Abstract
Background and purpose
In preclinical radiation studies, there is great interest in quantifying the radiation response of healthy tissues. Manual contouring has significant impact on the treatment-planning because of variation introduced by human interpretation. This results in inconsistencies when assessing normal tissue volumes. Evaluation of these discrepancies can provide a better understanding on the limitations of the current preclinical radiation workflow. In the present work, interobserver variability (IOV) in manual contouring of rodent normal tissues on cone-beam Computed Tomography, in head and thorax regions was evaluated.
Materials and methods
Two animal technicians performed manually (assisted) contouring of normal tissues located within the thorax and head regions of rodents, 20 cases per body site. Mean surface distance (MSD), displacement of center of mass (ΔCoM), DICE similarity coefficient (DSC) and the 95th percentile Hausdorff distance (HD95) were calculated between the contours of the two observers to evaluate the IOV.
Results
For the thorax organs, right lung had the lowest IOV (ΔCoM: 0.08 ± 0.04 mm, DSC: 0.96 ± 0.01, MSD:0.07 ± 0.01 mm, HD95:0.20 ± 0.03 mm) while spinal cord, the highest IOV (ΔCoM:0.5 ± 0.3 mm, DSC:0.81 ± 0.05, MSD:0.14 ± 0.03 mm, HD95:0.8 ± 0.2 mm). Regarding head organs, right eye demonstrated the lowest IOV (ΔCoM:0.12 ± 0.08 mm, DSC: 0.93 ± 0.02, MSD: 0.15 ± 0.04 mm, HD95: 0.29 ± 0.07 mm) while complete brain, the highest IOV (ΔCoM: 0.2 ± 0.1 mm, DSC: 0.94 ± 0.02, MSD: 0.3 ± 0.1 mm, HD95: 0.5 ± 0.1 mm).
Conclusions
Our findings reveal small IOV, within the sub-mm range, for thorax and head normal tissues in rodents. The set of contours can serve as a basis for developing an automated delineation method for e.g., treatment planning.
Keywords: Organ contouring, Interobserver variability, Preclinical, CT
1. Introduction
Preclinical radiation studies are essential in radiation therapy (RT) to understand the interaction of biological pathways with radiation exposure, to evaluate the long-term side effects of radiation and hence to enable clinical translation to humans [1], [2].Radiobiological researchers have shown great interest in obtaining information regarding the radiation response of normal tissues and tumors utilizing small animal models and image-guided precision radiotherapy [3]. Similarly, extensive work has been conducted on the development of advanced treatment-planning systems combined with high precision irradiation platforms mimicking human RT [4], [5], [6]. Even though there is extensive research in humans regarding the quantification of the interobserver variability (IOV) in organ segmentation [7], [8], similar research in small animals has only recently been started [9].
Assessment of organ delineation includes several applications, for instance, the contouring of organs for monitoring changes due to radiation toxicity such as lung fibrosis or monitoring adaptations of organ volumes longitudinally [10], [11], [12], [13], [14]. In the case of tumor presence, determination of its precise extent and position is essential to minimize the amount of radiation delivered to normal tissues besides the tumor target region translating in minimization of radiation-induced effects within the normal tissue volume [3]. The workflow followed in rodent studies with modern precision image-guided radiation platforms is shown in Supplementary Fig. 1 [11], [12], [13].
The workflow starts with the animal set-up followed by imaging, which is nowadays mostly X-ray cone-beam Computed Tomography (CBCT) imaging [6]. In the same step, image reconstruction is performed to obtain the CT volumes followed by delineation of the targeted areas. Next, the images are fed to the treatment-planning phase for quantitative analysis regarding the planned dose. This involves a separate process of tissue volume delineation, and possibly iterative steps in a feedback loop of irradiation preparation such as beam configuration and dose calculation until the optimal dose is chosen for each different tissue. Μanual contouring, i.e., segmentation of normal tissue in this case, is identified as one of the most time-consuming tasks which can be vastly improved using automated contouring algorithms [10], [15]. The latter commonly need sets of manual contours to develop the automated model. The time required for tissue segmentation is mainly dependent on anatomical complexities, introducing IOV per body site. Experimental settings such as the X-ray energy or the use of CT contrast media may also influence the degree of difficulty of manual contouring [16], [17]. Furthermore, the tedious task of contouring the tissues implies human bias, e.g., in experience level and therefore may compromise reproducibility and cause inaccurate dose delivery, especially combined with respiratory-induced motion [4], [18], [19].
The hypothesis of this work was that the IOV is within the order of sub-millimeters, and therefore negligible, on the conditions that clear contouring guidelines are provided. To demonstrate this, the IOV between two operators was quantified in manual contouring of normal organs for rodents in the thorax and head region for various organs.
2. Materials and methods
2.1. Dataset
In this work, two publicly available sets [20] of non contrast-enhanced and contrast-enhanced CBCT images (n = 20 each), acquired with an image-guided precision irradiator (X-RAD 225Cx, Precision X-Ray Inc., North Branford, USA) [21], were used retrospectively from two larger rodent radiation studies [22], [23]. Detailed information, such as the delineated organs and CT image acquisition information can be found in Supplementary Tables 1 and 2. For the thoracic organs, the dataset consists only of non tumor-bearing and non-treated mice. For the head region, data either without tumor or in an early stage of tumor growth (baseline) where it was not visible in the CT images were selected. The head data consists mainly of non-treated rats while 35% of them have been irradiated with a radiation dose of 20 Gy. For the head cases, the selection was performed to prevent side effects in anatomical structure such as neurotoxicity including only brain data from baselines and/or low radiation dose groups.
2.2. Organ segmentation
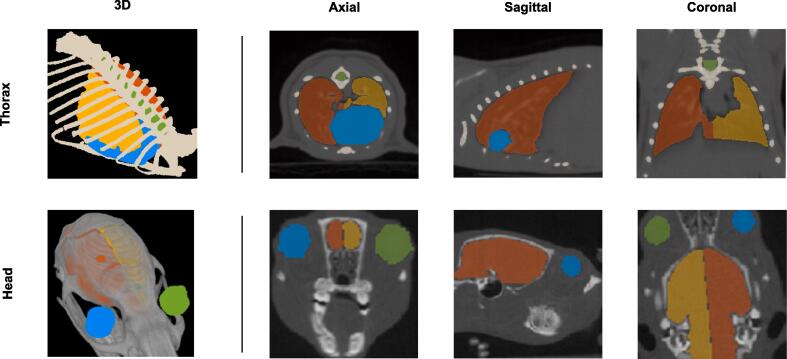
The thorax and head organs used in this work are depicted in Fig. 1, and a more in-depth overview of the delineation techniques can be found in Supplementary Table 3. The organs have been manually contoured, independently by two experienced animal technicians using the SmART-ATP software (Precision X-ray Inc., North Branford, CT & SmART Scientific Solutions BV, Maastricht, the Netherlands) [21]. In contrast to the Radiation Therapy Oncology Group guidelines for human organ delineation, there is no clear consensus for rodent organ delineations, even though their anatomy and atlases are known [9], [24]. The segmentation of the volumes consists of polygons with many vertices contoured in 2D slices of the CBCT scans. The orientation of the delineation plane, the time required, the selected drawing tool and the technique varies per organ due to anatomical complexities. The majority of the organs (i.e., lungs, spinal cord, thorax bone, brain hemispheres) were contoured semi-automatically assisted using 3D region growing, thresholding and slice interpolation followed by manual adjustments. For the eyes, 3D brushing was applied whereas the heart was contoured completely manually based on advice from an experienced radiation oncologist. Similar advice was used for the lung separation.
Fig. 1.
First row: mouse organs segmented in the thorax region depicted in 3D and 2D planes. Yellow: left lung; Orange: right lung; Green: spinal cord; Blue: heart; Beige: thorax bone. Second row: rat organs segmented in the head region depicted in 3D and 2D planes. Yellow: left brain hemisphere; Orange: right brain hemisphere; Green: left eye; Blue: right eye. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Evaluation metrics
For the evaluation of the IOV between the sets of contours from both annotators for each organ, four metrics were calculated: DICE similarity coefficient (DSC), mean surface distance (MSD), 95th percentile of Hausdorff distance (HD95) and the displacement of center of mass (ΔCoM) [10], [25]. The DSC computes the overlapping area between the contoured volumes of each organ, with values ranging from 0 (no overlap) to 1 (complete overlap). HD95 computes the maximum distance from a point in contour set A to the nearest point in contour set B. MSD calculates the average Euclidean distance between all points from contour set A with its corresponding nearest point of contour set B. Finally, the Cartesian metric ΔCoM stands for the distance between the centers of mass of volume A and volume B in a 3D space [26]. For each metric per organ, the results were calculated by comparing annotator A and B delineation IOV, for each rodent of the twenty thorax and head cases. The IOV is defined as the difference between the human expert delineations and the metrics are presented by calculating the mean (μ) and standard deviation (SD) of the studied sample per body site. Data was plotted using boxplots (with one inter-quartile range whiskers).
3. Results
3.1. Inter-observer disagreement analysis – thorax
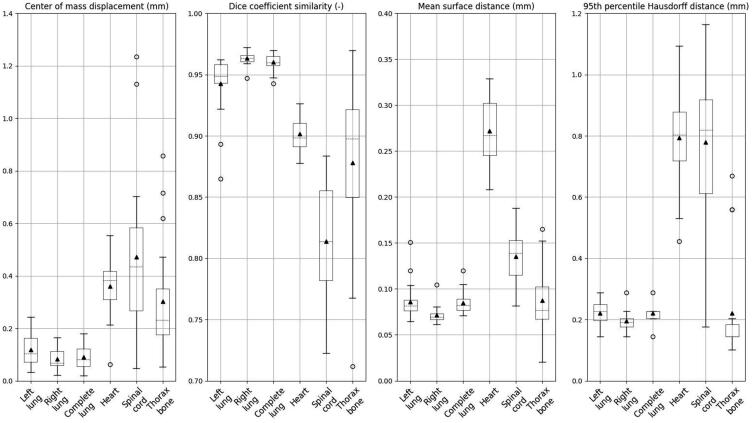
Quantitatively (Fig. 2), relatively small IOVs were observed for the lungs, with the lowest IOV for the right lung, which is more easily discerned from the heart (ΔCoM: 0.08 ± 0.04 mm, MSD: 0.07 ± 0.01 mm). The average HD95 for right lung was 0.20 ± 0.03 mm, the lowest standard deviation among all thoracic organs. The corresponding MSD and the HD95 presented submillimeter differences with an average of 0.09 ± 0.05 mm and 0.22 ± 0.03 mm, for the complete lung. Similarly, the DSC metric for the aforementioned organs showed high overlap for the two annotators’ delineations, with the highest DSC of 96% ± 1% founded in right lung, same as complete lung and slightly lower for the left lung. The thorax bone presented higher IOV for the two annotators, for all metrics, compared to the lung region. The findings show not only higher mean values but also larger deviations (ΔCoM: 0.33 ± 0.21 mm, DSC: 88% ± 7%, MSD: 0.09 ± 0.03 mm, HD95p: 0.22 ± 0.16 mm). Heart and spinal cord yielded the largest IOV for the thoracic site. For the spinal cord, the average ΔCoM was the highest measured with a value of 0.47 ± 0.29 mm, the highest standard deviation among thoracic organs. For the heart ΔCoM, a value of 0.36 ± 0.11 mm was obtained, 3.7 times higher than the average lungs IOV. The spinal cord DSC of 81% ± 5% was the lowest for the two annotators’ sets whereas the DSC of heart was 90% ± 1% indicating larger IOV than the lungs. Especially, the heart despite its small size had a mean MSD IOV of 0.27 ± 0.04 mm with the mean HD95 measured at 0.79 ± 0.15 mm. A complete overview is provided in Supplementary Table 4.
Fig. 2.
Boxplots of evaluation metrics for the different organs in the thorax region of 20 mice. The whiskers depict the 25th and 75th percentile; the dashed line indicates the median value, the triangle represents the mean value, and the circles represent outliers.
3.2. Inter-observer disagreement analysis – head
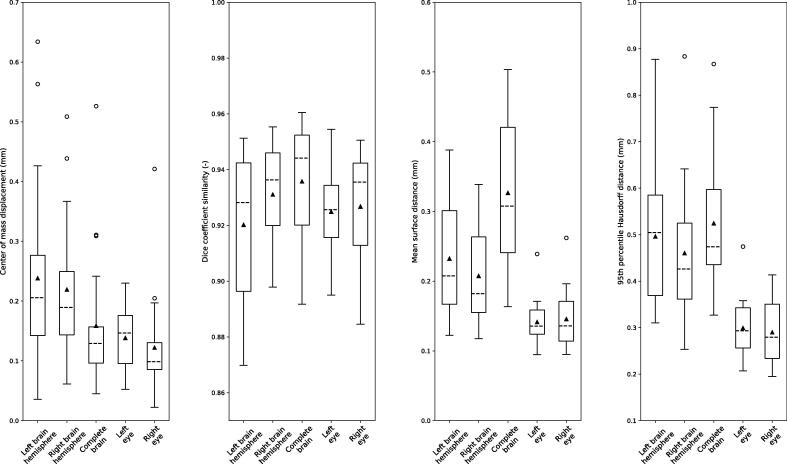
The quantitative analysis (Fig. 3) revealed submillimeter IOV for the eyes, while the right eye showed slightly less IOV for two metrics, with an average ΔCoM of 0.12 ± 0.08 mm and an average HD95:0.29 ± 0.07 mm. The highest IOV measured in ΔCoM was observed in the left brain hemisphere at 0.24 ± 0.15 mm. The average IOV, calculated for DSC, for complete brain was 94% ± 2% as the highest one, with small differences of 1–2% for the hemispheres, individually yielding good agreement in all cases, while the lowest DSC was found for the left eye. Similarly, the brain hemispheres presented higher IOV compared to eyes quantified with HD95 (left brain hemisphere: 0.47 ± 0.15 mm; right brain hemisphere: 0.46 ± 0.15 mm; complete brain: 0.52 ± 0.14 mm) and ΔCoM (left brain hemisphere: 0.24 ± 0.15 mm; right brain hemisphere: 0.22 ± 0.11 mm; complete brain: 0.16 ± 0.11 mm) with the highest degree of IOV found in left brain hemisphere. For the MSD, the lowest IOV was yielded for the right eye at 0.15 ± 0.04 mm while the annotators’ delineations varied more for the complete brain (MSD: 0.33 ± 0.10 mm).Last, the SD values of the hemispheres, individually compared to the complete brain, were twice larger than the corresponding one for the eyes. A complete overview is provided in Supplementary Table 4.
Fig. 3.
Boxplots of evaluation metrics for the different organs in the head region of 20 rats. The whiskers depict the 25th and 75th percentile; the dashed line indicates the median value, the triangle represents the mean value, and the circles represent outliers.
4. Discussion
This study investigated the IOV (n = 2) for manually (assisted) contouring of normal tissues located within the thorax and head rodents (n = 20 cases per body site). The analysis consisted of four metrics, MSD, ΔCoM, DSC and HD95 and revealed small IOV between the two independent annotators for all the metrics.
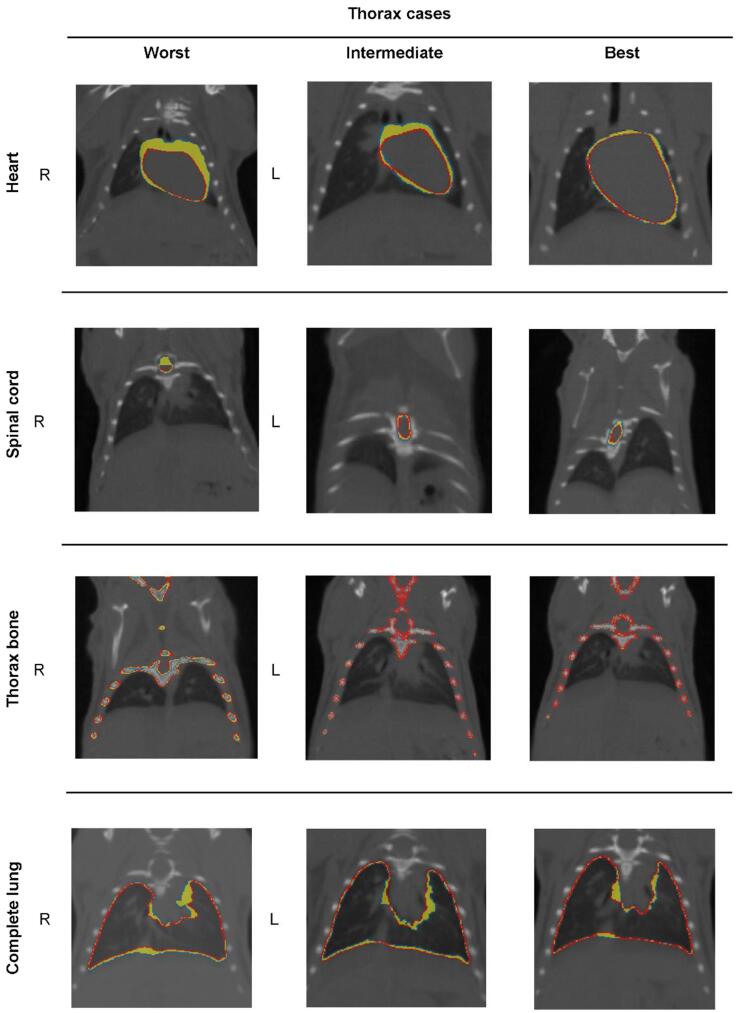
Regarding the thoracic region, the relatively low IOV values for all metrics found for both separate and complete lungs are in agreement with the qualitative analysis (Fig. 4 and Supplementary Fig. 2). The clear boundaries between lung tissue and ribs contributed to the low IOV values while in most cases, the largest IOV is detected close to the heart and in the lower parts of the lung (e.g., ‘worst’ and ‘intermediate’ thorax cases). In contrast, the spinal cord and heart yielded the largest IOV for the thoracic site. The disagreement between the two annotators was caused by different factors. For the heart, these were 1) the need for fully manual delineation and 2) the anatomical complexity consisting largely of soft tissue and the adjacent major blood vessels. For the spinal cord which showed similar IOV as the thorax bone, the annotators had differences in the longitudinal direction leading to uncertainties in the starting/ending slices.
Fig. 4.
Contoured organs by two annotators (blue: annotator #1; red: annotator #2) and region of IOV (yellow). Examples are shown for worst, intermediate and best cases per organ. First row: heart Second row: spinal cord; Third row: thorax bone; Forth row: complete lung. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
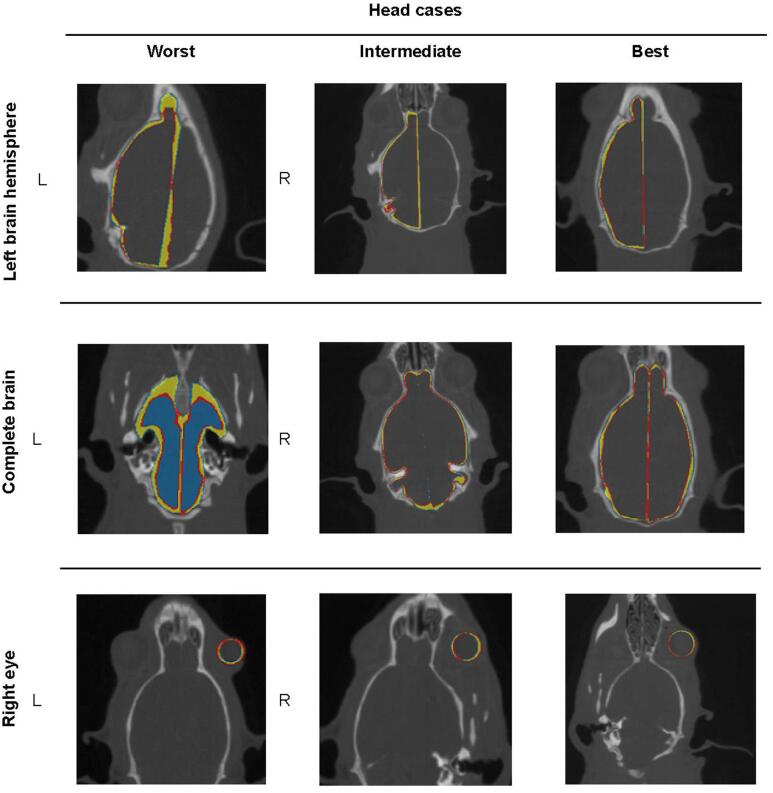
For the head region (Fig. 5), the qualitative analysis confirmed the quantitative results revealing the lowest IOV for the left and right eyes. This is connected to relatively simple anatomical complexity (i.e., spherical shape) and the smaller size compared to the other head organs. For the left and right brain hemispheres, the IOV were higher than for the eyes, mainly in the anterior-posterior direction, or in the lateral direction next to the brain cavity separating the two hemispheres, i.e., medial longitudinal fissure. The complete brain, derived from fusion of the two hemisphere contours, showed the highest IOV among all head organs, propagating the inconsistencies of the original separate brain contours. Specifically, in some cases subvolumes belonging to either olfactory bulbs or the posterior side of the brain have been insufficiently segmented because of poor visibility of the boundaries (Fig. 5 and Supplementary Fig. 3; i.e., ‘worst’ head cases).
Fig. 5.
Contoured organs by two annotators (blue: annotator #1; red: annotator #2) and region of IOV (yellow). Examples are shown for worst, intermediate and best cases per organ. First row: left hemisphere; Second row: complete brain; Third row: right eye.
To the best of our knowledge, the only available recent previous findings from Schoppe et al. using μCBCT, showed a similar DSC for lungs and heart as in this work [9]. More specifically, in our study both separate and complete lungs, as well as the eyes achieved the lowest IOV for most metrics, with the lungs showing the lowest IOV among all organs for the forty studied rodents of two different strains. In contrast, spinal cord, heart and complete brain showed larger IOV compared to other head and thorax organs. Spinal cord had the highest ΔCoM and the lowest DSC among all organs for the two body sites, even though IOV was in sub-mm level.
Our study used more quantitative and qualitative metrics. Schoppe et al. quantified the human IOV utilizing only the DSC metric and the volume size variability while in our study applied three additional metrics (ΔCoM, MSD, HD95) were studied. In addition, the IOV for the thorax bone or spinal cord cannot be compared directly to those of Schoppe et al. since the latter quantified IOV for the bony structure including other skeletal subvolumes different from the present study, based on three mouse atlases [9], [24]. Furthermore, in both studies, the skeletal volumes were delineated semi-automatically relying on the high voxel intensity in that area, thereby decreasing the likelihood for inconsistencies between the annotators. Regarding the head organs, there is no previous study. Thus, this work presents the first data for IOV in the head region in a preclinical rat study. Nonetheless, even though two annotators were used in both studies to investigate the annotator IOV, the amount of data used in the study of Schoppe et al. is larger (n = 220, mice) than the presented work (n = 40, mice and rats). The findings of this preclinical animal study differ from IOV for human contouring studies [7], [15]. For the latter, the IOVs in head and neck region are larger, especially adding more annotators to the analysis. While arguably the number of observers (n = 2) is small compared to five or more that are commonly used in human contouring studies, for preclinical studies it is hard to find multiple contouring experts. As shown in previous studies, the IOVs are even larger depending on the annotators experience or the guidelines resulting in uncertainties when delineating normal tissue [8], [15]. Human structures are also larger compared to the rodents and the findings in small animals with high spatial resolution during the CT acquisition need to be taken into account to compared to human studies, e.g., 10–50 times higher spatial CBCT resolution for rodents compared to humans. Similarly, in most cases, the animal studies present more homogeneous characteristics, such as sex, age, diet and health status.
In this work, a limited number of annotators from the same institution were used to study the IOV in preclinical CBCT images. The number of annotators is smaller compared to most human studies. In the only relevant published work of Schoppe et al., the authors also quantified the IOV using only two annotators. Furthermore, several differences, such as the imaging method or the evaluation metrics, exist between the two studies. However, the findings however from both studies are aligned for the organs which were in common (e.g., heart and lungs), validating our outcome. Likewise, presumably, evaluation using a third observer would not vastly alter the current outcome when they follow the same guidelines, although exploiting additional annotators from multi-institutional departments could be beneficial.
Finally, an important parameter to take into account for treatment planning is the time spent on the delineations. For the thoracic volumes, initially on average 60 min per rodent for the six organs was needed to segment the organs while a gain of 15 min per rodent was observed when the process was repeated multiple times. For the majority of the organs the time needed for delineation with assisted techniques including manual adjustments was 10 min per organ. The delineation of the heart was done completely manually. Finally, the time to delineate the five head organs was initially 40 min with a time gain of 20 min after some repetitions. In this study great care was taken to delineate accurately. In real-life studies, where animals are sedated while waiting to be irradiated, shorter delineation times may be used which may decrease the accuracy.
To standardize the treatment workflow, eliminate bias and the need for specialized annotators, and speed up the contouring vastly, fully automatic contouring methods can be developed. This could e.g., be based on artificial intelligence methods which can use datasets like the ones presented here as the gold standard for training the methods. Additionally, this dataset can serve as a basis to extract features for the studied organs enabling contouring for other body sites and organs exploiting transfer learning techniques [27].
The IOV found in this study is within the sub-mm range for thorax and head normal tissues in rodents’ treatment planning. Moreover, the IOV findings are well within the typically applied margins compensating for respiratory motion.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank prof. Dr. Dirk De Ruysscher for his cooperation in guidance regarding the anatomical guidelines for the tissue contouring. Moreover, the authors would like to thank the NVIDIA Corporation for the donation of the NVIDIA Quadro P6000 GPU used for this research. The OMEGA collaboration project is co-funded by the PPP allowance made available by Health-Holland, Top Sector Life Sciences&Health to stimulate public-private partnerships. This manuscript reflects the authors’ view only, and the Stichting LSH-TKI or the Ministry of Economic Affairs and Climate Policy (Netherlands) are not responsible for any use that may be made of the information it contains.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2022.01.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Schlaak R.A., Senthilkumar G., Boerma M., Bergom C. Advances in preclinical research models of radiation-induced cardiac toxicity. Cancers. 2020;12:415. doi: 10.3390/cancers12020415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koontz B.F., Verhaegen F., De Ruysscher D. Tumour and normal tissue radiobiology in mouse models: how close are mice to mini-humans? Br J Radiol. 2017;90:20160441. doi: 10.1259/bjr.20160441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iglesias VS, Hoof SJ van, Vaniqui A, Schyns LE, Lieuwes N, Yaromina A, et al. An orthotopic non-small cell lung cancer model for image-guided small animal radiotherapy platforms. 2018;92. https://doi.org/10.1259/BJR.20180476. [DOI] [PMC free article] [PubMed]
- 4.Verhaegen F, Granton P, Tryggestad E. Small animal radiotherapy research platforms. Phys Med Biol 2011;56. https://doi.org/10.1088/0031-9155/56/12/R01. [DOI] [PubMed]
- 5.Tillner F., Thute P., Bütof R., Krause M., Enghardt W. Pre-clinical research in small animals using radiotherapy technology - a bidirectional translational approach. Z Med Phys. 2014;24:335–351. doi: 10.1016/j.zemedi.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Verhaegen F., Dubois L., Gianolini S., Hill M.A., Karger C.P., Lauber K., et al. ESTRO ACROP: Technology for precision small animal radiotherapy research: Optimal use and challenges. Radiother Oncol. 2018;126:471–478. doi: 10.1016/j.radonc.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 7.van Baardwijk A., Bosmans G., Boersma L., Buijsen J., Wanders S., Hochstenbag M., et al. PET-CT-based auto-contouring in non-small-cell lung cancer correlates with pathology and reduces interobserver variability in the delineation of the primary tumor and involved nodal volumes. Int J Radiat Oncol Biol Phys. 2007;68:771–778. doi: 10.1016/j.ijrobp.2006.12.067. [DOI] [PubMed] [Google Scholar]
- 8.van der Veen J., Gulyban A., Willems S., Maes F., Nuyts S. Interobserver variability in organ at risk delineation in head and neck cancer. Radiat Oncol. 2021;16:1–11. doi: 10.1186/s13014-020-01677-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoppe O., Pan C., Coronel J., Mai H., Rong Z., Todorov M.I., et al. Deep learning-enabled multi-organ segmentation in whole-body mouse scans. Nat Commun. 2020;11 doi: 10.1038/s41467-020-19449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Heyden B., van de Worp W.R.P.H., van Helvoort A., Theys J., Schols A.M.W.J., Langen R.C.J., et al. Automated CT-derived skeletal muscle mass determination in lower hind limbs of mice using a 3D U-Net deep learning network. J Appl Physiol. 2020;128:42–49. doi: 10.1152/japplphysiol.00465.2019. [DOI] [PubMed] [Google Scholar]
- 11.Spiegelberg L., van Hoof S.J., Biemans R., Lieuwes N.G., Marcus D., Niemans R., et al. Evofosfamide sensitizes esophageal carcinomas to radiation without increasing normal tissue toxicity. Radiother Oncol. 2019;141:247–255. doi: 10.1016/j.radonc.2019.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Ruysscher D., Granton P.V., Lieuwes N.G., van Hoof S., Wollin L., Weynand B., et al. Nintedanib reduces radiation-induced microscopic lung fibrosis but this cannot be monitored by CT imaging: A preclinical study with a high precision image-guided irradiator. Radiother Oncol. 2017;124:482–487. doi: 10.1016/j.radonc.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Vande Velde G., Poelmans J., De Langhe E., Hillen A., Vanoirbeek J., Himmelreich U., et al. Longitudinal micro-CT provides biomarkers of lung disease that can be used to assess the effect of therapy in preclinical mouse models, and reveal compensatory changes in lung volume. Dis Models Mech. 2016;9:91–98. doi: 10.1242/DMM.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Worp W.R.P.H., van der Heyden B., Lappas G., van Helvoort A., Theys J., Schols A.M.W.J., et al. Deep learning based automated orthotopic lung tumor segmentation in whole-body mouse CT-scans. Cancers. 2021;13:4585. doi: 10.3390/cancers13184585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Veen J., Willems S., Deschuymer S., Robben D., Crijns W., Maes F., et al. Benefits of deep learning for delineation of organs at risk in head and neck cancer. Radiother Oncol. 2019;138:68–74. doi: 10.1016/j.radonc.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Chakraborty C., Richards P. Impact of CT window contouring on breast plans. Int J Med Res Health Sci. 2018;7:72–78. [Google Scholar]
- 17.Anderson BM, Lin E, Cardenas CE, Koay EJ, Odisio B, Brock KK. Automated contouring of contrast and non-contrast CT liver images with fully convolutional neural networks. Int J Radiat Oncol Biol Phys 2018;102:S55. https://doi.org/10.1016/j.ijrobp.2018.06.160.
- 18.Vaniqui A., van der Heyden B., Almeida I.P., Schyns L.EJR., van Hoof S.J., Verhaegen F. On the determination of planning target margins due to motion for mice lung tumours using a four-dimensional mOBY phantom. Br J Radiol. 2019;92 doi: 10.1259/bjr.20180445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heyden B van der, Hoof SJ van, Schyns LEJR, Verhaegen F. The influence of respiratory motion on dose delivery in a mouse lung tumour irradiation using the 4D MOBY phantom. 2016;90. https://doi.org/10.1259/BJR.20160419. [DOI] [PMC free article] [PubMed]
- 20.Verhaegen F. Head and thorax rodent data 2021. https://doi.org/10.34894/PJTTMJ.
- 21.van Hoof S.J., Granton P.V., Verhaegen F. Development and validation of a treatment planning system for small animal radiotherapy: SmART-Plan. Radiother Oncol. 2013;109:361–366. doi: 10.1016/j.radonc.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Granton P.V., Dubois L., van Elmpt W., van Hoof S.J., Lieuwes N.G., De Ruysscher D., et al. A longitudinal evaluation of partial lung irradiation in mice by using a dedicated image-guided small animal irradiator. Int J Radiat Oncol Biol Phys. 2014;90:696–704. doi: 10.1016/j.ijrobp.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Mowday AM, Lieuwes NG, Biemans R, Marcus D, Rezaeifar B, Reniers B, et al. Use of a luciferase-expressing orthotopic rat brain tumor model to optimize a targeted irradiation strategy for efficacy testing with temozolomide. Cancers 2020;12:1585. https://doi.org/10.3390/CANCERS12061585. [DOI] [PMC free article] [PubMed]
- 24.Akselrod-Ballin A., Dafni H., Addadi Y., Biton I., Avni R., Brenner Y., et al. Multimodal correlative preclinical whole body imaging and segmentation OPEN. Nature Publishing Group. 2016;6(1) doi: 10.1038/srep27940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaassen F., Hazelaar C., Vaniqui A., Gooding M., van der Heyden B., Canters R., et al. Evaluation of measures for assessing time-saving of automatic organ-at-risk segmentation in radiotherapy. Phys Imag Radiat Oncol. 2020;13:1–6. doi: 10.1016/j.phro.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deeley M.A., Chen A., Datteri R., Noble J.H., Cmelak A.J., Donnelly E.F., et al. Comparison of manual and automatic segmentation methods for brain structures in the presence of space-occupying lesions: A multi-expert study. Phys Med Biol. 2011;56:4557–4577. doi: 10.1088/0031-9155/56/14/021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karimi D., Warfield S.K., Gholipour A. Transfer learning in medical image segmentation: New insights from analysis of the dynamics of model parameters and learned representations. Artif Intell Med. 2021;116:102078. doi: 10.1016/j.artmed.2021.102078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.