Abstract
We present a genome assembly from an individual female Rana temporaria (the common frog; Chordata; Amphibia; Anura; Ranidae). The genome sequence is 4.11 gigabases in span. The majority of the assembly is scaffolded into 13 chromosomal pseudomolecules. Gene annotation of this assembly by the NCBI Eukaryotic Genome Annotation Pipeline has identified 23,707 protein coding genes.
Keywords: Rana temporaria, common frog, genome sequence, chromosomal
Species taxonomy
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Amphibia; Batrachia; Anura; Neobatrachia; Ranoidea; Ranidae; Rana; Rana temporaria Linnaeus 1758 (NCBI:txid8407).
Introduction
The common frog, Rana temporaria (Anura: Ranidae), is widely distributed throughout Europe. It has a biphasic life cycle that includes aquatic, benthic larvae and terrestrial (sometimes semi-aquatic) adults. In the United Kingdom, populations of R. temporaria breed as early as late January with most tadpoles metamorphosing in June or July, however, tadpoles occasionally overwinter ( Walsh et al., 2016). The common frog is an emerging model for the study of genetic sex determination, as different populations vary in their degree of sex chromosome differentiation (e.g. ( Phillips et al., 2020)).
The nuclear genome size of R. temporaria was previously estimated to be between 3.31 and 4.91 picograms (= 3.24 and 4.80 gigabases; ( Gregory, 2021)) which is consistent with our 4.11 gigabase assembly. The thirteen pseudomolecules in our assembly match the expected number of chromosomes in R. temporaria (2N = 26; five macro- and eight micro-chromosomes; ( Spasić-Bošković et al., 1997). This is the second nuclear genome sequence to be reported from a ranid anuran ( Hammond et al., 2017).
The R. temporaria reference genome sequence from a UK-collected individual will provide a useful resource for enhancing and further interpreting available datasets including transcriptomic data that document the immune response of R. temporaria to the amphibian diseases caused by Batrachochytrium dendrobatidis and Ranavirus ( Price et al., 2015).
Genome sequence report
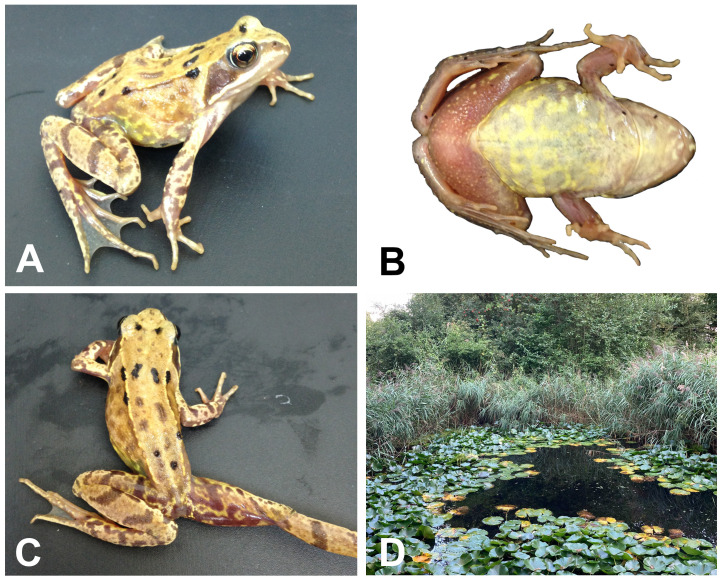
The genome was sequenced from one female R. temporaria ( Figure 1A–C) collected from The Natural History Museum Wildlife Garden, London, UK ( Figure 1D. A total of 63-fold coverage in Pacific Biosciences single-molecule long reads (N50 27 kb) and 51-fold coverage in 10X Genomics read clouds (from molecules with an estimated N50 of 25 kb) were generated. Primary assembly contigs were scaffolded with chromosome conformation Hi-C data. Manual assembly curation corrected 974 missing/misjoins and removed 22 haplotypic duplications, reducing the assembly length by 2.1% and the scaffold number by 42.4%, and increasing the scaffold N50 by 198.1%.
Figure 1. Images of the Rana temporaria specimen sequenced.
( A) Female voucher specimen of R. temporaria (BMNH 2013.483; Field ID, JWS 757; Snout–Vent Length 49.2 mm) from which the genome was sequenced. ( B) Ventral surface of NHMUK 2013.483. ( C) Dorsal and posterior thigh surfaces of NHMUK 2013.483. ( D) The individual was collected from the Natural History Museum Wildlife Garden, London, England.
The final assembly has a total length of 4.11 Gb in 555 sequence scaffolds with a scaffold N50 of 482 Mb ( Table 1). The majority, 99.5%, of the assembly sequence was assigned to 13 chromosomal-level scaffolds (numbered by sequence length) ( Figure 2– Figure 5; Table 2). The assembly has a BUSCO ( Simão et al., 2015) v5.1.2 completeness of 90.7% using the tetrapoda_odb10 reference set. However, a BUSCO (v4.0.2) score of 95.2% using the same reference set was obtained for the annotated gene set of the aRanTem1.1 assembly (see section Genome annotation), indicating that the assembly has a high level of completeness and that some genes were missed during BUSCO analysis of the whole genome assembly. The values obtained for this assembly are higher than for a previous transcriptome assembly ( Ma et al., 2018). While not fully phased, the assembly deposited is of one haplotype. Contigs corresponding to the second haplotype have also been deposited.
Table 1. Genome data for Rana temporaria, aRanTem1.1.
| Project accession data | |
|---|---|
| Assembly identifier | aRanTem1.1 |
| Species | Rana temporaria |
| Specimen | aRanTem1; NHMUK 2013.483 |
| NCBI taxonomy ID | NCBI:txid8407 |
| BioProject | PRJEB42239 |
| BioSample ID | SAMEA7521635 |
| Isolate information | Female, heart (genome assembly); kidney (Hi-C) |
| Raw data accessions | |
| PacificBiosciences SEQUEL I | ERR7012640-ERR7012642 |
| 10X Genomics Illumina | ERR6002771-ERR6002779, ERR6003050-
ERR6003052 |
| Hi-C Illumina | ERR6002780-ERR6002782 |
| BioNano | ERZ3003200 |
| Genome assembly | |
| Assembly accession | GCA_905171775.1 |
| Accession of alternate haplotype | GCA_905171725.1 |
| Span (Mb) | 4,111 |
| Number of contigs | 2,411 |
| Contig N50 length (Mb) | 6.26 |
| Number of scaffolds | 554 |
| Scaffold N50 length (Mb) | 482 |
| Longest scaffold (Mb) | 691 |
| BUSCO * genome score | C:90.7%[S:88.9%,D:1.8%],F:2.3%,M:6.9%,n:5310 |
| Genome annotation | |
| Number of genes | 36,124 |
| Number of protein-coding genes | 23,707 |
| Average length of gene (bp) | 52,818 |
| Average number of exons per gene | 14 |
| Average exon size (bp) | 273 |
| Average intron size (bp) | 9,757 |
| BUSCO annotation score ** | C:95.2%[S:92.8%,D:2.4%],F:0.6%,M:4.1%,n:5310 |
C= complete [S= single copy, D=duplicated], F=fragmented, M=missing, n=number of orthologues in comparison.
*BUSCO scores based on the terapoda_odb10 BUSCO set using v5.1.2, run on the aRanTem1.1 genome assembly using BlobToolKit. A full set of BUSCO scores is available at https://blobtoolkit.genomehubs.org/view/aRanTem1.1/dataset/CAJIMO01/busco.
**BUSCO scores based on the terapoda_odb10 BUSCO set using v4.0.2, run on the NCBI RefSeq annotation of the aRanTem1.1 genome assembly ( NCBI Rana temporaria Annotation Release 100).
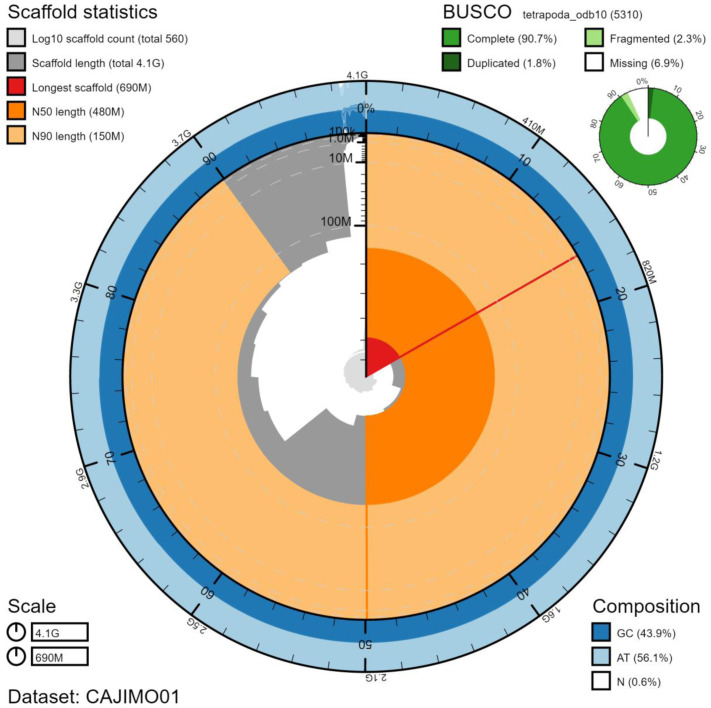
Figure 2. Genome assembly of Rana temporaria, aRanTem1.1: metrics.
The BlobToolKit Snailplot shows N50 metrics and BUSCO gene completeness. The main plot is divided into 1,000 size-ordered bins around the circumference with each bin representing 0.1% of the 4,111,445,260 bp assembly. The distribution of scaffold lengths is shown in dark grey with the plot radius scaled to the longest scaffold present in the assembly (690,654,357 bp, shown in red). Orange and pale-orange arcs show the N50 and N90 scaffold lengths (481,763,206 and 153,779,893 bp), respectively. The pale grey spiral shows the cumulative scaffold count on a log scale with white scale lines showing successive orders of magnitude. The blue and pale-blue area around the outside of the plot shows the distribution of GC, AT and N percentages in the same bins as the inner plot. A summary of complete, fragmented, duplicated and missing BUSCO genes in the tetrapoda_odb10 set is shown in the top right. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/aRanTem1.1/dataset/CAJIMO01/snail.
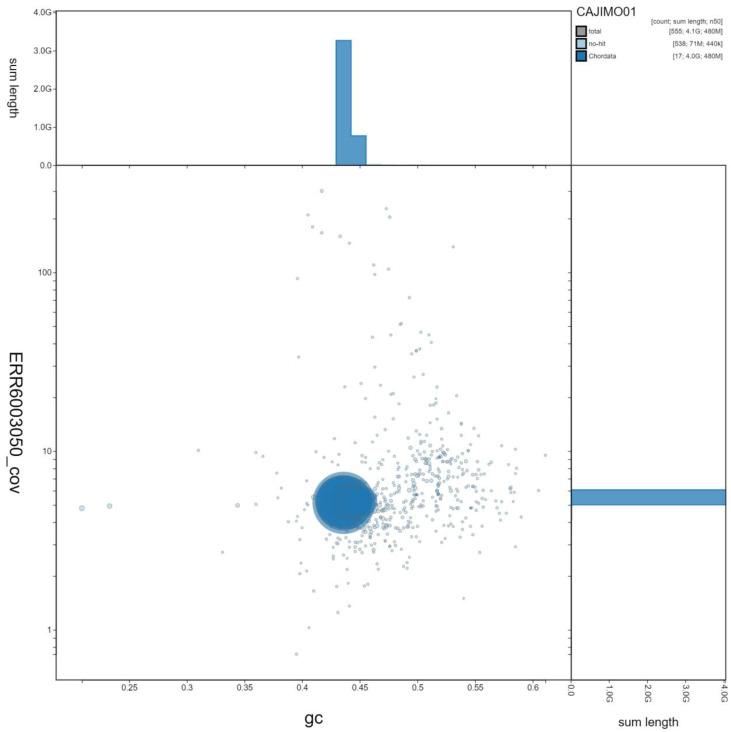
Figure 3. Genome assembly of Rana temporaria, aRanTem1.1: GC-coverage.
BlobToolKit GC-coverage plot. Scaffolds are coloured by phylum. Circles are sized in proportion to scaffold length. Histograms show the distribution of scaffold length sum along each axis. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/aRanTem1.1/dataset/CAJIMO01/blob.
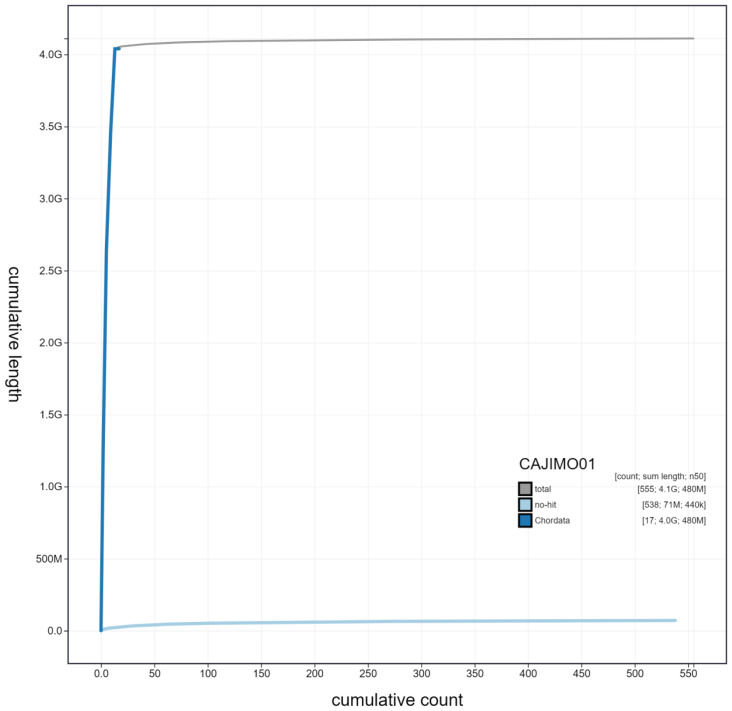
Figure 4. Genome assembly of Rana temporaria, aRanTem1.1: cumulative sequence.
BlobToolKit cumulative sequence plot. The grey line shows cumulative length for all scaffolds. Coloured lines show cumulative lengths of scaffolds assigned to each phylum using the buscogenes taxrule. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/aRanTem1.1/dataset/CAJIMO01/cumulative.
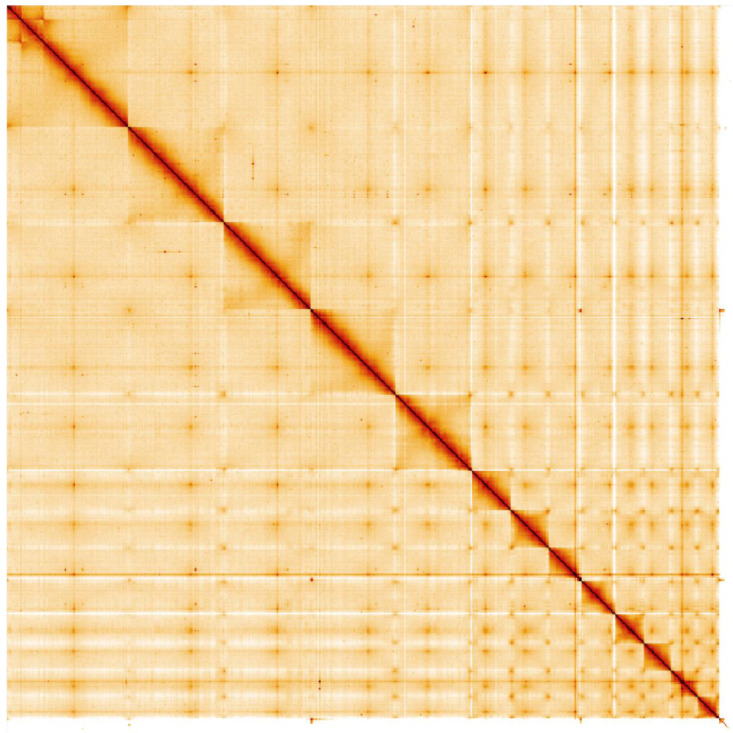
Figure 5. Genome assembly of Rana temporaria, aRanTem1.1: Hi-C contact map.
Hi-C contact map of the aRanTem1.1 assembly, visualised in HiGlass. Chromosomes are arranged in size order from left to right and top to bottom.
Table 2. Chromosomal pseudomolecules in the genome assembly of Rana temporaria, aRanTem1.1.
| INSDC accession | Chromosome | Size (Mb) | GC% |
|---|---|---|---|
| LR991680.1 | 1 | 690.65 | 43.8 |
| LR991681.1 | 2 | 541.44 | 43.7 |
| LR991682.1 | 3 | 495.42 | 44 |
| LR991683.1 | 4 | 481.76 | 43.8 |
| LR991684.1 | 5 | 429.35 | 43.9 |
| LR991685.1 | 6 | 224.82 | 44.4 |
| LR991686.1 | 7 | 212.59 | 44.7 |
| LR991687.1 | 8 | 190.44 | 44.4 |
| LR991688.1 | 9 | 184.30 | 44.4 |
| LR991689.1 | 10 | 153.78 | 44.7 |
| LR991690.1 | 11 | 164.33 | 44.8 |
| LR991691.1 | 12 | 148.93 | 45.4 |
| LR991692.1 | 13 | 121.98 | 45 |
| LR991693.1 | MT | 0.02 | 40.5 |
| - | Unplaced | 71.62 | 47.9 |
Genome annotation
The R. temporaria assembly was annotated by the NCBI Eukaryotic Genome Annotation Pipeline, an automated pipeline that annotates genes, transcripts and proteins on draft and finished genome assemblies. The annotation ( NCBI Rana temporaria Annotation Release 100; Table 1) was generated from transcripts and proteins retrieved from NCBI Entrez by alignment to the genome assembly, as described here ( Pruitt et al., 2014).
Methods
Sample acquisition
A single female R. temporaria was collected from a stable, isolated population in the NHM Wildlife Garden, London, UK (latitude 51.49586, longitude -0.178622, elevation 17 m) by Jeffrey W. Streicher on 1 July 2015 ( Figure 1D). The specimen of R. temporaria (NHMUK 2013.483, Field ID: JWS 757) was 49.2 mm snout–vent length (determined using a Miyamoto digital calliper to the nearest 0.1 mm). The specimen was collected with permission from the NHM Wildlife Garden management team and is part of a long-term monitoring project run by the Department of Life Sciences and the Angela Marmont Centre for UK Biodiversity. It was humanely euthanised using a saturated solution of tricaine mesylate (MS-222). Multiple tissues including heart, thigh muscle, liver, eyes, kidney, ovaries, and intestines were sampled and placed in an ammonium sulfate-based RNA + DNA preservation buffer. After ~24 hours of storage at 4°C, the tissues were transferred to -80°C until they were sent for genome sequencing. Sample tissue has been accessioned by the Natural History Museum Molecular Collections Facility (NHMUK 2013.483).
DNA extraction and sequencing
DNA was extracted from heart tissue in the Scientific Operations core of the Wellcome Sanger Institute using the Bionano Prep Animal Tissue DNA Isolation kit according to the manufacturer's instructions. Pacific Biosciences CLR long read and 10X Genomics read cloud sequencing libraries were constructed according to the manufacturers’ instructions. Hi-C data were generated from kidney tissue taken from the same animal using the Arima v2 Hi-C kit. Extraction and sequencing was performed by the Scientific Operations DNA Pipelines at the Wellcome Sanger Institute on Pacific Biosciences SEQUEL II (long-read) and Illumina HiSeq X (10X, Hi-C) instruments. DNA was labeled for Bionano Genomics optical mapping following the Bionano Prep Direct Label and Stain (DLS) Protocol and run on one Saphyr instrument chip flowcell.
Genome assembly
Assembly was carried out following the Vertebrate Genome Project pipeline v1.6 ( Rhie et al., 2021) with Falcon-unzip ( Chin et al., 2016), haplotypic duplication was identified and removed with purge_dups ( Guan et al., 2020) and a first round of scaffolding carried out with 10X Genomics read clouds using scaff10x. Hybrid scaffolding was performed using the BioNano DLE-1 data and BioNano Solve. Scaffolding with Hi-C data ( Rao et al., 2014) was carried out with SALSA2 ( Ghurye et al., 2019). The Hi-C scaffolded assembly was polished with arrow using the PacBio data, then polished with the 10X Genomics Illumina data by aligning to the assembly with longranger align, calling variants with freebayes ( Garrison & Marth, 2012) and applying homozygous non-reference edits using bcftools consensus. Two rounds of the Illumina polishing were applied. The mitochondrial genome was assembled using the mitoVGP pipeline ( Formenti et al., 2021). The assembly was checked for contamination and corrected using the gEVAL system ( Chow et al., 2016; Howe et al., 2021). Manual curation was performed using evidence from Bionano (using the Bionano Access viewer), using HiGlass ( Kerpedjiev et al., 2018) and Pretext, as described previously ( Howe et al., 2021). Figure 2– Figure 4 and BUSCO values were generated using BlobToolKit ( Challis et al., 2020). Table 3 includes a list of software tools used.
Table 3. Software tools used.
| Software tool | Version | Source |
|---|---|---|
| Falcon-unzip | falcon-kit 1.4.2 | Chin et al., 2016 |
| purge_dups | 1.0.0 | Guan et al., 2020 |
| SALSA2 | 2.2-14-g974589f | Ghurye et al., 2019 |
| scaff10x | 4.2 | https://github.com/wtsi-hpag/Scaff10X |
| Bionano Solve | 3.3_10252018 | https://bionanogenomics.com/downloads/bionano-solve/ |
| arrow | gcpp 1.9.0-SL-release-8.0.0+1-37-gd7b188d | https://github.com/PacificBiosciences/GenomicConsensus |
| longranger
align |
2.2.2 |
https://support.10xgenomics.com/genome-exome/software/
pipelines/latest/advanced/other-pipelines |
| freebayes | 1.3.1-17-gaa2ace8 | Garrison & Marth, 2012 |
| bcftools
consensus |
1.9-78-gb7e4ba9 | http://samtools.github.io/bcftools/bcftools.html |
| mitoVGP | Formenti et al., 2021 | |
| HiGlass | 1.11.6 | Kerpedjiev et al., 2018 |
| PretextView | 0.1 | https://github.com/wtsi-hpag/PretextView |
| gEVAL | N/A | Chow et al., 2016 |
| BlobToolKit | 2.6.1 | Challis et al., 2020 |
Ethical/compliance issues
The materials that have contributed to this genome note were supplied by a Tree of Life collaborator. The Wellcome Sanger Institute employs a process whereby due diligence is carried out proportionate to the nature of the materials themselves, and the circumstances under which they have been/are to be collected and provided for use. The purpose of this is to address and mitigate any potential legal and/or ethical implications of receipt and use of the materials as part of the research project, and to ensure that in doing so we align with best practice wherever possible.
The overarching areas of consideration are:
Ethical review of provenance and sourcing of the material;
Legality of collection, transfer and use (national and international).
Each transfer of samples is undertaken according to a Research Collaboration Agreement or Material Transfer Agreement entered into by the Tree of Life collaborator, Genome Research Limited (operating as the Wellcome Sanger Institute) and in some circumstances other Tree of Life collaborators.
Data availability
European Nucleotide Archive: Rana temporaria (common frog). Accession number PRJEB42239: https://identifiers.org/ena.embl:PRJEB42239
The genome sequence is released openly for reuse. The R. temporaria genome sequencing initiative is part of the Darwin Tree of Life (DToL) project and the Vertebrate Genomes Project. All raw sequence data and the assembly have been deposited in INSDC databases. Raw data and assembly accession identifiers are reported in Table 1.
Author information
Members of the Wellcome Sanger Institute Tree of Life programme collective are listed here: https://doi.org/10.5281/zenodo.5377053.
Members of Wellcome Sanger Institute Scientific Operations: DNA Pipelines collective are listed here: https://doi.org/10.5281/zenodo.4790456.
Members of the Tree of Life Core Informatics collective are listed here: https://doi.org10.5281/zenodo.5013542.
Members of the Darwin Tree of Life Consortium are listed here: https://doi.org/10.5281/zenodo.4783559.
Acknowledgements
JWS thanks Caroline Ware, Nicky Reilly, Naomi Lake, and the NHM Wildlife Garden Team for permitting specimen collection. JWS also thanks Donney Nicholson for assistance with tissue shipment as well as David Gower, Simon Loader (Department of Life Sciences), and John Tweddle (Angela Marmont Centre) for logistical assistance.
Funding Statement
This work was supported by the Wellcome Trust through core funding to the Wellcome Sanger Institute (206194) and the Darwin Tree of Life Discretionary Award (218328).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved]
References
- Challis R, Richards E, Rajan J, et al. : BlobToolKit - Interactive Quality Assessment of Genome Assemblies. G3 (Bethesda). 2020;10(4):1361–74. 10.1534/g3.119.400908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CS, Peluso P, Sedlazeck FJ, et al. : Phased Diploid Genome Assembly with Single-Molecule Real-Time Sequencing. Nat Methods. 2016;13(12):1050–54. 10.1038/nmeth.4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow W, Brugger K, Caccamo M, et al. : gEVAL - a Web-Based Browser for Evaluating Genome Assemblies. Bioinformatics. 2016;32(16):2508–10. 10.1093/bioinformatics/btw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formenti G, Rhie A, Balacco J, et al. : Complete Vertebrate Mitogenomes Reveal Widespread Repeats and Gene Duplications. Genome Biol. 2021;22(1):120. 10.1186/s13059-021-02336-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G: Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv [q-bio.GN]. arXiv,2012. Reference Source [Google Scholar]
- Ghurye J, Rhie A, Walenz BP, et al. : Integrating Hi-C Links with Assembly Graphs for Chromosome-Scale Assembly. PLoS Comput Biol. 2019;15(8):e1007273. 10.1371/journal.pcbi.1007273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR: Animal Genome Size Database.2021. Reference Source [Google Scholar]
- Guan D, McCarthy SA, Wood J, et al. : Identifying and Removing Haplotypic Duplication in Primary Genome Assemblies. Bioinformatics. 2020;36(9):2896–98. 10.1093/bioinformatics/btaa025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SA, Warren RL, Vandervalk BP, et al. : The North American Bullfrog Draft Genome Provides Insight into Hormonal Regulation of Long Noncoding RNA. Nat Commun. 2017;8(1):1433. 10.1038/s41467-017-01316-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Chow W, Collins J, et al. : Significantly Improving the Quality of Genome Assemblies through Curation. GigaScience. 2021;10(1):giaa153. 10.1093/gigascience/giaa153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, et al. : HiGlass: Web-Based Visual Exploration and Analysis of Genome Interaction Maps. Genome Biol. 2018;19(1):125. 10.1186/s13059-018-1486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma WJ, Veltsos P, Toups MA, et al. : Tissue Specificity and Dynamics of Sex-Biased Gene Expression in a Common Frog Population with Differentiated, Yet Homomorphic, Sex Chromosomes. Genes (Basel). 2018;9(6):294. 10.3390/genes9060294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips BC, Rodrigues N, van Rensburg AJ, et al. : Phylogeography, More than Elevation, Accounts for Sex Chromosome Differentiation in Swiss Populations of the Common Frog ( Rana Temporaria). Evolution. 2020;74(3):644–54. 10.1111/evo.13860 [DOI] [PubMed] [Google Scholar]
- Price SJ, Garner TW, Balloux F, et al. : A de Novo Assembly of the Common Frog ( Rana Temporaria) Transcriptome and Comparison of Transcription Following Exposure to Ranavirus and Batrachochytrium Dendrobatidis. PLoS One. 2015;10(6):e0130500. 10.1371/journal.pone.0130500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Brown GR, Hiatt SM, et al. : RefSeq: An Update on Mammalian Reference Sequences. Nucleic Acids Res. 2014;42(Database issue):D756–63. 10.1093/nar/gkt1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, et al. : A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159(7):1665–80. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, et al. : Towards Complete and Error-Free Genome Assemblies of All Vertebrate Species. Nature. 2021;592(7856):737–46. 10.1038/s41586-021-03451-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, et al. : BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics. 2015;31(19):3210–12. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Spasić-Bošković O, Tanić N, Blagojević J, et al. : Comparative Cytogenetic Analysis of European Brown Frogs: Rana Temporaria, R. Dalmatina and R. Graeca. Caryologia. 1997;50(2):139–49. 10.1080/00087114.1997.10797393 [DOI] [Google Scholar]
- Walsh PT, Downie JR, Monaghan P: Factors Affecting the Overwintering of Tadpoles in a Temperate Amphibian. J Zool. 2016;298(3):183–90. 10.1111/jzo.12296 [DOI] [Google Scholar]