Abstract
Irritable bowel syndrome (IBS) is a common clinical label for medically unexplained gastrointestinal symptoms, recently described as a disturbance of the microbiota-gut-brain axis. Despite decades of research, the pathophysiology of this highly heterogeneous disorder remains elusive. However, a dramatic change in the understanding of the underlying pathophysiological mechanisms surfaced when the importance of gut microbiota protruded the scientific picture. Are we getting any closer to understanding IBS’ etiology, or are we drowning in unspecific, conflicting data because we possess limited tools to unravel the cluster of secrets our gut microbiota is concealing? In this comprehensive review we are discussing some of the major important features of IBS and their interaction with gut microbiota, clinical microbiota-altering treatment such as the low FODMAP diet and fecal microbiota transplantation, neuroimaging and methods in microbiota analyses, and current and future challenges with big data analysis in IBS.
Keywords: Microbiota, Neurogastroenterology, Irritable bowel syndrome, Microbiota-gut-brain axis, Structural and functional magnetic resonance imaging, Machine learning, Big data analysis
Core Tip: Molecular biology, advanced neuroimaging and computer science is emerging to transform our understanding of the role of gut microbiota in irritable bowel syndrome (IBS). Herein, we provide an overview and discuss the role of gut microbiota in IBS, the clinical microbiota-altering interventions the low FODMAP diet and fecal microbiota transplantation, the role of brain-imaging and gut microbiota analyses, the importance of method selection, metadata, perspectives for improving microbiota role predictions, and big data analysis, in the seeking of understanding IBS pathology.
INTRODUCTION
Irritable bowel syndrome – a disturbance of the microbiota-gut-brain axis
Irritable bowel syndrome (IBS) is a chronic biopsychosocial disorder manifested by recurrent abdominal pain and alterations in stool form or frequency[1]. The condition affects 4%-10% of the global population and is associated with markedly reduced quality of life[2,3]. In addition to genetic predisposition, adverse life events, psychosocial factors, chronic and acute stress, and gastrointestinal (GI) infections[1], mounting evidence suggests the gut microbiota play a key role in IBS.
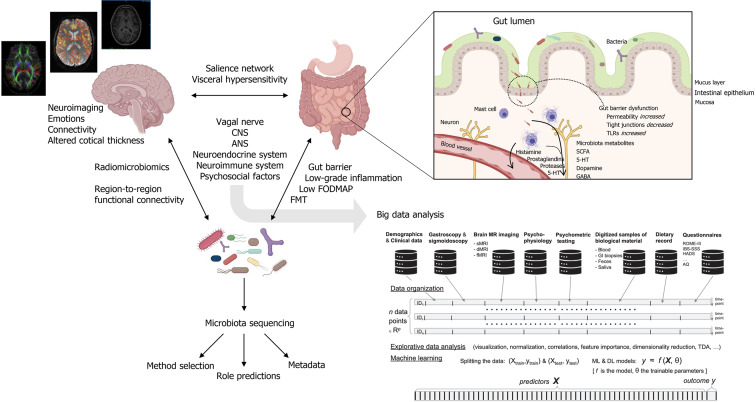
Because of its heterogeneity and unclear etiology, clear biomarkers and therapeutic targets for IBS have been difficult to identify. As a term, “IBS” is collective for medically unexplained disturbances of the bidirectional communication between the gut and the brain. These disturbances are multifactorial and include visceral hypersensitivity, low-grade inflammatory responses, intestinal motility disturbances, alterations of central nervous system (CNS) processing, and alterations in gut microbiota composition[1]. In the gut, a well-functioning microbiota is highly adapted to the host and carries out biochemical and metabolic processes that are important for host function. Signals coming from the gut microbiota modulate aspects of homeostasis through neural, endocrine and immune communication pathways between the gut and the brain[4,5]. Together, this has established the concept of the microbiota-gut-brain (MGB) –axis (Figure 1).
Figure 1.
Integration of multimodal and interdisciplinary approaches for big data analysis in irritable bowel syndrome. Created with BioRender.com. ANS: Autonomic nervous system; CNS: Central nervous system; FODMAP: Fermentable oligosaccharides, disaccharides, monosaccharides and polyols; FMT: Fecal microbiota transplant; 5-HT: 5-hydroxytryptamine (serotonin); SCFAs: Short chain fatty acids; GABA: γ-aminobutyric acid; TLRs: Toll-like receptors.
The vagus nerve serve as a major MGB pathway modulator. It is composed of somatic and afferent fibers (80%) and general and special visceral efferent fibers (20%). Under normal circumstances, the vagus nerve sense and is activated by diet-responsive gut microbes and metabolites such as short-chain fatty acids (SCFAs), or endocrine factors, enzymes, and neurotransmitters such as serotonin, dopamine, acetylcholine, glutamate, γ-aminobutyric acid (GABA), and noradrenaline[6-9]. Each of these factors are potentially affected by alterations in microbiota composition and are involved in IBS pathology, as shown in Figure 1. In the intestines, vagal endings synapse onto neurons of the enteric nervous system (ENS), which governs the function of muscular, neuro-hormonal, and secretory systems of the GI tract to generate patterns of functional digestion. In IBS, the pathophysiology implicates altered gut microbiota composition, impaired intestinal mucosal integrity, and low-grade inflammation[10]. In addition to pathways through the circulatory system, several of these factors may also trigger fluctuations in the activity of the ENS with subsequent effect on the brain. This relationship of reciprocal signals may be disturbed to the degree of chronic IBS. In the chronic IBS brain, efferent signals may be perceived as unpleasant or painful, potentially leading to chronic visceral discomfort or pain[11].
The heterogeneity of the “healthy gut microbiota” has made it difficult to identify a clear IBS microbial signature. Indeed, the composition of gut microbiota composition is influenced by multiple factors, e.g., geographic location, ethnicity, dietary choices, medication use, and pathogens, summarized by Adak et al[12]. Hence, the human gut microbiota composition is highly diverse. This heterogeneity makes is difficult to provide a clear definition to what a “healthy microbiota” is. Nevertheless, some features are considered important characteristics: a high level of diversity, a favorable amount of butyrate-producing bacteria, and resistance and resilience - the ability to withstand a disturbance promoting a shift in the composition and the attribute to return to its initial composition, functionally or taxonomical, following this disturbance[12,13]. On the contrary, in disease the microbiota composition is often associated with a decreased microbial diversity and loss of the typical balance between the host and the microorganisms[13], a so-called “dysbiosis” (a debated concept[14,15]), linked to several systemic and local human diseases.
Multiple studies have shown differences in the gut microbiota between IBS and healthy controls[16-20]. A comprehensive systematic review from 2019 showed that patients with IBS have increased levels of the bacterial families Enterobacteriaceae, Lactobacillaceae and Bacteroidales, whereas Bifidobacterium, Faecalibacterium, and Clostridiales were decreased compared to healthy controls[21]. On the contrary, Hugerth et al[22] recently reported no distinct microbiota signature of IBS in a random Swedish population of 3556 participants. Here, the between-sample divergence was higher in IBS compared to controls from the same population-sampling frame, but no clear biomarker of IBS was revealed. There are multiple individual reports on differences in distribution patterns of constipation predominant IBS (IBS-C), IBS with mixed constipation and diarrhea (IBS-M), and diarrhea predominant IBS (IBS-D), summarized by, among others, Liu et al[23], and Wang et al[24]. Pozuelo et al[25] found butyrate- and methane-producing bacteria were less abundant in IBS-D and IBS-M patients. However, Pittayanon et al[21] summarized six studies from 130 patients with IBS-M, demonstrating no significant difference between subtypes. Interestingly, intestinal bacterial composition has been reported to be highly dependent on sample type and regional localization. Also, mucosa-associated bacterial composition of the sigmoid colon differ between patients with IBS and healthy controls[26].
Indeed, the absence of a universal definition of what a “healthy microbiota” is, in addition to lack of consistency in sequencing methodology, study protocols, inter-individual variation that dominate intra-individual variation, definitions of “controls”, and different statistical methodologies being used have made the search for a common pathological IBS microbiota signature difficult. The importance of method selection, metadata, and perspectives for improving microbiota role predictions are discussed more thoroughly in section “Intestinal microbiota analyses”, below.
Another factor to consider is the impact of the circadian rhythm on gut microbiota variability. Both the level of host-derived autoantibodies and peptides and nutrient availability give fluctuations in the gut microbiota, and both are associated with circadian rhythm oscillations[6]. At least 10% of operational taxonomic units may oscillate due to the circadian rhythm, which is important to consider when collecting and analyzing fecal samples[27]. Thus, we might be in the mere beginning of understanding how alterations in gut microbiota may lead to the disruption of the intricate host-gut-microbiota-interaction. Is it a cause or a result of IBS pathology? In the last decade, much knowledge has been gained from clinical microbiota-altering interventions such as the low FODMAP-diet and fecal microbiota transplantation (FMT), which has emerged as debatably successful treatment strategies. However, their effects on the MGB-axis are still far from understood.
CLINICAL MICROBIOTA-ALTERING TREATMENT IN IBS – THE LOW FODMAP DIET AND FECAL TRANSPLANTATION
Dietary intervention
Diet is an environmental factor that is pivotal in shaping the architecture of gut bacteria. Although genetics have been assumed to be of great importance[28], a recent study shows that environmental factors, such as diet, are dominating[29]. In the symbiotic host/bacteria relationship, gut bacteria depend on host intake of complex polysaccharides to facilitate growth. As hosts, humans depend on gut bacteria to break down complex nutrients resistant to human GI metabolism and metabolites produced from fermentation, such as SCFAs. In IBS, foods play an important role among the contributing factors to symptom induction. In fact, the majority of patients with IBS experience increased symptom burden after food intake[30], despite the lack of objective evidence for food hypersensitivity or allergies[31]. Several underlying mechanisms generating symptoms are proposed to be involved[32]: (1) Local effects in the small and large intestine are caused by fermentable oligosaccharides, disaccharides, monosaccharides and polyols (FODMAPs). Intake of these short-chain carbohydrates have an osmotic effect in the gut lumen, increasing small intestinal water content and introduce undigested food particles to the gut bacteria who readily ferment them, causing gas production in the colon leading to abdominal pain as a consequence of a sensitive ENS i.e.: visceral hypersensitivity[33]. In 2017, Varjú et al[34] compared standard dietary therapy for IBS and the low FODMAP diet in 2017. Both diets showed to alleviate symptoms, but the low FODMAP diet showed a better therapeutic effect. However, with the available data on diet interventions in IBS, a low FODMAP diet has the greatest evidence of efficacy[35], with the most updated systematic review reporting significant improvements in GI symptoms and quality of life compared to control diets or habitual diets[36]; (2) Gut microbiota alterations and bacterial fermentation might have a role in food-related symptoms. Inexplicably, contradicting findings regarding different microbiota compositions between patients with IBS and healthy controls are often reported[21,22]. A recent matched case-control study from a Thai population reported no distinction in the gut microbiota between IBS-D and healthy subjects[37]. Here, the authors accredit the discordant results from those conducted in Western countries as an effect of different dietary lifestyles affecting the gut microbiota, suggesting that alterations in gut microbiota is not the main pathogenic mechanism of IBS-D in Thai patients[37]. In the Swedish random population, patients with IBS showed higher heterogeneity in microbiota composition compared to healthy individuals[22]. However, we need to keep in mind that bacterial fermentation capability may be more dependent on bacterial function rather than composition alone[38]. Nevertheless, differences in composition may still matter because it could result in differences in the effectiveness of a function[39]. Despite good documentation of a low FODMAP diet on symptom alleviation[36], FODMAP restriction is of concern due to possible unhealthy changes in gut microbiota composition with unknown consequences. Depriving the gut bacteria of carbohydrate and prebiotic substrates will shift the gut microbiota to ferment e.g., proteins and/or some amino acids, leading to production of potentially harmful compounds, summarized by Oliphant and Allen-Vercoe[40]. Desai et al[41] investigated gnotobiotic mice colonized with human gut microbiota fed a fiber-deprived diet. Because of fiber deficiency, the gut microbiota fed on the colonic mucosa layer that originally acted as a defense barrier against pathogens, leading to heighted pathogen susceptibility. Multiple features of alterations in the gut microbiota composition after a low FODMAP diet are reported, such as a lower total bacteria load, a lower absolute abundance of luminal Actinobacteria, Bifidobacteria, Clostridium cluster IV, Faecalibacterium prausnitzii, and a lower concentration of the SCFA butyrate[42-44]. These studies all report on short-term interventions compared to baseline or habitual diets. The inconsistency of the study results are intriguing. McIntosh et al[45] comparing a low and a high FODMAP diet found a higher bacteria richness and diversity of Actinobacteria, Firmicutes, and Clostridiales in patients with IBS-D/-M in the low FODMAP group, while a high FODMAP diet decreased the relative abundance of gas-producing bacteria[45]. The sparse documentation of the long-term consequences beyond 8-12 wk of FODMAP restrictions does evoke certain skepticism. The highly restrictive nature of the diet may lead to disordered eating habits and demands much effort and motivation from patients. These factors highlight the importance of reintroduction of FODMAPs after the strict phase in the clinical management of patients with IBS[46]. Inter-individual variability and high inconsistency between clinical findings have made the search for a microbiota signature to predict treatment outcome challenging[47]; and (3) Systemic immune and inflammation responses may also contribute in symptom generation in IBS. McIntosh et al[45] showed that urinary histamine levels were substantially reduced following a low FODMAP diet[45], leading to hypothesizing that a low FODMAP diet might be beneficial in a subset of patients with a particular microbiota profile leading to high histamine production, hence where histamine is being a pathophysiology modulator of importance[32,45]. Our group were the first to report reduced levels of pro-inflammatory IL-6 and IL-8, but not TNF-α, after a 3-wk low FODMAP diet. Simultaneously, selected bacteria associated with anti-inflammatory properties, e.g. Faecalibacterium prausnitzii and Bifidobacterium, total levels of SCFAs and n-butyric acid, decreased[43]. Others have reported that SCFAs may have anti-inflammatory and immunomodulatory effects, as summarized by Tan et al[48], indicating that our findings present a paradox. Indeed, the full connection between diet intake, gut microbiota and its metabolites, and immune and inflammatory responses remains elusive. Herein, the intestinal barrier, gut integrity and low-grade inflammation is further discussed in section “Intestinal barrier and gut integrity”, below.
Fecal microbiota transplantation
In fecal microbiota transplantation (FMT) screened stool from a healthy donor is transferred to a recipient with the purpose of altering the diversity of the gut microbiota. FMT is recommended as a therapeutic strategy in Clostridioides difficile (CDI) infection, and has also been demonstrated effective in inflammatory bowel disease and IBS[49]. There are multiple routes of FMT delivery available including colonoscopy, nasogastric tube, nasoduodenal tube, enema and oral capsules. Each of these modalities has been associated with varying clinical success. Additionally, whether the donor sample is fresh or frozen, or derived from a related or unrelated donor may result in different outcomes. Many recent randomized controlled trial (RCT) studies in IBS have been published, although with conflicting results. In a meta-analysis of five RCTs, overall FMT did not significantly improve IBS symptoms[50]. Here, the results were largely contradictive; one study showed amelioration of symptoms with FMT over placebo, while another study demonstrated superiority of placebo over FMT. The explanation for such contradictory results may be due to the heterogeneity of the disease. Another explanation may be the route of FMT administration. A recent double blinded RCT recruited 90 IBS patients and randomly assigned them to active treatment (n = 60) or placebo (n = 30) where fresh transplant was delivered with colonoscope to coecum[51]. FMT induced significant symptom relief in patients with IBS, compared to controls. In 2020, our group investigated the effect of a single FMT using different stool dosages (30 g and 60 g) of frozen feces, delivered to the distal duodenum through a gastroscope. Placebo was the patient’s own (autologous) feces. Here, patients responded best to the higher dosage. This study concluded that utilizing a well-defined donor with a normal dysbiosis index and a favorable specific microbial signature is important for a successful FMT[52]. Data on long-term follow-up post FMT in IBS have been sparse. However, 1-year effects were recently reported by Holvoet et al[53]. In a doubled blinded RCT of patients with treatment-refractory IBS with predominant bloating, patients were randomly assigned to single dose nasojejunal administration of donor stools or autologous stools[53]. Here, FMT relieved symptoms compared to placebo (autologous transplant), although the effects decreased over 1 year. A second FMT restored the response in patients with a prior response. Evidently, fecal samples from responders had higher microbiota diversity before administration of donor material than fecal samples from non-responders and distinct baseline composition, but unfortunately, no specific marker taxa were associated with response[53]. In addition, 5-year effects were recently reported in a retrospective analysis by Cui et al[54]. In this single-center retrospective study, patients with all subtypes of IBS were assigned to receive FMT through nasojejunal administration, colonoscopy administration, or freeze-dried capsules from healthy, screened donors[54]. Considering all patients, regardless of route of administration, 50% of patients reported gradual symptom improvement after one month to 70% and 75% after one and two years, respectively. After five years, 60% of patients experienced improvement. This decline suggests that repetitive FMT may be required for a sustained effect[54].
There are many hopes for the future in IBS treatment, and FMT capsules are one of them. Capsules are beneficial because the route of administration is much less demanding than endoscopy, they put much less stain on the patient, and can be orally administered by the patients themselves at home. Halkjær et al[55] performed a RCT study in patients with moderate-to-severe IBS. FMT resulted in altered gut microbiota composition, but patients in the placebo group experienced greater symptom relief compared to the treatment group after three months. Supporting this, Aroniadis et al[56] also found that placebo capsules did not induced symptom relief compared with placebo. Hence, the efficacy and safety of FMT in IBS is still under evaluation. Most researchers and clinicians strongly believe that more research is required before the FMT can become an openly available treatment option. A significant question that remains to be answered is whether the described dysbiosis in IBS is a consequence rather than cause of MGB-axis dysfunction in IBS. The varying abovementioned results may indicate that altering gut microbiota is not enough to obtain clinical improvement in IBS. FMT is a highly requested treatment among patients with IBS, and many practitioners find it difficult to refuse patients treatment that may be beneficial. However, researchers are calling for caution on FMT as a treatment of IBS[57]. With reference to safety, patients have reported adverse effects of abdominal pain, cramping or tenderness, diarrhea or constipation, in contrast to 2% in the placebo group[52]. In march 2020, the US Food and Drug Administration issued a warning of potential risk for serious infections due to FMT caused by enteropathogenic or Shigatoxin-producing E. coli that occurred following investigational use of an FMT product supplied by a stool bank, from pre-screened donors[58]. Hence, different study designs with larger cohorts are required to examine the efficacy and safety of FMT in IBS.
Indeed, the complex interplay between the host and gut microbiota is not fully elucidated, but certain features in IBS are documented to be involved, including luminal interactions and its pivotal role in the regulation of the immune system. Whether altered microbiota composition, function and abundance is the cause or a consequence of IBS, we need to understand more about their interplay with us as hosts, and importantly, the intestinal barrier and gut integrity in IBS.
INTESTINAL BARRIER AND GUT INTEGRITY
Molecular biology has revealed the presence of structural and functional alterations of the intestinal epithelial barrier and mild activation of the immune system both locally in the intestinal mucosa and systemically, in IBS. We now know that changes in intestinal permeability create a passage for microbiota and their metabolites from the lumen to the ENS, immune cells and systemic circulation, features that are associated with low-grade inflammation in IBS. Intestinal barrier dysfunction is present in a significant proportion of reported IBS studies, especially in the IBS-D and post-infectious subtype[59]. The association between impaired barrier function and symptoms in IBS are not fully understood but visceral hypersensitivity and pain is possibly explained by exposure of the submucosal neuronal and immune apparatus. Under normal conditions, the intestinal barrier consists of a monolayer of polarised epithelial cells, coated with a thick layer of mucus[60]. As a part of the host defence system, the mucus layer entraps pathogens and is inhabited by commensal microbes such as Bacteroides, Firmicutes, and Lactobacillus whose products, such as IgA, contribute to the prevention of pathogen colonization[61]. Microbiota also produce proteases and protease inhibitors that can modulate the host immune response. In IBS, dysbiosis-derived proteases are thought to contribute to loss of barrier function, immune activation, and symptom generation through activation of protease-activated receptors (PARs)[62]. Recently, higher levels of fecal proteolytic activity has been identified in IBS, particularly in patients suffering from post-infectious IBS[63]. Notably, this was associated with changes in microbiota composition, suggesting that specific microbes contribute to increased production or inadequate suppression of proteases and subsequent activation of PARs that may lead to intestinal barrier dysfunction.
In normal barrier permeability, the space between epithelial cells are sealed by tight junctions and maintained by a complex network of protein interactions. In IBS, the expression levels of tight junction proteins, such as Occludin, Zonula occludens-1, and Claudin-1, have been found to be reduced in the duodenum, jejunum and colon[59,64]. Interestingly, microbiota has been found to regulate the expression of tight junction proteins[65], and enhanced bacterial passage over the barrier has been observed[66].
In the intestinal mucosa, mast cells comprise 2%-3% of the immune cell pool of the lamina propria, and increased intestinal mast cell concentration or activation is one of the most consistent pathological findings in IBS[67-69]. Mast cells possess a great number of stimulatory molecules which allow interaction with a multitude of partners, including immune and non-immune cells. Activated mast cells release mediators such as histamine, serotonin, proteases, and prostaglandins, and they also secrete cytokines and chemokines. Their interactions are indeed complex. Some mast cells interact with both the commensal microbiota and the nervous system by signalling to enteric neurons though serotonin while being influenced by neurotransmitters such as substance P or noradrenalin[70]. In IBS, mast cell-induced activation of enteric neurons may contribute to visceral hypersensitivity[66,71]. Indeed, some bacteria specifically affect mast cells function and activation, but the role of dysbiosis-mast cell-interaction in IBS is yet to be elucidated.
A potential marker of low-grade inflammation in IBS is altered levels of cytokines. Multiple cytokine profiles of patients with IBS have been reported, but they are highly inconsistent. Some has found increased levels of circulating pro-inflammatory cytokines such as IL-6, IL-8, IL-17, and TNF-α[72-74], or reduced levels of the anti-inflammatory cytokine IL-10[72]. Other studies indicate no difference between patients with IBS and healthy controls [75,76]. Interestingly, associations between altered cytokine profiles and changes in gut microbiota have been observed. A study by Hustoft et al[43] found reduced levels of IL-6 and IL-8 and decreased levels of Actinobacteria, Bifidobacterium, and Faecalibacterium prausnitzii after three weeks of the low FODMAP diet. Changes in cytokine levels could thus indicate an abnormal mucosal immune response associated with changes in the gut microbiota.
Increased expression of Toll-like receptors (TLRs) is an interesting finding in patients with IBS[77,78]. These receptors are found on many different cells, including intestinal epithelial cells and immune cells, and they interact in close relation to neural and immune receptors that are involved in the homeostatic regulation in the gut mucosa[79]. TLRs recognise specific microbial components of both commensal and pathogenic bacteria and play a role in immunologic tolerance to commensals and defend against pathogens[80]. In association with increased levels of TLRs, changes in both cytokine profiles and gut microbiota have been observed in patients with IBS, suggesting that an altered microbiota profile may influence TLR expression and immune activation[78].
Indeed, dysbiosis may induce intestinal barrier loss and increased intestinal permeability that cause bacterial products and metabolites to permeate the epithelial barrier, thus triggering an inflammatory response[81]. Mast cells are ‘‘gate keepers’’, and they are not only involved in allergic reactions, but also in host defence including recruitment and activation of other immune cells which may evoke the symptom generation. We believe that further studies should be more focused on which triggering factors that are involved in the link between gut microbiota, intestinal permeability, and intestinal mucosal response in patients with IBS. Indeed, changes in intestinal permeability create a passage for microbiota and their metabolites from the lumen to the ENS, immune cells, and systemic circulation, features whose effect on the brain should also be investigated.
NEUROIMAGNING AND GUT MICROBIOTA IN IBS
The MGB-axis represent a paradigm shift in both neuroscience, gastroenterology and systems medicine. See Mayer et al[82] for a visionary and integrative systems-biology-based model approach to IBS. GI symptoms such as heartburn, indigestion, acid reflux, bloating, pain, constipation, and diarrhea can be triggered by emotional and psychosocial factors. Conversely, GI symptoms alter CNS processing in the absence of detectable organic disease and are implicated in neurological disorders and psychiatric conditions such as anxiety, depression, autism spectrum disorder (ASD), and Parkinson’s disease. Brain imaging modalities and techniques are valuable tools that can non-invasively extract both structure and function in the living brain at the millimeter scale and at a temporal resolution down to seconds. To study the IBS brain, or more generally, applying brain imaging to explore disorders of gut-brain interaction and relation to gut microbiota, the most important modality with whole brain coverage is likely magnetic resonance imaging (MRI)[83,84]. Among the plethora of MRI measurement techniques, there are (1) Structural MRI (sMRI), providing 3D images with high spatial resolution and various types of soft tissue contrast enabling quantitative assessment of brain morphometry such as volumes of different brain structures or regions, local and patch-wise cortical thickness and gyrification, and localized MR signal intensity patterns, e.g., radiomics[85]; (2) diffusion MRI (dMRI), measuring directional and tissue-dependent water diffusion at the microscopic scale enabling quantitative assessment of tissue microarchitecture, e.g., metrics such as fractional anisotropy (FA) derived from the voxel-wise diffusion tensor estimation, and large-scale structural connectivity between brain regions obtain by fiber-tracking algorithms; and (3) functional MRI (fMRI), based on blood-oxygen-level-dependent (BOLD) contrast imaging sensitive to local neuronal activity across the brain in situations where the brain is exposed to cognitive, emotional or sensory stimuli given under experimental control, or being in “resting state” where brain activity is assumed to be intrinsic due to spontaneous fluctuations in the paramagnetic BOLD signal and thereby detectable even in the absence of an externally prompted task or a specific sensory stimulus. A large proportion of neuroimaging studies, partly also targeting IBS, have focused on structural and functional brain connectivity using a combination of dMRI and/or fMRI recordings and topological network analysis based on graph theory[86-89], and more recently also deep learning methods and graph convolution approaches for functional annotation of cognitive states[90,91]. It is expected that these deep learning methodologies will also penetrate imaging and network-based analysis in IBS research[82].
Keeping the systems view on IBS, the term radiomicrobiomics was coined by De Santis, Moratal and Canals in their perspective paper on advancing along the gut-brain axis through big data analysis for diagnostic and prognostic purposes[92]. The term was introduced with reference to the efforts of combining microbiota sequencing data from the gut microbiota with imaging-based features that can be obtained from the conversion of brain images into mineable tabular data or graph representations in a network context. Interestingly, the gut microbiota seems to influence complex physiological systems other than the gut-brain axis and the pathophysiology of IBS, systems that are homeostatically regulated, partly involving CNS and ANS processing. These are blood pressure and the development and pathogenesis of hypertension, glycemic control, development of obesity and diabetes, mood regulation, and anxiety and depression[93-96]. In all these cases, neuroimaging and network analysis will be an important window to the brain and its interplay with microbiota composition and dynamics. More specifically, exploring the associations between neuroimaging parameters, such as brain regional volumes and gray matter densities assessed with sMRI, microstructural patterns assessed with dMRI and derived FA-values, or interregional functional connectivity in e.g. the salience network, visceroceptive, pain processing, or emotion-regulating networks assessed with resting state fMRI and specific gut microbiota signatures, has the potential of vastly enhancing our knowledge on gut-brain interactions in IBS. In neuroimaging of the IBS brain, one of the most consistent findings are alterations in the structure and function of key regions of the somatosensory network, including the globus pallidus, putamen, and caudate, composing the basal ganglia[83]. It has also been reported increased gray matter density (GMD) in the hypothalamus and decreased GMD in the prefrontal cortex in the IBS brain[97]. In rectal distention experiments, patients with IBS had a differential brain response in the pain matrix and default mode network[89]. Neuroimaging studies has also revealed gender-differences in IBS brain network alterations. Female IBS patients showed increased cortical thickness in the pre- and post-central gyrus and decreased thickness in the bilateral insula and the left subgenual anterior cingulate cortex (sgACC), compared to healthy female controls. Connecting emotions and altered brain function in IBS, patients with IBS and comorbid alexithymia have different brain responses to rectal distention in the right insula[98]. As a first in our field, Norlin et al[99] recently provided evidence that the vulnerability to fatigue in IBS is associated with connectivity within a mesocorticolimbic network as well as immune activation in the form of enhanced plasma levels of TNF-α, compared to controls. Indeed, there has been published a large series of papers on brain imaging in IBS, and now, evidence for disrupted subcortical and cortical regions mediated by gut microbial modulation are emerging. Labus et al[100] reporting associations between brain region-to-region functional connectivity and microbiota found a correlation between Clostridia and Bacteroidia with connectivity of the thalamus, the basal ganglia (caudate nucleus, putamen, pallidum, nucleus accumbens), the superior part of the precentral gyrus, the anterior insula and ventral prefrontal regions. Recently, the same lab also reported on fecal metabolites and resting state fMRI[101]. Here, the differences in histidine, cysteine, glycine, glutamate, spermidine, and anserine were significantly associated with the alteration in left dorsal part of the posterior cingulate gyrus to the left putamen. Also, the changes in histidine, tryptophan, uracil, 2-deoxyuridine, thymidine, and succinate were differentially associated with the alteration in the right superior frontal gyrus to the right putamen. Interestingly, this interaction may be mediated by aberrant tryptophan signaling in IBS, which is important because it is a substrate for serotonin synthesis.
In combining brain imaging data, molecular and genetic data, and metagenomic data for joint analysis, new challenges and opportunities arise in the attempt to elucidate the mechanisms and biomarkers of IBS. This endeavour is further described and discussed in the section “Big data analysis” below.
INTESTINAL MICROBIOTA ANALYSIS
Importance of method selection
Microbiome research is advancing rapidly, improving the precision of taxonomy and functional surveys and minimizing methodological limitations. After almost two decades of the earliest intestinal microbiota surveys in humans, we have advanced towards recommendation of quasi-standard methodological procedures to make next-generation sequencing (NGS) data comparable across studies. Notwithstanding, the complete implementation of standards is challenging, given the wide variety of commercial options for sampling, DNA extraction, amplicon generation, library preparation, and DNA sequencing that users fit at their own convenience, making cost-effectivity prevailing. In this regard, the International Human Microbiome Standards consortium has agreed that stool sampling requires minimal processing (no addition of preservation buffers and nuclease inhibitors) to maintain microbiota DNA and RNA integrity, thus facilitating sampling, storage and transport logistics by donors and patients. Besides, stool sub-sampling was revealed to produce minimal variation within individuals[102,103]. Among the multiple methodological steps, DNA extraction introduces the larger variation across experiments, and chemical (muralytic enzymes) or physical cell-wall disruption (bead-beating) methods during DNA extraction are recommended to gain the representation of some microbial species during massive and parallel sequencing and taxonomy data appraisals[103,104]. Taxonomy surveys via amplification and sequencing of bacterial 16S rRNA gene are widespread because of their cost-effectivity. A large collection of tools (e.g., QIIME2, Mothur, DADA2, etc.) and reference repositories (SILVA, RDP, Greengenes, GTDB) have been developed for such an aim. However, this methodology allows reliable identifications mostly at the family and genus levels. Also taxonomy classification depends on the region amplified[105,106] and, accordingly, inconsistencies have been found across studies. Currently, most of the studies are sequencing the V4 or V3-V4 hypervariable regions because of their larger genetic variation and discriminatory power, facilitating re-use and comparisons across different studies.
Metagenomic analysis, based on non-targeted massive DNA sequencing, outperforms the 16S rRNA gene hypervariable region sequencing, although it is much more expensive. Gigabase-level information normally recovered from individual samples permit inspection of microbial species present in human samples and other ecologically complex environments[107,108]. The discriminatory capability of this approach is constantly improved thanks to the existence of comprehensive genome catalogues[109], compiling a huge amount of microbial genetic variability and making it possible sample profiling at the strain level[110]. In addition to the detailed taxonomy surveys, metagenomics makes functional appraisals feasible via DNA read mapping strategies using curated and comprehensive repositories (e.g., KEGG, COG, eggnog databases). The functional analysis has also been developed for 16S rRNA gene amplicon sequencing data (namely PICRUSt). However, the predictions made with such an approach have a high degree of uncertainty due to the ambiguous taxonomic assignments of the 16S rRNA readouts and absence of functional variation information at the species level (minimal gene/functions shared on multiple genome examinations – pangenome – within a single species).
Metadata does matter
The interpretation of microbiome data require a proper control of covariates that many times are not available to be incorporated into the data analysis. This could lead to ambiguous (relying on the generally recognized strain-specific pathogenicity traits) and uncertain (plenty of false-positives) associations between the microbiota and health and disease states, largely influenced by confounding variables[111-113]. Of environmental factors, the intestinal microbiota is strongly influenced by the dietary patterns. Therefore, the use of dietary records around the sampling time are good strategies to integrate such information in the data analysis. The value of this type of information is even more important in microbiota-based biomarker discovery for example for IBS given the impact of food intake on disease symptoms[114]. Nevertheless, not all studies have found meaningful differences in microbiota when using dietary records[115,116]. There is growing evidence supporting a role of energy and macronutrient intake on the intestinal microbiota which could affect associations with health and disease. Body mass index (BMI), gender and age could affect both dietary habits and the intestinal microbiota and change through life differently in women and men[117-119]. By integrating gender, BMI, diet, and age information with microbiota data, the results are less influenced by the subjects' idiosyncratic variation and signals looking for links between gut microbes and health/disease states become more reliable. Moreover, pharmacological treatments given to IBS patients to tackle symptoms should be considered as they could bias the conclusions on the microbiota signatures correlated and playing a role in IBS. There is indeed extensive impact of non-antibiotic drugs on the composition and metabolic function of the gut microbiota[14,120]. In summary, good practices in microbiome research for clinical application undoubtedly involve a meticulous metadata recording covering a large set of individual and lifestyle information that permit uncover unquestionably the influence of gut microbes in our health.
Perspectives for improving microbiome role predictions
The integration of functional omics provides information on the potential role of intestinal microbes and the metabolic products resulting from host-diet-microbe interactions and allows generating human-data-driven hypotheses which could be latter validated in study models. These methods turn data processing more complex because of multidimensionality, but provide clues on the molecular mechanism driven by microbe-host interactions and underlying health and disease[121]. For instance, the correlations of gut microbes (metagenomics) with secondary bile acids (lipidomics), SCFAs (metabolomics), and pro-inflammatory molecules (proteomics/cytokine arrays) make it possible to distinguish microbial groups that plausibly explain disease states or the physiological response to a particular dietary components[122-124]. The high individual specificity and variability of the microbiome data also requires the application of statistical methods that minimize false-positives during biomarker discovery, permit an adequate covariate control and integrate other multidimensional datasets. In this context, the EU COST action ML4Microbiome represents an interesting initiative for advising, researching, and developing advanced statistical and machine learning approaches applied to microbiome research that could greatly contribute to standardizing and improving data analysis in this field[125,126].
Promising developments have also emerged to improve, for example, the accuracy of the dietary assessments. These are often based on self-reported data and, therefore, biased affecting the interpretation of its relationship with the microbiome and health outcomes. In this regard, metabarcoding of plant DNA has been proposed as a method to tacking human plant intake more accurately than using dietary questionnaires. Although this strategy has been only applied to gain information on plant components of the human diet, it looks promising to infer the dietary intakes and the resulting diet-microbe interactions[127].
Sequencing technological advances are also helping to improve the taxonomy resolution of targeted amplicon-based microbiota analysis. The emergence of single-molecule sequencing platforms (Oxford Nanopore Technologies and PacBio) has permitted to generate longer DNA reads, pivotal to increase the information of gene markers under inspection in microbial diversity assessments and to gain resolution at the species-level at a lower cost than metagenomics[128,129]. This methodology has already demonstrated good performance on microbiota surveys despite the modest per-base quality of its reads compared to the classical Sequencing-by-Synthesis (SBS)-based instruments (Illumina). Its potential is even more promising to infer strain-level variation pivotal to determine, for example, species engraftment after FMT[130], which, as mentioned above, is under investigation to treat IBS[52,131].
BIG DATA ANALYSIS
The enormous potential of big data, when harnessed efficiently by powerful statistical methods, mathematical models, and machine learning algorithms, often translates into deeper insights in multi-factorial dynamic systems, which are otherwise complicated to explore, describe and comprehend. The MGB-axis is a well-suited example of such a complex multivariate system and data science (i.e., machine learning algorithms and statistical analyses) is the method of choice to approach this problem. It has become highly evident that no single factor underlie the heterogeneous disorder of IBS. Its investigation requires analysis of large datasets collected from an array of clinical disciplines for a deeper understanding of pathophysiological mechanisms and pathways, and correlations with specific symptoms and symptom severity.
The large number of factors involved both from the brain and microbiota along with their continuous variability, yield large amount of information. These factors are probed using various clinical modalities across several points in time in longitudinal research. The number of resulting variables can range from few hundreds to tens of thousands and the magnitude of data can easily approach hundreds of thousands to several millions of data points, even for studies recruiting several tens of participants. This sheer scale of data combined with larger dimensionality and significant variability sets up a ripe case for employing sophisticated data science methods to study intricate relationships between brain and gut microbiota.
The MGB scientific community appear to already recognize and acknowledge the importance of data science in the field of our research. For example, in their review, De Santis et al[92] described the potential of large amount of information emerging from advanced neuroimaging systems and sophisticated microbiome sequencing techniques to probe complex interactions in the MGB-axis. Their proposal was to combine data from both of these domains to analyze quantitative features for intricate relationships using computational analysis methods, a process they termed “radiomicrobiomics”, which could potentially unveil novel biological information on the MGB-axis. Similar ideas were expressed in another critical review whose key note suggested that encoding microbial information along with other necessary variables into machine learning algorithms could excel our understanding on GI disorders, which are challenging to diagnose due to multifactorial nature of underlying pathology[132]. Mayer et al[83,133] had stressed the need of integrating large sets of host's multi-omics data and microbial data with machine learning techniques to reveal novel insights into the MGB-axis, independent of existing theories and hypothesis. Kaur et al[134] also emphasized the role of machine learning in multiomics data analysis to probe MGB relationship and discussed a framework to move beyond prediction to prevention and personalized therapeutics in MGB related disorders.
In line with these proposals, reports of initiatives and on-going work where the MGB-axis is being explored using the discipline of data science are beginning to emerge. In a recent study, statistical analyses were performed on combined brain and microbial datasets, acquired using resting state fMRI and genetic sequencing, respectively[135]. Probably for the first time, clear correlation-based associations were drawn between certain species of microbiota and corresponding brain regions affected by it, a step forward in the right direction. Wu et al[136] studied the association of gut microbiome in ASD using an array of statistical and machine learning-based analyses and realized presence of certain bacterial genera in ASD group, which could be used as a potential ASD biomarker. Stevens et al[137] studied association of depression phenotype with gut microbiome using microbial genetic information at single nucleotide resolution using multivariate analysis. Based on genetic data of microbiome, they were able to differentiate between depression and healthy cases. The Bergen brain-gut-microbiota study is a notable example of an on-going work that integrates data science with multiomics, where both brain and gut data is being collected from an IBS patient cohort and healthy controls, as shown in Figure 1[138]. BrainGutAnalytics, an advanced analysis project under the umbrella of the Bergen brain gut study, aims to apply sophisticated data science methods to locate IBS biomarkers in brain and peripheral organ systems[139].
Despite these rapidly growing applications of data science in investigating the MGB-axis, it appears that the full potential of data science is yet to be leveraged. Data science techniques, such as machine learning models, particularly thrive in scenarios where the sample size is high (i.e., in order of thousands or more), as it allows the models to adequately learn the underlying data structure by iterating over large number of observations. On the contrary, clinical studies are often limited by sample size albeit high dimensionality of data, as various studies report participant cohorts comprising a few tens to a few hundred subjects only[120-122]. This limited sample size, on one hand, impedes the development of reliable computer models and on the other, high feature to sample ratio could lead to overfitting of the model, which often result in misleading predictions[119]. One tangible way to address this problem is aggregation of several datasets coming from various small scale studies into a larger MGB-axis database, as also proposed by other researchers[80,118,121]. Such a collection will not only feed the needs of data starving computer models but will also represent diverse sectors of subject population in terms of demographics and genetic backgrounds, improving generalization and validation of analysis outcomes. However, such an initiative would only be meaningful if a highly controlled and uniform system of data collection could be developed and implemented across all participating studies. A merger of various datasets taken from isolated studies following their own highly customized protocols, based on variable inclusion and exclusion criteria, will not carry much scientific value. Similarly, an acceptable level of consistency in data management system across all studies is imperative for rapid data accessibility, interpretation and interoperability. The establishment of a larger MGB database would also facilitate much needed interdisciplinary collaboration in MGB research, as data scientists and clinicians must join forces together to solve this complex puzzle.
CONCLUSION
In recent years, research aiming to understand the influence of the gut microbiota on bidirectional interactions between the gut and brain has gained momentum. As described and discussed in this review, the role of microbiota in IBS is so multifaceted that it requires research approaches across disciplines and scientific fields, to reveal details of the complex interactions. Currently, most dietary or FMT interventions are limited to observations of transient microbial shifts within short time frames. Personalized responses of the host microbiota may explain some of the heterogeneity of research outcomes, but not all. Because IBS fluctuates between periods of remission and aggravation of symptoms, longitudinal sampling, multiple sample time-points, post-intervention follow-ups and washout periods for cross-over studies are needed to identify microbial changes that are missed when using cross-sectional sampling, will be of great importance in future studies. We know that gut microbiota profiles are significantly associated with alterations in intestinal gut integrity, brain microstructure, intrinsic neural activities, and cognitive function and mood. How this tremendously intricate symbiotic relationship works in IBS, remains to be unraveled. Multimodal and interdisciplinary clinical studies that include assessments of the gut microbiota composition and function in conjunction with neuroimaging and behavioral testing, such as the Bergen BrainGut microbiota-study[138] are necessary for verification of directionality and causality in the MGB-axis in IBS. Other important work to come are how probiotics influence gut microbiota and affect functional changes in the brain through gut microbiota[140]. As described in this review, the choice of method for analysis is important. We believe, that only through integration of multiple advanced techniques, such as metabolomics and neuroimaging, can we generate a complete picture of host and microbiota pathways in IBS.
Footnotes
Conflict-of-interest statement: No conflict of interest.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Peer-review started: April 17, 2021
First decision: June 3, 2021
Article in press: January 13, 2022
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Norway
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sales-Campos H S-Editor: Gong ZM L-Editor: A P-Editor: Gong ZM
Contributor Information
Eline Margrete Randulff Hillestad, Department of Clinical Medicine, University of Bergen, Bergen 5021, Norway; National Center for Functional Gastrointestinal Disorders, Department of Medicine, Haukeland University Hospital, Bergen 5021, Norway.
Aina van der Meeren, National Center for Functional Gastrointestinal Disorders, Department of Medicine, Haukeland University Hospital, Bergen 5021, Norway.
Bharat Halandur Nagaraja, Mohn Medical Imaging and Visualization Center, Department of Radiology, Haukeland University Hospital, Bergen 5021, Norway.
Ben René Bjørsvik, National Center for Functional Gastrointestinal Disorders, Department of Medicine, Haukeland University Hospital, Bergen 5021, Norway; Mohn Medical Imaging and Visualization Center, Department of Radiology, Haukeland University Hospital, Bergen 5021, Norway.
Noman Haleem, National Center for Functional Gastrointestinal Disorders, Department of Medicine, Haukeland University Hospital, Bergen 5021, Norway; Mohn Medical Imaging and Visualization Center, Department of Radiology, Haukeland University Hospital, Bergen 5021, Norway.
Alfonso Benitez-Paez, Host-Microbe Interactions in Metabolic Health Laboratory, Principe Felipe Research Center, Valencia 46012, Spain.
Yolanda Sanz, Microbial Ecology, Nutrition and Health Research Unit, Institute of Agrochemistry and Food Technology, National Research Council, Paterna-Valencia 46980, Spain.
Trygve Hausken, Department of Clinical Medicine, University of Bergen, Bergen 5021, Norway; National Center for Functional Gastrointestinal Disorders, Department of Medicine, Haukeland University Hospital, Bergen 5021, Norway.
Gülen Arslan Lied, National Center for Functional Gastrointestinal Disorders, Department of Medicine, Haukeland University Hospital, Bergen 5021, Norway; Center for Nutrition, Department of Clinical Medicine, University of Bergen, Bergen 5021, Norway.
Arvid Lundervold, Mohn Medical Imaging and Visualization Center, Department of Radiology, Haukeland University Hospital, Bergen 5021, Norway; Department of Biomedicine, University of Bergen, Bergen 5021, Norway.
Birgitte Berentsen, Department of Clinical Medicine, University of Bergen, Bergen 5021, Norway; National Center for Functional Gastrointestinal Disorders, Department of Medicine, Haukeland University Hospital, Bergen 5021, Norway. birgitte.berentsen1@helse-bergen.no.
References
- 1.Ford AC, Sperber AD, Corsetti M, Camilleri M. Irritable bowel syndrome. Lancet. 2020;396:1675–1688. doi: 10.1016/S0140-6736(20)31548-8. [DOI] [PubMed] [Google Scholar]
- 2.Cassar GE, Youssef GJ, Knowles S, Moulding R, Austin DW. Health-Related Quality of Life in Irritable Bowel Syndrome: A Systematic Review and Meta-analysis. Gastroenterol Nurs. 2020;43:E102–E122. doi: 10.1097/SGA.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 3.Sperber AD, Bangdiwala SI, Drossman DA, Ghoshal UC, Simren M, Tack J, Whitehead WE, Dumitrascu DL, Fang X, Fukudo S, Kellow J, Okeke E, Quigley EMM, Schmulson M, Whorwell P, Archampong T, Adibi P, Andresen V, Benninga MA, Bonaz B, Bor S, Fernandez LB, Choi SC, Corazziari ES, Francisconi C, Hani A, Lazebnik L, Lee YY, Mulak A, Rahman MM, Santos J, Setshedi M, Syam AF, Vanner S, Wong RK, Lopez-Colombo A, Costa V, Dickman R, Kanazawa M, Keshteli AH, Khatun R, Maleki I, Poitras P, Pratap N, Stefanyuk O, Thomson S, Zeevenhooven J, Palsson OS. Worldwide Prevalence and Burden of Functional Gastrointestinal Disorders, Results of Rome Foundation Global Study. Gastroenterology. 2021;160:99–114.e3. doi: 10.1053/j.gastro.2020.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Osadchiy V, Martin CR, Mayer EA. The Gut-Brain Axis and the Microbiome: Mechanisms and Clinical Implications. Clin Gastroenterol Hepatol. 2019;17:322–332. doi: 10.1016/j.cgh.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fülling C, Dinan TG, Cryan JF. Gut Microbe to Brain Signaling: What Happens in Vagus…. Neuron. 2019;101:998–1002. doi: 10.1016/j.neuron.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Leeming ER, Johnson AJ, Spector TD, Le Roy CI. Effect of Diet on the Gut Microbiota: Rethinking Intervention Duration. Nutrients. 2019;11 doi: 10.3390/nu11122862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bercik P, Park AJ, Sinclair D, Khoshdel A, Lu J, Huang X, Deng Y, Blennerhassett PA, Fahnestock M, Moine D, Berger B, Huizinga JD, Kunze W, McLean PG, Bergonzelli GE, Collins SM, Verdu EF. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neurogastroenterol Motil. 2011;23:1132–1139. doi: 10.1111/j.1365-2982.2011.01796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalile B, Vervliet B, Bergonzelli G, Verbeke K, Van Oudenhove L. Colon-delivered short-chain fatty acids attenuate the cortisol response to psychosocial stress in healthy men: a randomized, placebo-controlled trial. Neuropsychopharmacology. 2020;45:2257–2266. doi: 10.1038/s41386-020-0732-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mishima Y, Ishihara S. Molecular Mechanisms of Microbiota-Mediated Pathology in Irritable Bowel Syndrome. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21228664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enck P, Aziz Q, Barbara G, Farmer AD, Fukudo S, Mayer EA, Niesler B, Quigley EM, Rajilić-Stojanović M, Schemann M, Schwille-Kiuntke J, Simren M, Zipfel S, Spiller RC. Irritable bowel syndrome. Nat Rev Dis Primers. 2016;2:16014. doi: 10.1038/nrdp.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Oudenhove L, Crowell MD, Drossman DA, Halpert AD, Keefer L, Lackner JM, Murphy TB, Naliboff BD, Levy RL. Biopsychosocial Aspects of Functional Gastrointestinal Disorders. Gastroenterology. 2016 doi: 10.1053/j.gastro.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adak A, Khan MR. An insight into gut microbiota and its functionalities. Cell Mol Life Sci. 2019;76:473–493. doi: 10.1007/s00018-018-2943-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017;15:630–638. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 14.Brüssow H. Problems with the concept of gut microbiota dysbiosis. Microb Biotechnol. 2020;13:423–434. doi: 10.1111/1751-7915.13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hooks KB, O'Malley MA. Dysbiosis and Its Discontents. mBio. 2017;8 doi: 10.1128/mBio.01492-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maharshak N, Ringel Y, Katibian D, Lundqvist A, Sartor RB, Carroll IM, Ringel-Kulka T. Fecal and Mucosa-Associated Intestinal Microbiota in Patients with Diarrhea-Predominant Irritable Bowel Syndrome. Dig Dis Sci. 2018;63:1890–1899. doi: 10.1007/s10620-018-5086-4. [DOI] [PubMed] [Google Scholar]
- 17.Su T, Liu R, Lee A, Long Y, Du L, Lai S, Chen X, Wang L, Si J, Owyang C, Chen S. Altered Intestinal Microbiota with Increased Abundance of Prevotella Is Associated with High Risk of Diarrhea-Predominant Irritable Bowel Syndrome. Gastroenterol Res Pract. 2018;2018:6961783. doi: 10.1155/2018/6961783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagel R, Traub RJ, Allcock RJ, Kwan MM, Bielefeldt-Ohmann H. Comparison of faecal microbiota in Blastocystis-positive and Blastocystis-negative irritable bowel syndrome patients. Microbiome. 2016;4:47. doi: 10.1186/s40168-016-0191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung CS, Chang PF, Liao CH, Lee TH, Chen Y, Lee YC, Wu MS, Wang HP, Ni YH. Differences of microbiota in small bowel and faeces between irritable bowel syndrome patients and healthy subjects. Scand J Gastroenterol. 2016;51:410–419. doi: 10.3109/00365521.2015.1116107. [DOI] [PubMed] [Google Scholar]
- 20.Zhu S, Liu S, Li H, Zhang Z, Zhang Q, Chen L, Zhao Y, Chen Y, Gu J, Min L, Zhang S. Identification of Gut Microbiota and Metabolites Signature in Patients With Irritable Bowel Syndrome. Front Cell Infect Microbiol. 2019;9:346. doi: 10.3389/fcimb.2019.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pittayanon R, Lau JT, Yuan Y, Leontiadis GI, Tse F, Surette M, Moayyedi P. Gut Microbiota in Patients With Irritable Bowel Syndrome-A Systematic Review. Gastroenterology. 2019;157:97–108. doi: 10.1053/j.gastro.2019.03.049. [DOI] [PubMed] [Google Scholar]
- 22.Hugerth LW, Andreasson A, Talley NJ, Forsberg AM, Kjellström L, Schmidt PT, Agreus L, Engstrand L. No distinct microbiome signature of irritable bowel syndrome found in a Swedish random population. Gut. 2020;69:1076–1084. doi: 10.1136/gutjnl-2019-318717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu HN, Wu H, Chen YZ, Chen YJ, Shen XZ, Liu TT. Altered molecular signature of intestinal microbiota in irritable bowel syndrome patients compared with healthy controls: A systematic review and meta-analysis. Dig Liver Dis. 2017;49:331–337. doi: 10.1016/j.dld.2017.01.142. [DOI] [PubMed] [Google Scholar]
- 24.Wang L, Alammar N, Singh R, Nanavati J, Song Y, Chaudhary R, Mullin GE. Gut Microbial Dysbiosis in the Irritable Bowel Syndrome: A Systematic Review and Meta-Analysis of Case-Control Studies. J Acad Nutr Diet. 2020;120:565–586. doi: 10.1016/j.jand.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 25.Pozuelo M, Panda S, Santiago A, Mendez S, Accarino A, Santos J, Guarner F, Azpiroz F, Manichanh C. Reduction of butyrate- and methane-producing microorganisms in patients with Irritable Bowel Syndrome. Sci Rep. 2015;5:12693. doi: 10.1038/srep12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sundin J, Aziz I, Nordlander S, Polster A, Hu YOO, Hugerth LW, Pennhag AAL, Engstrand L, Törnblom H, Simrén M, Öhman L. Evidence of altered mucosa-associated and fecal microbiota composition in patients with Irritable Bowel Syndrome. Sci Rep. 2020;10:593. doi: 10.1038/s41598-020-57468-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, Kuperman Y, Biton I, Gilad S, Harmelin A, Shapiro H, Halpern Z, Segal E, Elinav E. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 28.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, Shilo S, Lador D, Vila AV, Zmora N, Pevsner-Fischer M, Israeli D, Kosower N, Malka G, Wolf BC, Avnit-Sagi T, Lotan-Pompan M, Weinberger A, Halpern Z, Carmi S, Fu J, Wijmenga C, Zhernakova A, Elinav E, Segal E. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 30.Böhn L, Störsrud S, Törnblom H, Bengtsson U, Simrén M. Self-reported food-related gastrointestinal symptoms in IBS are common and associated with more severe symptoms and reduced quality of life. Am J Gastroenterol. 2013;108:634–641. doi: 10.1038/ajg.2013.105. [DOI] [PubMed] [Google Scholar]
- 31.Monsbakken KW, Vandvik PO, Farup PG. Perceived food intolerance in subjects with irritable bowel syndrome-- etiology, prevalence and consequences. Eur J Clin Nutr. 2006;60:667–672. doi: 10.1038/sj.ejcn.1602367. [DOI] [PubMed] [Google Scholar]
- 32.Moayyedi P, Simrén M, Bercik P. Evidence-based and mechanistic insights into exclusion diets for IBS. Nat Rev Gastroenterol Hepatol. 2020;17:406–413. doi: 10.1038/s41575-020-0270-3. [DOI] [PubMed] [Google Scholar]
- 33.Whelan K, Martin LD, Staudacher HM, Lomer MCE. The low FODMAP diet in the management of irritable bowel syndrome: an evidence-based review of FODMAP restriction, reintroduction and personalisation in clinical practice. J Hum Nutr Diet. 2018;31:239–255. doi: 10.1111/jhn.12530. [DOI] [PubMed] [Google Scholar]
- 34.Varjú P, Farkas N, Hegyi P, Garami A, Szabó I, Illés A, Solymár M, Vincze Á, Balaskó M, Pár G, Bajor J, Szűcs Á, Huszár O, Pécsi D, Czimmer J. Low fermentable oligosaccharides, disaccharides, monosaccharides and polyols (FODMAP) diet improves symptoms in adults suffering from irritable bowel syndrome (IBS) compared to standard IBS diet: A meta-analysis of clinical studies. PLoS One. 2017;12:e0182942. doi: 10.1371/journal.pone.0182942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dionne J, Ford AC, Yuan Y, Chey WD, Lacy BE, Saito YA, Quigley EMM, Moayyedi P. A Systematic Review and Meta-Analysis Evaluating the Efficacy of a Gluten-Free Diet and a Low FODMAPs Diet in Treating Symptoms of Irritable Bowel Syndrome. Am J Gastroenterol. 2018;113:1290–1300. doi: 10.1038/s41395-018-0195-4. [DOI] [PubMed] [Google Scholar]
- 36.van Lanen AS, de Bree A, Greyling A. Efficacy of a low-FODMAP diet in adult irritable bowel syndrome: a systematic review and meta-analysis. Eur J Nutr. 2021;60:3505–3522. doi: 10.1007/s00394-020-02473-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jandee S, Chuensakul S, Maneerat S. No distinction in the gut microbiota between diarrhea predominant-irritable bowel syndrome and healthy subjects: matched case-control study in Thailand. Gut Pathog. 2021;13:16. doi: 10.1186/s13099-021-00406-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banerjee S, Schlaeppi K, van der Heijden MGA. Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol. 2018;16:567–576. doi: 10.1038/s41579-018-0024-1. [DOI] [PubMed] [Google Scholar]
- 39.Berg G, Rybakova D, Fischer D, Cernava T, Vergès MC, Charles T, Chen X, Cocolin L, Eversole K, Corral GH, Kazou M, Kinkel L, Lange L, Lima N, Loy A, Macklin JA, Maguin E, Mauchline T, McClure R, Mitter B, Ryan M, Sarand I, Smidt H, Schelkle B, Roume H, Kiran GS, Selvin J, Souza RSC, van Overbeek L, Singh BK, Wagner M, Walsh A, Sessitsch A, Schloter M. Microbiome definition re-visited: old concepts and new challenges. Microbiome. 2020;8:103. doi: 10.1186/s40168-020-00875-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliphant K, Allen-Vercoe E. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome. 2019;7:91. doi: 10.1186/s40168-019-0704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, Young VB, Henrissat B, Wilmes P, Stappenbeck TS, Núñez G, Martens EC. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167:1339–1353.e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Staudacher HM, Lomer MC, Anderson JL, Barrett JS, Muir JG, Irving PM, Whelan K. Fermentable carbohydrate restriction reduces luminal bifidobacteria and gastrointestinal symptoms in patients with irritable bowel syndrome. J Nutr. 2012;142:1510–1518. doi: 10.3945/jn.112.159285. [DOI] [PubMed] [Google Scholar]
- 43.Hustoft TN, Hausken T, Ystad SO, Valeur J, Brokstad K, Hatlebakk JG, Lied GA. Effects of varying dietary content of fermentable short-chain carbohydrates on symptoms, fecal microenvironment, and cytokine profiles in patients with irritable bowel syndrome. Neurogastroenterol Motil. 2017;29 doi: 10.1111/nmo.12969. [DOI] [PubMed] [Google Scholar]
- 44.Sun Q, Jia Q, Song L, Duan L. Alterations in fecal short-chain fatty acids in patients with irritable bowel syndrome: A systematic review and meta-analysis. Medicine (Baltimore) 2019;98:e14513. doi: 10.1097/MD.0000000000014513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McIntosh K, Reed DE, Schneider T, Dang F, Keshteli AH, De Palma G, Madsen K, Bercik P, Vanner S. FODMAPs alter symptoms and the metabolome of patients with IBS: a randomised controlled trial. Gut. 2017;66:1241–1251. doi: 10.1136/gutjnl-2015-311339. [DOI] [PubMed] [Google Scholar]
- 46.Gibson PR, Halmos EP, Muir JG. Review article: FODMAPS, prebiotics and gut health-the FODMAP hypothesis revisited. Aliment Pharmacol Ther. 2020;52:233–246. doi: 10.1111/apt.15818. [DOI] [PubMed] [Google Scholar]
- 47.Biesiekierski JR, Jalanka J, Staudacher HM. Can Gut Microbiota Composition Predict Response to Dietary Treatments? Nutrients. 2019;11 doi: 10.3390/nu11051134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- 49.D Goldenberg S, Merrick B. The role of faecal microbiota transplantation: looking beyond Clostridioides difficile infection. Ther Adv Infect Dis. 2021;8:2049936120981526. doi: 10.1177/2049936120981526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ianiro G, Eusebi LH, Black CJ, Gasbarrini A, Cammarota G, Ford AC. Systematic review with meta-analysis: efficacy of faecal microbiota transplantation for the treatment of irritable bowel syndrome. Aliment Pharmacol Ther. 2019;50:240–248. doi: 10.1111/apt.15330. [DOI] [PubMed] [Google Scholar]
- 51.Johnsen PH, Hilpüsch F, Cavanagh JP, Leikanger IS, Kolstad C, Valle PC, Goll R. Faecal microbiota transplantation versus placebo for moderate-to-severe irritable bowel syndrome: a double-blind, randomised, placebo-controlled, parallel-group, single-centre trial. Lancet Gastroenterol Hepatol. 2018;3:17–24. doi: 10.1016/S2468-1253(17)30338-2. [DOI] [PubMed] [Google Scholar]
- 52.El-Salhy M, Hatlebakk JG, Gilja OH, Bråthen Kristoffersen A, Hausken T. Efficacy of faecal microbiota transplantation for patients with irritable bowel syndrome in a randomised, double-blind, placebo-controlled study. Gut. 2020;69:859–867. doi: 10.1136/gutjnl-2019-319630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holvoet T, Joossens M, Vázquez-Castellanos JF, Christiaens E, Heyerick L, Boelens J, Verhasselt B, van Vlierberghe H, De Vos M, Raes J, De Looze D. Fecal Microbiota Transplantation Reduces Symptoms in Some Patients With Irritable Bowel Syndrome With Predominant Abdominal Bloating: Short- and Long-term Results From a Placebo-Controlled Randomized Trial. Gastroenterology. 2021;160:145–157.e8. doi: 10.1053/j.gastro.2020.07.013. [DOI] [PubMed] [Google Scholar]
- 54.Cui J, Lin Z, Tian H, Yang B, Zhao D, Ye C, Li N, Qin H, Chen Q. Long-Term Follow-Up Results of Fecal Microbiota Transplantation for Irritable Bowel Syndrome: A Single-Center, Retrospective Study. Front Med (Lausanne) 2021;8:710452. doi: 10.3389/fmed.2021.710452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Halkjær SI, Christensen AH, Lo BZS, Browne PD, Günther S, Hansen LH, Petersen AM. Faecal microbiota transplantation alters gut microbiota in patients with irritable bowel syndrome: results from a randomised, double-blind placebo-controlled study. Gut. 2018;67:2107–2115. doi: 10.1136/gutjnl-2018-316434. [DOI] [PubMed] [Google Scholar]
- 56.Aroniadis OC, Brandt LJ, Oneto C, Feuerstadt P, Sherman A, Wolkoff AW, Kassam Z, Sadovsky RG, Elliott RJ, Budree S, Kim M, Keller MJ. Faecal microbiota transplantation for diarrhoea-predominant irritable bowel syndrome: a double-blind, randomised, placebo-controlled trial. Lancet Gastroenterol Hepatol. 2019;4:675–685. doi: 10.1016/S2468-1253(19)30198-0. [DOI] [PubMed] [Google Scholar]
- 57.Camilleri M. FMT in IBS: a call for caution. Gut. 2021;70:431. doi: 10.1136/gutjnl-2020-321529. [DOI] [PubMed] [Google Scholar]
- 58.U.S. Food and Drug administration. Safety Alert Regarding Use of Fecal Microbiota for Transplantation and Risk of Serious Adverse Events Likely Due to Transmission of Pathogenic Organisms 2020. Available from: https://www.fda.gov/vaccines-blood-biologics/safety-availability-biologics/safety-alert-regarding-use-fecal-microbiota-transplantation-and-risk-serious-adverse-events-likely . [Google Scholar]
- 59.Hanning N, Edwinson AL, Ceuleers H, Peters SA, De Man JG, Hassett LC, De Winter BY, Grover M. Intestinal barrier dysfunction in irritable bowel syndrome: a systematic review. Therap Adv Gastroenterol. 2021;14:1756284821993586. doi: 10.1177/1756284821993586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30:492–506. doi: 10.1038/s41422-020-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ringel Y, Maharshak N. Intestinal microbiota and immune function in the pathogenesis of irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2013;305:G529–G541. doi: 10.1152/ajpgi.00207.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maharshak N, Huh EY, Paiboonrungruang C, Shanahan M, Thurlow L, Herzog J, Djukic Z, Orlando R, Pawlinski R, Ellermann M, Borst L, Patel S, Dotan I, Sartor RB, Carroll IM. Enterococcus faecalis Gelatinase Mediates Intestinal Permeability via Protease-Activated Receptor 2. Infect Immun. 2015;83:2762–2770. doi: 10.1128/IAI.00425-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edogawa S, Edwinson AL, Peters SA, Chikkamenahalli LL, Sundt W, Graves S, Gurunathan SV, Breen-Lyles M, Johnson S, Dyer R, Graham R, Chen J, Kashyap P, Farrugia G, Grover M. Serine proteases as luminal mediators of intestinal barrier dysfunction and symptom severity in IBS. Gut. 2020;69:62–73. doi: 10.1136/gutjnl-2018-317416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu H, Xiao X, Shi Y, Wu Y, Huang Y, Li D, Xiong F, He G, Chai Y, Tang H. Inhibition of miRNA-29a regulates intestinal barrier function in diarrhea-predominant irritable bowel syndrome by upregulating ZO-1 and CLDN1. Exp Ther Med. 2020;20:155. doi: 10.3892/etm.2020.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh R, Chandrashekharappa S, Bodduluri SR, Baby BV, Hegde B, Kotla NG, Hiwale AA, Saiyed T, Patel P, Vijay-Kumar M, Langille MGI, Douglas GM, Cheng X, Rouchka EC, Waigel SJ, Dryden GW, Alatassi H, Zhang HG, Haribabu B, Vemula PK, Jala VR. Enhancement of the gut barrier integrity by a microbial metabolite through the Nrf2 pathway. Nat Commun. 2019;10:89. doi: 10.1038/s41467-018-07859-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bednarska O, Walter SA, Casado-Bedmar M, Ström M, Salvo-Romero E, Vicario M, Mayer EA, Keita ÅV. Vasoactive Intestinal Polypeptide and Mast Cells Regulate Increased Passage of Colonic Bacteria in Patients With Irritable Bowel Syndrome. Gastroenterology. 2017;153:948–960.e3. doi: 10.1053/j.gastro.2017.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Casado-Bedmar M, Keita ÅV. Potential neuro-immune therapeutic targets in irritable bowel syndrome. Therap Adv Gastroenterol. 2020;13:1756284820910630. doi: 10.1177/1756284820910630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robles A, Perez Ingles D, Myneedu K, Deoker A, Sarosiek I, Zuckerman MJ, Schmulson MJ, Bashashati M. Mast cells are increased in the small intestinal mucosa of patients with irritable bowel syndrome: A systematic review and meta-analysis. Neurogastroenterol Motil. 2019;31:e13718. doi: 10.1111/nmo.13718. [DOI] [PubMed] [Google Scholar]
- 69.Krammer L, Sowa AS, Lorentz A. Mast Cells in Irritable Bowel Syndrome: A Systematic Review. J Gastrointestin Liver Dis. 2019;28:463–472. doi: 10.15403/jgld-229. [DOI] [PubMed] [Google Scholar]
- 70.De Zuani M, Dal Secco C, Frossi B. Mast cells at the crossroads of microbiota and IBD. Eur J Immunol. 2018;48:1929–1937. doi: 10.1002/eji.201847504. [DOI] [PubMed] [Google Scholar]
- 71.Buckley MM, O'Halloran KD, Rae MG, Dinan TG, O'Malley D. Modulation of enteric neurons by interleukin-6 and corticotropin-releasing factor contributes to visceral hypersensitivity and altered colonic motility in a rat model of irritable bowel syndrome. J Physiol. 2014;592:5235–5250. doi: 10.1113/jphysiol.2014.279968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choghakhori R, Abbasnezhad A, Hasanvand A, Amani R. Inflammatory cytokines and oxidative stress biomarkers in irritable bowel syndrome: Association with digestive symptoms and quality of life. Cytokine. 2017;93:34–43. doi: 10.1016/j.cyto.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 73.Seyedmirzaee S, Hayatbakhsh MM, Ahmadi B, Baniasadi N, Bagheri Rafsanjani AM, Nikpoor AR, Mohammadi M. Serum immune biomarkers in irritable bowel syndrome. Clin Res Hepatol Gastroenterol. 2016;40:631–637. doi: 10.1016/j.clinre.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 74.Bashashati M, Moradi M, Sarosiek I. Interleukin-6 in irritable bowel syndrome: A systematic review and meta-analysis of IL-6 (-G174C) and circulating IL-6 levels. Cytokine. 2017;99:132–138. doi: 10.1016/j.cyto.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 75.Bennet SM, Polster A, Törnblom H, Isaksson S, Capronnier S, Tessier A, Le Nevé B, Simrén M, Öhman L. Global Cytokine Profiles and Association With Clinical Characteristics in Patients With Irritable Bowel Syndrome. Am J Gastroenterol. 2016;111:1165–1176. doi: 10.1038/ajg.2016.223. [DOI] [PubMed] [Google Scholar]
- 76.Kamp KJ, Han C, Shulman RJ, Cain KC, Barney P, Opp MR, Chang L, Burr RL, Heitkemper MM. Cytokine Levels and Symptoms Among Women with Irritable Bowel Syndrome: Considering the Role of Hormonal Contraceptive Use. Biol Res Nurs. 2021;23:171–179. doi: 10.1177/1099800420941252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dlugosz A, Zakikhany K, Acevedo N, D'Amato M, Lindberg G. Increased Expression of Toll-Like Receptors 4, 5, and 9 in Small Bowel Mucosa from Patients with Irritable Bowel Syndrome. Biomed Res Int. 2017;2017:9624702. doi: 10.1155/2017/9624702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shukla R, Ghoshal U, Ranjan P, Ghoshal UC. Expression of Toll-like Receptors, Pro-, and Anti-inflammatory Cytokines in Relation to Gut Microbiota in Irritable Bowel Syndrome: The Evidence for Its Micro-organic Basis. J Neurogastroenterol Motil. 2018;24:628–642. doi: 10.5056/jnm18130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Assas BM, Miyan JA, Pennock JL. Cross-talk between neural and immune receptors provides a potential mechanism of homeostatic regulation in the gut mucosa. Mucosal Immunol. 2014;7:1283–1289. doi: 10.1038/mi.2014.80. [DOI] [PubMed] [Google Scholar]
- 80.de Kivit S, Tobin MC, Forsyth CB, Keshavarzian A, Landay AL. Regulation of Intestinal Immune Responses through TLR Activation: Implications for Pro- and Prebiotics. Front Immunol. 2014;5:60. doi: 10.3389/fimmu.2014.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Linsalata M, Riezzo G, Clemente C, D'Attoma B, Russo F. Noninvasive Biomarkers of Gut Barrier Function in Patients Suffering from Diarrhea Predominant-IBS: An Update. Dis Markers. 2020;2020:2886268. doi: 10.1155/2020/2886268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mayer EA, Labus JS, Tillisch K, Cole SW, Baldi P. Towards a systems view of IBS. Nat Rev Gastroenterol Hepatol. 2015;12:592–605. doi: 10.1038/nrgastro.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mayer EA, Labus J, Aziz Q, Tracey I, Kilpatrick L, Elsenbruch S, Schweinhardt P, Van Oudenhove L, Borsook D. Role of brain imaging in disorders of brain-gut interaction: a Rome Working Team Report. Gut. 2019;68:1701–1715. doi: 10.1136/gutjnl-2019-318308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu P, Peng G, Zhang N, Wang B, Luo B. Crosstalk Between the Gut Microbiota and the Brain: An Update on Neuroimaging Findings. Front Neurol. 2019;10:883. doi: 10.3389/fneur.2019.00883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging-"how-to" guide and critical reflection. Insights Imaging. 2020;11:91. doi: 10.1186/s13244-020-00887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 87.Yeh CH, Jones DK, Liang X, Descoteaux M, Connelly A. Mapping Structural Connectivity Using Diffusion MRI: Challenges and Opportunities. J Magn Reson Imaging. 2021;53:1666–1682. doi: 10.1002/jmri.27188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bassett DS, Khambhati AN, Grafton ST. Emerging Frontiers of Neuroengineering: A Network Science of Brain Connectivity. Annu Rev Biomed Eng. 2017;19:327–352. doi: 10.1146/annurev-bioeng-071516-044511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kano M, Grinsvall C, Ran Q, Dupont P, Morishita J, Muratsubaki T, Mugikura S, Ly HG, Törnblom H, Ljungberg M, Takase K, Simrén M, Van Oudenhove L, Fukudo S. Resting state functional connectivity of the pain matrix and default mode network in irritable bowel syndrome: a graph theoretical analysis. Sci Rep. 2020;10:11015. doi: 10.1038/s41598-020-67048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys. 2019;29:102–127. doi: 10.1016/j.zemedi.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 91.Zhang Y, Tetrel L, Thirion B, Bellec P. Functional annotation of human cognitive states using deep graph convolution. Neuroimage. 2021;231:117847. doi: 10.1016/j.neuroimage.2021.117847. [DOI] [PubMed] [Google Scholar]
- 92.De Santis S, Moratal D, Canals S. Radiomicrobiomics: Advancing Along the Gut-brain Axis Through Big Data Analysis. Neuroscience. 2019;403:145–149. doi: 10.1016/j.neuroscience.2017.11.055. [DOI] [PubMed] [Google Scholar]
- 93.Avery EG, Bartolomaeus H, Maifeld A, Marko L, Wiig H, Wilck N, Rosshart SP, Forslund SK, Müller DN. The Gut Microbiome in Hypertension: Recent Advances and Future Perspectives. Circ Res. 2021;128:934–950. doi: 10.1161/CIRCRESAHA.121.318065. [DOI] [PubMed] [Google Scholar]
- 94.Gérard C, Vidal H. Impact of Gut Microbiota on Host Glycemic Control. Front Endocrinol (Lausanne) 2019;10:29. doi: 10.3389/fendo.2019.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Simpson CA, Schwartz OS, Eliby D, Butler CA, Huang K, O'Brien-Simpson N, Callaghan BL, Dashper SG, Gooley PR, Whittle S, Haslam N, Simmons JG. Bugs and Brains, the Gut and Mental Health Study: a mixed-methods study investigating microbiota composition and function in anxiety, depression and irritable bowel syndrome. BMJ Open. 2021;11:e043221. doi: 10.1136/bmjopen-2020-043221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Margolis KG, Cryan JF, Mayer EA. The Microbiota-Gut-Brain Axis: From Motility to Mood. Gastroenterology. 2021;160:1486–1501. doi: 10.1053/j.gastro.2020.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Seminowicz DA, Labus JS, Bueller JA, Tillisch K, Naliboff BD, Bushnell MC, Mayer EA. Regional gray matter density changes in brains of patients with irritable bowel syndrome. Gastroenterology. 2010;139:48–57.e2. doi: 10.1053/j.gastro.2010.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kano M, Muratsubaki T, Yagihashi M, Morishita J, Mugikura S, Dupont P, Takase K, Kanazawa M, Van Oudenhove L, Fukudo S. Insula Activity to Visceral Stimulation and Endocrine Stress Responses as Associated With Alexithymia in Patients With Irritable Bowel Syndrome. Psychosom Med. 2020;82:29–38. doi: 10.1097/PSY.0000000000000729. [DOI] [PubMed] [Google Scholar]
- 99.Norlin AK, Walter S, Icenhour A, Keita ÅV, Elsenbruch S, Bednarska O, Jones MP, Simon R, Engström M. Fatigue in irritable bowel syndrome is associated with plasma levels of TNF-α and mesocorticolimbic connectivity. Brain Behav Immun. 2021;92:211–222. doi: 10.1016/j.bbi.2020.11.035. [DOI] [PubMed] [Google Scholar]
- 100.Labus JS, Osadchiy V, Hsiao EY, Tap J, Derrien M, Gupta A, Tillisch K, Le Nevé B, Grinsvall C, Ljungberg M, Öhman L, Törnblom H, Simren M, Mayer EA. Evidence for an association of gut microbial Clostridia with brain functional connectivity and gastrointestinal sensorimotor function in patients with irritable bowel syndrome, based on tripartite network analysis. Microbiome. 2019;7:45. doi: 10.1186/s40168-019-0656-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Osadchiy V, Mayer EA, Gao K, Labus JS, Naliboff B, Tillisch K, Chang L, Jacobs JP, Hsiao EY, Gupta A. Analysis of brain networks and fecal metabolites reveals brain-gut alterations in premenopausal females with irritable bowel syndrome. Transl Psychiatry. 2020;10:367. doi: 10.1038/s41398-020-01071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cardona S, Eck A, Cassellas M, Gallart M, Alastrue C, Dore J, Azpiroz F, Roca J, Guarner F, Manichanh C. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol. 2012;12:158. doi: 10.1186/1471-2180-12-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Santiago A, Panda S, Mengels G, Martinez X, Azpiroz F, Dore J, Guarner F, Manichanh C. Processing faecal samples: a step forward for standards in microbial community analysis. BMC Microbiol. 2014;14:112. doi: 10.1186/1471-2180-14-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Costea PI, Zeller G, Sunagawa S, Pelletier E, Alberti A, Levenez F, Tramontano M, Driessen M, Hercog R, Jung FE, Kultima JR, Hayward MR, Coelho LP, Allen-Vercoe E, Bertrand L, Blaut M, Brown JRM, Carton T, Cools-Portier S, Daigneault M, Derrien M, Druesne A, de Vos WM, Finlay BB, Flint HJ, Guarner F, Hattori M, Heilig H, Luna RA, van Hylckama Vlieg J, Junick J, Klymiuk I, Langella P, Le Chatelier E, Mai V, Manichanh C, Martin JC, Mery C, Morita H, O'Toole PW, Orvain C, Patil KR, Penders J, Persson S, Pons N, Popova M, Salonen A, Saulnier D, Scott KP, Singh B, Slezak K, Veiga P, Versalovic J, Zhao L, Zoetendal EG, Ehrlich SD, Dore J, Bork P. Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol. 2017;35:1069–1076. doi: 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- 105.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mizrahi-Man O, Davenport ER, Gilad Y. Taxonomic classification of bacterial 16S rRNA genes using short sequencing reads: evaluation of effective study designs. PLoS One. 2013;8:e53608. doi: 10.1371/journal.pone.0053608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Milanese A, Mende DR, Paoli L, Salazar G, Ruscheweyh HJ, Cuenca M, Hingamp P, Alves R, Costea PI, Coelho LP, Schmidt TSB, Almeida A, Mitchell AL, Finn RD, Huerta-Cepas J, Bork P, Zeller G, Sunagawa S. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat Commun. 2019;10:1014. doi: 10.1038/s41467-019-08844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Almeida A, Nayfach S, Boland M, Strozzi F, Beracochea M, Shi ZJ, Pollard KS, Sakharova E, Parks DH, Hugenholtz P, Segata N, Kyrpides NC, Finn RD. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat Biotechnol. 2021;39:105–114. doi: 10.1038/s41587-020-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Anyansi C, Straub TJ, Manson AL, Earl AM, Abeel T. Computational Methods for Strain-Level Microbial Detection in Colony and Metagenome Sequencing Data. Front Microbiol. 2020;11:1925. doi: 10.3389/fmicb.2020.01925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, Tito RY, Chaffron S, Rymenans L, Verspecht C, De Sutter L, Lima-Mendez G, D'hoe K, Jonckheere K, Homola D, Garcia R, Tigchelaar EF, Eeckhaudt L, Fu J, Henckaerts L, Zhernakova A, Wijmenga C, Raes J. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 112.Haro C, Rangel-Zúñiga OA, Alcalá-Díaz JF, Gómez-Delgado F, Pérez-Martínez P, Delgado-Lista J, Quintana-Navarro GM, Landa BB, Navas-Cortés JA, Tena-Sempere M, Clemente JC, López-Miranda J, Pérez-Jiménez F, Camargo A. Intestinal Microbiota Is Influenced by Gender and Body Mass Index. PLoS One. 2016;11:e0154090. doi: 10.1371/journal.pone.0154090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vujkovic-Cvijin I, Sklar J, Jiang L, Natarajan L, Knight R, Belkaid Y. Host variables confound gut microbiota studies of human disease. Nature. 2020;587:448–454. doi: 10.1038/s41586-020-2881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Spiller R. Impact of Diet on Symptoms of the Irritable Bowel Syndrome. Nutrients. 2021;13 doi: 10.3390/nu13020575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Koole JL, Bours MJL, Breedveld-Peters JJL, van Roekel EH, van Dongen MCJM, Eussen SJPM, van Zutphen M, van Duijnhoven FJB, Boshuizen HC, Weijenberg MP. Evaluating the Validity of a Food Frequency Questionnaire in Comparison with a 7-Day Dietary Record for Measuring Dietary Intake in a Population of Survivors of Colorectal Cancer. J Acad Nutr Diet. 2020;120:245–257. doi: 10.1016/j.jand.2019.09.008. [DOI] [PubMed] [Google Scholar]
- 116.Watanabe D, Nanri H, Yoshida T, Yamaguchi M, Sugita M, Nozawa Y, Okabe Y, Itoi A, Goto C, Yamada Y, Ishikawa-Takata K, Kobayashi H, Kimura M Kyoto-Kameoka Study Group KS. Validation of Energy and Nutrition Intake in Japanese Elderly Individuals Estimated Based on a Short Food Frequency Questionnaire Compared against a 7-day Dietary Record: The Kyoto-Kameoka Study. Nutrients. 2019;11 doi: 10.3390/nu11030688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Caine-Bish NL, Scheule B. Gender differences in food preferences of school-aged children and adolescents. J Sch Health. 2009;79:532–540. doi: 10.1111/j.1746-1561.2009.00445.x. [DOI] [PubMed] [Google Scholar]
- 118.Martin CK, Rosenbaum D, Han H, Geiselman PJ, Wyatt HR, Hill JO, Brill C, Bailer B, Miller BV 3rd, Stein R, Klein S, Foster GD. Change in food cravings, food preferences, and appetite during a low-carbohydrate and low-fat diet. Obesity (Silver Spring) 2011;19:1963–1970. doi: 10.1038/oby.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Song HJ, Simon JR, Patel DU. Food preferences of older adults in senior nutrition programs. J Nutr Gerontol Geriatr. 2014;33:55–67. doi: 10.1080/21551197.2013.875502. [DOI] [PubMed] [Google Scholar]
- 120.Vich Vila A, Collij V, Sanna S, Sinha T, Imhann F, Bourgonje AR, Mujagic Z, Jonkers DMAE, Masclee AAM, Fu J, Kurilshikov A, Wijmenga C, Zhernakova A, Weersma RK. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun. 2020;11:362. doi: 10.1038/s41467-019-14177-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project. Nature. 2019;569:641–648. doi: 10.1038/s41586-019-1238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Benítez-Páez A, Hess AL, Krautbauer S, Liebisch G, Christensen L, Hjorth MF, Larsen TM, Sanz Y MyNewGut consortium. Sex, Food, and the Gut Microbiota: Disparate Response to Caloric Restriction Diet with Fiber Supplementation in Women and Men. Mol Nutr Food Res. 2021;65:e2000996. doi: 10.1002/mnfr.202000996. [DOI] [PubMed] [Google Scholar]
- 123.Benítez-Páez A, Kjølbæk L, Gómez Del Pulgar EM, Brahe LK, Astrup A, Matysik S, Schött HF, Krautbauer S, Liebisch G, Boberska J, Claus S, Rampelli S, Brigidi P, Larsen LH, Sanz Y. A Multi-omics Approach to Unraveling the Microbiome-Mediated Effects of Arabinoxylan Oligosaccharides in Overweight Humans. mSystems. 2019;4 doi: 10.1128/mSystems.00209-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, Casero D, Courtney H, Gonzalez A, Graeber TG, Hall AB, Lake K, Landers CJ, Mallick H, Plichta DR, Prasad M, Rahnavard G, Sauk J, Shungin D, Vázquez-Baeza Y, White RA 3rd IBDMDB Investigators, Braun J, Denson LA, Jansson JK, Knight R, Kugathasan S, McGovern DPB, Petrosino JF, Stappenbeck TS, Winter HS, Clish CB, Franzosa EA, Vlamakis H, Xavier RJ, Huttenhower C. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marcos-Zambrano LJ, Karaduzovic-Hadziabdic K, Loncar Turukalo T, Przymus P, Trajkovik V, Aasmets O, Berland M, Gruca A, Hasic J, Hron K, Klammsteiner T, Kolev M, Lahti L, Lopes MB, Moreno V, Naskinova I, Org E, Paciência I, Papoutsoglou G, Shigdel R, Stres B, Vilne B, Yousef M, Zdravevski E, Tsamardinos I, Carrillo de Santa Pau E, Claesson MJ, Moreno-Indias I, Truu J. Applications of Machine Learning in Human Microbiome Studies: A Review on Feature Selection, Biomarker Identification, Disease Prediction and Treatment. Front Microbiol . 2021;12:634511. doi: 10.3389/fmicb.2021.634511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Moreno-Indias I, Lahti L, Nedyalkova M, Elbere I, Roshchupkin G, Adilovic M, Aydemir O, Bakir-Gungor B, Santa Pau EC, D'Elia D, Desai MS, Falquet L, Gundogdu A, Hron K, Klammsteiner T, Lopes MB, Marcos-Zambrano LJ, Marques C, Mason M, May P, Pašić L, Pio G, Pongor S, Promponas VJ, Przymus P, Saez-Rodriguez J, Sampri A, Shigdel R, Stres B, Suharoschi R, Truu J, Truică CO, Vilne B, Vlachakis D, Yilmaz E, Zeller G, Zomer AL, Gómez-Cabrero D, Claesson MJ. Statistical and Machine Learning Techniques in Human Microbiome Studies: Contemporary Challenges and Solutions. Front Microbiol. 2021;12:635781. doi: 10.3389/fmicb.2021.635781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Reese AT, Kartzinel TR, Petrone BL, Turnbaugh PJ, Pringle RM, David LA. Using DNA Metabarcoding To Evaluate the Plant Component of Human Diets: a Proof of Concept. mSystems. 2019;4 doi: 10.1128/mSystems.00458-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Benítez-Páez A, Portune KJ, Sanz Y. Species-level resolution of 16S rRNA gene amplicons sequenced through the MinION™ portable nanopore sequencer. Gigascience. 2016;5:4. doi: 10.1186/s13742-016-0111-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Benítez-Páez A, Sanz Y. Multi-locus and long amplicon sequencing approach to study microbial diversity at species level using the MinION™ portable nanopore sequencer. Gigascience. 2017;6:1–12. doi: 10.1093/gigascience/gix043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Benítez-Páez A, Hartstra AV, Nieuwdorp M, Sanz Y. Strand-wise and bait-assisted assembly of nearly-full rrn operons applied to assess species engraftment after faecal microbiota transplantation. bioRxiv . 2020:2020.09.11.292896. [Google Scholar]
- 131.Johnsen PH, Hilpüsch F, Valle PC, Goll R. The effect of fecal microbiota transplantation on IBS related quality of life and fatigue in moderate to severe non-constipated irritable bowel: Secondary endpoints of a double blind, randomized, placebo-controlled trial. EBioMedicine. 2020;51:102562. doi: 10.1016/j.ebiom.2019.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mukhtar K, Nawaz H, Abid S. Functional gastrointestinal disorders and gut-brain axis: What does the future hold? World J Gastroenterol. 2019;25:552–566. doi: 10.3748/wjg.v25.i5.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Martin CR, Osadchiy V, Kalani A, Mayer EA. The Brain-Gut-Microbiome Axis. Cell Mol Gastroenterol Hepatol. 2018;6:133–148. doi: 10.1016/j.jcmgh.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kaur H, Singh Y, Singh S, Singh RB. Gut microbiome-mediated epigenetic regulation of brain disorder and application of machine learning for multi-omics data analysis. Genome. 2021;64:355–371. doi: 10.1139/gen-2020-0136. [DOI] [PubMed] [Google Scholar]
- 135.Liu P, Jia XZ, Chen Y, Yu Y, Zhang K, Lin YJ, Wang BH, Peng GP. Gut microbiota interacts with intrinsic brain activity of patients with amnestic mild cognitive impairment. CNS Neurosci Ther. 2021;27:163–173. doi: 10.1111/cns.13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu T, Wang H, Lu W, Zhai Q, Zhang Q, Yuan W, Gu Z, Zhao J, Zhang H, Chen W. Potential of gut microbiome for detection of autism spectrum disorder. Microb Pathog. 2020;149:104568. doi: 10.1016/j.micpath.2020.104568. [DOI] [PubMed] [Google Scholar]
- 137.Stevens BR, Roesch L, Thiago P, Russell JT, Pepine CJ, Holbert RC, Raizada MK, Triplett EW. Depression phenotype identified by using single nucleotide exact amplicon sequence variants of the human gut microbiome. Mol Psychiatry. 2021;26:4277–4287. doi: 10.1038/s41380-020-0652-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Berentsen B, Nagaraja BH, Teige EP, Lied GA, Lundervold AJ, Lundervold K, Steinsvik EK, Hillestad ER, Valeur J, Brønstad I, Gilja OH, Osnes B, Hatlebakk JG, Haász J, Labus J, Gupta A, Mayer EA, Benitez-Páez A, Sanz Y, Lundervold A, Hausken T. Study protocol of the Bergen brain-gut-microbiota-axis study: A prospective case-report characterization and dietary intervention study to evaluate the effects of microbiota alterations on cognition and anatomical and functional brain connectivity in patients with irritable bowel syndrome. Medicine (Baltimore) 2020;99:e21950. doi: 10.1097/MD.0000000000021950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bergen BrainGut. Multiomics based analysis of brain-gut axis: A search for gastrointestinal disease phenotypes. 2021. Available from: https://cordis.europa.eu/project/id/895219 .
- 140.Liu L, Ni X, Tian T, Li X, Li F, Sun M, Chen J, Zhou S, Zhao L. Effect of regulating gut microbiota using probiotics on functional changes in the brain: protocol for a systematic review. BMJ Open. 2020;10:e037582. doi: 10.1136/bmjopen-2020-037582. [DOI] [PMC free article] [PubMed] [Google Scholar]