisms for rapid identification of Campylobacter and Arcobacter species. J Clin Microbiol. 1997;35:2401–2404. doi: 10.1128/jcm.35.9.2401-2404.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jalava K, Hielm S, Hirvi U, Hänninen M L. Evaluation of a molecular identification scheme based on 23S rRNA gene polymorphisms for differentiating canine and feline gastric Helicobacter spp. Lett Appl Microbiol. 1999;28:269–274. doi: 10.1046/j.1365-2672.1999.00527.x. [DOI] [PubMed] [Google Scholar]
- 6.Jalava K, Kaartinen M, Utriainen M, Happonen I, Hänninen M L. Helicobacter salomonis sp. nov., a canine gastric Helicobacter sp. related to Helicobacter felis and Helicobacter bizzozeronii. Int J Syst Bacteriol. 1997;47:975–982. doi: 10.1099/00207713-47-4-975. [DOI] [PubMed] [Google Scholar]
- 7.Jalava K, On S L W, Vandamme P A R, Happonen I, Sukura A, Hänninen M L. Isolation and identification of Helicobacter spp. from canine and feline gastric mucosa. Appl Environ Microbiol. 1998;64:3998–4006. doi: 10.1128/aem.64.10.3998-4006.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marshall S M, Melito P L, Woodward D L, Johnson W M, Rodgers F G, Mulvey M R. Rapid identification of Campylobacter, Arcobacter, and Helicobacter isolates by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene. J Clin Microbiol. 1999;37:4158–4160. doi: 10.1128/jcm.37.12.4158-4160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meining A, Kroher G, Stolte M. Animal reservoirs in the transmission of Helicobacter heilmannii—results of a questionnaire-based study. Scand J Gastroenterol. 1998;33:795–798. doi: 10.1080/00365529850171422. [DOI] [PubMed] [Google Scholar]
- 10.On S L. Identification methods for campylobacters, helicobacters, and related organisms. Clin Microbiol Rev. 1996;9:405–422. doi: 10.1128/cmr.9.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stolte M, Wellens E, Bethke B, Ritter M, Eidt H. Helicobacter heilmannii (formerly Gastrospirillum hominis) gastritis: an infection transmitted by animals? Scand J Gastroenterol. 1994;29:1061–1064. doi: 10.3109/00365529409094888. [DOI] [PubMed] [Google Scholar]